Abstract
NEDD8 (neural precursor cell expressed developmentally downregulated gene 8)-specific protease NEDP1 processes preNEDD8 to its mature form and deconjugates NEDD8 from substrates such as p53 and cullins. Although NEDD8 and ubiquitin are highly related in sequence and structure, their attachment to a protein leads to different biological effects. It is therefore critical that NEDP1 discriminates between NEDD8 and ubiquitin, and this requires remarkable precision in molecular recognition. To determine the basis of this specificity, we have determined the crystal structure of NEDP1 in isolation and in a transition state complex with NEDD8. This reveals that NEDP1 is a cysteine protease of the Ulp family. Binding of NEDD8 induces a dramatic conformational change in a flexible loop that swings over the C-terminus of NEDD8 locking it into an extended β-structure optimal for catalysis. Structural, mutational and biochemical studies have identified key residues involved in molecular recognition. A single-residue difference in the C-terminus of NEDD8 and ubiquitin contributes significantly to the ability of NEDP1 to discriminate between them. In vivo analysis indicates that NEDP1 mutants perturb deNEDDylation of the tumour suppressor p53.
Keywords: NEDD8, NEDP1, protease, structure, ubiquitin
Introduction
Ubiquitin and ubiquitin-like proteins are linked by an isopeptide bond to lysine side chains in target proteins (Johnson, 2002). In addition to ubiquitin, small ubiquitin-like modifier (SUMO) and neural precursor cell expressed developmentally downregulated gene 8 (NEDD8) (Kamitani et al, 1997) or related to ubiquitin 1 (Rub1) in yeast can be covalently coupled to target proteins with important functional consequences (Jentsch and Pyrowolakis, 2000). Thus, cells need appropriate mechanisms to ensure that the correct ubiquitin-like protein is added to substrates, and equally, proteases that carry out processing and deconjugation must recognise the correct ubiquitin-like protein. Ubiquitin-like proteins are conjugated to target proteins by enzymatic cascades that typically involve three activities: an E1 (activating enzyme); E2 (conjugating enzyme); and E3 (protein ligase) (Huang et al, 2004). Although NEDD8 is highly homologous to ubiquitin (57% identity), it has unique E1 and E2 enzymes that discriminate between ubiquitin and NEDD8. The NEDD8 E1 is a heterodimer composed of the amyloid precursor protein-binding protein (APP-BP1) and the Uba3 protein, while the NEDD8 E2 is Ubc12 (Lammer et al, 1998; Liakopoulos et al, 1998; Osaka et al, 1998; Pozo et al, 1998; Gong and Yeh, 1999). Genetic analysis in animals, plants and yeast has demonstrated the importance of the NEDD8 pathway in cell proliferation, viability and development. Until recently, the only known substrates for NEDD8 modification in mammals were the six members of the cullin family of proteins that are components of ubiquitin E3 ligase complexes. The cullins play an architectural role in a number of ubiquitin E3 ligase complexes, but the role of NEDD8 modification of cullins has been determined in SCF complexes, which also contain Rbx1 (Roc1), Skp1 (or homologue) and a substrate receptor protein that contains an F-box motif (Chiba and Tanaka, 2004). SCF-like complexes are responsible for the ubiquitination of proteins such as phosphorylated IκBα and hydroxylated HIF1α (Deshaies, 1999; Gray et al, 1999). Cullin protein modification is facilitated by the RING domain containing Rbx1, which may act as a NEDD8 E3 ligase (Kamura et al, 1999; Morimoto et al, 2003). Genetic experiments in yeast and plants indicate that Rub1 (NEDD8) modification is important for SCF ubiquitin ligase activity (Dharmasiri et al, 2003), while biochemical experiments demonstrated that NEDD8 modification of Cul-1 was responsible for recruitment of the Ubc4-ubiquitin thioester to the SCF complex (Kawakami et al, 2001; Wu et al, 2002). It is also thought that NEDD8 modification of cullins blocks binding of the SCF complex inhibitor CAND1 (Goldenberg et al, 2004). While a complete Rub1 (NEDD8) modification pathway is not required for the viability of Saccharomyces cerevisiae, it is required for viability of Schizosaccharomyces pombe. In ts41 hamster cells, a temperature-sensitive mutation in APP-BP1 results in cell cycle defects and indicates that NEDD8 modification is required for entry into mitosis and inhibition of entry into S phase (Handeli and Weintraub, 1992). Deletion of the Uba3 gene in mice leads to embryonic lethality and establishes an essential function of NEDD8 modification in higher eukaryotic cells (Tateishi et al, 2001). While NEDD8 modification of cullins is essential for SCF function, this does not appear to be the only role of NEDD8 modification and additional substrates have recently been detected. The von Hippel–Lindau (VHL) tumour suppressor is a component of an SCF-like E3 ligase complex that targets the hypoxia-inducible factor 1α (HIF-1α) for ubiquitination-mediated proteolysis and regulates fibronectin matrix assembly. NEDD8 modification of VHL is required for fibronectin matrix assembly but not for HIF-1α ubiquitination (Stickle et al, 2004). The p53 tumour suppressor has also been reported to undergo NEDD8 modification, and in this situation NEDDylation reduces the transcriptional activity of p53. In this case, the p53 ubiquitin ligase Mdm2 also functions as a NEDD8 E3 ligase and greatly stimulates NEDD8 modification of p53 in vivo (Xirodimas et al, 2004).
Ubiquitylation of p27Kip1 in cell extracts only proceeds in the presence of NEDD8 and NEDD8 modification enzymes. The continued requirement for NEDD8 conjugation during the ubiquitylation reaction suggests that the cell extract contains NEDD8-specific proteases that are capable of deconjugating NEDD8 from cullins (Podust et al, 2000). The COP9 signalosome (CSN) associates with cullin proteins both genetically and physically. CSN is present in all eukaryotes and is a multisubunit complex that has structural similarity to the lid of the proteasome and the eukaryotic translation initiation factor 3 (eIF3) complex (Seeger et al, 2001). A NEDD8-specific protease activity has been reported to be associated with the CSN, and while a metalloprotease motif in Jab1/Csn5 is required for this activity, the isolated protein is not active and only displays NEDD8 protease activity when part of the CSN complex (Cope et al, 2002). In Caenorhabditis elegans, it has been demonstrated that both NEDDylation and CSN-dependent deNEDDylation are required for SCF-mediated MEL-1 degradation (Pintard et al, 2003). Thus, it seems likely that multiple cycles of NEDDylation and deNEDDylation are required to allow SCF-mediated ubiquitin polymerisation onto target proteins.
Prior to conjugation, the primary translation products of ubiquitin, NEDD8 and SUMO are processed by specific proteases that expose the C-terminal glycine residue that will form the isopeptide bond with the lysine side chain of the target protein. Structures of ubiquitin and SUMO bound to their proteases have been determined and identify the basis of molecular recognition in each case (Johnston et al, 1999; Mossessova and Lima, 2000; Hu et al, 2002; Reverter and Lima, 2004). Although SUMO and ubiquitin are quite different, the high structural homology between ubiquitin and NEDD8 suggests that it is possible that deubiquitinating enzymes could carry out this processing. In budding yeast, the single member of the ubiquitin C-terminal hydrolase (UCH) family of deubiquitinating enzymes, Yuh1p, processes ubiquitin and the structure of ubiquitin bound to Yuh1p displays many specific interactions between the protease and its substrate (Johnston et al, 1999). However, Yuh1p also acts as a processing enzyme for Rub1 and strains deleted for Yuh1 fail to NEDDylate the cullin Cdc53 (Linghu et al, 2002). In mammals, the ubiquitin-specific protease USP21 catalyses the deNEDDylation of conjugated proteins (Gong et al, 2000) and the ubiquitin C-terminal hydrolase UCH-L3 can process both ubiquitin and NEDD8 (Wada et al, 1998), although UCH-L3−/− mice are viable with no defects in the NEDD8 pathway (Kurihara et al, 2000). Thus, it seems likely that additional NEDD8-specific proteases exist in mammals. Recently, a cysteine protease highly specific for NEDD8 has been identified in human cells. NEDP1 (Mendoza et al, 2003) or DEN1 (Gan-Erdene et al, 2003; Wu et al, 2003) is homologous to the SUMO proteases but shows remarkable specificity for NEDD8. It does not cleave SUMO and has a 60 000-fold preference for NEDD8 over ubiquitin (Gan-Erdene et al, 2003). Although not present in budding yeast, close homologues of NEDP1 are present in fission yeast, plants and animals (Mendoza et al, 2003). While this protein is capable of processing preNEDD8, it is also capable of deNEDDylating cullins and p53 and depolymerising polymeric chains of NEDD8 (Mendoza et al, 2003; Wu et al, 2003; Xirodimas et al, 2004). Clearly, the distinct functional consequences associated with ubiquitin and NEDD8 modification necessitate highly specific mechanisms that allow the cell to discriminate between these two highly related ubiquitin-like modifiers. This is achieved by uniquely specific enzymes that correctly bring about conjugation and deconjugation of each of these modifiers. Here, we determine the crystal structure of the NEDD8-specific protease NEDP1 in isolation and of a covalent thiohemiacetal transition state complex between NEDP1 and NEDD8. The structure reveals that NEDP1 is a cysteine protease of the Ulp family and undergoes a dramatic conformational change upon NEDD8 binding. Biochemical analysis has demonstrated how a single-residue change between NEDD8 and ubiquitin plays a significant role in discrimination by NEDP1.
Results
Structure determination
NEDP1 is a cysteine protease that can process preNEDD8 to the mature form that is competent for conjugation and it can deconjugate NEDD8 from modified substrates (Mendoza et al, 2003; Wu et al, 2003; Xirodimas et al, 2004). Despite the high degree of similarity between NEDD8 and ubiquitin, NEDP1 displays a 60 000-fold preference for NEDD8 over ubiquitin. To determine the basis for this remarkable specificity, we have determined the structure of NEDP1 alone and in a covalent complex with NEDD8. Sodium borohydride was used to reduce selectively the deacylation intermediate formed during proteolytic cleavage, yielding a chemically stable transition state analogue (Pickart and Rose, 1986; Mossessova and Lima, 2000). Thus, a complex containing a covalent thiohemiacetal linkage between the C-terminal glycine of NEDD8 (G76) and the active site cysteine (C163) of NEDP1 was prepared and crystallised. Phases for crystals of the complex were determined by a multiwavelength anomalous diffraction experiment on selenomethionine variants of the proteins. The structure was refined to 1.9 Å resolution on native protein and includes residues 1–211 of NEDP1 and residues 1–76 of NEDD8 (Table I). NEDP1 alone was crystallised and its structure solved to 2.0 Å resolution by molecular replacement using the coordinates of NEDP1 from the NEDP1–NEDD8 complex as a search model.
Table 1.
Crystallographic data
| NEDP1–NEDDP8 | NEDP1–NEDDP8 | NEDP1–NEDDP8 | NEDP1–NEDDP8 | NEDP1 | |
|---|---|---|---|---|---|
| Beamline | BM14 UK | BM14 UK | BM14 UK | ESRF ID14-3 | ESRF ID14-2 |
| Wavelength (Å) | 0.9792 | 0.9794 | 0.8984 | 1.008 | 0.934 |
| Cell | a=54.6 Å, b=74.2 Å, c=75.9 Å α=β=γ=90.0° | a=57.1 Å, b=57.5 Å, c=75.0 Å α=99.7° β=110.9° γ=92.4° | |||
| Space group | P212121 | P1 | |||
| Resolution (Å) (highest shell) | 47–2.8 (2.87–2.8) | 54–1.9 (1.95–1.9) | 69–2.0 (2.05–2.0) | ||
| Unique reflections | 17 003 | 16 291 | 16 287 | 25 411 | 54 875 |
| I/σ | 7.9 (2.1) | 8.6 (2.3) | 10 (3.0) | 5.5 (2.0) | 9.8 (3.0) |
| Multiplicity | 7.9 (8) | 5 (5.1) | 5.0 (5.1) | 6.9 (3.0) | 1.9 (2) |
| Data completeness (%) | 100 (100) | 100 (100) | 100 (100) | 97.8 (83.7) | 96.9 (95.7) |
| Rmerge (%) | 8.2 (31) | 7.1 (30.7) | 6.3 (23.2) | 8.9 (35.2) | 4.8 (25.3) |
| Refinement | |||||
| R-cryst | 16.9 (20.0) | 19.6 (22.0) | |||
| R-free | 20.9 (24.1) | 25.0 (28.8) | |||
| Bond r.m.s. | 0.017 | 0.013 | |||
| Angle r.m.s. | 1.569 | 1.590 | |||
| % Ramachandran most favourable | 93.1 | 91.2 | |||
| No. of atoms | 2292 | 6748 | |||
| No. of water molecules | 214 | 386 | |||
| PDB code | 2bkr | 2bkq | |||
NEDP1 structure
The structure of the 211-residue NEDP1 protease confirms that despite its low sequence homology (Figure 1) it is a member of the cysteine protease superfamily. These proteins are defined by the classic catalytic triad Cys, His and Asp. NEDP1 has a central five-stranded β-sheet in which the middle strand β-5 (Figures 1 and 2) is antiparallel to the other four. One face of the sheet packs against two helices (α-2 and α-7), and the axes of both helices are parallel to each other and to the β-strands, resulting in a narrow channel. Helix α-7 (also known as the central helix) is opposite strand β-5, and helix α-2 (also known as helix A) is opposite strand β-3. These two helices are sandwiched by helices α-1 and α-9, whose axes are aligned almost 90° to the direction of the β-strands (Figure 2). The other face of the β-sheet packs against α-6 that lies at an angle across the β-sheet and connects β-6 and β-7. Helices α-4 and α-5 are on this side of the molecule and form a U shape. Both helices are in the stretch of residues that connects β-3 and β-4. The long axes of these two helices are arranged antiparallel to each other and orthogonal to the face of the sheet. There are two small strands (β-1 and β-2) at the N-terminus and are in an antiparallel arrangement. A long loop connects β-4 and β-5 and a short turn β-5 and β-6. The key nucleophile Cys163 is located at the N-terminus of the central α-helix, while the required His102 is at the N-terminus of β-5 and Asp119 is on β-6. The central three strands (β-4, β-5 and β-6) and the central helix are found in all members of the protease superfamily. The closest structural homologue to the NEDP1 protease is the Ulp1 protease from yeast, which has 140 superimposable residues over which the root mean square deviation for C α-atoms is 1.3 Å. Ulp1 defined a subclass of cysteine proteases to which NEDP1 belongs. The most significant difference between the proteases appears to be that in NEDP1, the loop centred on Q96 that connects strands β-4 and β-5 is five residues longer (highlighted in Figure 2 and denoted as loop 1). A sequence alignment suggests that this insertion is conserved in homologues of NEDP1 from a diverse range of species but not in Ulp1 (Figure 1). There are four monomers of NEDP1 in the asymmetric unit and the monomers show substantial variability in the structure. The different packing arrangement of the monomers changes subtly the geometry of the catalytic triad. The N to S distance varies from 3.6 to 4.2 Å, which is too big to be an error and reflects a genuine variation. The estimated error in any distance measurement is 0.3 Å. Comparison of pairs of well-separated side-chain atoms within the central sheet shows a much smaller deviation (<0.4 Å) even when the separation is over 16 Å. The reason for this difference at the catalytic triad is probably small changes in the position of H102. H102 is the first residue of β-5 that is connected to loop 1, which as mentioned has very different conformations. The geometry of the triad is crucial for effective catalysis and it would appear that changes in loop 1 could perturb or in some manner regulate catalysis. An electrostatic analysis of the protein surface reveals it to be mainly negatively charged (acidic) (Figure 3).
Figure 1.

Sequence alignment of NEDD8 with ubiquitin and SUMO and NEDP1 with Senp2 and ULP1. Sequences were aligned using ClustalW (Thompson et al, 1994). Conserved residues are shaded in grey. (A) Sequence alignment of NEDD8 with ubiquitin and SUMO. An alanine marked with an asterisk in NEDD8 was mutated to arginine, while an arginine in ubiquitin was mutated to alanine. Secondary structure elements (in yellow) above the sequence are indicated for NEDD8, while those (blue) below the sequences are indicated for ubiquitin. (B) Sequence alignment of NEDP1 with ULP1 and SENP2. Residues of the catalytic triad are highlighted in red. Residues involved in direct NEDD8 and NEDP1 intermolecular interaction are marked with an asterisk. Mutations in these residues were created by alanine substitution. Residues marked with a triangle were deleted in a loop deletion. The secondary structure elements (yellow) above the sequences are indicated for NEDP1 and those (blue) below the sequences are indicated for SENP2. Sequences shown are from NCBI protein databases: NEDD8 (NP_006147), ubiquitin (AAA36787), SUMO (AAH66306), NEDP1 (AAA36787), SENP2 (AAH40609) and ULP1 (1EUVA).
Figure 2.

Structure of NEDP1 alone and in complex with NEDD8. (A) Monomers of NEDP1 from the apo (wheat) and covalent complex (slate). The loop centred around residue 100 is shown in green and is profoundly changed in the complex. (B) The final 2Fo−Fc electron density different map of the linkage contoured at 1σ. (C) Stereo diagram of the NEDP1–NEDD8 complex. NEDP1 is coloured slate and NEDD8 pink. The residues mutated are shown and labelled. (D) The β-sheet-like interaction between nep1 and NEDD8.
Figure 3.
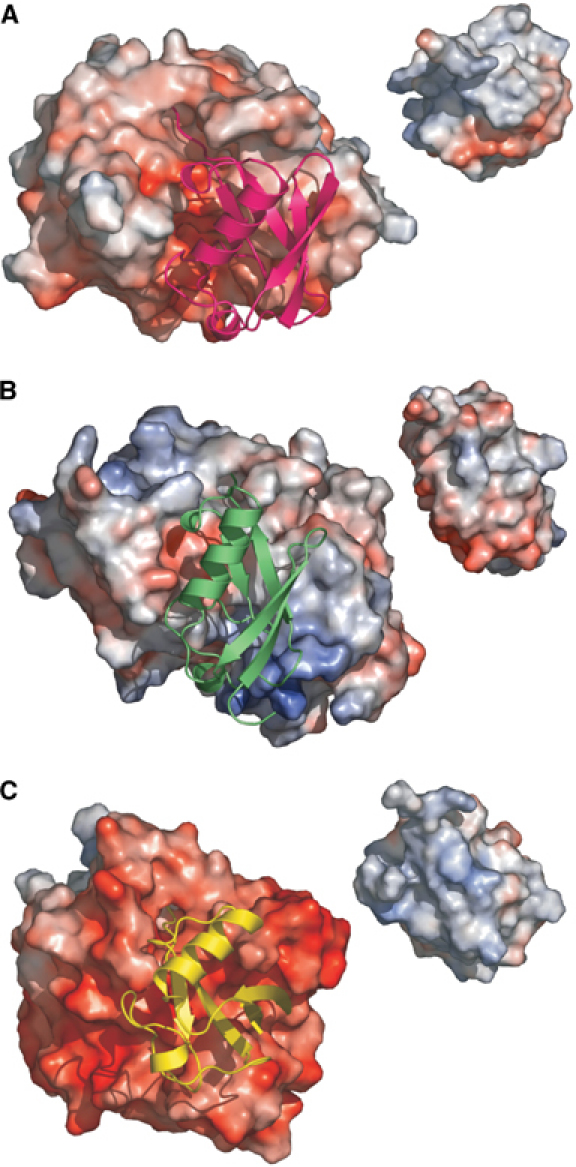
Comparison of protease-ubiquitin-like protein complexes. The complex between proteases, shown as electrostatic surfaces with the same scale in each and their target protein shown in a ribbon representation. Inset: The target turned to show its electrostatic surface that binds to the protease. (A) NEDP1–NEDD8. The protease interface is weakly acidic with no basic path. (B) Ulp1–SUMO. The strong positive patch at the bottom and a weaker smaller one at the top of protease are visible. (C) Yuh1–ubiquitin. The protease interface is much more acidic. In comparing ubiqutin and NEDD8, it can be seen that NEDD8 is less basic. This is consistent with changes in residues E53 and A72 (to Gly and Arg respectively in ubiquitin).
NEDD8–NEDP1 complex
In the complex of NEDP1 and NEDD8, the structure of NEDD8 is little changed from the previous report of the native protein (Whitby et al, 1998), the root mean square difference between C α-residues of the native and complexed forms is 0.56 Å. The protein has the classic ubiquitin fold of an antiparallel β-sheet with an α-helix packed on one face. The major difference occurs in residues 64–76, which are flexible in the native protein (Whitby et al, 1998) but are now found in a well-ordered extended conformation starting at residue Gly64 and finishing at Gly76. The gross fold of NEDP1 is unchanged from the native structure described above. However, there is a clear and striking difference at the loop centred on Gln96 (Figure 2). We have already highlighted that in the native structure, this loop is flexible. In the complex, the loop is well ordered and adopts a different arrangement from that seen in any of the four copies of the apo protease. There are several other small changes between the native and complex structure of NEDP1, which are likely to reflect differences in crystal packing.
The interaction surface between the proteins is substantial, burying nearly 2800 Å2. Within the interface, 44% of the residues are polar and 56% apolar. Interaction between the C-terminus of NEDD8 and the protease accounts for over 50% of the buried surface area. Visual inspection of the complex reveals that the C-terminus of NEDD8 (residues 72–76) sits in the narrow channel between the β-sheet and N-termini of helices α-2 and α-7 in the protease. The NEDD8 C-terminus makes backbone hydrogen bond contacts with NEDP1 residues 98 and 99 on one side of the channel and residues 27–29 on the other side of the channel, reminiscent of those seen between β-strands (Figure 2). Unsurprisingly, it is the positively charged face of NEDD8 that interacts with the negatively charged surface of NEDP1 (Figure 3). The interface can be decomposed into five segments, and we have mutated at least one protease residue in each segment and shown that efficiency of cleavage is reduced (Figures 2 and 4). Segment 1 comprises the N-terminus of NEDP1 and residues 45–56 in NEDD8. Segment 2 consists of the N-terminus of helix α-2 in the protease combined with the loop that connects to β-2 interacting with the C-terminus of NEDD8. The particularly negative charged patch of surface area of NEDP1 involves segments 1 and 2 (Figure 3). The third segment is formed by the U-shaped arrangement of helices α-4 and α-5 of the protease interacting with the loop connecting the first two β-strands of NEDD8. The fourth segment is the large loop (N91-T101) that changes conformation upon binding to the C-terminus of NEDD8. The final area of contact is between helix β-7 of NEDP1 and the C-terminus of NEDD8. The catalytic triad of residues (H102, D119 and C163) is consistent with the classic cysteine protease mechanism, and mutation of any of them effectively abolishes cleavage. The ordering of the loop and locking into a distinct conformation will perturb the position of H102 and may be important for efficient catalysis.
Figure 4.

Processing activities of NEDP1 mutant proteins. The substrate used in the assay is His-MBP-NEDD8-Ub and the assays are as detailed in Material and methods. NEDP1 and its mutants were mixed with the substrate and after incubation for 30 min at 37°C, the reaction was stopped by adding 6 × Laemmli loading buffer and boiled for 3 min. The processing products were fractionated using SDS–PAGE and the gel stained with 0.25% Coomassie blue. The designation of each mutant enzyme is indicated. CON: substrate only. The upper arrow indicates the substrate His-MBP-NEDD8-Ub, while the lower arrow indicates the cleavage product His-MBP-NEDD8. The released Ub has migrated off the bottom of the gel and is therefore not detected. Data presented are representative of three independent experiments.
Mutational analysis of NEDP1
To validate our structural analysis, we have employed site-directed mutagenesis to alter key residues in NEDP1 predicted to participate in substrate recognition and catalysis. To assay NEDD8 processing activity in vitro, proteins bearing altered residues were expressed in bacteria and purified to homogeneity. As substrate, a 6His-maltose binding protein-NEDD8-ubiquitin fusion (His-MBP-NEDD8-Ub) was expressed and purified to homogeneity. Incubation of His-MBP-NEDD8-Ub with NEDP1 releases ubiquitin, generating a His-MBP-NEDD8 product that has an increased electrophoretic mobility in SDS–PAGE. For each mutant, the extent of cleavage was determined by scanning of Coomassie blue-stained gels (Figure 4). While NEDP1 D10 forms a hydrogen bond with Y59 in NEDD8, the D10A version of NEDP1 has an activity that is indistinguishable from that of wild-type protein (Figure 4), suggesting that under the assay conditions employed, this residue is not a key determinant of NEDP1 activity. W26 in NEDP1 is highly conserved in the Ulp family of proteases and forms van der Vaals contacts with the C-terminal Gly–Gly of NEDD8. It sits directly above the protease catalytic site with its side chain locking the Gly–Gly into the active site cleft. Removal of the tryptophan side chain in the W26A mutation severely reduces the activity of the protease (Figure 4), thus underscoring the importance of this residue. Also conserved in all the Ulp proteins is D29 that forms a salt bridge with R42 in NEDD8. This appears to be a key interaction, as neither the D29A nor D29N versions of NEDP1 have detectable activity in the processing assay (Figure 4). N91 of NEDP1 forms a hydrogen bond with R74 of NEDD8 that immediately precedes the Gly–Gly motif. The importance of stabilising the C-terminus of NEDD8 in the NEDP1 active site is reflected in the lack of processing activity of the N91A NEDP1 mutant (Figure 4). Another highly conserved residue recognising the Gly–Gly motif is W103 that forms van der Waals interactions with G75 of NEDD8. Removal of this side chain in the W103A mutant results in a version of NEDP1 with severely reduced processing activity (Figure 4). Deletion of the large loop (LD, residues 93–99), which undergoes dramatic change in conformation upon NEDD8 binding, results in a protein with minimal NEDD8 processing activity (Figure 4). This is to be expected, as this region engages the NEDD8 C-terminus in a β-sheet-like arrangement with multiple interactions (Figure 2). The ordering of the loop and locking into a distinct conformation will perturb the location of the adjacent H102 and may be important for efficient catalysis. Analysis of the mutant protein by CD spectroscopy confirmed that it had not undergone any gross structural rearrangement (data not shown). Mutation of either of the residues that comprise the catalytic triad (H102, D119 and C163) abolishes processing activity (Figure 4), and this is entirely consistent with the proposed mechanism for this class of proteases. Although Q157 could interact with G76 of NEDD8, the Q157A mutation is without consequence (Figure 4) and indicates that this residue is probably not important for stabilisation of the NEDD8 C-terminus. Mutations V58A, F74A and P77A in helices α-4 and α-5 had little effect on processing activity (Figure 4).
While the assay employed above measures the processing activity of NEDP1 in vitro, it was also important to assay the deconjugating activity of the protease and to determine if the mutations altered the ability of the protease to discriminate between NEDD8 and ubiquitin. Using transfection-based assays to determine protease activity in vivo, we have previously determined that NEDP1 can deNEDDylate cullins (Mendoza et al, 2003) and the tumour suppressor p53 (Xirodimas et al, 2004). To evaluate the activity of the NEDP1 mutants in vivo, sequences encoding a representative selection of the mutant proteases were transferred into a eukaryotic expression vector and transfected into H1299 cells (genetically negative for p53) along with expression constructs for p53, its E3 ligase Mdm2 and either 6His-NEDD8 or 6His-Ub. After 48 h, a fraction of the cells was lysed under strongly denaturing conditions and 6His-containing proteins bound to Ni-agarose analysed by Western blotting using an antibody to p53. Prior to Ni-agarose chromatography, samples of whole-cell extracts were analysed by Western blotting to determine the levels of NEDP1. In the presence of Mdm2, there is a dramatic increase in the levels of NEDDylated and ubiquitinated p53 (Figure 5). When wild-type NEDP1 is coexpressed, p53 is completely deNEDDylated but there is no effect on the level of ubiquitinated p53, indicating that NEDP1 is specific for NEDD8 and has no activity against ubiquitin in vivo. In line with the in vitro data, mutants that have no activity in the processing assay are also defective in the in vivo deNEDDylation assay (Figure 5). None of the mutants have deubiquitinating activity, indicating that the mutations do not alter the specificity of the NEDP1 protease. Levels of NEDP1 did not display any substantial variation in expression levels that could explain the observed differences in in vivo deNEDDylation activity (Figure 5).
Figure 5.

Ability of NEDP1 mutants to deconjugate NEDDylated p53 in vivo. Top panel: H1299 cells were transfected with expression constructs for p53, mdm2, 6His-NEDD8 and NEDP1 mutants as indicated. At 36 h after transfection, cells were lysed in guanidine hydrochloride and 6His-NEDD8-conjugated species purified on Ni-NTA agarose as described in Materials and methods. NEDDylated p53 was detected by Western blotting with DO-1 anti-p53 monoclonal antibody. Total levels of NEDP1 are shown in the bottom panel. Middle panel: As in top panel, but 6His-ubiquitin was cotransfected rather than 6His-NEDD8.
Residues in NEDD8 and ubiquitin that allow discrimination by NEDP1
Many of the residues in NEDD8 that interact with NEDP1 are conserved in ubiquitin and therefore cannot be features that NEDP1 uses to discriminate between NEDD8 and ubiquitin. However, Glu53 and Ala72 are not conserved in ubiquitin where the equivalent residues are Gly and Arg respectively (Whitby et al, 1998). Glu53 appears to have only limited potential to make direct contacts with NEDP1, and in some species NEDD8 contains Asp at this position. In the ubiquitin-specific protease Yuh1, an Asp residue forms a salt bridge with R72 in ubiquitin and this interaction is important for catalysis (Johnston et al, 1999). Structural alignment of NEDD8 and ubiquitin in relation to their respective proteases indicates that NEDD8 and ubiquitin are oriented quite differently. The ubiquitin C-terminus is kinked at Arg72 by interaction with its cognate protease, whereas the NEDD8 C-terminus however contains Ala at this position and is straight (Figure 6A and B). We designed experiments to determine if the Arg/Ala difference is involved in discrimination between NEDD8 and ubiquitin. Fusions between NEDD8 and ubiquitin were generated that contained either wild-type NEDD8 or A72R NEDD8. These fusions were incubated with NEDP1 and the extent of cleavage determined over time. It is clear that the A72R mutant is cleaved significantly more slowly by NEDP1 than the wild-type NEDD8 (Figure 6C). We also made fusion proteins of ubiquitin connected to ubiquitin and connected to R72A ubiquitin. These constructs were incubated with NEDP1 and the products analysed by SDS–PAGE. As cleavage of ubiquitin by NEDP1 is inefficient, stoichiometric amounts of NEDP1 are required for cleavage to be evident (Figure 6D). However, incorporation of the R72A mutation into ubiquitin allows for an increased extent of cleavage by NEDP1 (Figure 6D). Conversely, the R72A ubiquitin construct is cleaved with a reduced efficiency compared to wild-type ubiquitin by the ubiquitin-specific protease HAUSP (Figure 6C). Thus, position 72 in NEDD8 and ubiquitin contributes to the discrimination of NEDD8 and ubiquitin by NEDP1.
Figure 6.

How NEDP1 discriminates between NEDD8 and ubiquitin. (A) Superposition of NEDD8 and ubiquitin, the key change is the backbone centred on residue 72. (B) The NEDP1–NEDD8 complex is shown coloured as before. Ubiquitin is shown as an yellow ribbon. The ubiquitin is positioned based on a superposition of NEDP1 and Yuh1. It is clear that ubiquitin has a very different spatial relationship to its protease. Residue 72 is shown and labelled; it is the kink at this position that is responsible for the dramatic difference in orientation. (C) Time course of NEDP1 protein on processing wild-type NEDD8 and NEDD8 A/R mutant. NEDP1 enzyme was incubated with NEDD8 and A72R mutant proteins at 37°C and the reaction was stopped at 0.5, 1, 2, 3, 4 and 6 min by adding an equal volume of 2 × Laemmli loading buffer and boiling for 3 min. The His-MBP-N8 was fractionated by SDS–PAGE and quantified by densitometric analysis using a multianalysis system (Doc-2000, Bio-Rad). The data presented are the mean of three independent experiments. Error bars represent the 95% confidence of the calculated means. (D) Activities of NEDP1 and HAUSP on processing of ubiquitin and R72A ubiquitin. Substrates used in the assay are His-MBP-N8-UB, His-MBP-N8(m)-UB (NEDD8 A72R mutant), HisUB-UB and HisUB9(m)-UB (ubiquitin R72A mutant). Substrates were mixed with NEDP1 or HAUSP and incubated for 30 min at 37°C. The processing products were fractionated in SDS–PAGE and stained with 0.25% Coomassie blue.
Discussion
We have determined the structure of the NEDD8-specific protease NEDP1 in isolation and of a covalent thiohemiacetal transition state complex between NEDP1 and NEDD8. Comparison of the structures of the bound and free forms of NEDP1 reveals that NEDD8 binding is accompanied by a dramatic change in the conformation of NEDP1. A flexible loop centred on Q96 in NEDP1 swings over the C-terminus of NEDD8, locking it into an extended β-structure and engaging residues important for NEDD8–ubiquitin discrimination. This loop is conserved in homologues of NEDP1 from other species (Mendoza et al, 2003), but is absent in other members of the Ulp family of proteases. After submission of this work, a structure was reported for the complex between NEDP1 (Den1) and NEDD8 (Reverter et al, 2005). Although the two structures for the complex are entirely compatible, the previous publication did not report on the structure of the free NEDP1 and thus structural changes that accompanied NEDD8 binding could not be identified.
In terms of the relative positioning of the two proteins in the complex, the complex between NEDD8 and NEDP1 is very similar to the complex between Ulp1 and SUMO (Mossessova and Lima, 2000). However, a detailed comparison of the interfaces in the Ulp1–SUMO complex with the NEDP1–NEDD8 complex reveals some very marked differences. The surfaces buried in the NEDP1–NEDD8 complex are much more extensive, principally because of loop 1 in NEDP1, which folds down over and makes hydrogen bonds with the C-terminus of NEDD8 (Figure 2). In Ulp1, a much shorter loop does not make such hydrogen bonds. Comparison of NEDP1–NEDD8 complex with Ulp1–SUMO complex reveals several differences in the position, size and charge of interacting residues. Examining the electrostatics of the complex (Figure 3) reveals that Ulp1 has two positively charged patches, whereas NEDP1 is uniformly negatively charged. These differences clearly explain the basis of the selectivity of Ulp1 for SUMO versus NEDD8 and NEDP1 for NEDD8 versus SUMO.
The key question, however, for NEDD8 biology is what drives the selectivity of NEDP1 for NEDD8 over ubiquitin, which is very similar in sequence (Figure 1), charge distribution (Figure 3) and in structure to NEDD8 (Figure 6). The complex of yeast Yuh1 covalently linked with ubiquitin aldehyde (Johnston et al, 1999) reveals the protease interface to be more acidic than the NEDP1 interface (Figure 3). Although the ubiquitin protease has no sequence similarity to NEDP1, a core of the secondary structural elements can be superimposed. The superpositions reveal that NEDD8 and ubiquitin are oriented quite differently with respect to their protease (Figure 6B) and there is no commonality in the recognition of ubiquitin and NEDD8. This gross difference is even more striking given that, of the very highly conserved NEDD8 residues that interact with NEDP1, only Ala72 and Glu53 are not equally well conserved in ubiquitin (found as Arg and Gly, respectively). Glu53 has limited contacts with NEDP1 and in some NEDD8 proteins, this residue is found as an Asp. The role of R72 in making a salt bridge with an aspartic acid in Yuh1 was highlighted as crucial in the formation of this complex and mutation of Arg severely affected binding. The structure of the complex between Ub and the deubiquitinylating enzyme (USP7/HAUSP) has also been reported (Hu et al, 2002). Although USP7/HAUSP has the same catalytic triad as NEDP1, there are very substantial differences in structure between the two enzymes, making a direct comparison of recognition impossible. USP7/HAUSP utilises a second domain (fingers), not found in YUH1 or NEDP1, to recognise ubiquitin. However, in this complex, Arg72 of ubiquitin was found to make a key salt bridge with a Glu residue in USP7/HAUSP. We have confirmed the importance of residue 72 by examining the cleavage of ubiquitin by the ubiquitin-specific protease HAUSP. While native ubiquitin was cleaved readily, the R72A mutant and NEDD8 were not susceptible to cleavage. Thus, the presence of an Ala residue at position 72 in NEDD8 and an Arg at the corresponding position in ubiquitin explains why ubiquitin-specific proteases cleave ubiquitin but not NEDD8. A more subtle question is why NEDP1 does not cleave ubiquitin, which is so similar to NEDD8. The most obvious difference between the two proteins is at position 72, which is Arg in ubiquitin and Ala in NEDD8. Using simple molecular modeling, it is possible to substitute an Arg residue at position 72 of NEDD8. However, in those conformations that avoid severe steric clashes, it is noticeable that the R72 would be close to either R74 or R42, both of which are conserved in NEDD8 and ubiquitin. R74 and R42 of both NEDD8 and ubiquitin are involved in recognition by their cognate proteases. The kink seen at the C-terminus in ubiquitin avoids this electrostatic repulsion between R72 and the two other Arg residues, but such a kink would destroy the interface with NEDP1. We suggest that an Arg will perturb the interface between NEDD8 and NEDP1 due to electrostatic and van der Waals repulsion with R42 and R74. In support of our structural analysis, biochemical experiments suggest a role for position 72 in discrimination between NEDD8 and ubiquitin by NEDP1. These conclusions are in contrast to those reached in the study reporting the structure of Den1 (NEDP1) with NEDD8. Based on the inability of R72A ubiquitin to inhibit the Den1 (NEDP1)-catalysed hydrolysis of ubiquitin-7-amido-4-methyl coumarin (AMC) (Reverter et al, 2005; data not shown), it was concluded that position 72 was not important for discrimination between NEDD8 and ubiquitin. While we feel that our structural arguments support a role for Ala72 in NEDD8–ubiquitin discrimination, the differences in interpretation may arise from the use of different biochemical analyses. In the experiments reported here, we generated four different protein substrates (NEDD8-Ub, A72RNEDD8-Ub, Ub-Ub and R72AUb-Ub) and tested their ability to be cleaved by NEDP1 and the ubiquitin-specific protease HAUSP (Figure 6). The biggest difference was observed in the assay where cleavage of NEDD8-Ub and A72RNEDD8-Ub by NEDP1 was compared. Our results indicated that A72RNEDD8-Ub was cleaved less efficiently than NEDD8-Ub by NEDP1. Conversely, R72AUb-Ub was cleaved more efficiently than Ub-Ub, although it should be noted that cleavage of N-terminal ubiquitin derivatives was rather inefficient and required stoichiometric amounts of NEDP1 for cleavage. In contrast, HAUSP cleaved R72AUb-Ub less efficiently than Ub-Ub. We therefore conclude that position 72 in NEDD8 and ubiquitin plays an important role in allowing NEDP1 to discriminate between these two highly related ubiquitin-like proteins. This is reminiscent of the situation in E1 enzymes where the NEDD8 E1 APPBP1-Uba3 uses a single conserved Arg residue to discriminate between Arg72 in ubiquitin and Ala72 in NEDD8 (Walden et al, 2003). In ubiquitin, the Arg72 clashes with the conserved Arg in the NEDD8 E1 and catalysis is blocked. However, when the cognate NEDD8 is present, there is no clash with the Ala at this position and catalysis is permitted. Thus, it is position 72 that appears to be responsible for the discrimination between ubiquitin and NEDD8 that is exerted when these Ubls both enter (via E1) and leave (via NEDP1) the modification cycle.
Materials and methods
Protein preparation
NEDP1 was cloned and expressed using vector pEHISTEV as an N-terminal His-tagged protein in Escherichia coli BL21(DE3) cells and purified using Nickel affinity chromatography (Ni-NTA-agarose, Qiagen). The His-tag was cleaved using TEV protease in 50 mM Tris–HCl (pH 8.0) 50 mM NaCl and 5 mM β-mercaptoethanol and removed from NEDP1 using Ni-NTA-agarose. NEDP1 was further purified by gel filtration chromatography. The purified NEDP1 was concentrated to ∼14 mg/ml in a buffer containing 20 mM Tris–HCl (pH 8.0) and 50 mM NaCl.
An N-terminal His-tagged full-length NEDD8 used for preparation of the NEDP1–NEDD8 complex was expressed and purified as described previously (Mendoza et al, 2003). Sel-Met-substituted NEDP1 and NEDD8 were generated as described previously (Guerrero et al, 2001).
All the mutants, including NEDD8(A72R), Ub(R72A) and NEDP1 mutants, were generated using a PCR-based mutagenesis (Brons-Poulsen et al, 1998).
Purified HAUSP generated in baculovirus-infected insect cells was a kind gift from Dr Roger Everett (MRC Virology Unit, Glasgow, UK).
Generation of NEDP1–NEDD8 complex
To generate the NEDP1–NEDD8 complex, NEDP1 was expressed as an N-terminal GST fusion and purified using glutathione Sepharose 4B column (Pharmacia). The covalent adduct was prepared using a molar ratio of 1:5 of GST-NEDP1 to His-tagged NEDD8 in a buffer containing 50 mM Tris–HCl (pH 8.0) 50 mM NaCl and 5 mM β-mercaptoethanol. A total of 10 aliquots of NaBH4 were added to the reaction mixture over 30 min to a final concentration of 30 mM. The GST-NEDP1-His NEDD8 complex was purified using glutathione Sepharose 4B column (Pharmacia) and Ni-NTA-agarose. After thrombin cleavage, GST was removed by glutathione affinity chromatography step and the NEDP1-His-NEDD8 complex purified by gel filtration chromatography. The complex was concentrated to ∼20 mg/ml in a buffer containing 20 mM Tris–HCl (pH 8.0) and 50 mM NaCl and used for crystallisation trials.
In vitro proteolysis assays
The His-MBP-NEDD8-Ub was expressed using vector pLous3 and purified using Ni-NTA-agarose and followed by gel filtration. An equal amount (20 ng) of the wild-type or mutant NEDP1 protein was incubated with 1 μg of His-MBP-NEDD8-Ub substrate at 37°C for 30 min in 15 μl of reaction buffer containing 50 mM Tris–HCl (pH 8.0), 50 mM NaCl and 5 mM β-mercaptoethanol. The reaction was stopped by the addition of 3 μl of 6 × SDS sample buffer and analysed by SDS–PAGE and Coomassie blue staining.
His-Ub-Ub and His-Ub(R72A)-Ub were expressed using pEHISTEV in E. coli BL21(DE3) and purified using nickel affinity and gel filtration chromatography.
To determine the effects of these mutations on processing by HAUSP and NEDP1, an equal amount (1.5 μg) of the His-Ub-Ub or His-Ub(R72A)-Ub was incubated with 1 μg of HAUSP or NEDP1 at 37°C in 15 μl of reaction buffer containing 50 mM Tris–HCl pH 8.0, 50 mM NaCl and 5 mM β-mercaptoethanol. The reaction was stopped by the addition of 3 μl of 6 × SDS sample buffer and analysed by SDS–PAGE.
In vivo analysis of NEDP1 activity
H1299 cells (genetically negative for p53) were transfected with expression constructs for p53, mdm2, NEDP1 and either 6His-NEDD8 or 6His-ubiquitin as described (Xirodimas et al, 2004). His-tagged proteins were isolated under denaturing conditions on Ni-agarose as described (Rodriguez et al, 1999).
Crystallography of the NEDP1–NEDD8 complex
Crystals were grown by setting-drop method by mixing the NEDP1–NEDD8 complex (20 mg/ml) with an equal volume of reservoir solution containing 20% PEG8000, 200 mM NaCl and 100 mM phosphate citrate pH 4.5. Crystals were equilibrated in a cryoprotectant buffer containing reservoir buffer plus 17.5% glycerol (v/v). Selenomethionine variants of both proteins were obtained using standard procedures (Doublie, 1997) and behaved essentially identically to the native form. The native and MAD data set was collected at ESRF beamline ID14 and BM14, respectively, and processed using DENZO/SCALEPACK (Otwinowski, 1997). There is one NEDP1 and one NEDD8 in each asymmetric unit. The crystals belong to the space group P212121 with cell dimensions of a=54.6 Å, b=74.2 Å, c=75.9 Å and α=β=γ=90°. The structure was determined by multiple anomalous dispersion. Data were collected at three wavelengths, treated with SOLVE (Terwilliger, 1997) and 10 selenium atoms were located. Initial phases, with a mean figure of merit of 0.60 at 2.8 Å resolution, were extended to 1.9 Å and improved with solvent flattening and histogram matching using DM (Collaborative Computational Project, 1994). A model was built using ARP/wARP (Morris et al, 2002), examined with the program O (Jones et al, 1991) and refined to a resolution of 1.9 Å with REFMAC5 (Murshudov et al, 1997, 1999).
Crystallisation and structure determination of NEDP1
Crystals were grown by setting-drop method by mixing NEDP1 (14 mg/ml) with an equal volume of reservoir solution containing 0.1 M bicine pH 9.0 and 30% PEG3000. Crystals were equilibrated in a cryoprotectant buffer containing reservoir buffer plus 15% glycerol (v/v). The native data set was collected at ESRF beamline ID14 and processed using DENZO/SCALEPACK (Otwinowski, 1997). The crystals belong to the space group P1 with cell dimensions a=57.1 Å, b=57.5 Å, c=75.0 Å and α=99.7°, β=110.9° and γ=92.4°. There are four NEDP1 monomers in each asymmetric unit. Attempts to merge the data in higher symmetry space groups failed. The structure was determined by molecular replacement, using the software AMoRe (Navaza, 1994). The coordinates of NEDP1 from the NEDP1–NEDD8 complex were used as a search model. All other calculations were carried out with programs in CCP4 suite (Collaborative Computational Project, 1994).
Acknowledgments
We thank Heidi Mendoza (University of Dundee) and Barbara Ink (Glaxo Smith Kline) for provision of the original NEDP1 expression constructs. We are indebted to Roger Everett (MRC Virology Unit) for providing us with purified HAUSP. We thank BM14UK and ESRF for data collection facilities. JHN is a BBSRC career development fellow. This work was supported by CRUK, Wellcome Trust and the BBSRC.
References
- Brons-Poulsen J, Petersen NE, Horder M, Kristiansen K (1998) An improved PCR-based method for site directed mutagenesis using megaprimers. Mol Cell Probes 12: 345–348 [DOI] [PubMed] [Google Scholar]
- Chiba T, Tanaka K (2004) Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr Protein Pept Sci 5: 177–184 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15: 435–467 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M (2003) The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J 22: 1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. Macromol Crystallogr A 276: 523–530 [PubMed] [Google Scholar]
- Gan-Erdene T, Kolli N, Yin L, Wu K, Pan ZQ, Wilkinson KD (2003) Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem 278: 28892–28900 [DOI] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N (2004) Structure of the Cand1–Cul1–Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119: 517–528 [DOI] [PubMed] [Google Scholar]
- Gong L, Kamitani T, Millas S, Yeh ET (2000) Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J Biol Chem 275: 14212–14216 [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET (1999) Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem 274: 12036–12042 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero SA, Hecht HJ, Hofmann B, Biebl H, Singh M (2001) Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl Microbiol Biotechnol 56: 718–723 [DOI] [PubMed] [Google Scholar]
- Handeli S, Weintraub H (1992) The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell 71: 599–611 [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Yao T, Wu JW, Gu W, Cohen RE, Shi Y (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Huang DT, Walden H, Duda D, Schulman BA (2004) Ubiquitin-like protein activation. Oncogene 23: 1958–1971 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Pyrowolakis G (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol 10: 335–342 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2002) Ubiquitin branches out. Nat Cell Biol 4: E295–E298 [DOI] [PubMed] [Google Scholar]
- Johnston SC, Riddle SM, Cohen RE, Hill CP (1999) Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J 18: 3877–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–118 [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET (1997) Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 272: 28557–28562 [DOI] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW (1999) The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev 13: 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, Suzuki H, Shimbara N, Hidaka Y, Osaka F, Omata M, Tanaka K (2001) NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J 20: 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Semenova E, Levorse JM, Tilghman SM (2000) Expression and functional analysis of Uch-L3 during mouse development. Mol Cell Biol 20: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M (1998) Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev 12: 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J 17: 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linghu B, Callis J, Goebl MG (2002) Rub1p processing by Yuh1p is required for wild-type levels of Rub1p conjugation to Cdc53p. Eukaryot Cell 1: 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, Hay RT (2003) NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem 278: 25637–25643 [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Nagayama Y, Yasuda H (2003) Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun 301: 392–398 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Perrakis A, Lamzin VS (2002) ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr D 58: 968–975 [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD (2000) Ulp1–SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe and automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev 12: 2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski ZMW (1997) Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol 275: 307–326 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Rose IA (1986) Mechanism of ubiquitin carboxyl-terminal hydrolase. Borohydride and hydroxylamine inactivate in the presence of ubiquitin. J Biol Chem 261: 10210–10217 [PubMed] [Google Scholar]
- Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B (2003) Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol 13: 911–921 [DOI] [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V (2000) A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci USA 97: 4579–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo JC, Timpte C, Tan S, Callis J, Estelle M (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280: 1760–1763 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2004) A basis for SUMO protease specificity provided by analysis of human Senp2–SUMO complex. Structure 12: 1519–1531 [DOI] [PubMed] [Google Scholar]
- Reverter D, Wu K, Gan-Erdene T, Pan Z-Q, Wilkinson KD, Lima CD (2005) Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J Mol Biol 345: 141–151 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J 18: 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Gordon C, Dubiel W (2001) Protein stability: the COP9 signalosome gets in on the act. Curr Biol 11: R643–R646 [DOI] [PubMed] [Google Scholar]
- Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG Jr, Ohh M (2004) pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol 24: 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Omata M, Tanaka K, Chiba T (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol 155: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (1997) Multiwavelength anomalous diffraction phasing of macromolecular structures: analysis of MAD data as single isomorphous replacement with anomalous scattering data using the MADMRG Program. Methods Enzymol 276: 530–537 [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T (1998) Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun 251: 688–692 [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL Jr, Holton JM, Schulman BA (2003) The structure of the APPBP1–UBA3–NEDD8–ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell 12: 1427–1437 [DOI] [PubMed] [Google Scholar]
- Whitby FG, Xia G, Pickart CM, Hill CP (1998) Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem 273: 34983–34991 [DOI] [PubMed] [Google Scholar]
- Wu K, Chen A, Tan P, Pan ZQ (2002) The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J Biol Chem 277: 516–527 [DOI] [PubMed] [Google Scholar]
- Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, Lee CG, Wei N, Wilkinson KD, Wang R, Pan ZQ (2003) DEN1 is a dual function protease capable of processing the C-terminus of Nedd8 deconjugating hyper-neddylated CUL1. J Biol Chem 278: 28882–28891 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP (2004) Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83–97 [DOI] [PubMed] [Google Scholar]


