Abstract
SCF-type (SCF: Skp1–Cullin–F-box protein complex) E3 ligases regulate ubiquitin-dependent degradation of many cell cycle regulators, mainly at the G1/S transition. Here, we show that SCFGrr1 functions during cytokinesis by degrading the PCH protein Hof1. While Hof1 is required early in mitosis to assemble a functional actomyosin ring, it is specifically degraded late in mitosis and remains unstable during the entire G1 phase of the cell cycle. Degradation of Hof1 depends on its PEST motif and a functional 26S proteasome. Interestingly, degradation of Hof1 is independent of APCCdh1, but instead requires the SCFGrr1 E3 ligase. Grr1 is recruited to the mother–bud neck region after activation of the mitotic-exit network, and interacts with Hof1 in a PEST motif-dependent manner. Our results also show that downregulation of Hof1 at the end of mitosis is necessary to allow efficient contraction of the actomyosin ring and cell separation during cytokinesis. SCFGrr1-mediated degradation of Hof1 may thus represent a novel mechanism to couple exit from mitosis with initiation of cytokinesis.
Keywords: cytokinesis, Grr1, Hof1/Cyk2, SCF, ubiquitin-dependent degradation
Introduction
Ubiquitin-dependent degradation of key proteins has emerged as an important mechanism to control numerous biological responses, including signal transduction, morphogenesis and cell cycle progression. Protein destruction is triggered by the covalent attachment of ubiquitin onto lysine residues of substrates, which targets them for degradation by the 26S proteasome (Ciechanover and Schwartz, 2002). Three classes of enzymes are required to mediate these reactions: the E1 ubiquitin-activating enzyme, the E2 ubiquitin-conjugating enzymes and the E3 ubiquitin ligases (Peters, 1998). The anaphase-promoting complex (APC) and Skp1–Cullin–F-box protein complex (SCF) are multiprotein E3 ligases that were discovered because of their essential roles in the regulation of the cell cycle (Peters, 1998). APC is thought to be involved in late cell cycle events including entry into anaphase and exit from mitosis. In budding yeast, entry into S phase requires SCFCdc4-dependent degradation of the cyclin-dependent kinase inhibitor Sic1 (Schwob et al, 1994; Verma et al, 1997). SCFCdc4 is composed of the cullin Cdc53, the E2-conjugating enzyme Cdc34, the ring-finger protein Hrt1 and Skp1, which binds to the F-box motif present in a large family of proteins (Deshaies, 1999). It is thought that F-box proteins function as adaptor subunits that recruit specific substrates to a core ligase complex composed of Cdc53, Cdc34, Hrt1/Rbx1 and Skp1 (Patton et al, 1998; Tyers and Jorgensen, 2000). Besides Sic1, SCFCdc4 is also required for the degradation of Far1, Cdc6 and Gcn4 (Drury et al, 1997; Henchoz et al, 1997; Skowyra et al, 1999; Meimoun et al, 2000). In contrast, ubiquitination of the transcription factor Met4 is dependent on SCFMet30 (Kaiser et al, 2000; Patton et al, 2000; Rouillon et al, 2000), while degradation of the G1 cyclins Cln1 and Cln2 (Barral et al, 1995; Seol et al, 1999; Skowyra et al, 1999) and the bud emergence protein Gic2 (Jaquenoud et al, 1998) is mediated by SCFGrr1.
Substrate phosphorylation and the subcellular localization of F-box proteins regulate temporal and spatial aspects of protein degradation. Many SCF substrates need to be phosphorylated for subsequent ubiquitination (Deshaies, 1997), and in some cases, phosphorylation of multiple sites is required to allow substrate binding to the F-box protein, thereby setting a threshold for kinase activity before abrupt degradation is triggered (Nash et al, 2001). While the shared core components of the SCF complex are found in the nucleus and cytoplasm (Blondel et al, 2000), the localization of F-box proteins provides an important determinant for the spatial control of substrate degradation. Cdc4 is localized and active only in the nucleus, thereby ensuring that cytoplasmic Far1 is stable during mating (Blondel et al, 2000). Likewise, the mammalian F-box protein Skp2 is nuclear and accumulates on centrosomes in human cells (Gstaiger et al, 1999). These results suggest that the subcellular localization of F-box proteins may provide important clues to their function and may help to identify potential substrates. The F-box protein Grr1 is found both in the nucleus and the cytoplasm. In addition, Grr1 also accumulates at the bud neck late in mitosis (Blondel et al, 2000), implying that it may play a role during cytokinesis by degrading an as yet unknown substrate.
Genetic experiments revealed two redundant pathways for cytokinesis in budding yeast. The first pathway involves the type II myosin Myo1 and the formin Bni1, and is required for the assembly of a functional actomyosin-based contractile ring. The second pathway controls cell separation and septum formation and involves a number of proteins including the formin-related protein Bnr1 and Hof1/Cyk2. Hof1 is a member of the PCH family of proteins (Lippincott and Li, 2000), which contain an N-terminal FER-CIP4 homology (FCH) domain flanked by coiled-coil domains, PEST sequences and one or more SH3 domains in their C-termini. Both Schizosaccharomyces pombe Cdc15 and Imp2 and Saccharomyces cerevisiae Hof1 localize to the actin ring and are important for the completion of cytokinesis. It has been suggested that Hof1 could play a role in coupling the actomyosin system to septum formation (Vallen et al, 2000). Alternatively, Hof1 could act by modulating the stability of the actomyosin ring during contraction (Lippincott and Li, 1998). Interestingly, overexpression of Hof1 in S. cerevisiae or Cdc15 in S. pombe has been shown to interfere with cell separation (Fankhauser et al, 1995; Lippincott and Li, 1998; Vallen et al, 2000). Similar results were obtained when overexpressing the mouse homolog PSTPIP in S. pombe (Spencer et al, 1997).
In this study, we have investigated a role of Grr1 during cytokinesis. We found that SCFGrr1 is required to degrade Hof1 on the cytokinesis ring. Degradation of Hof1 is important for efficient contraction of the actomyosin ring and cell separation during cytokinesis. Because localization of Grr1 on the mother–bud neck and degradation of Hof1 require activation of the mitotic-exit network (MEN), SCFGrr1 may couple mitotic exit and completion of cytokinesis.
Results
Accumulation of Grr1 at the bud neck requires activation of the MEN
We have shown previously that a functional Grr1-GFP fusion protein localizes to the cytoplasm and nucleus, and also accumulates at the mother–bud neck late in mitosis (Blondel et al, 2000). To determine when Grr1 is recruited to the mother–bud region, we performed time-lapse microscopy in wild-type cells expressing Grr1-GFP and the spindle marker CFP-Tub1. Grr1-GFP was absent from the mother–bud region during most of the cell cycle, and accumulated on the cytokinesis ring shortly after disassembly of the mitotic spindle (Figure 1A). Quantification of more than 10 cells in at least five independent movies revealed that Grr1-GFP transiently accumulated at the mother–bud neck shortly after disassembly of the mitotic spindle, but before cell separation as determined by DIC analysis (panel B). Consistent with these results, Grr1-GFP was not found at the mother–bud neck in cdc16-1 cells (data not shown), which arrest at the metaphase–anaphase transition. Grr1-GFP was also absent from the mother–bud neck in cdc14-3 (data not shown) and cdc15-1 arrested cells (panel C), suggesting that activation of the MEN may be necessary for its recruitment to the mother–bud neck. As a control, the localization of Hof1 at the mother–bud neck was normal, implying that the actin ring region is properly assembled under these conditions. Taken together, these results indicate that Grr1 is transiently recruited to the mother–bud neck region in a MEN-dependent manner, suggesting that Grr1 may degrade a substrate involved in cytokinesis.
Figure 1.
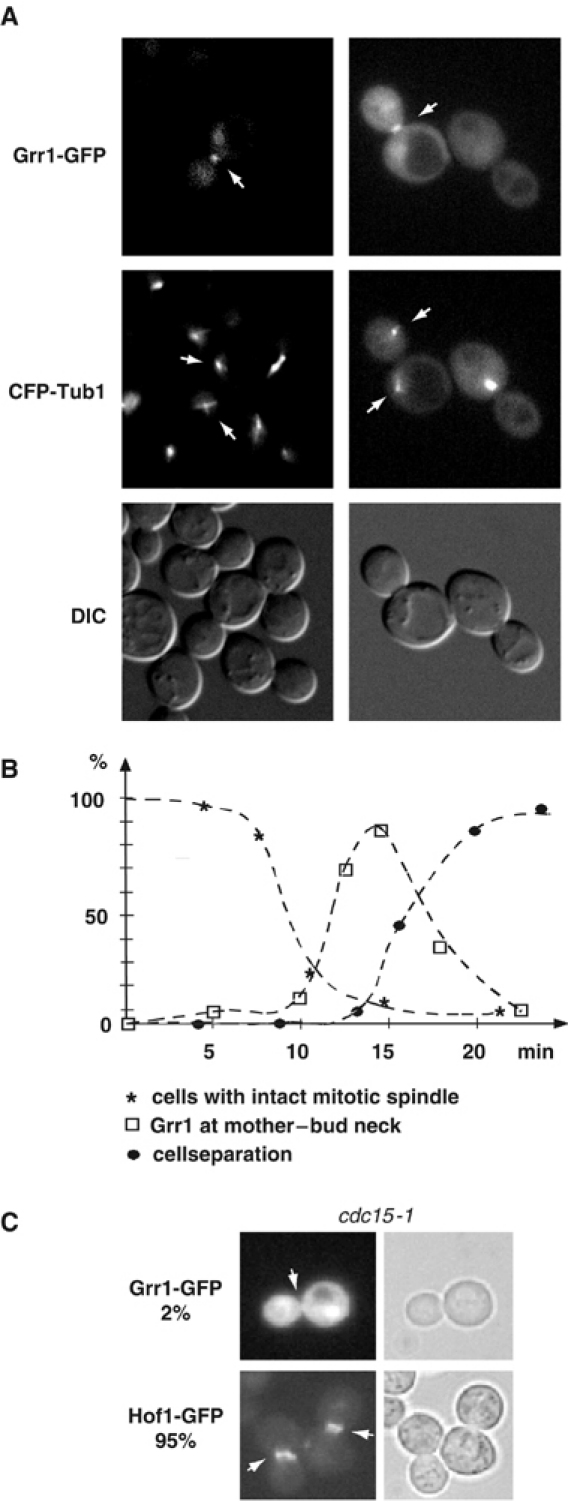
Accumulation of Grr1-GFP at the mother–bud neck occurs after spindle breakdown in a MEN-dependent manner. (A) Wild-type cells were analyzed microscopically for their localization of Grr1-GFP and the status of the mitotic spindle using CFP-Tub1. The arrows mark the position of Grr1 at the mother–bud neck and the mitotic spindle. (B) Time-lapse analysis of wild-type cells (n=10) expressing CFP-Tub1 and Grr1-GFP. Time 0 represents the onset of anaphase as determined by CFP-Tub1. Cell separation was determined by DIC microscopy, as the separated G1 cells slightly tilt after separation. (C) The Localization of Grr1-GFP (upper panel) and Hof1-GFP (lower panel) was analyzed in cdc15-1 cells at the restrictive temperature (37°C). The numbers indicate the percentage (%) of cells with Grr1-GFP (upper panel) or Hof1-GFP (lower panel) at the mother–bud neck region.
The localization of Grr1-GFP at the mother–bud requires a functional leucine-rich domain but not its F-box or nuclear localization signals
To determine the domains responsible for the different subcellular localizations of Grr1-GFP, we analyzed the localization of various mutant constructs as schematically indicated in Figure 2A. The primary sequence of Grr1 can be subdivided into four domains: an N-terminal domain (1–310), the F-box (311–361), the 12 leucine-rich repeat (LRR, 413–740) and a C-terminal domain (740–1150). Interestingly, neither an intact F-box nor the C-terminal domain was required for the subcellular localization of Grr1 (panel A). These Grr1 mutant proteins efficiently accumulated in the nucleus and were specifically targeted to the mother–bud region in late mitosis. In contrast, deletion of the N-terminal domain prevented the accumulation of Grr1-GFP in the nucleus. Grr1 deletion mutants lacking 179 or 311 amino-terminal amino acids (Figure 2A and B, and data not shown) were predominantly cytoplasmic throughout the cell cycle. However, these mutants still localized to the mother–bud neck region late in mitosis, indicating that the N-terminal domain is not required for ring localization of Grr1 but rather contains sequences necessary for its nuclear import. The N-terminal domain (amino acids 1–311) contains two potential bipartite nuclear localization signals (NLS) beginning at position 152 (RKQIHEFKLIVGKKIQEFLVVIEKRRKKE; Figure 2A). Indeed, the N-terminal domain of Grr1 was sufficient to target GFP to the nucleus (panel A), suggesting that the amino-terminal domain is necessary and sufficient for nuclear accumulation of Grr1.
Figure 2.
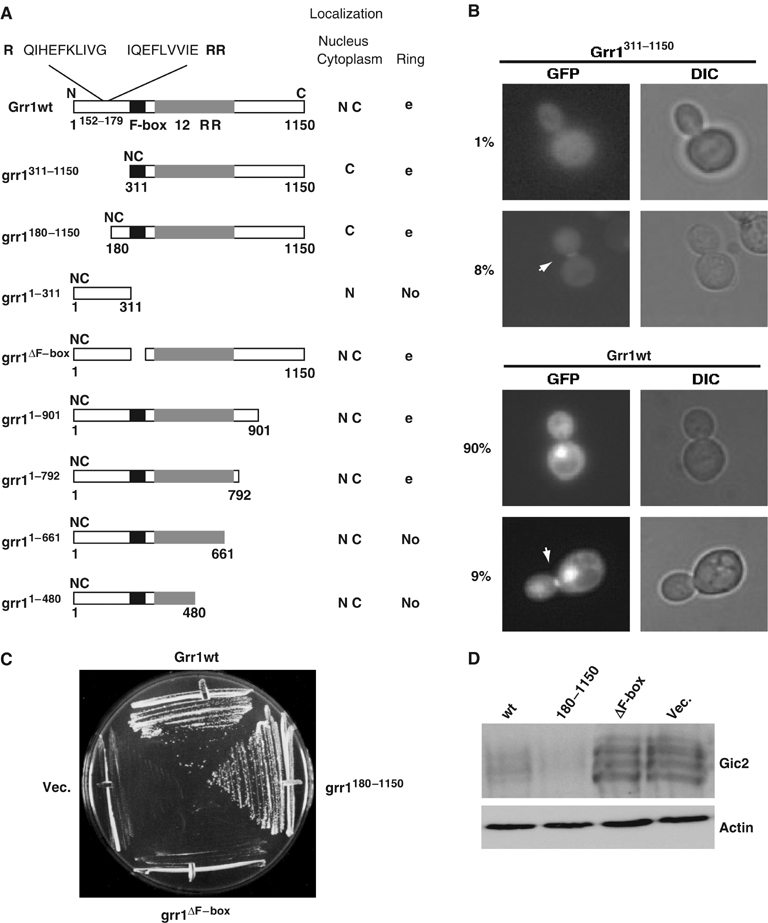
Grr1 domains required for function and subcellular localization. (A, B) Schematic representation of wild-type and various mutant forms of Grr1 (left panel). Black bar: F-box; gray bar: LRR. Their subcellular localization was determined microscopically using GFP fusions as indicated on the right. N: nuclear; C: cytoplasmic. The ability of wild-type and mutant GFP-Grr1 proteins to accumulate at the mother–bud neck region (ring) during cytokinesis was analyzed in at least 200 cells: ‘yes' indicates ring localization observed in 5–10% of cells in late mitosis, ‘no' indicates no accumulation of the GFP-Grr1 mutant protein detectable. The localization of wild-type Grr1-GFP and cytoplasmic Grr1311−1150-GFP is shown in panel B. The numbers indicate the percentage (%) of cells (n>200) with Grr1311−1150-GFP (upper panel) or wild-type Grr1-GFP (lower panel) with nuclear accumulation (upper images), or accumulation at the mother–bud neck region of cells in late mitosis as determined by DIC (lower images). The arrows mark the position of Grr1-GFP and Grr1311−1150-GFP at the mother–bud neck during cytokinesis. (C, D) Wild-type Grr1, Grr1-ΔF-box or cytoplasmic Grr1180−1150 expressed from the ADH promoter were analyzed for their ability to complement growth of grr1Δ cells overexpressing Gic2 (C), and degradation of Gic2 in grr1Δ cells by Western blot (D). An empty vector (Vec.) was included for negative control.
We also tested the importance of the 12 LRR for the subcellular localization of Grr1-GFP. This domain, encompassing amino acids 413 and 740, was previously described as the substrate interacting region of Grr1 and is predicted to fold into a characteristic horseshoe structure with a high density of positive charges on the concave surface (Hsiung et al, 2001). Based on structural predictions, deletion of a single LRR is expected to abolish its interactions with substrates (Hsiung et al, 2001). Interestingly, Grr1 mutant proteins lacking even small portions of the LRR domain failed to localize to the mother–bud region, while their nuclear accumulation was unaffected (Figure 2A, grr11−661 and grr11−480). We conclude that the putative substrate-interactive region of Grr1 is essential for its localization to the mother–bud neck region, suggesting that Grr1 could degrade one (or more) substrate(s) at the cytokinesis ring.
In addition to the subcellular localization, we also examined the function of the various Grr1 constructs in vivo (Figure 2C and D). grr1Δ cells exhibit pronounced morphological defects and accumulate as hyperpolarized, highly elongated cells (Blacketer et al, 1995), most likely due to their failure to degrade the G1 cyclins Cln1 and Cln2 (Barral and Mann, 1995). Likewise, grr1Δ cells fail to degrade the Cdc42 effector Gic2 after bud emergence (Jaquenoud et al, 1998), and as a result, overexpression of Gic2p is toxic in grr1Δ cells. As expected, both the Grr1 mutant lacking the F-box (grr1-ΔF) and the Grr1 mutant lacking LRR repeats were unable to complement these defects (Figure 2C and D, and data not shown). In contrast, cytoplasmic grr1-ΔN (grr1180−1150) was able to restore both growth of grr1Δ cells overexpressing Gic2 and degradation of Gic2 (panels C and D). Likewise, the morphology of grr1Δ cells expressing cytoplasmic Grr1-ΔN was normal (data not shown). These results imply that cytoplasmic Grr1 is sufficient to restore the function of Grr1 with respect to Gic2 degradation and its morphological functions, implying that ubiquitinylation of these targets may occur outside the nucleus.
Hof1 binds Grr1 and this interaction depends on its PEST domain
To confirm that Grr1 plays a role in cytokinesis, we searched for potential substrates located at the mother–bud neck. Interestingly, a genome-wide two-hydrid screen has previously detected an interaction between Grr1 and the PCH protein Hof1 (Ito et al, 2001). Indeed, Hof1 accumulates at the mother–bud neck shortly after bud emergence, and abruptly disappears at the end of mitosis concomitant with the appearance of Grr1 (Figure 4A; Vallen et al, 2000). We confirmed the two-hybrid interaction between Grr1 and Hof1 using two different two-hybrid systems (Figure 3A). To address the functional importance of this interaction, we attempted to identify a functional Hof1 mutant that fails to interact with Grr1. Several Grr1 targets including Cln1 and Cln2 contain functional PEST domains (Lanker et al, 1996), which are commonly found in unstable protein. Interestingly, the PEST-Find program (Rechsteiner and Rogers, 1996) revealed a high-scoring PEST domain (score of 11.85) between amino acids 418 and 438 of Hof1 (Figure 3A). To determine the significance of this PEST motif, we constructed a Hof1 mutant lacking this domain (hof1ΔPEST). This mutant was found to be functional as it was able to suppress the thermosensitivity of hof1Δ cells when expressed from the endogenous HOF1 gene promoter on a low-copy number plasmid (data not shown). Moreover, as for Hof1wt, overexpression of hof1ΔPEST from the inducible GAL1,10 promoter was able to block cytokinesis (Figure 6B). Importantly, the interaction between hof1ΔPEST and Grr1 was greatly reduced in both two-hybrid systems (Figure 3A). Taken together, these results confirm that Grr1 binds Hof1, and suggest that a functional PEST domain of Hof1 is required for this interaction.
Figure 4.
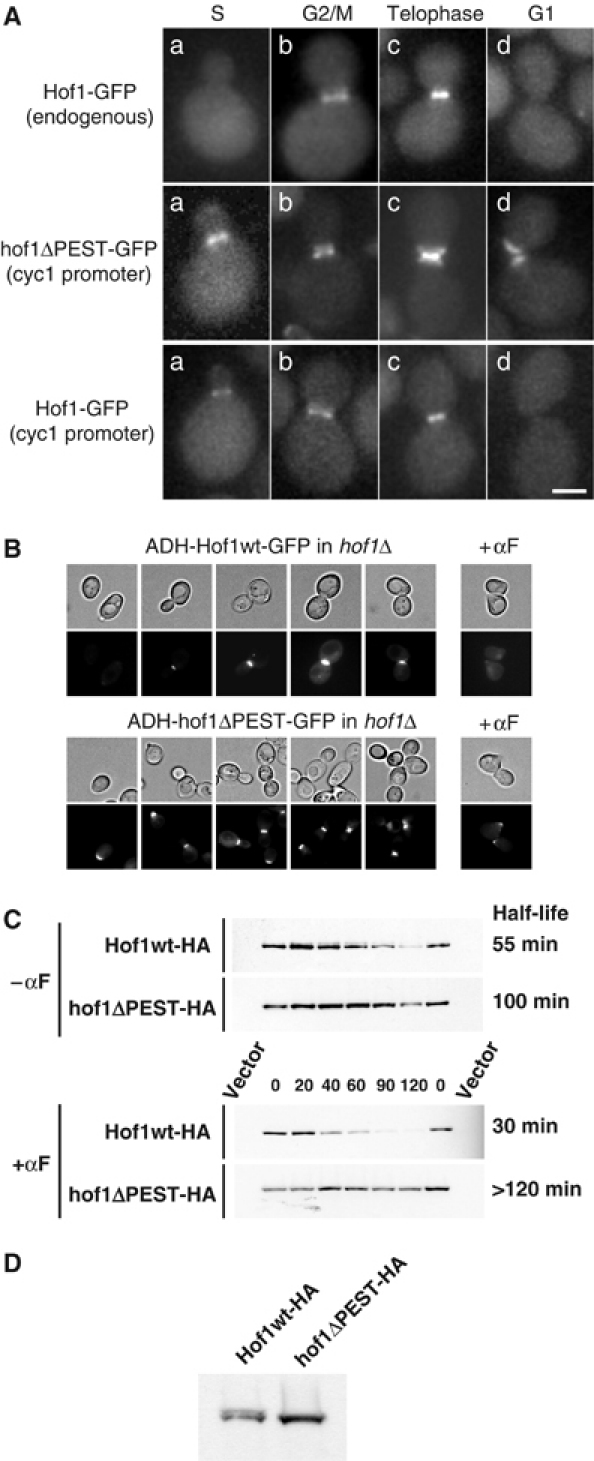
Cell cycle-dependent degradation of Hof1. (A, B) The localization of Hof1-GFP and hof1ΔPEST-GFP expressed from either the endogenous promoter or the constitutive CYC1 (A) or ADH promoters (B) was analyzed in hof1Δ cells (YSB100) by GFP microscopy at different cell cycle stages. The scale bar in panel A represents 2 μm. Where indicated, the cells were arrested with α-factor (+αF) for 90 min. (C) The half-life of Hof1-HA and hof1ΔPEST-HA in wild-type cells (K699) was determined by GAL shutoff experiments as described in Materials and methods. Times are listed in minutes after the addition of glucose (time 0) to turn off expression of Hof1-HA and hof1ΔPEST-HA. Where indicated, the cells have been arrested in G1 phase by α-factor (+αF). An empty vector was used to control for the specificity of the Western blot. The half-life (in minutes) on the right was averaged from at least three independent experiments. (D) The phosphorylation status of Hof1-HA and hof1ΔPEST-HA expressed from the ADH promoter was determined by Western blot analysis of extracts prepared from asynchronous hof1Δ cultures.
Figure 3.
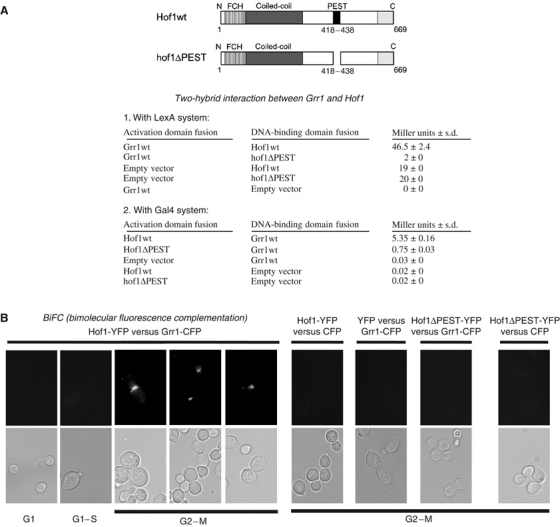
Interaction of Hof1 with Grr1 in a PEST-dependent manner. (A) Schematic representation of wild-type Hof1 and hof1ΔPEST. The various functional domains (FCH, coiled-coil, PEST and SH3) are indicated. Two-hybrid analysis between Grr1 and wild type and hof1ΔPEST using either the LexA (upper panel) or Gal4 system (lower panel). The numbers indicate Miller units (MU) with standard deviations (s.d.) of liquid β-galactosidase reporter assays determined from at least four independent experiments. (B) BiFC analysis of Grr1 and Hof1. Wild-type cells (K699) expressing Grr1 fused to CFP-C (Grr1-CFP) and Hof1 fused to YFP-N (Hof1-YFP) were analyzed by GFP microscopy. Cells expressing Grr1-CFP and YFP-N alone, and Hof1-YFP or hof1ΔPEST-YFP and CFP-C alone were included for control.
Figure 6.
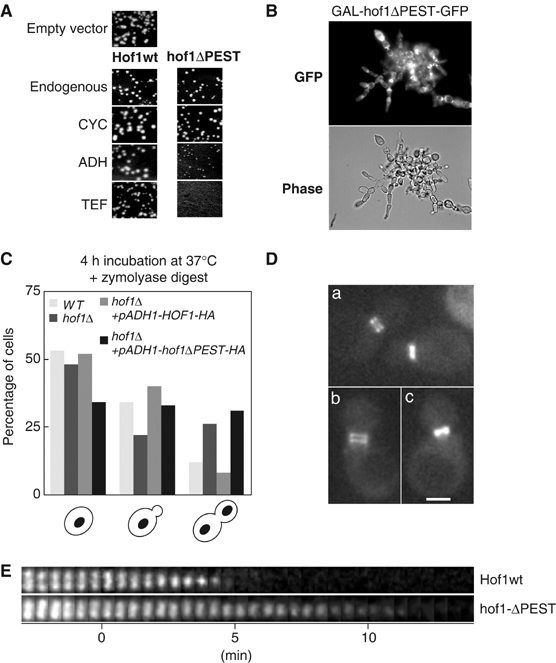
Stabilization of Hof1 inhibits efficient actomyosin contraction during cytokinesis. (A) Wild-type cells (K699) were transformed with the same quantity of an empty control vector (empty vector), or plasmids expressing either Hof1-GFP or hof1ΔPEST-GFP from the constitutive CYC (pBM360 and 365), ADH (pBM361 and 366) or TEF (pBM362 and 367) promoters. Sectors of the transformation plates were photographed after 3 days at 30°C. (B) Wild-type cells (K699) expressing hof1ΔPEST-GFP from the inducible GAL promoter were grown to early log phase in 2% raffinose, and photographed 24 h after addition of 2% galactose to induce expression of hof1ΔPEST-GFP. (C) Wild-type (S288C) or isogenic hof1Δ cells (DJ640), and hof1Δ cells with integrated plasmids expressing as indicated either Hof1-HA (DJ1600) or hof1ΔPEST-HA (DJ1608) from the ADH promoter were shifted to 37°C for 4 h, fixed, treated with zymolyase for 30 min and vortexed. Cell cycle stages were counted as shown in the graph. Note that expression of stabilized hof1ΔPEST-HA increases the number of cells arrested in cytokinesis. (D) The splitting of the septin ring was monitored using GFP-Cdc3 in cells expressing pADH1-HOF1-HA (a, DJ1600) cells and cells expressing pADH1-hof1ΔPEST-HA (b and c, DJ1608). The scale bar represents 2 μm. (E) Actomyosin ring (Myo1-CFP) assembly and contraction were analyzed in cells expressing pADH1-HOF1-HA (DJ1619, upper panel) and cells expressing pADH1-hof1ΔPEST-HA (DJ1618, lower panel). Cell cycle progression was monitored using a GFP-tagged septin (GFP-CDC3). Splitting of the septin ring marks the start point (0 min) and pictures were taken every 30 s.
To test whether Grr1 and Hof1 interact in vivo, we made use of the bimolecular fluorescence complementation (BiFC) system (Hu et al, 2002). Grr1 was fused to the amino-terminal half of YFP (termed YFP-N), while Hof1 was fused to the carboxy-terminal half of CFP (CFP-C). The GFP-chromophore is only restored if YFP-N and CFP-C are brought together by interaction of two proteins fused to the two respective halves. As shown in Figure 3B, coexpression of Grr1p-YFP-N and Hof1-CFP-C restored GFP fluorescence specifically on the mother–bud neck late in the cell cycle. This interaction was specific as no GFP signal was detected in cells expressing either YFP-N or CFP-C alone, or CFP-C fused to another protein unable to interact with Grr1 (not shown). Moreover, no GFP signal was observed after coexpression of Grr1p-YFP-N and hof1ΔPEST-CFP-C, further demonstrating that the interaction of Grr1 and Hof1 requires an intact PEST domain. Importantly, no GFP signal was observed during the G1, S and G2 phases of the cell cycle in cells coexpressing Grr1p-YFP-N and Hof1-CFP-C, demonstrating that Grr1 and Hof1 interact in vivo in a spatially and temporally controlled manner.
Cell cycle-dependent degradation of Hof1 but not hof1ΔPEST
Like other members of the CLB2 cluster (Spellman et al, 1998), HOF1 transcription gradually increases from S phase to mitosis and peaks around anaphase. Indeed, Hof1-GFP expressed from its endogenous promoter was absent during the G1 phase of the cell cycle, and assembled at the mother–bud neck only after S phase (Figure 4A; Vallen et al, 2000). Hof1-GFP expressed from the constitutive CYC1 or ADH promoters was also absent in unbudded G1 cells, while it assembled at the mother–bud region shortly after bud emergence (Figure 4A and B). The mother–bud neck signal increased in large budded cells and then abruptly disappeared during cytokinesis. The premature accumulation of Hof1-GFP expressed from the CYC1 promoter but not the endogenous promoter confirms that the transcriptional regulation of the HOF1 promoter contributes to its G2/M-specific expression in vivo. Importantly however, these data imply that the absence of Hof1 in late mitosis and the G1 phase of the cell cycle must be predominantly regulated at the post-transcriptional level. While wild-type Hof1-GFP was undetectable in most G1 cells, hof1ΔPEST-GFP expressed from the constitutive CYC1 or ADH promoters was present at all stages of the cell cycle and accumulated in a ring-like structure at the cell cortex of G1 cells (Figure 4A and B). This difference was also evident in α-factor-arrested G1 cells. Taken together, these data strongly suggest that Hof1, but not hof1ΔPEST-GFP, is degraded during the G1 phase of the cell cycle.
To measure directly the half-life (t1/2) of Hof1, we expressed HA- and GFP-tagged Hof1 and hof1ΔPEST from the GAL1,10 promoter, which allows a transcriptional shutoff by the addition of glucose. In asynchronous cells (−αf), HA- and GFP-Hof1 were degraded with a half-life of about 55 min (Figure 4C, and data not shown). Similar results were obtained when the half-life of HA-Hof1 expressed in the normal chromosomal context was determined after blocking protein translation by the addition of cycloheximide (CX, data not shown). When cells were synchronized in G1 by α-factor treatment (+αf), HA- and GFP-Hof1 were degraded with a half-life of about 30 min (Figure 4C, and data not shown). These results are consistent with the microscopy analysis presented above, and suggest that Hof1 is degraded between the end of mitosis and bud emergence. In contrast, GFP- and HA-hof1ΔPEST were stabilized at all cell cycle stages (Figure 4A–C), with a half-life of more than 120 min in α-factor-arrested cells. In asynchronous cells, slower migrating forms of HA-Hof1 corresponding to phosphorylated species were detectable (Figure 4C and D; Vallen et al, 2000). Such phosphorylated species were not observed with HA-hof1ΔPEST, suggesting that the PEST domain of Hof1 may be important for phosphorylation. Taken together, these results demonstrate that Hof1 is rapidly degraded during the G1 phase of the cell cycle by a PEST- and perhaps phosphorylation-dependent mechanism.
Degradation of Hof1 during G1 requires the 26S proteasome and SCFGrr1
Because Grr1 interacted with Hof1 in a PEST-dependent manner, we next examined the expression of wild-type Hof1-GFP in cells defective for components of the SCFGrr1 complex. Interestingly, we were unable to obtain grr1Δ cells expressing Hof1-GFP from the constitutive ADH promoter, probably because it is toxic without efficient degradation. However, although the cells grew very poorly, we were able to analyze grr1Δ cells expressing Hof1-GFP from the weaker CYC promoter. Interestingly, strong Hof1-GFP staining was observed in grr1Δ cells at all cell cycle stages including G1 (Figure 5A). Similarly, Hof1-GFP was present in G1 in cdc34-2 and cdc53-1 cells grown at the semipermissive temperature of 30°C (panel B, and data not shown). In contrast, Hof1-GFP was absent in G1 cells in cdc4-1 cells (panel C), which like cdc34-2 and cdc53-1 cells fail to degrade the Cdk inhibitor Sic1 at the G1/S-phase transition (Schwob et al, 1994). Half-life measurements confirmed that HA-Hof1 was stable in grr1Δ rgt1 and cdc34-2 cells (Figure 5D), while it was rapidly degraded in cdh1Δ cells, which are deficient for APC activity during the G1 phase of the cell cycle (Peters, 1999). Stabilization of HA-Hof1 expressed from its endogenous promoter was also observed in grr1Δ cells using CX chase (data not shown). HA-Hof1 but not HA-Hof1ΔPEST accumulated in its phosphorylated form in grr1Δ rgt1 and cdc34-2 cells, suggesting that the failure to degrade Hof1 is not caused by a defect in its phosphorylation. Hof1 was also stabilized in cim5-1 cells analyzed at the restrictive temperature (35°C) (half-life >120 min), implying that Hof1 is degraded by the 26S proteasome. Interestingly, a smear of high molecular mass HA-Hof1 species accumulated in these cells, which may represent ubiquitinylated species of Hof1. Consistent with this notion, such higher molecular weight Hof1 species were absent in grr1Δ rgt1 or cdc34-2 cells. Taken together, these results strongly suggest that Hof1 is ubiquitinylated by SCFGrr1, and specifically degraded by the 26S proteasome in vivo.
Figure 5.
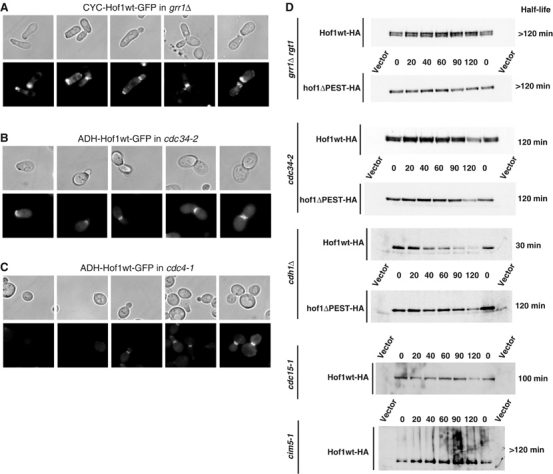
Hof1 degradation depends on SCFGrr1 and a functional MEN pathway. (A–C) grr1Δ rgt1 (YMJ166 (A)), cdc34-2 (YMT670 (B)) or cdc4-1 (YMT668 (C)) cells expressing Hof1-GFP from the constitutive CYC1 or ADH promoter were grown to early log phase in selective media at 30°C (semipermissive temperature), and analyzed by phase (upper panels) and GFP microscopy (lower panels). (D) The half-lives of HA-tagged Hof1 or hof1ΔPEST were determined by GAL shutoff experiments and the half-lives (in minutes) are shown on the right. The following strains were analyzed at 30°C (except where indicated): grr1Δ rgt1 (YMJ166), cdc34-2 (YMT670, 35°C), cdh1Δ (MJ1075), cdc15-1 (YMP690) and cim5-1 (YMP241, 35°C). Cells were treated with pheromones for 2 h (+αF).
To test whether activation of the MEN is necessary for Hof1 degradation, we determined the half-life of HA-Hof1 in cdc15-1 cells at semipermissive temperature (30°C). Interestingly, HA-Hof1 was stabilized (half-life approximately 100 min) and as expected accumulated in its unphosphorylated form (Vallen et al, 2000), suggesting that phosphorylation of Hof1 at the end of mitosis may trigger its degradation. Cdc15 and/or Dbf2 may directly phosphorylate Hof1 or, alternatively, another MEN-dependent kinase(s) may be responsible for these modifications.
Degradation of Hof1 is required for efficient actin ring contraction and cell separation during cytokinesis
To determine whether downregulation of Hof1 at the end of mitosis is important for cell viability, we expressed wild-type Hof1 or stabilized hof1ΔPEST from its endogenous promoter or under the control of the constitutive CYC, ADH or TEF promoters (Funk et al, 2002) in hof1Δ cells. Expression of Hof1 from the ADH promoter resulted in protein levels comparable to levels obtained from the endogenous promoter, while Hof1 is approximately three-fold overexpressed from the TEF promoter (data not shown). While constitutive expression of Hof1 in wild-type cells has no effect (Figure 6A), expression of hof1ΔPEST from the ADH or TEF promoters significantly diminished the growth rate. Likewise, grr1Δ cells expressing wild-type Hof1 from the constitutive ADH promoter were not viable (data not shown). Taken together, these results suggest that efficient inactivation of Hof1 at the end of mitosis is functionally important and is achieved both by transcriptional downregulation and ubiquitin-dependent proteolysis.
To determine why degradation of Hof1 is important for mitotic exit, we first analyzed the morphological phenotype of wild-type cells overexpressing hof1ΔPEST from the inducible GAL promoter. As shown in Figure 6B, these cells arrested as large groups of cells that remained attached to each other, characteristic for a defect during cytokinesis. Interestingly, after cell wall digestion by zymolyase, hof1Δ and hof1Δ cells expressing stabilized hof1ΔPEST from the ADH promoter exhibited an increased number of cells arrested in cytokinesis (Figure 6C), implying that cytokinesis could not be efficiently completed and that the cells continue to share the same cytoplasm. To determine more precisely what process during cytokinesis may require degradation of Hof1, we studied wild-type cells expressing stabilized hof1ΔPEST from the ADH promoter by time-lapse microscopy (Figure 6D and E). As expected, the cells had no significant defects at bud emergence, and initiated anaphase with normal kinetics (data not shown). The splitting of the septin ring as assayed by Cdc3-GFP occurred efficiently (panel D; Dobbelaere and Barral, 2004). Wild-type Hof1 was degraded at this stage of cytokinesis, while stabilized hof1ΔPEST abnormally accumulated between the two septin rings (data not shown). Interestingly, degradation of Hof1 was not required to assemble the actomyosin ring at the mother–bud neck (Figure 6E). However, time-lapse analysis of Myo1-CFP in cells expressing hof1ΔPEST revealed that initiation of actomyosin ring contraction was significantly delayed compared to cell expressing wild-type Hof1. Moreover, subsequent disassembly of the actomyosin ring was slower compared to wild-type cells. Taken together, these results strongly suggest that degradation of Hof1 occurs after splitting of the septin ring, and is required for efficient contraction of the actomyosin ring and proper cell separation during cytokinesis.
Discussion
We show here that Grr1 functions during cytokinesis by promoting degradation of the PCH protein Hof1, demonstrating that SCF-ubiquitin ligases also function during mitosis in budding yeast. Because degradation of Hof1 requires prior phosphorylation in a MEN-dependent manner, SCFGrr1 may be part of a mechanism to ensure that cytokinesis cannot occur until chromosome segregation has been completed.
Specific subcellular localization of F-box proteins provides information on their function
SCF-type E3 ligases target a large variety of substrates for degradation by the 26S proteasome. Not surprisingly therefore, the common core subunits of yeast SCF complexes including Cdc53, Skp1, Cdc34 and Hrt1 are distributed in the nucleus and cytoplasm (Blondel et al, 2000). In contrast, several F-box proteins localize to specific cellular sites. Cdc4 and Met30 are nuclear proteins (Choi et al, 1990; Blondel et al, 2000; Rouillon et al, 2000) and it has been shown that SCFCdc4 is active only in the nucleus, at least when tested for its substrates Far1, Sic1, Gcn4 and Cdc6 (Blondel et al, 2000; Pries et al, 2002; Luo et al, 2003). Interestingly, Grr1 has several substrates that localize to distinct cellular locations. The accumulation of Grr1 at the mother–bud neck region late in mitosis triggers degradation of Hof1, while the Cdc42 effector Gic2 is likely degraded at bud tips shortly after bud emergence (Jaquenoud et al, 1998). Because cytoplasmic Grr1 is able to complement the morphology defects of grr1Δ cells, it is likely that the G1 cyclins Cln1 and Cln2 are degraded in the cytoplasm (Miller and Cross, 2000). However, Grr1 is also found in the nucleus of cells throughout the cell cycle, and we have identified a specific region in the amino-terminus that contains functional NLS. Although these NLS sequences are not conserved in Grr1 orthologs from other species, we suspect that Grr1 in budding yeast may target an unknown nuclear protein for degradation. Grr1 is required for the repression of genes involved in glucose fermentation (Flick and Johnston, 1991), and it is possible that Grr1 may regulate a nuclear target involved in the regulation of gene expression.
Degradation of Hof1 by SCFGrr1 is required for efficient actomyosin contraction and cell separation during cytokinesis
Several lines of evidence strongly suggest that SCFGrr1 plays a role in cytokinesis. First, Grr1 is recruited to the mother–bud neck region just after completion of anaphase but before cell separation initiates. Second, this ring localization depends on its substrate interacting domain (LRR). Third, grr1Δ cells accumulate with a G2 DNA content and exhibit clear cell separation defects (Barral et al, 1995). Finally, we have identified the PCH protein Hof1 as a likely substrate of SCFGrr1 and the 26S proteasome during cytokinesis. Hof1 is degraded at the end of mitosis, and remains unstable during the entire G1 phase of the cell cycle. Hof1 degradation depends on its PEST motif, which is required for its interaction with the F-box protein Grr1. Cells expressing the stabilized mutant version hof1ΔPEST exhibit severe defects during cytokinesis. While assembly of the actomyosin structure and splitting of the septin ring occurred normally, contraction and subsequent ingression of the actomyosin ring were delayed and the cells were unable to separate their cytoplasm. This phenotype is reminiscent of the function of Imp2 in S. pombe (Demeter and Sazer, 1998), which is dispensable for actomyosin ring assembly but is required for proper contraction during cytokinesis. However, our data further suggest that SCFGrr1 may have substrates other than Hof1 during cytokinesis. For example, deletion of HOF1 in grr1Δ cells did not suppress the cell separation defect, but instead exacerbated the cytokinesis defects (data not shown). Moreover, although the substrate-binding domain of Grr1 was required for its localization to the mother–bud neck, Grr1 still accumulated on the cytokinetic ring in hof1Δ cells (data not shown). Additional SCFGrr1 substrates may include Bni1, which contains PEST domains, and the PCH protein Bzz1, which was also found to interact with Grr1 by comprehensive two-hybrid analysis (Ito et al, 2001). Finally, SCFGrr1 may also be involved in the degradation of Gic1 (M Blondel, unpublished observation), which localizes to the mother–bud neck during cytokinesis (Chen et al, 1997).
Dual regulation of Hof1 expression: cell cycle-dependent transcription and degradation
HOF1 transcription peaks in mitosis and is downregulated as cells exit from mitosis (Spellman et al, 1998). Our results show that this transcriptional regulation is not essential for cell viability as cells expressing the HOF1 gene under the control of the constitutive ADH promoter are viable and grow with wild-type rates. Likewise, decreasing the rate of Hof1 degradation alone does not have deleterious effects, as expression of hof1ΔPEST under the control of its normal promoter does not severely interfere with cell division. However, affecting both transcriptional regulation and proteolysis is toxic and blocks cytokinesis. These results indicate that yeast cells use two distinct and redundant pathways to downregulate Hof1 levels at the end of mitosis.
Hof1 degradation: a mechanism to couple cytokinesis to mitotic exit?
Binding of substrates to their F-box protein is often dependent on site-specific phosphorylation (Deshaies, 1997), providing the possibility to control protein degradation by regulating the kinase activity. Hof1 is heavily phosphorylated at the end of mitosis (Vallen et al, 2000), and this hyperphosphorylation depends on its PEST motif. PEST motifs are rich in proline, serine and threonine residues (Rechsteiner and Rogers, 1996), and are often directly phosphorylated. For example, the PEST motif of the G1 cyclin Cln2 is phosphorylated and regulates its interaction with SCFGrr1 (Lanker et al, 1996). Likewise, hof1ΔPEST was stable, most likely because the mutant protein fails to interact with Grr1. These results suggest that phosphorylation of Hof1 may regulate its binding to SCFGrr1. Available results suggest that Hof1 is phosphorylated by a kinase dependent on the MEN pathway. Hyperphosphorylation of Hof1 correlates with the activation of the MEN pathway, and Hof1 is strongly stabilized and accumulated in an unphosphorylated form in MEN mutants including cdc15-1 and dbf2-2 cells. Interestingly, Dbf2 also requires MEN activity for its accumulation at the mother–bud neck (Frenz et al, 2000), raising the possibility that Hof1 may be a direct substrate of Mob1/Dbf2. However, our results indicate that Hof1 is unstable in α-factor-arrested cells, and Mob1/Dbf2 is not thought to be active during the G1 phase of the cell cycle (Visintin and Amon, 2001). Nevertheless, it is possible that Mob1/Dbf2 may initiate Hof1 degradation as cells exit from mitosis, and that another kinase phosphorylates Hof1 during G1. Taken together, these results suggest that activation of the MEN pathway after completion of anaphase triggers phosphorylation of Hof1 (and maybe other substrates of SCFGrr1) at the mother–bud neck region. This allows its recognition by SCFGrr1 and subsequent degradation by 26S proteasomes. Because Hof1 inhibits actomyosin ring ingression, its degradation may trigger completion of cytokinesis. This mechanism may thus ensure that cell separation cannot be initiated as long as mitosis is not completed.
Degradation of PCH family members: a conserved mechanism to regulate cytokinesis?
PCH proteins constitute a conserved protein family including S. pombe Cdc15, Imp2 and YB65, S. cerevisiae Hof1 and Bzz1, Mus musculus PSTPIP, PSTPIP2, PACSIN and PACSIN 2 and Homo sapiens CIP4. Although these proteins share only around 20% sequence identity, they have the same domain structure. Most PCH family proteins contain an N-terminal FCH domain, coiled-coil domains followed by PEST sequences and finally one or more SH3 domains in their C-termini. Recent studies suggest that PCH proteins regulate actin-based processes and perhaps polarized secretion of membrane material during cytokinesis (Lippincott and Li, 2000). Several lines of evidence suggest that degradation of PCH proteins may be a widely conserved mechanism. First, all PCH members contain a conserved PEST domain. Second, transcriptional regulation of the S. pombe Hof1 ortholog Cdc15 is not essential (Utzig et al, 2000). However, overexpression of this protein is toxic and blocks cytokinesis, suggesting that an alternative pathway involving degradation of Cdc15 may also exist in S. pombe. Indeed, S. pombe Cdc15 is phosphorylated around the time of cytokinesis (Fankhauser et al, 1995) and these phosphorylations depend on its PEST domain (V Simanis, personal communication). Finally, overexpression of the M. musculus homolog PSTPIP2 is toxic in S. pombe cells (Spencer et al, 1997). Because Grr1 itself is a conserved protein among eukaryotes, it is possible that degradation of PCH proteins by SCFGrr1 or other SCF complexes may generally regulate cytokinesis in many organisms. However, although expression of the putative S. pombe Grr1 homolog Pof2 was able to at least partially complement the cryosensitivity of budding yeast grr1Δ cells, its inactivation in S. pombe did not result in any obvious cytokinesis phenotype (S Bamps and J Vandenhaute, unpublished observation).
At present, the molecular understanding of how PCH proteins first promote and then inhibit efficient actomyosin contraction and cell separation during cytokinesis is still unclear and will be an important goal for future experiments.
Materials and methods
Yeast strains and genetic manipulations
Yeast strains are described in Table I. Strains are derived from K699: Mat a, ade2-1, trp1-1, can1-100, leu2-3,112, his3-11,15, ura3, GAL+, psi+, ssd1-d2 (W303 background) or S288c: ade2-101, ura3-52, lys2-801, trp1-Δ1, his3Δ200, leu2-Δ. Standard yeast growth conditions and genetic manipulations were as described previously (Guthrie and Fink, 1991; Wach et al, 1994). The plasmids pDJ228, pDJ230, pDJ233 and pDJ234 were digested with StuI for integration at the URA3 locus of the hof1Δ strain DJ640. To analyze actomyosin ring behavior, the strains harboring an HA-tagged Hof1 construct were crossed with a strain containing Myo1-CFP (DJ1316).
Table 1.
Strain list
| Strain name | Relevant genotype | Background | Source |
|---|---|---|---|
| K699 | Mat a | W303 | Kim Nasmyth |
| YMP190 | Mat a cdc16-1 | W303 | Kim Nasmyth |
| YMP809 | Mat a cdc14-3 | W303 | Toni Hyman |
| YMP690 | Mat a cdc15-1 | W303 | Ray Deshaies |
| YMP1118 | Mat a dbf2-2 | W303 | Ray Deshaies |
| YSB100 | Mat a hof1∷KANMX | W303 | This study |
| YMP2957 | Mat a grr1∷LEU2 | W303 | Jaquenoud et al (1998) |
| YMJ166 | Mat a grr1∷LEU2 rgt1-101 | S288C | Carl Mann |
| YMT670 | Mat a cdc34-2 | W303 | Mike Tyers |
| YMT740 | Mat a cdc53-1 | W303 | Mike Tyers |
| YMT668 | Mat a cdc4-1 | W303 | Mike Tyers |
| MJ1075 | Mat a cdh1∷LEU2 | W303 | Angelika Amon |
| YMP241 | Mat a cim5-1 | S288C | Ghislain et al (1993) |
| YSB101 | Mat a bni1∷KANMX | W303 | This study |
| DJ640 | Mat α hof1∷HIS3 | S288C | Dobbelaere and Barral (2004) |
| DJ641 | Mat a HOF1-GFP∷His3MX6 | S288C | Dobbelaere and Barral (2004) |
| DJ1316 | Mat a MYO1-CFP∷Kan ura3-52 | S288C | Dobbelaere and Barral (2004) |
| DJ1601 | pCYC1-hof1-PEST-GFP∷URA3 hof1∷HIS3 | S288C | This study |
| DJ1602 | pCYC1-HOF1-GFP∷URA3 hof1∷HIS3 | S288C | This study |
| DJ1600 | pADH1-HOF1-HA∷URA hof1∷HIS3 | S288C | This study |
| DJ1608 | pADH1-hof1-PEST-HA∷URA hof1∷HIS3 | S288C | This study |
| DJ1618 | pADH1-HOF1-PEST-HA∷URA hof1∷HIS3 Myo1-CFP∷Kan | S288C | This study |
| DJ1619 | pADH1-HOF1-HA∷URA3hof1∷HIS3 Myo1-CFP∷Kan | S288C | This study |
| DJ1620 | hof1∷HIS3 Myo1-CFP∷Kan | S288C | This study |
DNA manipulations, Western blots and antibodies
Standard molecular biology techniques were used for plasmid constructions (Table II) and details will be provided upon request. PCR reactions were performed with the Expand polymerase kit as recommended by the manufacturer (Roche), and confirmed by sequencing. Standard procedures were used for yeast cell extract preparation and immunoblotting. Antibodies against GFP (Roche) or HA (Babco) were used as recommended by the manufacturers.
Table 2.
Plasmid list
| pBM200 | GAL GRR1wt-GFP URA3 CEN | Blondel et al (2000) |
| pJMG48 | GAL GRR1wt-GFP TRP1 CEN | Blondel et al (2000) |
| pMJ200 | GAL GFP TRP1 CEN | Jaquenoud et al (1998) |
| pBM210 | GAL grr1(1−901)-GFP TRP1 CEN | This study |
| pBM211 | GAL grr1(1−661)-GFP TRP1 CEN | This study |
| pBM213 | GAL grr1(1−311)-GFP TRP1 CEN | This study |
| pBM223 | GAL grr1(311–1150)-GFP TRP1 CEN | This study |
| pBM241 | GAL grr1(180–1150)-GFP TRP1 CEN | This study |
| pLC45 | GAL grr1(1–180)-GFP TRP1 CEN | This study |
| pLC50 | ADH GRR1wt LEU2 CEN | This study |
| pLC51 | ADH grr1(180–1150) LEU2 CEN | This study |
| pLC52 | ADH grr1ΔFbox LEU2 CEN | This study |
| pLC53 | ADH GRR1wt-GFP LEU2 CEN | This study |
| pLC13 | HOF1wt-GFP TRP1 CEN | This study |
| pBM320 | hof1Δ418–438-GFP TRP1 CEN | This study |
| pLC12 | GAL HOF1wt -GFP TRP1 CEN | This study |
| pBM321 | GAL hof1Δ418–438-GFP TRP1 CEN | This study |
| pBM325 | HOF1wt-GFP URA3 CEN | This study |
| pBM326 | hof1Δ418–438-GFP URA3 CEN | This study |
| pBM327 | GAL HOF1wt-GFP URA3 CEN | This study |
| pBM328 | GAL hof1Δ418–438-GFP URA3 CEN | This study |
| pBM351 | GAL grr1ΔFbox-GFP TRP1 CEN | This study |
| pBM352 | GAL grr1(1–792)-GFP TRP1 CEN | This study |
| pBM360 | CYC1 HOF1wt-GFP URA3 CEN | This study |
| pBM361 | ADH HOF1wt-GFP URA3 CEN | This study |
| pBM362 | TEF HOF1wt-GFP URA3 CEN | This study |
| pBM365 | CYC1 hof1Δ418–438-GFP URA3 CEN | This study |
| pBM366 | ADH hof1Δ418–438-GFP URA3 CEN | This study |
| pBM367 | TEF hof1Δ418–438-GFP URA3 CEN | This study |
| pBM370 | pEG203 HOF1 wt (from plasmid DNA) | This study |
| pBM371 | pEG203 hof1Δ418–438 | This study |
| pBM375 | pEG203 HOF1 wt (from genomic DNA) | This study |
| pBM385 | GAL HA3 TRP1 CEN | This study |
| pBM390 | GAL HOF1wt-HA3 TRP1 CEN | This study |
| pBM391 | GAL hof1Δ418–438-HA3 TRP1 CEN | This study |
| pBM400 | pACT2 HOF1wt | This study |
| pBM401 | pACT2 hof1Δ418–438 | This study |
| pBM402 | pGBT9 GRR1wt | This study |
| pBM405 | GAL HOF1wt-HA3 URA3 CEN | This study |
| pBM406 | GAL hof1Δ418–438-HA3 URA3 CEN | This study |
| pSOB1 | GAL linker-CFP(155–238) TRP1 CEN | This study |
| pSOB2 | GAL linker-YFP(1–173) URA3 CEN | This study |
| pSOB4 | GAL HOF1wt-linker-YFP(1–173) URA3 CEN | This study |
| pSOB11 | GAL GRR1wt-linker-CFP(155–238) TRP1 CEN | This study |
| pSOB15 | GAL hof1Δ418–438-linker-YFP(1–173) URA3 CEN | This study |
| pDJ63 | GFP-CDC3wt LEU2 CEN | Dobbelaere et al (2003) |
| pDJ228 | CYC1-HOF1wt-GFP URA3 | This study |
| pDJ230 | CYC1-hof1Δ418–438-GFP URA3 | This study |
| pDJ233 | ADH-HOF1wt-HA3 URA3 | This study |
| pDJ234 | ADH-hof1Δ418–438-HA3 URA3 | This study |
Microscopy
GFP-tagged proteins were visualized using a Chroma GFP4 filter (excitation 455–495 nm) on an Olympus BX61 microscope, photographed with a Spot RT cooled CCD camera and analyzed with Photoshop 5.0 software (Adobe). Cells expressing GFP fusion proteins from the inducible GAL1, 10 promoter were grown to early log phase at 25°C in selective media containing raffinose (2% final concentration), at which time galactose was added (2% final concentration) for 5 h as previously described (Blondel et al, 1999). The ability of wild-type and mutant Grr1-GFP proteins to accumulate at the mother–bud neck region (ring) was quantified by analyzing for each construct at least 200 cells in late mitosis (as judged by DIC). For the BiFC experiments, wild-type cells expressing either wild-type Hof1 or hof1ΔPEST fused to the N-terminal part of YFP (1–173) together with Grr1 fused to the C-terminal part of CFP (155–238) were analyzed using a Chroma GFP4 filter (Hu and Kerppola, 2003).
Time-lapse movies were acquired at room temperature unless noted otherwise, with a BX51 Olympus microscope connected to a Till Vision monochromator. All pictures/movies were the result of the projection of five different stacks. The examined strains were grown overnight on YPD plates, and then put on an agarose pad containing nonfluorescent medium (Dobbelaere et al, 2003). Analysis of septin ring behavior in both wild-type (S288C) and hof1ΔPEST (DJ1608) strains was performed after shifting the cells for 1 h to 35°C and then mounting them under a preheated microscope. These strains were transformed with a plasmid containing GFP-CDC3 (pDJ63). Actomyosin ring contraction was analyzed at 30°C in at least five different movies for each strain.
Cytokinetic assays
Cytokinetic assays were performed essentially as described previously (Dobbelaere et al, 2003). Briefly, S288C, DJ640, DJ1608 and DJ1600 were grown overnight at room temperature in YPD. Cells were diluted to OD=0.1 in fresh YPD and grown for 1 h to reach ±OD=0.2. Cells were shifted to 37°C and samples were taken every hour, fixed with ethanol and stained with DAPI. To analyze if cells were connected to each other, ethanol-fixed cells were treated with zymolyase (2 mg/ml) for 30 min at 30°C. Cells were vortexed extensively and then stained with DAPI. Cell cycle stages were determined microscopically and compared before and after zymolyase treatment.
Protein half-life determination
Half-lives were measured as described previously (Blondel et al, 2000) in strains that carry plasmids expressing HA-tagged Hof1 or hof1ΔPEST from the inducible GAL1,10 promoter. Half-life measurements by CX chase were performed as described (Galan and Peter, 1999).
Acknowledgments
We thank JM Galan and B André for providing reagents and helpful suggestions. Members of our laboratories are acknowledged for fruitful discussion, and V Simanis for communicating unpublished data and critical reading of the manuscript. MB is supported by the ‘Association pour la Recherche contre le Cancer' (ARC MB5812), a PRIR from the ‘Conseil Régional de Bretagne' and an ‘ACI Jeune Chercheur' fellowship from the French ‘Ministère de la Recherche'. MP is supported by the Swiss National Science Foundation (SNF) and the ETHZ. Part of this work was supported by an HFSP fellowship.
References
- Barral Y, Jentsch S, Mann C (1995) G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev 9: 399–409 [DOI] [PubMed] [Google Scholar]
- Barral Y, Mann C (1995) G1 cyclin degradation and cell differentiation in Saccharomyces cerevisiae. CR Acad Sci III 318: 43–50 [PubMed] [Google Scholar]
- Blacketer MJ, Madaule P, Myers AM (1995) Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics 140: 1259–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M, Alepuz PM, Huang LS, Shaham S, Ammerer G, Peter M (1999) Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev 13: 2284–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M, Galan JM, Chi Y, Lafourcade C, Longaretti C, Deshaies RJ, Peter M (2000) Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J 19: 6085–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GC, Kim YJ, Chan CS (1997) The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev 11: 2958–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WJ, Clark MW, Chen JX, Jong AY (1990) The CDC4 gene product is associated with the yeast nuclear skeleton. Biochem Biophys Res Commun 172: 1324–1330 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL (2002) Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology 35: 3–6 [DOI] [PubMed] [Google Scholar]
- Demeter J, Sazer S (1998) imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J Cell Biol 143: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ (1997) Phosphorylation and proteolysis: partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev 7: 7–16 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y (2004) Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305: 393–396 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Gentry MS, Hallberg RL, Barral Y (2003) Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell 4: 345–357 [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF (1997) The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J 16: 5966–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V (1995) The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell 82: 435–444 [DOI] [PubMed] [Google Scholar]
- Flick JS, Johnston M (1991) GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol 11: 5101–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz LM, Lee SE, Fesquet D, Johnston LH (2000) The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci 113 (Part 19): 3399–3408 [DOI] [PubMed] [Google Scholar]
- Funk M, Niedenthal R, Mumberg D, Brinkmann K, Ronicke V, Henkel T (2002) Vector systems for heterologous expression of proteins in Saccharomyces cerevisiae. Methods Enzymol 350: 248–257 [DOI] [PubMed] [Google Scholar]
- Galan JM, Peter M (1999) Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc Natl Acad Sci USA 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366: 358–362 [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Marti A, Krek W (1999) Association of human SCF(SKP2) subunit p19(SKP1) with interphase centrosomes and mitotic spindle poles. Exp Cell Res 247: 554–562 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press Inc. [Google Scholar]
- Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M (1997) Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev 11: 3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung YG, Chang HC, Pellequer JL, La Valle R, Lanker S, Wittenberg C (2001) F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol Cell Biol 21: 2506–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hu CD, Kerppola TK (2003) Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol 21: 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M, Gulli MP, Peter K, Peter M (1998) The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J 17: 5360–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Flick K, Wittenberg C, Reed SI (2000) Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102: 303–314 [DOI] [PubMed] [Google Scholar]
- Lanker S, Valdivieso MH, Wittenberg C (1996) Rapid degradation of the G(1) cyclin Cln2 induced by Cdk-dependent phosphorylation. Science 271: 1597–1601 [DOI] [PubMed] [Google Scholar]
- Lippincott J, Li R (1998) Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol 143: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R (2000) Involvement of PCH family proteins in cytokinesis and actin distribution. Microsc Res Tech 49: 168–172 [DOI] [PubMed] [Google Scholar]
- Luo KQ, Elsasser S, Chang DC, Campbell JL (2003) Regulation of the localization and stability of Cdc6 in living yeast cells. Biochem Biophys Res Commun 306: 851–859 [DOI] [PubMed] [Google Scholar]
- Meimoun A, Holtzman T, Weissman Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D (2000) Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell 11: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ME, Cross FR (2000) Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol 20: 542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Patton EE, Peyraud C, Rouillon A, Surdin-Kerjan Y, Tyers M, Thomas D (2000) SCF(Met30)-mediated control of the transcriptional activator Met4 is required for the G(1)–S transition. EMBO J 19: 1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet 14: 236–243 [DOI] [PubMed] [Google Scholar]
- Peters JM (1998) SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol 10: 759–768 [DOI] [PubMed] [Google Scholar]
- Peters JM (1999) Subunits and substrates of the anaphase-promoting complex. Exp Cell Res 248: 339–349 [DOI] [PubMed] [Google Scholar]
- Pries R, Bomeke K, Irniger S, Grundmann O, Braus GH (2002) Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot Cell 1: 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW (1996) PEST sequences and regulation by proteolysis. Trends Biochem Sci 21: 267–271 [PubMed] [Google Scholar]
- Rouillon A, Barbey R, Patton EE, Tyers M, Thomas D (2000) Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30 )complex. EMBO J 19: 282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev 13: 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, Elledge SJ, Harper JW (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284: 662–665 [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell 9: 3273–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, Simanis V, Lasky LA (1997) PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol 138: 845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Jorgensen P (2000) Proteolysis and the cell cycle: with this RING I do thee destroy. Curr Opin Genet Dev 10: 54–64 [DOI] [PubMed] [Google Scholar]
- Utzig S, Fankhauser C, Simanis V (2000) Periodic accumulation of cdc15 mRNA is not necessary for septation in Schizosaccharomyces pombe. J Mol Biol 302: 751–759 [DOI] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E (2000) Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell 11: 593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ (1997) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460 [DOI] [PubMed] [Google Scholar]
- Visintin R, Amon A (2001) Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol Biol Cell 12: 2961–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerecisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]


