Abstract
The spindle checkpoint ensures faithful chromosome segregation by linking the onset of anaphase to the establishment of bipolar kinetochore–microtubule attachment. The checkpoint is mediated by a signal transduction system comprised of conserved Mad, Bub and other proteins. In this study, we use live-cell imaging coupled with RNA interference to investigate the functions of human Bub1. We find that Bub1 is essential for checkpoint control and for correct chromosome congression. Bub1 depletion leads to the accumulation of misaligned chromatids in which both sister kinetochores are linked to microtubules in an abnormal fashion, a phenotype that is unique among Mad and Bub depletions. Bub1 is similar to the Aurora B/Ipl1p kinase in having roles in both the checkpoint and microtubule binding. However, human Bub1 and Aurora B are recruited to kinetochores independently of each other and have an additive effect when depleted simultaneously. Thus, Bub1 and Aurora B appear to function in parallel pathways that promote formation of stable bipolar kinetochore–microtubule attachments.
Keywords: Aurora B, Bub1, checkpoint, kinetochore, mitosis
Introduction
Chromosome segregation is a complex process during which paired sister chromatids attach to microtubules (MTs) of the mitotic spindle, align on the metaphase plate and then segregate into two equal sets. Chromosome–MT attachment is mediated by kinetochores, multiprotein complexes that assemble on centromeric DNA. Correct chromosome disjunction at anaphase requires bipolar attachment, in which one kinetochore on a chromatid pair binds to MTs emanating from one spindle pole and the other kinetochore binds to MTs from the opposite pole (for review, see Tanaka et al, 2002). Because kinetochores capture MTs through a stochastic search-and-capture process, the amount of time required for all kinetochores to make stable bipolar attachment varies from cell to cell. The spindle checkpoint ensures that anaphase onset is delayed in each cell until all chromatids are correctly MT-bound (for reviews, see Gorbsky, 2001; Hoyt, 2001; Musacchio and Hardwick, 2002; Cleveland et al, 2003). Loss-of-function mutations in checkpoint genes cause chromosome loss and genomic instability by allowing anaphase to proceed even if some kinetochores are incorrectly bound to spindle MTs (Draviam et al, 2004).
The spindle checkpoint is mediated by a set of highly conserved proteins that function as a signal transduction system and include Mad1, Mad2, Mad3/BubR1, Bub1, Bub3 and Mps1 (Hoyt et al, 1991; Li and Murray, 1991; Weiss and Winey, 1996). Prior to achieving bipolar MT attachment, kinetochores accumulate Mad and Bub proteins and act as upstream checkpoint signals (for review, see Cleveland et al, 2003). The downstream target of the checkpoint is Cdc20, an activator of the anaphase-promoting complex (APC/C). APC/C is an E3-ubiquitin ligase that marks key mitotic proteins for degradation and is the key regulator of the metaphase–anaphase transition (for review, see Nasmyth, 2002; Peters, 2002). Mad2 and BubR1 bind to Cdc20 and prevent it from activating APC/C (Sudakin et al, 2001; Tang et al, 2001; Fang, 2002). The molecular mechanisms by which checkpoint signals are generated by kinetochores and then sent to APC/C have not been elucidated in detail, but the deletion of any single Mad or Bub gene abolishes checkpoint control. Interdependencies among Mad and Bub proteins for kinetochore recruitment suggest that Bub1 and BubR1 kinases lie ‘upstream' of Mad1 and Mad2 (Sharp-Baker and Chen, 2001; Sironi et al, 2001; Chen, 2002; Martin-Lluesma et al, 2002; Liu et al, 2003; Gillett et al, 2004; Meraldi et al, 2004). Mad2, BubR1, Mps1 and Bub3 have also been shown to traffic on and off unattached kinetochores whereas Mad1 and Bub1 remain more firmly kinetochore-bound (Howell et al, 2004; Shah et al, 2004). The current consensus is that unattached and maloriented kinetochores activate Mad2 and BubR1, which then bind to and inhibit Cdc20, thereby turning APC off (Cleveland et al, 2003). In addition, Mad2 and BubR1 regulate the timing of mitotic progression in a kinetochore-independent manner (Meraldi et al, 2004).
The checkpoint processes monitoring chromosome–MT attachment are linked to error-correcting machinery that destabilizes incorrect MT links. The best-understood connection involves the dual-function mitotic kinase Aurora B/Ipl1. Aurora B/Ipl1p corrects syntely, in which both kinetochores on a pair of sister chromatids bind to MTs emanating from a single spindle pole (Tanaka et al, 2002; Hauf et al, 2003). Aurora B is also necessary for the spindle checkpoint to respond to reduced tension across sister kinetochores (Biggins and Murray, 2001; Ditchfield et al, 2003; Hauf et al, 2003). In this paper, we characterize the functions of human Bub1 using a combination of live-cell imaging and RNA interference (RNAi) and find that, like Aurora B, Bub1 has a dual function in mitosis. Human Bub1 is not only essential for the checkpoint response, but also required for the correct alignment of chromosomes on the metaphase spindle. Bub1 promotes the formation of stable end-on attachments necessary for correct sister chromatid disjunction at anaphase. Although similar in many ways, Aurora B and Bub1 appear to act independently of each other to regulate MT–kinetochore binding.
Results
Bub1 depletion abrogates the spindle checkpoint in human cells
To study the functions of human Bub1 during mitosis, HeLa cells expressing histone-H2B-GFP were transfected with one of six small interfering RNAs (siRNAs) directed against Bub1 mRNA, and chromosome movement was monitored by live-cell imaging (Kanda et al, 1998). Two siRNAs were found to target Bub1 efficiently. Following transfection of these siRNAs into cells, quantitative immunoblotting of extracts showed ∼25-fold depletion of Bub1 protein (Figure 1A and C), whereas levels of other spindle checkpoint proteins were unaltered. To ascertain the extent of Bub1 depletion from kinetochores, 3D deconvolution imaging was performed on multiple prometaphase cells stained with anti-Bub1 antibodies. Staining with CREST antisera, which is constant through the cell cycle, served as an intensity reference (Hoffman et al, 2001). Using this approach, we measured a 150- to 200-fold reduction in fluorescence associated with kinetochore-localized Bub1 in greater than 95% of cells (Figure 1B–D) arguing that Bub1 RNAi was very efficient. To investigate the consequences of Bub1 depletion on cell division, 150 Bub1-depleted or control cells were imaged every 3 min for 6 h and the key events of mitosis scored morphologically. Nuclear breakdown (NBD) was set as T=0 min in each cell. ‘Congression time' represented the point at which chromosomes had completed movement to the spindle equator and formed the metaphase plate. ‘Anaphase time' represented the onset of sister separation and toward-the-pole chromosome movement. Cells were followed until telophase so that errors in chromosome segregation that manifested themselves during anaphase could be quantitated.
Figure 1.
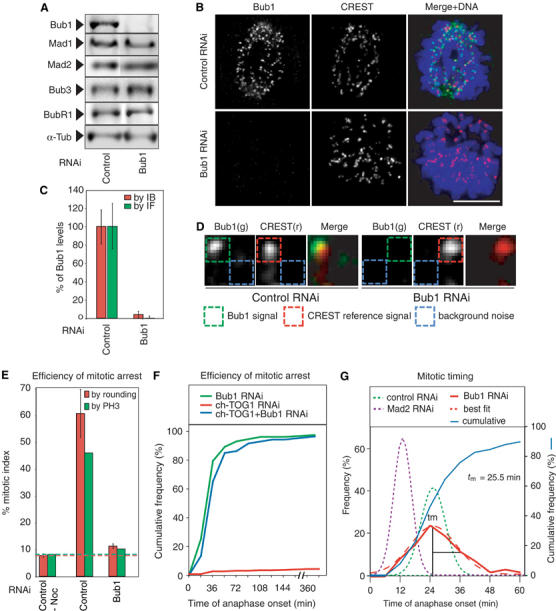
Bub1 depletion abrogates the spindle checkpoint in HeLa cells. (A) Immunoblots of lysates from cells transfected with siRNA against Lamin A (a negative control) or Bub1 and probed with antibodies as indicated. (B) Immunofluorescence images of prometaphase cells following transfection with Lamin A or Bub1 siRNA and stained with either Bub1 or CREST anti-sera. Scale bar: 10 μm. (C, D) Quantification of Bub1 levels in whole-cell lysates by immunoblotting (red bars) or on prometaphase kinetochores by immunofluorescence (green bars). Error bars show standard error of the mean (s.e.m.) from repeat determinations in multiple cells. Bub1 levels were determined from deconvolved 3D reconstructions. The Bub1 intensity (green) was determined relative to a CREST reference (red) and corrected for background noise (blue). See Supplementary data for details. Each box has a dimension of 0.25 × 0.25 μm. (E) Mitotic index of untransfected and siRNA-transfected cells, as indicated, treated with 100 ng/ml nocodazole for 16 h. The fraction of mitotic cells was determined by cell rounding under phase contrast microscopy (red) or FACS of phospho-histone H3-positive cells (green). Red and green dotted lines show comparable values from unperturbed cycling cells. Error bars represent s.e.m. (F) Cumulative frequency plots of anaphase times in siRNA-treated cells, as indicated, with NBD=T0, as determined from live-cell movies. Note that Bub1 depletion bypasses a mitotic arrest induced by ch-TOG1 depletion (blue lines). (G) Frequency distribution of anaphase times in siRNA-treated cells as indicated; tm denotes the peak time (mode) of the distribution. Mad2 data were derived from Meraldi et al (2004).
To monitor the status of the spindle checkpoint in cells, two assays were used. In a first set of experiments, Bub1 was depleted by RNAi and cells treated with the MT poison nocodazole for 16 h. Mitotic arrest was measured by cell rounding and anti-phospho-histone H3 staining followed by FACS. Following nocodazole treatment, control transfected cells had a high mitotic index (50–60%), whereas Bub1-depleted cells had a low mitotic index similar to that of normally growing cells (8–12%; Figure 1E). Thus, Bub1 depletion abolished the checkpoint-dependent cell cycle arrest provoked by nocodazole. In a second set of experiments, mitotic progression was monitored by live-cell imaging of individual cells following cotransfection with siRNA against ch-TOG1 and either Bub1 or a negative control (Lamin A). ch-TOG1 depletion perturbs kinetochore–MT attachment and provokes a robust metaphase arrest (Gergely et al, 2003; Meraldi et al, 2004) that is overcome by RNAi of Mad1, Mad2, Bub3 or BubR1 (Meraldi et al, 2004). When cells cotransfected with control and ch-TOG1 siRNA oligos were examined by live-cell imaging, >95% remained arrested for >6 h (Figure 1F; Meraldi et al, 2004) whereas ∼95% of cells cotransfected with Bub1 and ch-TOG1 siRNAs proceeded through mitosis (Figure 1F). Thus, Bub1 depletion over-rides the cell cycle arrest provoked by nocodazole treatment or depletion of ch-TOG1 as efficiently as Mad2 depletion (Meraldi et al, 2004). We therefore conclude that Bub1 is required for the spindle assembly checkpoint in human cells, in agreement with results from yeast (Hoyt et al, 1991) but contradicting recent reports based on RNAi of human Bub1 (Johnson et al, 2004; Kitajima et al, 2005; see below).
We have previously established that depletion of Mad and Bub proteins results in two distinct phenotypes with respect to mitotic timing. Depletion of Mad2 or BubR1, but not Mad1 or Bub3, abrogates the checkpoint and also accelerates mitotic progression dramatically (Meraldi et al, 2004). The time of anaphase onset in normally cycling HeLa cells exhibits a skew-normal distribution with a peak time (mode) of 26±1.5 min. In Mad2- or BubR1-depleted cells, the mode decreases to 12±2 min, whereas it is wild type in cells depleted of Mad1 or Bub3. When anaphase times were examined in 150 Bub1-depleted cells, we observed a broadening of the distribution but no significant change in the mode (25.5±1.5 min; Figure 1G). When both Bub1 and ch-TOG1 are depleted, the anaphase mode (27±2 min) was indistinguishable from the mode in cells transfected with Bub1 siRNA alone (Figure 1F). Thus, Bub1, like Mad1 and Bub3, is required for the spindle checkpoint but not for correct mitotic timing.
Bub1 is required for chromosome congression
Live-cell imaging reveals that, in cells depleted of Bub1, chromatids do not align correctly on the spindle equator prior to the onset of anaphase (Figure 2A and B). Whereas metaphase plates form correctly in the vast majority of control cells 6–8 min prior to the start of anaphase (i.e. by T=18 min), chromatids remained disorganized in 90% of Bub1-depleted cells until anaphase onset. To determine whether congression defects in Bub1-depleted cells were of a permanent nature or could be corrected by delaying anaphase onset, we treated cells with the proteasome inhibitor MG132 (Rock et al, 1994). MG132 blocks asynchronous cell populations at multiple points in the cell cycle, but when applied to early mitotic cells, leads to an arrest at the metaphase–anaphase transition independent of the spindle checkpoint. Cells were therefore synchronized using a double thymidine–aphidicolin block, transfected with siRNA, released from the block and treated with MG132 as they entered mitosis. Under these conditions, chromatid pairs congressed only very slowly to the metaphase plate so that 65% of Bub1 but only 2% of control cells contained maloriented chromosomes at T=60 min (Figure 2C). We therefore conclude that Bub1 depletion causes significant, long-lasting problems in chromosome congression.
Figure 2.
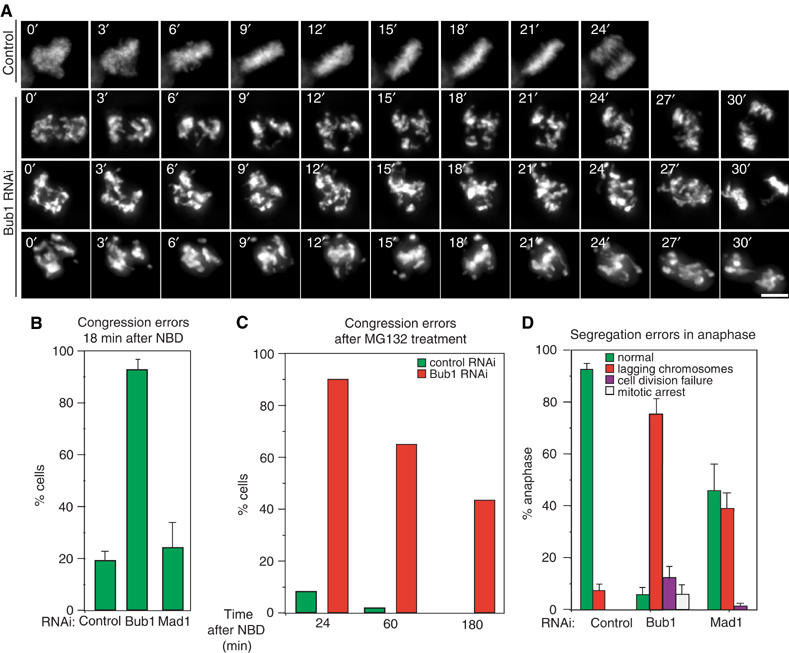
Bub1 depletion leads to errors in chromosome congression. (A) Successive frames every 3 min from live-cell movies of H2B-GFP-expressing HeLa cells transfected with siRNA as indicated. Scale bar: 10 μm. (B) Fraction of cells at T=18 min with uncongressed chromosomes in control and Bub1- and Mad1-depleted cells as determined visually. (C) Fraction of cells with uncongressed chromosomes at indicated time points after NBD in control and Bub1-depleted cells arrested at the metaphase–anaphase transition by MG132 treatment (see Materials and methods for details). (D) Fraction of cells with mitotic errors in anaphase. Green denotes morphologically normal anaphase, red lagging chromosomes, purple failure to divide into two chromosome masses and white sustained mitotic arrest at T=120 min. Error bars represent s.e.m. from four or more independent experiments.
Problems with sister chromatid congression of the type described above are not a simple consequence of spindle checkpoint disruption: unlike Bub1 depletion, Mad1 depletion did not increase the number of maloriented chromosomes at T=18 min as judged by live-cell imaging (when compared to control depletions; Figure 2B). However, to determine whether other Mad and Bub proteins are involved in congression, it was necessary to eliminate the effects of altered mitotic timing (Meraldi et al, 2004). Cells depleted of Mad or Bub proteins were therefore arrested at the metaphase–anaphase transition using MG132 and chromatids counted as incorrectly aligned if they were outside of a rectangular area encompassing the central 30% of the spindle (a volume that generously encompasses wild-type metaphase plates) or if the kinetochore-to-kinetochore axis was not parallel to the spindle axis (see the scheme in Figure 3A). This is a very conservative underestimate of chromosome misalignment. Nonetheless, 60% of Bub1-depleted cells contained 1–8 unaligned chromosomes by these criteria, as compared to 4% of control cells (Figure 3A and B). The depletion of Mad1, Mad2 or Bub3 did not increase the number of misaligned chromosomes relative to controls, whereas BubR1 depletion resulted in a small but significant increase (17%), consistent with previous reports (Figure 3B; Ditchfield et al, 2003; Lampson and Kapoor, 2005). As a positive control for these experiments, we also examined the effects of CENP-E depletion. CENP-E is a kinesin-like protein known to be required for chromosome congression. Intriguingly, it has been suggested that in Xenopus (Sharp-Baker and Chen, 2001; Vigneron et al, 2004) and humans (Johnson et al, 2004), the recruitment of CENP-E to kinetochores requires Bub1. When assayed using the MG132 arrest protocol described above, cells depleted of CENP-E had a similar number of misaligned chromatid pairs as compared to Bub1-depleted cells (Figure 3A and B). However, when levels of kinetochore-bound CENP-E were quantitated on a kinetochore-by-kinetochore basis (as described above), no significant differences were observed between control and Bub1-depleted cells (Figure 3C and D). Thus, congression defects caused by Bub1 depletion are similar in magnitude to those caused by depletion of the CENP-E motor, but the Bub1 phenotype in human cells cannot be explained by a failure to recruit CENP-E to kinetochores. Overall, these data demonstrate that Bub1 is unique among Mad and Bub proteins in being required for chromosome congression during metaphase.
Figure 3.
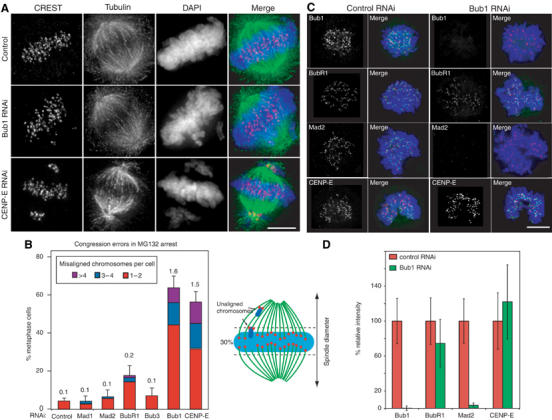
The role of Bub1 in chromosome congression is unique among checkpoint proteins. (A, B) Errors in chromosome congression in cells transfected with siRNA as indicated and arrested at the metaphase–anaphase transition for 1 h using MG132. Chromosomes in metaphase cells were counted as unaligned if they were located outside of the central 30% of the mitotic spindle (dotted lines in schematic) or if their kinetochores were aligned perpendicular to the spindle axis. (A) Immunofluorescence of arrested cells stained for CREST (red), tubulin (green) and DNA (blue) following RNAi as indicated. Scale bar: 10 μm. (B) Cumulative plot of number of fraction of cells with maloriented chromosomes; red: 1–2 maloriented chromosomes per cell; blue: 3–4; purple: >4. Numbers above each bar indicate the average number of misaligned chromosomes per cell. Error bars show s.e.m. (C, D) The Bub1, BubR1, Mad2 and CENP-E levels at kinetochores in control and Bub1-depleted cells as quantified using methods described in Figure 1. (C) Cells stained with DAPI (blue), CREST (red) and the indicated antibodies (green). Scale bar: 10 μm. (D) Quantification of protein levels (Bub1, BubR1, Mad2 and CENP-E) on individual kinetochores in control and Bub1-depleted cells as determined from images similar to those in (C). Error bars represent s.e.m. Note that CENP-E is not lost from human kinetochores following Bub1 depletion.
Bub1 depletion leads to defective MT–chromosome attachments
Chromosome misalignment can arise from several mechanistically distinct defects in chromosome–MT binding, including a complete absence of attachment, monopolar attachment (binding of only one kinetochore in a pair of sister chromatids to MTs), syntelic attachment (binding of both kinetochores to MTs emanating from a single spindle pole) and merotelic attachment (binding of a single kinetochore to MTs from opposite poles). No direct biochemical assays exist for these defects, but defects can be at least partially distinguished by monitoring chromosome dynamics during metaphase and anaphase, measuring interkinetochore distance (centromere stretching) as readout of the net force exerted on chromatid pairs and characterizing the levels of kinetochore-bound checkpoint proteins. Mad2 is typically used in the latter assay because it is present at high levels on unattached and maloriented kinetochores but at much lower levels on correctly attached kinetochores (Hoffman et al, 2001). Unfortunately, Bub1 is required for kinetochore binding by Mad2, precluding its use in our experiments (Meraldi et al, 2004; Figure 3C and D). However, BubR1 behaves much like Mad2 (Hoffman et al, 2001) and we have found that it is recruited to human kinetochores independent of Bub1 (Meraldi et al, 2004; Figure 3C and D). We therefore monitored chromosome dynamics, centromere stretching and BubR1 levels in CENP-E- and Bub1-depleted cells arrested at metaphase with MG132 for consistent differences between aligned and misaligned chromosomes.
Neither CENP-E nor Bub1 depletion caused chromatids to detach completely from MTs. Detached chromatids, such as those generated by Nuf2R or HEC1 depletion (DeLuca et al, 2002), are pushed rapidly out of the central spindle by astral ejection forces. While misaligned chromatid pairs in CENP-E-depleted cells exhibited interkinetochore distances (d=0.55±0.15 μm; Figure 4C) typical of detached chromatid pairs (d=0.5±0.15 μm), they remained close to the spindle poles (Figures 3A and 4A). Misaligned chromatids in Bub1-depleted cells were scattered throughout the spindle, moved rapidly back and forth along MTs (Figure 4B) and had interkinetochore distances (d=0.95±0.15 μm) midway between those of bound (d=1.8±0.35 μm) and detached chromatid pairs (Figure 4C). In CENP-E-depleted cells, BubR1 levels on unaligned metaphase kinetochores were high (as high as on unattached kinetochores in prometaphase cells) whereas, in Bub1-depleted cells, BubR1 levels on unaligned kinetochores were low (as low as on correctly aligned chromatid pairs; Figure 5A and B). Tight association of maloriented chromatid pairs with a single spindle pole, low centromere stretching and high levels of kinetochore-bound BubR1 are consistent with previous reports that CENP-E depletion leads to the accumulation of chromatids with monopolar MT attachments (Putkey et al, 2002). The defect in Bub1-depleted cells is clearly different from this. Following Bub1 depletion, both kinetochores on unaligned sister chromatids have low BubR1 levels and are partially stretched, leading us to infer that both are bound to MTs.
Figure 4.
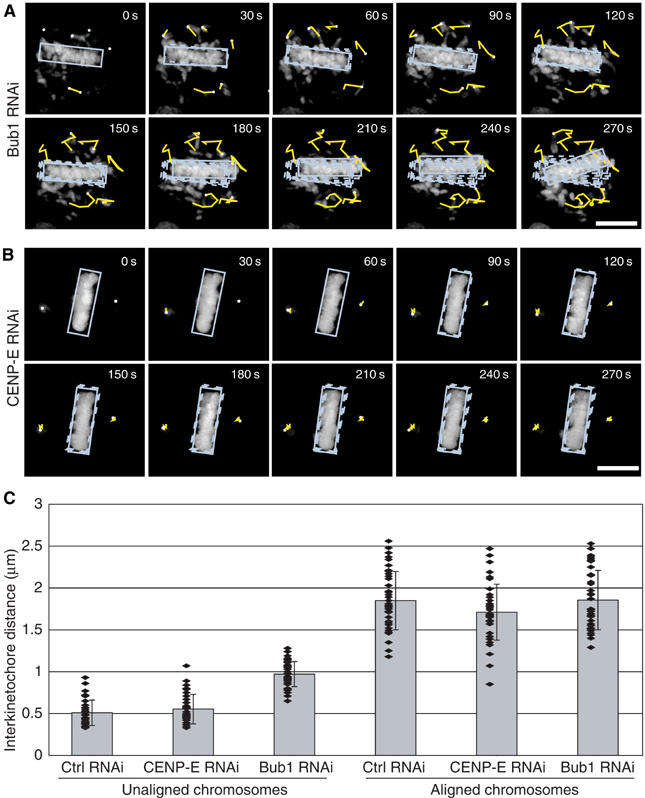
Rapid movement and reduced kinetochore stretching of maloriented sister chromatids generated by Bub1 depletion. (A, B) Approximate movement of the metaphase plate (blue box) and of maloriented chromosomes as monitored by 3D live-cell microscopy with one image stack every 30 s. Histone-GFP-expressing HeLa cells were transfected with Bub1 or CENP-E siRNA as indicated, arrested with MG132 and then monitored by 3D live-cell imaging. In each panel, yellow lines show the approximate path of chromosomes from time point to time point. (C) Interkinetochore distances of unaligned and aligned chromosomes from cells arrested at the metaphase–anaphase transition by MG132 for 1 h and transfected with siRNA as indicated. Interkinetochore distances were determined using the outer kinetochore marker BubR1. Each value was derived by measuring at least 40 kinetochore pairs in >8 cells. Values for unaligned chromosomes in control cells were obtained in prometaphase.
Figure 5.
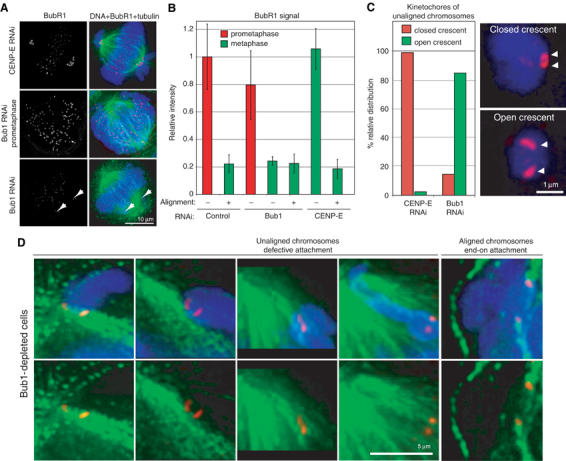
Bub1 depletion leads to defective MT–kinetochore attachments. (A) Immunofluorescence images of metaphase CENP-E-depleted (upper panel), prometaphase Bub1-depleted (middle panel) and metaphase Bub1-depleted (bottom panel) cells stained with DAPI (blue), anti-BubR1 antibodies (red) and tubulin antibodies (green). To arrest cells at metaphase, they were treated with MG132. Scale bar: 10 μm. Images of cells stained in parallel were acquired at the same time to assist comparisons across samples. Note that BubR1 levels on kinetochores in Bub1-depleted metaphase cells are low, implying that the kinetochores are MT-bound; high BubR1 levels in prometaphase demonstrate that BubR1 can bind kinetochores of unattached chromatids even when Bub1 is depleted. (B) Quantification of BubR1 levels on individual kinetochores from the data in (A). Error bars indicate s.e.m. Prometaphase and metaphase data are necessarily derived from different cells, but unaligned and aligned data for Bub1 and CENP-E depletions in metaphase were derived from the same cells. (C) Quantification of immunofluorescence images of unaligned chromosomes in Bub1- and CENP-E-depleted cells arrested for 1 h at the metaphase–anaphase transition with MG132, stained for DNA (blue) and BubR1 (red). Kinetochores were classified as having either a closed (upper panel) or open crescent morphology (lower panel). Scale bar: 1 μm. (D) Immunofluorescence images of unaligned chromosomes in Bub1-depleted cells arrested at the metaphase–anaphase transition for 1 h with MG132, stained with DAPI (blue), anti-CENP-E antibodies (red) and anti-β-tubulin antibodies (green). Note the MTs running past the open crescent-shaped kinetochores. As a comparison, end-on attached kinetochores of correctly aligned sister chromatids are shown. Scale bar: 5 μm.
To better visualize the mode of MT binding by maloriented chromatids in Bub1-depleted cells, we used high-resolution 3D imaging. When stained with BubR1 antibodies, kinetochores of unaligned chromatids in Bub1-depleted cells appeared as a pair of flattened crescents spaced about 0.9 μm apart, whereas those in CENP-E-depleted cells appeared as two tight crescents spaced less than 0.6 μm apart (Figure 5C, white triangle). The latter ‘closed' configuration is typical of a detached chromosome produced by nocodazole treatment (Hoffman et al, 2001). In Bub1-depleted cells, we frequently observed MTs running past pairs of flattened crescent-shaped kinetochores, suggesting binding to the walls of MTs (Figure 5D). With the exception of a failure to achieve end-on binding, the MT attachment defects in Bub1-depleted cells resemble aberrant syntelic attachments generated by Aurora B depletion: syntelic attachments are incompatible with correct chromosome congression and generate only intermediate levels of tension across sisters. We therefore speculate that maloriented sister kinetochores in Bub1-depleted cells are bound to MTs emanating from the same spindle pole, probably in a side-on configuration, although we have not proven that this is the case.
Independent regulation of congression by Bub1 and Aurora B
Bub1 and Aurora B share the fundamental similarity of having roles in both the spindle checkpoint and in correct chromosome–MT attachment (Hauf et al, 2003). Given this similarity, we wondered whether Bub1 and Aurora might be interdependent for kinetochore localization and activity. Indeed, several recent reports suggest that this might be the case in human (Hauf et al, 2003; Johnson et al, 2004) and Xenopus cells (Vigneron et al, 2004). To inactivate Aurora B, cells were transfected with one of three previously characterized siRNA oligos (Hauf et al, 2003; Honda et al, 2003; Johnson et al, 2004) and treated with a small molecule inhibitor of Aurora B (Straight et al, 2003). Aurora B was effectively inactivated under these conditions, as judged by two widely used criteria: (i) phosphorylation of histone H3, an Aurora B substrate, was reduced 15- to 20-fold as determined by anti-phospho-H3 staining of mitotic cells (see Hauf et al, 2003) and (ii) chromosome congression and anaphase B were disrupted (Ditchfield et al, 2003; Hauf et al, 2003; Figure 6A–D; data not shown). However, quantification of protein levels on individual kinetochores showed that levels of kinetochore-bound Bub1 were unchanged by Aurora B inhibition in both the presence and absence of nocodazole (Figure 6E–I). The reciprocal was also true: Aurora B remained on kinetochores at the same levels following Bub1 depletion (Figure 6E and G). Thus, Bub1 and Aurora B are recruited independently to kinetochores. If Bub1 and Aurora B lie in independent pathways regulating kinetochore–MT binding, we might expect an additive phenotype when both are inactivated. Consistent with this, the double-depletion phenotype was roughly twice as severe as either single-depletion phenotype (Figure 6J). We therefore conclude that Bub1 and Aurora B regulate chromosome attachment via separate pathways and are independently recruited to kinetochores.
Figure 6.
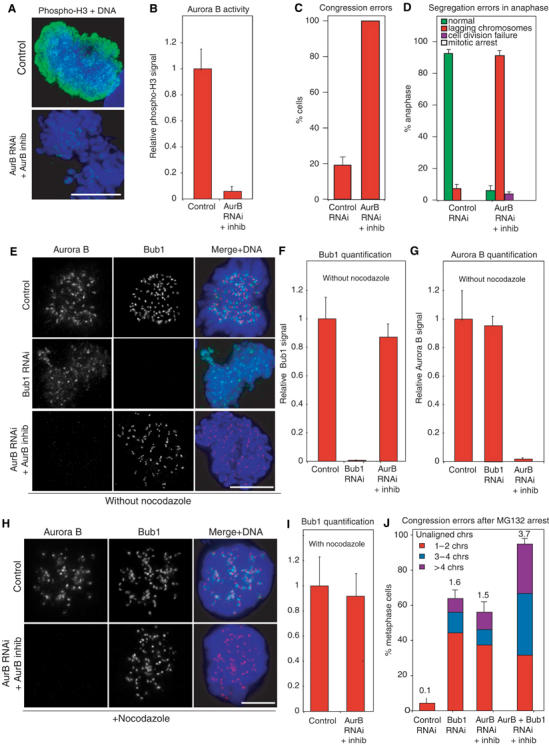
Bub1 and Aurora B act independently of each other. (A, B) Quantification of histone H3 phosphorylation following Aurora B inhibition, as a measure of Aurora B activity. Cells were stained with anti-phospho-H3 antibodies (green) and DAPI (blue) and the overall intensity measured in 10 prometaphase cells each. Error bars indicate s.e.m. Scale bar: 10 μm. (C, D) Fraction of cells with congression and segregation errors (see Figures 1 and 2) following Aurora B inhibition. (E) Immunofluorescence of cells following siRNA transfection as indicated and stained with DAPI (blue), anti-Bub1 antibodies (red) and anti-Aurora B antibodies (green). Scale bar: 10 μm. (F, G) Quantification by immunofluorescence of Aurora B and Bub1 levels on prometaphase kinetochores following RNAi (as in Figure 1). Error bars indicate s.e.m. (H) Immunofluorescence of nocodazole-treated cells following siRNA transfection as indicated, and stained as in (E). Scale bar: 10 μm. (I) Quantification of Bub1 levels on kinetochores of nocodazole-treated cells as in (F). (J) Fraction of cells with maloriented and misaligned chromatids following RNAi as assayed in MG132-arrested cells. Chromatids were scored as maloriented or misaligned as described in Figure 3.
Discussion
We have shown that human Bub1 is necessary for the spindle checkpoint and also for the correct congression of chromatids to the spindle plate during metaphase. In cells depleted of Bub1, a significant number of paired sister chromatids fail to move to the metaphase plate and the kinetochores on these chromatids appear to bind to the sides rather than the ends of MTs. However, some chromatid pairs do align on the metaphase plate in Bub1-depleted cells and become stretched, suggesting that normal attachment has been achieved. Thus, it appears that Bub1 is required for efficient establishment rather than maintenance of bipolar MT attachment. Bub1 is unique among Mad and Bub proteins in having this function: Mad1, Mad2, Bub3 and BubR1 have little if any role in congression. Given the high evolutionary conservation of spindle checkpoint proteins, and the requirement for Bub1 in checkpoint control in Saccharomyces cerevisiae, Schizosaccharomyces pombe and Xenopus laevis (Hoyt et al, 1991; Bernard et al, 1998; Sharp-Baker and Chen, 2001; Johnson et al, 2004; Meraldi et al, 2004), it is surprising that several recent reports (Johnson et al, 2004) have concluded that the depletion of Bub1 from human cells does not impair checkpoint control, and might even provoke cell cycle arrest (Tang et al, 2004). The two functional assays described in this paper are unambiguous in showing that human Bub1 is as important for the spindle checkpoint as Mad2 (although, unlike Mad2, Bub1 does not appear to play an important role in regulating the timing of mitosis; Meraldi et al, 2004). We believe that the primary difference between our data and those published by others involves the extent of Bub1 depletion. We have found that the presence of even low levels of kinetochore-localized Bub1 (2–5% of wild type) is sufficient to sustain a Mad2-dependent checkpoint response (Supplementary Figure S1) while leading to the accumulation of maloriented sister chromatids. We therefore encounter the paradoxical situation (described by Tang et al, 2004) that Bub1 can be depleted to levels at which congression is impaired but the checkpoint is functional, leading to cell cycle arrest. This situation is reminiscent of one we have previously described involving the kinetochore structural proteins HEC1 and Nuf2R (Meraldi et al, 2004). HEC1 and Nuf2R are required to assemble kinetochores capable of MT binding and also for Mad1 and Mad2 recruitment. Partial inactivation of HEC1 or Nuf2R engages the checkpoint, whereas complete inactivation abrogates it (Figure 7; Martin-Lluesma et al, 2002; Meraldi et al, 2004). Thus, to establish the loss-of-function phenotypes for kinetochore and checkpoint proteins, it is important to examine the effects of partial and complete RNAi and thereby determine the sensitivity to the degree of depletion. Having done this for all Mad and Bub proteins, our data clearly establish that in humans, as in other eukaryotes, inactivation of Bub1 by complete depletion abrogates the spindle checkpoint.
Figure 7.
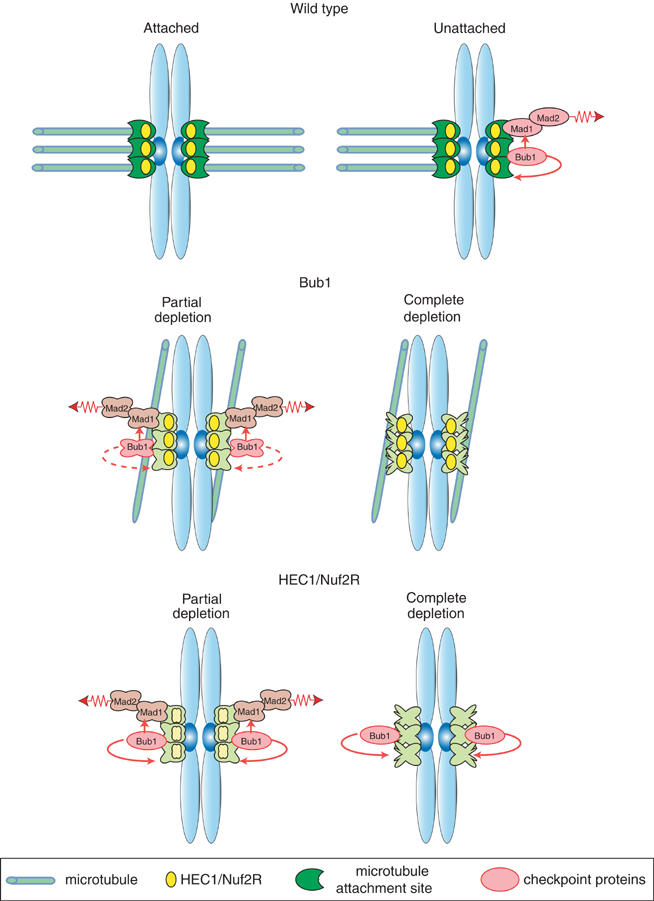
Speculative model for the effect of partial and complete Bub1 depletion on the spindle checkpoint. Schematics show the effect of partial and complete Bub1 depletion, and partial and complete HEC1/Nuf2R depletion on kinetochore-MT attachment and spindle checkpoint function as compared to correct operation of these processes in wild-type cells.
Bub1 promotes the correct attachment of kinetochores to MTs
Bub1 depletion is also associated with a dramatic increase in the number of chromatid pairs that fail to align correctly on the metaphase plate. The severity of the congression defect in Bub1-depleted cells is as great as in cells depleted of CENP-E, a kinesin-like motor known to play an active role in chromosome alignment (Putkey et al, 2002). However, our data establish that congression failure caused by Bub1 depletion is not a simple secondary consequence of checkpoint inactivation or altered mitotic timing (or of CENP-E loss from kinetochores), and is not observed when cells are depleted of other Mad or Bub proteins. These findings with human Bub1 are consistent with data from fungi. The deletion of S. pombe bub1+ increases the rate of chromosome mis-segregation (Bernard et al, 1998), and the depletion of either scBUB1 or scBUB3 in S. cerevisiae results in slow growth and elevated chromosome loss (Warren et al, 2002). The timing of scBub1p recruitment to budding yeast kinetochores has also been cited as evidence that scBub1p assists in the formation of mature end-on MT attachments (Gillett et al, 2004). One discrepancy between data from S. cerevisiae and humans is that the deletion of scBUB3 results in as severe a chromosome loss phenotype as scBUB1 deletion (Warren et al, 2002), whereas we find no role for huBub3 in congression in human cells. An important difference between yeast and human cells in this regard is that scBub3p is required for the recruitment of scBub1p to kinetochores, whereas, in human cells, huBub1 does not require huBub3 for kinetochore binding (Meraldi et al, 2004). Thus, we can conclude that, in organisms as diverse as yeast and human, Bub1 has a dual function in spindle checkpoint signaling and in promoting the establishment of correct chromosome–MT attachments.
Aurora B and Bub1 both contribute to correct chromosome–MT attachment
What is the nature of the MT-binding defect caused by Bub1 depletion? In Bub1-depleted cells, sister chromatids fail to achieve bipolar attachment and move toward the metaphase plate. Maloriented chromatids in Bub1-depleted cells are highly mobile and both kinetochores appear to contact MTs. The high density of MTs makes it difficult to image maloriented chromatids in HeLa cells, but our data suggest that the chromatids are displaced from the ends of MTs and make side-on attachments. If, as seems likely, many of these side-on attachments are to MTs emanating from a single spindle pole, they represent a state of syntely. Syntely in this case is related to, but distinct, from that caused by inactivation of the dual-function kinase Aurora B/Ipl1p. In Aurora B/Ipl1p-depleted cells, sister chromatids form end-on attachments to MTs emanating from a single spindle pole. This aberrant state of attachment is thought to represent a intermediate in the formation of correct bipolar attachment that must be actively corrected during prometaphase. In contrast, we have no evidence that the state of attachment observed in Bub1-depleted cells represents an intermediate in normal mitosis. Instead, we speculate that chromosome malorientation in the absence of Bub1 arises from defects in kinetochore assembly; Bub1 is known to be required for the binding of several proteins to kinetochores (Johnson et al, 2004; Tang et al, 2004; Kitajima et al, 2005). Shugoshin, for example, does not correctly assemble on kinetochores in Bub1-depleted cells, leading to a loss of centromeric cohesion (Tang et al, 2004; Kitajima et al, 2005). Whether the loss of centromeric cohesion is sufficient to explain chromosome misalignment in Bub1-depleted cells is not yet known.
Aurora B/Ipl1p and Bub1 are similar in both having functions in the spindle checkpoint and kinetochore–MT attachment. However, three findings argue that Bub1 functions independently of Aurora B/Ipl1p. First Bub1 and Aurora B are recruited independently of each other to kinetochores. In X. laevis, immunodepletion of Aurora B blocks kinetochore binding by Bub1 (Vigneron et al, 2004), and it has been suggested that this is also true in human cells (Hauf et al, 2003; Johnson et al, 2004). Differences between our data and previous results are not easily explained by incomplete inactivation of Aurora B on our part: the inhibition we use effectively blocks kinase activity in vivo, as measured using assays established by Hauf et al (2003). However, the published data are more ambiguous than might appear: following inhibition or Aurora B, Hauf et al (2003) reported only a slight decrease in the levels of kinetochore-bound Bub1 in one experiment of two (see Figure 9 of Hauf et al, 2003). The levels of Bub1 and Aurora B on kinetochores vary between early and late stages of mitosis and we have been careful to control this variable. Moreover, our conclusion that Bub1 and Aurora B function independently of each other is consistent with different loss-of-function phenotypes. Cells depleted of Bub1 have no detectable checkpoint response, whereas cells in which Aurora B has been inactivated or inhibited arrest in the presence of nocodazole (Ditchfield et al, 2003; Hauf et al, 2003). Finally, the phenotype of simultaneous Bub1 and Aurora B depletion is additive with respect to single depletions. We therefore believe that ambiguous data in previously published papers may have been misinterpreted. It follows that kinetochore–MT attachment is regulated by at least two related but independent dual-function kinases: one, based on the error-correcting Aurora B/Ipl1 kinase, acts to resolve syntelic attachment and the other, based on the Bub1 checkpoint kinase, promotes the formation of bipolar end-on attachment.
Conclusions
In human cells, Mad and Bub proteins fall into several distinct classes with regard to their loss-of-function phenotypes. (i) Mad1 and Bub3 depletion gives rise to a classical checkpoint defect in which the overall progress of mitosis is normal, but cells enter anaphase despite the presence of maloriented chromosomes (Meraldi et al, 2004). (ii) Mad2 and BubR1 depletion disrupts both the checkpoint response and the overall timing of mitosis, accelerating the entry into mitosis two- to three-fold (Meraldi et al, 2004). (iii) Bub1 depletion inactivates the checkpoint and disrupts the efficient congression of sister chromatids to the metaphase plate. We believe that this reflects a role for Bub1 in the correct assembly of kinetochores, and is similar in important ways to the role of the Aurora B/Ipl1 protein kinase. The two functions of Bub1 are also reminiscent of proteins in the DNA damage checkpoint that participate in both signaling and DNA repair (for review, see Rouse and Jackson, 2002). As a consequence, cells lacking Bub1 are particularly prone to genetic instability: not only is kinetochore–MT attachment defective in these cells but their ability to sense the errors is also lost.
Materials and methods
Cell culture, RNAi, MG132 treatment and Aurora B inhibitor
HeLa cells were grown as described (Meraldi et al, 2004). Bub1-targetting siRNA oligonucleotides (Dharmacon Research Inc.) were GAGUGAUCACGAUUUCUAU and CCUGAUAUUUCUGAUGACA; oligos against Mad1, Mad2, BubR1, Bub3, CENP-E, ch-TOG1 and Aurora B are described elsewhere (Martin-Lluesma et al, 2002; Ditchfield et al, 2003; Gergely et al, 2003; Hauf et al, 2003; Honda et al, 2003; Meraldi et al, 2004). HeLa cells were transfected as described (Elbashir et al, 2001) and analyzed 24–48 h after transfection. For MG132 experiments HeLa cells were synchronized using a double thymidine-aphidicolin block (Meraldi et al, 2004), transfected with siRNA 5 h prior to the aphidicolin treatment and treated for 1 h with 1 μM MG132 (Sigma) 13 h after the aphidicolin release. Aurora B inhibitor (kind gift of T Mitchison) was applied at 50 μM for 1 h before fixation (Straight et al, 2003).
Immunofluorescence microscopy
Cells were fixed, permeabilized and blocked as described (Kapoor et al, 2000). Antibodies used were as follows: affinity-purified goat anti-Bub1 (1 μg/1 μl; kind gift of S Taylor), goat anti-BubR1 (1:1000 dilution kind gift of S Taylor), anti-Aurora B (1:1000 dilution; Abcam), rabbit anti-Mad2 (1;500 dilution; Covance), rabbit anti-CENP-E (1:1500 dilution; Meraldi et al, 2004), rabbit anti-CENP-F (1:1000; Abcam), human anti-CREST (1:1500; kind gift of W Earnshaw) and mouse anti-β-tubulin monoclonal antibodies (1:400 dilution; Sigma clone Tub2.1). Cross-adsorbed secondary antibodies were used (Molecular Probes). Images were acquired as described (Martinez-Exposito et al, 1999). For quantification of kinetochore signals, the percentage of protein depletion at prometaphase kinetochores was quantified with SoftWorx (API) using the formula
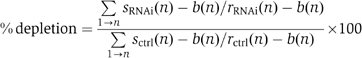
with s (signal), b (background) and r (reference signal). For each measurement, levels in at least eight cells (40 kinetochores) were determined. Interkinetochore distances were measured in individual focal plans of 3D deconvoluted microscopy images using the SoftWorx distance measurement tool. The distances were determined in at least eight cells (40 kinetochore pairs).
Immunoblotting
Whole-cell extracts were prepared by cell lysis in SDS sample buffer with 15% mercaptoethanol, resolved by SDS–PAGE and transferred to nitrocellulose membranes by semidry blotting (Hoefer). Membranes were blocked in blocking buffer (5% low-fat dried milk, PBS, 0.1% Tween 20) and incubated with goat anti-Bub1 (0.5 μg/ml kind gift of S Taylor), mouse anti-α-tubulin (1:10 000 dilution; Sigma clone B-1-5-2), goat anti-BubR1 antibodies (1:1000; kind gift of S Taylor), rabbit anti-Mad2 antibodies (1:500; Meraldi et al, 2004), rabbit anti-Mad1 antibodies (1:1000; Meraldi et al, 2004) or rabbit anti-Bub3 antibodies (1:1000; Meraldi et al, 2004) in blocking buffer. Anti-mouse and anti-sheep HRP-conjugated secondary antibodies (Amersham Pharmacia) were applied in blocking buffer and developed by enhanced chemiluminescence (Supersignal West Femto Maximum kit; Pierce).
Live-cell time-lapse imaging and analysis
Cells were imaged in ΔT 0.15 mm dishes (Bioptechs) in CO2-independent medium (GibcoBRL) at 37°C. Exposures (0.2 s) were acquired every 3 min for 6 h using a × 20 NA 0.75 objective on a Nikon Applied Precision Deltavision microscope equipped with a mercury 100 W lamp, GFP long-pass filter set (Chroma) and Coolsnap HQ camera. Point visiting was used to follow cells in multiple fields of view. Distributions of anaphase times were analyzed with MatLab. For high-resolution chromosome tracking, images of mitotic cells were recorded with × 60 NA 1.4 objective at 30 s intervals. For each time point, a stack of 40 images with 0.4 μm steps in the z-direction was obtained. The image stacks were subjected to a deconvolution algorithm (Applied Precision) and projected on a single 2D plane.
Nocodazole arrest
Cells were treated with 100 ng/ml nocodazole for 16 h and the fraction of rounded-up cells was determined by phase contrast microscopy. Alternatively, cells were methanol fixed, stained for anti-phospho-histone H3 and quantified using the FACSCALIBUR (Becton-Dickinson flow cytometer) as described by Meraldi et al (2004).
Supplementary Material
Supplementary Figure legends
Supplementary Figure S1
Acknowledgments
We thank S Taylor, W Earnshaw and T Mitchison for gifts of reagents, V Draviam for the FACS analysis and the members of the Sorger lab for helpful discussions. PM acknowledges receipt of an EMBO long-term fellowship. This work was supported by NIH grant CA84179.
References
- Bernard P, Hardwick K, Javerzat JP (1998) Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J Cell Biol 143: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW (2001) The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev 15: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH (2002) BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol 158: 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED (2002) Nuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam VM, Xie S, Sorger PK (2004) Chromosome segregation and genomic stability. Curr Opin Genet Dev 14: 120–125 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Fang G (2002) Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell 13: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW (2003) The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev 17: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett ES, Espelin CW, Sorger PK (2004) Spindle checkpoint proteins and chromosome–microtubule attachment in budding yeast. J Cell Biol 164: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ (2001) The mitotic spindle checkpoint. Curr Biol 11: R1001–R1004 [DOI] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM (2003) The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol 161: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Korner R, Nigg EA (2003) Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell 14: 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED (2004) Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol 14: 953–964 [DOI] [PubMed] [Google Scholar]
- Hoyt MA (2001) A new view of the spindle checkpoint. J Cell Biol 154: 909–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517 [DOI] [PubMed] [Google Scholar]
- Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS (2004) Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci 117: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM (1998) Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 8: 377–385 [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ (2000) Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol 150: 975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y (2005) Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol 15: 353–359 [DOI] [PubMed] [Google Scholar]
- Lampson MA, Kapoor TM (2005) The human mitotic checkpoint protein BubR1 regulates chromosome–spindle attachments. Nat Cell Biol 7: 93–98 [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW (1991) Feedback control of mitosis in budding yeast. Cell 66: 519–531 [DOI] [PubMed] [Google Scholar]
- Liu ST, Hittle JC, Jablonski SA, Campbell MS, Yoda K, Yen TJ (2003) Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat Cell Biol 5: 341–345 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- Martinez-Exposito MJ, Kaplan KB, Copeland J, Sorger PK (1999) Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc Natl Acad Sci USA 96: 8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK (2004) Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG (2002) The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol 3: 731–741 [DOI] [PubMed] [Google Scholar]
- Nasmyth K (2002) Segregating sister genomes: the molecular biology of chromosome separation. Science 297: 559–565 [DOI] [PubMed] [Google Scholar]
- Peters JM (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 9: 931–943 [DOI] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW (2002) Unstable kinetochore–microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell 3: 351–365 [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78: 761–771 [DOI] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2002) Interfaces between the detection, signaling, and repair of DNA damage. Science 297: 547–551 [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW (2004) Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol 14: 942–952 [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H, Chen RH (2001) Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J Cell Biol 153: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A (2001) Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J 20: 6371–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ (2003) Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299: 1743–1747 [DOI] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ (2001) Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K (2002) Evidence that the Ipl1–Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H (2001) Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 1: 227–237 [DOI] [PubMed] [Google Scholar]
- Tang Z, Sun Y, Harley SE, Zou H, Yu H (2004) Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA 101: 18012–18017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T (2004) Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell 15: 4584–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CD, Brady DM, Johnston RC, Hanna JS, Hardwick KG, Spencer FA (2002) Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell 13: 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M (1996) The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol 132: 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure legends
Supplementary Figure S1


