Abstract
Insect herbivores frequently cospeciate with symbionts that enable them to survive on nutritionally unbalanced diets. While ancient symbiont gain and loss events have been pivotal for insect diversification and feeding niche specialization, evidence of recent events is scarce. We examine the recent loss of nutritional symbionts (in as little as 1 MY) in sap-feeding Pariaconus, an endemic Hawaiian insect genus that has undergone adaptive radiation, evolving various galling and free-living ecologies on a single host-plant species, Metrosideros polymorpha within the last ∼5 MY. Using 16S rRNA sequencing, we investigated the bacterial microbiomes of 19 Pariaconus species and identified distinct symbiont profiles associated with specific host-plant ecologies. Phylogenetic analyses and metagenomic reconstructions revealed significant differences in microbial diversity and functions among psyllids with different host-plant ecologies. Within a few millions of years, Pariaconus species convergently evolved the closed-gall habit twice. This shift to enclosed galls coincided with the loss of the Morganella-like symbiont that provides the essential amino acid arginine to free-living and open-gall sister species. After the Pariaconus lineage left Kauai and colonized younger islands, both open- and closed-gall species lost the Dickeya-like symbiont. This symbiont is crucial for synthesizing essential amino acids (phenylalanine, tyrosine, and lysine) as well as B vitamins in free-living species. The recurrent loss of these symbionts in galling species reinforces evidence that galls are nutrient sinks and, combined with the rapidity of the evolutionary timeline, highlights the dynamic role of insect–symbiont relationships during the diversification of feeding ecologies. We propose new Candidatus names for the novel Morganella-like and Dickeya-like symbionts.
Keywords: galling, B vitamins, Pariaconus psyllids, symbiont loss, symbiosis
Introduction
Among the many and varied oceanic archipelagos in the Pacific, the Hawaiian Islands offer an unparalleled level of isolation, setting the stage for the evolution of extraordinary and diverse ecosystems. This remarkable isolation has made the Hawaiian archipelago a natural laboratory, where the phenomenon of adaptive radiations is observed more frequently compared to other environments (Gillespie et al. 2012; Shaw and Gillespie 2016). For example, sap-feeding insects known as psyllids from the endemic Pariaconus genus have been diversifying in the Hawaiian archipelago for several millions of years (MY) on a single endemic host plant, the ecologically dominant shrub ʻōhiʻa lehua, Metrosideros polymorpha (Percy 2017). This diversification on a single host-plant species is unusual for a monophyletic group, not only for psyllids but also for other plant-feeding insects (Joy and Crespi 2007; Hippee et al. 2021; Ward et al. 2022). Instead, the prototypical process of speciation in insect herbivores, including psyllids, is via geographic isolation or host-plant shifts (Percy 2003; Taylor et al. 2016; Bastin et al. 2024). For example, host-plant switching helped facilitate one of the largest radiations of insects on the Hawaiian Islands: the endemic Hawaiian leafhopper genus, Nesophrosyne, which has >200 species (Bennett and O’Grady 2012).
In contrast, Pariaconus has diversified on a single host plant, M. polymorpha, via a remarkable array of habitat and morphological shifts that have taken place within an insular radiation of more than 36 psyllid species over the span of just a few million years (Percy 2017). This sets Pariaconus apart from other insect study systems (Price 2005; Nyman 2010) as a unique group exhibiting evolutionary shifts, particularly between galling and nongalling ecological habits, on the same plant species. Although single plant species acting as a “superhost” for many different galling insects are not uncommon and such superhosts may increase local insect species richness (Araújo et al. 2019), examples of a single plant species acting as a superhost for a monophyletic clade of insects are much rarer. Some examples include oak gall wasps in Andricus (Hymenoptera; Cynipidae) (Cook et al. 2002) and gall midges in Asphondylia (Diptera: Cecidomyiidae) (Joy and Crespi 2007). However, these examples are not as diverse as Pariaconus in terms of the number of species found on a single host plant that exhibit different galling/nongalling host-plant ecologies. One factor that may contribute to Pariaconus speciation (Percy 2017) and possibly other insects associated with M. polymorpha (Gruner et al. 2005) is the extraordinarily high degree of phenotypic variation in the host plant. “Hypervariable” polymorphism in M. polymorpha is posited to result from an unexpectedly rich pool of ancestral genetic variation (Choi et al. 2021). An additional key factor that likely drives this psyllid radiation is the capacity exhibited by Pariaconus species to exploit different ecological niches, which span from free-living to different gall-inducing phenotypes on different parts of the host plant (Percy 2017). Furthermore, phylogenies of the host plant and the psyllids show these associated diversifications have occurred over similar time frames and in a similar fashion in that an initial colonization of Kauai was followed by colonization of the younger islands (Percy et al. 2008, 2018; Choi et al. 2021; Bastin et al. 2024).
The origins and diversification of many insect lineages with specialized, nutrient-poor diet niches are associated with intimate partnerships with mutualistic symbionts (Moran et al. 2005; Sudakaran et al. 2017; Cornwallis et al. 2023). In fact, 90% of insect species that feed on phloem, xylem, wood, and blood possess obligate symbionts that help them feed on these nutritionally unbalanced or recalcitrant diets (Cornwallis et al. 2023). Indeed, insect herbivores that possess obligate symbionts are associated with a 15-fold increase in the number of insect species compared to insect herbivore families without symbionts, further suggesting that these symbionts improve host fitness and foster diversification (Cornwallis et al. 2023). However, studies that show how different insect-feeding ecologies are associated with the gains and losses of symbionts at a finer evolutionary scale are limited because of poorly resolved species phylogenies (Moran et al. 2005; Sudakaran et al. 2017; Bell-Roberts et al. 2019; Cornwallis et al. 2023). In contrast, the Hawaiian Pariaconus psyllid radiation is highly resolved (Percy 2017).
Psyllids are globally distributed plant phloem feeders that harbor the primary obligate nutritional symbiont Candidatus Carsonella ruddii (hereafter Carsonella) (Thao et al. 2000). Carsonella's genome is dramatically reduced compared to its free-living relatives but nevertheless still encodes most genes for the essential amino acid pathways given the psyllids’ specialized sap diet is limited in these essential nutrients (Nakabachi et al. 2006). Gaps are still present in some of Carsonella's essential amino acid pathways, however, and co-symbionts (Sloan and Moran 2012; Dittmer et al. 2023) and/or psyllid host genes that were horizontally transferred from bacteria into the psyllid genome in an early psyllid ancestor are assumed to complement some of the enzymatic steps missing in Carsonella (Sloan et al. 2014; Kwak et al. 2023). Additional co-symbionts that are present in all individuals of a psyllid species and/or are cospeciating with related psyllid species (Hall et al. 2016; Morrow et al. 2017; Nakabachi et al. 2020, 2020; Kwak et al. 2021; Serbina et al. 2022) are hypothesized to be transitioning into a potential obligate role with their psyllid host by providing missing genes for Carsonella's essential amino acid and/or vitamin biosynthesis pathways.
Previous research on insect galling of plant tissue has shown that galls can act as nutrient sinks for essential amino acids and nitrogen in general (Forrest 1971; Larson and Whitham 1991; Inbar et al. 1995; Fay et al. 1996). Due to this potential concentration of otherwise limited nutrients in gall tissue, it was previously hypothesized that compared to free-living psyllids, gall-feeding psyllids most likely would not harbor additional bacterial nutritional symbionts other than their Carsonella, which is generally present in all psyllid species and cospeciated with their psyllid hosts (Spaulding and Von Dohlen 2001). Yet, to date, sequencing surveys have not revealed a clear pattern in the presence or absence of additional nutritional co-symbionts based on galling status (Sloan and Moran 2012; Morrow et al. 2017; Hammer et al. 2021). This lack of pattern can be due to low sample sizes and not examining symbiont gains and/or losses at a species-level resolution for a monophyletic group.
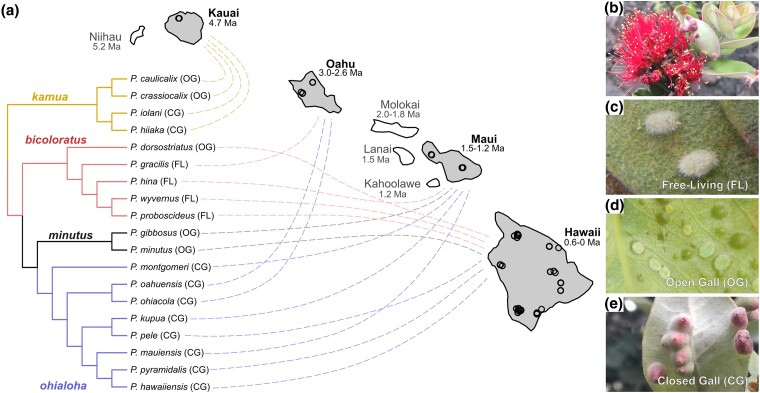
To investigate if shifts in host-plant ecologies are associated with gains or losses of nutritional symbionts at a species level, we examine the bacterial microbiomes of Pariaconus species that span the Hawaiian archipelago (Fig. 1a) and have evolved three main types of host-plant ecologies on M. polymorpha: (i) free-living; (ii) open-gall, living in a pit/cup gall that is open to the environment; and (iii) closed-gall, living in an enclosed gall that is not open to the external environment (Fig. 1b to e). We conduct 16S rRNA sequencing on 19 Pariaconus species from all four species groups (kamua, bicoloratus, minutus, and ohialoha) (Fig. 1a; supplementary Data Set S1, Supplementary Material online) across the Hawaiian archipelago and analyze their phylogenetic relationships with co-symbionts to explore associations with different galling ecologies. Additionally, we sequence metagenomes of coevolving symbionts and examine transcriptomic and genomic data from psyllids to investigate the role of horizontal gene transfer in complementing nutritional symbionts for essential amino acid and vitamin biosynthesis.
Fig. 1.
Hawaiian psyllid radiation spans multiple islands and lifestyles. a) Cladogram of Pariaconus psyllid species radiation and their sampling locations among the Hawaiian Islands of Kauai, Oahu, Maui, and Hawaii (black circles on gray islands). Psyllid relationships from the four species groups kamua, ohialoha, minutus, and bicoloratus are shown by the cladogram, which was modified from Percy (2017, 2018). Species names are linked to the island of specimen origin for study. Pariaconus gracilis is the only species found on multiple islands (Oahu, Molokai, and Maui). See supplementary Data Set S1, Supplementary Material online for more details on samples. The host-plant ecologies of each species are indicated next to their name as free-living (FL), open-gall (OG), or closed-gall (CG). Dating of the islands is presented as millions of years ago (Ma) and is based on dating from Bonacum et al. (2005). b to e) Pictures taken by D.M.P. and A.K.H. b) Single host-plant species of all Pariaconus psyllid species, ʻōhiʻa lehua, M. polymorpha. c) Nymph stage of P. proboscideus (FL), d) nymph stage of P. dorsostriatus (OG), and e) CG structure of P. pyramidalis.
Results
Influence of Island, Species Group, and Host-Plant Ecologies on Hawaiian Psyllid Microbiomes
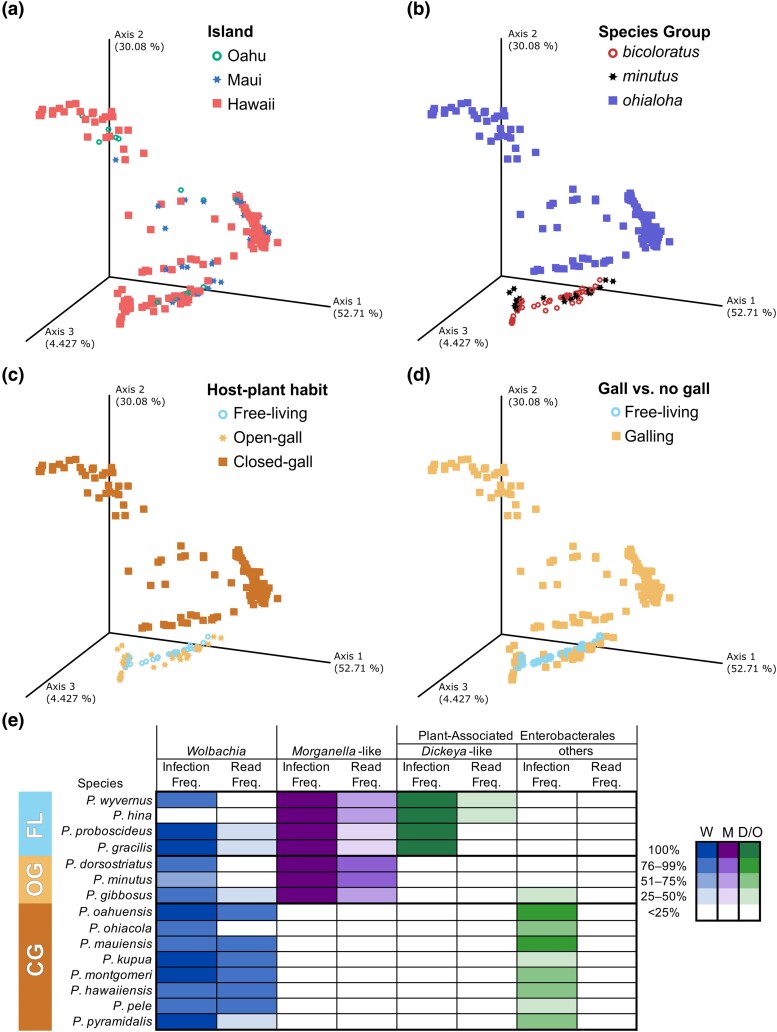
Over several MY Hawaiian Pariaconus psyllids have radiated across the islands and adopted distinct host-plant habits along the way (Percy 2017). To investigate how psyllid microbial communities have influenced psyllids or have been impacted by these processes, we conducted a principal coordinate analysis (PCoA) of the sample weighted UniFrac distances. This analysis identified trends in the similarity of microbial community composition among samples by island, species groups, host-plant ecologies, and galling types (Fig. 2a to d; supplementary Data Sets S1 and S2, Supplementary Material online). Rarefaction analyses of observed amplicon sequence variants (ASVs) further revealed that for both data sets, (i) all 319 samples/psyllid individuals and 15 psyllid species and (ii) a random subsample of the data set to standardize the number of samples/individuals per species (56 samples and 14 species), there was an asymptote between the sequence depth of 1,000 to 1,600 reads, suggesting that ASVs were saturated below our randomly sampled minimum read cutoff for all samples and species (supplementary fig. S1a to d, Supplementary Material online). To determine if these psyllid microbial communities are statistically associated with island, species groups, host-plant habits, and galling types, we conducted a PERMANOVA analyses, which accounts for species-level variation, using both the entire data set (n = 319 psyllid individuals/samples) and the randomly subsampled data set (n = 52 psyllid individuals/samples). For the PERMANOVA analyses, we found that island (Oahu vs. Maui vs. Hawaii) (Adonis, P = 0.001), species groups (ohialoha vs. bicoloratus vs. minutus) (Adonis, P = 0.001), host-plant ecologies (free-living vs. open-gall vs. closed-gall) (Adonis, P = 0.001), and galling type (free-living vs. open- and closed-gall) (Adonis, P = 0.001) were all significant factors associated with psyllid microbial communities, when using both data sets (n = 319 and n = 52).
Fig. 2.
Hawaiian psyllid microbiomes exhibit marked compositional differences. Weighted UniFrac PCoA plot representing the distances of the bacterial microbiomes of 319 psyllid samples (individuals) to one another and colored according to their island a), species group b), host-plant habit c), and gall/no-gall d) associations. e) High-frequency ASVs represent putative symbionts. A heatmap of the most prevalent high-frequency ASVs among sampled psyllid species, which belong to the Wolbachia-like, Morganella-like, and other plant-associated Enterobacterales, which includes the Dickeya-like ASVs. Infection frequency is the percent of psyllid individuals per species infected with one of the high-frequency taxa (>25%). Read frequency is the relative number of reads representing one of the high-frequency ASVs out of total reads for the species. Data represent 319 psyllid samples (individuals) where sample size per species ranges from 2 to 107 individuals; see Materials and Methods and supplementary Data Set S1, Supplementary Material online for more details on samples.
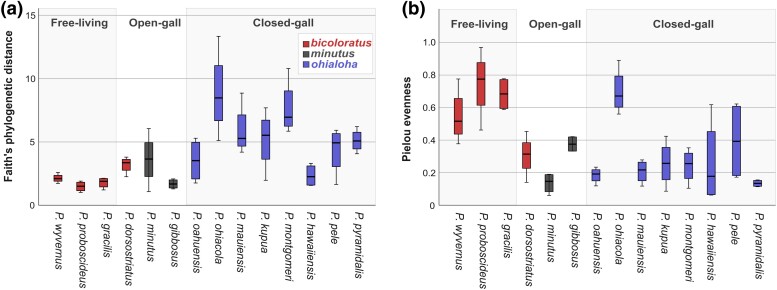
We also examined the patterns of within sample diversity. Using the subsampled data set, we calculated microbiome richness (Faith's phylogenetic distance) and evenness (Pielou) (Fig. 3). Faith's phylogenetic distance was not significantly associated with island (Oahu vs. Maui vs. Hawaii) (general linear model [GLM], F2,11 = 0.732, P = 0.503) (Fig. 3a). In contrast, Faith's phylogenetic distance was significantly associated with species groups (ohialoha vs. bicoloratus vs. minutus) (GLM, F2,11 = 6.664, P = 0.013) and host-plant ecologies (free-living vs. open-gall vs. closed-gall) (GLM, F2,11 = 8.018, P = 0.007). Post hoc tests revealed that Faith's phylogenetic distance was not significantly different between bicoloratus versus minutus groups (Bonferroni, P = 1.0) or free-living versus open-gall psyllids (Bonferroni, P = 0.098) (which largely reflects species groups); however, the other pairwise combinations of ohialoha versus bicoloratus groups (Bonferroni, P < 0.001), ohialoha versus minutus groups (Bonferroni, P < 0.001), closed-gall psyllids versus free-living psyllids (Bonferroni, P < 0.001), and closed-gall psyllids versus open-gall psyllids (Bonferroni, P < 0.001) were all significantly different. Together, these results suggest microbiome richness based on Faith's phylogenetic distance is on average higher in closed-gall psyllids (ohialoha group) compared to both free-living and open-gall psyllids (bicoloratus and minutus groups) (Fig. 3a). However, when all galling psyllids (open- and closed-gall) are compared to the free-living psyllids, galling psyllids display significantly higher microbiome richness compared to free-living psyllids (GLM, F1,12 = 7.551, P = 0.018) (Fig. 3a).
Fig. 3.
ASV diversity of Hawaiian Pariaconus psyllid microbiomes. Boxplots of ASV diversity per psyllid species for Faith's phylogenetic distance a) and Pielou's evenness b). Sample size is standardized to four individuals per species (see Materials and Methods for more details). Outer bars on species boxplots represent maximum and minimum diversity values. The box itself represents interquartile range from first to third quartile, and the bar inside the box represents the median.
Pielou evenness was not significantly associated with island (Oahu vs. Maui vs. Hawaii) (GLM, F2,11 = 0.859, P = 0.450) or species groups (ohialoha vs. bicoloratus vs. minutus) (F2,11 = 3.521, P = 0.066). In contrast, Pielou evenness was significantly associated with host-plant ecologies (e.g. free-living vs. open-gall vs. closed-gall) (F2,11 = 5.1, P = 0.027) where post hoc tests showed that evenness was significantly higher between free-living psyllids versus open-gall psyllids (Bonferroni, P < 0.001) and free-living psyllids versus closed-gall psyllids (Bonferroni, P < 0.001) (Fig. 3b). Pielou evenness was also significantly different between galling type (free-living vs. open- and closed-gall) (GLM, F1,12 = 7.551, P = 0.018) where psyllids with galls display lower microbiome evenness compared to free-living psyllids (Fig. 3b). In sum, free-living Hawaiian psyllids have lower phylogenetic diversity of ASVs and the relative abundances of the ASVs are more uniform while galling psyllids have greater phylogenetic diversity in their microbial community and are more biased in their relative abundance in/on the insects.
High-Frequency ASVs of Hawaiian Pariaconus Psyllid Species and Their Phylogenetic Origins
Closer analysis of the Hawaiian Pariaconus psyllid microbiomes identified recurrent, high-frequency ASVs within many of the psyllid species, where the frequency of ASVs is defined as the percent of psyllid individuals harboring the ASV within a psyllid species. Note we have defined high-frequency ASVs as those that occur in >25% of psyllid individuals within a psyllid species. Based on an understanding of insect-associated microbes, if a symbiont has a high frequency of occurrence within a psyllid species (Morrow et al. 2017), it may be involved in important biological roles for that psyllid species because of its high fidelity with the species either as an endosymbiont, exosymbiont, and/or environmental symbiont. We detected a variable number of these high-frequency ASVs in the psyllid host-plant ecologies free-living (x̅ = 3.7 ± 0.6 SD), open-gall (x̅ = 4 ± 1.0 SD), and closed-gall (x̅ = 15.1 ± 10.8 SD) (supplementary Data Set S2, Supplementary Material online). The detected number of high-frequency ASVs and the average observed ASVs per psyllid species (supplementary Data Set S2, Supplementary Material online) are consistent with the Faith's phylogenetic distance results above where free-living and open-gall psyllids have lower observed microbiome richness compared to closed-gall psyllids.
The highest frequency ASVs observed in these Hawaiian Pariaconus psyllids belong to Morganellaceae, plant-associated Enterobacterales, and Wolbachia (Fig. 2e; supplementary Data Set S3, Supplementary Material online). The ASVs from these three groups also possessed the highest relative read counts per psyllid species (if present in the species) compared to other high-frequency ASVs (Fig. 2e). We calculated the average relative read frequencies per psyllid species where the species contained reads for the ASV > 0% to be Wolbachia (x̅ = 54% ± 31 SD, n = 14), Morganellaceae (x̅ = 60% ± 17 SD, n = 7), and plant-associated Enterobacterales (x̅ = 15% ± 11 SD, n = 9) (Fig. 2e).
Putative Wolbachia Endosymbionts
The well-known insect endosymbiont Wolbachia was the most prevalent high-frequency ASV among Hawaiian Pariaconus psyllid species sampled except for one, Pariaconus hina. Additionally, in 6 out of 15 of the psyllid species, the Wolbachia ASVs were present at 100% infection frequency for sampled individuals/species. Further, these six psyllid species with 100% infection frequency represent both free-living and closed-gall psyllid species (Fig. 2e). To determine the phylogenetic affinity of the Wolbachia ASVs within the diverse Wolbachia genus, a phylogenetic tree was constructed (supplementary fig. S2a, Supplementary Material online). The Wolbachia taxa from all of the Pariaconus psyllids here appear to belong to Wolbachia subgroup B and cluster with other Wolbachia taxa from insects including other Hemipterans. Closer inspection reveals at least two closely related, but distinct lineages of Wolbachia taxa occur in five and nine of the Pariaconus psyllid species. Each of these lineages has members from all three plant ecologies (free-living, open-, and closed-gall) (supplementary fig. S2a, Supplementary Material online). Furthermore, three Wolbachia ASVs were shared by more than one psyllid species (supplementary fig. S2a, Supplementary Material online) and subsequent sequence analysis revealed these ASVs have 100% identity and coverage with existing Wolbachia sequences in NCBI from diverse insect species (grasshoppers, moths, beetles, aphids, psyllids, planthoppers, leafhoppers, spittlebugs, and whiteflies). This suggests this region of the Wolbachia 16S rRNA does not mutate rapidly compared to other bacterial taxa, precluding closer analysis of its population structure in these samples. As such, more gene markers other than 16S rRNA will be required to fully resolve the evolutionary relationships of these Wolbachia from Pariaconus psyllids (e.g. Baldo et al. 2006).
Putative Morganellaceae Endosymbionts
We identified ASVs initially assigned to the gammaproteobacterial family the Morganellaceae present in 100% of psyllid individuals from all free-living and open-gall psyllid species and therefore appear to be fixed in their populations (Fig. 2e). None of the closed-gall species were identified with high-frequency Morganellaceae ASVs (hereafter referred to as Morganella-like) (also see supplementary text S1, Supplementary Material online on the removal of contaminant ASVs). Further, phylogenetic reconstruction of these Morganella-like ASVs with outgroup 16S rRNA sequences from sequenced bacterial genomes with the greatest Blastn similarity suggests the Morganella-like microbes are distinct from sequenced insect endosymbionts (supplementary fig. S2b, Supplementary Material online).
Plant-Associated Enterobacterales Endo/Exosymbionts
Among the ASVs, those classified as Enterobacterales, which were not from the family Morganellaceae, represent the third most prevalent high-frequency ASV taxa among psyllid species and were detected in all host-plant ecologies (free-living, closed-gall, and open-gall) (Fig. 2e). Based on the phylogenetic analyses, two groups of the Enterobacterales ASVs clearly belong to two distinct plant-associated bacterial genera Rosenbergiella and Pantoea (supplementary fig. S2c, Supplementary Material online). The third group exhibited phylogenetic instability due to their divergent 16S sequences resulting in a floating long-branch that more frequently resolved near plant-associated bacterial genera Dickeya or Symbiopectobacterium (supplementary fig. S2d, Supplementary Material online). Interestingly, only free-living psyllid species are associated with microbes that belong to this Dickeya-like symbiont, and it is present and fixed at 100% infection frequency in all free-living species and at high relative read frequencies (Pariaconus wyvernus = 27%, P. hina = 34%, Pariaconus proboscideus = 24%, and Pariaconus gracilis = 19%) (Fig. 2e).
In contrast, the majority of closed-gall species are instead associated with a single Rosenbergiella ASV, with the exception to Pariaconus ohiacola, which is associated with an ASV assigned to the genus Pantoea (supplementary fig. S2c, Supplementary Material online). None of the ASVs for Rosenbergiella or Pantoea are found in 100% of psyllid individuals from any closed-gall species (x̅ = 61% ± 17% SD, n = 8), and relative read frequencies were low potentially suggesting an exosymbiont location (x̅ = 4.3% ± 5.2% SD, n = 8). Regarding phylogenetic patterns, similar to the Wolbachia ASVs above, the same Rosenbergiella ASV is associated with multiple closed-gall psyllid species. Based on Blastn, this Rosenbergiella ASV has 100% sequence identity and coverage with other Rosenbergiella sequences from NCBI (top 100) indicating that this region of the 16S rRNA is highly conserved for this bacterium as well.
Other Abundant Hawaiian Pariaconus Psyllid ASVs
Other high-frequency ASVs that were not fixed (e.g. 100%) in any species but were present in two or more psyllid species primarily belong to Gammaproteobacteria such as Pseudomonas (seven psyllid species), Enhydrobacter (four psyllid species), Acinetobacter (seven psyllid species), and Massilia (four psyllid species) taxa (supplementary Data Set S3, Supplementary Material online). Other Pseudomonadota taxa include those that belong to Alphaproteobacteria such as Sphingomonadaceae (five psyllid species), Beijerinckiaceae (eight psyllid species), and Acetobacteraceae (seven psyllid species). Other high-frequency ASVs among psyllid species belong to other bacterial phyla including Chryseobacterium from the phylum Bacteroidota (three psyllid species) and Cutibacterium from Actinobacteriota (four psyllid species) (supplementary Data Set S3, Supplementary Material online). Several of these ASVs appear specific to particular psyllid host-plant ecologies including Massilia, Sphingomonadaceae, Chryseobacterium, and Cutibacterium taxa, which were present only in several closed-gall psyllid species (supplementary Data Set S3, Supplementary Material online). None of the later taxa were present in all individuals of any of the closed-gall species (x̅ = 41% frequency ± 13.7% SD, n = 16) and were at relatively low read frequency (x̅ = 1.6% read frequency ± 3.9% SD, n = 16).
Cospeciation Patterns of Carsonella-, Morganella-, and Dickeya-like ASVs with Psyllids
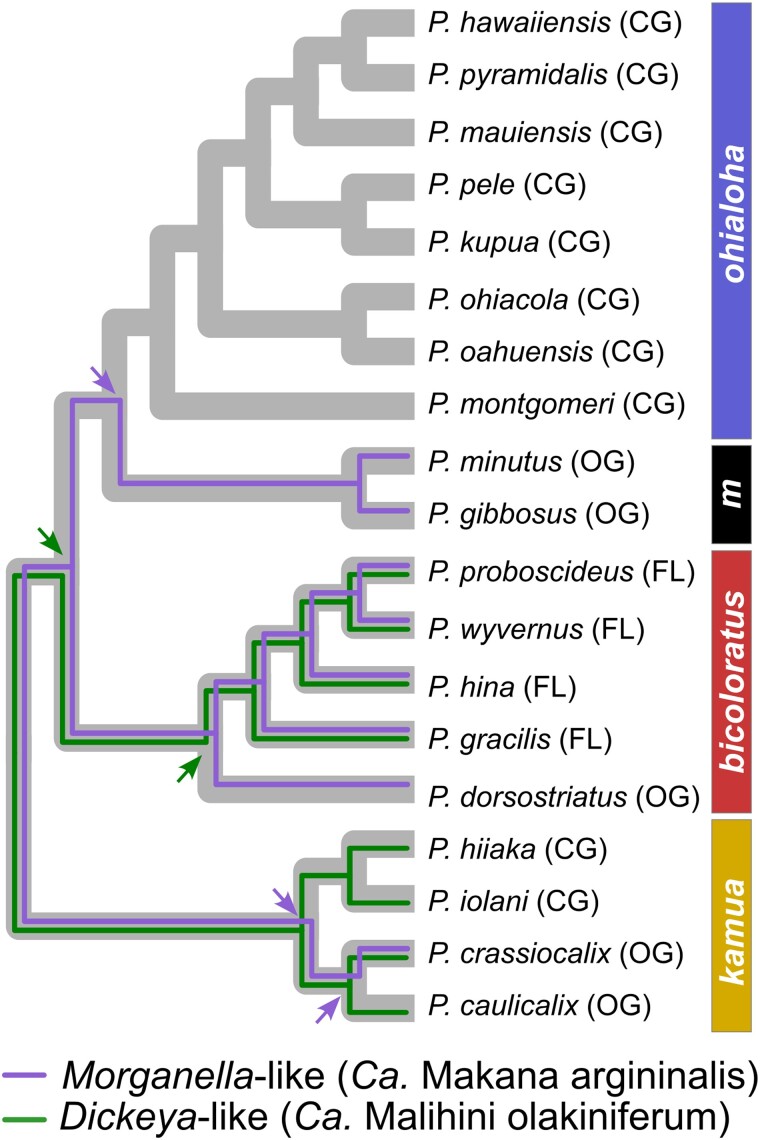
The Pariaconus high-frequency ASV groups represent phylogenetically distinct lineages of the Pseudomonadota (supplementary fig. S2a to d, Supplementary Material online). However, to obtain more information on the ancestral state of the potentially coevolved Morganella-like and Dickeya-like symbionts, we obtained additional 16S sequence data for four additional psyllid species (Pariaconus hiiaka, Pariaconus iolani, Pariaconus crassiorcalix, and Pariaconus caulicalix) from the kamua species group from Kauai (Fig. 1). Previous data indicate that the closed-gall state is derived, evolving twice, once on Kauai and again on the younger islands (Percy 2017). There are no extant free-living taxa on Kauai; therefore, only closed-gall (P. hiiaka and P. iolani) and open-gall (P. crassiorcalix and P. caulicalix) psyllid species were obtained for this post hoc 16S rRNA analysis. Screening the 16S rRNA clones identified two to six unique bacterial taxa from each of the insect species (n = 2 to 3 individuals/species). We detected the Dickeya-like symbiont and Wolbachia at variable frequencies (4.7% to 65%) in all four Kauai Pariaconus species, and the Morganella-like symbiont was only detected in the open-gall species P. crassiorcalix (supplementary Data Set S4, Supplementary Material online). Additional unique 16S rRNA clones were detected in P. hiiaka (n = 2) and P. caulicalix (n = 4) (supplementary Data Set S4, Supplementary Material online).
Using 16S rRNA sequences from all four psyllid species groups, we compared the phylogenetic histories of Carsonella-, Morganella-, and Dickeya-like symbionts with the established Pariaconus species phylogeny (Percy 2017, 2018) and found compelling evidence for a history of cospeciation for all three symbionts (Fig. 4; supplementary fig. S2e, Supplementary Material online). Individual phylogenies for each of the three symbionts were not significantly different from the psyllid phylogeny (approximately unbiased [AU] test, P > 0.05) (supplementary fig. S2e, Supplementary Material online). This result is expected for the obligate nutritional endosymbiont Carsonella; however, the Morganella-like and Dickeya-like microbes exhibiting the same pattern were surprising and suggest a long-term coevolved relationship with their psyllid hosts that is not the result of horizontal transfer. Given the free-living or open-gall state is hypothesized to be the ancestral host-plant ecology of Pariaconus psyllid species, it appears that the Morganella-like symbiont was independently lost twice, in tandem with the convergent evolution of the closed-gall ecology, i.e. in both the kamua group and the ohialoha group (Fig. 4). Interestingly, the Dickeya-like symbiont is present in both open- and closed-gall psyllids species in the kamua group and then is lost from all galling species after the Pariaconus lineage left the oldest island, Kauai, thereafter only remaining in free-living species (Fig. 4). These losses of the Morganella-like and Dickeya-like symbionts associated with galling host-plant ecologies and island colonization events appear to be fixed and nonreversible. We do note that our sampling and sequencing depth of the kamua group is not as extensive as for the other three species groups. Therefore, additional sampling will be important to test these observations.
Fig. 4.
Hawaiian psyllid radiation harbors additional coevolved symbiont lineages. Coevolutionary history of Pariaconus symbionts is traced onto the insect species cladogram. Individual sequence phylogenies for Morganella-like (Ca. Makana arginalis) and Dickeya-like (Ca. Malihini olakiniferum) 16S ASV symbiont sequences are not significantly different from the insect phylogeny (AU test, P > 0.05) (see also supplementary fig. S2, Supplementary Material online). As such, inferred evolutionary events of Morganella- and Dickeya-like symbiont loss are shown by arrows near the relevant ancestral node. Note cospeciation of Carsonella is not shown, but it is in all Pariaconus psyllids and mirrors the insect species cladogram (supplementary fig. S2, Supplementary Material online). Species groups are shown to the right of the tree, with minutus indicated by an “m.”
Metagenomic Assembly of Hawaiian Psyllid Endosymbionts
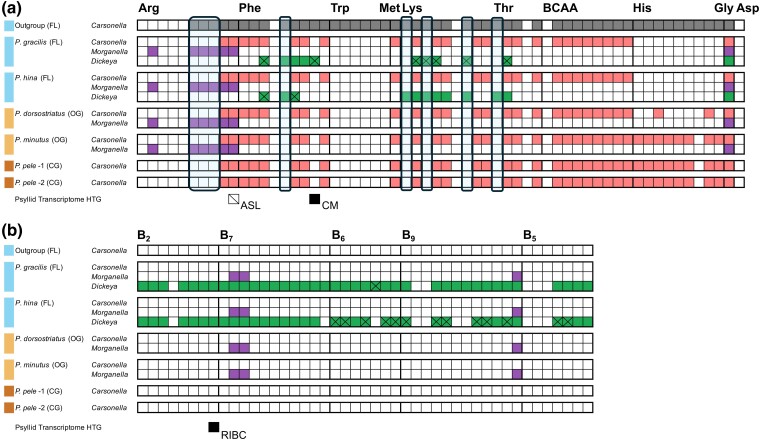
To better illuminate the impact of long-term coevolution and changes in host-feeding ecologies on these endosymbionts, we sequenced six metagenomes from psyllids representing each host-plant ecology, which includes closed-gall species (Pariaconus pele f pele 1 and P. pele f pele 2), open-gall species (P. minutus and P. dorsostriatus), and free-living species (P. gracilis and P. hina). Illumina sequencing resulted in an average of ∼220,364,824 high-quality reads (SE ± 25,382,186; n = 6) remaining after quality trimming (supplementary Data Set S5, Supplementary Material online). Reads from the primary nutritional endosymbiont Carsonella were de novo assembled into one contig/chromosome for all six samples with an average depth of coverage of ∼1,055 (SE ± 408; n = 6) (supplementary Data Set S6, Supplementary Material online). A high degree of synteny was detected among all of the Carsonella strains with only a single small rearrangement observed in P. gracilis involving the clsA gene (supplementary fig. S3a, Supplementary Material online). Also, a high similarity was observed in %GC (x̅ = ∼14%; SE ∼0.11%), genome length (x̅ = ∼152 kb; SE ∼1.6 kb), and average pairwise nucleotide identity (ANI) of 92.3% (SE ± 0.32) (supplementary Data Sets S6 and S7, Supplementary Material online). Gene content was also largely the same among the Carsonella genomes except for the presence and absence of coding sequences that were lineage specific (supplementary fig. S3b, Supplementary Material online). Notably, the lineage within the species group bicoloratus composed of both free-living and open-gall psyllids lost six genes within the His pathway and the two free-living species sequenced further lost two additional genes within the His pathway (supplementary fig. S3b, Supplementary Material online). When Hawaiian Carsonella was compared to the free-living outgroup Carsonella from the same psyllid family (Triozidae: Bactericera cockerelli) (Riley et al. 2017), several genes within five essential amino acids were lost (Arg, Phe, Lys, Trp, and His) (Fig. 5). All Hawaiian Carsonella genomes had the same genes lost from the Arg, Phe, Lys, and Trp pathways indicating an early loss that is ancestral to the three species groups bicoloratus, minutus, and ohialoha. In contrast, gene loss within the His pathway was extremely variable and was based on species group affiliation and lineage, with free-living psyllids losing all nine genes within the His pathway (Fig. 5).
Fig. 5.
Metabolic reconstructions of putative long-term symbionts of Pariaconus psyllid species. Matrices of gene presence, absence, or inactivation in assembled Pariaconus symbiont genomes and an outgroup from same psyllid family (B. cockerelli) for several critical a) amino acid and b) B vitamin biosynthesis pathways. Gene absences (not encoded) are white boxes and gene presence (encoded) are colored boxes corresponding to the Carsonella-, Morganella- (Ca. Makana arginalis), and Dickeya-like (Ca. Malihini olakiniferum) symbiont assemblies. Boxes with an “X” indicate inactivated pseudogene copies. The presence of horizontally transferred genes/transcripts from P. gracilis and P. montgomeri for homologs and/or same enzymes in corresponding symbiont pathways are indicated as detected (black box) or not detected (outlined box with cross).
Morganella-like symbiont genomes were only identified in the free-living and open-gall psyllid species samples, consistent with 16S rRNA analyses. De novo assemblies reconstructed single circular contigs (chromosomes) for each sample with an average depth of coverage of ∼1,218 (SE ± 615; n = 4) (supplementary Data Set S6, Supplementary Material online). Similar to Carsonella, a high degree of synteny was observed among all Morganella-like strains with no observed rearrangements (supplementary fig. S3c, Supplementary Material online). Also, Morganella-like taxa were highly similar in %GC (x̅ = ∼19.4%; SE∼0.003%), highly reduced sizes (x̅ = ∼279.6 kb; SE ± ∼2.5 kbp), ANI (87.3%; SE ± 0.15), and shared gene content (supplementary Data Sets S6 and S7 and fig. S3d, Supplementary Material online). Morganella-like symbionts only encode six genes involved in the essential amino acid biosynthesis pathways and these genes are all involved within the Arg pathway, and all Hawaiian psyllid Carsonella taxa do not encode four of these genes (Fig. 5). Importantly, three of these genes are downstream from the intermediate ornithine, which is often provided by the host psyllid via nuclear-encoded genes (Hansen and Moran 2014; Kwak et al. 2023) and the outgroup Carsonella still encodes these downstream genes (Fig. 5). In turn, the free-living and open-gall psyllids, which possess Morganella-like symbionts, are assumed to be able to produce the essential amino acid Arg through the complementation of genes involved in Arg biosynthesis from both the Morganella-like symbiont and Carsonella.
Again, consistent with the 16S rRNA analyses, the Dickeya-like endosymbiont genomes were only detected in the metagenomes of the two free-living psyllid species sampled. The genomes were de novo assembled into eight and ten contigs with an average depth of coverage of ∼410× and ∼86× for Dickeya-gracilis and hina, respectively (supplementary Data Set S6, Supplementary Material online). Unlike Carsonella- and Morganella-like genomes, genomes of the Dickeya-like strains have experienced multiple small- and large-scale rearrangements (supplementary fig. S4a, Supplementary Material online). The Dickeya-like genomes are also substantially larger than Carsonella and the Morganella-like symbiont (Dickeya-gracilis: 1.1 Mb and Dickeya-hina: 1.3 Mb) and have greater %GC compositions (Dickeya-gracilis: ∼44.4% and Dickeya-hina: ∼38%). Analysis of the gene annotations revealed surprisingly low coding density in both genomes (43% to 59%) that upon further inspection is the result of rampant pseudogene formation (Dickeya-gracilis: 270 and Dickeya-hina: 184) (supplementary fig. S4b, Supplementary Material online). Furthermore, despite the synteny between flanking genes, many long intergenic regions were found to be highly diverged, providing evidence that ancestral genes have been completely eroded by mutation. As a result, only 486 intact, orthologous protein-coding genes were detected between both Dickeya-like de novo genomes. This left 32% to 35% of predicted CDS in each genome as unique (Dickeya-gracilis: 227 and Dickeya-hina: 261). As indicated above, all Hawaiian Carsonella are missing two key enzymes in Phe biosynthesis and three key enzymes for Lys biosynthesis. Although both Dickeya-like taxa appear to have lost most of their genes for essential amino acid biosynthesis, there are exceptions in the Lys and Phe pathways (Fig. 5). Dickeya-gracilis can complement both enzymes for Phe biosynthesis while Dickeya-hina can only complement one. This complementation is largely reversed for Lys, as Dickeya-hina can complement all three enzymatic steps and these genes are missing or pseudogenes in Dickeya-gracilis. In addition, both Dickeya-like genomes encode intact B vitamin biosynthetic pathways for riboflavin (B2) and biotin (B7). However, only Dickeya from P. gracilis has intact pyridoxal 5′-phosphate (B6), folate (B9), and CoA (pantothenic acid) (B5) pathways. Fifty percent or more of the genes for these three pathways have been inactivated in Dickeya-hina (Fig. 5).
Phylogenetic Origins of Novel Pariaconus Symbionts
To further determine if Pariaconus symbionts are closely related to other insect symbionts, we conducted phylogenetic analyses with additional sequence data from NCBI. Initial naïve Bayes taxonomy classifier analysis placed the Morganella-like symbiont in the family Morganellaceae; however, the initial ASV phylogeny did not conclusively place this group of symbionts (supplementary fig. S2b, Supplementary Material online). Therefore, additional phylogenetic analyses were employed to determine if it was related to any of the other known insect bacterial symbionts from this family of microbes. An array of identified Morganellaceae and Enterobacteriaceae symbiont 16S rRNA sequences from both psyllids and other insects was analyzed with those from the Pariaconus symbiont. This revealed that the Pariaconus Morganella-like symbionts are a monophyletic group that is diverged from the other known symbionts (supplementary fig. S5a, Supplementary Material online). As discussed above, short 16S rRNA ASVs can sometimes encode limited phylogenetic information. Due to the apparent sequence divergence (long-branch artifact), the 16S ASVs for the Dickeya-like symbiont exhibited phylogenetic instability at times branching near or within the genus Dickeya or with Symbiopectobacterium. To resolve this, we leveraged the whole-genome sequences to recover a core set of 330 protein genes conserved in the symbiont along with representative Dickeya, Symbiopectobacterium, and Pectobacterium species. The resulting phylogenetic estimation places the Dickeya-like symbiont as sister to the genus Symbiopectobacterium (rather than within the genus) with 100% bootstrap support (supplementary fig. S5b, Supplementary Material online). Further comparisons of their overall shared proteomes strongly suggest the distinction of the Dickeya-like symbionts from all three genera. Within genus, Percentage of COnserved Proteins values (Qin et al. 2014) averaged between 63% and 83%, whereas between genus, values were overlapping but always lower, ranging between 22% and 69% (supplementary Data Set S8, Supplementary Material online). Together, these data suggest both Pariaconus symbionts represent novel Candidatus genera.
Rates of Evolution in Coevolving Symbiont Genomes
Given the varied combinations of symbionts found among the Hawaiian Pariaconus species, it is conceivable that natural selection may have left its mark on the genes of one or more of the symbionts. Therefore, rates of molecular evolution were estimated among the Carsonella-, Morganella-, and Dickeya-like genomes. We found that in all symbiont lineages, the overwhelming majority of orthologous genes have experienced purifying selection (tree dN/dS < 1; 176/178, 242/242, and 486/486, respectively). Note that the few exceptions appear to be driven by gene comparisons where dS ∼ 0 due either to recently diverged lineages and/or conserved hypothetical genes that are incredibly short < 130 bp. Regardless, whole-gene estimates of dN/dS can mask positive selection that may have acted at individual codons within genes or along specific lineages; therefore, a nested approach was used to determine if there is any evidence of such selection (Álvarez-Carretero et al. 2023). Only a minority of genes met the significance threshold of encoding potentially positive selected codon positions (2Δl > χ21,5%), including rpsP, rpsM, and def in Carsonella, rpsA, rpsO, and secE in Morganella and rplN, rpmC and 12 other genes in Dickeya. Furthermore, only two genes in the Morganella-like symbiont show evidence of lineage-specific positive selection (rpsA, secE), and in both instances, greater dN/dS values were detected on the branch leading to Morganella-minutus.
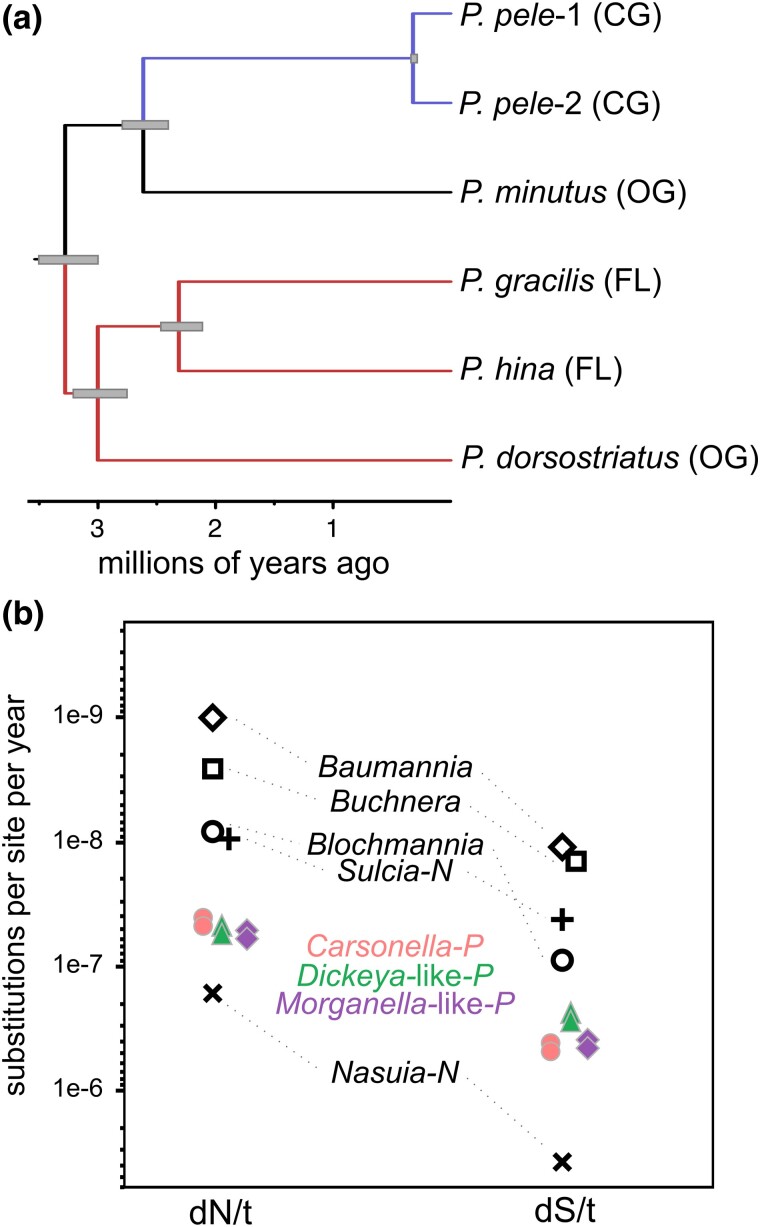
Like many insect-associated endosymbionts, Carsonella-, Morganella-, and Dickeya-like symbionts from Pariaconus spp. are on long phylogenetic branches indicative of elevated rates of nucleotide substitutions compared to free-living Bacteria (supplementary figs. S2 and S5, Supplementary Material online). In addition, it has been previously estimated that the Pariaconus bicoloratus and minutus lineages emerged ∼3 to 3.5 MYA (Percy 2017). This enabled us to calibrate the rate of substitutions per year of these three symbionts (Fig. 6; supplementary Data Set S9, Supplementary Material online). Remarkably, the rates of dN/t and dS/t of all three symbionts are all within 2-fold of one another. Furthermore, their overall substitutions rates are ∼3 to 38× faster than similar estimates for symbionts of aphids, ants, sharpshooters, and leafhoppers. Despite these elevated rates, we note that the Pariaconus symbionts are still 9 to 15 times slower than the incredibly fast-evolving symbiont Nasuia of Hawaiian leafhoppers (Vasquez and Bennett 2022).
Fig. 6.
Rapidly evolving Pariaconus symbiont genomes. a) Calibrated Carsonella ultrametric tree based on 153 single-copy orthologs. Gray bars represent estimated age ranges for each node based on a fixed divergence time at the root node of 3 or 3.5 MY. b) Genome-wide estimates of time-resolved rates of nonsynonymous (dN) and synonymous (dS) divergence in Pariaconus symbiont genomes compared to other published symbionts rates (Vasquez and Bennett 2022). Two points are shown for Carsonella- (filled circles), Morganella- (filled diamonds; Ca. Makana arginalis), and Dickeya-like (triangles; Ca. Malihini olakiniferum) based on upper (3.5 MY) and lower (3 MY) bound time estimates.
Presence of Known HTG in Hawaiian Pariaconus Psyllids
Genomic and transcriptomic data from P. gracilis (free-living) and Pariaconus montgomeri (closed-gall), respectively (supplementary Data Set S10, Supplementary Material online), were analyzed for evidence of psyllid-acquired, horizontally transferred genes (Sloan et al. 2014; Kwak and Hansen 2023). We identified alleles of all characterized HTGs in both species except for argininosuccinate lyase (ASL, EC 4.3.2.1), which was not found in either species and 16S rRNA methyltransferase (RSMJ), which was found in P. gracilis but not P. montgomeri. The missing transcript/gene of ASL is important because it carries out the last step of Arg biosynthesis in collaboration with Carsonella (Sloan et al. 2014; Kwak and Hansen 2023). Carsonella taxa, including all the taxa sequenced in this study, also encode a redundant terminal step enzyme (argH, EC 4.3.2.1). Nevertheless, this HTG and its paralogs, in addition to other HTGs that appear to be transferred early in the psyllid lineage, appear to still be maintained in three previously studied divergent psyllid species (Sloan et al. 2014; Kwak and Hansen 2023). Given that we do not have a fully sequenced genome for the two Pariaconus psyllid species, we cannot discount that this gene is still encoded and/or just not expressed highly compared to other HTG. Based on a previous whole-body adult transcriptome analysis of another triozid psyllid species (B. cockerelli), all three ASL copies were detected between 48% and 58% relative expression compared to other genes expressed in the psyllid's genome (Kwak et al. 2023). In addition to ASL, the only other HTG involved in the essential amino acid pathways with Carsonella is chorismate mutase (CM, EC 5.4.99.5), in which all three copies were detected in both Hawaiian species; for reference, these three copies were expressed in B. cockerelli whole-body adult samples between 35% and 93% relative gene expression compared to other expressed genes in the whole-body transcriptome (Kwak et al. 2023). The HTG CM is thought to be involved in Phe biosynthesis with Carsonella, and, similar to ASL, Carsonella taxa encode a redundant enzyme function (Sloan and Moran 2012; Kwak et al. 2023). The HTG RIBC involved in the last step of riboflavin biosynthesis (B2) was also detected in both Hawaiian species (P. gracilis and P. montgomeri) (Fig. 5b).
Discussion
Pariaconus-Nutritional Symbionts as a Model System
Dynamic evolutionary systems are, arguably, better captured and more tractable for hypothesis testing in island systems than anywhere else. In the Hawaiian Islands during the last ∼3.5 MY, Pariaconus psyllids rapidly lost two coevolving nutritional symbionts, the Morganella-like and Dickeya-like symbionts, and this is associated with shifts in their galling ecologies. In contrast, studies that show how different insect-feeding ecologies are associated with the gains and losses of symbionts are often looking at changes that may have occurred over 10 to 100 s of millions of years (Moran et al. 2005; Sudakaran et al. 2017; Bell-Roberts et al. 2019). Whereas, in Pariaconus psyllids, the Morganella-like symbiont appears to have been lost independently twice, and the two occasions coincide with the convergent evolution of the closed-gall state. These gall shifts occurred within both the basal kamua species group (∼4.7 MY based on island dating) and the derived ohialoha group (∼3.6 MY based on island dating) (Bastin et al. 2024). The Dickeya-like symbiont was lost in all sampled open- and closed-gall Pariaconus after the psyllid lineage left the oldest major aerial island of the Hawaiian archipelago (Kauai) (∼5 to 3 MY based on island dating) and appears to remain only in free-living psyllids, which are currently known to be present on all the major Hawaiian Islands except Kauai (Percy 2017) (Fig. 1). This loss of the coevolved Morganella-like and Dickeya-like symbionts, which encode missing genes from Carsonella's essential amino acid and B vitamin biosynthesis pathways, supports a previous hypothesis suggesting that galling insects should harbor fewer nutritional co-symbionts compared to nongalling insects, because the galling habit creates a nutritional sink for certain insect hosts (Spaulding and Von Dohlen 2001). To our knowledge, the loss of coevolved insect symbionts concurrent with the evolution of the galling habit has never been systematically observed before in insects especially for a monophyletic species-level phylogeny. Pariaconus psyllids are therefore one of the first in situ model systems with which to observe this phenomenon. This endemic Hawaiian radiation with a diversity of galling/nongalling ecologies on the same host-plant species across differently aged islands has created a natural “common garden” host-plant species design for testing symbiosis evolutionary hypotheses within a time-calibrated phylogenetic framework. In other psyllid genera that have multiple species occurring on the same host-plant species (e.g. in genera Arytainilla, Bactericera, Cacopsylla, Calophya, Mitrapsylla, and Queiroziella), there are generally fewer than ten species on the same host (e.g. Burckhardt 2021), with fewer distinct host-plant ecologies, and they are all continental genera in which processes of speciation have not been explicitly tested for monophyly, sister taxon relationships, or sympatry. In turn, observing psyllid symbioses in association with galling habit is more difficult in these systems.
The loss of coevolved nutritional symbionts has been observed previously at the family level or higher for specialized diet shifts in insects (Cornwallis et al. 2023). Examples of diet shifts that are associated with symbiont losses include the loss of Sulcia, an ancient nutritional symbiont, which is associated with specialized diet transitions from xylem to phloem as well as shifts to parenchyma feeding in Auchenorrhyncha insects (Moran et al. 2005; Bell-Roberts et al. 2019). An example of this type of diet switch includes Typhlocybides planthoppers that lost their obligate symbiont when they switched from a plant sap to a parenchyma diet, which is more nutritionally enriched (Buchner 1965; Bell-Roberts et al. 2019). In another example, the mealybug genus Hippeococcus lost its symbionts when shifting from an ancestral sap-feeding habit to a myrmecophilous lifestyle, where it requires nutrient provisioning by ants. Indeed, Hippeococcus female ovaries and embryos only develop after they have been carried off into ant nests (Buchner 1956).
Carsonella Functional Roles
Pariaconus Carsonella taxa from all three host-plant ecologies did not differ significantly in gene presence or absence except for several housekeeping genes involved in ribosomal and tRNA processes, NAD kinase, PRPP biosynthesis, and genes involved in the biosynthesis of the essential amino acid His (Fig. 5; supplementary fig. S3b, Supplementary Material online). Some of these genes may have been lost in a cascading fashion due to their interdependence on each other within interacting metabolic pathways, thereby resulting in a relaxation of purifying selection and subsequent gene loss. For example, within the bicoloratus lineage, which encompasses both free-living and open-gall psyllid species, six genes involved in histidine biosynthesis are lost, with three additional genes lost exclusively in the free-living psyllid species. Furthermore, the loss of ribose-phosphate diphosphokinase (prs) in the bicoloratus lineage, involved in PRPP biosynthesis, could be linked to the loss of genes within the His biosynthesis pathway given PRPP is important for the biosynthesis of two amino acids, His and Trp (Hove-Jensen et al. 2017), both of which Carsonella in the bicoloratus group do not synthesize anymore (Fig. 5). The complete loss of the His and Trp pathways in Carsonella has been observed previously. In free-living (Heteropsylla) and closed-gall (Pachypsylla) psyllid species, the co-symbionts are not always present to aid in His or Trp biosynthesis (Sloan and Moran 2012; Hansen and Moran 2014). These results indicate that for some free-living and closed-gall psyllids, their diets are sufficient in His and Trp. Interestingly, in three independent Carsonella lineages that cannot synthesize His anymore, the loss of purA also co-occurs in these Carsonella taxa from Heteropsylla and Pachypsylla (Sloan and Moran 2012) and the Pariaconus members of the bicoloratus group in this study (Fig. 5). The interaction of purA with the His biosynthesis pathway has been observed previously in free-living bacteria given ATP is needed for His biosynthesis and purA is the first step for ATP biosynthesis (Hartman 1956; Shedlovsky and Magasanik 1962).
In comparison to Carsonella from the free-living psyllid B. cockerelli (which belongs to the same psyllid family as Pariaconus), all Pariaconus Carsonella genomes sequenced here, representing all three host-plant ecologies, are missing the same genes within the Arg, Phe, and Lys biosynthesis pathways (Fig. 5). This suggests that the loss of these genes in the Hawaiian radiation occurred either once in the Carsonella ancestor of the colonizing free-living or open-gall psyllid or independently multiple times after the psyllid lineages (bicoloratus, minutus, and ohialoha) split off from the basal Pariaconus lineage (kamua), which only resides on Kauai, the oldest major aerial island of the archipelago. Based on metabolic reconstructions and 16S rRNA and phylogenetic analyses, the symbiotic role we predict for the Morganella-like symbiont—as a coevolved symbiont of free-living and open-gall psyllids—is for complementing Carsonella's gaps for Arg biosynthesis using host-derived ornithine as an intermediate (Fig. 5). Interestingly, the HTG ASL, which is conserved and expressed in divergent psyllid species and redundant with Carsonella's terminal step for Arg biosynthesis (Sloan et al. 2014; Kwak et al. 2023) along with Morganella-like symbiont here, is not expressed or encoded in either closed-gall or free-living Pariaconus species analyzed here. This suggests, based on parsimony, the loss or reduction of expression levels of this HTG in the ancestor of the Pariaconus lineage. Potentially, the Morganella-like symbiont, instead of the host HTG ASL, collaborates with Carsonella in this terminal step for Arg biosynthesis in open-gall and free-living psyllids, but this requires further examination.
Arginine is generally either encoded in Carsonella and/or its co-symbiont from all psyllid metagenomes sequenced to date (Sloan and Moran 2012; Dittmer et al. 2023). The one exception is the free-living psyllid Ctenarytaina spatulata (Sloan and Moran 2012) and now the closed-gall Pariaconus psyllids here. Potentially, the host-plant diets of C. spatulata and closed-gall Pariaconus psyllids are not deficient in the essential amino acid Arg due to these psyllids modifying their host-plant environment to elevate Arg concentrations. Indication that the host-plant feeding environment of Pariaconus closed galls is dramatically different compared to Pariaconus open galls was found recently in Amada et al. (2020) where closed-gall leaf tissue has much higher moisture stress compared to open-gall leaf tissue. Bailey et al. (2015) corroborate these findings that Pariaconus closed galls create moisture stress in M. polymorpha leaves by showing that drought genes are upregulated only in Pariaconus galling leaf tissue compared to nongalled leaf tissue on the same plant. Given that drought stress in addition to other stressors can increase nitrogen availability to psyllids and other sap-feeding insects (White 1984), potentially, closed galls are more enriched in some essential amino acids, such as Arg, compared to both free-living and open-gall psyllids. For the free-living psyllid C. spatulata, it could potentially induce an elevation in Arg concentrations in its host plant even though it is not a gall former. This is because it is known to cause leaf senescence, necrotic lesions, and the proliferation of lateral shoots on certain tree species, especially when they are drought stressed (Queiroz et al. 2010). Leaf senescence that can be initiated through stressors including wounding and feeding by certain psyllids, such as the free-living psyllid Cardiaspina near densitexta, has been shown to increase the levels of Arg in addition to other essential amino acids such as Ile, Leu, Lys, Thr, Trp, and Val when causing leaf damage to Eucalyptus moluccana (Steinbauer et al. 2014). In turn, the loss of obligate co-symbionts from psyllid lineages may not just be associated with gall formation but other factors as well where psyllid species reliably induce plant stress (e.g. moisture stress) by manipulating their host-plant environment thereby elevating essential amino acid concentrations when feeding and developing on their host plants.
Coevolved Nutritional Symbionts
The Morganella-like symbiont displays hallmarks of a long-term obligate nutritional co-endosymbiont given that it (i) is present in all psyllid individuals screened from open-gall and free-living psyllid species after leaving Kauai, (ii) is present in these individuals at high 16S rRNA relative read frequencies compared to other putative co-symbionts per individual, (iii) appears to be cospeciating with psyllid species based on phylogenetic analyses, and (iv) is highly reduced in genome size (∼280 kb), AT rich, and highly syntenic. Compared to the Morganella-like symbiont, the Dickeya-like symbiont's genome is not as reduced in genome size or AT rich suggesting that this symbiont has a more recent association to Pariaconus psyllids. Nevertheless, similar to the Morganella-like symbiont, the Dickeya-like symbiont also appears to infect all individuals sampled from free-living psyllid species and has coevolved with these psyllids before the Pariconous lineage split into four species groups (Fig. 4). These latter results support the hypothesis that a free-living psyllid was the ancestral colonizer of the Pariconous lineage on Kauai instead of the other plausible alternative, an open-gall psyllid, based on prior phylogenetic reconstructions (Percy 2017). The Dickeya-like symbiont's genome lost many genes for the provisioning of essential amino acids except only for several genes that may complement Carsonella's gaps for Phe and Lys biosynthesis (Fig. 5). These results suggest that free-living Pariconous psyllids may need these additional essential amino acids compared to open- and closed-gall psyllids. Observations suggest that both open and closed galls may alter their host-plant tissue environment. This is evident from the fact that galls from a variety of Pariaconus species are occasionally reddish in color and chlorotic (Percy 2017). These leaf phenotypes have previously been linked to other psyllid species, attributed to psyllid-induced leaf senescence, which can mobilize nitrogen nutrients to psyllid feeding sites (Steinbauer et al. 2014). However, the development of this reddish leaf color, associated with high concentrations of anthocyanins, does not always occur during psyllid-induced leaf senescence. Its occurrence depends on abiotic conditions (Steinbauer et al. 2014).
The Dickeya-like symbiont's genome encodes several partial and intact B vitamin pathways, which can be beneficial to an insect host. Like essential amino acids, insects cannot produce their own B vitamins de novo and rely on their diet and/or microbiome to provide these coenzymes for their central metabolic pathways (Douglas 2017). Insects that switch to diets with elevated B vitamin concentrations have previously been associated with symbiont loss, highlighting the importance of nutritional symbionts for B vitamin provisioning when feeding on diets with low B vitamin concentrations (Cornwallis et al. 2023). Symbiont taxa of insects that feed on low B vitamin diets belong to a diversity of bacterial lineages demonstrating convergent evolution for these important symbiotic associations (Cornwallis et al. 2023; Serrato-Salas and Gendrin 2023). Sap-feeding insects such as aphids, whiteflies, planthoppers, and leafhoppers rely on symbiont-provisioned B vitamins for postembryonic development, longer lifespans, and/or egg production (Serrato-Salas and Gendrin 2023). In psyllids, the primary nutritional symbiont Carsonella generally does not encode genes for B vitamin biosynthesis; however, psyllids, including the species sampled here, are known to express/encode the HTG RIBC, which is the end step of the riboflavin biosynthesis pathway (Sloan et al. 2014; Kwak et al. 2023). These results indicate that psyllids must obtain an intermediate from either their host-plant diet and/or co-symbiont for riboflavin biosynthesis. Psyllid co-symbionts that have been found to encode B vitamin pathways include Profftella of Diaphorina host species and Psyllophila of Cacopsylla host species (Nakabachi et al. 2020; Dittmer et al. 2023). These latter psyllid co-symbionts encode all genes for the riboflavin pathway except for the end step, which Diaphorina citri is known to upregulate (RIBC) in symbiotic cells (bacteriomes) compared to other body tissues (Kwak and Hansen 2023). Other Hawaiian hemipterans that harbor a co-symbiont that encodes B vitamin biosynthesis genes include seed-feeding Nysius leafhoppers (Stever et al. 2021) and phloem-feeding planthoppers that live both aboveground feeding on ferns and in lava tubes feeding on roots of M. polymorpha (Gossett et al. 2023). The co-symbiont Purcelliella from lava tube-dwelling planthoppers, which feeds on the same host plant (M. polymorpha) as Pariaconus psyllids, encodes nearly all the genes within the B vitamin pathways of biotin (B7), riboflavin (B2), and pyridoxin (B6) similar to the Dickeya-like symbiont of Pariaconus psyllids. It will be of interest of future studies to measure essential amino acids and B vitamin concentrations in different M. polymorpha plant parts, in addition to plant tissues that are galled and ungalled to have a greater understanding of how symbiont genome metabolisms relate to essential nutrient concentrations in phloem from these different plant tissues.
New Candidatus Species Designation for co-symbionts
Given our results of distinct genomic, phylogenetic, 16S rRNA read depth and frequency patterns, and metabolic and ecological traits described above, we propose new Candidatus names for the Morganella-like and Dickeya-like co-symbionts discovered here. We propose the name “Candidatus Makana argininalis” for the new Morganella-like lineage. The generic name comes from the Hawaiian term for “gift,” makana, and the specific epithet refers to the role in providing the arginine metabolic pathway (+ -alis, Latin suffix for “pertaining to”). The genus “Ca. Makana” is classified in Bacteria, Pseudomonadota, Gammaproteobacteria, and Enterobacterales. We also propose the name “Candidatus Malihini olakiniferum” for the new Dickeya-like lineage. The generic name comes from the Hawaiian term for “guest,” malihini (treated as indeclinable neuter), and the specific epithet from the Hawaiian term for “health” referring to the nutritive role (+ -iferum, Latin suffix for “bearing”), hence “health-bearing guest.” The genus “Ca. Malihini” is classified in Bacteria, Pseudomonadota, Gammaproteobacteria, Enterobacterales, and Enterobacteriaceae.
Coevolved Symbiont Genome Paradigm
Many reduced genome symbionts experience recurrent genetic bottlenecks and exhibit coincident patterns of elevated mutation rates, compositional biases, and genome-wide purifying selection. Indeed, all three symbiont lineages in this study align to this model, exhibiting genome-wide AT biases and strong evidence of purifying selection acting on intact protein-coding genes. Notably, we did detect a minority of genes with evidence of positively selected amino acid positions, although not nearly as many genes as detected in some other insect symbionts (e.g. Vasquez and Bennett 2022). Several of the genes with signatures of positive selection (ribosomal protein rpsA and def) have previously been identified as experiencing positive selection in the planthopper symbiont Sulcia as well (Gossett et al. 2023). While it is not immediately clear what effect the selected amino acid substitutions may have on these proteins, it is possible that these substitutions may modulate translational activity in these symbionts.
Remarkably, we found highly similar estimated rates of synonymous and nonsynonymous substitutions per year in all three symbionts (Fig. 6). This is despite the marked differences in the genes they retain for DNA repair and replication and the considerable differences in their genome sizes overall (supplementary Data Set S9b, Supplementary Material online). For instance, both Carsonella and C. Makana argininalis (Morganella-like symbiont) have diminutive genomes (<300 kb), yet C. Makana argininalis unlike many insect symbionts of similar size has retained a complete DNA recombination complex (recBCD) and DNA mismatch repair gene (mutS). While C. Malihini olakiniferum (Dickeya-like symbiont) genomes have similar substitution rates, their genomes are 4 to 6× larger and show a genome-wide trend of gene inactivation. These traits coincide with genome trends in “recently” acquired endosymbionts (McCutcheon and Moran 2012). Despite the coevolutionary history of both C. Makana argininalis and C. Malihini olakiniferum with Hawaiian Pariaconus species, and given the dramatic differences in their genomes, we speculate that C. Makana argininalis (Morganella-like symbiont) may share a longer history with Pariconous prior to the arrival of these insects in the Hawaiian Islands, whereas C. Malihini olakiniferum (Dickeya-like symbiont) potentially was acquired shortly after arriving. Regardless, it appears that the C. Malihini olakiniferum (Dickeya-like symbiont) continues to experience relaxed purifying selection across large swaths of its genome, leading to gradual decay of unnecessary gene functions (supplementary fig. S4, Supplementary Material online). Similar patterns appear to have affected several members of the sister genus Symbiopectobacterium, which also appear to have transitioned from free-living microbes to host-associated symbionts (Martinson et al. 2020).
Symbiont Role in Gall Formation
Bacteria are known to induce galls by producing metabolites within cytokine and auxin pathways (Barash and Manulis-Sasson 2009); however, the role of insect symbionts in gall formation is unclear currently. For instance, a previous study found no systematic differences in insect microbiome diversity between galling and nongalling insects when examined at a high phylogenetic resolution, suggesting that microbial associates of insects may not play a significant role in gall formation (Hammer et al. 2021). Nevertheless, other studies examining galling sawflies and flies have found potential correlations between specific insect microbiome taxa and galling (Bansal et al. 2014; Michell and Nyman 2021). Gall induction on M. polymorpha from Pariaconus closed-galling psyllids is hypothesized to be due to an auxin response based on transcriptional evidence from psyllid galled M. polymorpha plant tissue compared to nongalled tissue on the same individual host plants (Bailey et al. 2015). Auxin and cytokine production has been previously observed in insects that produce galls (Yamaguchi et al. 2012; Suzuki et al. 2014). For example, Suzuki et al. (2014) characterized the de novo pathway for auxin via tryptophan both in divergent galling and nongalling insects suggesting that endogenous auxin pathways are present in insects in general. Given Carsonella and the co-symbionts from Pariaconus psyllids do not encode a pathway for Trp, it is expected that these psyllids, especially closed-gall psyllids, must obtain this essential amino acid from their diet and/or other microbial associates. Bacterial taxa that were associated only with Pariaconus closed-gall psyllid species (Cutibacterium, Chryseobacterium, and Massilia) and open-gall species (Enhydrobacter) were not present in all individuals of these galling species and were present at low read frequencies compared to Wolbachia. Wolbachia is a known insect endosymbiont that is generally horizontally transferred in insects (supplementary fig. S2a, Supplementary Material online) and potentially fixed in some populations (Fig. 2); however, its effects on psyllid hosts is not well known (Kwak et al. 2021) and does not seem to have an association with host-plant ecologies in Pariaconus psyllids. The taxa found exclusively in closed-gall Pariaconus psyllids, Cutibacterium, Chryseobacterium, and Massilia are known to be plant-associated bacteria, where Chryseobacterium species (Chryseobacterium tagetis sp. nov.) are known to produce antibiotics and plant growth promoting abilities such as auxin (Campisano et al. 2014; Chhetri et al. 2022; Francioli et al. 2022; Jeon et al. 2023). Only three closed-gall species were associated with Chryseobacterium taxa, however, and 25% to 49% of the individuals sampled from those species were infected with this microbe. Alternatively, if galling is associated with microbiome members, multiple bacterial taxa that share functionally redundant genes for Trp and/or auxin or cytokine-producing metabolites would be needed, as we did not find any clear patterns of one particular taxon only associated with a galling species that was fixed at 100% infection frequency. Thus, more evidence is needed from metagenomic sequencing to determine if ASVs associated with gall formers here encode genes that are associated with auxin production.
Conclusion
In conclusion, our study provides compelling evidence that Pariaconus psyllids serve as an exemplary model for understanding the rapid loss of coevolved nutritional symbionts in association with shifts in galling ecologies. The independent losses of C. Makana argininalis (Morganella-like) and C. Malihini olakiniferum (Dickeya-like) across different lineages within the Pariaconus radiation highlight a complex interaction between insect hosts and their symbiotic partners potentially driven by ecological factors such as diet and habitat. Since animals, including insects, cannot synthesize their own essential amino acids, these amino acids must come from symbionts or their plant diet. Therefore, we argue that these different nutritional symbiont coevolutionary patterns and their metagenomes predict changes in the psyllids’ requirements from the diet. This phenomenon underscores the dynamic nature of symbiotic relationships and their potential role in facilitating ecological diversification over rapid evolutionary timescales. Our findings also support the hypothesis that galling insects harbor fewer nutritional co-symbionts due to the nutritional modifications caused by their galling behavior. Future research should focus on elucidating the specific mechanisms by which these psyllids manipulate their host plants and how this impacts their nutritional environment. Ultimately, understanding the intricate balance between host-plant manipulation and symbiont loss will enhance our comprehension of insect–plant–microbe interactions and their evolutionary implications.
Materials and Methods
Psyllid Sample Collection, Identification, and DNA Extraction Methods/Details
Specimens were field collected by DP between 2002 and 2014. Field sampled material was collected by sweep netting individual plants, aspirating directly from the host plant, or in the case of immatures removing directly from galls and leaf surfaces with tweezers; specimens were transferred live into 90–95% ethanol and stored at −20 °C. In a few cases, adults were raised from bagged galled leaf material before preservation in ethanol. Nondestructive DNA extractions, similar to Kwak et al. (2021), were conducted using whole individual psyllid specimens using the Qiagen Blood and Tissue Kit (Qiagen) or QIAamp UCP DNA Micro Kit (Qiagen). DNA voucher specimens were preserved in ethanol (70% to 85%) and retained in DP personal collection (DMPC, University of British Columbia).
The final sample size per species (e.g. individual psyllids/species) ranged from 2 to 107 individuals/species. Samples were removed if (i) minimum reads/sample not met or (ii) if species validation via CO1 primers (Simon et al. 1994) and/or mitogenome analysis could not be confirmed. See 16S rRNA analysis methods below and supplementary Data Sets S1 and S2, Supplementary Material online and Fig. 1 for more sample details. The sample site map image was generated using the mapview function in R package version 2.11.0 (Appelhans et al. 2024).
Sample Preparation and Illumina miSeq
Illumina sequencing of 16S rRNA was carried out as in Kwak et al. (2021). Briefly custom 16S rRNA primers were synthesized by Integrated DNA Technologies, Inc. (San Diego, CA, USA) using a dual barcode design with 25 primer pairs using the forward primer, 16S-341F = 5′-CCTACGGGNGGCWGCAG-3′, and the reverse primer, 16S-805R = 5′-GACTACHVGGGTATCTAATCC-3′ (Morrow et al. 2017). These latter primers span the V3 to V4 region of the 16S rRNA gene resulting in a 464b-p amplicon. Library preparation directly follows the pipeline detailed in Kwak et al. (2021), and 16S rRNA amplicons were multiplexed in a single lane for dual index sequencing on the Illumina MiSeq PE300 at the DNA Technologies and Expression Analysis Core Laboratory at the University of California, Davis.
The quality of raw reads was verified with FASTQC v.0.11.3 (Andrews 2010). FASTQC results revealed that R2 reads are of better overall quality than R1 reads. In turn, given that single-end reads are sufficient to observe the same relationships among samples that are revealed with paired-end reads (Caporaso et al. 2012) for diversity and statistical analyses, we analyzed the R2 reads for microbiome data analyses (see below). When possible, we used the paired-end data for phylogenetic analyses (see below) to provide greater sequence information.
Processing and Statistical Analysis of 16S rRNA Reads
The analysis of 16S rRNA read data was performed using the Qiime2/v2023.5 pipeline (Bolyen et al. 2019) to demultiplex and denoise quality filter reads into ASV tables.
Single-end reads were imported, demultiplexed (q2-demux), and then denoised using DADA2 (q2-dada2; Callahan et al. 2016) and trimmed to 200 nt in length. Separately, paired-end reads were first joined (q2-vsearch; Rognes et al. 2016), followed by quality filtering (q-score-joined), then denoising with DEBLUR (q2-deblur; Amir et al. 2017) and trimming to 400 nt in length. To assign taxonomy to the ASVs, the classify-sklearn naïve Bayes taxonomy classifier (q2-feature-classifier; Bokulich et al. 2018) was used against the SILVA database (silva-138-99-nb-classifier) (Quast et al. 2012). Single- and paired-end ASV tables were exported from Qiime, and contaminating sequences were identified and filtered out of the data sets using the prevalence of ASVs in the five negative control samples with the decontam R package (threshold = 0.05; Davis et al. 2018). The final ASV tables were filtered to remove all contaminating reads; insect mitochondrial and plant chloroplast sequences were also filtered out from the data sets using Silva classifications before subsequent analyses.
Rarefaction and diversity analyses of the single-end data were conducted in Qiime2/v2023.5, and libraries were rarified to 2,000 reads per sample. Samples under 2,000 reads (n = 13) were removed from the data set. Ordination analyses of the data were conducted in Qiime2/v2023.5 by using a weighted UniFrac PCoA (Lozupone and Knight 2005). A PERMANOVA (ADONIS) was conducted in Qiime2/v2023.5 using the weighted UniFrac distances and nested to account for species-level variation where the significance level was set to P < 0.05. This analysis was conducted both on the entire data set (n = 319 psyllid individuals/samples) and a randomly subsampled data set that standardized the number of individuals/samples per psyllid species where species with ≥4 samples/psyllid were retained resulting in a random subsampling of four individuals per species (n = 52 psyllid individuals/samples and 14 species total). This subsampled data set was rarified to 2,750 reads per sample based on the sample with the lowest number of reads. Alpha diversity measures of richness (Faith's phylogenetic distance) and evenness (Pielou evenness) were calculated in Qiime2/v2023.5. These values were normalized by Log10 transformation and analyzed using a factorial ANOVA with a univariate GLM where species was included as a random factor and the significance level was set to 0.05, and Bonferroni post hoc tests were carried out using IBM SPSS 29 (IBM 2024). The alpha diversity analyses were conducted only on the subsampled data set to standardize the number of samples per species.
Additional sequencing of 16S rRNA sequences for post hoc analyses was obtained from limited sample material from Kauai (supplementary Data Set S1, Supplementary Material online) where DNA extract was used for prior phylogenetic analyses (Percy 2017, 2018). All DNA extracts available were used per species per sample/individual for 16S rRNA amplification using the same primers as above for the 16S amplicon Illumina sequencing following the same PCR concentrations and conditions. PCR amplicons were cloned and Sanger sequenced using the same conditions, kits, and platforms as in Kwak et al. (2021) and Kwak and Hansen (2023). A minimum of ten clones were picked per individual extract except for one sample where DNA availability did not allow additional cloning. Additional screening of samples for Morganella-like symbiont was conducted with MorgF—5′-GCACAATGGGGGAAACCCTG-3′ and 16S-805R-GACTACHVGGGTATCTAATCC-3′. PCR conditions were the following: 95 °C for 3 min, 35 cycles of 30 s at 95 °C, 30 s at 60 °C, and 30 s at 68 °C, with a final extension of 5 min at 68 °C.
ASV Phylogenetic Analyses
Representative ASV and 16S rRNA clone sequences of interest were retrieved and aligned with appropriate outgroups using MUSCLE v3.8.1551 (Edgar 2004) (supplementary Data Set S11, Supplementary Material online). The outgroup 16S rRNA gene sequences were identified by BLASTn using the NCBI preformatted “ref_prok_rep_genomes” database (download September 21, 2023) and from Maruyama et al. (2023) (supplementary Data Set S11, Supplementary Material online). Sequences were retrieved from the database, trimmed to lengths corresponding to the ASVs and aligned as above. Aligned sequences were then subjected to approximately maximum likelihood tree estimation with FastTree v2.1.11 (Price et al. 2010). The nucleotide alignments were analyzed with a generalized time-reversible model of evolution under a discrete gamma model with 20 rate categories, and Shimodaira-Hasegawa-like local supports are reported.
To test if symbiont and psyllid phylogenies were congruent, unrooted, “best” phylogenies for Carsonella-, Morganella-, and Dickeya-like ASVs were generated with FastTree (-nt -gtr -gamma) (Price et al. 2010). In addition, the Carsonella ASV tree was reestimated using a constraint tree representing the species “subfamilies” as previously identified (Percy et al. 2018). These phylogenies were then compared to corresponding pared-down topologies based on the mitogenome tree from Percy et al. (2018) with the AU test (Shimodaira 2002) using the software IQ-TREE v2.2.2.6 (Nguyen et al. 2015). The AU test was run using the GTR + F + R3 model of evolution with weighted tests and 10,000 RELL replicates.
Metagenomic Methods
Six metagenomes were sequenced from one psyllid individual per psyllid species that represent each host-plant ecology that includes closed-gall species (P. pele f pele 1 and P. pele f pele 2), open-gall species (P. minutus and P. dorsostriatus), and free-living species (P. gracilis and P. hina) (supplementary Data Set S5, Supplementary Material online). The same DNA extraction method as above for 16S rRNA libraries was used here. Standard Illumina library preparation and sequencing were conducted by the University of California, Riverside Genomics Core facility. Metagenomic libraries were constructed using the New England Biolabs (Ipswich, MA, USA) FS DNA Library Prep Kit (E7805, E6177) with inputs of ≤100 ng of DNA. The libraries for the psyllid samples were pooled and sequenced as paired-end, 300-bp reads on three lanes of NextSeq 2000 using the Illumina P2 reaction kit.
The quality of raw reads for each sample run was verified with FASTQC v0.11.9 (Andrews 2010), and low-quality reads and adapter sequences were removed using Trimmomatic v.0.36 (Bolger et al. 2014) with quality parameters LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:100 or MINLEN:50 dependent on the run. Symbiont reads were assembled into contigs using MEGAHIT (v1.2.9) (Li et al. 2015) and binned using Anvi’o (v8.0) (Eren et al. 2020) with manual curation guided by PFAM (v33.1) and TIGRFAM (v15.0) databases. Manual curation of assemblies was further performed using Mauve (v2.4.0) (Darling et al. 2004) and Tablet (v1.21.02.08) (Milne et al. 2013). Draft genome completeness and quality were examined by computing the N50 using QUAST (v5.2) (Gurevich et al. 2013) and Busco (v5.4.4) (Simão et al. 2015). The percent nucleotide similarity between symbiont assemblies was conducted using JSpeciesWS (v4.1.1) (Richter et al. 2016). Initial gene prediction and annotation were carried out with PROKKA (Seemann 2014). Intergenic spacers from genomes with low gene density were reevaluated with BlastX to identify the presence of potential pseudogenes. These predictions were manually cross-referenced with additional pseudogene predictions made by Pseudofinder using default parameters (Syberg-Olsen et al. 2022). Final pseudogene predictions were marked when intact CDS represented <2/3 the length of known homologs and the presence of >1 disruptive mutation.
Phylogenetic Analyses of Dickeya-Like Symbiont
The origins of the Dickeya-like symbiont were determined by identifying single-copy orthologous proteins among a set of Symbiopectobacterium, Pectobacterium, and Dickeya genomes using OrthoVenn3 (Sun et al. 2023) (supplementary Data Set S8, Supplementary Material online). Alignments of the amino acid sequences for individual genes were performed using MUSCLE v3.8.1551 (Edgar 2004) and then combined into a single matrix. A maximum likelihood phylogeny was reconstructed using RAxML v8.2.12 using the JTT model of protein evolution (Stamatakis 2014) on the CIPRES server (Miller et al. 2010). Bootstrapping was allowed to be automatically stopped using MRE-based bootstrapping criteria threshold of 0.03 (autoMRE).
Molecular Evolution Analyses of Symbiont Genomes
Orthologous gene clusters and whole-genome species trees for each symbiont lineage were identified and generated by OrthoVenn3 (Sun et al. 2023). Nucleotide sequences for all gene clusters (n ≥ 2) were individually aligned with Muscle v3.8.1551 (Edgar 2004) based on their amino acid translations and then trimmed to remove all gaps and stop codons. Corresponding unrooted phylogenies for each gene were assigned based on the genome species tree. Rates of synonymous (dS) and nonsynonymous (dN) substitutions were then estimated with CodeML v4.9 using the strategies to detect positive selection with heterogeneous dN/dS rates across sites and branches in Álvarez-Carretero et al. (2023). This included testing five nested site models (m0, m1a, m2a, m7, and m8) and nested branch models allowing for two distinct dN/dS rate categories along tagged branches (foreground vs. background branches). Calculated maximum likelihood scores were tested between appropriate nested models using a likelihood ratio test, and significance was determined based on the appropriate χ2 critical values for a 5% cutoff. Following Vasquez and Bennett (2022), the only genes reported with potentially positive selected codon positions were significant in comparisons of both m1a → m2a and m7 → m8 models. Subsequent statistical analysis of dS, dN, and dN/dS was performed in JMP v17.
To estimate absolute substitution rates, the ages of internal nodes for OrthoVenn3 generated whole-genome phylogenies were determined using r8s (v1.8) (Sanderson 2003). Ages were estimated by “fixing” the age of the divergence between the bicoloratus and minutus clades at 3.5 or 3.0 MYA based on consideration of the rate of divergence identified in Percy (2017), phylogenetic topology (Percy et al. 2018), and island ages. Estimates were calculated for both Carsonella- and Morganella-like symbionts; however, in the final calculations, the appropriate ages from the resulting Carsonella ultrametric tree were then used as divisors to determine the average rates of dS and dN per year (dS/t and dN/t). Genes with saturated dS values (dS ≥ 3) were excluded.
Detecting Psyllid HTG
Genomic data obtained from Johnson et al. (2018) originated from ten adult whole bodies pooled for the closed-gall psyllid P. montgomeri's transcriptome (PRJNA295750) and six adult whole bodies pooled for the free-living psyllid P. gracilis's DNA shotgun assembly (PRJNA389892). DNA and RNA extractions were prepared and sequenced prior by Johnson et al. (2018). Raw RNA sequence data were assembled into contigs using Trinity v2.15.1 (Grabherr et al. 2011). Trinity transcripts were merged using SuperTranscripts (Davidson et al. 2017) to substitute for the lack of reference genomes. Alternative assemblies were also generated to have equal read depths for both species to determine if read depth influences the presence and absence of specific genes by randomly selecting paired-end reads.
RNA/DNA contigs were then screened for horizontally transferred genes (HTG) in psyllids using BlastX (% similarity > 60%, E-value cutoff < 10−5) using amino acid sequences of HTG previously identified from both B. cockerelli and D. citri (Kwak et al. 2023). Corresponding contigs were manually inspected further if they were below the Blast threshold % similarity parameter and Blast was then used against the NCBI nr and B. cockerelli database in addition to all symbiont assemblies to ensure the best blast hit was a psyllid species. To evaluate the completeness of both the psyllid transcriptomes and DNA assemblies BUSCO (v5.4.4) (Simão et al. 2015), scores were calculated using the insect_odb10 database.
Supplementary Material
Acknowledgments
We would like to the thank University of California, Riverside Entomology Department and the Microbiology and Plant Pathology Department (A.K.H. and P.H.D., respectively) for funding for this project. We also would like to thank Doug Perry for the help the with initial preliminary analyses. We would also like to thank Kevin Johnson for access to psyllid transcriptome and shotgun sequencing data, which was supported by NSF award DEB-1239788 to Kevin P. Johnson, Christopher H. Dietrich, and Hugh M. Robertson. We are grateful to Cerise Chen who performed a subset of psyllid DNA extractions supported by an NSF Dimensions of Biodiversity award (DEB 1241253) to the University of California, Berkeley, that also supported fieldwork by D.M.P. in the Hawaiian Islands; additional support that assisted in fieldwork is gratefully acknowledged from a Smithsonian Institution Postdoctoral Fellowship (2004 to 2005). D.M.P is grateful to Quentin Cronk for the use of laboratory facilities at the University of British Columbia.
Contributor Information
Allison K Hansen, Department of Entomology, University of California, Riverside, CA, USA.
Jacob A Argondona, Department of Entomology, University of California, Riverside, CA, USA.
Sen Miao, Department of Entomology, University of California, Riverside, CA, USA.
Diana M Percy, Department of Botany, University of British Columbia, Vancouver, BC, Canada.
Patrick H Degnan, Department of Microbiology and Plant Pathology, University of California, Riverside, CA, USA.
Data Availability
The amplicon and metagenomic data have been deposited under the BioProject accession PRJNA1125431. The amplicon data are submitted into the NCBI Sequence Read Archive database under BioSample accession SAMN41896535. The metagenome-assembled genome samples are available under BioSample numbers SAMN41940397 to SAMN41940408. Author annotated versions of the metagenome-assembled genomes are available on GitHub (https://github.com/phdegnan/Pariaconus-Symbiont-Genomes).
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
A.K.H. and D.M.P. designed the research; S.M. and D.M.P. produced the data; A.K.H., J.A.A., and P.H.D. analyzed the data; and A.K.H., D.M.P., and P.H.D. wrote the paper.
Funding
This research was supported by funding from the University of California, Riverside Entomology Department and the Microbiology and Plant Pathology Department (A.K.H. and P.H.D.), the National Science Foundation (NSF) award DEB-1239788 to D.M.P., Kevin P. Johnson, Christopher H. Dietrich, and Hugh M. Robertson, the NSF Dimensions of Biodiversity award (DEB 1241253) to the University of California, Berkeley and D.M.P., and a Smithsonian Institution Postdoctoral Fellowship (2004 to 2005) to D.M.P.
References
- Álvarez-Carretero S, Kapli P, Yang Z. Beginner's guide on the use of PAML to detect positive selection. Mol Biol Evol. 2023:40(4):msad041. 10.1093/molbev/msad041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amada G, Kobayashi K, Izuno A, Mukai M, Ostertag R, Kitayama K, Onoda Y. Leaf trichomes in Metrosideros polymorpha can contribute to avoiding extra water stress by impeding gall formation. Ann Bot. 2020:125(3):533–542. 10.1093/aob/mcz196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017:2(2):e00191–e00116. 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data—FastQC a quality control tool for high throughput sequence data. 2010. [accessed 2022 Mar 16]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Appelhans T, Detsch F, Reudenbach C, Woellauer S. mapview: interactive viewing of spatial data in R. R package version 2.11.2.9000. 2024. [accessed 2024 Feb 2]. https://github.com/r-spatial/mapview.
- Araújo M, Falcão B, Fagundes F, Guilherme G. Superhost plants alter the structure of plant–galling insect networks in neotropical savannas. Plants (Basel). 2019:8(10):369. 10.3390/plants8100369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Percy DM, Hefer CA, Cronk QCB. The transcriptional landscape of insect galls: psyllid (Hemiptera) gall formation in Hawaiian Metrosideros polymorpha (Myrtaceae). BMC Genomics. 2015:16(1):943. 10.1186/s12864-015-2109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MCJ, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006:72(11):7098–7110. 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal R, Hulbert S, Reese J, Whitworth R, Stuart J, Chen M-S. Pyrosequencing reveals the predominance of Pseudomonadaceae in gut microbiome of a gall midge. Pathogens. 2014:3(2):459–472. 10.3390/pathogens3020459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash I, Manulis-Sasson S. Recent evolution of bacterial pathogens: the gall-forming Pantoea agglomerans case. Annu Rev Phytopathol. 2009:47(1):133–152. 10.1146/annurev-phyto-080508-081803. [DOI] [PubMed] [Google Scholar]
- Bastin S, Reyes-Betancort JA, De La Rosa FS, Percy DM. Origins of the central Macaronesian psyllid lineages (Hemiptera; Psylloidea) with characterization of a new island radiation on endemic Convolvulus floridus (Convolvulaceae) in the Canary Islands. PLoS One. 2024:19(1):e0297062. 10.1371/journal.pone.0297062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Roberts L, Douglas AE, Werner GDA. Match and mismatch between dietary switches and microbial partners in plant sap-feeding insects. Proc Biol Sci. 2019:286(1902):20190065. 10.1098/rspb.2019.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, O’Grady PM. Host–plants shape insect diversity: phylogeny, origin, and species diversity of native Hawaiian leafhoppers (Cicadellidae: Nesophrosyne). Mol Phylogenet Evol. 2012:65(2):705–717. 10.1016/j.ympev.2012.07.024. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 2018:6(1):90. 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014:30(15):2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019:37(8):852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacum J, O’Grady PM, Kambysellis M, DeSalle R. Phylogeny and age of diversification of the planitibia species group of the Hawaiian Drosophila. Mol Phylogenet Evol. 2005:37(1):73–82. 10.1016/j.ympev.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Buchner P. Endosymbiosestudien an schildläusen IV. Hippeococcus, eine myrmekophile pseudococcine 1. Zeitschrift für Morphologie und Ökologie der Tiere. 1956:45(4):379–410. 10.1007/BF00407704. [DOI] [Google Scholar]
- Buchner P. Endosymbiosis of animals with plant microorganisms. Rev. Eng. ed. New York: Interscience Publishers; 1965. p. 1–909. [Google Scholar]
- Burckhardt D. Queiroziella gen. nov., a new genus of jumping plant-lice (Hemiptera, Psyllidae) from southern Brazil associated with Mimosa scabrella (Leguminosae). Zootaxa. 2021:4927(3):zootaxa.4927.3.3. 10.11646/zootaxa.4927.3.3. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016:13(7):581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisano A, Ometto L, Compant S, Pancher M, Antonielli L, Yousaf S, Varotto C, Anfora G, Pertot I, Sessitsch A, et al. Interkingdom transfer of the acne-causing agent, Propionibacterium acnes, from human to grapevine. Mol Biol Evol. 2014:31(5):1059–1065. 10.1093/molbev/msu075. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012:6(8):1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri G, Kim I, Kim J, So Y, Seo T. Chryseobacterium tagetis sp. nov., a plant growth promoting bacterium with an antimicrobial activity isolated from the roots of medicinal plant (Tagetes patula). J Antibiot (Tokyo). 2022:75(6):312–320. 10.1038/s41429-022-00525-7. [DOI] [PubMed] [Google Scholar]
- Choi JY, Dai X, Alam O, Peng JZ, Rughani P, Hickey S, Harrington E, Juul S, Ayroles JF, Purugganan MD, et al. Ancestral polymorphisms shape the adaptive radiation of Metrosideros across the Hawaiian Islands. Proc Natl Acad Sci U S A. 2021:118(37):e2023801118. 10.1073/pnas.2023801118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JM, Rokas A, Pagel M, Stone GN. Evolutionary shifts between host oak sections and host-plant organs in Andricus gallwasps. Evolution. 2002:56(9):1821–1830. 10.1111/j.0014-3820.2002.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Cornwallis CK, van ‘t Padje A, Ellers J, Klein M, Jackson R, Kiers ET, West SA, Henry LM. Symbioses shape feeding niches and diversification across insects. Nat Ecol Evol. 2023:7(7):1022–1044. 10.1038/s41559-023-02058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004:14(7):1394–1403. 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NM, Hawkins ADK, Oshlack A. SuperTranscripts: a data driven reference for analysis and visualisation of transcriptomes. Genome Biol. 2017:18(1):148. 10.1186/s13059-017-1284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018:6(1):226. 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer J, Corretto E, Štarhová SL, Michalik A, Nováková E, Schuler H. Division of labor within psyllids: metagenomics reveals an ancient dual endosymbiosis with metabolic complementarity in the genus Cacopsylla. mSystems. 2023:8(5):e00578-23. 10.1128/msystems.00578-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. The B vitamin nutrition of insects: the contributions of diet, microbiome and horizontally acquired genes. Curr Opin Insect Sci. 2017:23:65–69. 10.1016/j.cois.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004:32(5):1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, Fink I, Pan JN, Yousef M, Fogarty EC, et al. Community-led, integrated, reproducible multi-omics with Anvi’o. Nat Microbiol. 2020:6(1):3–6. 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay PA, Preszler RW, Whitham TG. The functional resource of a gall-forming adelgid. Oecologia. 1996:105(2):199–204. 10.1007/BF00328547. [DOI] [PubMed] [Google Scholar]
- Forrest JMS. THE GROWTH OF APHIS FABAE AS AN INDICATOR OF THE NUTRITIONAL ADVANTAGE OF GALLING TO THE APPLE APHID DYSAPHIS DEVECTA. Entomologia Exp Applicata. 1971:14(4):477–483. 10.1111/j.1570-7458.1971.tb00187.x. [DOI] [Google Scholar]
- Francioli D, Cid G, Hajirezaei M-R, Kolb S. Leaf bacterial microbiota response to flooding is controlled by plant phenology in wheat (Triticum aestivum L.). Sci Rep. 2022:12(1):11197. 10.1038/s41598-022-15133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie RG, Baldwin BG, Waters JM, Fraser CI, Nikula R, Roderick GK. Long-distance dispersal: a framework for hypothesis testing. Trends Ecol Evol. 2012:27(1):47–56. 10.1016/j.tree.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Gossett JM, Porter ML, Vasquez YM, Bennett GM, Chong RA. Genomic comparisons reveal selection pressure and functional variation between nutritional endosymbionts of cave-adapted and epigean Hawaiian planthoppers. Genome Biol Evol. 2023:15(3):evad031. 10.1093/gbe/evad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011:29(7):644–652. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner DS, Taylor AD, Forkner RE. The effects of foliar pubescence and nutrient enrichment on arthropod communities of Metrosideros polymorpha (Myrtaceae). Ecol Entomol. 2005:30(4):428–443. 10.1111/j.0307-6946.2005.00710.x. [DOI] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013:29(8):1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AAG, Morrow JL, Fromont C, Steinbauer MJ, Taylor GS, Johnson SN, Cook JM, Riegler M. Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts. Environ Microbiol. 2016:18(8):2591–2603. 10.1111/1462-2920.13351. [DOI] [PubMed] [Google Scholar]
- Hammer TJ, De Clerck-Floate R, Tooker JF, Price PW, Miller DG, Connor EF. Are bacterial symbionts associated with gall induction in insects? Arthropod Plant Interact. 2021:15(1):1–12. 10.1007/s11829-020-09800-6. [DOI] [Google Scholar]
- Hansen AK, Moran NA. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol. 2014:23(6):1473–1496. 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- Hartman PE. Linked loci in the control of consecutive steps in the primary pathway of histidine synthesis in Salmonella typhimurium. Genetic Studies with Bacteria. 1956:612:35–61. [Google Scholar]
- Hippee AC, Beer MA, Bagley RK, Condon MA, Kitchen A, Lisowski EA, Norrbom AL, Forbes AA. Host shifting and host sharing in a genus of specialist flies diversifying alongside their sunflower hosts. J Evol Biol. 2021:34(2):364–379. 10.1111/jeb.13740. [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B, Andersen KR, Kilstrup M, Martinussen J, Switzer RL, Willemoës M. Phosphoribosyl diphosphate (PRPP): biosynthesis, enzymology, utilization, and metabolic significance. Microbiol Mol Biol Rev. 2017:81(1):e00040–e00016. 10.1128/MMBR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM . SPSS statistics 29.0.0. 2024. [accessed 2024 Jan 4]. https://www.ibm.com/docs/en/spss-statistics/29.0.0.
- Inbar M, Eshel A, Wool D. Interspecific competition among phloem-feeding insects mediated by induced host-plant sinks. Ecology. 1995:76(5):1506–1515. 10.2307/1938152. [DOI] [Google Scholar]
- Jeon D, Jiang L, Kim K-H, Peng Y, Cho D, Jeong R-D, Kim CY, Jeong JC, Lee J. Bioplastic (poly-3-hydroxybutyrate)-producing Massilia endophytica sp. nov., isolated from Cannabis sativa L. ‘Cheungsam.’. Sci Rep. 2023:13(1):17767. 10.1038/s41598-023-44976-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KP, Dietrich CH, Friedrich F, Beutel RG, Wipfler B, Peters RS, Allen JM, Petersen M, Donath A, Walden KKO, et al. Phylogenomics and the evolution of hemipteroid insects. Proc Natl Acad Sci U S A. 2018:115(50):12775–12780. 10.1073/pnas.1815820115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy JB, Crespi BJ. Adaptive radiation of gall-inducing insects within a single host-plant species. Evolution. 2007:61(4):784–795. 10.1111/j.1558-5646.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Kwak Y, Argandona JA, Degnan PH, Hansen AK. Chromosomal-level assembly of Bactericera cockerelli reveals rampant gene family expansions impacting genome structure, function and insect-microbe-plant-interactions. Mol Ecol Resour. 2023:23(1):233–252. 10.1111/1755-0998.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y, Hansen AK. Unveiling metabolic integration in psyllids and their nutritional endosymbionts through comparative transcriptomics analysis. iScience. 2023:26(10):107930. 10.1016/j.isci.2023.107930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y, Sun P, Meduri VR, Percy DM, Mauck KE, Hansen AK. Uncovering symbionts across the psyllid tree of life and the discovery of a new Liberibacter species, “Candidatus“ Liberibacter capsica. Front Microbiol. 2021:12:739763. 10.3389/fmicb.2021.739763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson KC, Whitham TG. Manipulation of food resources by a gall-forming aphid: the physiology of sink–source interactions. Oecologia. 1991:88(1):15–21. 10.1007/BF00328398. [DOI] [PubMed] [Google Scholar]
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015:31(10):1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005:71(12):8228–8235. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson VG, Gawryluk RMR, Gowen BE, Curtis CI, Jaenike J, Perlman SJ. Multiple origins of obligate nematode and insect symbionts by a clade of bacteria closely related to plant pathogens. Proc Natl Acad Sci U S A. 2020:117(50):31979–31986. 10.1073/pnas.2000860117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama J, Inoue H, Hirose Y, Nakabachi A. 16S rRNA gene sequencing of six psyllid species of the family Carsidaridae identified various vacteria including Symbiopectobacterium. Microb Environ. 2023:38(3):ME23045. 10.1264/jsme2.ME23045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012:10(1):13–26. 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Michell CT, Nyman T. Microbiomes of willow-galling sawflies: effects of host plant, gall type, and phylogeny on community structure and function. Genome. 2021:64(6):615–626. 10.1139/gen-2020-0018. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 gateway computing environments workshop (GCE). New Orleans, LA, USA: IEEE. p. 1–8. Available from: http://ieeexplore.ieee.org/document/5676129/. [Google Scholar]
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013:14(2):193–202. 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005:71(12):8802–8810. 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow JL, Hall AAG, Riegler M. Symbionts in waiting: the dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome. 2017:5(1):58. 10.1186/s40168-017-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Piel J, Malenovský I, Hirose Y. Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: providing toxins, vitamins, and carotenoids. Genome Biol Evol. 2020:12(11):1975–1987. 10.1093/gbe/evaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006:314(5797):267–267. 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman T. To speciate, or not to speciate? Resource heterogeneity, the subjectivity of similarity, and the macroevolutionary consequences of niche-width shifts in plant-feeding insects. Biol Rev Camb Philos Soc. 2010:85(2):393–411. 10.1111/j.1469-185X.2009.00109.x. [DOI] [PubMed] [Google Scholar]
- Percy DM. Radiation, diversity, and host-plant interactions among island and continental legume-feeding psyllids. Evolution. 2003:57(11):2540–2556. 10.1111/j.0014-3820.2003.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Percy DM. Making the most of your host: the Metrosideros-feeding psyllids (Hemiptera, Psylloidea) of the Hawaiian Islands. Zookeys. 2017:649:1–163. 10.3897/zookeys.649.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy DM, Crampton-Platt A, Sveinsson S, Lemmon AR, Lemmon EM, Ouvrard D, Burckhardt D. Resolving the psyllid tree of life: phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Syst Entomol. 2018:43(4):762–776. 10.1111/syen.12302. [DOI] [Google Scholar]
- Percy DM, Garver AM, Wagner WL, James HF, Cunningham CW, Miller SE, Fleischer RC. Progressive island colonization and ancient origin of Hawaiian Metrosideros (Myrtaceae). Proc Biol Sci. 2008:275(1642):1479–1490. 10.1098/rspb.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PW. Adaptive radiation of gall-inducing insects. Basic Appl Ecol. 2005:6(5):413–421. 10.1016/j.baae.2005.07.002. [DOI] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010:5(3):e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q-L, Xie B-B, Zhang X-Y, Chen X-L, Zhou B-C, Zhou J, Oren A, Zhang Y-Z. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014:196(12):2210–2215. 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012:41(D1):D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz DL de, Zanol KMR, Oliveira EB, dos Anjos N, Majer J. Feeding and oviposition preferences of Ctenarytaina spatulata Taylor (Hemiptera, Psyllidae) for Eucalyptus spp. and other Myrtaceae in Brazil. Rev Bras Entomol. 2010:54(1):149–153. 10.1590/S0085-56262010000100023. [DOI] [Google Scholar]
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016:32(6):929–931. 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley AB, Kim D, Hansen AK. Genome sequence of “Candidatus Carsonella ruddii” strain BC, a nutritional endosymbiont of Bactericera cockerelli. Genome Announc. 2017:5(17):e00236-17. 10.1128/genomeA.00236-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016:4:e2584. 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. R8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003:19(2):301–302. 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014:30(14):2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Serbina LŠ, Gajski D, Pafčo B, Zurek L, Malenovský I, Nováková E, Schuler H, Dittmer J. Microbiome of pear psyllids: a tale about closely related species sharing their endosymbionts. Environ Microbiol. 2022:24(12):5788–5808. 10.1111/1462-2920.16180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato-Salas J, Gendrin M. Involvement of Microbiota in insect physiology: focus on B vitamins. mBio. 2023:14(1):e02225–e02222. 10.1128/mbio.02225-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KL, Gillespie RG. Comparative phylogeography of oceanic archipelagos: hotspots for inferences of evolutionary process. Proc Natl Acad Sci U S A. 2016:113(29):7986–7993. 10.1073/pnas.1601078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlovsky AE, Magasanik B. A defect in histidine biosynthesis causing an adenine deficiency. J Biol Chem. 1962:237(12):3725–3730. 10.1016/S0021-9258(19)84515-5. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002:51(3):492–508. 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015:31(19):3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994:87(6):651–701. 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- Sloan DB, Moran NA. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 2012:29(12):3781–3792. 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, Moran NA. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 2014:31(4):857–871. 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding AW, Von Dohlen CD. Psyllid endosymbionts exhibit patterns of co-speciation with hosts and destabilizing substitutions in ribosomal RNA. Insect Mol Biol. 2001:10(1):57–67. 10.1046/j.1365-2583.2001.00231.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014:30(9):1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauer MJ, Burns AE, Hall A, Riegler M, Taylor GS. Nutritional enhancement of leaves by a psyllid through senescence-like processes: insect manipulation or plant defence? Oecologia. 2014:176(4):1061–1074. 10.1007/s00442-014-3087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stever H, Eiben J, Bennett GM. Hawaiian Nysius insects rely on an obligate symbiont with a reduced genome that retains a discrete nutritional profile to match their plant seed diet. Genome Biol Evol. 2021:13(9):evab189. 10.1093/gbe/evab189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran S, Kost C, Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017:25(5):375–390. 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Sun J, Lu F, Luo Y, Bie L, Xu L, Wang Y. OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023:51(W1):W397–W403. 10.1093/nar/gkad313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Yokokura J, Ito T, Arai R, Yokoyama C, Toshima H, Nagata S, Asami T, Suzuki Y. Biosynthetic pathway of the phytohormone auxin in insects and screening of its inhibitors. Insect Biochem Mol Biol. 2014:53:66–72. 10.1016/j.ibmb.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Syberg-Olsen MJ, Garber AI, Keeling PJ, McCutcheon JP, Husnik F. Pseudofinder: detection of pseudogenes in prokaryotic genomes. Mol Biol Evol. 2022:39(7):msac153. 10.1093/molbev/msac153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GS, Fagan-Jeffries EP, Austin AD. A new genus and twenty new species of Australian jumping plant-lice (Psylloidea: Triozidae) from Eremophila and Myoporum (Scrophulariaceae: Myoporeae). Zootaxa. 2016:4073(1):1–84. 10.11646/zootaxa.4073.1.1. [DOI] [PubMed] [Google Scholar]
- Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 2000:66(7):2898–2905. 10.1128/AEM.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez YM, Bennett GM. A complex interplay of evolutionary forces continues to shape ancient co-occurring symbiont genomes. iScience. 2022:25(8):104786. 10.1016/j.isci.2022.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AKG, Bagley RK, Egan SP, Hood GR, Ott JR, Prior KM, Sheikh SI, Weinersmith KL, Zhang L, Zhang YM, et al. Speciation in Nearctic oak gall wasps is frequently correlated with changes in host plant, host organ, or both. Evolution. 2022:76(8):1849–1867. 10.1111/evo.14562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TCR. The abundance of invertebrate herbivores in relation to the availability of nitrogen in stressed food plants. Oecologia. 1984:63(1):90–105. 10.1007/BF00379790. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Tanaka H, Hasegawa M, Tokuda M, Asami T, Suzuki Y. Phytohormones and willow gall induction by a gall-inducing sawfly. New Phytol. 2012:196(2):586–595. 10.1111/j.1469-8137.2012.04264.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The amplicon and metagenomic data have been deposited under the BioProject accession PRJNA1125431. The amplicon data are submitted into the NCBI Sequence Read Archive database under BioSample accession SAMN41896535. The metagenome-assembled genome samples are available under BioSample numbers SAMN41940397 to SAMN41940408. Author annotated versions of the metagenome-assembled genomes are available on GitHub (https://github.com/phdegnan/Pariaconus-Symbiont-Genomes).