Abstract
SU(VAR)3–9 like histone methyltransferases control heterochromatic domains in eukaryotes. In Arabidopsis, 10 SUVH genes encode SU(VAR)3–9 homologues where SUVH1, SUVH2 and SUVH4 (KRYPTONITE) represent distinct subgroups of SUVH genes. Loss of SUVH1 and SUVH4 causes weak reduction of heterochromatic histone H3K9 dimethylation, whereas in SUVH2 null plants mono- and dimethyl H3K9, mono- and dimethyl H3K27, and monomethyl H4K20, the histone methylation marks of Arabidopsis heterochromatin are significantly reduced. Like animal SU(VAR)3–9 proteins SUVH2 displays strong dosage-dependent effects. Loss of function suppresses, whereas overexpression enhances, gene silencing, causes ectopic heterochromatization and significant growth defects. Furthermore, modification of transgene silencing by SUVH2 is partially transmitted to the offspring plants. This epigenetic stability correlates with heritable changes in DNA methylation. Mutational dissection of SUVH2 indicates an implication of its N-terminus and YDG domain in directing DNA methylation to target sequences, a prerequisite for consecutive histone methylation. Gene silencing by SUVH2 depends on MET1 and DDM1, but not CMT3. In Arabidopsis, SUVH2 with its histone H3K9 and H4K20 methylation activity has a central role in heterochromatic gene silencing.
Keywords: DNA methylation, gene silencing, heterochromatin, histone methylation
Introduction
Epigenetically established changes in chromatin structure define the gene expression potential during development. These processes are controlled by complex DNA and histone modification systems. Modifications at the highly conserved N-terminal histone tails include acetylation, methylation, phosphorylation and ubiquitination, and show characteristic differences between active and repressed chromatin states (Stahl and Allis, 2000; Jenuwein and Allis, 2001). Lysine methylation at H3K4, H3K36 and H3K79 marks transcriptionally active chromatin, whereas methylation of H3K9, H3K27 and H4K20 defines repressed chromatin domains (Lachner et al, 2001; Fischle et al, 2003). Mono-, di- and trimethylation states of histone lysine residues extend the coding potential of the ‘epigenetic histone code' and specific states of H3K9, H3K27 and H4K20 methylation define repressed heterochromatic domains in mouse and Drosophila (Peters et al, 2003; Schotta et al, 2004). In mouse H3K9 trimethylation, H3K27 monomethylation and H4K20 trimethylation index pericentric heterochromatin, whereas in Drosophila these regions show H3K9 di- and tri-, H3K27 mono-, di- and tri-, and H4K20 trimethylation (Ebert et al, 2004).
Plant development in contrast to animals is rather plastic and considerably affected by environmental factors. Therefore, subtle changes in chromatin structure might be required for fine-tuning of gene expression and probably for this reason multi-gene families for DNA and histone modification systems are found in plants. In Arabidopsis, three different classes of DNA methylases (Cao and Jacobsen, 2002), 12 putative methylcytosine-binding proteins (Zemach and Grafi, 2003), 37 SET domain proteins (Baumbusch et al, 2001), 18 putative histone deacetylases and 12 putative histone acetylases have been identified (Arabidopsis Genome Initiative, 2000; Pandey et al, 2002).
A predominant role in establishment of epigenetically stable active or repressed chromatin domains is attributed to histone lysine methylation by SET domain proteins. These proteins can be assigned to four groups typified by their Drosophila homologues E(Z), TRX, ASH1 and SU(VAR)3–9 (Jenuwein et al, 1998). In Arabidopsis, several of these genes were identified by developmental phenotypes of mutations. Like animal E(Z), CLF (Goodrich et al, 1997) and MEA (Grossniklaus et al, 1998) act as negative regulators and are involved in control of flower and endosperm development, respectively. The homeotic effects of clf and the function of MEA in imprinting of paternal genes first indicated an important role of SET domain proteins for chromatin regulation and epigenetic inheritance in plants (Goodrich et al, 1997; Vielle-Calcada et al, 1999). Similar to TRITHORAX in animals, Arabidopsis ATX1 acts as an activator of homeotic genes (Alvarez-Venegas et al, 2003).
Epigenetically stable transmission of a condensed and transcriptionally inert chromatin state is characteristic for heterochromatin and heterochromatic gene silencing. With the evolutionary conserved SU(VAR)3–9 SET domain protein (Tschiersch et al, 1994; Ivanova et al, 1998; Aagaard et al, 1999) and demonstration of its function in histone H3K9 methylation (Rea et al, 2000), a basic factor of heterochromatin formation has been identified. In plants, in general, a large fraction of the genome is heterochromatic and extensive heterochromatic silencing processes are found. Involvement of histone H3K9 and DNA methylation in heterochromatin formation and heterochromatic gene silencing was documented in studies of the SU(VAR)3–9 homologue SUVH4 (KYP) and DNA methylation defective mutations of the MET1, CMT3, DRM and DDM1 genes (Cao and Jacobsen, 2002; Gendrel et al, 2002; Jackson et al, 2002; Tariq et al, 2003). In contrast to mammals, plants contain a large number of SU(VAR)3–9 homologues (Baumbusch et al, 2001). In Arabidopsis, 10 different SU(VAR)3–9 homologous SUVH proteins are found and we studied the function of SUVH1, SUVH2 and SUVH4, representative members of three subgroups of Arabidopsis SU(VAR)3–9 homologues.
Here, for the first time, we show the heterochromatin association of the Arabidopsis SU(VAR)3–9 homologues SUVH1 and SUVH2 and present evidence that SUVH proteins differentially control heterochromatic histone methylation marks. Our data define SUVH2 as a central function in heterochromatin formation and heterochromatic gene silencing in Arabidopsis. In suvh2 null mutations all heterochromatin-specific histone methylation marks are significantly reduced, which is connected with strong suppression of transcriptional gene silencing (TGS). After overexpression, SUVH2 shows ectopic nuclear distribution and causes extensive heterochromatization, accompanied with an increase of DNA and heterochromatic histone methylation. SUVH2-dependent suppression or enhancement of TGS shows partial epigenetic stability connected with heritable changes in symmetric and nonsymmetric DNA methylation over consecutive generations. Mutational dissection of SUVH2 revealed a role of its N-terminus in control of nuclear protein distribution and a function of its YDG domain in directing DNA methylation to the target sequences. We also show that this DNA methylation does not cause silencing but rather is a prerequisite for consecutive SUVH2-dependent histone H3 and H4 methylation to establish heterochromatic silencing.
Results
Heterochromatin association of SUVH proteins in Arabidopsis
The heterochromatin-associated SU(VAR)3–9 proteins control repressive chromatin structures by histone H3K9 methylation (Rea et al, 2000; Nakayama et al, 2001; Schotta et al, 2002). In contrast to animals and fungi, which have one or two Su(var)3–9 homologues, in Arabidopsis 10 SUVH genes encode SU(VAR)3–9 homologous proteins (Baumbusch et al, 2001). Tree reconstruction of 21 SUVH protein sequences from different plant species results in four clearly distinct subgroups of SUVH proteins in angiosperms (Figure 1A). Branching of both moss and fern SUVH fragments at the root of the angiosperm SUVH subgroups argues for a phylogenetic split of SUVH genes during early evolution of seed plants. The SUVH4-like genes are more distant to all groups of SUVH genes. Only SUVH4-like genes contain introns and retrotransposition might have been involved in evolution of the different SUVH gene families. Branching of gymnosperm PtaSUV1f with the SUVH4 subgroup suggests that functional differentiation of SUVH proteins had already occurred before the angiosperm–gymnosperm split.
Figure 1.
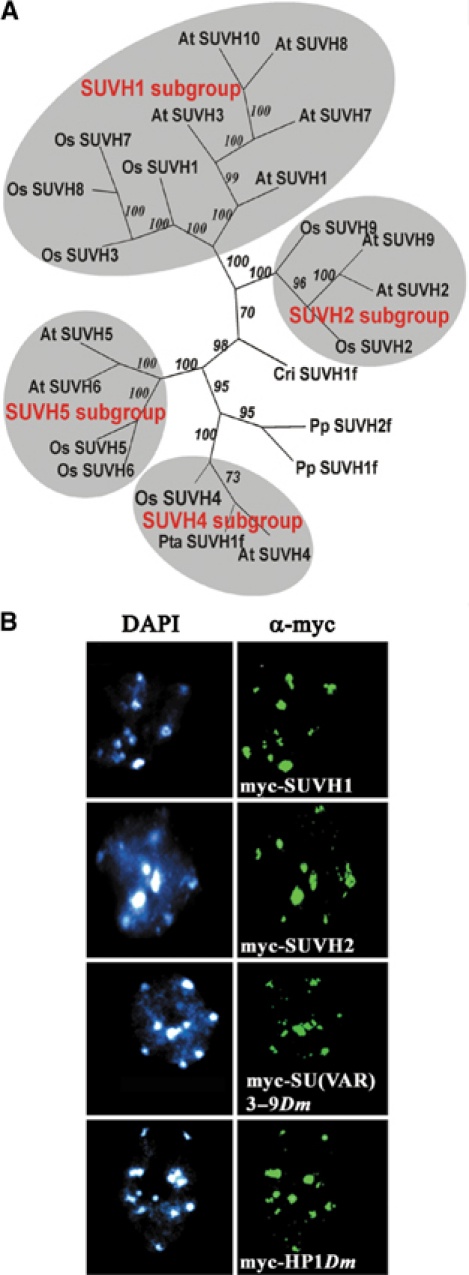
SU(VAR)3–9 homologous proteins in plants. (A) Four conserved groups of SUVH genes are found in angiosperms. Phylogenetic analysis of 21 SUVH protein sequences from Arabidopsis (AtSUVH), rice (OsSUVH), Pinus taeda (Pta), Physcomitrella patens (Pp) and Ceratopteris richardii (Cri). (B) Immunostaining of plants expressing myc fusion protein of SUVH1, SUVH2 and Drosophila SU(VAR)3–9 and HP1 in Arabidopsis interphase nuclei with α-myc. Heterochromatin association is found for the SUVH1 and SUVH2 proteins as well as for Drosophila SU(VAR)3–9 and HP1.
To identify chromatin targets of the Arabidopsis SU(VAR)3–9 homologues SUVH1 and SUVH2 proteins, we studied their nuclear distribution in transgenic Arabidopsis plants expressing myc or EGFP fusion proteins. The SUVH1 and SUVH2 fusion proteins associate with the DAPI bright regions of interphase nuclei (Figure 1B), which represent pericentromeric heterochromatin in Arabidopsis (Fransz et al, 2000; Soppe et al, 2002). We also tested nuclear distribution of the Drosophila SU(VAR)3–9 and HP1 heterochromatin proteins in Arabidopsis. The Drosophila proteins in Arabidopsis also associate with heterochromatin (Figure 1B), indicating conserved mechanisms of heterochromatin association of these proteins in plants and animals. SUVH1 and SUVH2 proteins are both heterochromatin associated and could comprise redundant functions. Alternatively, these proteins could control distinct heterochromatic histone methylation marks as well as their combinatorial interplay.
Differential control of heterochromatic histone methylation by SUVH proteins
We analysed the effect of SUVH1, SUVH2 and SUVH4 on heterochromatic histone H3 and H4 methylation marks using specific histone methylation antibodies (Peters et al, 2003). In these studies, we used loss-of-function T-DNA insertion mutations or antisense lines (cf. Supplementary Figures 1 and 2). None of the insertional mutations or the antisense lines show a significant phenotypic defect in homozygous constitution.
Arabidopsis heterochromatin is enriched in methylated DNA and mono- and dimethylated histone H3K9 (Tariq et al, 2003; Figure 2A). Other heterochromatic histone methylation marks in Arabidopsis are mono- and dimethyl H3K27 and monomethyl H4K20 (Figure 2A). In contrast, trimethyl H3K9, trimethyl H3K27, and di- and trimethyl H4K20 are found together with methylated H3K4 and H3K36 in Arabidopsis euchromatin (Figure 2A and B). In our studies loss-of-function mutations of SUVH1 and SUVH4 appear to show only weak reduction of mono- and dimethyl H3K9 in pericentromeric heterochromatin (Figure 2A and Supplementary Figure 1). In contrast, SUVH2 loss-of-function mutations strongly reduce all heterochromatin-specific histone and DNA methylation marks (Figure 2A and C). The most dramatic effect of SUVH2 is on H4K20 monomethylation. Reduction of DNA methylation at heterochromatic sequences in SUVH2 null plants was further quantified by bisulphite sequence analysis of Athila transposons (Supplementary Figure 3). In vitro analysis shows that SUVH2 is a nucleosome-dependent HMTase and methylates histone H3 and H4 in recombinant nucleosomes (Figure 2D).
Figure 2.
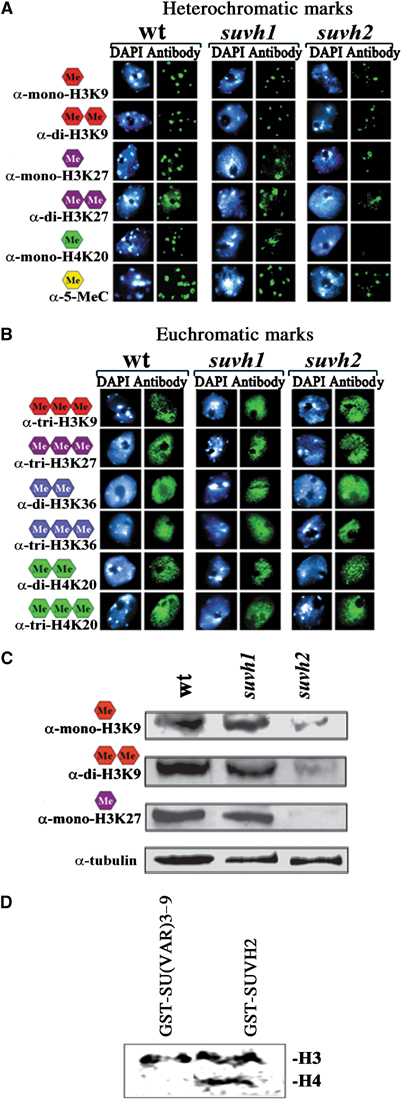
Differential effects of suvh1 and suvh2 mutations on histone and DNA methylation. (A, B) Immunohistochemical staining of wild-type, suvh1 and suvh2 interphase nuclei with antibodies recognizing specific histone and DNA methylation marks. In suvh2, but not in suvh1, mono- and dimethyl H3K9, mono- and dimethyl H3K27, monomethyl H4K20 and 5-methylcytosine (heterochromatic marks) are significantly reduced (A). No effects are found on the trimethyl H3K9, trimethyl H3K27, di- and trimethyl H3K36 and di- and trimethyl H4K20 euchromatic marks (B). (C) Western analysis of nuclear extracts of wild-type, suvh1 and suvh2 mutant plants. Only in suvh2 significant reduction of mono- and dimethyl H3K9 and monomethyl H3K27 is found. (D) In vitro recombinant SUVH2 shows H3 and H4 HMTase activity in assays with reconstituted nucleosomes.
Drosophila Su(var)3–9 is a haplo- and triplo-dosage-dependent modifier of heterochromatic gene silencing (Schotta et al, 2002). In order to study the dosage-dependent effects of Arabidopsis SUVH1 and SUVH2, we constructed transgenes with the 35S* promotor (Mindrinos et al, 1994) or used a glucocorticoid-mediated transcriptional system, which allows controlled induction of gene expression by dexamethasone treatment (Aoyama and Chua, 1997). Overexpression of SUVH1 has no significant effect on pericentromeric heterochromatin and the protein remains heterochromatin associated (Figure 3A). In contrast, after overexpression, SUVH2 shows dispersed nuclear distribution, resulting in ectopic heterochromatization. By electron microscopic analysis, additional blocks of heterochromatic material can be detected (Figure 3A). Immunocytological analysis of SUVH2 overexpression plants reveals a significant increase in mono- and dimethyl H3K9, mono- and dimethyl H3K27, and monomethyl H4K20, as well as cytosine methylation (Figure 3B). We have quantified the dosage-dependent effects of SUVH2 on five histone methylation marks by Western analysis of bulk histones. In suvh2 mutant plants, heterochromatic H3K9 mono- and dimethylation, H3K27 monomethylation and H4K20 monomethylation are significantly reduced (Figures 2C and 3C), whereas SUVH2 overexpression causes enhancement of H3K9 dimethylation and H4K20 monomethylation (Figure 3B and C). In contrast, trimethylation of H3K9, which is a euchromatic histone methylation mark in Arabidopsis, is significantly reduced after SUVH2 overexpression (Figure 3C and D). Significant reduction in immunostaining for other euchromatic histone modification marks like dimethyl H3K4, acetyl H3K9 and trimethyl H3K27 is also found in SUVH2 overexpression plants (Figure 3D).
Figure 3.
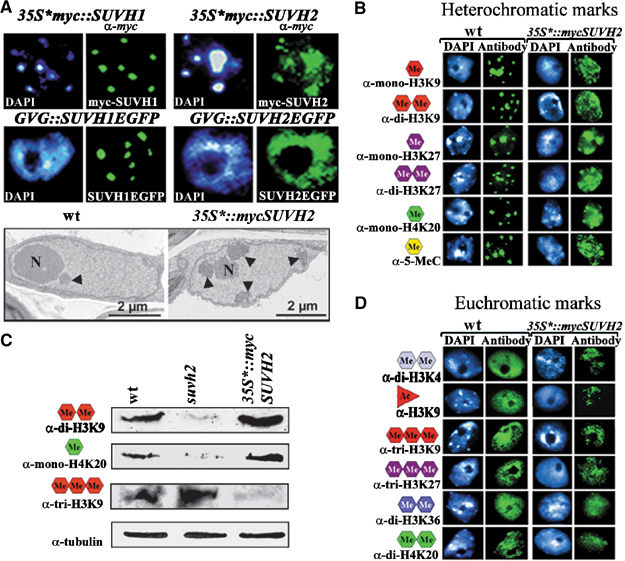
Ectopic heterochromatization in SUVH2 overexpression lines. (A) Immunostaining of nuclei from 35S*∷mycSUVH1, 35S*∷mycSUVH2 with α-myc and GFP fluorescence analysis in dexamethasone-treated GVG∷SUVH1EGFP and GVG∷SUVH2EGFP plants. Only SUVH2 shows ectopic distribution. Electron microscopic analysis of nuclei from 35S*∷mycSUVH2 plants shows ectopic heterochromatin (arrowheads). (B) Immunocytological analysis of heterochromatic histone and 5-methylcytosine methylation in SUVH2 overexpression plants. Enhanced staining for all heterochromatic marks (mono- and dimethyl H3K9, mono- and dimethyl H3K27, monomethyl H4K20 and 5-methylcytosine) is found. (C) Western analysis of suvh2 mutant and 35S*∷mycSUVH2 overexpression plants. In suvh2, dimethyl H3K9 and monomethyl H4K20 are reduced. In 35S*∷mycSUVH2 overexpression plants, heterochromatic H3K9 dimethyl and H4K20 monomethyl are enriched, whereas euchromatic H3K9 trimethyl is reduced. (D) Immunostaining for euchromatic histone modification marks in SUVH2 overexpression plants. Staining for dimethyl H3K4, acetyl H3K9, trimethyl H3K27, dimethyl H3K36 and dimethyl H4K20 is significantly reduced.
SUVH2-dependent growth defects and the mutational dissection of its molecular function
None of the 24 35S*∷mycSUVH1 overexpression lines nor the GVG∷SUVH1-EGFP lines after dexamethasone treatment show any phenotypic defects (not shown). In contrast, four of 24 independent 35S*∷mycSUVH2 transgenic lines show significant growth reduction (mini-plant phenotype) and a curled cotyledon phenotype (Figure 4A and B). Western blot analysis shows direct correlation between the extent of SUVH2 overexpression and the strength of mini-plant growth defects (Figure 4C). Line 35S*∷mycSUVH2#4 with lower amount of additional SUVH2 is only weakly affected, whereas the three lines 35S*∷mycSUVH2#5, #6 and #22 with higher amount of SUVH2 manifest a strong mini-plant phenotype. Similarly, the GVG∷SUVH2-EGFP lines manifest a mini-plant phenotype only after dexamethasone treatment (Figure 4A). Significant rescue of SUVH2 overexpression phenotypes is found in suvh2/+; 35S*∷mycSUVH2#6/+ plants by introducing a suvh2 null allele (Figure 4B).
Figure 4.
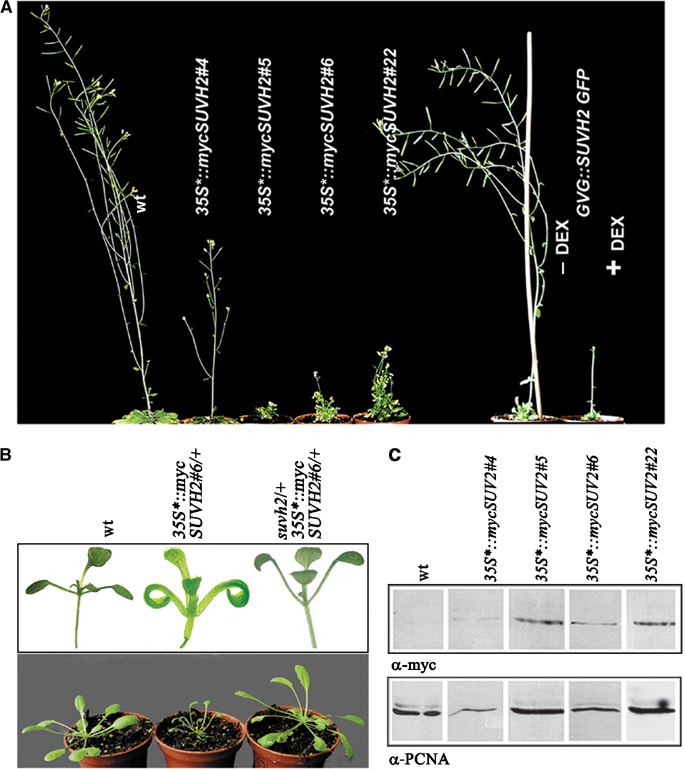
Growth and developmental defects in SUVH2 overexpression plants. (A) SUVH2 overexpression causes mini-plant phenotype in 35S*∷mycSUVH2#5, #6, #22 and dexamethasone-treated GVG∷SUVH2EGFP lines. (B) SUVH2 overexpression seedlings show a curled cotyledon phenotype (upper panel). By introducing a suvh2 null allele, the mini-plant and curled cotyledon phenotypes are significantly rescued in suvh2/+; 35S*∷mycSUVH2#6/+ plants (lower panel). (C) Western analysis of extracts from 35S*∷mycSUVH2 lines with α-myc. The amount of mycSUVH2 protein correlates with the strength of growth defects. 35S*∷mycSUVH2#4 plants with a weak growth reduction expresses a lower amount of additional SUVH2 as compared to lines with a mini-plant phenotype.
For functional dissection of SUVH2, we isolated mutations within the 35S*∷mycSUVH2#5 transgene after EMS mutagenesis. These mutations were identified as dominant suppressors of the curled cotyledon phenotype in M1 seedlings. All confirmed dominant suppressors rescued the mini-plant growth defect. In M3 two classes of mutant lines could be established. In 224 lines the suppressor mutation and the SUVH2 transgene segregate independently, whereas in 96 lines complete linkage is found. The latter class represents 35S*∷mycSUVH2#5 transgene mutations and we studied the functional consequence of seven such mutations (Figure 5). All are missense mutations located either in the N-terminus (5–1), the YDG (5–2) or the SET domain (5–3 to 5–7) of SUVH2 (Figure 5A). Only the N-terminus mutation 5–1 interferes with ectopic nuclear distribution of SUVH2 (Figure 5B). Mutations 5–2 and 5–3 located in the YDG domain and the region between the YDG domain and the preSET region, respectively, cause loss of ectopic 5-methylcytosine, H3K9 dimethylation and H4K20 monomethylation, although ectopic distribution of SUVH2 is not affected (Figure 5B and C). Mutations in the SET domain (5–3 to 5–7) eliminate ectopic H3K9 and H4K20 methylation, but leave ectopic DNA methylation unaffected.
Figure 5.
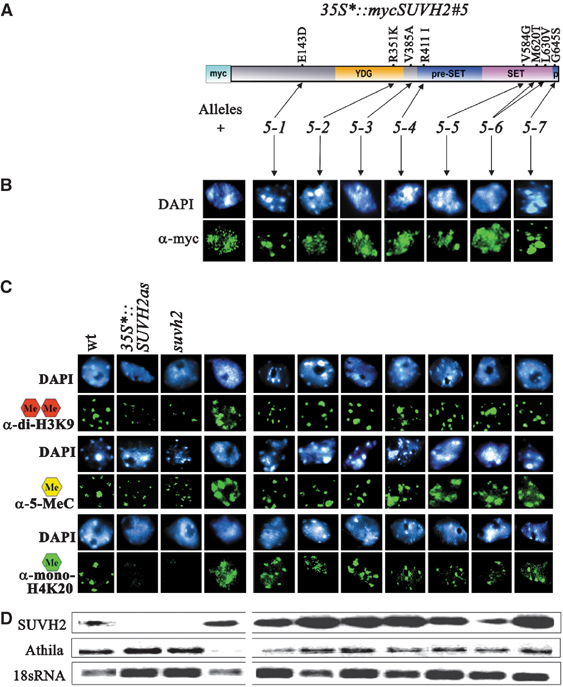
Functional dissection of SUVH2 by transgene mutations. (A) Molecular nature of 35S*∷mycSUVH2#5 transgene mutations. Structure of SUVH2 with the conserved YDG, preSET, SET and postSET (p) domains. (B) Immunostaining with α-myc shows that only the N-terminus mutation 5-1 eliminates ectopic distribution of SUVH2. (C) All mutations eliminate ectopic H3K9 and H4K20 methylation. The N-terminus mutation 5-1, the YDG mutation 5-2 and mutation 5-3 eliminate ectopic DNA methylation, whereas in plants with the SET domain mutations 5-4, 5-5, 5-6 and 5-7 ectopic DNA methylation is observed. (D) Silencing of Athila transposons by SUVH2 overexpression is rescued by all transgene mutations independent of DNA hypermethylation (RT–PCR) (cf. Supplementary Figure 3 for bisulphite data).
In order to study SUVH2 overexpression effects on heterochromatic silencing of endogenous sequences, we analysed expression of Athila transposons. Overexpression of SUVH2 causes strong repression of Athila, whereas all SUVH2 transgene mutations rescue this silencing effect (Figure 5D). Immunocytological and bisulphite sequence analysis shows that none of the SET domain mutations reduce DNA hypermethylation at Athila although its silencing is released (Figure 5D and Supplementary Figure 3).
Taken together, our mutant analysis resolves a sequence of molecular events connected with SUVH2-induced heterochromatic gene silencing. Mutation 5–1 indicates a possible role of the SUVH2 N-terminus in target sequences recognition, whereas the YDG domain region appears to be involved in directing DNA methylation to these sequences. DNA methylation alone is not sufficient for silencing, but rather functions as a mark directing SUVH2-dependent histone methylation to sequences subjected to silencing. Moreover, our data suggest that SUVH2-mediated DNA methylation precedes histone methylation.
Dosage dependence and epigenetic maintenance of SUVH2-induced transgene silencing
To study dosage-dependent effects of SUVH proteins on repeat-induced TGS, we used a new type of transgene constructs that contain four tandem copies of the luciferase gene (cf. Materials and methods). The LUC2 transgene shows moderate luciferase silencing, which is significantly enhanced by SUVH2 (Figure 6) but not SUVH1 overexpression (data not shown). Significant enhancement of TGS is already found in crosses with the 35S*∷mycSUVH2#4 line showing only weak growth defects (Figure 5), indicating that TGS is already efficiently enhanced by low levels of SUVH2 overexpression. The loss-of-function effect of SUVH2 on TGS was studied using a specific antisense line. Out of 25 independent 35S*∷SUVH2as antisense lines, we selected line SUVH2as#11 causing complete elimination of SUVH2 transcript, whereas other tested SUVH genes are not affected (cf. Supplementary Figure 1). After a cross of SUVH2as#11/SUVH2as#11 with LUC2 homozygous plants, all SUVH2as#11/+; LUC2/+ offspring show strong suppression of TGS (Figure 6). Significant suppressor effects are also seen with antisense lines only partially eliminating the SUVH2 transcript (not shown). Our experiments reveal the dosage-dependent modifier effect of SUVH2 on TGS. Overexpression enhances whereas loss-of-function suppresses TGS. We further studied the epigenetic stability of SUVH2 modified LUC2 transgene silencing in progeny plants, lacking SUVH2 overexpression or antisense constructs which were generated by a backcross of the F1 progeny with wild-type plants. Partial epigenetic stability of modified LUC2 transgene silencing is found in the offspring plants for the enhanced as well as suppressed state of LUC2. Reciprocal crosses produced identical results (data not shown). Reversion to the level typical for control plants is found in offspring of the second backcross generation (Figure 6A and B). To study whether epigenetic maintenance of SUVH2-modified TGS is based on stable transmission of changes in DNA methylation, we performed bisulphite sequence analysis of the LUC2 transgene. In plants produced by the first backcross (BC1) to wild type, the LUC2 transgene inherited either from SUVH2 overexpression or SUVH2 antisense plants still shows increased and decreased DNA methylation, respectively (Figure 6A and C). The data suggest that DNA methylation is involved in maintenance of the SUVH2-induced epigenetic effects.
Figure 6.
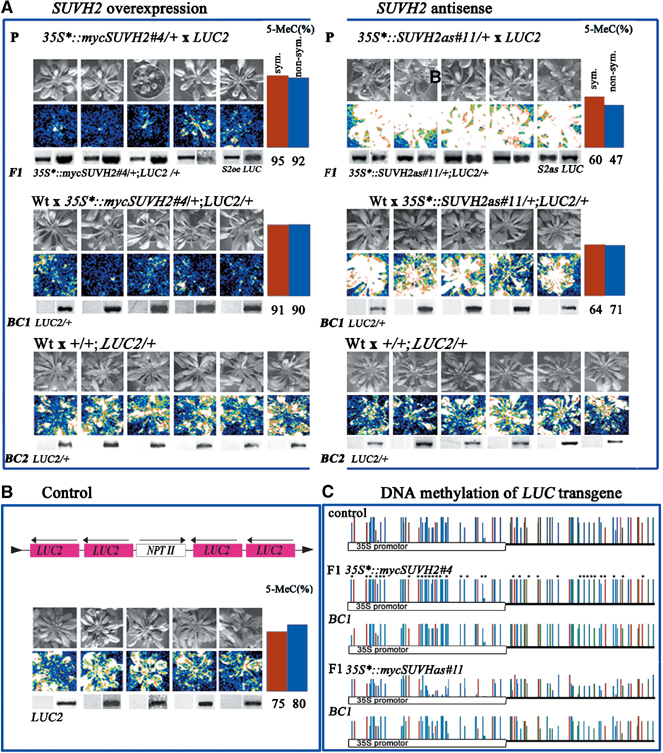
Dosage-dependent modifier effects of SUVH2 on LUC2 transgene silencing. (A–C) Crosses of 35S*∷mycSUVH2#4/+ and 35S*∷SUVH2as#11/+ with LUC2 homozygous plants and backcrosses (BC) of F1 and F2 LUC2/+ plants to wild type (A). Structure and activity of the LUC2 repeated transgene in control plants (B). In 35S*∷mycSUVH2#4/+; LUC2/+ plants with SUVH2 overexpression, LUC2 silencing is enhanced, whereas in 35S*∷SUVH2as#11/+; LUC2/+ plants without SUVH2 LUC2 silencing is strongly released. The repressed or activated state of the LUC2 is maintained after a backcross of 35S*∷mycSUVH2#4/+;LUC2/+ and 35S*∷SUVH2as#11/+;LUC2/+ with wild-type plants, respectively. In LUC2/+ offspring from the second backcross generation, reversion of transgene silencing to the control level is found. Symmetric (red bars) and nonsymmetric (blue bars) DNA methylation at LUC2 transgenes was studied by bisulphite sequencing (A and B). Bisulphite sequence analysis of control LUC2/+, F1 35S*∷mycSUVH2#4/+; LUC2/+ and 35S*∷SUVH2as#11/+; LUC2/+ as well as LUC2/+ BC1 progeny plants. (C) Stars denote significantly changed symmetric CpG (red) and CpNpG (green) and nonsymmetric CpNpN (blue) cytosine residues (N, no G).
Modification of TGS by SUVH2 is connected with complex DNA methylation pattern
We analysed the effect of SUVH2 on DNA methylation at the LUC2 transgene, GUS transgene repeats and Athila transposon sequences (Figure 6 and Supplementary Figure 3). All these sequences show already in wild-type a consistent amount of DNA methylation. In our studies, we compared CpNpN (N, no G) nonsymmetric with CpG and CpNpG symmetric DNA methylation. Loss or overexpression of SUVH2 affects both symmetrical and nonsymmetrical DNA methylation at all studied sequences. In suvh2 mutant as well as SUVH2as#11 antisense plants, strongest reduction of nonsymmetrical methylation at CpC is observed (Supplementary Figure 3).
Since DNA methylase MET1 is suggested to function in maintenance of cytosine methylation (Finnegan and Kovac, 2000), we studied at the completely silenced LUC7 transgene the effects of a newly isolated strong met1-h1 mutation on SUVH2-induced transgene silencing (Figure 7A and B). Furthermore, we also studied interaction between SUVH2 and a newly isolated strong cmt3-h1 CHROMOMETHYLASE mutation (Figure 7C). In LUC7 met1-h1 and LUC7 cmt3-h1 plants, silencing of the LUC7 transgene is significantly released (Figure 7B and C). LUC7 transgene silencing is differentially affected in 35S*∷mycSUVH2#5/+; LUC7/LUC7; met1-h1/met1-h1 and 35S*∷mycSUVH2#5/+; LUC7/LUC7; cmt3-h1/cmt3-h1 plants. The suppressor effect of met1-h1 dominates the enhancer effect of SUVH2 overexpression, whereas the enhancer effect of SUVH2 dominates the suppressor effect of cmt3-h1. These results demonstrate that transgene silencing by SUVH2 depends on MET1, but not significantly on CMT3.
Figure 7.
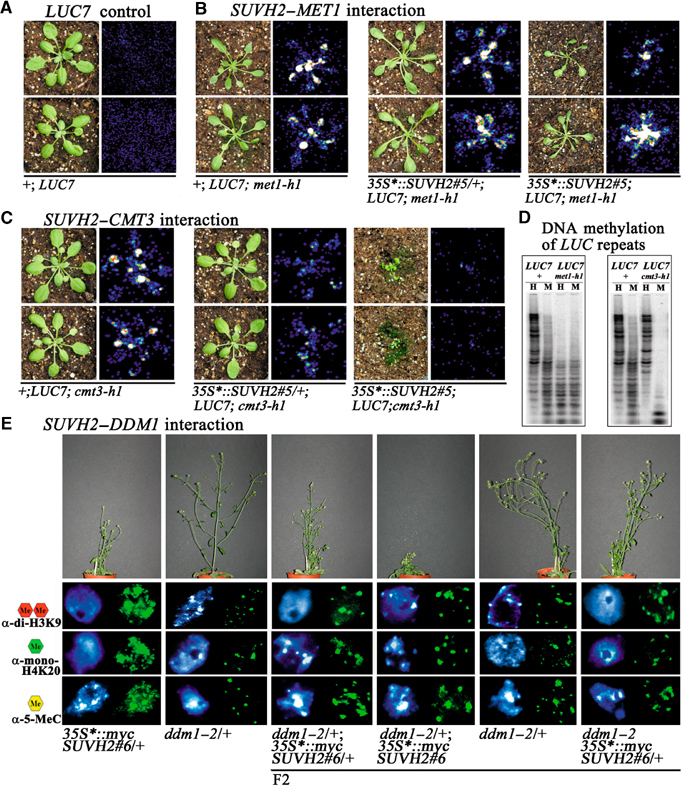
Genetic interaction of SUVH2 with MET1, CMT3 and DDM1. (A) The LUC7 transgene shows complete repression of LUC activity (no fluorescence). (B) Relaxation of gene silencing by met1-h1 dominates the enhancer effect of SUVH2 overexpression and significantly rescues the SUVH2 overexpression mini-plant phenotype. (C) The silencing enhancer effect of SUVH2 overexpression dominates the suppressor effect of cmt3-h1 on LUC7 silencing. (D) The met1-h1 and cmt3-h1 mutations confer reduced CpG and CpNpG methylation, respectively. HpaII (H) and MspI (M) restriction map of LUC7 with LUC as a probe. (E) Suppression of SUVH2-dependent mini-plant phenotype in 35S*∷SUVH2#6/+; ddm1–2 plants and rescue of ectopic dimethyl H3K9, monomethyl H4K20 and 5-methylcytosine methylation.
In addition to DNA methylases, the DDM1 protein, which shows homology to the SWI2/SNF2 family of chromatin remodelling factors, controls DNA methylation independent of their sequence context (Mittelsten Scheid et al, 1998; Jeddeloh et al, 1999). In 35S*∷mycSUVH2#6/+; ddm1–2/ddm1–2 plants, the mini-plant phenotype caused by SUVH2 overexpression as well as ectopic dimethyl H3K9, monomethyl H4K20 and 5-methylcytosine are all significantly suppressed (Figure 7E). Our results suggest that the function of SUVH2 in heterochromatin formation and gene silencing depends on both MET1 and DDM1. Genetic interaction of SUVH2 with different silencing factors also demonstrate that the clear-cut SUVH2 overexpression phenotypes allow systematic genetic dissection of functions involved in the control of heterochromatic gene silencing in Arabidopsis.
Discussion
SUVH proteins function differentially in control of heterochromatin formation
SU(VAR)3–9 proteins are evolutionary conserved and their H3K9 HMTase activity controls heterochromatin formation in eukaryotic organisms (Nakayama et al, 2001; Peters et al, 2001; Tamaru and Selker, 2001; Jackson et al, 2002; Schotta et al, 2002). In Arabidopsis 10 SUVH genes encode SU(VAR)3–9 homologues and sequence analysis suggests that these genes evolved via gene duplications (Baumbusch et al, 2001). It is likely that a duplicated gene, which does not adopt a distinct function, tends to become lost by mutation. Functional difference could be reached by either having different expression patterns or by acquiring distinct functions. As SUVH genes are expressed in all tissues, it is likely that they have adopted at least partially nonoverlapping functions. The first SUVH protein analysed is SUVH4 (KYP), which shows in vitro H3K9 HMTase activity and in vivo is required for CMT3-dependent CpNpG DNA methylation of several transposon and repeat sequences (Jackson et al, 2002; Malagnac et al, 2002; Lindroth et al, 2004). Our analysis could not confirm the report that dimethyl H3K9 is lost from the chromocentre of kyp-2 plants (Supplementary Figure 2; Jasencakova et al, 2003). Analysis of suvh1 and suvh4 null mutations shows only weak reduction of total histone H3K9 dimethylation (Figure 2 and Supplementary Figure 2), whereas all other histone methylation marks or DNA methylation in chromocentres are not significantly changed. This suggests that both SUVH1 and SUVH4 have no primary role in overall heterochromatin organization. Loss-of-function and overexpression of SUVH1 do not affect silencing of repeated LUC or GUS transgenes and therefore these proteins might only affect a restricted number of heterochromatic sequences.
In contrast, the SUVH2 protein has a strong impact on heterochromatin formation and gene silencing. The protein shows dosage-dependent nuclear distribution and affects all heterochromatin-specific histone methylation marks. In vitro SUVH2 methylates H3 and H4, and our data demonstrate that in vivo SUVH2 is one of the major mono- and dimethyl H3K9 and monomethyl H4K20 HMTase. SUVH2 also significantly affects mono- and dimethylation of H3K27. As none of the heterochromatic histone methylation marks are completely lost in the suvh2 null mutant line, other SUVH proteins also contribute to the establishment of these marks. Whether this is mainly connected with target site specificity of different SUVH protein complexes remains to be studied. Finally, differences in HMTase activities, target sequence recognition specificity, differential recruitment of DNA methylases as well as functional interdependence could all contribute to the functional complexity of SUVH protein complexes. In mammals and Drosophila, functional interdependence between the H3K9 HMTase SU(VAR)3–9 and the H4K20 trimethyl specific HMTase SUV4–20 was recently shown (Schotta et al, 2004). In these organisms, binding of HP1 to heterochromatin depends on H3K9 di- and trimethylation by SU(VAR)3–9. HP1 not only binds SU(VAR)3–9 but also recruits the SUV4–20 HMTase to heterochromatin, and therefore a loss of SU(VAR)3–9 activity excludes HP1 heterochromatin association and successive recruitment of the SUV4–20 HMTase.
Proteins binding specific heterochromatic histone methylation marks in vivo are still unknown in plants. In vitro, a high affinity of a CMT3 chromodomain homodimer to lysine 9 and lysine 27 double methylated histone H3 tails was recently shown (Lindroth et al, 2004), indicating that in plants also proteins with differential binding activities to histone methylation marks exist. Complex interactions between such proteins, various SUVH proteins, other SET domain proteins and DNA methylases could finally result in a highly complex network of regulatory interactions.
The interplay between DNA and histone methylation in heterochromatic silencing
Histone H3K9 and DNA methylation represent interrelated marks of repressed chromatin (Martienssen and Colot, 2001; Selker, 2002). Recently, two alternative models are discussed. Either DNA methylation is triggered by histone methylation (Tamaru and Selker, 2001; Jackson et al, 2002; Malagnac et al, 2002; Lindroth et al, 2004) or vice versa histone methylation is initiated after DNA methylation (Johnson et al, 2002; Soppe et al, 2002; Tariq et al, 2003). Our analysis with SUVH2 transgene mutations clearly favours the latter hypothesis. Overexpression of SUVH2 results in silencing of Athila transposons. This effect is completely released with an HMTase inactive SUVH2 protein although Athila sequences remain hypermethylated. Mutant analysis of a SUVH2 transgene revealed an interesting hierarchic sequence of molecular events. N-terminal regions of SUVH2 appear to be involved in target sequence recognition. Specificity of these processes might also depend on a variety of unknown interactors. The YDG domain and possibly a region N-terminal to the preSET region of SUVH2 appear to be involved in recruitment of DNA methylation to target sequences, which is a prerequisite for histone methylation by SUVH2. Preference for CpNpG symmetric methylation is shown for SUVH4-dependent silencing (Jackson et al, 2002; Lindroth et al, 2004), whereas in silencing processes induced by SUVH2 both symmetric and nonsymmetric DNA methylation is involved. This difference is also reflected by dependence of SUVH4-induced silencing on CMT3, whereas SUVH2 is largely independent of CMT3. The epistatic effect of met1-h1 mutation on SUVH2-induced silencing and suppression of ectopic SUVH2 distribution in ddm1–2, 35S*∷mycSUVH2#5 mutant plants shows that SUVH2-induced gene silencing requires both MET1 and DDM1 but not CMT3. Furthermore, in plants a large number of genes encoding MBD proteins is found and these proteins could also be necessary to convey differentially cytosine methylation to histone methylation. Interaction with HDAC complexes was demonstrated for AtMBD5, AtMBD6 and AtMBD7 (Zemach and Grafi, 2003), and MBD proteins might be involved in control of euchromatic as well as heterochromatic chromosomal subdomains. Genetic dissection of gene-silencing processes which depend either on SUVH2 or other SUVH proteins with suppressor and enhancer mutations should contribute substantially to understanding of complex regulatory networks involved in chromatin regulation, heterochromatin formation and gene silencing in plants.
Materials and methods
Plant material and growth conditions
The 35S*∷mycSUVH1, 35S*∷mycSUVH2, 35S*∷mycDmSU(VAR)3–9 and 35S*∷mycDmHP1 Arabidopsis transgenes were introduced with the pBI1-4tr vector. SUVH1 and SUVH2 ORFs were PCR-amplified from genomic DNA using primers Su1start-Su1stop and Su2start-Su2stop. All primers are listed in Supplementary Table 1. SalI fragments (SUVH1 and SUVH2) and the Drosophila BamHI (DmSU(VAR)3–9) and EcoRI–BamHI (DmHP1) fragments (Schotta et al, 2002) were cloned with 3xmyc in frame in sense orientation and in antisense orientation without 3xmyc (35S*∷SUVH1as and 35S*∷SUVH2as transgenes) into pBI1-4tr, which was derived from pBI121 (Clontech) after insertion of a 1.4 kb XbaI–SacI fragment of pKEx4tr (Baumbusch et al, 2001) containing the 35*S promotor (Mindrinos et al, 1994).
For controlled ectopic expression, GVG∷SUVH1EGFP, GVG∷SUVH2EGFP transgenic plants were generated. SalI–NheI fragments of SUVH1, SUVH2 and GFP were cloned in frame into the XhoI–SpeI site of pTA7002 (Aoyama and Chua, 1997). The SalI–NheI EGFP fragment was PCR-amplified from CD3-327 (ABRC Stock Center) with primers GFP-Nhestop and GFPstart. Expression of the SUVH1–GFP and SUVH2–GFP fusion proteins was induced by 0.01 mM dexamethasone (Sigma) treatment. All constructs were confirmed by sequencing using an ABI 377 sequencer.
For studies of repeat-dependent TGS, the pGPTV 2xLUC Kan 2xLUC (LUC2 and LUC7 lines) vector was constructed. The NcoI–XbaI LUC fragment from pSP-LUC (Promega) was cloned into pRT100 (Töpfer et al, 1987) and, subsequently, the LUC PstI fragment from pRT100LUC cloned into the PstI site of partially PstI digested pRT100, resulting in pRT100-2xLUC. After HindIII digestion, the 2xLUC repeat was cloned into pGPTV-Kan, the resulting construct was digested with PstI–HindIII and the PstI–HindIII LUC fragment cloned into the PstI–HindIII site of pGEM-3zf+ (Promega), resulting in pGEM-2xLUC. KpnI–HindIII digestion of pGEM-2xLUC produces a 2xLUC fragment, which was cloned into pBC-pAnos, resulting in pBC-pAnos-2xLUC. After EcoRI–Ecl136II digestion of pBC-pAnos-2xLUC, the 2LUC-nos-ter fragment was inserted into the EcoRI–SmaI site of pBluescript KS (Stratagene). The BamHI fragment of pBluescript 2xLUC-nos-ter was cloned into the BamHI site of pGPTV-Kan 2xLUC producing the pGPTV 2xLUC Kan 2xLUC vector.
The GUS27 line was produced with a pGPTV-3xuidA-Kan vector containing three tandem copies of the uidA gene, provided by Dr Bettina Tschiersch (Institute of Plant Biochemistry, Halle).
All binary vectors were transferred to the Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) and used to transform Arabidopsis ecotype Columbia (Col) plants by vacuum infiltration. T1 seeds were plated on MS medium supplemented with kanamycin or hygromycin. The plates were left at 4°C in the dark for 1 day and then transferred to a growth chamber and incubated at 23°C under continuous light. Plants were transferred to soil after 2 weeks and grown in a chamber with 16-h-light/8-h-dark cycle at 23°C.
The sequence-indexed Arabidopsis T-DNA insertion mutants of SUVH1, SUVH2 and SUVH4 (SALK 003675, 079574, 105816, 130630) were kindly provided by the Salk Institute Genomic Analysis Laboratory. The new cmt3-h1 (P715L) and met1-h1 (E1272K) mutant alleles were selected as suppressors of LUC7 transgene silencing after EMS treatment of LUC7 homozygous plants (I Hofmann, unpublished). The alleles strongly reduce CpNpG and CpG DNA methylation, respectively. The ddm1–2 mutant allele is described in Vongs et al (1993). All mutant material is available upon request.
Genetic analysis
35S*∷mycSUVH2#6 and antisense 35S*∷SUVH2as#11 plants were used as female parent in all crosses with LUC2, LUC7 and GUS27 homozygous lines. Genotypes in offspring were determined with the specific primers 35S-1HindIII, 2RNAi3BamH1, Lu/1626B and Lu8F, Gus-start and Gus-stop or S2-1631B and mycSal1 (Supplementary Table 1). Luciferase activity was monitored with a CCD camera system employing Argus 50 Software (Hamamatsu Photonic Deutschland) after spraying of plants with 1.3mM Luciferin (Molecular Probes). GUS histochemical staining was performed as previously described (Baumbusch et al, 2001).
Functional interaction of SUVH2 with MET1 and CMT3 in transcriptional silencing of the LUC7 transgene was studied in crosses of the 35S*∷mycSUVH2#5 overexpression line with homozygous LUC7; met1-h1 and LUC7; cmt3-h1 mutant plants. F1 35S*∷mycSUVH2#5/+; LUC7/+; met1h-1/+ and 35S*∷mycSUVH2#5/+; LUC7/+; cmt3-h1/+ plants were selfed over two generations and genotypes in F3 plants originated from LUC7 homozygous F2 plants by using the derived cleaved amplified polymorphic sequence primers Met1-6f and Met1-6r or CMT3-eF and CMT3-eR (Supplementary Table 1). Amplified fragments were digested with Eco57I (met1-h1) or BspI (cmt3-h1). In crosses with ddm1–2, the F2 offspring originated from 35S*∷mycSUVH2#6/+; ddm1–2/+ F1 plants were analysed using primers DDM1f and ddm1–2dRsaI (Neff et al, 1998). The 35S*∷mycSUVH2#6 transgene was identified with primers 35S-1HindIII and S2RNAi3X/Bstop.
To screen for SUVH2 transgene mutations, seeds of the 35S*∷mycSUVH2#5 overexpression line were mutagenized with ethylmethane sulphonate (EMS, 90 mM). M2 progeny seedlings were grown on agar medium with kanamycin and scored after 10 days for a non-curled cotyledon phenotype. Putative mutants were transferred to soil and genomic DNA PCR-amplified with SUVH2 transgene-specific primers 35S-1HindIII and S2RNAi3X/Bstop. The SUVH2 transgenes were sequenced using primers S2RNAi3X/Bstop, 2RNAi5-BamH1F, 2RNAi5X B and 2RNAi3BamH1.
Immunocytology and EM analysis
For all immunocytological analyses, young rosette leaves from Arabidopsis ecotype Columbia plants were used. Leaf pieces were fixed in 4% formaldehyde in PBS on a glass slide, chopped, covered with coverslips and squashed. Slides were freezed in liquid nitrogen and transferred immediately into PBS after removing the coverslips. The slides were pre-incubated for 30 min at 37°C in 1% BSA in PBS and incubated with the following antibodies (1:100): α-mono-, α-di- and α-trimethyl H3K9, α-mono-, α-di- and α-trimethyl H3K27, α-di- and α-trimethyl H3K36, α-di- and α-trimethyl H4K20, α-dimethyl H3K4, α-acetyl H3K9, α-monomethyl H4K20 (Upstate). Detection of antibodies was performed with Alexa-488 conjugated secondary antibodies (1:200; Molecular Probes).
For detection of myc-tagged fusion proteins and 5-methylcytosine, leaves were fixed for 1 h in cold methanol:acetic acid (3:1), washed in citrate buffer (0.01 M citric acid monohydrate, 0.01 M trisodium citrate-2-hydrate, pH 4.8) and digested with 20% pectinase (Onozuka R-10) and 2% cellulase (Merk) in citrate buffer at 37°C for 30 min. The preparations were washed for 30 min in citrate buffer, squashed and coverslips were removed after freezing on dry ice.
For detection of myc-tagged fusion proteins, slides were washed in PBS, preincubated for 30 min at 37°C in 5% dry milk/PBS and incubated with mouse monoclonal α-myc (1:30; Labvision).
For detection of 5-methylcytosine, slides were baked after enzyme treatment at 60°C for 30 min, denatured in a 70% formamide, 2 × SSC, 50 mM sodium phosphate buffer (pH 7.0), washed in ice-cold PBS and preincubated in 1% BSA in PBS for 30 min at 37°C, followed by incubation with mouse monoclonal α-5-methylcytosine (1:400; Eurogenetech). For antibody detection Alexa-488 conjugated secondary antibodies (1:200; Molecular Probes) were used. All preparations were washed in PBS, stained with DAPI in mounting solution and examined with Zeiss Axioscop fluorescence microscope.
For electron microscopic analysis, plant material was processed as described previously (Hause and Hahn, 1998). Ultrathin sections were observed with an EM900 transmission electron microscope (Carl Zeiss NTS).
In vitro histone methyltransferase assay
GST constructs used for the assays were Arabidopsis SUVH2 amino acids 291–651 and Drosophila SU(VAR)3–9 amino acids 305–635 (Schotta et al, 2002). Generation and purification of the GST fusion proteins and histone methyltransferase assay were performed as described previously (Rea et al, 2000), with about 2 μg recombinant protein and 1 μg reconstituted recombinant nucleosomes.
Bisulphite sequencing
Cytosine methylation was assayed by the bisulphite genomic sequencing (Grunau et al, 2001) with 2 μg of EcoRI digested and alkali denatured genomic DNA. Denatured DNA was incubated in 1.2 ml freshly prepared 3.1 M sodium bisulphite/0.5 mM hydoquinone (Sigma), pH 5.0 at 95°C for 1 h, followed by desalting (QIAaexII, Quiagen), desulphonation and neutralization. DNA was precipitated, resuspended in 100 μl of 1 mM Tris, pH 8.0 and stored at −20°C. Gene fragments were PCR amplified and cloned into pGEM (Promega) for sequencing. For each genotype, 10–15 independent clones were analysed.
Protein isolation and Western blot analysis
Proteins from approximately 1 g leaf tissue were isolated as described (Tariq et al, 2003), separated on SDS–15% polyacrylamide gels and visualized by Coomassie Brilliant Blue staining or transferred to nitrocellulose membranes.
Membranes were used for immunodetection with α-dimethyl H3K9, and α-monomethyl-H4K20 (Upstate). Detection of primary antibodies was performed with peroxidase-conjugated secondary antibodies using the ECL kit (Amersham).
Supplementary Material
Supplementary Material
Acknowledgments
We thank Dr G Schotta for helpful comments on the manuscript. We are grateful to Dr M Lachner for nucleosome preparations, C Fiedler and A Thuemmler for their help in genotyping of plants, K Kittlaus and R Franke for excellent technical assistance. This work was supported by grants from SFB363 of the Deutsche Forschungsgemeinschaft.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T (1999) Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13: 627–637 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen R (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionary conserved classes. Nucleic Acid Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G (2004) Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev 18: 2973–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA (2000) Plant DNA methyltransferases. Plant Mol Biol 43: 189–201 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003) Extending the histone code: modification cassettes and switches. Nature 425: 475–479 [DOI] [PubMed] [Google Scholar]
- Fransz P, Armstrong S, de Jong JH, Parnell LD, van Drunen C, Dean C, Zabel P, Bisseling T, Jones GH (2000) Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere regions. Cell 100: 367–376 [DOI] [PubMed] [Google Scholar]
- Gendrel A-V, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomiee P, Martin M, Long D, Meyerowitz E, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998) Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Grunau G, Clark SJ, Rosenthal A (2001) Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acid Res 29: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause G, Hahn H (1998) Cytological characterization of multicellular structures in embryogenic microspore cultures of Brassica napus L. Bot Acta 111: 204–211 [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJS (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet 19: 192–195 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jasencakova Z, Soppe WJJ, Meister A, Gernand D, Turner B, Schubert I (2003) Histone modification in Arabidopsis—high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33: 471–480 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ (1999) Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G (1998) SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 54: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Vao X, Jacobsen SE (2002) Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promotor of TL-DNA gene 5 controls the tissue specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lindroth MA, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE (2004) Dual histone H3 methylation marks at lysine 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4146–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J (2002) An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Colot V (2001) DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070–1074 [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Asubel FM (1994) The A. thaliana disease resistance gene RPS encodes a protein containing nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Asfar K, Paszkowski J (1998) Release of epigenetic silencing by trans-acting mutations in Arabidopsis. Proc Natl Acad Sci USA 95: 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JD, Strahl BD, Allis CD, Grewal SIS (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE (1998) DCAPS a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acid Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AHFM, Kubicek S, Mechtler K, O'Sullivan J, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JHA, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Peters AHFM, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39h histone methyltransferase impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun Z-W, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Reuter G (2002) Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 tri-methylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU (2002) Repeat-induced gene silencing in fungi. Adv Genet 46: 439–450 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz P (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Tariq M, Saze H, Probst A, Lichota J, Habu Y, Paszkowski J (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res 15: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J 13: 3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calcada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U (1999) Maintenance of genomic imprinting at the Arabidopsis MEDEA locus requires zygotic DDM1 activity. Genes Dev 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Matienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Zemach A, Grafi G (2003) Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J 34: 565–572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


