Abstract
Selective gene amplification is associated with normal development, neoplasia, and drug resistance. One class of amplification events results in large arrays of inverted repeats that are often complex in structure, thus providing little information about their genesis. We made a recombination substrate in Saccharomyces cerevisiae that frequently generates palindromic duplications to repair a site-specific double-strand break in strains deleted for the SAE2 gene. The resulting palindromes are stable in sae2Δ cells, but unstable in wild-type cells. We previously proposed that the palindromes are formed by invasion and break-induced replication, followed by an unknown end joining mechanism. Here we demonstrate that palindrome formation can occur in the absence of RAD50, YKU70, and LIG4, indicating that palindrome formation defines a new class of nonhomologous end joining events. Sequence data from 24 independent palindromic duplication junctions suggest that the duplication mechanism utilizes extremely short (4-6 bp), closely spaced (2-9 bp), inverted repeats to prime DNA synthesis via an intramolecular foldback of a 3′ end. In view of our data, we present a foldback priming model for how a single copy sequence is duplicated to generate a palindrome.
Keywords: DNA palindrome, gene amplification, MR complex, SAE2, Saccharomyces cerevisiae, BFB
Gene amplification is the process whereby a cell can increase the copy number of a gene or genomic region. In some organisms this is an integral part of normal development, as in the expansion of rDNA genes during macronuclear development in Tetrahymena (Butler et al. 1995) or of chorion genes required for eggshell formation in Drosophila oocytes (Claycomb et al. 2004). Gene amplification is also associated with abnormal processes, such as increased drug resistance (Schimke et al. 1978; Paquin et al. 1992) and the onset and progression of tumorigenesis (Lengauer et al. 1998). Several mechanisms of gene amplification have been described, including the formation and overreplication of extrachromosomal circular elements (Thomas et al. 2004), in situ endoreplication (Botchan and Levine 2004), unequal sister chromatid exchange (Axelrod et al. 1994), and palindromic amplification.
Palindromic amplicons are comprised of in loco arrays of inverted repeats corresponding to at least a part of the amplicon (Ford and Fried 1986). The mechanism leading to palindromic amplification is poorly understood because the end products are usually complex and provide little information about the initiating events. The currently favored model for palindromic amplification, initially proposed by McClintock (1941) is that replication of a chromosome with a spontaneous double-strand break (DSB) leads to the formation of two broken sister chromatids that can fuse to form a dicentric chromosome. Mitotic segregation of the centromeres to opposite poles can then lead to new chromosome breaks and additional rounds of amplification via breakage-fusion-bridge (BFB) events. Eventually, the chromosome is stabilized by the acquisition of a telomere (see Ciullo et al. 2002).
Several lines of evidence support the conclusion that the initiating lesion is a DSB. Treatment of cells with reagents that promote DSB formation enhances the induction of amplification (Yunis et al. 1987; Paulson et al. 1998). Also, the introduction of a site-specific DSB has been shown to lead to amplification near the break site (Pipiras et al. 1998; Difilippantonio et al. 2002; Tanaka et al. 2002; Zhu et al. 2002). Chromosomal fragile sites (CFSs), which represent sites at which chromosomes are often broken when cells are exposed to conditions of limiting replication (Sutherland and Richards 1995; Lemoine et al. 2005), are associated with amplification events in several types of tumors (Stark et al. 1989). Importantly, the boundary of the amplified region often maps to the fragile site (Ciullo et al. 2002; Hellman et al. 2002).
There is abundant cytological evidence for sister chromatid fusion and subsequent BFB cycles with eventual healing by the acquisition of a telomeric cap (McClintock 1941; Windle et al. 1991). However, these studies do not address the fusion mechanism. Some mechanistic details have been suggested by molecular studies. For example, the first products of SV40 amplification events induced by carcinogens are hairpins that involve multiple rounds of replication initiation from the SV40 origin (Cohen et al. 1994). Also, in Tetrahymena, 42-bp inverted repeats adjacent to a break site promote palindrome amplification of the rDNA by foldback between the inverted repeats that are used to prime DNA synthesis. (Butler et al. 1995). These latter studies were extended to show that artificially constructed substrates with short (≥28 bp) inverted repeats adjacent to DSBs can promote palindrome formation in yeast (Butler et al. 2002), and in CHO cells (Tanaka et al. 2002).
We previously found that the repair of a site-specific DSB near an inverted repeat (separated by >1 kb) (Fig. 1A) in Saccharomyces cerevisiae often resulted in palindromic duplications in cells deficient in SAE2, but not in wild-type cells (Rattray et al. 2001). The function of Sae2p is only beginning to be elucidated. It is a poorly conserved protein with no obvious motifs. Experimental evidence suggests that it is likely to function by regulating the nuclease activity of the Mre11p/Rad50p/Xrs2p (MRX) complex (referred to as the MRN complex in mammalian cells where Nbs1p is the mammalian ortholog of Xrs2). A deletion of SAE2 has a phenotype indistinguishable from that of certain hypomorphic alleles of MRE11 and RAD50, termed “s” for separation of function (McKee and Kleckner 1997; Prinz et al. 1997; Rattray et al. 2001; Neale et al. 2002). Importantly, we have shown that in strains bearing our substrate, a single amino acid change resulting in a nuclease-deficient mre11 allele (H125N) led to a similarly increased frequency of palindrome formation (Rattray et al. 2001). Recently it has been shown that Mre11p is constitutively phosphorylated in the absence of Sae2p (Baroni et al. 2004), suggesting that Sae2p could function by inhibiting a negative regulator of the Mre11p nuclease activity.
Figure 1.
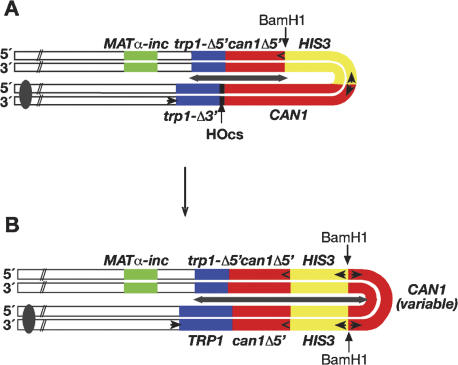
HO endonuclease induction of DSB repair events in Mush18/21. (A) The substrate consists of inverted repeats comprised of two truncated but overlapping alleles of TRP1, a full-length CAN gene, and a truncated can1-5′Δ allele, separated by a full-length HIS3 gene. The extent of the inverted repeat sequence is marked by the gray double-headed arrow. Between the trp1-3′Δ allele and the full-length CAN1 gene, there is a unique recognition sequence for the HO endonuclease. The normal HO recognition sequence at the MAT locus has been mutated (MATα-inc). The substrate is inserted near the MATα locus on chromosome III. (B) Structure of the palindromic duplications. Note that the can1-5′Δ, HIS3, and a variable amount of the 3′ end of the CAN1 gene have been duplicated. The increase in the extent of the inverted repeat is indicated by the gray double-headed arrow.
Mre11p and Rad50p are highly conserved with homologs from bacteria (SbcCD) to mammals (MR) (Connelly and Leach 2002). This complex is also functionally conserved, having a preference for cleaving at single-strand/double-strand DNA junctions, including cleavage of hairpin DNA molecules at both the loop and at the bottom of the stem (Lewis et al. 2004). The MRX/N complex is important for many different DNA metabolic processes in the cell, including telomere maintenance, nonhomologous end joining (NHEJ), meiotic DSB formation and processing, and homologous recombination (D'Amours and Jackson 2002).
NHEJ is the process by which broken ends are repaired in the absence of significant homology. In yeast, NHEJ usually requires the MRX complex, the Ku complex (Yku70p/Yku80p), the Lig4p complex (Lig4p/Lif1p), as well as Nej1p/Lif2p (Dudasova et al. 2004). The molecular details leading to NHEJ are not well defined, and it is likely that it can occur by more than one pathway. However, in general, Ku binds to and protects the broken ends (Boulton and Jackson 1996) and may also serve to recruit MRX/N (Goedecke et al. 1999). MRX probably has multiple functions during NHEJ. Rad50p is responsible for tethering the DNA ends together via its SMC/cohesin-like structure and zinc hooks (de Jager et al. 2001; Hopfner et al. 2002), and the Mre11p nuclease may be necessary to clean up the ends in preparation for ligation. Since MRX/N interacts with Lig4p/Lif4p (Chen et al. 2001), it may also recruit the ligase for the final joining step.
Here we show that the palindromic duplications observed in sae2Δ cells are made efficiently in the absence of Rad50p, Yku70p, and Lig4p, thus indicating that these functions are not required for their formation. In order to gain some insight into the mechanism by which the palindromic duplications were made, we sequenced the novel junction at the tip of the palindromes. Our data provide compelling support for a mechanism by which very short (4-6 bp) inverted repeats serve as primers for foldback DNA synthesis.
Results
The starting substrate used in this study is shown in Figure 1A. In wild-type cells we previously found that the introduction of a site-specific DSB by a galactose-regulated HO endonuclease gene resulted in a large induction of Trp+ recombinants (∼6000-fold). Among these, 0.4% were also His+ Canr (Table 1). Physical analysis of both the Trp+ His+ Cans and Trp+ His+ Canr events indicated that almost all of the DSBs were repaired via a recombinational gap repair mechanism (Rattray et al. 2001). In contrast, when DSBs were introduced in sae2Δ cells, we found a similar level of induction to Trp+, but now 12% were also His+ Canr (Table 1). From a physical analysis of the Trp+ His+ Canr events by Southern blots, we determined that 48% of the events had a palindromic duplication of the substrate as shown in Figure 1B (Table 1). Notably, there was a duplication of the can1-5′Δ allele, the HIS3 gene, and a variable amount of the 5′ end of the full-length CAN1 gene, accompanied with a deletion of the remainder of the full-length CAN1 gene. The duplicated sequences were organized as a palindrome, with a novel junction fragment that varied in size between different isolates (Rattray et al. 2001).
Table 1.
Role of NHEJ functions on palindrome formation
| Strain | Relevant genotype | Induced frequency Trp+ (×103) | No. His+ Canr/total analyzed (%) | Palindromes/total analyzed |
|---|---|---|---|---|
| GRY1654a | Wild type | 24 | 6/1500 (0.4) | 1/44 |
| YAR523a | sae2Δ | 18 | 61/523 (12) | 27/56 |
| YAR771 | rad50Δ | 10 | 14/150 (9) | 14/24 |
| YAR774 | rad50Δ sae2Δ | 9 | 16/200 (8) | 13/24 |
| YAR896 | lig4Δ | 19 | 2/489 (0.4) | 0/24 |
| YAR897 | lig4Δ sae2Δ | 43 | 33/314 (11) | 11/22 |
| YAR720 | yku70Δ | 21 | 5/238 (2.1) | 0/22 |
| YAR775 | yku70Δ sae2Δ | 22 | 14/142 (10) | 9/20 |
Data from Rattray et al (2001).
Palindrome formation does not require known NHEJ functions.
From the deduced structure of the palindromes, we proposed that the palindromic duplication events were formed by invasion of the trp1-3′Δ allele into the uncut trp1-5′Δ allele (to produce TRP1), followed by break-induced replication (BIR) toward the broken end of the inverted repeat, and subsequent joining of the ends by some type of NHEJ (Rattray et al. 2001). To test this prediction, we have analyzed the role of genes required for NHEJ on palindrome formation.
We previously found that mre11-H125N and rad50-K81I cells promoted palindrome formation as efficiently as do sae2Δ cells (Rattray et al. 2001) and, hence, the formation of a novel junction at the center of the palindromes. Since sae2Δ cells and mre11-H125N cells are proficient for NHEJ (Moreau et al. 1999; A.J. Rattray and J.N. Strathern unpubl.), rad50-K81I may also be NHEJ proficient. Therefore, we have analyzed a rad50Δ strain (deficient in NHEJ) and find that it promotes palindromic duplications as efficiently as a sae2Δ mutant (Table 1). Furthermore, the double mutant rad50Δ sae2Δ, is similar to either single mutant. Therefore, the NHEJ function of Rad50p is not required for palindrome formation. In fact, these data indicate that MRX is involved in inhibiting the formation of palindromic duplications.
Cells deleted for LIG4 or YKU70 promote gap repair to Trp+ as efficiently as a wild-type strain (Table 1). Significantly, the double mutants lig4Δ sae2Δ, or yku70Δ sae2Δ are as efficient at promoting palindrome formation as is the sae2Δ single mutant. That is, all three strains show a large increase in the proportion of His+ Canr events among the Trp+ recombinants, and approximately half of these have a palindromic structure (Table 1). Therefore, we conclude that neither Lig4p nor Yku70p is required for palindrome formation. Thus it appears that the process involved in forming the junction during palindrome formation is a novel pathway of end joining that is independent of the known NHEJ functions
Physical analysis of palindromic events from sae2Δ cells
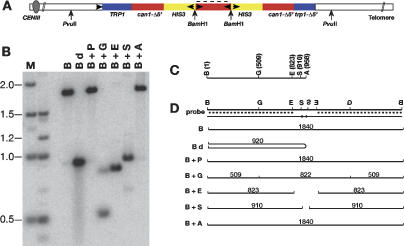
From the Southern blot analysis of the gene amplification events depicted in Figure 1B, we concluded that there was a novel BamH1 fragment of variable size that harbored the palindromic junction (Fig. 2A). In order to gain some insight into how the palindromic junctions are formed, we have done a more detailed Southern blot analysis of the BamH1 fragment from several independent isolates. One example is shown in Figure 2. The probe used in this blot is specific to the unique 5′ sequence of the full-length CAN1 gene (indicated by the dotted lines in Fig. 2A,D) and thus can only hybridize with sequences within the BamH1 fragment. Our interpretation of the blot shown in Figure 2B is depicted in Figure 2D. Genomic DNA was first digested with BamH1, and divided into seven aliquots for further processing. In Figure 2B, DNA that had been digested with BamH1 alone (lane B), or with BamH1 and PvuII (lane B + P) migrates with a molecular size of ∼1.8 kb, consistent with the interpretation that the novel band is created by duplication of the BamH1 site (the next closest BamH1 sites are outside of the PvuII sites). Denaturation by boiling of the BamH1 cleaved DNA prior to electrophoresis results in a tight band that migrates with an apparent size of ∼900 bp (Fig. 2B, lane Bd). This result is expected from a palindromic molecule, due to formation of an intramolecular snap-back molecule (Fig. 2D, Bd). The other sites within the CAN1 gene used for secondary digestion are shown in Figure 2C. Our results are consistent with a palindromic structure up to and including the BstEII site. Notably, although the AvaII site is only 48 bp beyond the BstEII site in the native CAN1 sequence, the DNA digested with BamH1 and AvaII (Fig. 2B, lane B + A) migrates identically to that digested with BamH1 alone (Fig. 2B, lane B), indicating that the AvaII site is not present. Therefore, the tip of the palindrome is located between the BstEII site, and the AvaII site.
Figure 2.
Southern blot analysis of a palindromic gene duplication event. (A) Schematic of the palindromic molecule showing the location of the BamH1 (B) and PvuII (P) sites. (B) Southern blot analysis of BamH1-digested DNA. (Bd) BamH1-digested DNA that was denatured by boiling prior to electrophoresis; (G) BglI; (E) EcoR1; (S) BstEII; (A) AvaII. (A,D) The blot was probed with unique sequences from the 3′ end of the CAN1 gene (dotted lines). (C) Location of restriction sites in relevant region of the native CAN1 gene. (D) Schematic showing interpretation of the Southern blot shown in panel B.
Sequence analysis of palindromic events
Having identified the approximate location of the palindromic junctions, we analyzed the junction sequence of 24 independently generated palindromes (Fig. 3). As noted above (Fig. 2B,D, lane Bd), the palindromic DNAs have a propensity to form intramolecular snap-back molecules upon denaturation. Thus it is impossible to directly sequence the junctions, since no amount of exogenously added oligonucleotide can compete with the local concentration provided by the adjacent repeat. To overcome this problem, we used sodium bisulfite modification of the DNA that deaminates cytosine, converting it to uracil and thus reducing intrastrand pairing (Rattray 2004 and references therein). The deaminated DNA can then be PCR amplified using primers specific for the deaminated DNA (Supplementary Table S1). Due to the incomplete modification during the deamination procedure, it is necessary to clone the PCR products prior to sequencing. Although Escherichia coli cannot tolerate palindromic molecules (Leach 1994), bisulfite modification disrupts the palindromic structure sufficiently to permit cloning and plasmid purification from E. coli (Rattray 2004).
Figure 3.
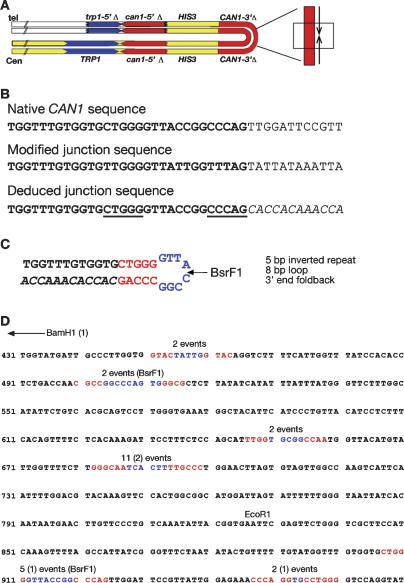
DNA sequence analysis of palindromic junctions. (A) Schematic of a palindromic event with the location of the region sequenced shown by the boxed region. (B) Sequence analysis of the same event analyzed in Figure 2. (Top) Sequence of the relevant region of the native CAN gene. (Center) Bisulfite-modified sequence of the junction. (Bottom) Deduced junction sequence. Sequence in italics diverges from the native sequence. Underlined sequences indicate short inverted repeats preceding the point of divergence. (C) Interpretation of sequence analysis with inverted repeats shown in red and loop region shown in blue. BsrF1 indicates the site used for vectorette PCR analysis. (D) Summary of sequence from 24 independent junctions. Sequence shown is a portion of the CAN1 gene, where the BamH1 site of the starting substrate is at position 1 (not shown). The end point of each different palindromic junction is shown with the stem regions highlighted in red and the loop region highlighted in blue. For each junction the homology ends at the second inverted repeat sequence followed immediately by the complement to the sequence preceding the first inverted repeat (as drawn in C). The shortest palindrome is the first sequence highlighted and the longest palindrome is the last sequence highlighted. The number of independent events mapping to each of the six junctions is shown above the sequence. Junction sequences from rad50Δ cells are noted in parentheses, the remaining 20 events are derived from sae2Δ cells. EcoR1 defines the site at which homology to the can1-5′Δ allele begins. BsrF1 indicates the location of the sites used for verifying the relevant events by vectorette PCR.
The sequence analysis of the tip of the same palindrome analyzed in Figure 2 is shown in Figure 3A-C. Figure 3B shows the alignment of the native CAN1 sequence (top), the sequence obtained by sodium bisulfite sequencing (middle), and the deduced junction sequence (bottom). Notably, there is a short inverted repeat of 5 bp (Fig. 3B, underlined and separated by 8 bp) immediately preceding the point at which the sequences diverge. Also, the diverged sequence is complementary (Fig. 3B, in inverted orientation) to the sequence preceding the inverted repeat. This arrangement suggests that the DNA folded back upon itself intramolecularly and used the 3′ end to prime DNA synthesis (Fig. 3C; see Discussion).
Since the sequence analysis relies on our interpretation of sodium bisulfite-modified DNA, we took advantage of a BsrF1 restriction site found at the junction to further verify the sequence of unmodified genomic DNA (Fig. 3C). The DNA was cleaved with BsrF1 and ligated to vectorette bubble linkers. The DNA is then amplified with a vectorette primer and a fixed primer within the sequence being analyzed (see Materials and Methods). Vectorette PCR analysis on palindromic DNA is expected to generate two different junction sequences. One sequence is identical to the native CAN1 locus (upper strand shown in Fig. 3C) and the other derives from the novel junction (Fig. 3C, lower strand). Sequencing of the vectorette clones confirmed our interpretation of the bisulfite sequence data.
The sequence analyses of all 20 independent junctions analyzed from a sae2Δ strain and four independent junctions from a rad50Δ strain are summarized in Figure 3D, embedded within the native CAN1 sequence. The 24 junctions map to six different positions within the CAN1 gene. The terminus of each palindrome is marked by the highlighted sequences, with the inverted repeats shown in red and the intervening sequence shown in blue. For each junction the sequence aligns with the native CAN1 sequence through the second inverted repeat, and immediately diverges. Remarkably, in every case, the divergent sequence is complementary to the sequence immediately preceding the first inverted repeat (as shown in Fig. 3C). This sequence analysis implicates that all of the junctions were formed by a hairpin that provided a replication primer (see Discussion).
Further confirmation of the sequence analysis was obtained by vectorette PCR of the unmodified DNA for two additional independent isolates of the same locus as that described above (position 915, Fig. 3D: one derived from a sae2Δ strain and one derived from a rad50Δ strain) as well as for the DNA from two independent sae2Δ-derived events from a second locus with a BsrF1 site at the junction (position 501, Fig. 3D).
It is worth noting that although we are only presenting the sequence at the tips of the palindromic junctions, the size of the palindromes are constrained to at least 2.4 kb/arm since we are selecting His+ events. The palindromes are also constrained to a maximum of 3.4 kb/arm since we are selecting Canr events, and thus the full-length copy of the CAN1 gene must be disrupted. Palindromic events that extend more than ∼300 bp into the region of the CAN1 gene homologous to the can1-5′Δ allele (beginning at the EcoR1 site noted in Fig. 3) are expected to break down by either homologous recombination or single strand annealing to produce a wild-type CAN1 gene and therefore are excluded by our selection (Rattray et al. 2001).
Instability of palindromic amplicons
Lobachev et al. (2002) found that palindromic molecules (with either 0 or 12 intervening base pairs) are highly unstable when inserted within the LYS2 gene in SAE2 cells but are significantly stabilized in sae2Δ cells. The instability of palindromes in SAE2 cells provides us with a genetic assay to help distinguish palindromes from nonpalindromes.
To monitor the stability of the chromosome with the rearranged substrate in SAE2 cells, we crossed MATα sae2Δ (strain YAR523) Trp+ His+ Canr events that either were normal recombinants or had a palindrome to a MATa strain (GRY1509; Materials and Methods) that has a URA3 gene located at the same position as our substrate on chromosome III. We monitored the stability of the locus in the MATa/α diploid cells by measuring loss of the HIS3 gene (two copies of which are located within the palindrome), loss of the adjacent MATα locus (Fig. 1B), and loss of the MATa and URA3 loci. Whereas MATa/α diploids do not normally mate, they are able to mate if they lose one MAT allele. All 48 zygotes colonies resulting from crosses with nonpalindromic events remained His+, showed no illegitimate mating, and did not produce Ura- cells (measured as 5-FOA resistance), indicating that both chromosomes were stable. In contrast, two of the 71 zygote colonies derived from crosses with palindromic events were His- (3%). More strikingly, 54 (76%) of these colonies showed illegitimate mating with the MATα tester strain, indicating that the cells were losing all or part of the chromosome bearing the palindrome near the MATα locus. Notably, the MATa chromosome (marked with URA3) did not appear to be unstable as monitored by the lack of appearance of 5-FOA-resistant cells. We note that all of the cells in a patch have to lose both copies of the HIS3 gene in order to become His-, but only a proportion (∼5%-10%) of the cells need to lose either MAT locus in order to show significant illegitimate mating. Our findings indicate that the MATα chromosome, bearing the palindrome, becomes unstable when crossed with a SAE2 strain.
Discussion
Palindromic amplicons have been linked with tumor progression and prognosis for many different types of cancers. Thus, understanding the molecular mechanisms by which they arise or are destabilized could lead to novel cancer therapeutic agents. The correlation between the presence of palindromic sequences and the genomic instability of cancer cells was recently elegantly demonstrated by taking advantage of the propensity of palindromic sequences for intrastrand annealing (Tanaka et al. 2005). In the study by Tanaka et al. (2005) isolated snap-back DNA from either normal cells or tumor-derived cells and found several palindromic loci only in the tumor cells. This study provides an insight into some of the loci that are subject to amplification, but it does not provide any information as to how the events arise. Perhaps the most difficult step of palindromic amplification to envision is how a single copy of a sequence becomes duplicated to produce a palindrome. Here we present data from S. cerevisiae that provide that mechanistic insight. Our data indicate that the initial palindromic duplication event does not require canonical NHEJ functions and further suggest that the palindromic duplications are initiated by a foldback replication between extremely short inverted repeats near the site of a DSB.
When we sequenced the centers of the palindromes, we found that they were derived from short (4-6 bp), closely spaced (2-9 bp spacer), inverted repeats. For two reasons we reject the hypothesis that these junctions derive from microhomology-mediated NHEJ pathways. First, we have shown that they are formed in cells that lack functions required for NHEJ in other studies. That is, palindrome formation does not require RAD50, YKU70, and LIG4. In fact, they are formed at an elevated frequency in rad50Δ cells. Second, there is the statistical argument that while there are hundreds of microhomologies that could have been used to join two copies of the CAN1 gene made by BIR (Table 2), all of the junctions that we identified had a short inverted repeat structure. For example, in the relevant region of the CAN1 gene there were 101 6-bp inverted repeats, but only the two with the short inverted repeat structures were used. Among the 24 events sequenced, one of these was used 11 times, and the other two times (Fig. 3). Similarly, although there are 446 different 5-bp microhomologous sequences that could have been used by NHEJ, we found that the same junction was used on five independent occasions. This sequence had a foldback structure. Finally, the three different junctions that we found that involved 4-bp microhomologies were each found twice despite the fact that there are 1452 such possible 4-bp sequences. Those three junctions each have the property that they derive from a sequence that is a closely spaced inverted repeat. Thus, this requirement for a hairpin structure leads us to suggest that the junctions are formed by foldback DNA synthesis primed by these short inverted repeats.
Table 2.
Distribution of inverted repeats in unique CAN1 sequence
| Size of inverted repeat | No. of repeats in size class | No. of hairpins (loop of ≤9 bp) | No. of hairpins used for amplification | No. of GC residues in stem at used sites |
|---|---|---|---|---|
| 9 bp | 1 | 0 | 0 | NA |
| 8 bp | 9 | 0 | 0 | NA |
| 7 bp | 21 | 1 | 0 | NA |
| 6 bp | 101 | 2 | 2 | 5, 4 |
| 5 bp | 446 | 9 | 1 | 4 |
| 4 bp | 1452 | 31 | 3 | 4, 2, 2 |
NA indicates not applicable.
The parameters that lead to stable DNA hairpins with such short stems are not entirely understood, but some of the characteristics are being defined (Varani 1995). For example, it is clear that the presence of a GC base pair on the loop side provides an unexpected amount of stability (Moody and Bevilacqua 2003). Notably all three of the 4-bp stem hairpins found in our sequence analysis share this feature.
The formation of palindromes via short inverted repeats has been shown in several other organisms, including bacteria (Qin and Cohen 2000; Lin et al. 2001), fission yeast (Albrecht et al. 2000), protozoans (Ouellette et al. 1991; Butler et al. 1995), and mammalian cells (Tanaka et al. 2002). Indeed, the ciliated protozoan Tetrahymena uses short-inverted repeats near a DSB as part of its normal mechanism of rDNA amplification during macronuclear development (Butler et al. 1995). Presumably, cells that use palindromy as a normal mechanism for gene amplification have acquired characteristics that allow them to tolerate palindromes.
Model for palindromic gene amplification
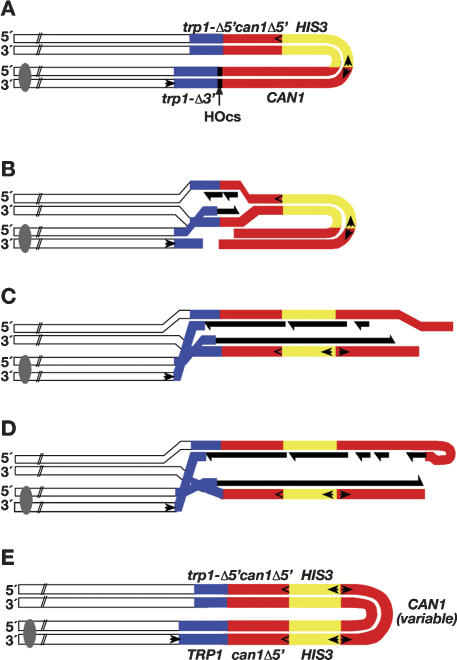
A model based on our observations is presented in Figure 4. After the introduction of a DSB in G1 cells (Fig. 4A), homology-mediated invasion (Fig. 4B) leads to BIR toward the end of the molecule (Fig. 4C). The product of BIR is still inviable, since the telomere and centromere are not joined. Occasionally the unreplicated 3′ end of the molecule can fold back upon itself via short inverted repeats (perhaps requiring processing to expose the repeats) (Fig. 4D). This foldback structure is used to prime DNA synthesis. The resulting product has one strand with both a centromere and a telomere. Branch migration of the intermediate then results in a single-strand gap that can be filled in to produce the molecule shown in Figure 4E. Given that a hairpin sequence as short as 4 bp is apparently sufficient for the foldback priming shown in Figure 4D, we wonder what DNA polymerase can extend such a primer. We note that a similar type of foldback replication was recently shown to be utilized for stabilizing the ends of chromosomes from yeast cells lacking both telomerase and recombinational repair functions provided they are also exo1 deficient (Maringele and Lydall 2004).
Figure 4.
Model for palindrome formation. (A) DSB repair substrate. (B) Recombination is induced by turning on the expression of the HO endonuclease. (C) After invasion, DNA synthesis proceeds by BIR toward the end of the molecule. (New DNA synthesis is shown in black.) (D) A free 3′ end folds back intramolecularly via short (4-6 bp), closely spaced (2-9 bp), inverted repeats. The paired repeats function to prime DNA synthesis. Eventually, the newly primed strand becomes ligated to the other newly synthesized DNA. (E) Branch migration of the intermediate leads to a palindromic molecule with a gap that is subsequently filled in. The Watson and Crick strands of the DNA molecules are indicated by 5′ and 3′.
Palindromic amplicons are often, but not always associated with translocations. For example, the PIP gene duplication associated with the T47D breast carcinoma cell line involves a palindromic duplication internal to the chromosome (Ciullo et al. 2002). Ciullo et al. (2002) suggest that the amplification was derived by the following steps: breakage at the FRA7I CFS in G1, replication and fusion of the sister chromatids, breakage of the resulting dicentric chromosome during mitotic chromosome segregation, and, finally, recapture of the chromosome fragment bearing the telomere. Two questions remain unaddressed in this model: the mechanism of sister chromatid fusion and the mechanism of telomere recapture. We suggest that perhaps palindromic amplifications similar to the PIP gene duplication are derived from a mechanism akin to that outlined in Figure 4. Sequence analysis of the novel junction at the center of such palindromes may identify junctions associated with a short inverted repeat.
Although our starting substrate consists of an inverted repeat (separated by >1 kb), the entire inverted repeat is probably not necessary for palindrome formation. We recently replaced the can1-5′Δ allele with a URA3 gene and found that this substrate also promotes efficient palindrome formation (A. Welcker and J. Strathern, unpubl.). The homology at trp1 is presumably important for initiating BIR. As few as 72 bp appear to be sufficient to initiate BIR events that can replicate several hundred kilobases (Bosco and Haber 1998). Therefore, repetitive dispersed repeats found in most eukaryotic cells may be able to serve the same role in initiating BIR as the trp1 sequences provide here (see below).
Is BIR itself necessary for palindrome formation? We suspect that it may play an important role based on the following observations. Several years ago Kunes et al. (1985) found that transformation of linear DNA into S. cerevisiae resulted in the formation of circular head-to-head dimer molecules. They found that two molecules were required for dimer formation, unless the plasmid was linearized within an inverted repeat (Kunes et al. 1990). This result suggests that a similar mechanism, involving invasion via homology, followed by BIR is likely to be involved in inverted dimer formation. Butler et al. (2002) also found that two molecules were required for producing large inverted palindromes from broken plasmid DNA molecules containing inverted repeats on the same side of the break site. In their system it is not obvious why the inverted repeats cannot simply fold back upon themselves to initiate DNA replication in a unimolecular reaction. Butler et al. (2002) concluded these events must require an intermolecular end-joining event to produce the palindromes. The data presented here suggest that intermolecular end joining is not used to generate palindromes, since it would not be expected to require the closely spaced inverted repeats that we found here. The largest spacer we found was of 9 nucleotides, even though hundreds of inverted repeats with much larger spacers are clearly present within the appropriate region of the CAN1 gene (Table 2). Furthermore, large central spacers are expected to be more stable and thus should be favored over shorter spacers. Therefore, we suggest that the requirement for two DNA molecules in the experiments of Butler et al. (2002) and Kunes et al. (1990) reflects a requirement for BIR in palindrome formation. One role for BIR could be that it serves to deposit the necessary replication complex near the end of the DNA molecule.
The absence of Sae2p is vital in the recovery of our palindromic duplications. This is presumably because when it is present it licenses the hairpin cleaving activity of the MRX/N nuclease. We do not know if the role of Sae2p (or MRX/N) is to inhibit palindrome formation to begin with, to inhibit palindrome maintenance, or both. We suspect that Sae2p functions at both stages. Although palindromes are unstable in SAE2 cells (Lobachev et al. 2002; and data presented here), they can still be maintained (Lobachev et al. 1998, 2002), indicating that palindrome breakdown is not rampant. However, we only detected one DSB repair event that could be consistent with a palindrome from the analysis of Trp+ His+ Canr events from wild-type cells (representing <0.2% of all Trp+ Canr events) (Table 1). Since the frequency of palindromes in sae2Δ cells is >30-fold higher, perhaps their formation is inhibited in wild-type cells.
We note that Lig4p complex has also been shown to not be required for palindromic gene amplification in mice. Whereas Lig4-/- or XRCC4-/- (mammalian Lif4) mice are embryonic lethal, they are rescued by a p53 mutation. However, the doubly mutated mice undergo a rapid onset of genome rearrangements including palindromic amplification, leading to aggressive lymphomas. (Difilippantonio et al. 2002; Zhu et al. 2002).
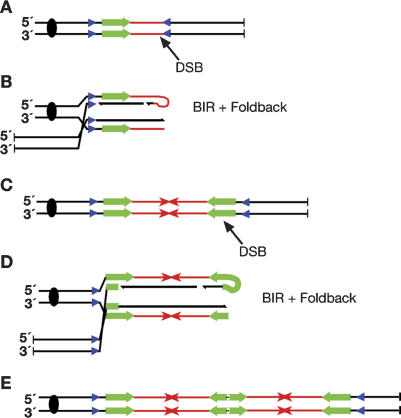
A model based on our data, depicted with dispersed inverted repeats used to initiate BIR, is shown in Figure 5. After a spontaneous break occurs in an unreplicated chromosome (Fig. 5A), an inverted repeat can invade and initiate BIR. Once BIR has extended toward the break site, foldback priming (Fig. 5B) would lead to palindromic fusion of the duplicated region (Fig. 5C). A second break within the duplicated region would lead to further cycles of BIR and foldback replication (Fig. 5D,E), resulting in further amplification of the array.
Figure 5.
Model for palindromic amplification using dispersed repeats. (A) A spontaneous chromosomal DSB can lead to invasion at an inverted dispersed repeat (blue triangles), which initiates BIR. (B) Extension by BIR toward the broken end followed by foldback priming can lead to palindrome formation. (C-E) Subsequent breakage cycles, possibly induced by the presence of the palindrome itself, can lead to further amplification of the array.
We suggest that once a palindrome is formed, the palindrome itself leads to chromosome instability and drives further amplification. Indeed, amplification of sequences that initiate as palindromes has been documented (Zhou et al. 2001; Lobachev et al. 2002; Tanaka et al. 2002). We suggest that it is the modulation of the MRX/N nuclease activity by a factor like Sae2p that drives amplification. Although breakage of a palindrome at the tip inhibits palindrome formation, breakage at the stem may provide the substrate necessary for subsequent rounds of amplification. Thus amplification may require turning the nuclease activity on and off. It will be interesting to determine whether cells somewhat deficient in the MRX/N complex, such as in ATLD and NBS syndromes (Taylor 2001) have a greater propensity for palindromic gene amplification events.
Materials and methods
Strains, plasmids, and general methods
For E. coli transformations, we used chemically competent Top10 F cells (Invitrogen) that were grown in LB supplemented with ampicillin and X-gal where necessary.
S. cerevisiae cells were grown in YEPD (Sherman et al. 1986) or the appropriate AA-synthetic drop-out media. AA drop-out media is similar to SD media described by Sherman et al. (1986) except that all amino acids, including myo-inositol, are at 85 μg/mL, except for leucine, which is at 170 μg/mL, and para-aminobenzoic acid, which is at 17 μg/mL. 5-FOA media was made as described (Boeke et al. 1987).
The strains used in this study are all derivatives of GRY1647, (MATα-inc::mush18/21 can1Δ::hisG his3-Δ200 leu2-Δ1 lys2Δ::hisG trp1Δ:: hisG ura3-52 tyr7-1 [pGalHO]), or YAR523 (a sae2Δ::KanMX derivative of GRY1647) whose construction has been described in detail (Rattray et al. 2001). The rad50Δ allele was created by transplacement with the appropriate fragment from pNKY83 resulting in strain YAR585 (Alani et al. 1987) the yku70Δ::KanMX allele was created by replacement of the wild-type gene with a PCR fragment derived from the S. cerevisiae knockout collection strain (Open Biosystems). The lig4::HphMX allele was first created by replacing KanMX from the lig4Δ::KanMX allele from the knockout collection with the HphMX allele as described (Malagon et al. 2004), and subsequently transferred into our strain background by PCR replacement. Double mutants were obtained by crosses.
Twenty of the palindromic events used in this study are all Trp+ His+ Canr derivatives of strain YAR523. Four additional mutants are Trp+ His+ Canr derived from YAR585. Palindrome stability was tested by crossing either palindromic or nonpalindromic Trp+ His+ Canr events from strain YAR523 (sae2Δ) with strain GRY1509 (SAE2) MATa-inc::URA3 ade2-101 his3-Δ200, leu2-Δ1 lys2Δ::hisG trp1Δ::hisG ura3-52 (McGill et al. 1993). Instability of the palindromic locus (adjacent to the MAT locus) was monitored by the ability of the resulting MATa/α diploid cells (by definition nonmaters) to acquire the ability to mate with tester strains. The mating tester strains were DC14 (MATa his1) and DC17 (MATα his1; Cold Spring Harbor Laboratory).
The oligonucleotides used in this study were synthesized by Invitrogen, and the sequences are provided in Supplementary Table S1. Oligonucleotides used to isolate the appropriate PCR fragment from the knockout collection are as described at the Saccharomyces Genome Deletion Project (http://www.sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). All constructions were verified by Southern blot analysis.
Fluctuation tests and Southern blot analysis
Fluctuation tests were done as previously described with a minimum of 11 independent colonies for each strain (Rattray et al. 2001). DNA was isolated from at least 20 independent Trp+ His+ Canr events from each strain and monitored for palindrome formation by Southern blot analysis as previously described (Rattray et al. 2001).
Sodium bisulfite modification and sequence analysis
Sodium bisulfite modification was done as described (Rattray 2004), except that we used ∼50 μg of genomic DNA as the starting material and doubled the volume of the reagents for the modification reaction. Sodium bisulfite DNA was PCR amplified with oligos specific for the modified DNA (Supplementary Table S1). After Topoisomerase-mediated cloning the products into pCR2.1 (Invitrogen), at least four independent clones were sequenced for each PCR product. The sequencing was done by the Laboratory of Molecular Technology (SAIC Corp.).
DNA sequences were read using the PHRED base-calling program. Sequence reads from each clone were assembled into Contigs using PHRAP (Ewing et al. 1998). The resulting contigs were then aligned using Sequencher software (GeneCodes Corp.). The deduced sequence of the palindromic junctions was then determined as described (Rattray 2004).
Vectorette PCR (Riley et al. 1990) was used for further confirmation of the sequence analysis. Unmodified genomic DNA from five independent palindromic junctions was first digested with BsrF1 and ligated to the BsrF1 vectorette bubble primers (Supplementary Table S1). The DNA was then PCR amplified with the primer “bubble amplify” and the appropriate unmodified oligonucleotides (Supplementary Table S1) as previously described (Rattray 2004).
Analysis of the inverted repeat sequences present in the 1.1-kb relevant region of the CAN1 gene was done using the EMBOSS PALINDROME program (http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html).
Acknowledgments
We thank P. Bullock, S. Lewis, and F. Malagon for their critical review of this manuscript, and M. Mills and D. Naugle for their administrative support. This work was sponsored by the National Cancer Institute, Department of Health and Human Services.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1315805.
References
- Alani E., Cao, L., and Kleckner, N. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116: 541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht E.B., Hunyady, A.B., Stark, G.R., and Patterson, T.E. 2000. Mechanisms of sod2 gene amplification in Schizosaccharomyces pombe. Mol. Biol. Cell 11: 873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D.E., Baggerly, K.A., and Kimmel, M. 1994. Gene amplification by unequal sister chromatid exchange: Probabilistic modeling and analysis of drug resistance data. J. Theor. Biol. 168: 151-159. [DOI] [PubMed] [Google Scholar]
- Baroni E., Viscardi, V., Cartagena-Lirola, H., Lucchini, G., and Longhese, M.P. 2004. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 24: 4151-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart, J., Natsoulis, G., and Fink, G.R. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164-175. [DOI] [PubMed] [Google Scholar]
- Bosco G. and Haber, J.E. 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150: 1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M. and Levine, M. 2004. A genome analysis of endoreplication in the Drosophila ovary. Dev. Cell 6: 4-5. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. and Jackson, S.P. 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15: 5093-5103. [PMC free article] [PubMed] [Google Scholar]
- Butler D.K., Yasuda, L.E., and Yao, M.C. 1995. An intramolecular recombination mechanism for the formation of the rRNA gene palindrome of Tetrahymena thermophila. Mol. Cell. Biol. 15: 7117-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D.K., Gillespie, D., and Steele, B. 2002. Formation of large palindromic DNA by homologous recombination of short inverted repeat sequences in Saccharomyces cerevisiae. Genetics 161: 1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Trujillo, K., Ramos, W., Sung, P., and Tomkinson, A.E. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8: 1105-1115. [DOI] [PubMed] [Google Scholar]
- Ciullo M., Debily, M.A., Rozier, L., Autiero, M., Billault, A., Mayau, V., El Marhomy, S., Guardiola, J., Bernheim, A., Coullin, P., et al. 2002. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: The model of PIP gene duplication from a break at FRA7I. Hum. Mol. Genet. 11: 2887-2894. [DOI] [PubMed] [Google Scholar]
- Claycomb J.M., Benasutti, M., Bosco, G., Fenger, D.D., and Orr-Weaver, T.L. 2004. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev. Cell 6: 145-155. [DOI] [PubMed] [Google Scholar]
- Cohen S., Hassin, D., Karby, S., and Lavi, S. 1994. Hairpin structures are the primary amplification products: A novel mechanism for generation of inverted repeats during gene amplification. Mol. Cell. Biol. 14: 7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.C. and Leach, D.R. 2002. Tethering on the brink: The evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27: 410-418. [DOI] [PubMed] [Google Scholar]
- D'Amours D. and Jackson, S.P. 2002. The Mre11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3: 317-327. [DOI] [PubMed] [Google Scholar]
- de Jager M., van Noort, J., van Gent, D.C., Dekker, C., Kanaar, R., and Wyman, C. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8: 1129-1135. [DOI] [PubMed] [Google Scholar]
- Difilippantonio M.J., Petersen, S., Chen, H.T., Johnson, R., Jasin, M., Kanaar, R., Ried, T., and Nussenzweig, A. 2002. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 196: 469-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudasova Z., Dudas, A., and Chovanec, M. 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28: 581-601. [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred, I: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Ford M. and Fried, M. 1986. Large inverted duplications are associated with gene amplification. Cell 45: 425-430. [DOI] [PubMed] [Google Scholar]
- Goedecke W., Eijpe, M., Offenberg, H.H., van Aalderen, M., and Heyting, C. 1999. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat. Genet. 23: 194-198. [DOI] [PubMed] [Google Scholar]
- Hellman A., Zlotorynski, E., Scherer, S.W., Cheung, J., Vincent, J.B., Smith, D.I., Trakhtenbrot, L., and Kerem, B. 2002. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 1: 89-97. [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Craig, L., Moncalian, G., Zinkel, R.A., Usui, T., Owen, B.A., Karcher, A., Henderson, B., Bodmer, C.T. McMurray, C.T., et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418: 562-566. [DOI] [PubMed] [Google Scholar]
- Kunes S., Botstein, D., and Fox, M.S. 1985. Transformation of yeast with linearized plasmid DNA: Formation of inverted dimers and recombinant plasmid products. J. Mol. Biol. 184: 375-387. [DOI] [PubMed] [Google Scholar]
- ____. 1990. Synapsis-mediated fusion of free DNA ends forms inverted dimer plasmids in yeast. Genetics 124: 67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.R. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893-900. [DOI] [PubMed] [Google Scholar]
- Lemoine F.J., Degtyareva, N.P., Lobachev, K., and Petes, T.D. 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587-598. [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler, K.W., and Vogelstein, B. 1998. Genetic instabilities in human cancers. Nature 396: 643-649. [DOI] [PubMed] [Google Scholar]
- Lewis L.K., Storici, F., Van Komen, S., Calero, S., Sung, P., and Resnick, M.A. 2004. Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166: 1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.T., Lin, W.H., Lyu, Y.L., and Whang-Peng, J. 2001. Inverted repeats as genetic elements for promoting DNA inverted duplication: Implications in gene amplification. Nucleic Acids Res. 29: 3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K.S., Shor, B.M., Tran, H.T., Taylor, W., Keen, J.D., Resnick, M.A., and Gordenin, D.A. 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K.S., Gordenin, D.A., and Resnick, M.A. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183-193. [DOI] [PubMed] [Google Scholar]
- Malagon F., Tong, A.H., Shafer, B.K., and Strathern, J.N. 2004. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation-elongation transition. Genetics 166: 1215-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L. and Lydall, D. 2004. Telomerase- and recombination-independent immortalization of budding yeast. Genes & Dev. 18: 2663-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill C.B., Shafer, B.K., Derr, L.K., and Strathern, J.N. 1993. Recombination initiated by double-strand breaks. Curr. Genet. 23: 305-314. [DOI] [PubMed] [Google Scholar]
- McKee A.H. and Kleckner, N. 1997. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146: 797-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody E.M. and Bevilacqua, P.C. 2003. Thermodynamic coupling of the loop and stem in unusually stable DNA hairpins closed by CG base pairs. J. Am. Chem. Soc. 125: 2032-2033. [DOI] [PubMed] [Google Scholar]
- Moreau S., Ferguson, J.R., and Symington, L.S. 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19: 556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M.J., Ramachandran, M., Trelles-Sticken, E., Scherthan, H., and Goldman, A.S. 2002. Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell 9: 835-846. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Hettema, E., Wust, D., Fase-Fowler, F., and Borst, P. 1991. Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J. 10: 1009-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C.E., Dorsey, M., Crable, S., Sprinkel, K., Sondej, M., and Williamson, V.M. 1992. A spontaneous chromosomal amplification of the ADH2 gene in Saccharomyces cerevisiae. Genetics 130: 263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson T.G., Almasan, A., Brody, L.L., and Wahl, G.M. 1998. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol. Cell. Biol. 18: 3089-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipiras E., Coquelle, A., Bieth, A., and Debatisse, M. 1998. Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J. 17: 325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Amon, A., and Klein, F. 1997. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146: 781-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z. and Cohen, S.N. 2000. Long palindromes formed in Streptomyces by nonrecombinational intra-strand annealing. Genes & Dev. 14: 1789-1796. [PMC free article] [PubMed] [Google Scholar]
- Rattray A.J. 2004. A method for cloning and sequencing long palindromic DNA junctions. Nucleic Acids Res. 32: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A.J., McGill, C.B., Shafer, B.K., and Strathern, J.N. 2001. Fidelity of mitotic double-strand break repair in Saccharomyces cerevisiae: A role for SAE2/COM1. Genetics 158: 109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J., Butler, R., Ogilvie, D., Finniear, R., Jenner, D., Powell, S., Anand, R., Smith, J.C., and Markham, A.F. 1990. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18: 2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R.T., Kaufman, R.J., Alt, F.W., and Kellems, R.F. 1978. Gene amplification and drug resistance in cultured murine cells. Science 202: 1051-1055. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink, G., and Hicks, J. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Stark G.R., Debatisse, M., Giulotto, E., and Wahl, G.M. 1989. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57: 901-908. [DOI] [PubMed] [Google Scholar]
- Sutherland G.R. and Richards, R.I. 1995. The molecular basis of fragile sites in human chromosomes. Curr. Opin. Genet. Dev. 5: 323-327. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Tapscott, S.J., Trask, B.J., and Yao, M.C. 2002. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. 99: 8772-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Bergstrom, D.A., Yao, M.C., and Tapscott, S.J. 2005. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 37: 320-327. [DOI] [PubMed] [Google Scholar]
- Taylor A.M. 2001. Chromosome instability syndromes. Best. Pract. Res. Clin. Haematol. 14: 631-644. [DOI] [PubMed] [Google Scholar]
- Thomas L., Stamberg, J., Gojo, I., Ning, Y., and Rapoport, A.P. 2004. Double minute chromosomes in monoblastic (M5) and myeloblastic (M2) acute myeloid leukemia: Two case reports and a review of literature. Am. J. Hematol. 77: 55-61. [DOI] [PubMed] [Google Scholar]
- Varani G. 1995. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 24: 379-404. [DOI] [PubMed] [Google Scholar]
- Windle B., Draper, B.W., Yin, Y.X., O'Gorman, S., and Wahl, G.M. 1991. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes & Dev. 5: 160-174. [DOI] [PubMed] [Google Scholar]
- Yunis J.J., Soreng, A.L., and Bowe, A.E. 1987. Fragile sites are targets of diverse mutagens and carcinogens. Oncogene 1: 59-69. [PubMed] [Google Scholar]
- Zhou Z.H., Akgun, E., and Jasin, M. 2001. Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl. Acad. Sci. 98: 8326-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Mills, K.D., Ferguson, D.O., Lee, C., Manis, J., Fleming, J., Gao, Y., Morton, C.C., and Alt, F.W. 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109: 811-821. [DOI] [PubMed] [Google Scholar]