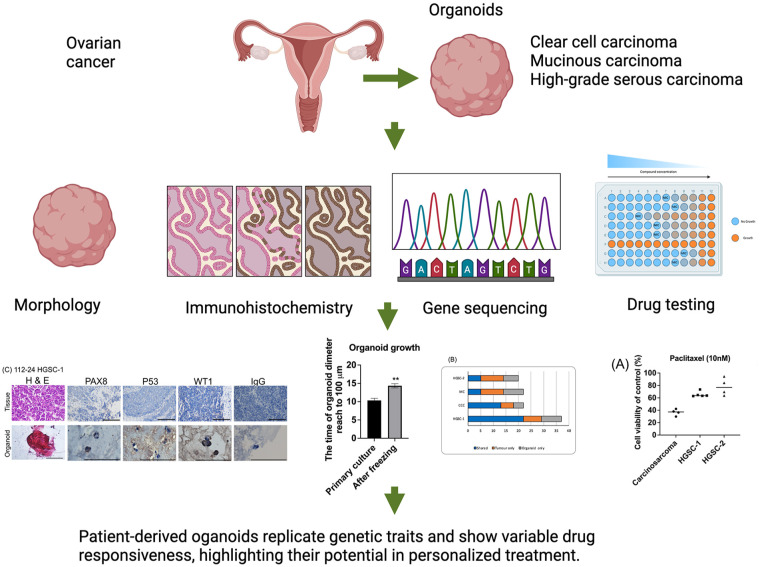
Abstract
This study aimed to explore the role of ovarian cancer patient-derived organoids (PDOs) in their replicating genetic characteristics and testing drug responsiveness. Ovarian cancer PDOs were cultured in Matrigel with a specialized medium. The successful rate and proliferation rate were calculated. Morphology, histology, and immunohistochemistry (IHC) (PAX8, P53, and WT1) were used to identify the tumor characteristics. Gene sequencing, variant allele frequency (VAF), and copy number variation were used to explore the mutation profile. The sensitivity to chemodrugs (carboplatin, paclitaxel, gemcitabine, doxorubicin, and olaparib) was conducted. Successful generation of organoids occurred in 54% (7/13) of attempts, encompassing 4 high-grade serous carcinomas (HGSC), 1 mucinous carcinoma (MC), 1 clear cell carcinoma (CCC), and 1 carcinosarcoma. The experiments used six organoids (3 HGSC, 1 CCC, 1 MC, and 1 carcinosarcoma). The derived organoids exhibited spherical-like morphology, and the diameter ranged from 100 to 500 μm. The histology and IHC exhibited the same between organoids and primary tumors. After cryopreservation, the organoid’s growth rate was slower than the primary culture (14 days vs 10 days, P < 0.01). Targeted sequencing revealed shared DNA variants, including mutations in key genes, such as BRCA1, PIK3CA, ARID1A, and TP53. VAF was similar between primary tumors and organoids. The organoids maintained inherited most copy number alterations. Drug sensitivity testing revealed varying responses, with carcinosarcoma organoids showing higher sensitivity to paclitaxel and gemcitabine than HGSC organoids. Our preliminary results showed that ovarian cancer PDOs could be successfully derived and histology, mutations, and diverse copy numbers of genotypes could be faithfully captured. Drug testing could reveal the individual PDO’s responsiveness to drugs. PDOs might be as valuable resources for investigating genomic biomarkers for personalized treatment.
Keywords: organoids, ovarian cancer, high-grade serous carcinoma, clear cell carcinoma, endometrioid adenocarcinoma, mucinous carcinoma
Graphical Abstract.
Introduction
Ovarian cancer, the deadliest gynecological malignancy 1 , is often detected at advanced stages, leading to metastasis in over 80% of cases 2 . High-grade serous carcinoma (HGSC) is a significant contributor to ovarian cancer–related mortality and poses a challenge to gynecological oncology care. Primary treatment involves tumor-debulking surgery and adjuvant chemotherapy. Following treatment, 70%–80% of patients experience tumor relapse and the development of chemoresistance 3 . In addition to chemotherapy, therapeutic approaches include antiangiogenic agents, poly (adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors, growth factor inhibitors, and immunotherapy 4 .
Ovarian cancer is classified into two types: Type I (e.g., endometrioid and clear cell) with mutations in the MMR, Wnt-catenin, ARID1A, and PI3K pathways and Type II (e.g., HGSC and carcinosarcoma) originating from fallopian tube intraepithelial carcinomas with p53 pathway inactivation, genomic variation, and CCNE1 activation5–8.
Understanding the mechanisms underlying ovarian cancer pathobiology remains incomplete, leading to limited improvements in treatment efficacy and patient survival 9 . Many cancer studies rely on cancer cell lines that inadequately recapitulate the histopathological and molecular features of ovarian cancer and its origin, resulting in limited clinical relevance 10 . Consequently, organoid research systems are emerging as an alternative.
Ovarian cancer organoids are three-dimensional, miniature models of ovarian tumors derived from cancer cells, which closely resemble the original tumor architecture 11 . This patient-specific approach ensures that the organoids carry genetic mutations and characteristics unique to an individual’s cancer type 12 . One of the primary applications of ovarian cancer organoids is drug sensitivity and resistance testing 13 . They also allow scientists to study the biology of ovarian cancer, investigate disease mechanisms, and test potential therapies using controlled and representative in vitro systems 14 .
Nevertheless, patient-derived organoids (PDOs) have raised several unresolved questions. First, the overall efficiency of organoid generation could be improved. Second, even in the same patient, different organoid lines can exhibit distinct behaviors, potentially leading to variability in drug screening results.
In this study, we aimed to establish ovarian cancer PDO from different histologies effectively, compare genetic features between organoids and tissues, and use them for chemo-drug sensitivity testing.
Materials and Methods
Ethics
This study was approved by the Research Ethical Committee of Hualien Tzu Chi Hospital, Hualien, Taiwan (IRB111-011-A). Informed consent was obtained from all patients.
Establishing Organoids From Ovarian Cancer Specimens
Biopsies of epithelial ovarian cancer specimens following primary or interval debulking surgeries were performed at Hualien Tzu Chi Hospital. Cancer specimens (1–3 cm3) were placed in specimen boxes soaked in normal saline and immediately sent to the laboratory. Each sample was cut into four pieces for organoid culture, fixed in formalin, stored in RNAlater solution, and embedded in optimal cutting temperature (OCT) compound (Thermo Fisher Scientific, MA, USA). For organoid culture, tissue fragments were rinsed with Ca2+/Mg2+-free phosphate-buffered saline (PBS, Thermo Fisher Scientific). Dispase (2 U/mL; Thermo Fisher Scientific), 1 mg/mL collagenase from clostridium histolyticum (C9407, Sigma, St. Louis, MO, USA), 5 μM Y27632 (Merk Millipore, MA, USA), and 10 μg/mL Dnase I (Thermo Fisher) in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Thermo Fisher) was used for tissue digestion for 1–2 h at 37°C. DMEM/F12 supplemented with 10% fetal bovine serum (FBS, Sigma) inhibited enzyme digestion. The resulting solution was centrifuged at 300 × g for 5 min at 4°C, and pellets were formed. The pellet was re-suspended in the growth factor–reduced Matrigel (BD Bioscience, Frank Lakes, NJ, USA)/DMEM/F12 (70%/30%) containing rho-associated protein kinase inhibitor (ROCK) inhibitor (Y-27632, 10 μM). The drop (20 μL/30,000 cells) was solidified on prewarmed 48-well plates at 37°C/5% CO2 for 20 min. The prewarmed culture medium was then added. The medium was changed every 2–3 days. The basal medium was called AdDF+++ media and was composed of DMEM/F12 with 1× Glutamax (Sigma), 10 mM HEPES (Sigma), and penicillin/streptomycin (Sigma).
The organoid culture media consisting of AdDF+++ supplemented with 1× B27 supplement (Life Technologies, Paisley, UK), 1.25 mM N-acetyl-l-cysteine (Sigma-Aldrich, MO, USA), 50 ng/mL WNT3A (R&D Systems, MN, USA), 250 ng/mL R-spondin1 (R&D Systems), 100 ng/mL Noggin (R&D Systems), 5 mM nicotinamide (Sigma-Aldrich), 50 ng/mL recombinant human EGF (epidermal growth factor, R&D Systems), 100 ng/mL recombinant human FGF10 (R&D Systems), 10 μM forskolin (R&D Systems), 5 μM A8301 (Tocris Bioscience, Bristol, UK), 500 ng/mL hydrocortisone (Sigma-Aldrich), 37.5 ng/mL Heregulinβ-1 (R&D Systems), 100 nM β-Estradiol (Sigma-Aldrich), and 10 μM Y27632 (Merck Millipore). Cell viability was assessed using Cell Titer Glow 3D Reagent (Promega, WI, USA).
Passage of Organoids and Cryopreservation
Organoid passage was performed after 2–4 weeks of culture, depending on the proliferation rate of the organoids. Organoids in Matrigel were dissociated using TrypLE Express (Thermo Fisher Scientific), containing ROCK inhibitor, at 37°C for 5–10 min, depending on the density of the organoids. TrypLE Express was inactivated by 1:1 medium dilution, and the suspension was centrifuged at 300 × g (4°C for 5 min). Subsequently, mechanical titration into single cells was performed by intense pipetting. The cells were centrifuged again at the same specifications and plated onto Matrigel as described above. For cryopreservation, the dissociated cells were resuspended in DMEM/F12, FBS, and dimethyl sulfoxide (DMSO; Sigma-Aldrich) [60%: 30%: 10%] and stored at −80°C overnight and subsequently moved to liquid nitrogen until further processing.
Immunohistochemistry
Matrigel-embedded organoids were gathered in a 15-mL tube with Cell Recovery Solution (354253, Corning, 500 µL/well). To ensure complete dissolution, the organoid suspension underwent gentle pipetting at intervals for 30 min on ice. Subsequently, the tube was placed on ice to allow the organoids to sediment. Following removing the supernatant, the organoids were washed with a small quantity of cold PBS. iPGell (FNK-PG20-1, Funakoshi Co., Tokyo, Japan) was used to solidify the organoids, and the organoid blocks were then fixed.
Tissues and organoids were fixed in 4% paraformaldehyde at 4°C overnight and at room temperature for 1 h, respectively. The tissue and organoids were paraffin-embedded and sectioned to 5-μm thickness. The sections sent for histology were stained with hematoxylin and eosin (H&E). Immunohistochemistry (IHC) was performed using antibodies against ovarian cancer markers (p53, Dako, Glostrup, Denmark). Antigen retrieval was carried out using citrate-based buffer (10 mM trisodium-citrate in H2O, pH 6; Merck) at 95°C for 30 min, and permeabilization was carried out using 1% Triton-X in PBS (PBT). Blocking was performed with 0.15% glycine/2 mg/mL bovine serum albumin in PBT. For blocking, immunofluorescence staining using 10% donkey serum (Sigma-Aldrich) was performed before incubation with primary antibodies at 4°C overnight. Anti-rabbit or anti-mouse horseradish peroxidase-labeled polymers (Dako) were then incubated at room temperature for 30 min. For immunofluorescence, fluorescein isothiocyanate (FITC)-labeled antibodies were incubated for 10 min. As a negative control, the primary antibodies were omitted, in which case no signals were detected. All immunostained slides were stained with hematoxylin or DAPI (4ʹ,6-diamidino-2-phenylindole). Images were captured using a Zeiss LSM 900 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany). The proportion of immunoreactive cells was counted in at least three replicates using the ImageJ software version 8 (https://imagej.net/ImageJ).
DNA Quantification and Qualification
The degree of DNA degradation and potential contamination was assessed using 1% agarose gel electrophoresis. DNA quantification was conducted using the Qubit® dsDNA Assay Kit (Thermo Fisher) on a Qubit 4.0 Fluorometer (Thermo Fisher).
Library Preparation
For sequencing experiments, passage 1 organoids were used. For library preparation, 1 μg of DNA per sample served as the input material. Sequencing libraries were generated according to the manufacturer’s recommendations using the xGenTM Exome Research Panel V2 (Integrated DNA Technologies, NC, USA), and unique index codes were incorporated to assign individual sequences to each sample. The library quality was evaluated using a Qubit 4.0 Fluorometer and the Agilent bioanalyzer system. Subsequently, the libraries underwent sequencing on an Illumina NovaSeq 6000 platform, producing paired 150 bp reads.
Data Analysis
The paired-end reads underwent thorough analysis for alignment to the human reference genome, utilizing the Burrows-Wheeler Aligner (v2.2.1) with default settings 15 . Following alignment, the Genome Analysis ToolKit (GATK) (v4.2.1.0) was employed for read processing 16 , incorporating MarkDuplicate and base quality score recalibration. Variant calling for single-nucleotide variants (SNVs) and small insertion/deletion (INDEL) events were conducted using HaplotypeCaller, with the GATK Variant Quality Score Recalibration tool utilized to obtain high-confidence variants.
Comprehensive annotation of these variants was carried out with ANNOVAR (v2020) 17 , SnpEff (v4.3) 18 , and VEP (v100.4) 19 . Data integration was effectively managed using an in-house algorithm. For the identification of copy number variations (CNVs) and structural variations (SVs), Control-FREEC (v11.6), and Manta (v1.6.0)20,21 were employed for CNV and SV detection, respectively. Annotation of these variants was performed using AnnotSV (v3.0.9) 22 . Genes exhibiting a variant allele frequency (VAF) of <20% in tumors and organoids were excluded from the analysis.
Copy Number Detection and Visualization
Tissue and organoid CNV analysis utilized the CNV kit software toolkit (v0.9.8). This software detected and visualized CNVs based on whole-exome sequencing data 23 . The analysis entailed comparing the read depths of on-target sequencing reads to provide copy number estimates for the entire genome within target regions. The results are visualized as a scatter plot displaying normalized copy ratios.
Drug Screening
Cells derived from Matrigel-embedded organoids were collected and then dissociated into single cells through trypLE treatment and mechanical dispersion. The cell suspension was reconstituted in Matrigel/cancer organoid medium (70%/30%), and 2000 cells/3 μL drop were seeded per well of a 96-well plate. Passage 1 organoids were used in drug testing experiments. Culture medium was added, and the organoids were allowed to grow for 1–2 weeks. Chemodrugs, such as paclitaxel (Formoxol, Yung Shin Pharm. Ind., Co. Ltd., Taichung, Taiwan), carboplatin (Abiplatin, ABIC Ltd., Netanya, Israel), doxorubicin (Adriblastina, Pfizer, Kent, NJ, USA), gemcitabine (Lilly France, Fegersheim, Frace), and Olaparib (Sigma), and vehicle (DMSO) were tested. These drugs are commonly used for the treatment of ovarian cancer patients 24 . A fixed concentration of each drug was applied to the organoid culture depending on the proliferation rate of the organoids. The initial chemodrug concentrations were paclitaxel (10–15 nM), carboplatin (1–20 μM), doxorubicin (100 nM–800 μM), gemcitabine (100–800 nM), and Olaparib (1 μM) 25 . Each drug’s concentration was determined using the IC50 concentration reported in the previous study 13 . Cell viability was assessed using the CellTiter-Glo 3D assay (Promega) on the sixth day.
Statistical Analysis
Statistical analysis was conducted using GraphPad Prism (v8, San Diego, CA, USA), with statistical significance indicated by a P-value of <0.05. Multiple comparisons were performed by one-way analysis of variance (ANOVA) and post hoc tests (Dunnett’s multiple comparison test with controls) for drug sensitivity tests. Results are presented as mean ± standard deviation, based on a minimum of three biological replicates for each experiment.
Results
Establishing Primary Ovarian Cancer Organoids
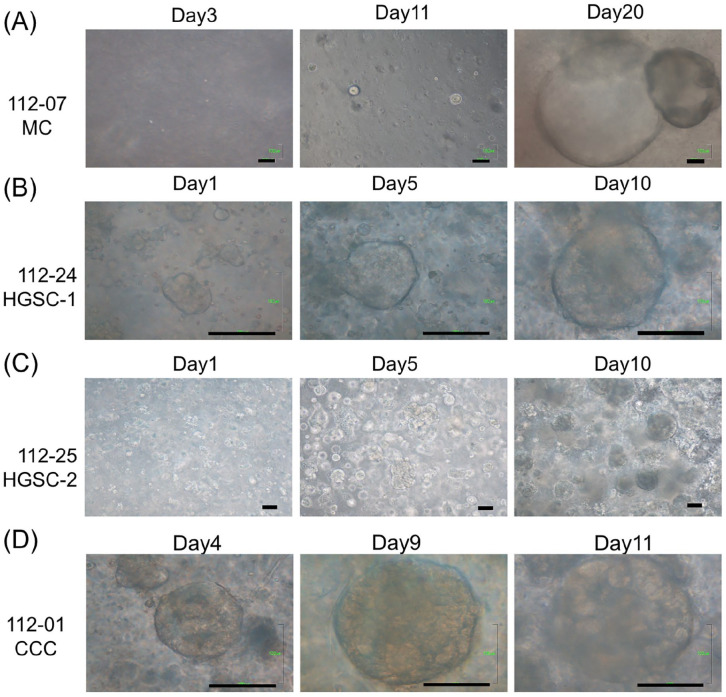
Single cells dissociated from primary ovarian cancer were first cultured and expanded. By cultivating these dissociated single tumor cells in Matrigel with a niche factor cocktail that included WNT-3A, R-spondin, and others, we successfully generated ovarian cancer organoids from patients with stage I–III ovarian cancer of various histologic subtypes (112-24 HGSC-1, 112-25 HGSC-2, 112-07 MC [mucinous carcinoma], and 112-01 CCC [clear cell carcinoma]) within 1–3 weeks (as depicted in Fig. 1 and outlined in Table 1). The derived organoids exhibited spherical-like morphology, and the diameter ranged from 100 to 500 μm (Fig. 1). The MC organoids grew more slowly than other types of cancer organoids, taking an additional 10 days to reach the same size as the other cancer organoids (Fig. 1).
Figure 1.
Primary ovarian cancer organoids derived from patients from day 1 to day 20. Brightfield microscopy images of the organoid lines on different days of growth. Scale bars 100 µm. (A) MC: mucinous carcinoma. (B-C) HGSC: high-grade serous carcinoma. (D) CCC: Clear cell carcinoma.
Table 1.
The Characteristics of Patients Whose Tissues Were Utilized for Organoid Derivation.
| Number | Case | Age at diagnosis | Stage | NAC | Debulking status | Observation time (m) | Recurrence | Time to recurrence after chemo | Status |
|---|---|---|---|---|---|---|---|---|---|
| 112-25 | HGSC-2 | 34 | IIIC | None | Optimal | 9 | No | NA | NED |
| 112-01 | CCC | 43 | IA | None | Complete | 7 | No | NA | NED |
| 112-24 | HGSC-1 | 48 | IIIC | None | Optimal | 9 | No | NA | NED |
| 112-07 | MC | 57 | IC | None | Complete | 3 | No | NA | NED |
NA: not available, NED: no evidence of disease.
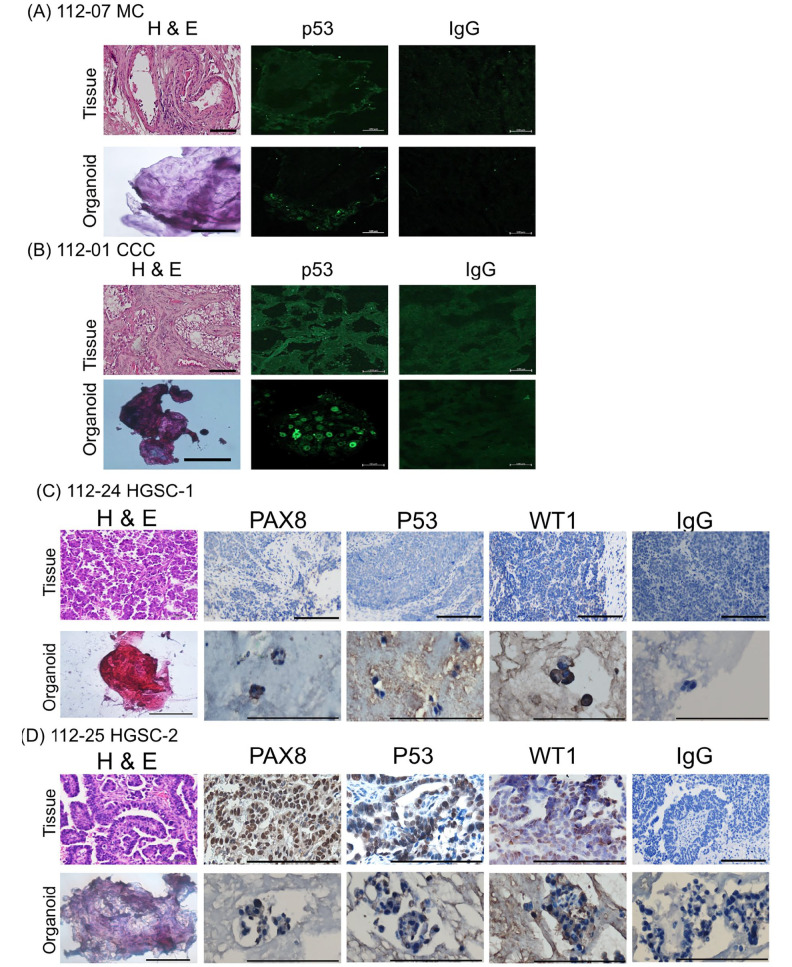
We introduce organoids cultured with the cocktail medium (low WNT3A), showcasing the superior efficacy for multi-tissue-type cultivation. The primary organoid culture achieved a success rate of 54%, with 7 out of 13 attempts yielding positive results (Table 2). Six organoids were derived from tumor tissue, and one HGSC was derived from omentum cake. These established organoids, MC (Fig. 2A), CCC (Fig. 2B), HGSC-1 (Fig. 2C), and HGSC-2 (Fig. 2D), accurately mirrored the histological characteristics and exhibited p53 positivity observed in the primary tumors. In HGSC organoids, PAX8 and WT1 were also positive (Fig. 2C–D).
Table 2.
The Success Rate of Organoid Culture From Each Histologic Subtype of Ovarian Cancer.
| Number of cases | Successful numbers of organoid culture | Successful rate (%) | Organoid origin | |
|---|---|---|---|---|
| HGSC | 6 | 4 | 66.7 | Tumor (3), omentum cake (1) |
| EM | 2 | 0 | 0 | 0 |
| CCC | 2 | 1 | 50 | Tumor (1) |
| MC | 2 | 1 | 50 | Tumor (1) |
| Others | 1 | 1 | 100 | Tumor (1) |
Other: carcinosarcoma; HGSC: high-grade serous carcinoma; CCC: clear cell carcinoma; MC: mucinous carcinoma; EM: endometrioid carcinoma.
Figure 2.
Haematoxylin and eosin (H&E) staining and p53, PAX8, and WT1 immunohistochemistry of primary ovarian tumors and derived organoids. It is noteworthy that organoids recapture the histologic features of the primary tumors. (A) mucinous ovarian (MC). (B) clear cell carcinoma (CCC). (C-D) high-grade serous (HGSC), and p53 staining pattern. HGSC-2 and CCC expressed p53. MC and HGSC-1 did not express p53. Scale bar = 100 µm. Scale bar = 50 µm in immunofluorescence pictures of organoids (nuclear staining of p53).
Investigate whether cryopreservation affects the growth rate of the organoids by comparing the growth rate of cryopreserved organoids with those that have never been cryopreserved. Both 112-24 HGSC-1 and 112-25 HGSC-2 were evaluated. Primary derivation organoids reaching 100 μm in diameter took 10 days, and subsequent culture time was the same as the primary derivation time. After cryopreservation, the growth rate to 100 μm in diameter was slower than the primary culture and took approximately 14 days (P < 0.01, Fig. 3).
Figure 3.
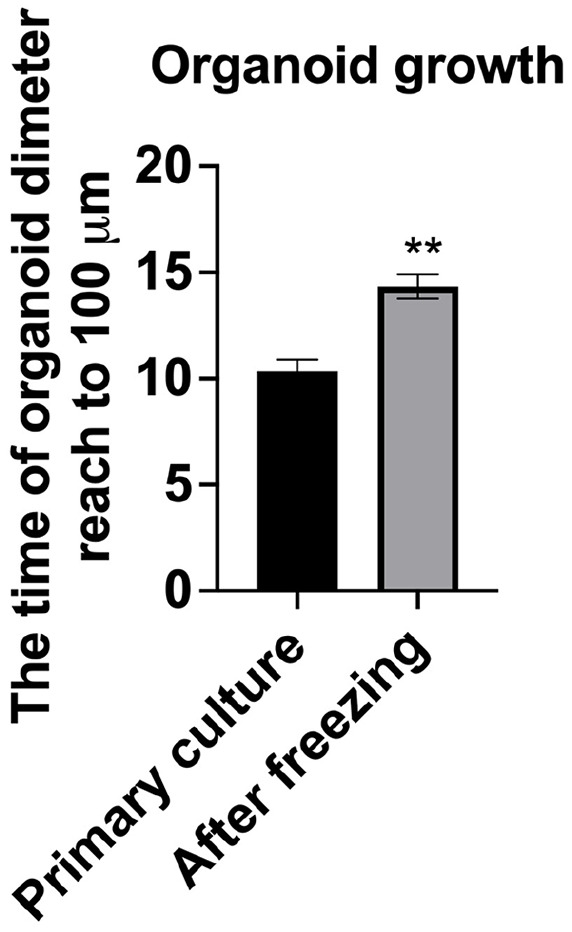
Organoid growth rate before and after freezing. The time the organoid size reached 100 µm was evaluated in organoids derived from high-grade serous carcinoma (n = 3 each in 112-24 HGSC-1 and 112-25 HGSC-2). **P < 0.01.
Capturing Primary Tumor Genomic Characteristics in Organoids
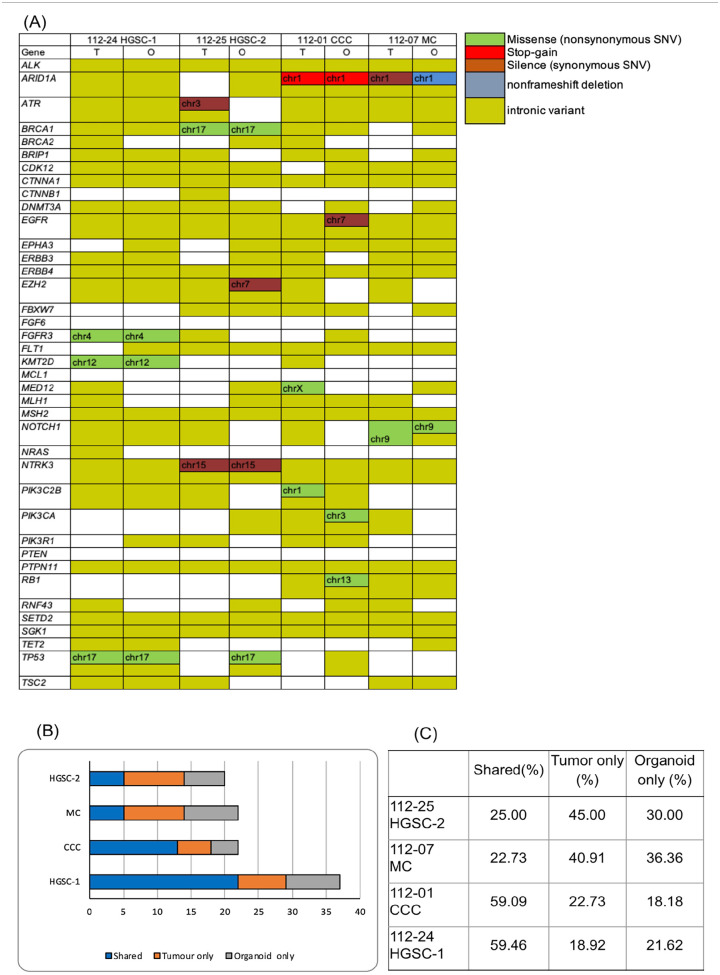
For a comprehensive analysis of genomic features between the original tumors and the derived organoids, targeted capture sequencing of 1,053 cancer-related genes was performed in four pairs of organoids and primary tumors spanning of 112-24 HGSC-1, 112-25 HGSC-2, 112-01 CCC, and 112-07 MC.
The genes in Fig. 4A were selected based on their well-established roles in promoting cancer (oncogenes) or inhibiting it (tumor suppressor genes). This includes commonly mutated genes in ovarian and other cancers, such as TP53, BRCA1, and BRCA2. In addition, genes identified as frequently mutated in ovarian cancer by previous studies, such as those from The Cancer Genome Atlas (TCGA), were also included.
Figure 4.
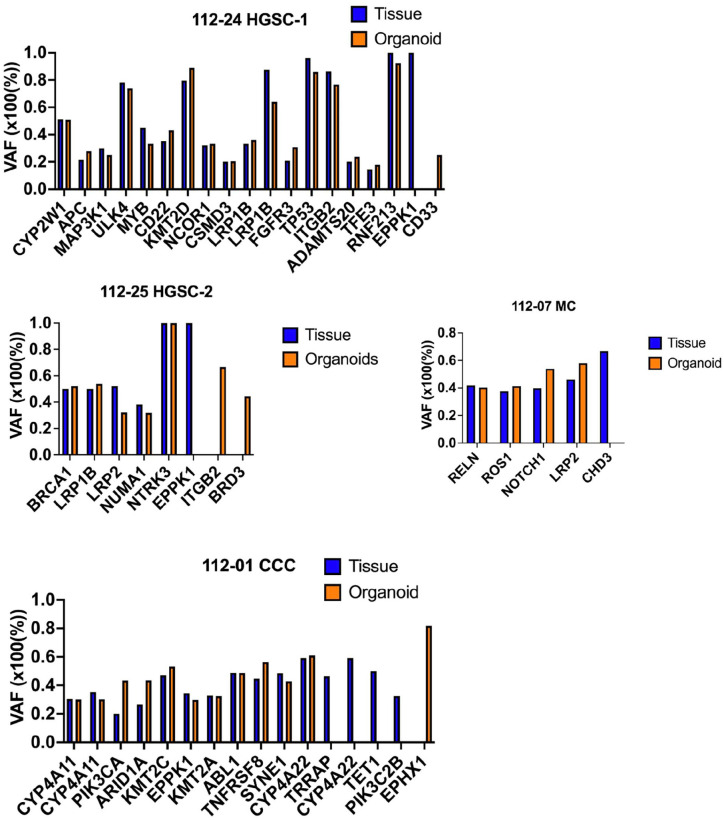
Organoids maintain the genetic alteration present in the original tumor. (A) A cancer-related set of variants was detected in both organoids and primary tumors with the mutation type indicated in the legend. Gene variants in the tumor and organoid from the same patient exhibit identical. T: tumor, O: organoid. HGSC: high-grade serous carcinoma, CCC: clear cell carcinoma, MC: mucinous carcinoma. The preservation of genetic alteration from the original tumor is evident in organoids. (B) Stacked bar graphs depict the number of mutations per patient sample identified in cancer tissues and derived organoids, organoid only and tumor only. (C) The percentage of shared, organoid-only, and tumor-only. Primary tumors and organoids exhibited a shared variation percentage ranging from 22% to 59%.
Our study revealed that these pairs share crucial DNA variants, including those in BRCA1, ARID1A, PIK3CA, and TP53 (Fig. 4).
The 112-25 HGSC-2 cells harbored a missense mutation in BRCA1 (p.N862I, pathogenic). At the same time, 112-01 CCC cells displayed stop-gain mutations in ARID1A (p.Y471*). The 112-07 MC cells contained silent (p.G786G) and non-frameshift deletion (p.A88_G93del) mutations in ARID1A, which were inconsistent in organoids and tumors.
Fig. 4 illustrates the mutations shared between the organoids and tissues. Overall, 59% of the gene mutations were shared between the organoids and primary tumors in 112-24 HGSC-1 and 112-01 CCC. However, only 25% of shared gene mutations were noted between organoids and primary tumors in 112-25 HGSC-2. The 112-07 MC was responsible for only 22.73% of the shared gene mutations.
VAF in the Tumor and Organoid
The VAF was similar between primary tumors and organoids (Fig. 5). The TP53 variant (c.G347A) had a VAF of 96% in the tumor tissue of 112-24 HGSC-1 and 85% in the organoids. The NOTCH1 variant (c.G4368C) had a VAF of 39.8% in the tumor tissue of 112-07 MC and 53.1% in the organoids. The PIK3CA variant (c.G1624A) had a VAF of 20% in the tumor tissues of 112-01 CCC and 43.4% in the organoids. The VAF elevation suggests a wild-type allele loss during organoid derivation.
Figure 5.
The variant allele frequency (VAF) was assessed in the occurrence of subclonal populations. Genes displaying a VAF of <20% in both tumor and organoid were excluded.
In summary, the observed difference in VAF between tumor and tumor-derived organoids is likely influenced by tumor heterogeneity, clonal selection during organoid culture, genomic instability, selective pressures, technical considerations, and potential evolutionary changes during organoid establishment. Further investigation and validation, possibly through additional sequencing and experimental analyses, are necessary to understand the underlying genetic dynamics.
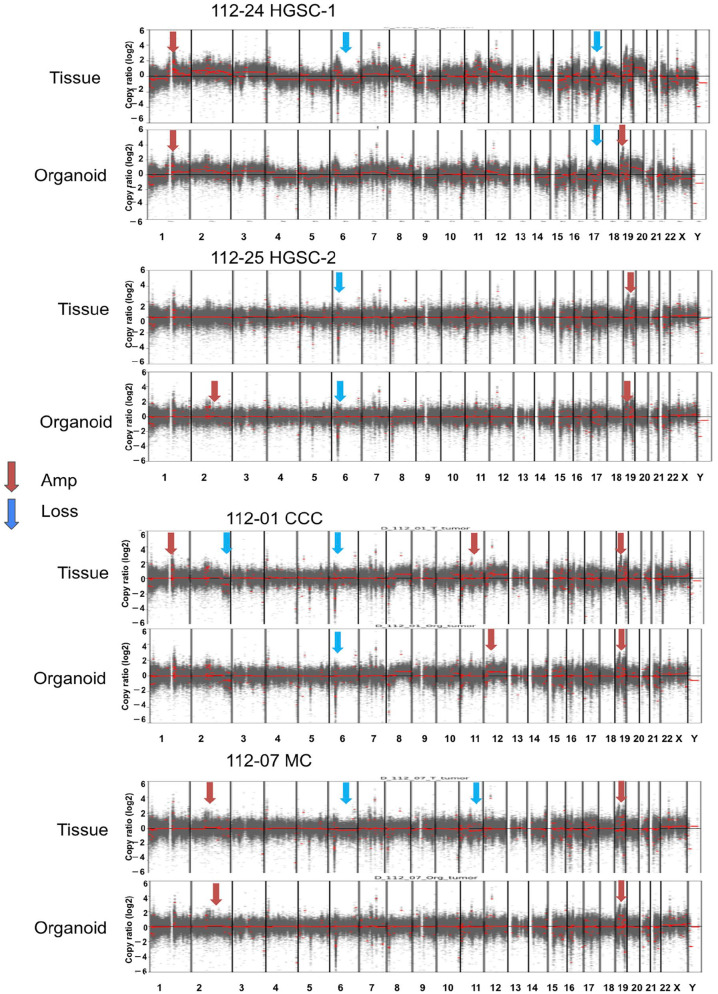
Copy Number Variants in Ovarian Organoids and Tumors
The four pairs of ovarian organoids and primary tumors exhibited consistent patterns of amplification and loss across their chromosomes (Fig. 6). Both 112-24 HGSC-1 and 112-25 HGSC-2 had limited CNVs, indicating non-homologous recombination deficiency (HRD). In the 112-01 CCC cases, parental tumors and organoids exhibit chromosome 19 amplification; however, chromosome 2 loss is exclusive to the primary tumor. Notably, organoids inherited most copy number alterations observed in tumors. However, 112-24 HGSC-1 and 112-25 HGSC-2 cells acquired new chromosomal aberrations.
Figure 6.
We created copy number variation (CNV) profiles for four instances and assessed the correlations (Pearson’s r) between tumor tissue and organoid samples. These CNV profiles were generated using a comprehensive cancer panel covering 1,053 genes. In the plot, regions with amplification are depicted in red, while areas with loss are shown in blue. T: tumor, O: organoid samples. HGSC: high-grade serous carcinoma, CCC: clear cell carcinoma, MC: mucinous carcinoma.
Utilizing Organoids for Personalized Drug Sensitivity Testing
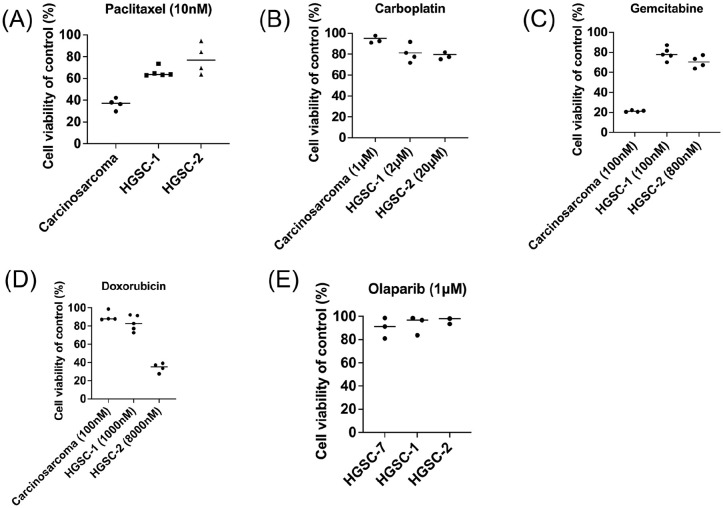
Finally, we tested the drug sensitivity of our samples carcinosarcoma, 112-24 HGSC-1, 112-25 HGSC-2, and HGSC-7 to drug paclitaxel, carboplatin, gemcitabine, doxorubicin, and olaparib. The clinical treatment concentration in serum for paclitaxel was 5.1 μM and for gemcitabine was 32–70 μM26,27.
After paclitaxel (10 nM) treatment, the carcinosarcoma organoids revealed 40% survival, and 2 HGSC organoids revealed 60%–80% cell survival (10 nM, Fig. 7A). After carboplatin (1–20 μM) treatment, the carcinosarcoma organoids revealed 90% survival (1 μM), and 2 HGSC organoids revealed 80% cell survival (2–20 μM, Fig. 7B). After gemcitabine treatment, the carcinosarcoma organoids revealed 20% survival (100 nM), and 2 HGSC organoids revealed 70%–80% cell survival (100–800 nM, Fig. 7C). After doxorubicin treatment, the carcinosarcoma organoids revealed 85% survival (100 nM), and 2 HGSC organoids revealed 36%–82% cell survival (1000–8000 nM, Fig. 7D). After the olaparib treatment (1 μM), the 3 HGSC organoids revealed 80%–90 % cell survival (Fig. 7E).
Figure 7.
Ovarian cancer organoids as a platform for drug screening. Cell viability was expressed as a percentage of control. (A) Paclitaxel (10nM). (B) Carboplatin (1μM to 20μM). (C) Gemcitabine (100–800 nM). (D) Doxorubicin (100–8000 nM). (E) Olaparib (1μM). Chemodrugs were used for testing cell survival after treating 72 hours (n = 4 each). HGSC was more resistant to chemodrugs than carcinosarcoma. HGSC: high-grade serous carcinoma.
Table 3 lists the chemodrugs used in these patients compared to the tested drugs. No recurrence was noted in two patients with HGSC observed for 9 months. Decreased tumor markers (CA125) were faster in 112-24 HGSC-1 than in 112-25 HGSC-2 (Table 4). The decreasing trend of CA125 in patients was correlated with drug testing.
Table 3.
The Chemodrugs Used and Outcomes in These Patients Were Compared.
| Age (years) | Tested drugs | Chemotherapy drugs | Clinical response | Observation time (m) | |
|---|---|---|---|---|---|
| Carcinosarcoma, stage IV | 68 | Paclitaxel (10 nM), carboplatin (1 μM), gemcitabine (100 nM), doxorubicin (100 nM) | None | dead | 18 days |
| 112-24 HGSC-1, stage IIIC | 48 | Paclitaxel (10 nM), carboplatin (2 μM), gemcitabine (100 nM), doxorubicin (1 μM) | Paclitaxel (175 mg/m2), carboplatin (AUC = 5), adding Avastin and 7.5 mg/kg5 since the third course, will be continued for 22 course | NED | 9 m |
| 112-25 HGSC-2, stage IIIC | 34 | Paclitaxel (15 nM), carboplatin (20 μM), gemcitabine (800 nM), doxorubicin (8 μM) | Paclitaxel (135 mg/m2), carboplatin (AUC = 5) | NED | 9 m |
NED: no evidence of disease, m: month, AUC: area under curve.
Table 4.
Change of the Tumor Marker (CA125: U/mL) During the Treatment Course.
| Course | before | 1 | 2 | 3 | 4 | 5 | 6 | post-6-1m | post-6-2m |
|---|---|---|---|---|---|---|---|---|---|
| 112-24 HGSC-1 | 1618 | 435 | 104 | 54.1 | 34.2 | 21.5 | 16.2 | 13.4 | 10.2 |
| 112/25 HGSC-2 | 302 | 302 | 288.4 | 164 | 98.6 | 63 | 48 | 41 | 19.9 |
HGSC: high-grade serous carcinoma, m: month
Discussion
This study successfully established primary ovarian cancer organoids that closely resembled the original tumors’ morphology, histology, and genomic characteristics. These organoids can be utilized for personalized drug sensitivity testing to reveal variations in drug responses among different histological subtypes of ovarian cancer, which might be valuable for tailoring treatment approaches. The patient demographic and pathological characteristics, organoid derivation success rate, and drug responsiveness are summarized in Table 5.
Table 5.
The Patient Demographic and Pathological Characteristics, Organoid Derivation Success Rate, and Drug Responsiveness.
| Organoid | Age (year) | Parity | BMI (kg/m2) | DM | H/T | Histology | Tumor IHC status | Stage | Organoid-derivation success rate (%) | Drug testing |
|---|---|---|---|---|---|---|---|---|---|---|
| 112-01 | 43 | 0 | 25.78 | - | + | CCC | ER (−), PR (−). Napsin A (+), p53 (+, wild type), WT-1 (−), PAS (+), PAS-D (−). | IA | 100 | - |
| 112-07 | 57 | 0 | 34.21 | - | + | MC | CK20 (−), CK7 (+), PMS2 (+), MSH6 (+) and WT-1 (−). | IC | 100 | - |
| 112-25 | 34 | 0 | 17.6 | - | + | HGSC-2 | CK7(+), CK20(−), p53 (mutational overexpression) MSH6: normal, PMS2: normal, ER: Positive (strong, 60%), PR: Positive (weak, 5%). | IIIC | 100 | T: +, C: +, G: +, D: +, Pi: + |
| 112-24 | 48 | 2 | 20.11 | - | + | HGSC-1 | WT-1 (diffuse +), p53 (totally negative), napsin A (-), ER (2+, 50%), PR (2+, <5%). | IIIC | 100 | T: +, C: +, G: +, D: +, Pi: + |
| 113-07 | 68 | 2 | 21.41 | - | + | HGSC-7 | CK7 (+), CK20 (−), GATA3 (−), NapsinA (-) and PAX8 (+), WT-1 weak and focal. | IIIC | 100 | Pi: + |
| 112-12 | 68 | 3 | 25.91 | - | + | Carcinosarcoma | PAX8 (−), CK (focal +), vimentin (diffuse +), S100 (focal +), WT-1 (−), Desmin (−), Myogenin (−), CD34 (focal +), c-kit (−) | IV | 100 | T: +++, C: +, G: +++, D: + |
CCC: clear cell carcinoma, MC: mucinous carcinoma, HGSC: high-grade serous carcinoma, ER: estrogen receptor, PR: progesterone receptor, CK: cytokeratin, PAS: Periodic acid-Schiff, PAS-D: PAS with predigestion with diastase, T: taxol, C: carboplatin, G: gemcitabine, D: doxorubicin, Pi: PARP inhibitor, BMI: body mass index, DM: diabetes mellitus, H/T: hypertension, IHC: immunohistochemistry, +: small response, +++: good response.
Establishing robust preclinical tumor models is important for advancing the therapeutic discovery of ovarian HGSC 28 . The previous study has explored ovarian cancer organoids as models of genomic instability 29 . As organoid culture has gained broader acceptance, short- and long-term ovarian cancer models have been created30–33. However, there is a need for more consistency in the available data regarding success rates and survival durations in culture. In our study, the success rate of organoid derivation was 54%, which was comparable with other studies. After gaining experience in organoid culture, the success rate would be increased.
Alterations in copy number can affect the expression of highly abundant human genes, suggesting a global sensitivity to gene dosage 34 . It is important to note that this relationship does not consistently result in proportional effects owing to the presence of transcriptional adaptive mechanisms 35 . In this study, organoids and primary tumors displayed consistent amplification and loss across chromosomes. The organoids faithfully inherited most of the copy number alterations observed in the tumors. However, some organoids have acquired new chromosomal aberrations. Sequencing cancer-related genes through targeted capture unveiled genomic insights within organoids and primary tumors that shared crucial DNA variants, including BRCA1, ARID1A, PIK3CA, and TP53 mutations. Notably, some organoids and tumors exhibited shared mutations.
The genetic changes in ovarian cancer are exceptionally diverse. Consequently, achieving personalized treatments requires the creation of genomically annotated patient-specific characteristics for therapeutic interventions 36 . Previous studies have also explored the use of ovarian cancer organoids in therapeutic drug screening11,13,29. This study found that the response to chemotherapy varied among the different organoids. Carcinosarcoma organoids showed greater sensitivity to paclitaxel and gemcitabine than HGSC organoids. The HGSC organoids exhibited lower sensitivity to these drugs, even at higher dosages.
The composition of the cancer organoid culture medium differed among the studies (Table 6). Matrigel and a niche factor cocktail containing WNT-3A and R-spondin were important for organoid culture. The concentrations of WNT3A and RSPO1 differed between the studies. We used 50 and 250 ng/mL of WNT3A and RSPO1, respectively. We did not use FGF2 but used a high concentration of FGF10 (100 ng/mL). In addition, our approach did not employ IGF1 (insulin-like growth factor 1) and HGF (hepatocyte growth factor). The organoid derivation efficiency was 54% in our study, which was lower than in other studies (80% in Nanki et al., 45%–90% in Maru et al.)13,37. This study uses unique combinations and concentrations of growth factors and supplements, potentially influencing organoid growth and behavior differently than other studies. Higher concentrations of estradiol and FGF10 could impact hormonal signaling pathways, crucial for ovarian cancer biology. The exclusion of specific factors like IGF1, HGF, and p38-inhibitor might reduce certain proliferative and survival signals, leading to different growth dynamics. These differences highlight the potential for this study to provide novel insights and optimizations in the development of ovarian cancer organoids.
Table 6.
The Composition of Ovarian Cancer Organoid Medium.
| Product | Current study | Kopper et al. 30 | Maenhoudt et al. 33 | Nanki et al. 13 |
|---|---|---|---|---|
| DMEM/F12 | Yes | Yes | Yes | Yes |
| L-glutamine | 1x | 1x | 1x | 1x |
| Penicillin/streptomycin | 1x | 0.2% primocin | 1x | 200 U/mL |
| A83-01 | 5 μM | 0.5 μM | 0.25–0.5 μM | 0.5 μM |
| Nicotinamide | 5 mM | 10 mM | 1–5 mM | / |
| N2 | / | / | 1x | / |
| B27+vitamin A | 1x | 1x | 1x | 1x |
| N-acetylcysteine | 1.25 mM | 1.25 mM | 1.25 mM | 1 mM |
| Estradiol | 100 nM | 100 nM | 10 nM | / |
| p38i (SB203580) | / | / | 1–10 μM | / |
| EGF | 50 ng/mL | 5 ng/mL | 50 ng/mL | / |
| bFGF | / | / | 2 ng/mL | 50 ng/mL |
| FGF10 | 100 ng/mL | 10 ng/mL | 0–10 ng/mL | / |
| Noggin | 100 ng/mL | 1% | 10% | 100 ng/mL |
| RSPO1 | 250 ng/mL | 10% | 20%, 50 ng/mL | 1 µg/mL |
| IGF1 | / | / | 0–20 ng/mL | 100 ng/mL |
| HGF | / | / | 0–10 ng/mL | / |
| Heregulinβ-1 | 37.5 ng/mL | 37.5 ng/mL | 50 ng/mL | / |
| WNT3A | 50 ng/mL | 20% | / | 20% |
| Forskolin | 10 μM | 10 μM | / | / |
| Leu15-Gastrin I | / | / | / | 10 nM |
| Hydrocortisone | 500 ng/mL | 500 ng/mL | / | / |
| Y27632 | 10 μM | 5 μM | 10 μM | 10 μM |
The previous study by Senkowski et al. 28 indicated that the organoid passage time ranges from 17 to 20 days, with passage criteria based on organoid number. Earlier research reported long-term expansion success rates of 23% to 38% using fresh primary cells and 53% with cryopreserved cells28,32,33,38. This suggests that long-term culture is feasible for ovarian cancer organoids 28 . In this study, we compared the growth time for organoids to reach a diameter of 100 μm before and after freezing. Our findings showed that organoids required 10 days to reach 100 μm before freezing and 14 days after freezing. This suggested the proliferation time of organoids after freezing became slower than in the primary culture.
This study had certain limitations. First, only four of seven organoids were studied for genomic characteristics, and six of seven were studied for drug responsiveness. This limited the gene mutation profile used to predict drug responsiveness. Second, the follow-up time of the patients was not long enough to see the accuracy of the drug responsiveness test. Third, this study did not subtract/normalize the sequence coming from the tumor-associated cells, such as fibroblasts and immune cells, from the actual cancer cell sequence of the tumor.
Conclusion
Our preliminary results showed that ovarian cancer PDOs could be successfully derived. The organoid’s histology, mutations, and diverse copy numbers of genotypes could be faithfully captured. Drug testing could reveal the individual PDO’s responsiveness to drugs. PDOs might be as valuable resources for investigating genomic biomarkers for personalized treatment.
Acknowledgments
The authors would like to thank the National Science and Technology Council (grant nos: NSTC 111-2314-B-303-019 and 112-2314-B-303-010-MY3).
Footnotes
Author Contributions: Conceptualization: D.C.D.; Methodology: K.H.W.; software, D.-C.D.; validation, D.-C.D., K.-H.W., and K.-C. W., and Y.-H.C.; formal analysis: D. C. D., K.-H.W., and K.-C. W., and Y.-H.C.; interpretation of data: K.-C.W. and D.-C.D.; resources: D.-C.D. and Y.-H.C.; data curation: Y.-H.C. and D.-C.D.; writing: D.-C.D., K.-H.W., and K.-C. W., and Y.-H.C.; original draft preparation: K.-H.W., Y.-H.C., and D.-C.D.; review and editing: D.-C.D.; supervision: D.-C.D. All authors have read and agreed to the published version of this manuscript.
Data Availability Statement: The datasets generated and/or analyzed in the current study are available from the corresponding author upon reasonable request.
Ethical Approval: This study was approved by the Ethics Committee of Hualien Tzu Chi Hospital (reference no. IRB111-011-A).
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by the National Science and Technology Council (grant nos: NSTC 111-2314-B-303-019 and 112-2314-B-303-010-MY3).
ORCID iD: Dah-Ching Ding  https://orcid.org/0000-0001-5105-068X
https://orcid.org/0000-0001-5105-068X
References
- 1. Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2018;144(8):1941–53. [DOI] [PubMed] [Google Scholar]
- 2. Narod S. Can advanced-stage ovarian cancer be cured? Nat. Rev Clin Oncol. 2016;13(4):255–61. [DOI] [PubMed] [Google Scholar]
- 3. Pignata SC, Cecere S, Du Bois A, Harter P, Heitz F. Treatment of recurrent ovarian cancer. Ann. Oncol. 2017;28:viii51–56. [DOI] [PubMed] [Google Scholar]
- 4. Cortez AJ, Tudrej P, Kujawa KA, Lisowska KM. Advances in ovarian cancer therapy. Cancer Chemother Pharmacol. 2018;81(1):17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Koshiyama M, Matsumura N, Konishi I. Recent concepts of ovarian carcinogenesis: type I and type II. Biomed Res Int 2014;2014:934261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurman RJ, Shih I-M. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186(4):733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shih I-M, Wang Y, Wang T-L. The origin of ovarian cancer species and precancerous landscape. Am J Pathol. 2021;191(1):26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu N-YY Fang C, Huang H-S, Wang J, Chu T-Y. Natural history of ovarian high-grade serous carcinoma from time effects of ovulation inhibition and progesterone clearance of p53-defective lesions. Mod Pathol. 2020;33(1):29–37. [DOI] [PubMed] [Google Scholar]
- 9. Timmermans M, Sonke GS, Van de Vijver KK, van der Aa MA, Kruitwagen RFPM. No improvement in long-term survival for epithelial ovarian cancer patients: A population-based study between 1989 and 2014 in the Netherlands. Eur J Cancer. 2018;88:31–37. [DOI] [PubMed] [Google Scholar]
- 10. Lengyel E, Burdette JE, Kenny HA, Matei D, Pilrose J, Haluska P, Nephew KP, Hales DB, Stack MS. Epithelial ovarian cancer experimental models. Oncogene. 2014;33(28):3619–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yee C, Dickson K-A, Muntasir MN, Ma Y, Marsh DJ. Three-dimensional modelling of ovarian cancer: from cell lines to organoids for discovery and personalized medicine. Front Bioeng Biotechnol. 2022;10:836984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang J, Huang S, Cheng S, Jin Y, Zhang N, Wang Y. Application of ovarian cancer organoids in precision medicine: key challenges and current opportunities. Front Cell Dev Biol. 2021;9:701429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nanki Y, Chiyoda T, Hirasawa A, Ookubo A, Itoh M, Ueno M, Akahane T, Kameyama K, Yamagami W, Kataoka F, Aoki D. Patient-derived ovarian cancer organoids capture the genomic profiles of primary tumours applicable for drug sensitivity and resistance testing. Sci Rep. 2020;10(1):12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liang J, Li X, Dong Y, Zhao B. Modeling human organ development and diseases with fetal tissue-derived organoids. Cell Transplant. 2022;31:9636897221124481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28(3):423–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–22. [DOI] [PubMed] [Google Scholar]
- 22. Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, Muller J. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34(20):3572–74. [DOI] [PubMed] [Google Scholar]
- 23. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. Plos Comput Biol. 2016;12(4):e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baert T, Ferrero A, Sehouli J, O’Donnell DM, González-Martín A, Joly F, van der Velden J, Blecharz P, Tan DSP, Querleu D, Colombo N, et al. The systemic treatment of recurrent ovarian cancer revisited. Ann Oncol. 2021;32(6):710–25. [DOI] [PubMed] [Google Scholar]
- 25. Cao Q, Li L, Zhao Y, Wang C, Shi Y, Tao X, Cai C, Han X-X. PARPi decreased primary ovarian cancer organoid growth through early apoptosis and base excision repair pathway. Cell Transplant. 2023;32:9636897231187996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stage TB, Bergmann TK, Kroetz DL. Clinical pharmacokinetics of paclitaxel monotherapy: an updated literature review. Clin Pharmacokinet. 2018;57(1):7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ciccolini J, Serdjebi C, Peters GJ, Giovannetti E. Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: an EORTC-PAMM perspective. Cancer Chemother Pharmacol. 2016;78(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Senkowski W, Gall-Mas L, Falco MM, Li Y, Lavikka K, Kriegbaum MC, Oikkonen J, Bulanova D, Pietras EJ, Voßgröne K, Chen Y-J, et al. A platform for efficient establishment and drug-response profiling of high-grade serous ovarian cancer organoids. Dev Cell. 2023;58(12):1106–11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vias M, Morrill Gavarró L, Sauer CM, Sanders DA, Piskorz AM, Couturier D-L, Ballereau S, Hernando B, Schneider MP, Hall J, Correia-Martins F, et al. High-grade serous ovarian carcinoma organoids as models of chromosomal instability. eLife. 2023;12:e83867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Witte CJ, Espejo Valle-Inclan J, Hami N, Lõhmussaar K, Kopper O, Vreuls CPH, Jonges GN, van Diest P, Nguyen L, Clevers H, Kloosterman WP, et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 2020;31(11):107762. [DOI] [PubMed] [Google Scholar]
- 31. Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ, Jr, Feltmate CM, Nucci MR, Swisher EM, Nguyen H, Yang C, et al. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 2018;8(11):1404–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffmann K, Berger H, Kulbe H, Thillainadarasan S, Mollenkopf H-J, Zemojtel T, Taube E, Darb-Esfahani S, Mangler M, Sehouli J, Chekerov R, et al. Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. EMBO J. 2020;39(6):e104013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maenhoudt N, Defraye C, Boretto M, Jan Z, Heremans R, Boeckx B, Hermans F, Arijs I, Cox B, Van Nieuwenhuysen E, Vergote I, et al. Developing organoids from ovarian cancer as experimental and preclinical models. Stem Cell Reports. 2020;14(4):717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao X, Lv N, Liao J, Long J, Xue R, Ai N, Xu D, Fan X. Copy number variation is highly correlated with differential gene expression: a pan-cancer study. BMC Med Genet. 2019;20(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sui Y, Peng S. A Mechanism leading to changes in copy number variations affected by transcriptional level might be involved in evolution, embryonic development, senescence, and oncogenesis mediated by retrotransposons. Front Cell Dev Biol. 2021;9:618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hollis RL, Gourley C. Genetic and molecular changes in ovarian cancer. Cancer Biol Med. 2016;13(2):236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maru Y, Tanaka N, Itami M, Hippo Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol Oncol. 2019;154(1):189–98. [DOI] [PubMed] [Google Scholar]
- 38. Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H, van Wijk LM, et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25(5):838–49. [DOI] [PubMed] [Google Scholar]