Abstract
MicroRNAs (miRNAs) represent a family of small, regulatory, noncoding RNAs that are found in plants and animals. Here, we describe the miRNA profile of the zebrafish Danio rerio resolved in a developmental and cell-type-specific manner. The profiles were obtained from larger-scale sequencing of small RNA libraries prepared from developmentally staged zebrafish, and two adult fibroblast cell lines derived from the caudal fin (ZFL) and the liver epithelium (SJD). We identified a total of 154 distinct miRNAs expressed from 343 miRNA genes. Other experimental/computational sources support an additional 10 miRNAs encoded by 19 genes. The miRNAs can be classified into 87 distinct families. Cross-species comparison indicates that 81 families are conserved in mammals, 17 of which also have at least one member conserved in an invertebrate. Our analysis reveals that the zygotes are essentially devoid of miRNAs and that their expression begins during the blastula period with a zebrafish-specific family of miRNAs encoded by closely spaced multicopy genes. Computational predictions of zebrafish miRNA targets are provided that take into account the depth of evolutionary conservation. Besides miRNAs, we identified a prominent class of repeat-associated small interfering RNAs (rasiRNAs).
Keywords: Development, microRNA, rasiRNA, zebrafish
The zebrafish is an important model organism to study vertebrate development (Grunwald and Eisen 2002), and its genome sequence is close to completion (http://www.sanger.ac.uk/Projects/D_rerio). The current zebrafish genome assembly (Zv4 June 2004, ftp://ftp.ensembl.org/pub/assembly/zebrafish/Zv4release) contains 1.56 × 109 base pairs (Gbp), which corresponds to about half the size of the available human genome sequence. The number of predicted genes, which is ∼24,000, is nearly identical for the zebrafish and human assemblies (http://www.ensembl.org). Recently, an abundant noncoding RNA gene family has been discovered in plants and animals, whose members are known as microRNAs (miRNAs) (for review, see Ambros 2004; Bartel 2004; He and Hannon 2004). miRNAs regulate gene expression post-transcriptionally and are expressed in a developmental and cell-type-specific manner. The developmental regulation of miRNAs has only been studied systematically in the invertebrates Caenorhabditis elegans (Lau et al. 2001; Lee and Ambros 2001; Ambros et al. 2003; Lim et al. 2003b) and Drosophila melanogaster (Lagos-Quintana et al. 2001; Aravin et al. 2003; Lai et al. 2003; Sempere et al. 2003), but very little information is available concerning vertebrate miRNAs. The miRNA profile during the development from oocyte to tadpole stage was recently studied in Xenopus laevis using a combination of stage-specific small RNA cloning and Northern analysis (Watanabe et al. 2005). This study only identified 28 distinct miRNAs, three of which were novel miRNA genes. The number of currently validated miRNA genes in zebrafish is of similar size (Lim et al. 2003a), and expression was not resolved as a function of development. In rat, miRNA expression changes were noticed during brain development (Krichevsky et al. 2003). In mouse embryos, the spatial expression patterns of let-7b and let-7c, miR-1, miR-196a, and miR-10a have been examined during development (Mansfield et al. 2004).
Rather than examining the specific expression of miRNAs, it is possible to assess the general contribution of miRNAs during development by knocking out components of the RNA silencing machinery. Dicer RNase III knockout in mouse causes early embryonic lethality (Bernstein et al. 2003). Dicer-deficient zebrafish arrest during larval stage development only at around day 10, because maternally contributed Dicer maintains miRNA maturation during the early development of the homozygous mutant (Wienholds et al. 2003). However, if the maternal Dicer contribution is eliminated, defects appear much earlier during gastrulation, brain formation, somitogenesis, and heart development (Giraldez et al. 2005).
In order to obtain a comprehensive picture of the total number of miRNAs expressed during development of a vertebrate model organism, we recorded the miRNA profiles during the development of zebrafish. In contrast to previous studies, we have cloned and sequenced miRNAs at a larger scale that permitted the identification of 154 distinct miRNAs, 10 of which have not been previously identified in either zebrafish or other species. These 154 miRNAs map to 343 unique miRNA precursors. Phylogenetic conservation analysis of experimentally confirmed miRNAs in mammals and previous PCR-based specific amplification of zebrafish miRNA candidates (Lim et al. 2003a) supports the presence of another 10 low-abundance miRNAs encoded by 19 miRNA genes. The cloning approach also revealed the presence of repeat-associated small interfering RNAs (rasiRNAs) in fish as a distinct size class of small RNAs. To facilitate biochemical analysis of miRNA function in zebrafish, we provide zebrafish target predictions explicitly including conservation analysis.
Results and Discussion
Characterization of zebrafish miRNAs
In order to identify the zebrafish miRNAs, we cloned and sequenced small RNA libraries prepared from total RNA isolated from zebrafish at different developmental stages and some selected zebrafish cell lines (Pfeffer et al. 2003). The stages we examined corresponded to the early zygote period (0 h), the blastula period (4 h), the segmentation period (12 h), the pharyngula period (24 h), the hatching period (48 h), and several months old male and female adults (Kimmel et al. 1995). In addition, two fibroblast cell lines derived from caudal fin (ZFL) and liver epithelium (SJD) were examined.
From 7835 small RNA clones sequenced, 4658 (59%) could be annotated as miRNAs (Supplementary Table 2). The majority of the remaining small RNAs correspond to fragments of rRNA, tRNA, mRNA, and repeat-annotated sequences from zebrafish. Eight percent of the small RNA clones could not be functionally annotated but mapped to the currently available zebrafish genomic sequences, and 5.5% of the clones could not be mapped to the zebrafish genome, but we noticed that some of these sequences matched bacterial genomic sequences.
We have identified by cloning 154 distinct miRNAs (Supplementary Table 1). Four other miRNAs have been previously validated in zebrafish using a miRNA-selective PCR-based amplification method (Lim et al. 2003a), and another six miRNAs can be identified as homologs of cloned mammalian miRNAs. Taken together, these miRNAs can be classified into 87 distinct families (Supplementary Table 3). Relying on the currently available sequence information for human, mouse, rat, chicken, pufferfish, frog, fruitfly, and the nematode Caenorhabditis elegans, the families can be subdivided by the pattern of evolutionary conservation. Eighty-one families are conserved in mammals, out of which 17 include conservation in at least one invertebrate. Four families are found only in chicken, fish, and frog; one family is found only in fish and frog; and one miRNA appears specific only to zebrafish. The zebrafish miRNA precursor sequences are distributed over 362 unique genomic locations (Supplementary Table 4). Sixty-eight of the distinct miRNAs are potentially transcribed from multicopy miRNA genes, unless some of these copies are transcriptionally inactive pseudogenes.
miRNAs are frequently organized in gene clusters (Lagos-Quintana et al. 2001; Lau et al. 2001). We considered pre-miRNAs as clustered if they mapped <20 kb apart and had the same direction of transcription. The 20-kb cutoff was selected to incorporate genomic segments encoding the spliced clusters of miR-125-let-7-miR-99/100 (Aravin et al. 2003; Rodriguez et al. 2004). We obtained 53 clusters, which together contain 218 of the 362 precursors (Supplementary Table 5). The two largest clusters contain 58 and 33 of sequence-related pre-miRNAs, and their genomic regions are 16 and 30 kb in size, respectively. The remaining clusters are much smaller and contain on average 2.5 pre-miRNAs, with a mean cluster size of 3163 bp (SD = 4714 bp) (see also Supplementary Fig. 2). Pairs of clustered miRNAs have higher alignment scores than random pairs of zebrafish miRNAs, consistent with the hypothesis that gene duplication is responsible, at least in part, for the emergence of these clusters (Supplementary Fig. 1).
To identify repeat-derived small RNAs that may play a role in regulating chromatin structure (for review, see Lippman and Martienssen 2004), we extracted sequences from the small RNA pool that were annotated as “repeat” or “none” and that mapped perfectly to at least five genomic loci (Supplementary Table 2). We had a total of 291 such sequences, of which 250 were distinct. We observed that they frequently map to the genome in a clustered manner within kilobases of each other, and that some of these clusters contain small RNAs mapped to the plus and minus strand of nearby genomic regions (Supplementary Table 6), consistent with the hypothesis that they originated from processing of longer dsRNAs. Seventy-two percent of these small RNAs have a U nucleotide at the first position, their mean length is 24.6 nucleotides (nt) (SD = 3.2 nt), and their frequency is highest in the 0-h sample (15% of the sequences, compared with 1%-5% in other tissues and developmental stages) (Supplementary Table 7). Although the degree of completion of the zebrafish genome sequence and annotation is much less than those that were available for the fruit fly, these data suggest that zebrafish, similar to fruit fly, processes repeat transcripts into a class of rasiRNAs (Aravin et al. 2003).
miRNAs are essentially absent from the early zygote stage as indicated by a mere 3% miRNA content of the small RNA library, and undetectable amounts for the cloned miRNAs by Northern blotting (Fig. 2, below). A similar observation was made in a study characterizing miRNAs during development of Xenopus laevis, and also showed extremely low levels of miRNA in the fertilized egg (Watanabe et al. 2005). In general, for miRNAs with relatively high cloning frequency (>20 independent clones), the relative expression can be correlated with the signal intensities in the Northern blot, except for the 0-h time points at which very few miRNAs were present at very low levels. To remedy this problem for miRNA profile display, we normalized clone numbers, taking into account the relative fraction of miRNAs identified within the total pool of cloned small RNAs of a given RNA sample (see legend to Fig. 1).
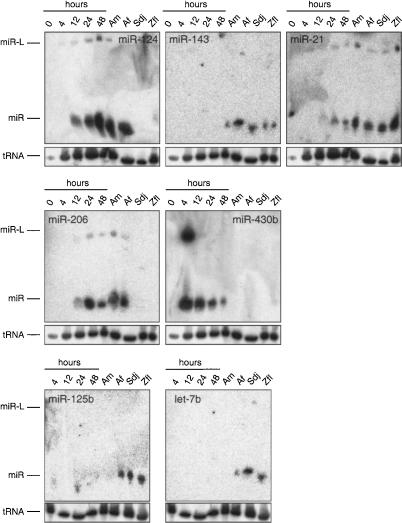
Figure 2.
Northern blot analysis of selected zebrafish miRNAs. The embryonic developmental stages are indicated in hours (h). Adult male (Am), adult female (Af), and fibroblast cell lines Sdj and Zfl are also examined. miR-L indicates the foldback dsRNA precursor form; miR refers to the mature predominantly 22-nt form. tRNA indicates the band detected with a probe complementary to tRNAval to monitor equal loading of the samples.
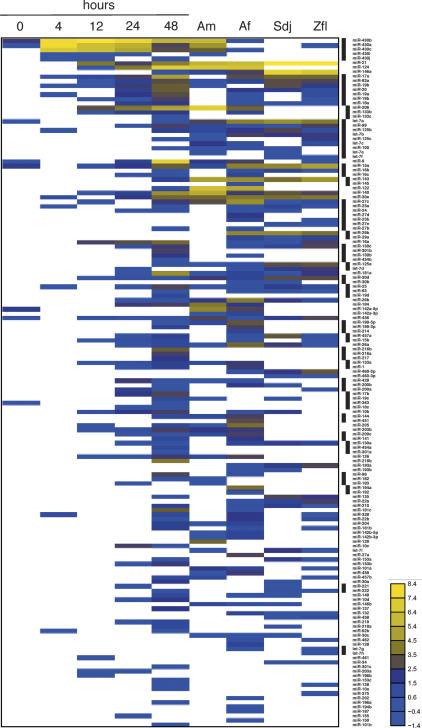
Figure 1.
Graphic representation of the miRNA profile. The profile is based on clone numbers and is scaled such that the total number of clones from each RNA sample (Supplementary Table 2) is the same and arbitrarily set at 1000 clones. This adjustment corrects for the difference in the number of clones at different stages assuming that the total small RNA composition of each pool is constant. The profile shows log2 of normalized miRNA clone numbers. The black bars to the right identify miRNAs that reside in miRNA gene clusters (Supplementary Table 5), and they were grouped together because they are assumed to be coexpressed.
The picture changed 4 h post-fertilization (hpf), when zygotic transcription is initiated, and a zebrafish-specific miR-430 family composed of five members was expressed. This miRNA family is very unusual in that it has ∼100 gene copies distributed over two large clusters of 30 and 17 kb within unassembled genome sequence, and a very small (500 bp) cluster of three miRNAs positioned on chromosome 13 (Supplementary Fig. 3). The genomes of Fugu rubripes and Tetraodon nigroviridis also appear to contain multiple copies of sequences that are either identical or closely related to zebrafish miR-430a, although the triple-repeat structure of the miR-430 miRNA clusters does not appear to be conserved in the other fish. This miRNA family is also related in sequence to the human and mouse embryonic stem (ES) cell-specific miRNAs (miR-291 to miR-295, miR-302, miR-371 to miR-373), which also occur in gene clusters (Houbaviy et al. 2003; Suh et al. 2004).
The expression of the miR-430 family clusters peaked at the 4-h stage, dominated the miRNA profile up to the 24-h-stage miRNA, and then decreased (Figs. 1, 2; Supplementary Table 1). This family was recently examined in zebrafish zygotic Dicer mutants, and injection of the processed form of a member of this miRNA family was able to rescue the brain morphogenesis phenotype (Giraldez et al. 2005). A miRNA related in sequence and expression level to zebrafish miR-430b was also discovered in X. laevis, and it peaked in expression during the blastula period (Watanabe et al. 2005), which roughly corresponds to the 4-h zebrafish developmental stage.
Another strongly expressed miRNA emerging early in development, at the 12-h stage, was miR-206, a member of the universally conserved miR-1 family, which was first shown to be specifically expressed in the adult mouse or human heart (Lee and Ambros 2001; Lagos-Quintana et al. 2002). miR-206 is ∼15 times more abundant in zebrafish than miR-1, and its timing of expression is similar in X. laevis (Watanabe et al. 2005).
At the 24-h stage, when segmentation and much of brain development have already taken place, we found that miR-9 and miR-124, both of which were shown to be specifically expressed in mouse brain (Lagos-Quintana et al. 2002), had accumulated to 5% of the miRNA pool, and this fraction increased to ∼30% at the 48-h stage. The relative increase in miR-124 is due to an increase in absolute expression level, as determined by Northern blotting (Fig. 2).
miR-122, which was shown to be specifically expressed in mouse liver (Lagos-Quintana et al. 2002), emerged at the 48-h stage. At this stage, the liver is a coherent mass of cells rostral to the intestine containing hepatocytes, but biliary function has not yet been established (Lorent et al. 2004).
The developmentally regulated and in nematodes best-characterized let-7 and lin-4/miR-125 family members (for review, see Ambros 2004) showed very low expression levels, and individual members were only detectable by Northern analysis in the adult female zebrafish and were enriched in the fibroblast cell lines (Fig. 2). In X. laevis, expression of let-7 members was also only detectable late in development, in the tailbud and tadpole stage (Watanabe et al. 2005).
The miRNA profiles of the fibroblast cell lines closely resemble each other, even though the cell lines were established independently from different tissue sources, liver and caudal fin. Their miRNA profile is dominated by the expression of miR-21, which accounted for ∼40% of all cloned miRNAs, followed by miR-146a, which accounted for ∼15% of all miRNA clones. The remaining 45% of miRNA clones are subdivided between 40 additional miRNA families, where the miR-15 and the let-7 families each represented ∼5%. The absolute amount of miR-21 in fibroblast cell lines, however, was comparable to the amount present in adult fish as determined by Northern blotting (Fig. 2).
Predicted miRNA target genes and sites
To date, predicted miRNA targets have only been provided for an incomplete set of zebrafish miRNAs (John et al. 2004). Another early study predicted miRNA targets conserved between mammals and the pufferfish F. rubripes, but does not present specific zebrafish targets (Lewis et al. 2003). To provide an overview of the potential control of gene regulation by miRNAs and for convenience in planning experiments, we provide several tables of miRNA targets at different levels of cross-species conservation (Supplementary Tables 8-10). Additional details about the predicted targets, such as sequence context of sites on aligned UTRs, are available at http://www.microrna.org/zebrafish. In general, target genes are ranked by the total alignment score (S), which reflects the sum over all sites for all miRNAs that may cooperatively target the gene if they are coexpressed (John et al. 2004). Note that one miRNA typically targets more than one gene (multiplicity), and one gene can be targeted by more than one miRNA (cooperativity) (Enright et al. 2003). The targets of the miRNAs that are highly expressed during early development (<48 h) (miR-430, miR-124, miR-206, and miR-9) are particularly interesting candidates for initial experimental validation. We note that many genes implicated in the Wnt and TGFβ/Nodal signaling pathways important during development (Schier 2003) are predicted to be regulated by the strong and early expressed miR-430 family (Supplementay Table 8; see also http://www.microrna.org/zebrafish).
The total number of predicted targets at a given score cutoff and chosen level of species conservation provides a rough estimate of the breadth of gene regulation by miRNAs within one species and the extent of cross-species conservation of specific miRNA-target relationships. Interestingly, there are strong differences in the specificities of individual miRNAs. For example, in D. rerio, the number of predicted target genes per miRNA, called target multiplicity (Enright et al. 2003), ranges from 0 to 99, with an average of 28 target genes per miRNA. The cooperativity of the miRNAs (the number of target sites per gene) ranges from 1 to 11, with an average of 2.5 miRNA target sites per gene (numbers based on targets conserved in fish and frog). In planning validation experiments, we think it is particularly important to take into account the cooperative or combinatorial control of gene expression by a group of miRNAs (Enright et al. 2003; Bartel and Chen 2004; Hobert 2004; John et al. 2004), which has recently also been called “coordinate” control (Krek et al. 2005).
Current computational target prediction methods applied to vertebrates differ in their emphasis on qualitative aspects. Some miRNA target prediction methods require a perfect match between miRNA positions 2-8 and its complementary target site (Rajewsky and Socci 2004; Lewis et al. 2005). Our method places a higher weight on this region in a dynamic programming method over the entire 21-residue length. Similarly, cooperativity is reflected in our method by numerical addition of S scores for all target sites by one or more miRNAs on a given gene, after removal of overlap. Such differences between algorithms tend to disappear for very high scoring targets. For example, 47% of the targets in Supplementary Table 8 have a perfect match with miRNA positions 2-8. We expect further improvements in target prediction methods to come from large-scale experimental validation.
Material and methods
RNA isolation
To collect total RNA from zebrafish at different developmental stages, zebrafish embryos were raised at 28°C under standard conditions in 0.3× Danieau's solution (Westerfield 1995). The following stages were examined: zygote period (0 hpf), blastula period (4 hpf), segmentation period (12 hpf), pharyngula period (24 hpf), hatching period (48 hpf), and male and female adult (6 mo old). Total RNA was isolated from ∼1500 embryos per stage or six male or six female fish. To facilitate sample processing only 500 embryos or two adult fish were homogenized at a time with 5 mL of extraction solution, which was freshly prepared by mixing 1 volume of solution A (4 M guanidinium isothiocyanate, 25 mM sodium citrate, 0.5% N-lauroylsarcosinate, 50 mM β-mercaptoethanol) with 1 volume of acidic phenol (equilibrated with water at pH 4) containing 0.5% 5-hydroxychinolin (Chomczynski and Sacchi 1987). Five-hundred microliters of chloroform:isoamyl alcohol (24:1) and 250 μL of 3 M sodium acetate (pH 4.2) were added, and homogenization continued until a white and turbid solution was obtained. The solution was transferred to 15-mL glass centrifuge tubes and centrifuged for 10 min at 10,000 rpm (Sorvall SS34 rotor) at 4°C. The upper phase was transferred to a new tube, extracted with 1/2 volume of acidic phenol:chloroform:isoamyl alcohol (24:24:1), and centrifuged again for 10 min at 10,000 rpm at 4°C. The extraction of the aqueous phase was repeated until a clear interphase was observed. The RNA was precipitated from the aqueous solution by the addition of 2.5 volumes of ethanol and incubation at -20°C overnight. The RNA was collected by centrifugation for 30 min at 12,000 rpm at 4°C. The supernatant was discarded, and the pellet was redissolved in 1 mL of solution A. The RNA was precipitated by the addition of 1 mL of isopropanol and incubation for 15 min at room temperature. The RNA was then collected by centrifugation for 30 min at 10,000 rpm at 4°C, and the pellet was washed twice with 5 mL of 70% ethanol at room temperature. The dried pellet was dissolved in formamide to a concentration of ∼1 mg/mL.
Zebrafish cell lines
Zebrafish ZFL and SJD cell lines were purchased from LGC Promochem, catalog numbers CRL-2643 and CRL-2296, respectively. Cells were grown in standard culture medium (Dulbecco's Modified Eagle's medium supplemented with 15% fetal calf serum) at 28°C and 5% CO2 in a humidified incubator. Cells (3 × 108) were trypsinized, collected, and transferred to a small glass homogenizer. The RNA was isolated following the protocol described above starting with 3 mL of solution A.
Small RNA cloning and Northern blotting
Small RNA cloning was performed according to the alternate protocol (Pfeffer et al. 2003), using chemically adenylated 3′ adapter and the truncated T4 RNA ligase 2, Rnl2(1-249) (Ho et al. 2004; Meister et al. 2004). As size markers for the RNA fractionation, the following 19- and 24-nt RNAs were used: 5′-32pCGUACGCGGGUUUAAACGA-3′ and 5′-32pCGUACGCGGAAUAGUUUAAACUGU-3′. Two-hundred micrograms of total RNA was used as starting material for library preparation. The PCR product was digested with PmeI restriction endonuclease to avoid cloning and sequencing of size markers before the second PCR step. Cloning and sequencing were performed as described (Pfeffer et al. 2004). On average, 300 clones containing concatamerized PCR products were sequenced per library. Northern blot analysis was performed as described (Pfeffer et al. 2004) loading 30 μg of total RNA per lane and using 5′32P-radiolabeled 21- or 22-nt oligodeoxynucleotides complementary to the predominantly cloned miRNA sequence. To monitor equal loading of total RNA, the blots were reprobed with 5′-TGGTGTTTCCGCCCGGTTT-3′ to detect tRNAval.
Sequence analysis
We used the most recent version, Zv4 June 2004, of the zebrafish genome assembly produced by the Zebrafish Sequencing Group at the Sanger Institute (ftp://ftp.ensembl.org/pub/assembly/zebrafish/Zv4release). The genome assembly and some functional annotation are available from the genome browser at the University of California at Santa Cruz (http://genome.cse.ucsc.edu). For our functional annotation, we assembled a database of rRNA, tRNA, snRNA, snoRNA, and mRNA sequences by querying GenBank (http://www.ncbi.nih.gov/Genbank/index.html), with the appropriate feature key and species (zebrafish, other fish including Barbus barbus, Carassius carassius, Cynoscion nebulosus, Cyprinus carpio, Gobio gobio, Notropis hudsonius, Pimephales promelas, Rutilus rutilus, Oncorhynchus mykiss, Salvelinus alpinus, Salmo trutta, mouse, and human). We additionally used a data set of human tRNA sequences (http://rna.wustl.edu/GtRDB/Hs/Hs-seqs.html), a data set of human and mouse sn/snoRNA sequences (http://condor.bcm.tmc.edu/smallRNA/smallrna.html), the Rfam database of miRNAs (http://www.sanger.ac.uk/Software/Rfam/mirna), and repeat annotations from University of California at Santa Cruz. We also queried GenBank for species, Rattus norvegicus, and for feature, scRNA (small cytoplasmic RNA). Finally, we incorporated the snoRNA database (http://www-snorna.biotoul.fr) as well as tables of predicted miRNAs (Lim et al. 2003a; Berezikov et al. 2005; Legendre et al. 2005) into our own database of noncoding RNA.
All small RNAs obtained by cloning were compared with functionally annotated sequences using the Washington University implementation (http://blast.wustl.edu, W. Gish, 1996-2004) of BLAST (Altschul et al. 1990) as well as in-house sequence alignment programs. For each small RNA, the best alignments to a functionally annotated sequence (up to at most three errors) were used to assign a functional category to the small RNA. In cases where multiple functional annotations were possible, we used the one reflecting the relative abundance of RNAs in the cell and the selection imposed by the cloning protocol (rRNA > tRNA > miRNA > sn/snoRNA > miscRNA > mRNA).
Prediction of miRNA target genes and sites
We used the miRanda method (Enright et al. 2003; John et al. 2004; software version 2.0 as available at http://www.microrna.org/miranda) to detect potential target sites for the zebrafish miRNA sequences (Supplementary Table 1) on any of the 23,524 zebrafish 3′-UTR sequences retrieved from the ENSEMBL (build 29_4c) database (Birney et al. 2004). Cut-off conditions for reported target sites: match score S ≥ 140 and duplex free energy ΔG ≤ -10 kcal/mol. Other parameters and conditions: scale factor w = 4.0 for complementary nucleotide match score in positions 2-8, counting from the miRNA 5′-end; not more than one non-Watson-Crick base pair at positions 2-8 and less than four G:U base pairs at positions 9-21. These parameters were chosen to reflect current knowledge as derived from a relatively small number of experimentally validated miRNA-target relationships and an even smaller number of validated miRNA-target sites (John et al. 2004).
Evolutionary conservation of candidate miRNA-target relationships was tested between zebrafish and several organisms (F. rubripes, T. nigroviridis, Xenopus tropicalis, Gallus gallus, Mus musculus, Rattus norvegicus, and Homo sapiens). Homologous miRNAs were defined as the most sequence-similar after cross-species alignment. Homologous gene pairs were retrieved using Ensmart (Kasprzyk et al. 2004). Sequence similarity of target sites, that is, of the mRNA subsequences after optimal alignment, was computed in terms of a weighted normalized sum (C) of the number of identical residues, with a weight, w = 4.0 on miRNA positions 2-8 and w = 1.0 elsewhere, reflecting nonuniform functional constraints along a target site. Target sites were considered conserved if C ≥ 0.85 between D. rerio and each of F. rubripes, T. nigroviridis, and X. tropicalis. Similarly, in mammals (human vs. mouse or rat), we required C ≥ 0.9. Between fish and mammals, only the miRNA-target relationship was required to be conserved, with no additional cutoff in C.
Acknowledgments
We thank Erez Raz for support with zebrafish, S. Pfeffer and M. Lagos-Quintana for introduction to miRNA cloning, P. Landgraf for assistance in developing the miRNA annotation tools, M. Pack for discussion, S. Shuman for providing Rnl2 ligase, and members of the laboratory for critical reading of the manuscript. We also thank M. Wilson for the development of the Web interface to miRNA targets. The work was financially supported by NIH grant P01 GM073047-01 and the Bundesministerium für Bildung und Forschung (BMBF), Biofuture grant number 0311856.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1310605.
Corresponding authors.
References
- Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Ambros V. 2004. The functions of animal microRNAs. Nature 431: 350-355. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807-818. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337-350. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. and Chen, C.Z. 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5: 396-400. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H., and Cuppen, E. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21-24. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn, H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., and Hannon, G.J. 2003. Dicer is essential for mouse development. Nat. Genet. 35: 215-217. [DOI] [PubMed] [Google Scholar]
- Birney E., Andrews, D., Bevan, P., Caccamo, M., Cameron, G., Chen, Y., Clarke, L., Coates, G., Cox, T., Cuff, J., et al. 2004. Ensembl 2004. Nucleic Acids Res. 32: D468-D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156-159. [DOI] [PubMed] [Google Scholar]
- Enright A.J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:RI: 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli, R.M., Glasner, M.E., Enright, A.J., Thomson, M.J., Baskerville, S., Hammond, S.M., Bartel, D.P., and Schier, A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308: 833-838. [DOI] [PubMed] [Google Scholar]
- Grunwald D.J. and Eisen, J.S. 2002. Headwaters of the zebrafish—Emergence of a new model vertebrate. Nat. Rev. Genet. 3: 717-724. [DOI] [PubMed] [Google Scholar]
- He L. and Hannon, G.J. 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5: 522-531. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Wang, L.K., Lima, C.D., and Shuman, S. 2004. Structure and mechanism of RNA ligase. Structure (Camb.) 12: 327-339. [DOI] [PubMed] [Google Scholar]
- Hobert O. 2004. Common logic of transcription factor and microRNA action. Trends Biochem. Sci. 29: 462-468. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell-specific MicroRNAs. Dev. Cell 5: 351-358. [DOI] [PubMed] [Google Scholar]
- John B., Enright, A.J., Aravin, A., Tuschl, T., Sander, C., and Marks, D. 2004. Human miRNA targets. PLoS Biol. 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk A., Keefe, D., Smedley, D., London, D., Spooner, W., Melsopp, C., Hammond, M., Rocca-Serra, P., Cox, T., and Birney, E. 2004. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 14: 160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard, W.W., Kimmel, S.R., Ullmann, B., and Schilling T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253-310. [DOI] [PubMed] [Google Scholar]
- Krek A., Grün, D., Poy, M.N., Wolf, R., Rosenberg, L., Epstein, E.J., MacMenamin, P., da Piedade, I., Gunsalus, K.C., Stoffel, M., et al. 2005. Combinatorial microRNA target prediction. Nat. Genet. 37: 495-500. [DOI] [PubMed] [Google Scholar]
- Krichevsky A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9: 1274-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853-858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735-739. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tomancak, P., Williams, R.W., and Rubin, G.M. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862-864. [DOI] [PubMed] [Google Scholar]
- Legendre M., Lambert, A., and Gautheret, D. 2005. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics 21: 841-845. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih, I., Jones-Rhoades, M.W., Bartel, D.P., and Burge, C.B. 2003. Prediction of mammalian microRNA targets. Cell 115: 787-798. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge, C.B., and Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15-20. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. 2003a. Vertebrate microRNA genes. Science 299: 1540. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003b. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z. and Martienssen, R. 2004. The role of RNA interference in heterochromatic silencing. Nature 431: 364-370. [DOI] [PubMed] [Google Scholar]
- Lorent K., Yeo, S.Y., Oda, T., Chandrasekharappa, S., Chitnis, A., Matthews, R.P., and Pack, M. 2004. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multiorgan defects compatible with an Alagille syndrome phenocopy. Development 131: 5753-5766. [DOI] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe, B.D., Nissen, R., Obenauer, J., Srineel, J., Chaudhuri, A., Farzan-Kashani, R., Zuker, M., Pasquinelli, A.E., Ruvkun, G., et al. 2004. MicroRNA-responsive `sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 36: 1079-1083. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., and Tuschl, T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15: 185-197. [DOI] [PubMed] [Google Scholar]
- Pfeffer S., Lagos-Quintana, M., and Tuschl, T. 2003. Cloning of small RNA molecules. In Current protocols in molecular biology (eds. F.M. Ausubel et al.), pp. 26.4.1-26.4.16. John Wiley and Sons, New York. [DOI] [PubMed]
- Pfeffer S., Zavolan, M., Grasser, F.A., Chien, M., Russo, J.J., Ju, J., John, B., Enright, A.J., Marks, D., Sander, C., et al. 2004. Identification of virus-encoded microRNAs. Science 304: 734-736. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. and Socci, N.D. 2004. Computational identification of microRNA targets. Dev. Biol. 267: 529-535. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones, S., Ashurst, J.L., and Bradley, A. 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res. 14: 1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A.F. 2003. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19: 589-621. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Sokol, N.S., Dubrovsky, E.B., Berger, E.M., and Ambros, V. 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 259: 9-18. [DOI] [PubMed] [Google Scholar]
- Suh M.R., Lee, Y., Kim, J.Y., Kim, S.K., Moon, S.H., Lee, J.Y., Cha, K.Y., Chung, H.M., Yoon, H.S., Moon, S.Y., et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270: 488-498. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Takeda, A., Mise, K., Okuno, T., Suzuki, T., Minami, N., and Imai, H. 2005. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 579: 318-324. [DOI] [PubMed] [Google Scholar]
- Westerfield M. 1995. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). University of Oregon Press, Eugene.
- Wienholds E., Koudijs, M.J., Van Eeden, F.J., Cuppen, E., and Plasterk, R.H. 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35: 217-218. [DOI] [PubMed] [Google Scholar]