Abstract
TrkA, the high affinity receptor for nerve growth factor (NGF), is essential for the development of nociceptive sensory and sympathetic neurons. The zinc finger transcription factor Klf7 interacts with an important cis element of the TrkA minimal enhancer and is coexpressed with TrkA in these neurons. We show that Klf7 binds to the endogenous TrkA minimal enhancer and can activate transcription from the TrkA minimal enhancer in a sequence-dependent manner. In Klf7-/- newborn mice, we find a significant reduction in sensory neurons due to increased apoptosis. The neuronal loss is restricted to nociceptive neurons that normally depend on TrkA for neurotrophic support, while other populations of somatosensory neurons appear normal. The reduction of TrkA expression in sensory neurons is a direct effect of Klf7 gene ablation, rather than a secondary effect of cell death. As a result, Klf7-/- mice have deficient response to noxious stimuli. Finally, removal of one TrkA allele exacerbates the loss of TrkA(+) neurons in Klf7-/- mice. Thus, Klf7 specifically regulates TrkA gene expression and is required for the development of a subset of nociceptive sensory neurons.
Keywords: Klf7, TrkA, nociception, transcriptional regulation, sensory neuron, neurotrophin
The vertebrate somatosensory system carries information from external and internal receptors to the central nervous system (CNS), where the information is processed to guide movement. The sensory pathways can be divided into exteroceptive and proprioceptive: The former mediates sensation of touch, temperature, and pain, while the latter conveys positional information. Vertebrate sensory neurons are located in dorsal root ganglia (DRG) and cranial ganglia. While different cranial ganglia receive contributions from the neural crest, neural placodes, or both, the DRG is entirely derived from the neural crest (Serbedzija et al. 1992, 1994).
In the peripheral nervous system (PNS), the NGF family of neurotrophins and their Trk receptors regulate cell survival, proliferation, the fate of neural precursors, and axon and dendrite growth and patterning (Bibel and Barde 2000; Huang and Reichardt 2001, 2003). Among the Trk receptors, TrkA expression is required for nociceptive sensory and sympathetic neurons in the PNS (Martin-Zanca et al. 1990; Holtzman et al. 1992; Smeyne et al. 1994; Lei et al. 2001). TrkB is expressed broadly in both the PNS and CNS and is required for a subset of mechanoreceptive neurons (Klein et al. 1989, 1990, 1993). TrkC is also expressed broadly in both the PNS and CNS and is required for proprioceptive sensory neurons (Tessarollo et al. 1993; Klein et al. 1994; Liebl et al. 1997).
To gain understanding of TrkA gene regulation, we previously identified a minimal enhancer that drives the expression of a β-galactosidase transgene specifically in TrkA(+) sensory and sympathetic neurons in vivo (Ma et al. 2000). A detailed site-directed mutagenesis study revealed that nine highly conserved cis elements within the 457-bp minimal enhancer are important for the enhancer function. One of these cis elements is a canonical binding site for the zinc finger transcription factor Ikaros, which is expressed in the immune system (Georgopoulos et al. 1992, 1997).
Expression cloning from a mouse embryonic DRG cDNA library identified the Kruppel-like zinc finger transcription factor Klf7 as able to interact with the Ikaros site of the TrkA minimal enhancer (Lei et al. 2001). Klf7 is coexpressed with TrkA in sensory and sympathetic neurons in the PNS. Consistent with Klf7 being a regulator of TrkA gene expression in these neurons, the onset of Klf7 expression is prior to that of TrkA expression (Lei et al. 2001).
In the present work we investigate the physiological relevance of our preceding study on the regulation of the TrkA enhancer by Klf7. Our data demonstrate an in vivo role for the regulation of the endogenous TrkA gene expression and sensory neuron survival by Klf7.
Results
Klf7 activates the TrkA minimal enhancer in a sequence-dependent manner
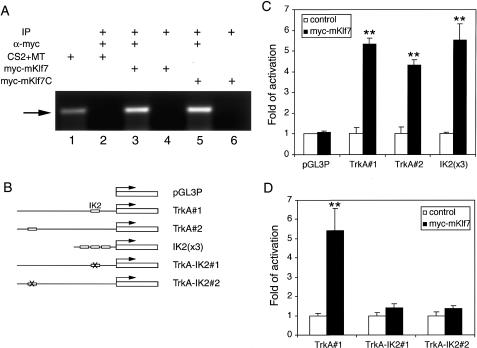
Using an in vitro gel mobility shift assay, we previously showed that the C-terminal DNA-binding domain of Klf7 binds to a site within the TrkA minimal enhancer that is required for in vivo enhancer activity (Lei et al. 2001). To determine whether this interaction was sustained under more physiological conditions, we probed whether Klf7 can bind to the endogenous TrkA minimal enhancer using chromatin immunoprecipitation (ChIP) assays (Fig. 1A). The results of the ChIP analysis showed that the myc-epitope-tagged mKlf7 (myc-mKlf7) binds specifically to the DNA fragment containing nucleotides +252-424 of the TrkA minimal enhancer in cell culture (Fig. 1A, lanes 3,4). This fragment of the TrkA minimal enhancer contains the Klf7-binding site (Ma et al. 2000; Lei et al. 2001). myc-mKlf7C, containing the C-terminal DNA-binding domain (amino acids 202-301) of Klf7, also binds specifically to the TrkA minimal enhancer (Fig. 1A, lanes 5,6).
Figure 1.
Klf7 specifically activates transcription from the TrkA minimal enhancer by binding its cognate site IK2. (A) ChIP analysis. Lane 1 is from empty vector (CS2 + MT) transfected cells without immunoprecipitation. Lane 2 is from CS2 + MT transfected cells after immunoprecipitation with an anti-Myc antibody. Lanes 3 and 4 are from myc-mKlf7 transfected cells following immunoprecipitation with (lane 3) and without (lane 4) an anti-Myc antibody. Lanes 5 and 6 are from myc-mKlf7C transfected cells following immunoprecipitation with (lane 5) and without (lane 6) an anti-Myc antibody. After ChIP, PCR was carried out using primers specific for the TrkA minimal enhancer to detect the precipitated DNA fragments. The arrow indicates the 173-bp PCR product. (B) Reporter constructs used in the luciferase assays. pGL3P is the empty vector pGL3-Promoter containing the firefly luciferase coding region under the control of a SV40 minimal promoter (Promega). TrkA#1 and TrkA#2 contain the wild-type TrkA minimal enhancer upstream of the SV40 minimal promoter in opposite orientations. TrkA-IK#1 and TrkA-IK#2 contain the TrkA minimal enhancer with a mutated IK2 core sequence, in opposite orientations. IK2(x3) contains three copies of the wild-type IK2 site in tandem. (C) Klf7 activates transcription from reporter constructs containing either the TrkA minimal enhancer or just its cognate site IK2 in PC12 cells. (D) Klf7 does not activate transcription from reporter constructs containing the TrkA minimal enhancer with a mutated IK2 site in PC12 cells. In both C and D, the empty vector CS2 + MT was used as a negative control. (★★) p < 0.01, t-test.
We also tested the ability of Klf7 to activate the TrkA minimal enhancer in PC12 cells. Reporter constructs containing the wild-type TrkA minimal enhancer, the TrkA minimal enhancer with a mutated Klf7-binding site, or three copies of the Klf7-binding site, were generated and analyzed in dual-luciferase assays (Fig. 1B). As shown in Figure 1C, Klf7 activates transcription from the reporters containing the full-length TrkA minimal enhancer in either orientation. Klf7 also activated transcription from the reporter containing only three copies of Klf7-binding sites, demonstrating that the interaction of Klf7 and its cognate site is sufficient for transcriptional activation by Klf7. In contrast, mutation of the Klf7-binding site abolished the response of the TrkA minimal enhancer to Klf7 (Fig. 1D). Taken together, these results indicate that the Klf7-binding site is necessary and sufficient for Klf7 to activate the TrkA minimal enhancer in cell culture. These data are in agreement with previous experiments showing that a TrkA minimal enhancer harboring the same cis-element mutation had limited capacity to drive expression of a β-galactosidase transgene in vivo (Ma et al. 2000). Therefore, Klf7 binds a physiologically important cis-regulatory element of the TrkA minimal enhancer and activates this enhancer in a sequence-dependent manner.
Decreased TrkA expression in Klf7-/- sensory neurons
To examine whether Klf7 is required for TrkA expression in vivo, we analyzed Klf7-null mice (Laub et al., 2005). Klf7+/+, Klf7+/-, and Klf7-/- mice are born in Mendelian ratio from Klf7+/- intercrosses, and >97% of Klf7-/- mice die within 2 d after birth. The Klf7-null pups have little or no milk in their stomachs, suggesting that starvation may be the cause of death.
We examined TrkA mRNA and protein expression throughout embryonic development in Klf7-/- DRG neurons. As shown in Figure 2A and B, at embryonic day 12.5 (E12.5), TrkA expression is significantly reduced in the mutant DRG. One day later (E13.5), at the onset of NGF/TrkA dependence, the mutant DRG exhibit fewer TrkA(+) neurons than wild-type DRG (Fig. 2C-F), and this difference is maintained through post-natal day 0 (P0) (Fig. 2G,H). We also examined TrkB and TrkC mRNA expression in P0 DRG by in situ hybridization and found no obvious difference in their expression between wild-type and Klf7-/- DRG (see Supplementary Fig. S1A-D). Ret, a receptor tyrosine kinase expressed in a subset of TrkA(-) DRG neurons, was also unaffected in Klf7-/- mice (see Supplementary Fig. S1E,F). Real-time RT-PCR analysis of TrkA, TrkB, and TrkC at E11.5, E12.5, and E13.5 provided consistent results in which only TrkA levels exhibit a significant reduction in Klf7-null mutants (Fig. 2I). Therefore, among the different populations of sensory neurons, only TrkA(+) neurons are affected in Klf7-/- mice.
Figure 2.
Reduced expression of TrkA in Klf7-/- DRG. (A-F) Protein expression of TrkA in E12.5 DRG (A,B) and E13.5 DRG (C-F) of wild-type (A,C,E) and mutant (B,D,F) embryos assayed by anti-TrkA immunohistochemistry. (G,H) In situ hybridization of wild-type (G) and mutant (H) P0 DRG using a TrkA antisense RNA probe. (I) Real-time RT-PCR analysis of TrkA, TrkB, and TrkC at E11.5, E12.5, and E13.5. The RNA levels of TrkA, TrkB, and TrkC were normalized against the level of β-actin. The numbers represent the ratios of the relative RNA level of the mutant embryos to that of the wild-type embryos. N = 3 for each embryonic stage and genotype. Bars: A-D,G,H, 100 μm; E,F, 200 μm.
Proliferation and apoptosis in Klf7-/- DRG
The size and cellularity of lumbar mutant DRG appear normal at E12.5 (see Supplementary Fig. S2A,B). However, by E13.5, the mutant DRG appear smaller than wild-type counterparts (see Supplementary Fig. S2C,D). BrdU incorporation was examined in E12.5 DRG, and no detectable differences were found between the wild-type and mutant DRG (see Supplementary Fig. S2E-G) (p = 0.716). Thus, the reduced size of the mutant DRG is not caused by proliferation defects in Klf7-/- neuronal precursor cells.
For murine DRG neurons, the period of neurotrophin dependence and developmental cell death spans the gestational ages E13.5 to E17.5 (White et al. 1996, 1998). As shown in Figure 3, TUNEL assays demonstrate a comparable level of apoptosis in E12.5 DRG of the wild-type and mutant embryos (Fig. 3A,B,G). However, at E13.5, there is a threefold increase in TUNEL(+) cells in Klf7-/- DRG compared with wild-type DRG (Fig. 3C,D,G). Therefore, the reduction in DRG neurons seen at E13.5 appears to be the consequence of increased apoptosis in Klf7-/- DRG. TUNEL and TrkA double-labeling experiments demonstrate that the increase in apoptosis occurs in cells in which we cannot detect TrkA (Fig. 3E,F). We counted equal numbers of lumbar DRG sections (n = 10) of wild-type or Klf7-/- embryos for TUNEL(+) and TrkA(+) cells and found that 95% (304/320) of TUNEL(+) cells in Klf7-/- DRG are TrkA(-), while 46% (41/90) of TUNEL(+) cells in wild-type DRG are TrkA(-). This demonstrates that low levels of TrkA expression correlate with TUNEL reactivity in Klf7-/- DRG. Therefore, a subset of Klf7-/- DRG neurons loses TrkA expression and undergoes apoptosis.
Figure 3.
Increased apoptosis in Klf7-/- DRG at E13.5. (A,B) DRG sections of E12.5 wild-type (A) and mutant (B) embryos stained with the TUNEL method for the detection of cells undergoing apoptosis. (C,D) DRG sections of E13.5 wild-type (C) and mutant (D) embryos stained with the TUNEL method for the detection of cells undergoing apoptosis. (E,F) DRG sections of E13.5 wild-type (E) and mutant (F) embryos double-labeled with TUNEL (green) and anti-TrkA antibodies (red). (G) Quantification of TUNEL(+) cells showing comparable numbers of apoptotic cells at E12.5 between the wild-type and mutant DRG but a marked increase of apoptotic cells in mutant DRG at E13.5. Shown are the average numbers of TUNEL(+) cells per section from the wild-type (N = 3) and mutant (N = 3) embryos. Twenty sections of lumbar DRG of each embryo were used for counting TUNEL(+) cells. E12.5, p = 0.192; E13.5, p = 2.2 × 10-9, t-test. Bars: A-D, 50 μm; E,F, 35 μm.
Loss of NGF-dependent DRG neurons in culture
To examine directly the consequence of Klf7 loss on the function of Trk receptors, we cultured E13.5 DRG neurons from Klf7-/- and wild-type embryos in the presence of 10 ng/mL NGF, BDNF, or NT3. These culture conditions enrich for TrkA(+), TrkB(+), or TrkC(+) neurons, respectively (Klesse and Parada 1998). As shown in Figure 4A, Klf7-/- and wild-type neurons survive to the same extent in the presence of BDNF or NT-3, while Klf7-/- neurons exhibit progressive cell loss in the presence of NGF. These data indicate that the increased apoptotic neurons in Klf7-/- embryos include TrkA(+) neurons. We next cultured wild-type E13.5 DRG neurons in the presence of NGF, BDNF, or NT3 and infected the neurons with recombinant adenoviruses expressing the full-length Klf7 or the C-terminal DNA-binding domain of Klf7 (Fig. 4B). The C-terminal DNA-binding domain of Klf7 can inhibit the ability of the full-length Klf7 to activate transcription of target genes through competitive binding of the DNA cis element and therefore functions as a dominant-negative form of Klf7 (DN-Klf7) (data not shown). To control against effects induced by the recombinant adenovirus system, we also infected neurons with adenoviruses expressing Xraf, an inactive and inert form of raf (Klesse and Parada 1998; Klesse et al. 1999). As shown in Figure 4C, DN-Klf7 inhibits the survival of NGF-dependent neurons in a dose-dependent manner, while it has no effect on BDNF or NT3-dependent neurons. Therefore, the survival of cultured sensory neurons mediated by NGF/TrkA depends on Klf7 expression and function.
Figure 4.
A significant loss of NGF-dependent primary DRG neurons in culture. (A) Primary DRG neurons were isolated from E13.5 wild-type or Klf7-/- embryos and cultured in the presence of 10 ng/mL NGF, BDNF, or NT3. The percentages of neuronal survival after 48 and 72 h in culture are shown. N = 8 for each genotype. (★★) p < 0.01, t-test. (B) Diagram showing the DNA constructs of the full-length Klf7 and dominant-negative Klf7 (DN-Klf7). The number of amino acids is shown. AD represents the transactivation domain, and DBD represents the DNA-binding domain of Klf7. (C) DN-Klf7 inhibits the survival of NGF-dependent wild-type DRG neurons in a dose-dependent manner. DRG neurons from E13.5 wild-type embryos were cultured in the presence of 10 ng/mL NGF, BDNF, or NT3 and infected with recombinant adenoviruses expressing an inactive form of raf (Xraf), Klf7, or DN-Klf7, respectively. The multiplicity of infection (MOI) was 50 for Xraf, Klf7, and DN-Klf7★, and 10 for DN-Klf7. The percentage of neuronal survival 72 h after the infection is shown. (★★) p < 0.01, t-test.
Loss of TrkA-specific sensory neurons in Klf7-/- DRG
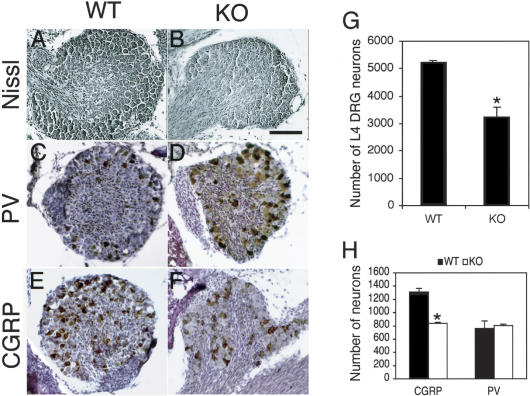
Newborn mice lacking TrkA or NGF by gene knockout have small DRG due to selective apoptosis of small-diameter nociceptive sensory neurons (Crowley et al. 1994; Smeyne et al. 1994). Klf7-/- newborn pups also have small DRG compared with wild-type littermates (Fig. 5A,B). We counted lumbar 4 (L4) DRG neurons from Klf7-/- and wild-type newborn littermates (Fig. 5G). Consistent with the smaller ganglia, the neuron number in L4 DRG of the mutants is reduced by 37.8% (p = 0.023).
Figure 5.
Loss of TrkA-specific neurons in Klf7-/- DRG. (A,B) Representative images of L4 DRG of wild-type (A) and Klf7-/- (B) P0 pups. (C,D) Parvalbumin (PV) immunostaining of L4 DRG of wild-type (C) and Klf7-/- (D) P0 pups. (E,F) CGRP immunostaining of L4 DRG of wild-type (E) and Klf7-/- (F) P0 pups. (G) Neuron counts of L4 DRG. N = 5 for each genotype. (★) p < 0.05, t-test. (H) Quantification of CGRP(+) or PV(+) neurons in L4 DRG showing a significant reduction of CGRP(+) neurons in Klf7-/- DRG but similar numbers of PV(+) neurons in wild-type and Klf7-/- DRG. Serial sections of spinal columns of wild-type (N = 3) and Klf7-/- (N = 3) P0 pups were stained with anti-CGRP or anti-PV antibodies, and the numbers of immunoreactive neurons were counted. CGRP, p = 0.015; PV, p = 0.776, t-test. Bar: A-F, 100 μm.
Within DRG neurons, the calcitonin-gene-related peptide (CGRP) is coexpressed with TrkA in small-diameter nociceptive neurons, while parvalbumin (PV) is coexpressed with TrkC in large-diameter proprioceptive neurons (Liebl et al. 1997). To further establish whether the loss of sensory neurons in vivo is specific to the TrkA(+) population, we counted CGRP(+) and PV(+) neurons of L4 DRG by immunohistochemistry (Fig. 5C-F,H). While there is no difference in PV(+) neurons (p = 0.776), there is a 35.4% reduction (p = 0.015) of CGRP(+) neurons in Klf7-/- DRG. Therefore, consistent with the in vitro culture data (Fig. 4), a significant proportion of TrkA(+) neurons is lost in Klf7-/- DRG.
Removal of one TrkA allele exacerbates the neuronal loss in Klf7-/- mice
Mutational analysis of the Klf7-binding element on the TrkA enhancer had demonstrated a significant reduction in enhancer activity (Ma et al. 2000). We therefore tested whether further reduction of TrkA expression by removal of one functional allele would enhance the neuronal phenotype. We crossed the Klf7 mutant allele into the TrkA+/- background and counted the number of total neurons and TrkA(+) neurons in the P0 trigeminal ganglion (TG) of various genotypes (Fig. 6). Although there is no difference in both total neuron numbers and TrkA(+) neuron numbers between Klf7+/-, TrkA+/-, and wild-type mice, there is a small but statistically significant reduction in Klf7+/- TrkA+/- mice (Fig. 6A-H,Q). Similar to DRG, there is a severe reduction of both total neuron numbers and TrkA(+) neuron numbers in Klf7-/- TG (Fig. 6I,J,Q). In fact, the neuronal loss in Klf7-/- TG can be entirely accounted for by the loss of TrkA(+) neurons (Fig. 6Q). Interestingly, the loss of both total neurons and TrkA(+) neurons is more severe in Klf7-/- TrkA+/- mice than in Klf7-/- mice (Fig. 6I-L,Q). These data strengthen the model that the level of TrkA expression is attenuated in Klf7-/- mice, resulting in reduced neuronal survival.
Figure 6.
Neuronal loss in TrkA+/- or Bax-/- background. (A,C,E,G,I,K,M,O) Nissl staining of P0 TG sections of indicated genotypes. (B,D,F,H,J,L,N,P) Anti-TrkA immunostaining of P0 TG sections of indicated genotypes. (Q) Quantification of total neurons and TrkA(+) neurons in P0 TG of indicated genotypes. N = 4 for each genotype. (★) p < 0.05, t-test. Keys: (K) Klf7; (A) TrkA; (B) Bax. Bars: A,C,E,G,I,K,M,O, 200 μm; B,D,F,H,J,L,N,P, 50 μm.
Reduced TrkA expression in Klf7-/- mice is independent of cell death
To further examine whether the reduced expression of TrkA in Klf7-/- mice is a direct effect of Klf7 gene ablation or a secondary effect of cell death, we crossed the Klf7 mutant allele into the Bax-null background. It is known that naturally occurring cell death is virtually eliminated in peripheral ganglia of Bax-/- mice during embryonic life (White et al. 1998; Patel et al. 2000). Therefore, Bax-/- mice provide a unique system in which to separate the survival requirement for Klf7 from other possible functions. Compared with wild-type TG, there is a marked increase of total neurons and TrkA(+) neurons in Bax-/- TG (Fig. 6M,N,Q). Importantly, while there is a similar increase of total neurons in Klf7-/- Bax-/- TG, the number of TrkA(+) neurons in the double knockouts is significantly smaller than in wild-type mice (Fig. 6A,B,M-Q). Therefore, the reduced expression of TrkA in Klf7-/- mice is the direct effect of Klf7 gene ablation rather than a secondary effect of apoptosis.
Reduced axonal growth and target innervation
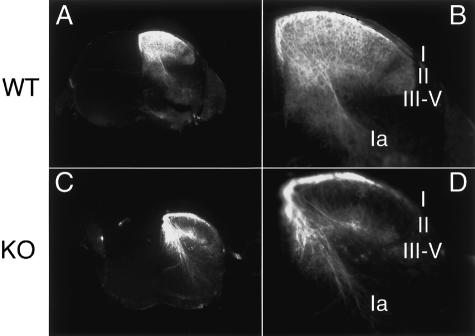
NGF/TrkA signaling is important for axonal growth and target innervation of DRG neurons (Patel et al. 2000; Tucker et al. 2001). DRG neurons project centrally to specific layers of the spinal cord depending on their specialized function (Ozaki and Snider 1997). The small-diameter TrkA(+) neurons innervate laminae I and II and synapse on dorsal horn interneurons, while the large-diameter TrkC(+) neurons innervate the deep lamina and make contacts with motoneurons in the ventral horn (Klein et al. 1994; Liebl et al. 1997). We applied DiI particles to lumbar 4-6 (L4-L6) DRG and analyzed the dorsal root afferent projection to the spinal cord in Klf7 mutants and wild-type littermates. The DiI tracing indicates that as in TrkA knockouts (Liebl et al. 1997), while the Ia afferent from the TrkC(+) neurons is normal, the innervation of lamina II by TrkA(+) neurons is greatly reduced in Klf7-/- newborn pups (Fig. 7). The innervation of laminae III-V by low-threshold mechanoreceptors is also normal in Klf7-/- spinal cord. Thus, consistent with a reduction in TrkA(+) nociceptive neurons, Klf7 mutant mice have deficient nociceptive afferent innervation into the spinal cord.
Figure 7.
Reduced nociceptive innervation of spinal cord in Klf7-/- P0 pups. DiI tracing of the spinal cord of the wild-type (A,B) and Klf7-/- (C,D) P0 pups. Sections shown are at the same level from the middle of the spinal cord segment. N = 8 for each genotype. Laminae I-V are labeled numerically, and Ia denotes the Ia afferent from TrkC(+) neurons.
We next examined the cutaneous innervation of the hindlimbs in Klf7-/- and wild-type P0 pups by staining for the axonal marker NF200 (neurofilament 200). NF200 staining was found in the major nerve trunks in the hind limb of P0 wild-type mice (see Supplementary Fig. S3A) and revealed the extensive collateral branching of axons in the superficial skin (see Supplementary Fig. S3C). In Klf7-/- mice, axons in the major nerve trunks were stained; however, collateral innervation to the superficial skin was substantially reduced (see Supplementary Fig. S3B,D).
Defects in pain sensitivity in Klf7-/- mice
The sensory anatomical deficits seen in Klf7 mutants would be expected to result in behavioral deficits in response to noxious stimuli. Consistent with this prediction, Klf7-/- mutant pups have reduced response to cold temperature (0°C), hot temperature (50°C), and tail pinch, as compared with wild-type littermates (data not shown).
While most Klf7-/- mice die as neonates, 3% of the mutant mice survived to adulthood and showed no obvious spontaneous behavioral abnormalities. To experimentally assess for nociceptive function, we performed a battery of behavioral tests on these adult Klf7-/- mice and wild-type littermates (N = 6 for each genotype) (Fig. 8). The Klf7-/- survivors displayed significantly longer latency in response to the hot plate test for thermal sensitivity (Fig. 8E). In addition, they exhibited reduced response in the test of chemical nociception using 5% formalin (Fig. 8F). Because TrkB- and TrkC-expressing sensory neurons remain intact in Klf7 mutant sensory ganglia, mechanoreception and proprioception should remain functional in these mice. Accordingly, Klf7-/- mice performed equivalently to wild-type littermates to innocuous mechanical stimulation (Fig. 8G) and to walking on a grid (Fig. 8H). In the adult mice, the innervation of the superficial spinal cord by nociceptive sensory neurons can be divided into laminae I, IIo, and IIi. Laminae I and IIo are CGRP(+), while lamina IIi binds a plant lectin IB4(+) (Snider and McMahon 1998). Consistent with the behavior tests, the innervation of laminae I, IIo, and IIi was significantly reduced in the adult Klf7-/- spinal cords (Fig. 8A-D).
Figure 8.
Reduced sensitivity to noxious stimuli in adult Klf7-/- mice. (A-D) The nociceptive innervation of the dorsal spinal cord is greatly reduced in adult Klf7-/- mice. A and B show anti-CGRP immunostaining of laminae I and IIo, while C and D show IB4 staining of lamina IIi. Both types of innervation are significantly reduced. (E) Test of thermal sensitivity. The response latency of Klf7-/- mice is significantly longer than that of wild-type littermates; (★★★) p < 0.001, t-test. (F) Test of chemical nociception. Injection of a 5% formalin solution into the hind paw evokes two phases of paw licking: Phase I reflects direct activation of primary nociceptors; phase II reflects peripheral inflammation and sensitization of dorsal horn neurons. Both phases of licking are significantly reduced in Klf7-/- mice; (★★) p < 0.01; (★★★) p < 0.001, t-test. (G) Test of sensitivity to innocuous mechanical stimulation using calibrated von Frey filaments. The threshold weight (in grams) necessary to produce a response is similar for Klf7-/- and wild-type mice (p = 0.979, t-test). (H) Proprioception as determined by a grid-walking test is not significantly different between wild-type and Klf7-/- mice (p = 0.081, t-test). N = 6 for each genotype in all tests.
Discussion
The formation of neural circuits regulating pain sensation is an important issue in neural development. However, little is known about the molecular mechanisms that control this process. In this study, we demonstrate that the loss of Klf7 function reduces the sensation of pain as determined by response to noxious stimuli.
The large majority of the small-caliber cutaneous sensory fibers are nociceptors (Snider and McMahon 1998). These nerve fibers respond to mechanical, chemical, and thermal stimuli applied to their peripheral receptors. During embryonic development, these fibers extend from neurons that express TrkA and require NGF for survival. These sensory neurons innervate laminae I and II of the superficial dorsal horn of the spinal cord. TrkA-/- and NGF-/- mutant mice lack nociceptive sensory neurons and their projections and are defective in response to noxious stimuli (Crowley et al. 1994; Smeyne et al. 1994; Liebl et al. 1997). In humans, the autosomal recessive genetic disorder congenital insensitivity to pain with anhidrosis (CIPA), characterized by loss of pain and temperature sensation and defects in thermal regulation, can arise as a consequence of loss-of-function mutations in the TrkA gene (Indo et al. 1996; Alberti et al. 2003). The present study implicates Klf7 as a possible target for disruption in CIPA.
The transcription factor DRG11 has also been implicated in nociception. DRG11 is a homeodomain transcription factor expressed in all DRG neurons and in the dorsal horn of spinal cord (Saito et al. 1995; Chen et al. 2001). In DRG11-/- mice, although the expression of Trk receptors in DRG is normal, the central projections of DRG neurons are altered and the mice show defects in nociception (Chen et al. 2001). Therefore DRG11 functions in a different but equally important aspect of nociception from that of TrkA and Klf7.
Klf7 specifically regulates expression of TrkA
The loss of TrkA(+) nociceptive sensory neurons in Klf7 mutant embryos is preceded by a reduction in TrkA gene expression. This indicates that the reduced expression of TrkA is a direct effect of Klf7 gene ablation, rather than a secondary effect of cell death. This is further strengthened by the result of fewer TrkA(+) neurons in Klf7-/- Bax-/- TG as compared with the wild-type TG, despite a marked increase of total neurons in the double-knockout TG. Considering the essential function of TrkA in sensory neuron survival, on the basis of the present study we propose that the reduced expression of TrkA leads to an increased apoptosis and therefore the loss of TrkA(+) neurons. Klf7 is thus the first transcription factor identified that specifically regulates the survival of a subset of nociceptive sensory neurons during development. Additional transcription factors remain to be identified that regulate the survival of the remaining nociceptive sensory neurons. One candidate is the Brn3a POU domain transcription factor that apparently contributes to the regulation of all Trk receptor family genes (Huang et al. 1999; Ma et al. 2003).
Although Klf7 is expressed in all DRG and TG sensory neurons, not all TrkA(+) neurons are lost in Klf7-/- mice. This is consistent with our previous functional analysis of the TrkA minimal enhancer (Ma et al. 2000). Within the TrkA minimal enhancer, nine highly conserved cis elements contribute to the enhancer function in transgenic reporter assays. When the Klf7-binding site in the TrkA minimal enhancer was mutated, the binding of Klf7 was abolished in vitro and the resulting transgenic embryos harboring the mutated TrkA minimal enhancer have diminished but not extinguished β-galactosidase transgene expression in vivo (Ma et al. 2000; Lei et al. 2001). Therefore, the Klf7-binding site is an important cis element that cooperates with additional DNA elements to attain full TrkA transcriptional activity.
It is possible that the loss of Klf7 causes reduction in TrkA expression in vivo. Subthreshold reduction causes death, while neurons that retain sufficient TrkA molecules survive. Alternatively, Klf7-related proteins may compensate for Klf7 in regulating TrkA expression in the remaining neurons. Twenty members of Kruppel-like factors have been identified in mammals and all share a similar DNA-binding domain characterized by the presence of three C2H2-type zinc fingers (Turner and Crossley 1999; Black et al. 2001). Klf6 is closely related to Klf7, and is also expressed in DRG during development (Laub et al. 2001b) and remains expressed in Klf7-/- DRG (data not shown).
Over the past decade, multiple transcription factors have been implicated in the development of sensory neurons. These include Ngn1, Ngn2, NeuroD, NSCL1, NSCL2, Brn3a, Islet1, Sox10, Pax3, and CREB (Anderson 1997; Lonze et al. 2002). Although the expression patterns and physiological functions of these transcription factors are well documented, few downstream target genes have been described. In this study, we identify Klf7 as an important transcription factor for the development of the nociceptive sensory sublineage.
In the CNS, TrkA expression is limited to the basal forebrain cholinergic neurons, while Klf7 has a broad expression pattern throughout development (Laub et al. 2001a; Lei et al. 2001). In a more thorough and broad analysis of the Klf7 mutant mice, several additional abnormalities have been found in the CNS that have no direct relation to TrkA function or gene expression (Laub et al. 2005). These include deficits in olfactory bulb development and corpus callosum. We cannot rule out that the behavioral deficits described in this study may be in part caused by as-yet-undetermined CNS defects.
Materials and methods
Breeding and genotyping of Klf7, TrkA, and Bax mutant mice
Klf7-null mice were generated by homologous recombination (Laub et al. 2005). Genotyping of Klf7 mutant mice was done by PCR using the following primers: for the wild-type allele, 5′-TTTCCTGGCAGTCATCTGCAC-3′ and 5′-GGGTCTGTTTGTTTGTCAGTCTGTC-3′; for the mutant allele, 5′-GCAGTCATCTGCACTGTACACG-3′ and 5′-GCGGCTGCATACGCTTGATCCGGC-3′. The PCR conditions are 94°C for 2 min; 94°C for 1 min, 59°C for 1 min, and 72°C for 2 min, 35 cycles. Genotyping of Bax mutant mice was done as previously described (Ma et al. 2003). Genotyping of TrkA mutant mice was done by PCR using primers P1 (5′-TGTACGGCCATAGATAAGCAT-3′), P2 (5′-GATTGCATAACTGTGTATTTCAC-3′), and P3 (5′-CGCCTTCTTGACGAGTTCTTCTGA-3′). The PCR conditions are 94°C for 2 min, 94°C for 45 sec, 55°C for 45 sec, and 72°C for 75 sec, 35 cycles. The wild-type allele was amplified with P1-P2 (164-bp PCR product), and the mutant allele was amplified using P3-P2 (600-bp PCR product). Klf7+/- mice were crossed with Bax+/- mice to generate Klf7+/- Bax+/- mice. Klf7+/- Bax+/- mice were then mated to generate Klf7-/- Bax-/- mice. Similarly, Klf7+/- mice were crossed with TrkA+/- mice to generate Klf7+/- TrkA+/- mice. Klf7+/- TrkA+/- mice were then mated to generate Klf7-/- TrkA+/- mice. Mice were housed and maintained according to the UTSW IACUC guidelines.
Generation of recombinant adenoviruses
Recombinant adenoviruses expressing the full-length Klf7 or the C-terminal DNA-binding domain (amino acids 202-301) of Klf7 (i.e., DN-Klf7) were generated and titered as described (He et al. 1998).
Neuron counts
To count the number of DRG and TG neurons, P0 pups were perfused with 1× PBS followed by 4% PFA. The heads were decapitated, and the spinal columns were dissected out; both were fixed in 4% PFA overnight, and then decalcified in CalRite (Richard-Allan Scientific) for 48 h at 4°C. The heads and spinal columns were then processed and embedded in paraffin. Eight-micron continuous sagittal sections were collected and stained using Nissl substance. The lumbar 4 (L4) DRG was identified using the ribs and pelvic bone as landmarks. Neurons were identified by their large round or oval nuclei (Francis et al. 1999). The images of every fourth section of L4 DRG or every tenth section of TG were used to count the number of neurons manually. Neuron numbers for whole ganglia were calculated by summing counts from the individual sections and multiplying by 4 for DRG or 10 for TG.
Immunohistochemistry
Immunohistochemistry was carried out as described (Ma et al. 2003). The primary antibodies included TrkA (Advanced Targeting System; 1:500), Ret (IBL; 1:100), NF200 (Sigma; 1:1000), CGRP (Sigma; 1:2000), and parvalbumin (Swant; 1:1000). Embryos and P0 pup tissues were embedded directly in OCT (Statlab). Fourteen-micron sections were cut on a cryostat (Leica), and sections were mounted directly onto Superfrost(+) slides (Statlab) and kept in a -80°C freezer until use. Before each experiment, the sections were dried on a slide warmer at 50°C for 15 min and fixed in 4% paraformaldehyde (PFA) for 1 h. After post-fixation, the slides were washed three times for 5 min in 1× PBS, blocked for 30 min in 1× PBS containing 0.3% Triton X-100 and 3% normal goat serum, and incubated in 1× PBS containing 0.3% Triton X-100, 3% normal goat serum, and appropriate primary antibodies overnight at 4°C. Next day, the slides were washed three times for 5 min in 1× PBS, and incubated with fluorescence dye or biotin conjugated secondary antibodies for 45 min. All secondary antibodies were used at 1:200 dilution. For fluorescence dye conjugated secondary antibodies, the slides were then washed with 1× PBS four times for 5 min and cover-slipped. For biotin-conjugated secondary antibodies, the slides were processed using the ABC kit (Vector Laboratories), and the signals were detected using the DAB reagent (Signet Pathology Systems) as instructed.
For counting the number of CGRP(+) or parvalbumin(+) neurons in DRG and TrkA(+) neurons in TG, 8-μm paraffin sections of P0 heads or spinal columns were used. The sections were deparaffinized and rehydrated, blocked, and then incubated with anti-CGRP, anti-parvalbumin, or anti-TrkA antibodies overnight. The sections were then washed and incubated with biotin-conjugated secondary antibodies for 45 min, and processed using the ABC kit. Signals were detected using the DAB reagent. The numbers of immunoreactive neurons in L4 DRG or TG were counted.
For analyzing the peripheral innervation of DRG neurons, 20-μm sagittal sections of P0 hindlimbs were incubated with anti-NF200 antibodies and visualized using the ABC kit and DAB. For analyzing the central innervation, 8-μm transverse sections of adult spinal cords were incubated with anti-CGRP antibodies or biotin-conjugated isolectin IB4 (10 μg/mL; Molecular Probes) and visualized using the ABC kit and DAB.
DRG primary neuron culture
DRG primary neuron culture was carried out as described (Klesse and Parada 1998). DRG were isolated from E13.5 embryos, and dissociated dorsal root ganglion neurons were cultured in the presence of 10 ng/mL NGF (NGF-7S; Sigma), BDNF (Sigma), or NT-3 (Sigma) on poly-ornithine and laminin-coated plates. The numbers of DRG neurons were counted at 6 h, 48 h, and 72 h after plating on an Olympus Optical IMT-2 inverted microscope under phase contrast. The percentage of neuronal survival was calculated by dividing the neuron numbers at 48 h and 72 h by the numbers at 6 h. The values for the mutants were normalized against the values for the wild-type neurons, which were set at 100%.
For adenovirus infection, 4 h after the initial plating of DRG neurons, recombinant adenoviruses were added to the cultures at the indicated multiplicity of infection (MOI). Neurons were counted every 24 h, and the percentage of neuronal survival at 72 h was reported. To control against effects induced by the recombinant adenovirus system, an inactive and inert form of raf, Xraf, was used (Klesse and Parada 1998; Klesse et al. 1999).
Cell proliferation assays
Pregnant female mice were injected with 50 μg/g of body weight 2′-bromo-5′-deoxyuridine (BrdU) 2 h before sacrifice. Embryos were embedded directly in OCT. Fourteen-micron transverse sections were made and used in immunohistochemistry using anti-BrdU antibodies (Novacastra; 1:100) following the instruction of the manufacturer. The numbers of BrdU(+) cells per section in lumbar DRG were counted.
TUNEL assays
Cryostat sections were processed for the end-labeling of nicked DNA (TUNEL) using the NeuroApop kit (FD NeuroTechnologies) following the instruction of the manufacturer. After TUNEL, sections were counterstained using methyl green. The numbers of TUNEL(+) cells per section in lumbar DRG were counted. For the combination of TUNEL staining with anti-TrkA immunohistochemistry, sections from E13.5 embryos were reacted according to the kit protocol and later incubated with fluorescein-conjugated anti-digoxigenin antibodies along with anti-TrkA antibodies.
In situ hybridization
The digoxigenin (DIG) nonisotopic in situ hybridization was carried out as described (Dong et al. 2001). The antisense probes include TrkA, TrkB, and TrkC.
DiI tracing
DiI tracing was done as described (Liebl et al. 1997). Briefly, P0 pups were perfused and fixed overnight in 4% PFA. DiI particles (Molecular Probes) were placed into the L4, L5, and L6 DRG. Two weeks later, the spinal cords were dissected out and the lumbar regions of the spinal cords were sectioned transversely at 50 μm. The sections were mounted onto glass slides and the images were taken using a Cy3 filter and a fluorescence microscope (Olympus).
Behavior tests
A plantar test was used to assess thermal sensitivity (Romero et al. 2000). The response latency to hind paw licking or jumping was recorded. Chemical nociception was assessed by the formalin test (Chen et al. 2001). Mice received a 20-μL intraplantar injection of 5% formalin, and the licking behavior of the injected paw was observed over 40 min. Mechanical sensitivity was assessed by a set of calibrated von Frey filaments (Chen et al. 2001). Proprioception was assessed by a grid-walking test (Romero et al. 2000). The mice were first trained to walk on a grid runway. After the training, the mice underwent two trials in which the numbers of misplaced steps were counted. The average of the two trials is considered representative for each animal. In all tests, six animals of wild-type or Klf7-/- were used. Analysis of Klf7+/- animals (N = 4) indicated no difference from wild-type animals in all of the behavioral tests performed.
Quantitative RT-PCR
Total RNA was isolated from E11.5 embryos, E12.5 trunks, and E13.5 trunks using the TRIzol reagent (Invitrogen). cDNA was synthesized using the Superscript II reverse transcriptase and oligo(dT)12-18 primers (Invitrogen). Real-time PCR reactions were done in duplicates using an ABI Prism 7000 machine (Applied Biosystems) and SYBR Green according to the manufacturer's instruction. Each 25-μL reaction contains 12.5 μL of 2× SYBR Green master mix, 2 μL of primers at 5 μM, 5 μL of diluted cDNA template, and 5.5 μL of ddH2O. The PCR conditions are 94°C for 15 sec, 60°C for 1 min, 40 cycles. The primers are for β-actin, 5′-ACAGTCCGCCTAGAAGCACT-3′ and 5′-T CCGATGCCCTGAGGCTCTT-3′; for TrkA, 5′-ATATCTAGCCAGCCTGCACTTTGT-3′ and 5′-TGCTCATGCCAAAGTCTCCA-3′; for TrkB, 5′-AAGGCTCTGTTGGTAGCGAGTC-3′ and 5′-CTTGTGTGGGTCTAATGCAAGG-3′; for TrkC, 5′-CATTCAAGGTTCTGGACCTTCCT-3′ and 5′-CAAGTTCACCTGGCTCACATTTC-3′. Each amplicon is 100 bp. The relative RNA levels of TrkA, TrkB, and TrkC were calculated and normalized against the level of β-actin. The ratios of the knockout values to the wild-type values (KO/WT) for TrkA, TrkB, and TrkC were reported.
ChIP assay
ChIP was done by using a commercial kit (Upstate Biotech) following the manufacturer's protocol. NIH3T3 cells were transfected with 20 μg of the myc-tagged vector CS2 + MT, expression vector myc-mKlf7, or myc-mKlf7C. myc-mKlf7 and myc-mKlf7C were generated by cloning the entire coding region or the region encoding the C-terminal DNA-binding domain (amino acids 202-301) of Klf7 into the EcoRI and XhoI sites of the vector CS2 + MT so that the Klf7 coding regions were inframe with the myc epitope tag on CS2 + MT. After immunoprecipitation using an anti-Myc antibody (Santa CruZ Biotechnology), DNA was phenol-chloroform-purified, isopropanol precipitated, and dissolved in 25 μL of water. PCR was carried out using 5 μL of the sample DNA as the template and using the primers 5′-CGAGAGGTCTCCACTTTCCAATC-3′ and 5′-TTAAGAGGGTGTGCATGTCACTTG-3′. Both primers are specific for the mouse TrkA minimal enhancer and the amplified PCR product corresponds to the nucleotides +252-424 of the TrkA minimal enhancer. This region of the TrkA minimal enhancer contains the Klf7-binding site (Ma et al. 2000; Lei et al. 2001). The PCR conditions are 94°C for 2 min, 94°C for 1 min, 60°C for 1 min, 72°C for 1 min, 35 cycles. Amplified products were resolved by standard 2% agarose gel electrophoresis.
Reporter constructs and luciferase assays
The core sequence of the Ikaros2 (IK2) site in the TrkA minimal enhancer was mutated from TCCC to CTTT by site-directed mutagenesis (Ma et al. 2000). The TrkA minimal enhancer containing such a mutation was partially defective in driving expression of a β-galactosidase transgene in DRG and TG sensory neurons (Ma et al. 2000). To generate the reporter constructs wild-type IK2-forward (TrkA#1), wild-type IK2-reverse (TrkA#2), mutant IK2★-forward (TrkA-IK#1), or mutant IK2★-reverse (TrkA-IK#2), the 457-bp TrkA minimal enhancer fragment with either a wild-type Ikaros2 site (IK2) or a mutated Ikaros2 site (IK2★) was released from the vector TA-IK2 or TA-IK2★ by EcoRI digestion, blunted, and subcloned into the SmaI site of the vector pGL3-Promoter (Promega). Plasmids containing the insert in either orientation were confirmed by restriction digestion and DNA sequencing. In addition, two complementary oligonucleotides containing three copies of the wild-type IK2 site (5′-GACTCTTCTCTCCCACTATTTTTC-3′) in the same orientation were annealed and subcloned into the SmaI site of the vector pGL3-Promoter to generate the reporter plasmid IK2(x3). The core sequence TCCC was highlighted. The resulting plasmid was confirmed by DNA sequencing. The same DNA fragment was used as a probe in identifying Klf7 by expression cloning and Klf7 binds specifically to a single IK2 site in vitro (Lei et al. 2001).
To analyze the transcriptional activity of Klf7, transient transfection and dual-luciferase assays were used (Promega). Briefly, PC12 cells were grown in DMEM containing 10% heat-inactivated horse serum and 5% heat-inactivated fetal bovine serum to near confluency. The day before transfection, PC12 cells were split and plated onto six-well plates. For transfection, individual reporter plasmid (1 μg), expression vector myc-mKlf7 (5 μg) or empty vector CS2 + MT (5 μg), and pCMV-Renila (internal control, 100 ng) was mixed with Fugene 6 reagent (Roche) and serum-free medium for 30 min, and the mixture was added into appropriate wells. All transfections were done in triplicates. Forty-eight hours after transfection, cell lysates were collected and both firefly and Renilla luciferase activities were measured using a fluorometer (Rosys Anthos Inc.). The activity of the firefly luciferase was divided by that of the Renilla luciferase to obtain an arbitrary unit of transcription.
Acknowledgments
We thank Drs. Steve Kernie and Jane Johnson for critically reading the manuscript. L.L. is a recipient of a National Research Service Award from the NIH. This work was supported by NIH grant R37 NS033199 to L.F.P. F.R. acknowledges support from NIH-2R37 AR38648.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1227705.
References
- Alberti L., Carniti, C., Miranda, C., Roccato, E., and Pierotti, M.A. 2003. RET and NTRK1 proto-oncogenes in human diseases. J. Cell Physiol. 195: 168-186. [DOI] [PubMed] [Google Scholar]
- Anderson D.J. 1997. Cellular and molecular biology of neural crest cell lineage determination. Trends Genet. 13: 276-280. [DOI] [PubMed] [Google Scholar]
- Bibel M. and Barde, Y.A. 2000. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes & Dev. 14: 2919-2937. [DOI] [PubMed] [Google Scholar]
- Black A.R., Black, J.D., and Azizkhan-Clifford, J. 2001. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 188: 143-160. [DOI] [PubMed] [Google Scholar]
- Chen Z.F., Rebelo, S., White, F., Malmberg, A.B., Baba, H., Lima, D., Woolf, C.J., Basbaum, A.I., and Anderson, D.J. 2001. The paired homeodomain protein DRG11 is required for the projection of cutaneous sensory afferent fibers to the dorsal spinal cord. Neuron 31: 59-73. [DOI] [PubMed] [Google Scholar]
- Crowley C., Spencer, S.D., Nishimura, M.C., Chen, K.S., Pitts-Meek, S., Armanini, M.P., Ling, L.H., MacMahon, S.B., Shelton, D.L., Levinson, A.D., et al. 1994. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell 76: 1001-1011. [DOI] [PubMed] [Google Scholar]
- Dong X., Han, S., Zylka, M.J., Simon, M.I., and Anderson, D.J. 2001. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 106: 619-632. [DOI] [PubMed] [Google Scholar]
- Francis N., Farinas, I., Brennan, C., Rivas-Plata, K., Backus, C., Reichardt, L., and Landis, S. 1999. NT-3, like NGF, is required for survival of sympathetic neurons, but not their precursors. Dev. Biol. 210: 411-427. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., Moore, D.D., and Derfler, B. 1992. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258: 808-812. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., Winandy, S., and Avitahl, N. 1997. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 15: 155-176. [DOI] [PubMed] [Google Scholar]
- He T.C., Zhou, S., da Costa, L.T., Yu, J., Kinzler, K.W., and Vogelstein, B. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. 95: 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D.M., Li, Y., Parada, L.F., Kinsman, S., Chen, C.K., Valletta, J.S., Zhou, J., Long, J.B., and Mobley, W.C. 1992. p140trk mRNA marks NGF-responsive forebrain neurons: Evidence that trk gene expression is induced by NGF. Neuron 9: 465-478. [DOI] [PubMed] [Google Scholar]
- Huang E.J. and Reichardt, L.F. 2001. Neurotrophins: Roles in neuronal development and function. Annu. Rev. Neurosci. 24: 677-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Trk receptors: Roles in neuronal signal transduction. Annu. Rev. Biochem. 72: 609-642. [DOI] [PubMed] [Google Scholar]
- Huang E.J., Zang, K., Schmidt, A., Saulys, A., Xiang, M., and Reichardt, L.F. 1999. POU domain factor Brn-3a controls the differentiation and survival of trigeminal neurons by regulating Trk receptor expression. Development 126: 2869-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indo Y., Tsuruta, M., Hayashida, Y., Karim, M.A., Ohta, K., Kawano, T., Mitsubuchi, H., Tonoki, H., Awaya, Y., and Matsuda, I. 1996. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat. Genet. 13: 485-488. [DOI] [PubMed] [Google Scholar]
- Klein R., Parada, L.F., Coulier, F., and Barbacid, M. 1989. trkB, a novel tyrosine protein kinase receptor expressed during mouse neural development. EMBO J. 8: 3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R., Martin-Zanca, D., Barbacid, M., and Parada, L.F. 1990. Expression of the tyrosine kinase receptor gene trkB is confined to the murine embryonic and adult nervous system. Development 109: 845-850. [DOI] [PubMed] [Google Scholar]
- Klein R., Smeyne, R.J., Wurst, W., Long, L.K., Auerbach, B.A., Joyner, A.L., and Barbacid, M. 1993. Targeted disruption of the trkB neurotrophin receptor gene results in nervous system lesions and neonatal death. Cell 75: 113-122. [PubMed] [Google Scholar]
- Klein R., Silos-Santiago, I., Smeyne, R.J., Lira, S.A., Brambilla, R., Bryant, S., Zhang, L., Snider, W.D., and Barbacid, M. 1994. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements [see comments]. Nature 368: 249-251. [DOI] [PubMed] [Google Scholar]
- Klesse L.J. and Parada, L.F. 1998. p21 ras and phosphatidylinositol-3 kinase are required for survival of wild-type and NF1 mutant sensory neurons. J. Neurosci. 18: 10420-10428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesse L.J., Meyers, K.A., Marshall, C.J., and Parada, L.F. 1999. Nerve growth factor induces survival and differentiation through two distinct signaling cascades in PC12 cells. Oncogene 18: 2055-2068. [DOI] [PubMed] [Google Scholar]
- Laub F., Aldabe, R., Friedrich Jr., V., Ohnishi, S., Yoshida, T., and Ramirez, F. 2001a. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev. Biol. 233: 305-318. [DOI] [PubMed] [Google Scholar]
- Laub F., Aldabe, R., Ramirez, F., and Friedman, S. 2001b. Embryonic expression of Kruppel-like factor 6 in neural and non-neural tissues. Mech. Dev. 106: 167-170. [DOI] [PubMed] [Google Scholar]
- Laub F., Lei, L., Sumiyoshi, H., Kajimura, D., Dragomir, C., Smaldone, S., Puche, A.C., Petros, T.J., Mason, C., Parada, L.F., et al. 2005. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the central and peripheral nervous system. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Lei L., Ma, L., Nef, S., Thai, T., and Parada, L.F. 2001. mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons. Development 128: 1147-1158. [DOI] [PubMed] [Google Scholar]
- Liebl D.J., Tessarollo, L., Palko, M.E., and Parada, L.F. 1997. Absence of sensory neurons before target innervation in brain-derived neurotrophic factor-, neurotrophin 3-, and TrkC-deficient embryonic mice. J. Neurosci. 17: 9113-9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonze B.E., Riccio, A., Cohen, S., and Ginty, D.D. 2002. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron 34: 371-385. [DOI] [PubMed] [Google Scholar]
- Ma L., Merenmies, J., and Parada, L.F. 2000. Molecular characterization of the TrkA/NGF receptor minimal enhancer reveals regulation by multiple cis elements to drive embryonic neuron expression. Development 127: 3777-3788. [DOI] [PubMed] [Google Scholar]
- Ma L., Lei, L., Eng, S.R., Turner, E., and Parada, L.F. 2003. Brn3a regulation of TrkA/NGF receptor expression in developing sensory neurons. Development 130: 3525-3534. [DOI] [PubMed] [Google Scholar]
- Martin-Zanca D., Barbacid, M., and Parada, L.F. 1990. Expression of the trk proto-oncogene is restricted to the sensory cranial and spinal ganglia of neural crest origin in mouse development. Genes & Dev. 4: 683-694. [DOI] [PubMed] [Google Scholar]
- Ozaki S. and Snider, W.D. 1997. Initial trajectories of sensory axons toward laminar targets in the developing mouse spinal cord. J. Comp. Neurol. 380: 215-229. [PubMed] [Google Scholar]
- Patel T.D., Jackman, A., Rice, F.L., Kucera, J., and Snider, W.D. 2000. Development of sensory neurons in the absence of NGF/TrkA signaling in vivo. Neuron 25: 345-357. [DOI] [PubMed] [Google Scholar]
- Romero M.I., Rangappa, N., Li, L., Lightfoot, E., Garry, M.G., and Smith, G.M. 2000. Extensive sprouting of sensory afferents and hyperalgesia induced by conditional expression of nerve growth factor in the adult spinal cord. J. Neurosci. 20: 4435-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Greenwood, A., Sun, Q., and Anderson, D.J. 1995. Identification by differential RT-PCR of a novel paired homeodomain protein specifically expressed in sensory neurons and a subset of their CNS targets. Mol. Cell. Neurosci. 6: 280-292. [DOI] [PubMed] [Google Scholar]
- Serbedzija G.N., Bronner-Fraser, M., and Fraser, S.E. 1992. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development 116: 297-307. [DOI] [PubMed] [Google Scholar]
- ____. 1994. Developmental potential of trunk neural crest cells in the mouse. Development 120: 1709-1718. [DOI] [PubMed] [Google Scholar]
- Smeyne R.J., Klein, R., Schnapp, A., Long, L.K., Bryant, S., Lewin, A., Lira, S.A., and Barbacid, M. 1994. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature 368: 246-249. [DOI] [PubMed] [Google Scholar]
- Snider W.D. and McMahon, S.B. 1998. Tackling pain at the source: New ideas about nociceptors. Neuron 20: 629-632. [DOI] [PubMed] [Google Scholar]
- Tessarollo L., Tsoulfas, P., Martin-Zanca, D., Gilbert, D.J., Jenkins, N.A., Copeland, N.G., and Parada, L.F. 1993. trkC, a receptor for neurotrophin-3, is widely expressed in the developing nervous system and in non-neuronal tissues. Development 118: 463-475. [DOI] [PubMed] [Google Scholar]
- Tucker K.L., Meyer, M., and Barde, Y.A. 2001. Neurotrophins are required for nerve growth during development. Nat. Neurosci. 4: 29-37. [DOI] [PubMed] [Google Scholar]
- Turner J. and Crossley, M. 1999. Mammalian Kruppel-like transcription factors: More than just a pretty finger. Trends Biochem. Sci. 24: 236-240. [DOI] [PubMed] [Google Scholar]
- White F.A., Silos-Santiago, I., Molliver, D.C., Nishimura, M., Phillips, H., Barbacid, M., and Snider, W.D. 1996. Synchronous onset of NGF and TrkA survival dependence in developing dorsal root ganglia. J. Neurosci. 16: 4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White F.A., Keller-Peck, C.R., Knudson, C.M., Korsmeyer, S.J., and Snider, W.D. 1998. Widespread elimination of naturally occurring neuronal death in Bax-deficient mice. J. Neurosci. 18: 1428-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]