Abstract
Human immunodeficiency virus type 1 (HIV-1) entry is triggered by the interaction of the gp120 envelope glycoprotein with a cellular chemokine receptor, either CCR5 or CXCR4. We have identified different mutations in human CXCR4 that prevent efficient infection by one HIV-1 strain (NDK) but not another (LAI) and sought to define these strain-dependent effects at the gp120 level. The lack of activity toward the NDK strain of the HHRH chimeric CXCR4 in which the second extracellular loop (ECL2) derived from the rat CXCR4 and of CXCR4 with mutations at an aspartic acid in ECL2 (D193A and D193R) was apparently due to the sequence of the third variable loop (V3) of gp120, more precisely, to its C-terminal part. Indeed, substitution of the LAI V3 loop or only its C-terminal part in the NDK gp 120 context was sufficient to restore usage of the HHRH, D193A, and D193R receptors. The same result was achieved upon mutation of a single lysine residue of the NDK V3 loop to alanine (K319A) but not to arginine (K319R). These results provide a strong case for a direct interaction between the gp120 V3 loop and the ECL2 domain of CXCR4. By contrast, V3 substitutions had no effect on the inability of NDK to infect cells via a mutant CXCR4 in which the amino-terminal extracellular domain (NT) is deleted. In experiments with a set of chimeric NDK-LAI gp120s, the V1/V2 region from LAI gp120 was both necessary and sufficient for usage of the NT-deleted CXCR4. Different variable domains of gp120 can therefore cooperate for a functional interaction with CXCR4.
The envelope glycoproteins of human immunodeficiency virus type 1 (HIV-1) consist of trimeric complexes of a surface (gp120) and a transmembrane (gp41) subunit and mediate fusion of the viral envelope with the target cell membrane (reviewed in references 21, 37, and 57). To achieve this goal, the gp120-gp41 complex must undergo conformation changes which are not yet understood in their molecular details but are known to be triggered, in most situations, by the interaction of gp120 with two components of the cell membrane, CD4 and a chemokine receptor, either CCR5 or CXCR4 (reviewed in references 2, 18, and 44). These chemokine receptors can therefore be viewed as CD4-associated coreceptors for HIV-1. Viral strains dependent on CCR5 or CXCR4 or capable of using either of them are termed R5, X4, and R5X4, respectively. While R5 strains can be isolated at all stages of HIV-1 infection in vivo, R5X4 and X4 strains generally emerge at later stages, marked by a decline of the immune defenses (17, 38, 45). Several types of CCR5 and CXCR4 ligands can prevent the interaction with gp120 and block HIV-1 infection (4, 15, 24, 39, 49). To pursue this type of antiviral strategy, it is important to gain a better understanding of the interaction of gp120 with chemokine coreceptors and of its consequences on the HIV-1 entry process. Among other means, this can be achieved by functional studies involving mutant forms of chemokine receptors and HIV-1 envelope proteins.
There is relatively limited information on how gp120 binds chemokine receptors. Elucidation of the crystal structure of the gp120 core (30) allowed definition of a CCR5-binding site formed by juxtaposition of residues from different conserved domains (43). It is not known if these conserved residues of gp120 also participate in the interaction with CXCR4. Regions of gp120 that display extreme genetic variation among HIV-1 strains, in particular the third hypervariable loop (V3), seem to contribute in an important way in the interaction of gp120 with the chemokine receptors. First, the CCR5-binding site seems to be masked by variable loops V3 and V2 until gp120 has interacted with CD4 (43). Second, there was no detectable binding to CCR5 of a recombinant gp120 with a V3 loop deletion (56). Finally, there are numerous observations that the primary sequence of the V3 and to a lesser extent the V1/V2 loops can determine the selectivity of HIV-1 strains for CCR5 or CXCR4 (13, 26, 27, 35, 47, 51); likewise, it determines biological properties, such as tropism for macrophages or cell lines and ability to induce syncytia in infected cell cultures, which are largely dependent on coreceptor choice (reviewed in references 2 and 25). On the cellular side, the absence of information on the spatial structure of chemokine receptors is a major obstacle to determining their interaction with gp120. Indirect information was mainly gathered from functional studies performed with mutant forms of CXCR4 or CCR5. For both receptors, the amino-terminal domain (NT) and the second extracellular loop (ECL2) seem to play a critical role in the interaction with HIV-1, as evidenced by the effect of mutations or domain swapping with other chemokine receptors (reviewed in references 3 and 44). These effects often vary between HIV-1 strains, indicating a certain flexibility in the interaction.
Here we took advantage of such strain-dependent effects of mutations in CXCR4 to address its interaction with HIV-1. Cells expressing ECL2 mutants or an NT-deleted form of CXCR4 could be infected by HIV-1 strain LAI but not by HIV-1 strain NDK (6, 8, 41). We found that the inability of the NDK strain to use the ECL2 mutants resulted from differences with the LAI strain in the level of the V3 loop, while the lack of infection via the NT-deleted CXCR4 was due to differences in the V1/V2 region. These results suggest that the variable domains of gp120 can both contribute to the interaction with CXCR4.
MATERIALS AND METHODS
Cell lines and viruses.
The CD4+ human astroglioma cell line U373MG-CD4, stably transfected with the Escherichia coli β-galactosidase gene (lacZ) under transcriptional control of the HIV-1 long terminal repeat (LTR) (23), and derivatives stably expressing the human or rat CXCR4 (31) have been described. U373MG-CD4 cells were cotransfected with the D193A or D193R human CXCR4 expression vectors (see below) and a vector allowing selection with puromycin. Individual clones were screened for their ability to fuse with cells expressing HIV-1 envelope proteins as described previously (42). Viral stocks of HIV-1 strains LAI (40), NDK (52), and 89.6 (16), as well as LAI and NDK derivatives with mutant env genes (see below), were produced by transient transfection of HeLa cells with molecularly cloned proviruses. Stocks of HIV-1 strains GUN-1 (48) (a gift from R. Weiss). OUA, and ATE (50) were supernatants of acutely infected T cells. All infectious titers were determined by scoring β-galactosidase-positive cells 24 h after infection of LTRlacZ HeLa-P4 cells (14).
Plasmid vectors.
All forms of CXCR4 were expressed from the cytomegalovirus immediate-early promoter. The vectors allowing expression of the wild-type (WT) human (H) and rat (R) CXCR4 (41), the HHRH and RRHR chimeras, and the Δ4-36 CXCR4 (8), D193A, and D193R human CXCR4 mutants (6) have been described. All HIV-1 envelope proteins (Env) were expressed in an LAI provirus with a gag-pol deletion (46). The WT LAI Env and chimeric LAI-V3NDK Env vectors have been described (32). The NDK Env vector (33) actually allows expression of a chimeric Env with gp120 and the gp41 ectodomain from NDK and the rest of gp41 from LAI (see Fig. 6A for the structure of the chimeric Env). A ClaI restriction site created in the V3-encoding region of LAI env at nucleotide (nt) 6471 (provirus sequence) was used to derive the LAI-V3LN and LAI-V3NL constructs by ligating SalI (nt 5320)-ClaI and ClaI-BamHI (nt 8068) PCR fragments amplified from LAI env or from LAI-V3NDK env, respectively. Restriction sites flanking V1/V2 (PstI, nt 6192, and SpeI, nt 6409) and V3 (MluI, nt 6706, and SmaI, nt 6818) created in the LAI env gene allowed exchanges with PCR fragments amplified from the NDK env. Site-directed mutagenesis was performed on an NDK env subclone with MluI and SmaI sites flanking V3 (32), allowing substitution of mutant NDK V3 into either NDK or LAI Env expression vectors. For infection assays, LAI and NDK proviruses with V3 substitutions (LAI-V3NDK and NDK-V3LAI) have been described (41). All other mutant env were subcloned in an LAI provirus (40).
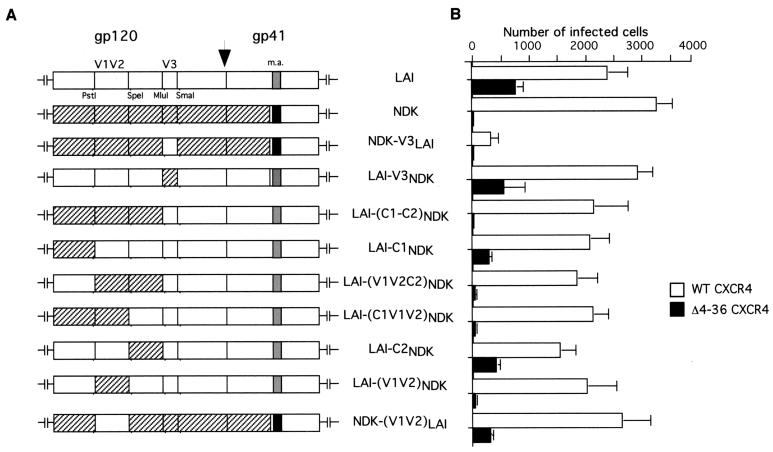
FIG. 6.
Effect of exchanges between the envelope proteins of LAI and NDK on usage of the NT-deleted CXCR4. (A) Schematic organization of chimeric env genes, with restriction sites created to allow substitutions. m.a., membrane anchor domain of gp41. (B) Infections of cells stably expressing human CXCR4 or the Δ4-36 mutant with LAI or NDK virus or recombinant LAI viruses bearing the indicated env gene, performed and scored as in Fig. 5.
Infection assays.
Infections of subconfluent U373MG-CD4 cells were performed in 12-well trays with approximately 103 infectious units (I.U.) per well 24 h after transfection with WT or mutant CXCR4 expression vectors as described (41). Cells were washed and fixed in 0.5% glutaraldehyde 48 h after infection and stained for β-galactosidase activity with the X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) substrate as described previously (19). Blue-stained foci were scored under 20× magnification. Cell counts of >200 were obtained by extrapolation from randomly selected fields.
Cell fusion assays.
Subconfluent monolayers of HeLa cells in six-well trays (∼2 × 105 cells per well) were transfected with Env expression vectors by calcium phosphate precipitation. An equivalent amount of U373MG-CD4 cells stably expressing WT or mutant human or rat CXCR4 freshly detached by trypsinization was added 20 h after transfection. After overnight coculture, adherent cells were fixed in 0.5% glutaraldehyde and stained with X-Gal. Blue-stained foci were scored under 20× magnification.
RESULTS
V3 loop and ECL2 substitutions.
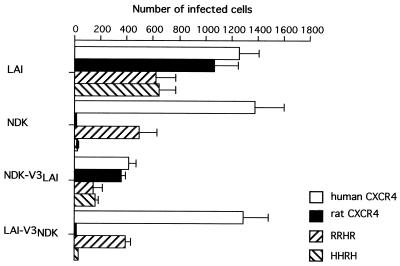
The HIV-1 coreceptor activity of different forms of CXCR4, i.e., their ability to allow infection of CD4+ cells, was tested in the human astroglioma cell line U373MG-CD4, which is naturally resistant to HIV-1 entry and to fusion with cells expressing the HIV-1 envelope proteins (23). Cells transfected to express the human (H) or the rat (R) CXCR4 or the derived HHRH and RRHR chimeric receptors corresponding to reciprocal ECL2 substitutions could be infected by the LAI HIV-1 strain, while infection by the NDK HIV-1 strain was detected only for cells expressing human CXCR4 or the RRHR chimera (Fig. 1). Previous studies have shown that these different forms of CXCR4 were expressed at a similar level at the surface of transfected cells (6). In agreement with our earlier observations (41), substitution of the NDK V3 loop in the LAI gp 120 context yielded a chimeric virus (LAI-V3NDK) infectious for cells expressing the human but not the rat CXCR4, while the chimeric NDK-V3LAI virus (NDK gp 120 with the V3 loop from LAI) could infect cells via the human or the rat CXCR4 (Fig. 1). The V3 substitutions also modified the ability of LAI or NDK to infect cells via the chimeric forms of CXCR4, HHRH, and RRHR (Fig. 1). The efficiency of infection by the NDK-V3LAI virus was relatively low whether cells expressed chimeric or wild-type receptors. This suggests that this V3 substitution impairs the function of the gp120-gp41 complex in a way that is independent of CXCR4. In these experiments, the NDK V3 loop determined a strict dependence for receptors with the human CXCR4 ECL2, while the V3 loop of LAI was required for usage of receptors with the rat CXCR4 ECL2 and could confer this property on the gp120 of a genetically divergent strain (LAI and NDK belong to clades B and D, respectively).
FIG. 1.
Infection of U373MG-CD4 cells expressing human or rat CXCR4 or the derived chimeric receptors HHRH and RRHR by HIV-1 strains LAI and NDK or derivatives with substitutions of the gp120 V3 loop. Cells were infected with approximately 1,000 I.U. per well (12-well plates) and stained with X-Gal after another 48 h. Blue-stained cells, representing HIV-1-infected cells, were scored in duplicate wells. Bars represent means of three independent transfections.
This analysis was refined by substituting the N-terminal or the C-terminal part of the V3 loop of LAI with the homologous region from the NDK V3, yielding the LAI-V3NL and LAI-V3LN chimeric Envs, respectively (Fig. 2). Both forms of Env mediated HIV-1 infection or syncytium formation with U373MG-CD4 cells expressing human CXCR4, but only the LAI-V3NL Env allowed infection and fusion when cells expressed the rat CXCR4 (Fig. 3A and B). Sequence differences in the distal part of the V3 loop therefore seem to account for the phenotypic differences between LAI and NDK in usage of the rat CXCR4 as a coreceptor.
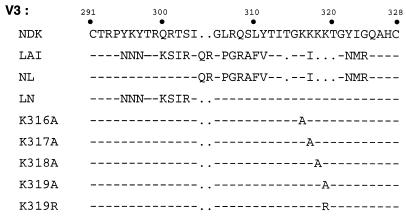
FIG. 2.
Alignment of the amino acid sequence of gp120 V3 loops of NDK and LAI strains and derived mutants. Numbering is according to the NDK gp120 sequence (52).
FIG. 3.
Effect of V3 substitutions and mutations on usage of rat and human CXCR4. (A) Infections of U373MG-CD4 cells stably expressing human or rat CXCR4 were performed and scored as described in the legend to Fig. 1. (B) Syncytium formation assays between these cell lines and HeLa cells transfected with Env expression vectors with the indicated type of gp120. Cocultures were performed in six-well plates, and cells were stained with X-Gal after 24 h. Blue-stained foci representing syncytia were scored in duplicate wells. Bars represent means of three independent transfections.
Mutagenesis of NDK V3.
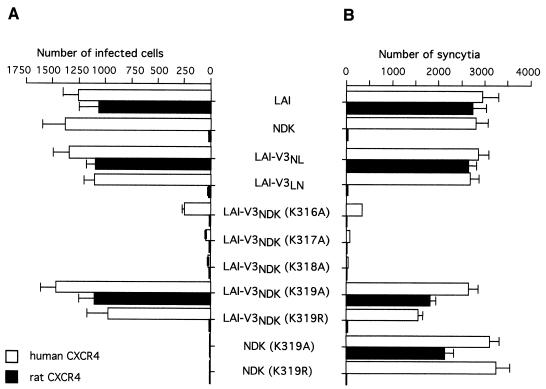
A striking feature of the NDK V3 loop in its distal moiety is a cluster of four adjacent lysine residues (positions 316 to 319), while only one lysine is found in the corresponding region of the LAI V3 loop (Fig. 2). Each of these four lysine residues of the NDK V3 was replaced with an alanine, and the resulting mutant V3 was inserted in the LAI gp120 context. The function of these chimeric envelope proteins was tested in HIV-1 infection (Fig. 3A) and cell fusion (Fig. 3B) assays. The K317A and K318A mutations were apparently not compatible with these Env functions. The K316A mutation markedly reduced the efficiency of infection and fusion with cells expressing human CXCR4 and did not seem to confer ability to use the rat CXCR4. In contrast, the K319A mutation was compatible with efficient infection or fusion with cells expressing the human CXCR4 and also with cells expressing the rat CXCR4. When the lysine residue was replaced with an arginine (K319R), the corresponding chimeric LAI-V3NDK gp120 could only mediate HIV-1 infection and fusion with cells expressing human CXCR4, not rat CXCR4. In the NDK gp120 context, the K319A and K319R mutations resulted in a complete loss of function in infection assays (Fig. 3A) but not in syncytium formation assays (Fig. 3B). In that case, the K319A but not K319R mutation restored usage of the rat CXCR4 by the NDK gp120. The presence of a positively charged residue at a defined position in the V3 loop of gp120 apparently explained why the NDK HIV-1 strain cannot functionally interact with the rat CXCR4. Why the mutant NDK Env can mediate cell-cell fusion but not HIV-1 infection has not been determined, but mutations in the V3 loop can reduce the stability of the gp120-gp41 complex (55). This can result in gp120 shedding from virions and loss of infectivity. But even unstable gp120-gp41 complexes can probably mediate syncytium formation, provided they reach the cell surface.
Usage of ECL2 mutants.
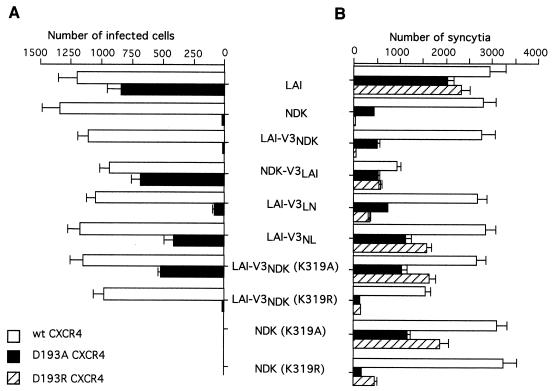
We have previously shown that a negatively charged aspartic acid residue (Asp193) in the ECL2 of the human CXCR4 was critical for a functional interaction with the NDK strain (6). The presence of a neutral asparagine residue at the corresponding position in the ECL2 of the rat CXCR4 could therefore explain its lack of coreceptor activity for NDK. We have tested the ability of LAI and NDK gp120 with V3 loop substitutions or mutations at the Lys319 residue to mediate infection of U373MG-CD4 cells stably expressing human CXCR4 with mutation of the Asp193 residue to alanine (D193A) (Fig. 4A). This mutant CXCR4 enabled infection by the LAI and NDK-V3LAI viruses, but not the NDK or LAI-V3NDK virus. Also, infection via the D193A receptor was markedly more efficient for the LAI-V3NL virus, in which the first part of the V3 loop derived from NDK and the second from LAI, than for the LAI-V3LN virus. The distal moiety of the LAI V3 therefore seemed important for usage of human CXCR4 with the Asp193 mutation, likewise for usage of the rat CXCR4 and HHRH chimera.
FIG. 4.
Effect of V3 substitutions and mutations on usage of human CXCR4 and the derived ECL2 mutants D193A and D193R. Infections (A) and syncytium formation assays (B) were performed and scored as described in the legend to Fig. 3.
Cells expressing the D193A mutant CXCR4 could be infected by a chimeric LAI-V3NDK virus with the K319A but not K319R mutation in the NDK V3 loop (Fig. 4A). Similar results were obtained in syncytium formation assays performed with cells expressing these different HIV-1 envelope proteins and U373MG-CD4 cells stably expressing human CXCR4 with a D193A or D193R mutation (Fig. 4B). The latter CXCR4 mutant was used because it yielded clearer results. Indeed, cells expressing the D193A mutant were not completely resistant to fusion mediated by the NDK or LAI-V3NDK Env. If a negative charge at position 193 in ECL2 is important for function, it was not unexpected that substitution with a positively charged residue (D193R) would have a stronger effect than substitution with a neutral residue (D193A). In cell fusion assays, we could also monitor the effect of the K319A and K319R mutations in the NDK Env context (Fig. 4B). Again, the inability to use the D193A or D193R mutant CXCR4 appeared to be related to the presence of a positively charged residue at position 319 in the V3 loop of the NDK strain.
Usage of NT-deleted CXCR4.
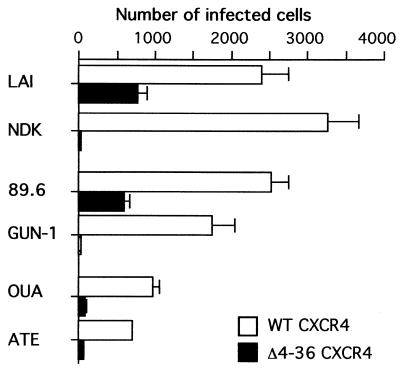
The deletion of 32 of the 39 residues of the amino-terminal domain (NT) of CXCR4 is compatible with coreceptor activity for the LAI HIV-1 strain (8). The efficiency of infection mediated by this mutant (Δ4-36) or the WT CXCR4 was in proportion of their levels of cell surface expression (8), suggesting that the NT domain of CXCR4 is dispensable for a functional interaction with the gp120 of the LAI strain. Cells stably expressing the Δ4-36 CXCR4 could be infected by the HIV-1 strain 89.6 (R5X4) but were resistant to infection by the NDK (X4) and GUN-1 (R5X4) strains and almost resistant to infection by the primary X4 isolates OUA and ATE (Fig. 5). Among this limited set of HIV-1 strains, there was no apparent correlation between the ability to use the NT-deleted CXCR4 and a strictly X4 or R5X4 phenotype, or with the genetic subtype, since LAI, GUN-1, and 89.6 all belong to env subtype B.
FIG. 5.
Effect of a deletion in the amino-terminal domain (NT) of CXCR4 on its coreceptor activity for different HIV-1 strains. Infections of U373MG-CD4 cells stably expressing human CXCR4 or the Δ4-36 mutant were performed in 12-well plates with approximately 2,000 I.U. per well. Cells were stained with X-Gal 48 h later, and blue-stained cells were scored in duplicate wells.
The V3 loop sequence had no apparent role in the ability of LAI to use the NT-deleted form of CXCR4, since cells expressing the Δ4–36 mutant could be infected with similar efficiency by the LAI and NDK-V3LAI viruses and were equally resistant to infection by the NDK and LAI-V3NDK viruses (Fig. B). We sought to determine whether a defined region of gp120 (or the gp41 ectodomain) was responsible for the ability of the LAI strain to use the NT-deleted form of CXCR4, likewise V3 for the ECL2 mutants. We tested the ability of a set of LAI/NDK chimeric Envs (Fig. 6A) to mediate HIV-1 infection of U373MG-CD4 cells stably expressing either WT or Δ4–36 CXCR4 (Fig. 6B). Results with the LAI-(C1-C2)NDK chimera showed that the region of LAI gp120 which is N-terminal to V3, i.e., the C1 and C2 conserved domains and the interventing V1/V2 variable loops, was required for usage of the NT-deleted CXCR4. Replacing the C1 or C2 domain of LAI gp120 with the corresponding NDK domain (LAI-C1NDK and LAI-C2NDK, respectively) reduced the efficiency of infection of cells expressing Δ4–36 CXCR4 but did not abolish it. In contrast, there was no detectable infection of these cells when the V1/V2 region of LAI gp120 was absent, for example, with the LAI-(C1-V1V2)NDK or the LAI-(V1V2)NDK chimera. We next found that substitution of the V1/V2 region from LAI into the NDK gp120 context (NDK-V1V2LAI chimera) was sufficient to restore detectable infection of cells expressing the Δ4–36 CXCR4. The V1/V2 region of the LAI gp120 was both necessary and sufficient to confer usage of the NT-deleted CXCR4 in the genetically distant context of the NDK gp120. Other regions of gp120 or the gp41 ectodomain apparently had no role in the phenotype difference between LAI and NDK.
DISCUSSION
Mutations or substitutions in the extracellular domains of the CCR5 and CXCR4 chemokine receptors can reduce or abolish their HIV-1 coreceptor activity, that is, their ability to mediate HIV-1 entry in CD4+ cells. The effects of such mutations can be very different in different HIV-1 strains, suggesting that not all gp120 envelope glycoproteins have the same requirements for a functional interaction with their coreceptors. This may represent an obstacle to antiviral approaches but can also be a source of information on the gp120-coreceptor interaction. Here we have used two CXCR4-dependent (X4) HIV-1 strains, LAI and NDK, that have relatively divergent gp120s and display clear-cut phenotypic differences in usage of certain forms of CXCR4. The NDK strain cannot infect CD4+ cells via (i) the rat CXCR4 (41), (ii) the HHRH chimeric human CXCR4, in which the second extracellular loop (ECL2) derives from the rat CXCR4 (8), (iii) human CXCR4 with mutations at position 193 in ECL2 (D193A or D193R) (6), or (iv) human CXCR4 with an almost complete deletion of the amino-terminal extracellular domain (NT) (7). These four types of CXCR4 were functional coreceptors for the LAI strain. We had already observed that the ability to use the rat CXCR4 as a coreceptor could be conferred on NDK by replacing the third variable loop (V3) with the homologous region of LAI gp120 (41). Here we found that this V3 substitution was also sufficient to allow usage of the HHRH chimeric receptor and the D193A and D193R CXCR4 mutants, but not the NT-deleted CXCR4. It confirmed the importance of the V3 loop of gp120 for a functional interaction with CXCR4 and showed that other domains of gp120 contribute to this interaction.
The role of the V3 loop was further studied by exchanging fragments of V3 between the gp120 of LAI and NDK and by site-directed mutagenesis in the NDK V3 loop. In the LAI gp120 context, only the C-terminal part of V3 appeared to be necessary for infection of cells via the rat CXCR4, the HHRH chimera, and the D193A and D193R mutants. The importance of the C-terminal part of V3 for usage of CXCR4 is in agreement with a recent study based on chimeric gp120 derived from X4 and R5 HIV-1 strains (27). The usage of these three types of mutant CXCR4 coreceptors was restored by the mutation of a lysine residue of the NDK V3 loop to alanine (K319A) but not to arginine (K319R). These results were consistent with some form of interaction between the V3 loop of the NDK gp120 and the ECL2 domain of CXCR4. The LAI V3 loop could have different requirements for its interaction with ECL2; in particular, it could be independent of the Asp193 residue. Alternatively, the LAI V3 loop could allow an interaction with CXCR4 that is totally independent of the ECL2 domain. We consider the second possibility less likely, given the negative effect of other ECL2 mutations on HIV-1 infection (6, 7) and its blocking by reagents targeting the ECL2 domain of CXCR4, such as the AMD3100 bicyclam (31) and the 12G5 monoclonal antibody (8).
There is evidence for the accumulation of basic amino acids in the V3 loop of X4 HIV-1 strains (20, 29). These positively charged residues could allow electrostatic interactions with the negatively charged residues of CXCR4 that are apparently involved in HIV-1 coreceptor activity and located in the NT or ECL2 domain (10, 11, 28, 54). In line with this view, it could be proposed that contact between the Lys319 residue of the NDK V3 loop and the Asp193 residue of ECL2 is critical for a functional interaction of gp120 and CXCR4. However, for the effect of the K319A mutation to fit with this model, one also has to envision that the basic residue Lys319 in the NDK V3 loop is detrimental to the interaction with CXCR4 in the absence of a negatively charged Asp193 residue in ECL2, which is the case for the rat CXCR4 (Ser). Alternatively, the K319A mutation could indirectly affect the structure of the NDK V3 loop, allowing it to cope with mutations at the Asp193 residue, likewise the LAI V3 loop.
The V3 loop sequence had no apparent role in the ability of the LAI strain to infect cells via an NT-deleted form of CXCR4. This property was apparently linked to the first variable loops V1/V2 of the gp120, which suggested their role in the functional interaction of gp120 with CXCR4. This is in apparent contradiction to the observed binding to CXCR4 of a recombinant gp120 deleted in the V1/V2 region (36). Two types of explanations can be proposed. First, the monomeric gp120 used in direct binding assays may behave differently from the trimeric gp120 complexed to gp41 involved in functional assays. Second, the V1/V2 region could have a minor role in terms of binding of gp120 to CXCR4, as it can be physically measured but still be important for a correct positioning and hence a functional interaction. In the case of the NDK gp120, it can be postulated either that the V1/V2 region directly interacts with the NT region of CXCR4 or that it has no role in the interaction with CXCR4. We lack experimental evidence to sort out these possibilities, but the former is indirectly supported by several reports involving the V1 or V2 region of gp120 in the tropism of different HIV-1 strains for T-cell lines and hence their adaptation to the CXCR4 coreceptor (1, 5, 9, 22). More recently, substitution of either the V1/V2 or the V3 region from a dualtropic strain (R5X4) in the context of a macrophage-tropic (R5) strain was found sufficient to confer ability to utilize the CXCR4 coreceptor (12). It can therefore be proposed that the V1/V2 and V3 variable loops of gp120 cooperate for interaction with the extracellular loops of CXCR4 (LAI strain) or with the loops and NT domain (NDK strain).
An important issue that could not be addresed in this study is the possible contribution to the interaction with CXCR4 of the conserved gp120 residues forming the putative CCR5 binding site (43). Does this site contribute to the interaction with CXCR4, particularly in R5X4 strains, or is it dispensable, particularly for strictly X4 strains? These questions will probably be answered by functional assays with a set of gp120 mutants. More generally, it can be wondered if the ability of CCR5 or CXCR4 — and other chemokine receptors in certain experimental conditions — to mediate HIV-1 entry is due only to their ability to bind gp120, itself dependent on the structure of their extracellular domains, or is also dependent on other features, for example, the colocalization of chemokine receptors with CD4 (53) or their monomeric versus dimeric status (34).
Differences in the interaction of HIV-1 strains with CXCR4 could represent an obstacle to the development of CXCR4 ligands as antiviral agents. The NT domain of CXCR4 contains a high affinity binding site for the SDF-1 chemokine, but signaling requires binding to a conformational site formed by residues of the extracellular loops and membrane-spanning domains (7). Natural or synthetic CXCR4 ligands binding to the NT domain of CXCR4 may have limited antiviral efficacy for HIV-1 strains such as LAI and 89.6. On the other hand, ligands targeting the extracellular loops of CXCR4 could have a broader spectrum of antiviral activity but be more likely to interfere with the natural signaling function. This should be kept in view for the selection or design of inhibitors of the CXCR4 coreceptor.
ACKNOWLEDGMENTS
We thank F. Letourneur (ICGM) for assistance with DNA sequencing.
This work was supported by the Agence Nationale de Recherche sur le SIDA.
REFERENCES
- 1.Andeweg A C, Leeflang P, Osterhaus A D, Bosch M L. Both the V2 and V3 regions of the human immunodeficiency virus type 1 surface glycoprotein functionally interact with other envelope regions in syncytium formation. J Virol. 1993;67:3232–3239. doi: 10.1128/jvi.67.6.3232-3239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Berson J F, Doms R W. Structure-function studies of the HIV-1 coreceptors. Semin Immunol. 1998;10:237–248. doi: 10.1006/smim.1998.0130. [DOI] [PubMed] [Google Scholar]
- 4.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 5.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brelot A, Heveker N, Adema K, Hosie M J, Willett B, Alizon M. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J Virol. 1999;73:2576–2586. doi: 10.1128/jvi.73.4.2576-2586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736–23744. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 8.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo A, Ratner L. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J Virol. 1996;70:1301–1309. doi: 10.1128/jvi.70.2.1301-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chabot D J, Chen H, Dimitrov D S, Broder C C. N-linked glucosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J Virol. 2000;74:4404–4413. doi: 10.1128/jvi.74.9.4404-4413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabot D J, Zhang P F, Quinnan G V, Broder C C. Mutagenesis of CXCR4 identifies important domains for human immunodeficiency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J Virol. 1999;73:6598–6609. doi: 10.1128/jvi.73.8.6598-6609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 16.Collman R. Human immunodeficiency virus type 1 tropism for human macrophages. Pathobiology. 1992;60:213–218. doi: 10.1159/000163725. [DOI] [PubMed] [Google Scholar]
- 17.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrov D S, Xiao X, Chabot D J, Broder C C. HIV coreceptors. J Membr Biol. 1998;166:75–90. doi: 10.1007/s002329900450. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Hazan U, Alizon M. Detection of cell fusion mediated by the envelopes of human retroviruses by transactivation of a reporter gene. Methods Mol Genet. 1995;7:218–236. [Google Scholar]
- 20.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freed E O, Martin M A. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 22.Groenink M, Fouchier R A, Broersen S, Baker C H, Koot M, van't Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 23.Harrington R D, Geballe A P. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J Virol. 1993;67:5939–5947. doi: 10.1128/jvi.67.10.5939-5947.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heveker N, Montes M, Germeroth L, Amara A, Trautmann A, Alizon M, Schneider-Mergener J. Dissociation of the signalling and antiviral properties of SDF-1-derived small peptides. Curr Biol. 1998;8:369–376. doi: 10.1016/s0960-9822(98)70155-1. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman T L, Doms R W. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol Membr Biol. 1999;16:57–65. doi: 10.1080/096876899294760. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type 1 envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung C S, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J Virol. 1999;73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kajumo F, Thompson D A, Guo Y, Dragic T. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by negatively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology. 2000;271:240–247. doi: 10.1006/viro.2000.0308. [DOI] [PubMed] [Google Scholar]
- 29.Kuiken C L, de Jong J J, Baan E, Keulen W, Tersmette M, Goudsmit J. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J Virol. 1992;66:4622–4627. doi: 10.1128/jvi.66.7.4622-4627.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrosse B, Pleskoff O, Sol N, Jones C, Henin Y, Alizon M. Resistance to a drug blocking human immunodeficiency virus type 1 entry (RPR103611) is conferred by mutations in gp41. J Virol. 1997;71:8230–8236. doi: 10.1128/jvi.71.11.8230-8236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrosse B, Treboute C, Alizon M. Sensitivity to a nonpeptidic compound (RPR103611) blocking human immunodeficiency virus type 1 Env-mediated fusion depends on sequence and accessibility of the gp41 loop region. J Virol. 2000;74:2142–2150. doi: 10.1128/jvi.74.5.2142-2150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapham C K, Zaitseva M B, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. . (Erratum, 5:590.) [DOI] [PubMed] [Google Scholar]
- 35.Lee M K, Heaton J, Cho M W. Identification of determinants of interaction between CXCR4 and gp120 of a dual-tropic HIV-1 DH12 isolate. Virology. 1999;257:290–296. doi: 10.1006/viro.1999.9686. [DOI] [PubMed] [Google Scholar]
- 36.Mondor I, Moulard M, Ugolini S, Klasse P J, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau Q J. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology. 1998;248:394–405. doi: 10.1006/viro.1998.9282. [DOI] [PubMed] [Google Scholar]
- 37.Moore J P, Jameson B A, Weiss R A, Sattentau Q J. The HIV-cell fusion reaction. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press Inc.; 1993. pp. 233–89. [Google Scholar]
- 38.Mosier D E. Virus and target cell evolution in human immunodeficiency virus type 1 infection. Immunol Res. 2000;21:253–258. doi: 10.1385/IR:21:2-3:253. [DOI] [PubMed] [Google Scholar]
- 39.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. . (Erratum, 384:288.) [DOI] [PubMed] [Google Scholar]
- 40.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 41.Pleskoff O, Sol N, Labrosse B, Alizon M. Human immunodeficiency virus strains differ in their ability to infect CD4+ cells expressing the rat homolog of CXCR-4 (fusin) J Virol. 1997;71:3259–3262. doi: 10.1128/jvi.71.4.3259-3262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 43.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 44.Ross T M, Bieniasz P D, Cullen B R. Role of chemokine receptors in HIV-1 infection and pathogenesis. Adv Virus Res. 1999;52:233–267. doi: 10.1016/s0065-3527(08)60300-0. [DOI] [PubMed] [Google Scholar]
- 45.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz O, Alizon M, Heard J M, Danos O. Impairment of T cell receptor-dependent stimulation in CD4+ lymphocytes after contact with membrane-bound HIV-1 envelope glycoprotein. Virology. 1994;198:360–365. doi: 10.1006/viro.1994.1042. [DOI] [PubMed] [Google Scholar]
- 47.Shieh J T, Martin J, Baltuch G, Malim M H, Gonzalez-Scarano F. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J Virol. 2000;74:693–701. doi: 10.1128/jvi.74.2.693-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu N, Takeuchi Y, Naruse T, Inagaki M, Moriyama E, Gojobori T, Hoshino H. Six strains of human immunodeficiency virus type 1 isolated in Japan and their molecular phylogeny. J Mol Evol. 1992;35:329–336. doi: 10.1007/BF00161170. [DOI] [PubMed] [Google Scholar]
- 49.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 50.Sol N, Ferchal F, Braun J, Pleskoff O, Treboute C, Ansart I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spire B, Sire J, Zachar V, Rey F, Barre-Sinoussi F, Galibert F, Hampe A, Chermann J C. Nucleotide sequence of HIV1-NDK: a highly cytopathic strain of the human immunodeficiency virus. Gene. 1989;81:275–284. doi: 10.1016/0378-1119(89)90188-1. [DOI] [PubMed] [Google Scholar]
- 53.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q J. HIV-1 gp120 induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 54.Wang Z X, Berson J F, Zhang T Y, Cen Y H, Sun Y, Sharron M, Lu Z H, Peiper S C. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism: unmasking of activity with M-tropic Env glycoproteins. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 55.Willey R L, Theodore T S, Martin M A. Amino acid substitutions in the human immunodeficiency virus type 1 gp120 V3 loop that change viral tropism also alter physical and functional properties of the virion envelope. J Virol. 1994;68:4409–4419. doi: 10.1128/jvi.68.7.4409-4419.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 57.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]