Abstract
To gain insight into splicing regulation, we developed a microarray to assay all annotated alternative splicing events in Drosophila melanogaster and identified the alternative splice events controlled by four splicing regulators: dASF/SF2, B52/SRp55, hrp48, and PSI. The number of events controlled by each of these factors was found to be highly variable: dASF/SF2 strongly affects >300 splicing events, whereas PSI strongly affects only 43 events. Pairwise analysis also revealed many instances of splice site usage affected by multiple factors and provides the framework to understand the network controlling the alternatively spliced mRNA isoforms that compose the Drosophila transcriptome.
Keywords: Alternative splicing, SR proteins, hnRNP proteins, microarray, Drosophila melanogaster
Higher eukaryotes exploit alternative pre-mRNA splicing to diversify their proteome and to regulate gene expression with developmental stage-and tissue-specificity (Maniatis and Tasic 2002). Therefore, a comprehensive understanding of an organism's gene expression program must include an understanding of alternative splicing (Black 2003). Toward this end, a well-conserved core of pre-mRNA splicing regulatory factors has been identified in all metazoan organisms (Moore et al. 1993; Jurica and Moore 2003). However, the majority of the interactions between these regulators and their target pre-mRNAs remain unknown.
Historically, it has been challenging to identify genes specifically regulated by individual splicing factors. Despite tremendous effort, several years separated the initial identification of the hnRNP proteins PSI and hrp48 as regulators of the P-transposase pre-mRNA (Siebel et al. 1994) and the identification of a single additional targeted cellular gene (Burnette et al. 1999; Labourier et al. 2002). Recent advances in microarray technology now permit monitoring of various aspect of RNA processing and maturation (Lee and Roy 2004). In particular, high-density microarrays have been successfully used to monitor pre-mRNA processing events in yeast (Clark et al. 2002; Burckin et al. 2005), to identify new instances of alternative splicing in human and Drosophila (Hu et al. 2001; Johnson et al. 2003; Wang et al. 2003; Stolc et al. 2004) and to monitor alternative splicing levels of cassette exons in different mouse and human tissues (Pan et al. 2004; Relogio et al. 2005). Here we describe the development of a new Drosophila microarray platform and its use to monitor all the annotated pre-mRNA splicing junctions specifically controlled by four canonical splicing regulators, the hnRNPs PSI and hrp48 as well as the argine/serine-rich (SR) proteins dASF/SF2 and dSRp55/B52. This study identified tens to hundreds of distinct splice events modulated by each of these splicing factors and reveals the amount of coregulation and antagonism between each.
Results
A microarray for monitoring alternative splicing in Drosophila melanogaster
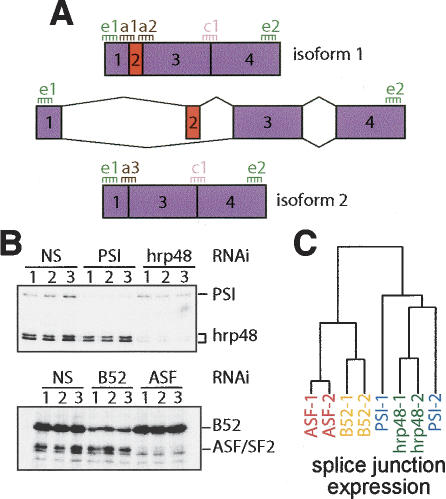
In order to rapidly and efficiently identify target genes and specific splicing events regulated by specific splicing factors, we have developed a microarray for monitoring changes of all the known alternatively spliced transcripts in Drosophila melanogaster. From the 13,472 genes in the GadFly 3.2 Drosophila genome annotation (http://flybase.bio.indiana.edu/annot/download_sequences.html), 2931 were found to have cDNA (EST) evidence of alternative splicing and generate 8315 different alternatively spliced mRNAs (Celniker and Rubin 2003). In order to monitor the complete set of annotated alternatively spliced transcripts, the single custom microarray contains probes spanning all the Drosophila annotated alternative splice junctions regardless of the specific alternative splicing pattern (probes labeled “a” in Fig. 1A; 9868 probes), and, up to two probes for constitutive splice junctions (probes labeled “c” in Fig. 1A; 4377 probes) for each of the alternatively spliced genes. Since it is known that there are many alternative mRNA isoforms yet to be annotated as such (Stolc et al. 2004), some of the junctions labeled constitutive may actually be alternative. Two common exon probes spanning segments present in all isoforms of each gene (probes labeled “e” in Fig. 1A; 5650 probes) were also selected for monitoring overall expression levels of the alternatively spliced mRNAs. This feature of the design allows potential changes in transcription level, or secondary effects, to be separated from effects on splicing patterns for a given gene.
Figure 1.
Experimental design and clustering results. (A) Thirty-six-mer probes were selected for all alternatively spliced junctions from the GadFly 3.2 Drosophila genome annotation (“a” probes). For each gene, two exonic probes were selected from regions common to all isoforms to gauge total gene expression (“e” probes). Up to two constant constitutive junction probes were also selected (“c” probes). (B) Immunoblot analysis confirmed effective RNAi knockdown of the hnRNP proteins PSI and hrp48 and of the SR proteins B52/SRp55 and dASF/SF2. (C) Hierarchical clustering using average log expression ratios from all splice junction probes was performed to assess the global affects of biological replicates of each splicing factor RNAi knockdown and to compare between splicing factors. This analysis indicates that the dASF/SF2, B52/SRp55, and hrp48 experiments produce a characteristic splicing response. The PSI results, however, were more variable (see text). The global splicing response to dASF/SF2 or B52/SRp55 knockdown includes more similarities than either does to hrp48 or PSI knockdown.
Genome-wide monitoring of alternative splicing
Using our array, we monitored splicing profile changes in Drosophila SL2 cells following RNAi knockdown of four splicing regulators: the SR proteins dASF/SF2 and B52/SRp55, and the hnRNP proteins PSI and hrp48 (Supplementary Fig. 1). Each of these four well-characterized splicing regulators is highly expressed in SL2 cells, and several of them have known pre-mRNA targets. Following treatment with double-stranded RNA (dsRNA) against each splicing factor, efficient protein reduction was confirmed by immunoblot analysis using antibodies specific for each protein (Fig. 1B). RNAi knockdown of each of these splicing factors generated no obvious morphological or growth phenotype in SL2 cells, despite the fact that in Drosophila PSI, hrp48, and B52/SRp55 are essential and dASF/SF2 is likely to be essential (Wang et al. 1996, 1998; Longman et al. 2000). From each RNAi-treated sample and from control cells treated with nonspecific dsRNA, total RNA was extracted, cDNA prepared, and labeled using a protocol developed to give good coverage over the entire length of all mRNAs (Castle et al. 2003; Johnson et al. 2003). Following standard hybridization, scanning, and data extraction, each experiment and each probe signal were filtered for consistency and RNAi target specificity (for a description of the filters, see Supplemental Material). Expression ratios (red/green ratios) of RNAi knockdown of each splicing factor versus no knockdown control were computed for each probe.
Biochemical experiments demonstrate that B52/SRp55 and PSI associate with, and presumably modulate splicing of, at least dozens and perhaps hundreds of distinct pre-mRNAs (Labourier et al. 2002; Kim et al. 2003). This is also likely the case for dASF/SF2 and hrp48. Therefore, reduction of any of these factors may impede or deregulate pre-mRNA processing severely and inconsistently, rendering the array data undecipherable or irreproducible. To address this possibility, we carried out multiple RNAi knockdown experiments for each splicing factor and compared the effect of experiments using simple hierarchical clustering (Eisen et al. 1998). Clustering experiments using data aggregated for each locus, each isoform, or each splice junction (see Materials and Methods) generated nodes specific for the dASF/SF2, B52/SRp55, and hrp48 experiments (Fig. 1C; data not shown). This indicates that the global expression patterns and splicing responses assayed on this array are largely distinct for the splicing factor that has been knocked down. The results of this high-level analysis support the notion that knockdown of each of these splicing factors results in a characteristic, interpretable, and reproducible splicing response. Interestingly, the PSI replicates were more variable in this analysis. This is likely due to the relatively small number of PSI-affected splicing events (see below). Despite this variability, PSI experimental replicates did identify several mRNAs that were previously found to be associated with PSI in an embryonic nuclear RNP fraction and whose expression was deregulated in PSI-mutant flies (Supplementary Table 1; Labourier et al. 2002).
RNAi knockdown of each splicing factor causes a general decrease in processed mRNA
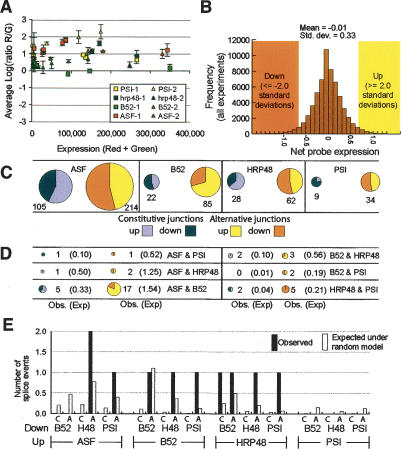
Five distinct, exogenous and heterologous, in vitro-transcribed, positive control RNAs were amplified and labeled along with the SL2 total cellular RNA to provide an independent assessment of any global changes in gene expression that would otherwise be masked by the normalization procedure. Following a standard assumption, we normalized the expression data from each array so that the average log expression ratio is 0; i.e., no net change in expression (see Materials and Methods). Interestingly, after normalization, the average log expression ratio of RNAi to control of nearly all positive control RNAs in each sample, across the range of expression intensities, was positive (Fig. 2A). Since identical amounts of the positive control RNAs were introduced in both the reference and experimental samples, these observations indicate an average reduction of the detectable, expressed genes on the array following knockdown of any of these four splicing factors. It is unclear to what extent this represents a biologically direct effect or a secondary consequence of target gene transcript reduction. Anecdotally, a previous screen identifying four B52/SRp55 target pre-mRNAs showed that the predominant effect of B52/SRp55 deletion was a reduction in target gene mRNA levels (Kim et al. 2003).
Figure 2.
Global microarray analysis. (A) Plot of total expression values versus log expression ratio of each positive control probe reveals a strong positive skew. Equal amounts of each positive control RNA were added to control and experimental samples. (B) The distribution of the net expression for each splice junction probe across all experiments is shown in the histogram. Cut-offs for up- or down-regulation were set at 2 standard deviations from unchanged. (C) Size of the sets of alternative and constitutive junctions that are strongly and consistently up- and down-regulated following knockdown of each splicing factor. dASF/SF2 affects the largest number of splicing events; PSI affects the smallest number. (D) Number of splicing events strongly and consistently affected by RNAi knockdown of each of two splicing factors. In parentheses are the expected sizes of each set assuming that each splicing factor affects in-dependent sets of splicing events. For each combination except B52 and PSI, the number of affected events is larger than expected by chance. (E) Antagonistically affected splice junctions. The numbers of splice events that were increased following knockdown of one splicing factor and decreased following knockdown of another are shown. Also shown is the number expected under the random model in which each splicing factor's affects are independent of the affects of the other splicing factors.
Each splicing factor affects a distinct set of splice junctions
Following data extraction and normalization, we computed log expression ratios for each probe in experimental versus control experiments. In order to determine which splicing events are affected by knockdown of which splicing factors, we calculated a value we call the net expression for each splicing junction (see Materials and Methods). The net expression for each splicing junction is the log expression ratio for that splice junction minus the average log expression ratio for all other probes on the isoforms containing that junction. This value indicates the increase or decrease in abundance of the specific splicing junction in question above and beyond any increase or decrease in abundance of the isoforms that contain the splice junction, and is designed to highlight individual splice junction changes by removing any differences in isoform expression or overall RNA levels. Because each splice junction is considered separately, this strategy should be minimally affected by incomplete data about which isoforms exist. The net expression value was calculated for all alternative and all constitutive junctions using the same formula. The distribution of the net expression values in all experiments (Fig. 2B) was used to generate cut-offs for classifying the affect of knockdown of each splicing factor on each splice junction. In order to find pre-mRNA splicing events that were strongly and consistently affected following knockdown of each splicing factor, net expression differences for each junction were compared in the biological replicates (see Materials and Methods; Supplementary Table 1).
The splicing events that were strongly (≥2 standard deviations) and consistently (in both replicates) affected by knockdown of each splicing factor were identified (Supplementary Table 1). The knockdown of dASF/SF2 affected the largest number of splicing events (319 events), while PSI affected the smallest number of splicing events (43 events) (Fig. 2C). This result is consistent with the notions that dASF/SF2 is a general regulator of alternative splicing, affecting a large number of targets, and that PSI is a more specialized regulator of alternative splicing. B52/SRp55 and hrp48 affected intermediate numbers of splice events (107 and 90 events, respectively). Interestingly, several of the splicing events detected for B52/SRp55 and for PSI were on genes previously found to interact with these splicing regulators (Supplementary Table 2).
The lists of splicing events consistently affected by each splicing factor were examined across experiments to determine which are significantly affected by knockdown of more than one of these splicing factors (Fig. 2D). The number of such splicing events was then compared with the number that would be expected by chance under a random model; i.e., assuming the splice junctions affected by one splicing factor are chosen independently of the splice junctions affected by any other splicing factor. The combination of dASF/SF2 and B52/SRp55 produced the most striking result: 22 splicing events were similarly affected by knockdown of either SR protein compared with 1.87 expected by chance (P-value ≤ 0.00001). This result is particularly interesting considering that, in vitro, SR proteins can complement one another for activity on several RNA targets (Fu et al. 1992; Zahler et al. 1992). Since dASF/SF2 and B52/SRp55 are the closest Drosophila SR protein paralogs (data not shown), this functional overlap is perhaps not surprising. However, the unique character of each SR protein is demonstrated by the fact that the majority of the splicing events strongly and consistently affected by RNAi of either dASF/SF2 or B52/SRp55 alone are not strongly or consistently affected by the other alone (22 shared targets out of 319 dASF/SF2 and 127 B52/SRp55 affected targets, respectively) (Fig. 2C,D).
PSI and hrp48 are known to coregulate alternative splicing of the P-element transposase pre-mRNA by binding to an exonic splicing silencer (Siebel et al. 1994, 1995). This analysis suggests their partnership may extend beyond P-element splicing. Of the 43 consistent and strong PSI targets, seven were found to also be strongly and consistently under the control of hrp48, whereas only 0.257 would be expected under the random model of independent effect (P-value ≤ 0.00017). Furthermore, for the 25 splice events that were consistently and strongly decreased following PSI knockdown, 21 were reduced following hrp48 knockdown by an average of -1.93 standard deviations (Supplementary Fig. 3). Similarly, for the 18 splice events that were consistently and strongly increased following PSI knockdown, 14 were increased following hrp48 knockdown by an average of 1.30 standard deviations (Supplementary Fig. 3). Therefore, nearly all of the splicing events under the control of PSI are similarly controlled by hrp48, suggesting that hrp48 may be an obligate partner for PSI. Interestingly, PSI does not appear to be an obligate partner for hrp48 as there are many hrp48 splicing events not similarly affected by PSI (Supplementary Fig. 3).
Traditionally, SR proteins and hnRNP proteins have been viewed as antagonistic partners, regulating in opposite directions, many of the same alternative splicing units (Smith and Valcarcel 2000; Black 2003). One of the best-studied models of antagonistic regulation is the HIV pre-mRNA in which binding of hnRNP proteins to cis-acting splicing silencer elements can be counter-acted by binding of SR proteins to nearby enhancer elements to regulate utilization of adjacent splice sites (Caputi et al. 1999; Jacquenet et al. 2001; Zhu et al. 2001; Caputi and Zahler 2002; Zahler et al. 2004). We analyzed antagonistic regulations by looking at splicing events that were increased following knockdown of one splicing factor and decreased following knockdown of a different splicing factor (Fig. 2E). Surprisingly, very few splicing events were found to be consistently and strongly regulated in an antagonistic relationship by any combination of these splicing factors. At a cut-off of 2.0 standard deviations, only dASF/SF2 and hrp48, which are the Drosophila homologs of the two canonical antagonistic splicing factors ASF/SF2 and hnRNP A1 (Mayeda and Krainer 1992; Cáceres et al. 1994; for review, see Black 2003), are found in more than a single antagonistic splicing event (Fig. 2E). At a more permissive cut-off, 1.5 standard deviations, more such antagonistic affects can be seen (Supplementary Fig. 4). These data indicate that antagonism between SR proteins and hnRNPs appears to be highly specific for both interacting partners (Supplementary Fig. 3) and somewhat uncommon.
RT-PCR validates the microarray results
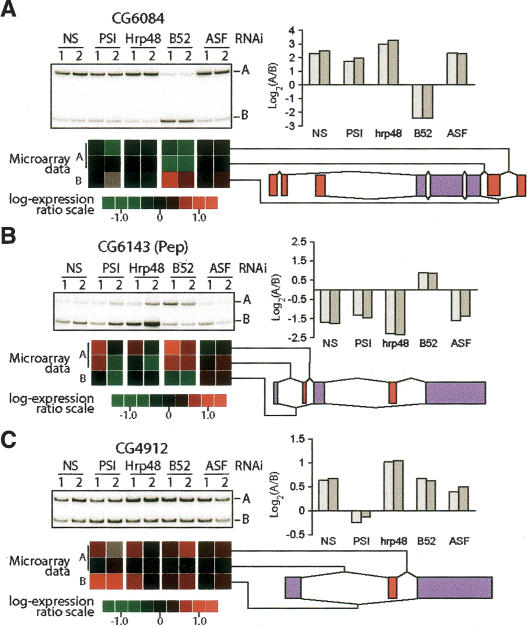
A validation of the microarray results was performed on several individual genes by RT-PCR designed to amplify the different affected mRNA isoforms. From the analysis described above, six genes were chosen on the basis that they were previously unknown targets of any of the four factors tested and their structures were amenable to RT-PCR analysis using a single pair of primers for each target (Fig. 3; Supplementary Fig. 2). All the selected targets confirmed the microarray results with differences in expressed isoforms ranging from less than twofold (PSI-specific target CG4912) (Fig. 3C) to 25-fold (B52/SRp55-specific target CG6084) (Fig. 3A).
Figure 3.
RT-PCR validation of selected targets. Shifts in alternatively spliced isoforms predicted from the microarray analysis were monitored by RT-PCR for three different targets using oligonucleotides flanking the affected alternative splice sites. (Bottom panel) Together with the RT-PCR gel analysis, the alternative spliced junction expression computed from the microarray data are shown. The densitometry of the gels are shown on the left expressed as a log2 ratio of the two measured isoforms. (A) CG6084 is a predicted target of the SR protein B52/SRp55. B52/SRp55 knockdown promoted skipping of the alternative cassette exon. (B) CG6143 (PEP) is a predicted B52/SRp55 target whose cassette exon is included upon knockdown of B52/SRp55. (C) CG4912 is a predicted target of the hnRNP protein PSI. The knockdown of PSI promotes skipping of the alternative cassette exon.
B52-binding sites are overrepresented around the 5′ splice site of the B52-affected splicing junctions
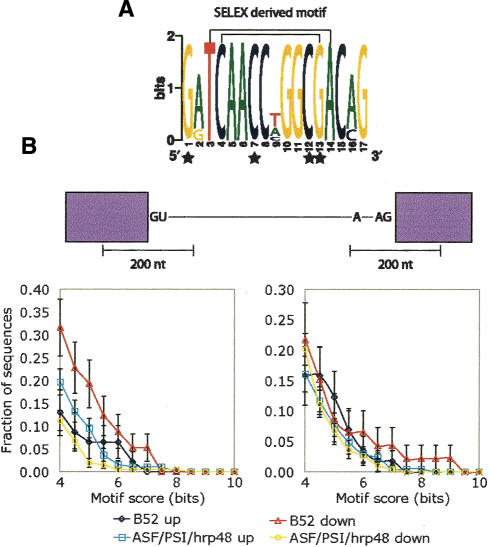
Any observed change in pre-mRNA splicing in the experiments may be due to a direct interaction between the splicing factor and pre-mRNA in question or to an indirect effect. One feature of direct targets for each splicing factor may be the presence of a cis-acting element within the pre-mRNA near the affected splice site. The assembled lists of splicing events specifically affected by reduction of each of these four splicing factors were used to identify potential binding elements for these factors. While many SELEX and other biochemical studies have been conducted to characterize the RNA binding preferences for splicing factors, the results have been limited in their ability to predict in vivo splicing affects on specific targets. However, one particularly informative study identified a sequence that likely forms a stem-loop structure that binds Drosophila B52/SRp55 tightly in vivo (Shi et al. 1997). A position weight matrix model of this sequence was used to search for similar sequences in a database composed of sequences around either 3′ or 5′ splice sites that were either reduced or increased specifically upon B52/SRp55 knockdown (Fig. 4; see Materials and Methods). For comparison, a search was performed using a database of 3′ or 5′ splice site regions found to be affected by RNAi against any of the other three (non-B52/SRp55) splicing factors, but not affected by B52 knockdown. The fraction of 3′ splice site regions that contain two strong matches to this sequence motif is similar to the fraction found in the comparison database search (Fig. 4B). However, the regions around the 5′ splice site in the B52/SRp55 knockdown-reduced junctions were specifically enriched for pairs of this motif compared with the corresponding non-B52/SRp55 set (Fig. 4B). A similar search using the SELEX-defined motif recognized by the human ASF/SF2 homolog (Tacke and Manley 1995) also shows an overrepresentation near the 5′and 3′ splice site regions regulated by the Drosophila ASF/SF2 homolog in our experiments (Supplementary Fig. 5). Although suggestive, this analysis has the caveat of being performed using human-derived SELEX sites. Although the human and Drosophila orthologs are very similar (62% identical with 10% additional similarity; data not shown), their RNA binding specificities may have diverged. Similar analyses using known binding site motifs for hrp48 and PSI failed to show enrichment around the affected splice junctions (data not shown). However, these negative results may be due to imprecise or inaccurate definition of the PSI- and hrp48-binding site motifs: Both proteins bind very degenerate sites derived from very limited binding data (Supplementary Table 3).
Figure 4.
B52/SRp55-binding motifs near B52/SRp55 uniquely affected splice junctions. (A) Sequence logo of the previously identified B52/SRp55-binding motif (Shi et al. 1997). Lines connecting residues indicate predicted base-pairing interactions. Stars underneath residues indicate B52/SRp55 footprint contacts (Shi et al. 1997). (B) Pairs of motifs similar to the previously identified B52/SRp55-binding motif (Shi et al. 1997) are overrepresented around 5′ splice sites that are down-regulated when B52/SRp55 is knocked-down relative to the 5′ splice sites affected in the other experiments. No significant difference is seen in 5′ splice site regions around up-regulated junctions or in either up- or down-regulated 3′ splice site regions. Error bars are determined analytically using the binomial distribution and correspond to 1 standard deviation: sqrt[np(1 - p)]/n, where n is the number of sequences searched and p is the observed probability of having sites of given score.
Discussion
This study represents the first genome-wide identification of alternative splicing events modulated by the four splicing factors, dASF/SF2, B52/SRp55, hrp48, and PSI. Traditionally, thorough genetic or in vitro biochemical analyses have been required to identify splicing events controlled by specific splicing factors (Black 2003). This difficulty accounts for the paucity of well-defined systems for studying alternative splicing. Although genome-wide analyses involve substantial risk of false positives and false negatives, this new splice junction platform provides the ability to rapidly identify many splicing events regulated by individual splicing factors and provides the basis for more focused searches for corresponding RNA regulatory motifs.
Analysis of the array results allows us to characterize the extent to which alternative splicing events require multiple splicing factors. The significant overlap between dASF/SF2 targets and B52/SRp55 targets reinforces earlier biochemical characterizations that indicate partial functional overlap between these factors (Fu et al. 1992; Zahler et al. 1992). However, as many splicing events were found to be uniquely affected by either factor individually, these data also demonstrate their discrete characteristics. It remains to be determined whether the observed functional overlap is due to similar RNA binding specificities, the presence of unique binding sites for dASF/SF2 and B52/SRp55 in target pre-mRNAs, or other properties of these factors such as interaction with a common binding partner already present on target pre-mRNAs.
hrp48 and PSI were also found to regulate many of the same splicing events. As nearly every identified target of PSI was similarly affected by hrp48 knockdown, these data suggest that hrp48 may be an obligate partner for PSI action. However, since many of the hrp48-affected splicing events were not similarly affected by PSI knockdown, the reverse does not appear to be the case; i.e., hrp48 does not appear to require PSI to regulate splicing.
Several antagonistic relations were also defined in this analysis, especially between dASF/SF2 and the hnRNPA1 homolog, hrp48. However, the absence of an overall negative correlation between dASF/SF2 knockdown and hrp48 knockdown supports a model in which their antagonism is mediated through cis elements present in target pre-mRNAs rather than through direct interaction between these proteins. That is, dASF/SF2 and hrp48 appear to be antagonistic only for a subset of splice sites that bind both factors and apparently most target pre-mRNAs of both do not.
While computational searches for the high-affinity B52/SRp55 SELEX motif (Shi et al. 1997) showed an enrichment near some of the affected junctions on the microarray, searches for the Drosophila PSI SELEX motif (Amarasinghe et al. 2001), which can be found in a very large fraction of pre-mRNAs (data not shown), showed no enrichment near the 43 PSI-affected splice junctions. This observation is reminiscent of the well-known splicing factor Sex-lethal (Sxl). whose binding site can be found in all known pre-mRNAs (R. Singh, pers. comm.) but controls alternative splicing of only three known pre-mRNAs (Maniatis and Tasic 2002). Our results suggest that the mere presence of strong SELEX-defined RNA-binding sites is generally not sufficient to predict regulation of nearby splice sites in a physiological setting. Functional SELEX has been performed in vitro and in vivo to identify both splicing enhancers and silencers (Coulter et al. 1997; Liu et al. 1998; Wang et al. 2004). The identified motifs were very short and presumably regulated by binding of a single protein, arguing that single factors can control individual alternative splicing events. However, it is also known that several specific regulated splice sites require the formation of large, multiprotein complexes compatible with the requirement for a higher order of complexity, rather than a single RNA-protein interaction (Siebel et al. 1992; Lynch and Maniatis 1996). As has been the case for transcriptional regulation via DNA-binding proteins, a combination of genome-wide methods to identify target genes together with bioinformatics searches using protein-binding site information, may prove to be the only way to validate in vivo the activity of putative cis-acting pre-mRNA elements controlled by specific regulatory proteins.
Current evidence indicates that the number of alternative splice junctions in Drosophila is at least 10,000 (Celniker and Rubin 2003) and may be as high as 40,000 (Stolc et al. 2004). Based on the present study using an arbitrary cut-off of 2 standard deviations, each splicing factor regulates a few hundred (43 for PSI to 319 for dASF/SF2) alternative splicing events in a given cell type. Since there are ∼200 putative splicing factors in the Drosophila genome (Celniker and Rubin 2003; Jurica and Moore 2003), the observed range of splicing junctions regulated by individual splicing factors is within the expected order of magnitude to account for the level of splicing complexity of the fly transcriptome. These splicing microarray experiments demonstrate on a genomic scale the unique character of each of these four splicing factors and give a first glimpse into the network of interactions regulating alternative splicing. Similar experiments using this technology should lead to an understanding of how the genes involved in RNA processing interact to regulate the tens of thousands of alternative splicing events in metazoans (Brett et al. 2002).
Materials and methods
Drosophila melanogaster alternative splicing array design
All transcript sequences from the GadFly 3.2 Drosophila melanogaster genome annotation (dmel_all_transcript_r3.2.0.fasta) (Celniker and Rubin 2003) were mapped to the masked genome sequence (whole_genome_masked_genomic_dmel_RELEASE3-1.FASTA) using Spidey version 1.40 (Wheelan et al. 2001). Transcripts that overlapped on the same strand were clustered into loci. Only loci with multiple, unique transcripts (alternatively spliced loci) were considered further. For each of these loci, each splice junction was labeled “constitutive” if it was found in all transcripts from that locus and “alternative” otherwise. Note that some alternative isoforms contain only alternative transcriptional initiation or termination regions and, therefore, may not be alternatively spliced in the strict sense. For each locus, junction probes were selected that are complementary to the 18 exon nucleotides on each side of the junction. Junction probes were selected for each alternative junction and up to two constitutive junctions per locus. Two exon probes for each locus were selected such that they avoid splice junctions and have at least three mismatches when compared with any other transcribed Drosophila sequence. Five unique probes from each of five exogenous genes were included as positive controls and the same number as negative controls. All probe features were included on two locations on the array for consistency checking.
RNAi, RNA extraction, RNA labeling, and array hybridization
Production of dsRNA and RNAi was as described (Clemens et al. 2000). RNAi were done for 4 d by incubating 10 μg of the different dsRNA with 0.5 × 106 serum-free adapted Schneider SL2 cells (Invitrogen) with addition of 10 μg of dsRNA after 48 h. At day 4, 10% of the cells were recovered and lysed in protein gel loading buffer, while the remaining cells were used for total RNA extraction using the Qiagen mRNA Easy purification kit with on column DNAse digestion following the manufacturer's protocol. In vitro transcribed RNAs of the human U17, U19, and 7Sk small RNA, as well as the human and tetrahymena telomerase RNA (300, 100, 10, 2.5, and 25 fmol each, respectively), were added as internal quality and sensitivity control to 10 μg of total RNA and were amplified, labeled, and hybridized as described (Castle et al. 2003) on a 44-k custom Agilent oligonucleotide array.
Array analysis
After hybridization, arrays were scanned and images analyzed following the manufacturer's recommendation (Agilent). Linear-LOWESS dye normalization was performed using all probes except for negative controls. The Pearson correlation coefficient was computed for all probe expression values using the two instances of each probe on the array as the x and y values and found to be >0.97 for all experiments. One B52/SRp55 experiment was removed from the analysis on the basis of a nonconsistent effect in the red channel (Supplementary Fig. 1A). Array data from each experiment were then analyzed to determine the extent of specific knockdown of each target. Experiments that failed to yield a strongly negative log expression ratio for probes of each RNAi target (log-ratio ≤ -0.2) or that also yielded a strongly negative log expression ratio for probes of a different RNAi target were discarded (Supplementary Fig. 1B). This removed the first PSI experiment, the second hrp48 experiment, and the first ASF/SF2 experiment. Remaining for further analysis were two experiments of each splice factor target. Since each probe occurs twice on the array, a single expression average was computed. Any probe whose expression at one position in either red or green channels was >150% of its expression in that channel at its other position was removed from the analysis. Hierarchical clustering of experiments using average linkage clustering was performed. A single average expression value for each locus, each isoform, or each junction was generated. Locus and isoform averages were computed by taking the average log expression ratio of all probes from each locus or from each isoform.
To separate gene-level or isoform-level expression changes from splicing changes, the average log expression ratio of all junctions for each isoform was subtracted from the expression ratio of each junction. This value, which we call the net expression of each junction, is given by the formula log expression ratio (x) - average of log expression ratios (y), where x is the splice probe in question and y is the set of all probes that are on all isoforms that contain junction x. This approach deemphasizes changes in splicing events that are correlated with other splicing events of the same transcript. However, it makes minimal assumptions about the set of isoforms present for a given locus. Therefore, it is unharmed by missing transcript isoform models since each splicing event is considered individually. The distribution of the net expression was found for each experiment and for all experiments and was found to be similar (data not shown). Therefore, we used the data compiled for all experiments to generate statistical cut-offs.
For each splicing junction probe, the net expression was compared between biological replicates of each splice factor knockdown experiment. Bona fide targets of each splicing factor are expected to be consistently affected in biological replicates whereas noise is expected to vary among replicates. Therefore, splicing junction net expression values were used to filter for those within 2 standard deviations of the net-expression value of each other. We compiled lists of splicing events that were strongly (net expression value deviated >2 standard deviations from 0) and consistently (in both biological replicates) affected by knockdown of each splicing factor (Supplementary Table 1). Comparisons of each pair of lists were used to determine overlap in splicing factor targets. The statistical significance was determined using the hypergeometric distribution with Bonferonni correction for three observations on each splicing factor. These results are given in Supplementary Table 4.
RT-PCR
For each experiment, 5 μg of RNA was extracted and primed with a dT16 oligonucleotide and reverse-transcribed using SuperScript II (Invitrogen) following the manufacturer's protocol. Amplification was performed following standard conditions for 21 cycles using one-twentieth of the cDNA reaction in the presence of 5 μCi of α32P-dCTP (3000 Ci/mmol). Oligonucleotide sequences can be obtained upon request. RT-PCR products were fractionated on a 6% acrylamide-bisacrylamide gel run in 1× TBE buffer. All gels were dried, exposed, and scanned on a Typhoon phosphorimager (Amersham-Pharmacia). Densitometry of unsaturated exposure was performed using ImageQuant (Amersham-Pharmacia).
Sequence motif search
Position weight matrix motifs of ASF/SF2-, B52/SRp55-, PSI-, and hrp48-binding sites were generated using previously published binding data from a variety of sources (Supplementary Table 3). Databases of regions 100 nucleotides upstream and downstream of 5′ and 3′ splice sites were constructed using the splice junctions found to be uniquely affected for each splicing factor knockdown. The comparison databases for each factor were composed of the sequences uniquely affected by any of the three other splicing factors not under examination. The motif was used to scan each position on each sequence in the database, and high-scoring positions were counted and shown in Figure 4.
Acknowledgments
We thank G. Crooks, M. Levine, and M. Eisen for helpful comments, as well as B. Hogg, D. Fu, C. O'Connor, and D. Cunningham for providing constructs for the in vitro transcribed control RNAs. This work was supported by a U.S. National Institutes of Health grant (R01GM61987). M.B. was the recipient of a Human Frontier Science Program long-term fellowship.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1314205.
References
- Amarasinghe A.K., MacDiarmid, R., Adams, M.D., and Rio, D.C. 2001. An in vitro-selected RNA-binding site for the KH domain protein PSI acts as a splicing inhibitor element. RNA 7 1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72 291-336 [DOI] [PubMed] [Google Scholar]
- Brett D., Pospisil, H., Valcarcel, J., Reich, J., and Bork, P. 2002. Alternative splicing and genome complexity. Nat. Genet. 30 29-30. [DOI] [PubMed] [Google Scholar]
- Burckin T., Nagel, R., Mandel-Gutfreund, Y., Shiue, L., Clark, T.A., Chong, J.L., Chang, T.H., Squazzo, S., Hartzog, G., and Ares Jr., M. 2005. Exploring functional relationships between components of the gene expression machinery. Nat. Struct. Mol. Biol. 12 175-182. [DOI] [PubMed] [Google Scholar]
- Burnette J.M., Hatton, A.R., and Lopez, A.J. 1999. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics 151 1517-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres J.F., Stamm, S., Helfman, D.M., and Krainer, A.R. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265 1706-1709. [DOI] [PubMed] [Google Scholar]
- Caputi M. and Zahler, A.M. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21 845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Mayeda, A., Krainer, A.R., and Zahler, A.M. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18 4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle J., Garrett-Engele, P., Armour, C.D., Duenwald, S.J., Loerch, P.M., Meyer, M.R., Schadt, E.E., Stoughton, R., Parrish, M.L., Shoemaker, D.D., et al. 2003. Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biol. 4 R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S.E. and Rubin, G.M. 2003. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 4 89-117. [DOI] [PubMed] [Google Scholar]
- Clark T.A., Sugnet, C.W., and Ares Jr., M. 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296 907-910. [DOI] [PubMed] [Google Scholar]
- Clemens J.C., Worby, C.A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B.A., and Dixon, J.E. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. 97 6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter L.R., Landree, M.A., and Cooper, T.A. 1997. Identification of a new class of exonic splicing enhancers by in vivo selection Mol. Cell Biol. 17 2143-2150. [ [DOI] [PMC free article] [PubMed] [Google Scholar]; Published erratum appears in Mol. Cell Biol. 1997. 17 3468]. [Google Scholar]
- Eisen M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D., Mayeda, A., Maniatis, T., and Krainer, A.R. 1992. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl. Acad. Sci. 89 11224-11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G.K., Madore, S.J., Moldover, B., Jatkoe, T., Balaban, D., Thomas, J., and Wang, Y. 2001. Predicting splice variant from DNA chip expression data. Genome Res. 11 1237-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquenet S., Mereau, A., Bilodeau, P.S., Damier, L., Stoltzfus, C.M., and Branlant, C. 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 276 40464-40475. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., and Shoemaker, D.D. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302 2141-2144. [DOI] [PubMed] [Google Scholar]
- Jurica M.S. and Moore, M.J. 2003. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12 5-14. [DOI] [PubMed] [Google Scholar]
- Kim S., Shi, H., Lee, D.K., and Lis, J.T. 2003. Specific SR protein-dependent splicing substrates identified through genomic SELEX. Nucleic Acids Res. 31 1955-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labourier E., Blanchette, M., Feiger, J.W., Adams, M.D., and Rio, D.C. 2002. The KH-type RNA-binding protein PSI is required for Drosophila viability, male fertility, and cellular mRNA processing. Genes & Dev. 16 72-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. and Roy, M. 2004. Analysis of alternative splicing with microarrays: Successes and challenges. Genome Biol. 5 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.X., Zhang, M., and Krainer, A.R. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes & Dev. 12 1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Johnstone, I.L., and Caceres, J.F. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19 1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K.W. and Maniatis, T. 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes & Dev. 10 2089-2101. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418 236-243. [DOI] [PubMed] [Google Scholar]
- Mayeda A. and Krainer, A.R. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68 365-375. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Query, C.C., and Sharp, P.A. 1993. Splicing of precursors to messenger RNAs by the spliceosome. In The RNA world (eds. R.F. Gesteland and J.F. Atkins), pp. 303-357. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Pan Q., Shai, O., Misquitta, C., Zhang, W., Saltzman, A.L., Mohammad, N., Babak, T., Siu, H., Hughes, T.R., Morris, Q.D., et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 16 929-941. [DOI] [PubMed] [Google Scholar]
- Relogio A., Ben-Dov, C., Baum, M., Ruggiu, M., Gemund, C., Benes, V., Darnell, R.B., and Valcarcel, J. 2005. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin lymphoma cells. J. Biol. Chem. 280 4779-4784. [DOI] [PubMed] [Google Scholar]
- Shi H., Hoffman, B.E., and Lis, J.T. 1997. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 17 2649-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C.W., Fresco, L.D., and Rio, D.C. 1992. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: Multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes & Dev. 6 1386-1401. [DOI] [PubMed] [Google Scholar]
- Siebel C.W., Kanaar, R., and Rio, D.C. 1994. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes & Dev. 8 1713-1725. [DOI] [PubMed] [Google Scholar]
- Siebel C.W., Admon, A., and Rio, D.C. 1995. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes & Dev. 9 269-283. [DOI] [PubMed] [Google Scholar]
- Smith C.W. and Valcarcel, J. 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25 381-388. [DOI] [PubMed] [Google Scholar]
- Stolc V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M.F., Rifkin, S.A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P.E., et al. 2004. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science 306 655-660. [DOI] [PubMed] [Google Scholar]
- Tacke R. and Manley, J.L. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14 3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Takagaki, Y., and Manley, J.L. 1996. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 10 2588-2599. [DOI] [PubMed] [Google Scholar]
- Wang J., Xiao, S.H., and Manley, J.L. 1998. Genetic analysis of the SR protein ASF/SF2: Interchangeability of RS domains and negative control of splicing. Genes & Dev. 12 2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hubbell, E., Hu, J.S., Mei, G., Cline, M., Lu, G., Clark, T., Siani-Rose, M.A., Ares, M., Kulp, D.C., et al. 2003. Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics 19 (Suppl. 1): i315-i322. [DOI] [PubMed] [Google Scholar]
- Wang Z., Rolish, M.E., Yeo, G., Tung, V., Mawson, M., and Burge, C.B. 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119 831-845. [DOI] [PubMed] [Google Scholar]
- Wheelan S.J., Church, D.M., and Ostell, J.M. 2001. Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 11 1952-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A.M., Lane, W.S., Stolk, J.A., and Roth, M.B. 1992. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 6 837-847. [DOI] [PubMed] [Google Scholar]
- Zahler A.M., Damgaard, C.K., Kjems, J., and Caputi, M. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 279 10077-10084. [DOI] [PubMed] [Google Scholar]
- Zhu J., Mayeda, A., and Krainer, A.R. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8 1351-1361. [DOI] [PubMed] [Google Scholar]