Abstract
Rab7 regulates vesicle traffic from early to late endosomes, and from late endosomes to lysosomes. The crystal structure of Rab7-GTP in complex with the Rab7 binding domain of RILP reveals that Rab7 interacts with RILP specifically via two distinct areas, with the first one involving the switch and interswitch regions and the second one consisting of RabSF1 and RabSF4. Disruption of these interactions by mutations abrogates late endosomal/lysosomal targeting of Rab7 and RILP. The Rab7 binding domain of RILP forms a coiled-coil homodimer with two symmetric surfaces to interact with two separate Rab7-GTP molecules, forming a dyad configuration of Rab7–RILP2–Rab7. Mutations that disrupt RILP dimerization also abolish its interactions with Rab7-GTP and late endosomal/lysosomal targeting, suggesting that the dimeric form of RILP is a functional unit. Structural comparison suggests that the combined use of RabSF1 and RabSF4 with the switch regions may be a general mode of action for most Rab proteins in regulating membrane trafficking.
Keywords: membrane trafficking, Rab proteins, Rab7, Rab–effector complex, RILP
Introduction
Rab proteins, as general regulators of intracellular vesicle transport, constitute the largest GTPase family with more than 60 members in mammalian systems. Rab proteins are intrinsically cytosolic proteins and behave as membrane-associated molecular switches to regulate vesicle transport (Pfeffer, 2001; Zerial and McBride, 2001). Similar to other GTPases, only the GTP-bound form can recruit diverse effectors and associate with the membrane. Newly synthesized GDP-bound Rab proteins are bound by Rab escort protein (REP) and delivered to Rab geranylgeranyl transferase for prenylation (Andres et al, 1993). After prenylation, REP delivers Rab proteins to membrane for use in vesicular traffic (Alory and Balch, 2000). The later activation, inactivation and transition are regulated by guanine nucleotide exchange factors (GEF), GTPase-activating proteins (GAP) and GDP dissociation inhibitors (GDI), respectively.
Rab proteins share specific and conserved Rab family motifs (RabF1–RabF5) clustered in and around the switch I and switch II regions (Pereira-Leal and Seabra, 2000). These motifs, combined with the conserved PM/G motifs (phosphate/Mg2+ and guanine binding motifs; Valencia et al, 1991) and C-terminal double-cysteine prenylation, allow the classification of a Rab GTPase. In addition, Rab proteins are classified into subfamilies by specific motifs, called RabSF motifs (RabSF1–RabSF4). These motifs are unique characteristics for Rab subfamily (Pereira-Leal and Seabra, 2000).
Rab7 functions in the endocytic pathway of mammalian cells by regulating traffic from early to late endosomes and then to lysosomes (Feng et al, 1995; Meresse et al, 1995; Press et al, 1998; Bucci et al, 2000). Rab7 is recently shown to participate in growth factor-regulated cell nutrition and apoptosis (Edinger et al, 2003). Two effectors have been identified for Rab7: Rab7-interacting lysosomal protein (RILP) and Rabring7 (Cantalupo et al, 2001; Mizuno et al, 2003). RILP is recruited onto late endosomal and lysosomal membranes by Rab7 (Cantalupo et al, 2001), and it represents a downstream effector of Rab7. Recruitment of RILP by Rab7 is important for phagosome maturation and fusion with late endosomes and lysosomes (Harrison et al, 2003; Harrison et al, 2004; Marsman et al, 2004). RILP is a 45 kDa protein and contains a domain comprising two coiled-coil regions, and is mainly found in the cytosol (Cantalupo et al, 2001). Studies on chimeric proteins containing sequences from RILP and RILP-like proteins (RLPs) showed that a 62-residue unique region (amino acids 272–333) in RILP is responsible for its specific role in regulating lysosomal morphology as well as interacting with Rab7-GTP (Wang et al, 2004). However, the molecular mechanism by which Rab7 interacts with RILP to regulate late endosomal/lysosomal morphogenesis remains elusive.
Rab proteins are multifunctional and have multiple effectors often unrelated to each other (Pfeffer, 2001; Zerial and McBride, 2001). A key question concerns the molecular mechanism by which Rab GTPases generate specificity for a diverse spectrum of effectors and regulatory factors. Several hypervariable regions that play an important role in determining functional specificity were shown for Rab proteins (Brennwald and Novick, 1993; Stenmark et al, 1994). Consistently, crystal structure of the Rab3A–Rabphilin3A complex (Ostermeier and Brunger, 1999) revealed that both switch regions and three hypervariable regions (RabSF1, RabSF3 and RabSF4) are involved in the complex interface, supporting the hypothesis that RabSFs may also determine the specificity of Rab–effector interactions. However, the newly determined crystal structure of Rab5 in complex with the effector domain of Rabaptin5 revealed that only the switch and interswitch regions of Rab5 are involved in the interactions (Zhu et al, 2004).
To investigate how Rab7-GTP binds to RILP, we determined the crystal structure of Rab7-GTP complexed with the Rab7 binding domain of RILP. The structure shows that the Rab7 binding domain of RILP forms a tightly associated homodimer with two surfaces for symmetric binding to two separate Rab7 molecules simultaneously. In addition to switch and interswitch regions, RabSF motifs (RabSF1 and RabSF4) of Rab7 are also involved in the interactions with the Rab7 binding domain of RILP. These observations are similar to those observed in the Rab3A–Rabphilin3A complex but different from those in the crystal structure of Rab5 in complex with Rabaptin5. Structural comparison combined with mutational analysis provides a framework for understanding the molecular basis of recruitment of RILP by Rab7-GTP, and how Rab GTPases recognize their effectors specifically in general.
Results
Structure determination
Human full-length Rab7 (GTP-restricted mutant Rab7Q67L) and the Rab7 binding domain of RILP (residues 241–320; denoted hereafter as RILPe) were expressed in Escherichia coli. The complex was formed by mixing the purified Rab7 and RILPe and subsequently purified by gel filtration chromatography. The Rab7–RILPe complex was eluted with a Rab7/RILPe molar ratio of 2:2, suggesting the existence of a heterodimeric dimer in solution (data not shown). The structure was solved by molecular replacement method using Rab7-GTP (PDB code: 1T91; M Wu and H Song, unpublished results) as a search model. The current model has been refined at a resolution of 3.0 Å to working and free R factors of 26.8 and 27.8%, respectively, with good statistics and stereochemistry. A representative portion of the initial electron density map in the region of the Rab7 binding domain of RILP is shown in Figure 1A. All protein main-chain torsion angles are located in the energetically allowed regions for L-amino acids. Two regions are not visible in the electron density map and are assumed to be disordered, namely residues 186–207 in the C-terminus of Rab7 and residues 309–320 in the C-terminal segment of RILPe. Statistics of structure determination and refinement are summarized in Table I (see Materials and methods).
Figure 1.
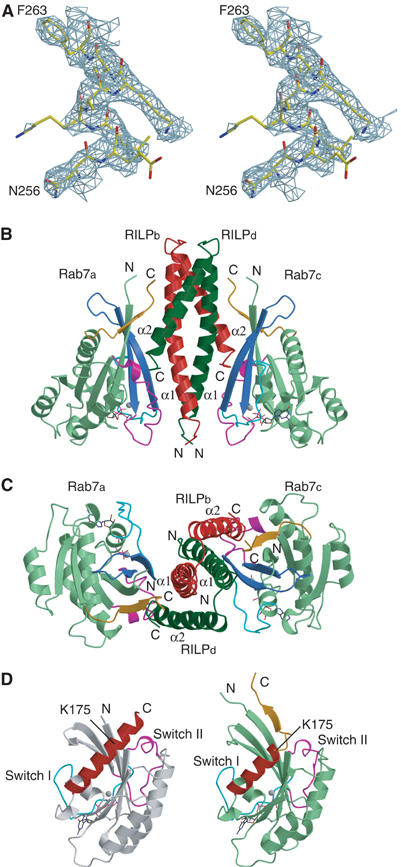
Structure of the Rab7–RILPe complex. (A) Stereo view of a representative portion of the Fo−Fc electron density map (contoured at 1.6σ) covering residues 256–263 of helix α1 in the Rab7 binding domain of RILP. The map was calculated with phase from Rab7 molecule only. (B) Ribbon diagram of the Rab7–RILPe complex. Two Rab7 molecules a and c are colored light green whereas two RILPe molecules b and d are red and dark green, respectively. The switch I, interswitch and switch II regions are colored cyan, royal blue and magenta, respectively. GTP molecule is shown in stick model and Mg2+ as cyan gray sphere. Residues 175–185 containing RabSF4 of Rab7 are shown in orange. (C) Top view of the Rab7–RILPe complex. The molecules are rotated 90° along a horizontal axis relative to the view in (B). (D) Comparison of Rab7-GTP in a free form and in complex with RILPe. Free Rab7-GTP (left panel) and that in complex with RILPe (right panel) are colored white and light green, respectively. Helix α5 of Rab7 is highlighted in red. The coloring scheme for the switch regions, GTP, Mg2+ and residues 175–185 of Rab7 in complex with RILPe is as in (B). Figures 1, 2 and 6 were generated using Molscript (Kraulis, 1991).
Table 1.
Data collection and refinement statistics
| Crystal | Rab7–RILP complex |
| Data collection | |
| Space group | P6522 |
| Unit cell dimension | |
| a/b/c (Å) | 93.13/93.13/132.83 |
| α/β/γ (deg) | 90.00/90.00/120.00 |
| Resolution range (Å) | 50–3.0 |
| Completeness (%) | 100 (100) |
| Unique reflections (N) | 7285 |
| Redundancy | 11.3 (11.4) |
| Rmerge (%)a | 9.7 (42.9) |
| I/σ | 6.6 (1.8) |
| Refinement | |
| Resolution (Å) | 20–3.0 |
| Used reflections (N) | 6904 |
| Total atoms (N) | 2082 |
| Protein atoms | 2015 |
| Nucleotide atoms | 32 |
| Mg2+ | 1 |
| Water molecules | 34 |
| Rwork (%)b | 26.8 (28.0) |
| Rfree (%)c | 27.8 (35.0) |
| R.m.s. deviation from ideal values | |
| Bond distance (Å) | 0.015 |
| Bond angle (deg) | 1.682 |
| Values in parentheses indicate the specific values in the highest resolution shell (3.1–3.0 Å). | |
| Rmerge=∑∣Ij−〈I〉∣/∑Ij, where Ij is the intensity of an individual reflection and 〈I〉 is the average intensity of that reflection. | |
| Rwork=∑∣∣Fo∣−∣Fc∣∣/∑∣Fc∣, where Fo denotes the observed structure-factor amplitude and Fc denotes the structure-factor amplitude calculated from the model. | |
| Rfree is as for Rwork but calculated with 5.0% of randomly chosen reflections omitted from the refinement. | |
Overall structure
The ribbon diagram of the Rab7–RILPe complex is shown in Figure 1B and C. There is one Rab7-GTP plus one RILPe molecule in the asymmetric unit. The structure of Rab7 in the complex contains a central six-strand β-sheet (β1–6) flanked by five α-helices (α1–5) and is in common with other Ras-like small GTPases. The fold of Rab7 in the complex is very similar to that of Rab7-GTP (Figure 1D) in which the conformation of switch II region is an extended loop rather than a well-defined helical structure present in most solved structures of Ras-like GTPase superfamily so far. Despite such high similarity, substantial structural differences are observed in the N- and C-terminal regions of Rab7. In the structure of Rab7-GTP, the N-terminal (residues 1–6) and C-terminal regions (183–207) are disordered, consistent with the observations that the hypervariable regions of Rab7 do not have a well-defined secondary structure in solution (Neu et al, 1997). In contrast, in the Rab7–RILPe complex structure, the N- and C-terminal regions extend to residues 3 and 185, respectively. Furthermore, the usual helical structure of residues 175–185 is melted into a small β-strand (residues 179–182) to form an antiparallel β-sheet with β1 (Figure 1D). This small β-sheet is involved in interactions with RILPe and may have important functional implications (see below).
RILPe is folded into a long helix α1 and a short helix α2 connected by a very tight loop. Since only helix α1 contacts Rab7 molecule in the asymmetric unit whereas helix α2 interacts with the crystallographic two-fold symmetry-related Rab7, the biologically meaningful complex was generated by the application of a crystallographic two-fold operation, which can be best described as Rab7–RILP2–Rab7 (Figure 1B and C). In this complex, one RILPe molecule interacts with its crystallographic two-fold symmetry-related counterpart to form a four helices homodimer in which both helices α1 and α2 are involved in dimerization. Such a homodimer binds to two separate Rab7-GTP molecules on opposite sides, with both helices involved in the interaction with Rab7. In the complex interface, although each Rab7-GTP interacts with both helices of RILPe, these two helices are contributed by two different molecules, with helix α1 coming from one protomer and helix α2 from the other protomer.
Rab7–RILP interaction
In the structure of the complex, there are two identical Rab7–RILPe interfaces (Figure 1B and C). For simplicity, we discuss only the interface formed between chain A of Rab7 molecules and the RILPe homodimer (referred to as chains B and D). Rab7-GTP and RILPe share an extensive interface with a buried accessible surface area of 2273 Å2, most of which involves hydrophobic interactions with some additional hydrogen bonds.
There are two distinct contact areas in the interface between Rab7 and RILPe. The first area involves the switch and interswitch regions of Rab7 as observed in other Ras-like small GTPases complexed with their effectors, and the N-terminal half of helix α1 and the C-terminus of helix α2 of RILPe (Figure 2A). Most residues involved in these interactions are highly conserved among members of the Rab7 subfamily. Lys38 (Lys82 in Rab34), a conserved residue in Rab7 and Rab34 (Figure 3A) in switch I region, which has been shown to be essential for interaction of Rab34 with RILP (Wang and Hong, 2002), makes van der Waals contacts with Glu247d and Gln250d. The invariant hydrophobic triad Phe45, Trp62 and Phe77 (Figure 3A), which has been implicated in effector binding for Rab5 subfamily members (Merithew et al, 2001), makes extensive hydrophobic interactions with RILPe (Figure 2A). Phe45 makes hydrophobic contacts with the methylene groups of both Lys259b and Asn256b. Ile41 makes hydrophobic interactions with the side chain of Phe248b, while Phe70 makes van der Waals contacts with the side chain of Arg245d. The methylene groups of both Thr47 and Thr58 in the interswitch region make contacts with aromatic ring from Phe263b. The side chains of Trp62 and Leu73 contact those of Leu306d and Leu252b, respectively, via hydrophobic interactions. Moreover, the side chain of Asp44 is hydrogen-bonded to the NZ group of Lys259b and the NH2 group of Arg255b, while the hydroxyl group of Asp82 forms a salt bridge with the NZ group of Lys304d.
Figure 2.
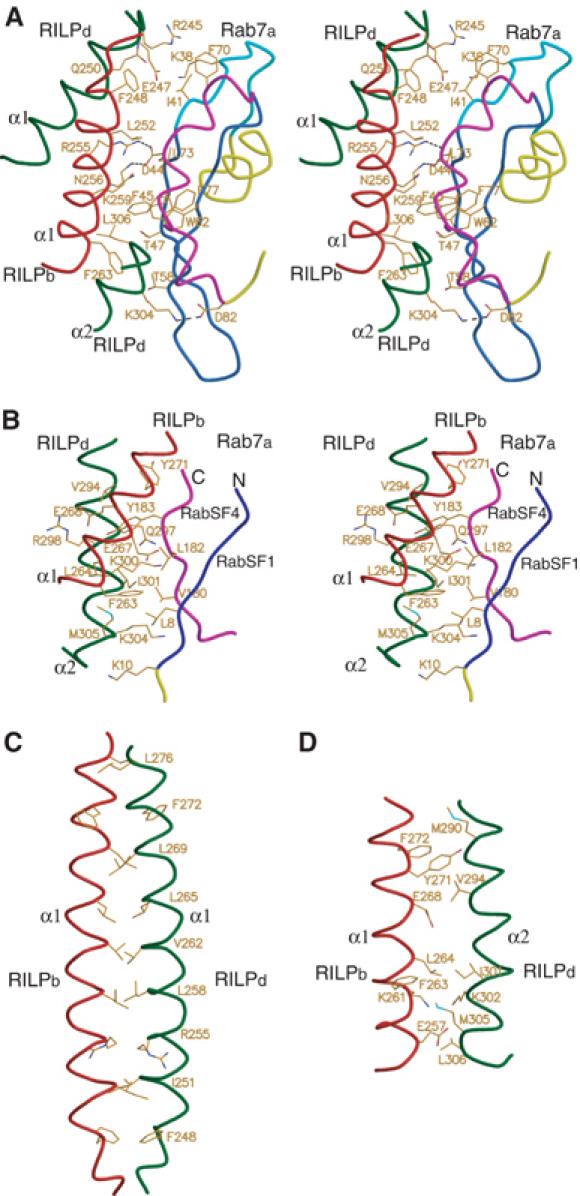
RILP–RILP and Rab7–RILP interfaces. (A) Stereo diagram of the first Rab7–RILP interface between the switch and interswitch regions of Rab7 and RILPe. The coloring scheme for the switch and interswitch, and RILPe molecules is as in Figure 1B, and the rest of Rab7 is yellow. (B) Stereo diagram of the second Rab7–RILP interface between the N- and C-terminal regions of Rab7 and RILPe. The N- and C-terminal regions of Rab7 are blue and magenta, respectively, with the rest of Rab7 in yellow. The coloring scheme for RILPe is as in (A). (C) The RILP–RILP interface showing the interactions of two long helices α1 from chain b (red) and d (dark green). For simplicity, only pairwise located residues involved in dimer interface are shown in stick model. (D) The RILP–RILP interface showing the interactions of the long helix α1 from chain b and the short helix α2 from chain d. The coloring scheme is as in (C). All residues involved in the interactions are shown in stick model.
Figure 3.
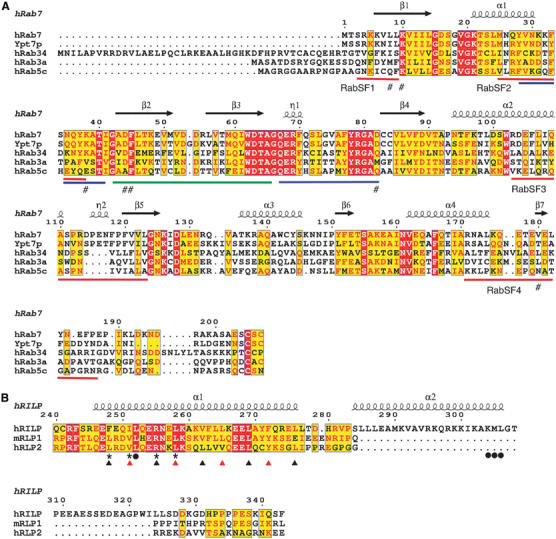
Sequence alignment of Rab GTPases, and the Rab7 binding domain of RILP with RLPs. (A) Sequence alignment of human Rab7, yeast Ypt7p, human Rab34, human Rab3a and human Rab5c. The secondary structures of human Rab7 in complex with RILPe are shown at top. Switch and interswitch regions are marked with blue and green lines, respectively. RabSF motifs are marked with red lines. Mutated residues involved in interaction with RILP are marked with #. (B) Sequence alignment of human RILP, mouse RLP1 and human RLP2. The secondary structures of RILPe are shown at top. Mutated residues involved in RILP dimerization are marked with * and those involved in interactions with Rab7 are marked with •. Residues showed in Figure 2C in positions a and d of the coiled-coil helix α1 are marked with black and red triangles, respectively.
The second contact area consists of parts of the N- and C-terminal segments of Rab7, which correspond to two hypervariable regions referred to as RabSF1 and RabSF4, respectively, and the C-terminal half of both helix α1 and α2 of RILPe (Figure 2B). Residues Leu8, Val180 and Leu182 combined with the methylene groups of Thr47 and Thr58 from the interswitch region described above form a hydrophobic surface to interact with a hydrophobic surface in RILPe, which is formed by the aliphatic group of K304d and the side chains of M305d, I301d, Leu264b and Phe263b. In the N-terminal region of Rab7, Leu8 makes multiple hydrophobic contacts with Lys304d, Met305d, Ile301d and Phe263b, while the side chain of Lys10 makes a hydrogen bond with the main-chain oxygen of Lys304d and van der Waals interactions with the main chains of both Met305d and Gly307d. In the C-terminal end of Rab7, the side chain of Val180 contacts that of Ile301d and the methylene groups of Lys300d and Lys304d via hydrophobic interactions, while Leu182 makes hydrophobic interactions with the aliphatic groups of Ile301d and Glu267b. Interestingly, the side chain of Tyr183 fits into a hydrophobic pocket formed by the side chains of Leu264b, Glu267b, Glu268b, Val294d, Gln297d, Arg298d and Ile301d and makes multiple hydrophobic contacts with these residues.
RILP dimer interface
The structure shows that two RILPe molecules form a tight homodimer, which is held together predominantly by the parallel long helix α1 (Figure 1B and C). In addition, the short helix α2 also interacts with helix α1 from the symmetry-related RILPe molecule. Thus, the four helices hold together to form the dimer interface dominated mainly by extensive hydrophobic interactions with a buried accessible surface area of 3680 Å2.
Two long helices α1 forming the core of the dimer interface is a typical short coiled-coil protein structure that consists of two identical strands of amino-acid sequences that wrap around each other in a gradual left-handed superhelical manner (Figure 1B). The amino-acid sequences in this coiled-coil structure are characterized by a heptad repeat denoted as a-b-c-d-e-f-g, in which the hydrophobic residues generally locate at positions a and d (O'Shea et al, 1991; Figure 3B). There are four heptad repeats in the coiled-coil region of RILPe dimer. All residues at positions a and d except Arg255 are hydrophobic in nature. Two sets of pairwise residues (Phe248, Ile251, Arg255, Leu258, Val262, Leu265, Leu269, Phe272 and Leu276) contact with each other to form the core of the dimer interface (Figure 2C). The spatial arrangements of these residues in the dimer interface are quite similar to those observed in the leucine zipper motif (O'Shea et al, 1991). Helix α2 also contributes to the dimerization of RILPe with its C-terminal segment contacting that of helix α1 from the symmetry-related RILP molecule via predominantly hydrophobic interactions (Figure 2D).
To verify the existence of the RILP homodimer in vitro and in vivo, a chemical crosslinking experiment was carried out using the recombinant RILPe, and the products were detected using SDS–PAGE. This result combined with that from analytical gel filtration confirmed that the Rab7 binding domain of RILP forms a homodimer in solution (data not shown). These results are further supported by deletion mutations and yeast two-hybrid interaction studies (Figure 4A). Both RILP199–401 and RILP241–310, which contain the intact helices α1 and α2, can self-interact well. Deletion of the C-terminal segment of helix α2 in RILP241–294 affected neither its self-interaction nor its interaction with RILP241–310. However, RILP261–310, a construct with the N-terminal half of helix α1 deleted, abolished its self-interaction and interactions with RILP241–310. These results suggest that the N-terminal half of helix α1 is essential for RILP dimer formation, whereas helix α2 is dispensable for dimerization.
Figure 4.

Effects of mutations on RILP dimerization and Rab7–RILP interaction. (A) RILP forms homodimer via self-interaction in a manner that is dependent on residues (241–260) in its coiled-coil region. Wild type (WT) refers to full-length RILP. (B) Effects of Ala point mutations of RILP on its dimerization. L287A serves as a positive control. (C) Effects of truncation or Ala point mutations in Rab7Q67L on its interaction with RILP. Rab7Q67L and E185A mutant serve as positive controls. Rab7ΔN contains residues 11–207, Rab7ΔC1 contains residues 1–176 and Rab7ΔC2 contains residues 1–185. (D) Effects of mutations of RILP on its interaction with Rab7Q67L. Wild-type (WT) RILP and RILPe serve as positive controls.
Mutagenesis and cellular localization
To examine the role of residues involved in the formation of RILP homodimer and in the interfaces between Rab7-GTP and RILP homodimer, Rab7 and RILP variants were created by site-directed mutagenesis. The resulting mutant Rab7 and RILP proteins were examined for their roles in Rab7–RILP interaction and/or RILP self-interaction by yeast two-hybrid interaction assay. The cellular localization of these mutants was also examined by immunofluorescence microscopy in HeLa cells expressing these mutants. Mutation of Leu8 to Ala (L8A) in Rab7 disrupted the binding of Rab7 to RILP (Figure 4C), and substantially reduced its late endosomal/lysosomal targeting (data not shown), while removal of the N-terminal 10 residues (Rab7ΔN; residues 11–207) or substitution of Lys10 with Ala (K10A) abrogated both interaction with RILP and its late endosomal/lysosomal targeting (Figures 4C and 5A). Mutations of some residues in the C-terminal region of Rab7 also affected its binding to RILP and cellular localization. Substitution of Val180 with Ala (V180A) abolished the binding of Rab7 to RILP (Figure 4C) and dramatically reduced its colocalization with RILP in the clustered late endosomes/lysosomes (Figure 5A), whereas mutation of Leu182 or Tyr183 or both to Ala had no effect on its RILP binding or cellular localization (data not shown). Moreover, removal of residues 177–207 (Rab7ΔC1) of Rab7 abolished its interaction with RILP, whereas deletion of residues 186–207 (Rab7ΔC2) had no effect on its interaction with RILP (Figure 4C). The C-terminal hypervariable region of Rab proteins has been shown to act as one of the main signals for subcelluar targeting (Chavrier et al, 1991; Stenmark et al, 1994; Ali et al, 2004). These results suggest that both RabSF1 and RabSF4 regions of Rab7 act together as the structural determinants for RILP binding and subcellular targeting. In support of our observations, residues 19–22 of the RabCDR (RabSF1) of Rab3A has been found to be essential for Rab3A–rabphilin3A complex formation (Ostermeier and Brunger, 1999). Consistent with a role of switch and interswitch regions, Ala substitutions of Asp44, Phe45 and Asp82 of Rab7 also disrupted its interaction with RILP and reduced substantially or abolished its targeting to the clustered late endosomes/lysosomes marked by co-overexpressed RILP (Figures 4C and 5A).
Figure 5.
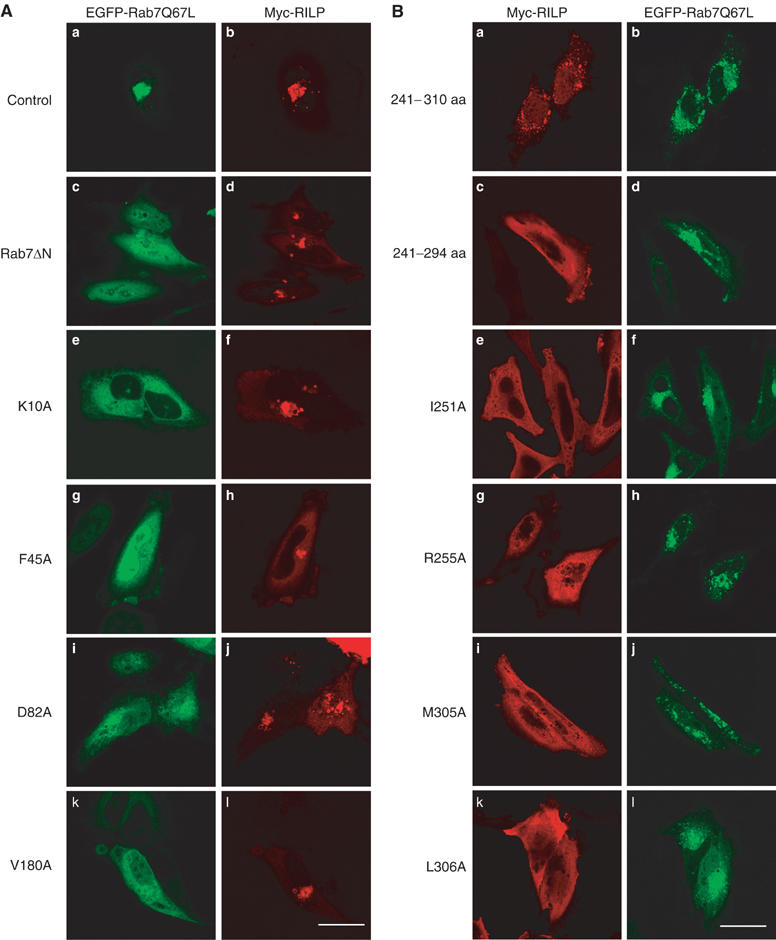
Effects of mutations of Rab7 and RILP on their cellular localization and membrane recruitment. (A) Representative site-directed mutants (panels e, g, i and k) or a truncated form (Rab7ΔN; panel c) of Rab7Q67L defective in interaction with RILP are mistargeted to the cytosol (and nucleus for F45A) (as revealed by GFP attached to the N-terminus of these proteins) and are not detected in the clustered lysosomes marked by coexpressed RILP (panels d, f, h, j and l as revealed by Myc tag). EGFP-Rab7Q67L (panel a) and Myc-RILP (panel b) serve as the positive control. Bar, 20 μm. (B) Representative mutants of RILP (panels c, e, g, i and k, revealed by Myc tag) defective in interaction with Rab7 are mistargeted to the cytosol and did not associate with punctuate late endosomes/lysosomes marked by coexpressed EGFP-Rab7Q67L (panels b, d, f, h, j and l, viewed by GFP). The fragment encompassing residues 241–310 of RILPe is peripheral distributed, but still can be efficiently recruited to the punctuate structures marked by EGFP-Rab7Q67L (panels a and b). Bar, 20 μm.
We also mutated residues of RILP that are predicted to participate in Rab7 interactions. Mutations of Leu252, Lys304, Met305 and Leu306 of RILP to Ala abrogated its interaction with Rab7, and these mutants failed to colocalize with Rab7 in late endosomes/lysosomes (Figures 4D and 5B). Moreover, deletion of residues 295–306 in the C-terminal half of helix α2 led to defect in late endosomal/lysosomal targeting (Figure 5B). Since Lys304, Met305 and Leu306 are located in a 62-residue unique region of RILP (residues 272–333), which has been shown to distinguish it from RLPs in regulation of lysosomal morphology and interactions with Rab7 (Wang et al, 2004), these observations confirm our previous results and explain why RILP rather than RLPs can interact with Rab7 and regulate lysosomal morphology. Collectively, these results suggest that the specific interactions between Rab7 and RILPe are essential for targeting of Rab7 and RILP to late endosomes/lysosomes.
The extensive dimer interface of RILPe and the existence of such a dimer in vitro and in vivo suggest that dimerized RILPe may function as a structural unit for interaction with Rab7. We mutated residues that are located in the core of RILPe dimer interface. Consistent with the observation that the N-terminal half of helix α1 is essential for RILP dimerization (Figure 4A), Ala substitutions of Ile251 and Arg255 (I251A and R255A) abolished both the dimerization of RILPe and targeting to late endosomes/lysosomes (Figures 4B and 5B), while mutations of Phe248 and Leu258 to Ala (F248A and L258A) reduced dramatically the self-interaction of RILPe (Figure 4B), and substantially reduced late endosomal/lysosomal targeting (data not shown). Moreover, mutations of Ile251 and Arg255 to Ala abolished the interaction of RILP with Rab7, while substitution of Leu258 for Ala substantially reduced its binding to Rab7 (Figure 4D). Residues Ile251, Arg255 and Leu258 make no direct contacts with Rab7-GTP, but substitutions of these residues with Ala still abolished or dramatically reduced the interaction with Rab7 and targeting of RILP to late endosomes/lysosomes. These results suggest that mutations disrupting the RILPe dimerization also abolish indirectly its interaction with Rab7-GTP and its late endosomal/lysosomal targeting. Combined with the observation that the Rab7 binding domain of RILP exists as a homodimer, these results suggest that the dimerized Rab7 binding domain of RILP is the structural and functional unit for interaction with two Rab7 molecules. In this structural unit, helix α1 is essential for both dimerization and late endosomal/lysosomal targeting, whereas helix α2 is dispensable for dimerization but is absolutely required for late endosomal/lysosomal targeting.
Structural diversity of Rab–effector recognition
Numerous studies have established that Rab proteins are distributed in distinct intracellular compartments and function in concert with multiple effectors to regulate transport between organelles (Zerial and McBride, 2001). One important issue about the Rab–effector interaction is how the GTP-bound form of Rab protein recognizes its effectors specifically. To understand the structural basis of the Rab–effector recognition in general, we compared the crystal structures of our Rab7–RILPe complex, the Rab3A–Rabphlin3A complex (Ostermeier and Brunger, 1999) and the recently reported Rab5–Rabaptin5 complex (Zhu et al, 2004). Comparison of these complexes revealed some similarities and differences as well. Although in all three complexes, Rab molecules interact with their respective effectors using the relative conserved switch and interswitch regions, the overall binding modes of three complexes are markedly different (Figure 6). First, Rabphilin3A does not form a homodimer whereas the effector domain of both Rabaptin5 and RILP forms a homodimer for interaction with two Rab molecules. Second, only switch and interswitch regions of Rab5 in the Rab5–Rabaptin5 complex are involved in the interactions with the effector domain of Rabaptin5 in a manner similar to those observed in Arf/Sar GTPase family, whereas RabSF1 and RabSF4 of Rab7, and RabSF1, RabSF3 and RabSF4 of Rab3A are also involved in the interactions with their respective effector molecules. The fact that the RabSF1 region of Rab7 is absolutely important for interaction with RILP and its late endosomal/lysosomal targeting suggests that this region plays a determining role in interaction with RILP. Moreover, although in all three complexes, the effector molecules contact their respective Rab proteins using an N-terminal long helix and a C-terminal short helix, the specific recognition between Rab proteins and their effectors is achieved in a remarkably different way (Figure 6). In the Rab3A–Rabphilin3A complex, the N-terminal segment of the long helix of Rabphilin3A contacts the switch and interswitch regions of Rab3A, while the C-terminal helix and the adjacent SGAWFF structural element of Rabphlin3A fit into a hydrophobic pocket formed by RabSF1, RabSF3, RabSF4 and loop α2–β4 (Ostermeier and Brunger, 1999). In the Rab5–Rabaptin5 complex, the effector domain of Rabaptin5 contacts Rab5 using only the N-terminal long helices of both protomers in the center of the homodimer, with its C-terminal short helix playing no role in binding of Rab5 (Figure 6). The structure of the Rab7–RILP complex (Figures 1B and 6) revealed that RILPe contacts Rab7 using the N-terminal long helix α1 of one protomer and the C-terminal short helix α2 of the other protomer. Furthermore, both helices α1 and α2 of RILPe contact the RabSF1 and RabSF4 regions and the switch and interswitch regions, with helix α1 contacting more with the switch and interswitch regions of Rab7 and helix α2 making more interactions with RabSF1 and RabSF4. Finally, the unique feature in our structure is that the small β-sheet formed by the RabSF1 and RabSF4 of Rab7 is involved in effector binding (Figures 1D and 6). The structural diversity of Rab–effector recognition and involvement of the RabSF motifs of Rab7 and Rab3A in effector binding suggest that each Rab protein interacts with its effector in a highly specific manner, presumably using the combination of the RabSF motifs in addition to the switch mechanism, which is conserved for all small GTPases.
Figure 6.
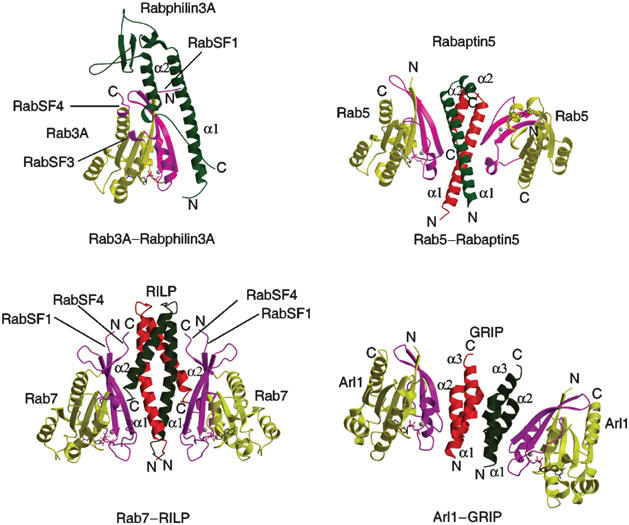
Structural comparison of the complexes of Rab3A–Rabphilin3A, Rab5–Rabaptin5, Rab7–RILP and Arl1–GRIP. The regions of GTPases involved in interactions with their effectors are highlighted in magenta. The N- and C-termini of GTPases and their effectors are labeled.
Discussion
Structure of the Rab7-GTP in complex with the Rab7 binding domain of RILP shows that Rab7-GTP interacts with RILPe in a highly specific manner with the switch and interswitch regions conferring the main binding affinity and the RabSF1 and RabSF4 motifs providing the additional affinities that are absolutely required for the interaction measured by yeast two-hybrid assays. Structural and mutational studies explain why Rab7 but not other Rab proteins interact with RILPe since some key residues of Rab7 involved in RILPe binding are unique to Rab7 proteins across species but not in other Rab proteins (Figure 3A). Moreover, the fact that the helix α2 of RILPe interacts specifically with the RabSF1 and RabSF4 regions of Rab7 confirmed our earlier studies on RILP, showing that a unique region of RILP consisting of helix α2 is essential for regulation of late endosomal/lysosomal morphology and interactions with Rab7 and Rab34 (Wang et al, 2004). Sequence comparison between Rab7 and Rab34 showed that most residues of Rab7 involved in interactions with RILP are conserved in Rab34 (Figure 3A). This observation probably accounts for the fact that RILP is a common effector for Rab7 and Rab34.
In the Rab7–RILPe complex, RILPe forms a homodimer, which acts as a structural and functional unit that creates two symmetric surfaces on the two opposite sides of the dimer to interact with two separate Rab7-GTP molecules. The mode of action of RILPe dimer in Rab7 binding is reminiscent of GRIP in Arl1 binding (Panic et al, 2003; Wu et al, 2004) but with markedly different binding mode in several details (Figure 6). First, all three helices of GRIP are indispensable for dimerization, whereas only the long helix α1 of RILPe is essential for dimerization. Second, GRIP interacts with Arl1 using its first two helices (α1and α2) from the same protomer, while RILPe contacts Rab7 using the long helix α1 from one protomer and the short helix α2 from the other protomer. Third, GRIP interacts predominantly with the switch II region of Arl1, while RILPe contacts not only the switch and interswitch regions but also the RabSF1 and RabSF4 regions of Rab7. So far, three small GTPases in complex with their effectors, the Rab7–RILP complex, the Arl1–GRIP complex (Panic et al, 2003; Wu et al, 2004) and the Rab5–Rabaptin5 complex (Zhu et al, 2004), exhibit a dyad-symmetric binding mode. For Golgin-245 and Rabaptin5, the dimerization of the C-terminal domain may facilitate their anchoring on the membrane. However, the Rab7 binding domain of RILP is located in the middle of its sequence. Although RILPe dimerization is necessary for interaction with Rab7 as well as for its membrane targeting, the significance for its Rab7 binding domain being positioned in the middle of the molecule remains to be examined.
Structural comparison of three Rab–effector complexes revealed some common features of effector binding with some striking differences as well. The use of switch and interswitch regions for interaction with effectors or regulators seems to be a common characteristic for all small GTPases (Hanzal-Bayer et al, 2002; Panic et al, 2003; Shiba et al, 2003; Wu et al, 2004). These GTPases use their switch mechanism to sense the state of the bound nucleotide, thereby binding to their respective effectors or regulators. For example, Rab7 in GDP form binds to REP-1 (Rak et al, 2004) while in GTP form it binds to RILPe using the common switch and interswitch regions. Rab proteins constitute a unique family of small GTPase in terms of their diverse roles in membrane trafficking and the heterogeneity of their interaction with effectors. The crystal structure of Rab3A–Rabphilin3A (Ostermeier and Brunger, 1999) revealed for the first time that Rab3A, unlike other Ras-like small GTPases, interacts with its effector using combination of its switch mechanism and its RabCDRs, and led to a conclusion that RabCDRs are probably the key determinants for the regulation of vesicle traffic. Our structural study reported here supports this conclusion by revealing that RabCDRs (RabSF1 and RabSF4) of Rab7 are additional and required structural determinants for effector binding. Although RabSF3 (α3–β5 loop) has been shown to be involved in the specific interaction with Rabphilin3A (Ostermeier and Brunger, 1999), it does not contribute to the Rab7–RILP interaction. These observations suggest that the α3–β5 loop may be just specific to Rab3A subfamily and not a general structural determinant for effector binding, despite the finding that α3–β5 loop is important for the function of other Rab proteins including Ypt1p and Rab5 (Brennwald and Novick, 1993; Stenmark et al, 1994). Instead, the RabSF1 and RabSF4 motifs of Rab GTPases play more important roles in effector recognition (Chavrier et al, 1991; Steele-Mortimer et al, 1994; Ostermeier and Brunger, 1999). The observation that only the switch and interswitch regions of Rab5 are involved in Rabaptin5 binding is probably not due to the truncation of the C-terminal end of Rab5 used for crystallization (Zhu et al, 2004), as the full-length and C-terminal truncated Rab5 bind equally well to the N-terminus of early endosome antigen 1 (Merithew et al, 2003). Additional Rab5–Rabaptin5 interactions might exist, as the N-terminus of Rab5 has been suggested to be a structural determinant for effector binding (Stenmark et al, 1994). In our structure, upon RILPe binding, the C-terminal region of Rab7 undergoes a strikingly structural transition and part of helix α5 transforms into a β-sheet with the extended N-terminus, thereby interacting with RILP (Figure 1). The involvement of the RabSF motifs, particularly the N- and C-terminal RabSFs, may be a general feature for most Rab–effector interaction. The combination of the RabSFs with the conserved switch and interswitch regions allows Rab proteins to bind a wide range of effectors in a specific manner and participate in distinct trafficking pathways.
In conclusion, we present evidences that Rab7-GTP interacts with the Rab7 binding domain of RILP with high specificity, and disrupting these interactions abolishes late endosomal/lysosomal targeting for both Rab7 and RILP. Similar to the GRIP domain of Golgin-245, the Rab7 binding domain of RILP forms a homodimer, which serves as a structural and functional unit for Rab7 binding and thereafter membrane targeting. This conclusion raises an intriguing possibility that Rab7 and RILP are targeted simultaneously to the membrane as Rab7–RILP2–Rab7 complex. With regard to membrane targeting, Rab7 and RILP may influence each other, rather than one regulates the other. The combined use of the switch and interswitch regions with the RabSF1 and RabSF4 motifs for effector binding as shown here for Rab7 and Rab3A (Ostermeier and Brunger, 1999) may be a general mode of action for most Rab proteins.
Materials and methods
Protein expression and purification
Both full-length human Rab7 and the Rab7 binding domain of RILP (RILPe; residues 241–320) were cloned into pGEX-6p-1 (Amersham) and expressed as GST fusion protein in E. coli BL-21 cells. The GTP-restricted Rab7Q67L was constructed using the site-directed mutagenesis kit (Stratagene). Cells were grown to OD600 nm=0.6 at 37°C and then induced by 0.2 mM IPTG at 28°C for 6 h. After centrifugation, cells expressing RILPe were resuspended in the lysis buffer (30 mM Tris–HCl pH 8.0, 500 mM NaCl, 2 mM DTT, 2 mM MgCl2, 0.1 mM PMSF, 2 mM benzamidine and 1 mg/ml lysozyme) for 30 min, and lysed by sonication. The clarified cell lysate was loaded onto glutathione-Sepharose 4B column (Amersham). GST fusion protein was eluted by glutathione and cleaved by PreScission protease (Amersham) overnight at 4°C. After desalting, cleaved protein was loaded on glutathione-Sepharose 4B column and further purified by MonoQ ion-exchange and Superdex 75 (Amersham) gel filtration columns. Cells expressing Rab7Q67L were lysed in the lysis buffer containing 1 mM GTP. Rab7Q67L was purified by glutathione-Sepharose 4B column as described above and further purified by Superdex 75 gel filtration column (Amersham). Purified Rab7Q67L and RILPe were mixed together in the presence of 2 mM GTP in a buffer containing 30 mM Tris–HCl (pH 8.0), 50 mM NaCl, 2 mM DTT and 2 mM MgCl2. The complex of Rab7Q67L and RILPe was further purified by Superdex 75 gel filtration column. Eluted complex-containing fractions were concentrated to 10 mg/ml for crystallization.
Crystallization, data collection and structure determination
The crystals of the complex were grown at 20°C using the hanging-drop method. Equal volume of protein solution was mixed with the precipitant solution containing 2 M ammonium sulfate, 0.5% polyvinylpyrrolidone K15 and 100 mM HEPES, pH 7.6. Crystals were cryoprotected using 2.5 M malonic acid (pH 7.4) and flash-frozen in liquid nitrogen (Holyoak et al, 2003).
Diffraction data were collected at 100 K on beamline BW7A at DESY (Hamburg, Germany). Data were processed using MOSFLM and intensities were reduced and scaled using SCALA (CCP4, 1994). The structure was solved using AMoRe (Navaza and Saludjian, 1997) with GTP-bound Rab7Q67L (MS Wu and H Song, unpublished results) as a search model. The subsequent model rebuilding was carried out using the program O (Jones et al, 1991). Refinement was performed using CNS (Brunger et al, 1998) and the solvent molecules were included automatically and manually edited with electron densities. The final round of the refinement was carried out with REFMAC5 (Murshudov et al, 1997). The average B-factors for Rab7, RILP, GTP and water molecules are 22.2, 48.0, 44.8 and 43.2 Å2, respectively. The real space correlation coefficient per residue based on a simulated annealed composite omit map for ordered residues is in the range of 0.70–0.97. All data statistics are shown in Table I.
Yeast two-hybrid and cellular localization
Protein–protein interaction analysis was carried out by yeast two-hybrid assays as described previously (Wang and Hong, 2002; Wu et al, 2004). Ala point mutations or truncation of wild-type RILP and Rab7Q67L were generated using standard PCR-mediated mutagenesis. Wild-type RILP and its mutants, and Rab7Q67L and its mutants were inserted into pGBKT7 or pGADT7 vectors (BD Clontech). The self-interaction (dimerization) of RILP was tested using the yeast clones transformed with wild-type protein and its truncation and some Ala point variants in QDO (without tryptophan, leucine, histidine and adenine) agar plates. The interaction between Rab7 and RILP was checked using the yeast clones transformed with Rab7Q67L and its variants cloned into pGADT7 vector and the wild-type RILP and its variants cloned into pGBKT7 vector. The zygotes were grown in QDO plates as described above.
For immunofluorescence microscopy, the wild-type and mutated RILP were cloned into pDMYCneo vector (Wang et al, 2004). The Rab7Q67L and its mutants were cloned into pEGFP-C1 vector (BD Clontech). The indicated RILP and the Rab7 constructs were cotransfected into HeLa cells. The transfected cells were fixed and immunostained with Myc-tag antibody (9E10, ATCC). Microscopy analysis was performed by using Carl Zeiss Axioplan II confocal microscope.
Acknowledgments
We are grateful to the beamline scientists at BW7A, DESY (EMBL, Hamburg) for their kind help during our data collection. This work is financially supported by the Agency for Science, Technology and Research (A* Star) in Singapore (to WH and HS). The coordinates and structure-factor amplitudes of the Rab7–RILPe complex have been deposited in the Protein Data Bank with ID code 1YHN.
References
- Ali BR, Wasmeier C, Lamoreux L, Strom M, Seabra MC (2004) Multiple regions contribute to membrane targeting of Rab GTPases. J Cell Sci 117: 6401–6412 [DOI] [PubMed] [Google Scholar]
- Alory C, Balch WE (2000) Molecular basis for Rab prenylation. J Cell Biol 150: 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73: 1091–1099 [DOI] [PubMed] [Google Scholar]
- Brennwald P, Novick P (1993) Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature 362: 560–563 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B (2000) Rab7: a key to lysosome biogenesis. Mol Biol Cell 11: 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C (2001) Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J 20: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project No. 4) (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chavrier P, Gorvel JP, Stelzer E, Simons K, Gruenberg J, Zerial M (1991) Hypervariable C-terminal domain of rab proteins acts as a targeting signal. Nature 353: 769–772 [DOI] [PubMed] [Google Scholar]
- Edinger AL, Cinalli RM, Thompson CB (2003) Rab7 prevents growth factor-independent survival by inhibiting cell-autonomous nutrient transporter expression. Dev Cell 5: 571–582 [DOI] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A (1995) Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol 131: 1435–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC (2002) The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J 21: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RE, Brumell JH, Khandani A, Bucci C, Scott CC, Jiang X, Finlay BB, Grinstein S (2004) Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol Biol Cell 15: 3146–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S (2003) Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol 23: 6494–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak T, Fenn TD, Wilson MA, Moulin AG, Ringe D, Petsko GA (2003) Malonate: a versatile cryoprotectant and stabilizing solution for salt-grown macromolecular crystals. Acta Crystallogr D 59: 2356–2358 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Marsman M, Jordens I, Kuijl C, Janssen L, Neefjes J (2004) Dynein-mediated vesicle transport controls intracellular Salmonella replication. Mol Biol Cell 15: 2954–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meresse S, Gorvel JP, Chavrier P (1995) The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J Cell Sci 108 (Part 11): 3349–3358 [DOI] [PubMed] [Google Scholar]
- Merithew E, Hatherly S, Dumas JJ, Lawe DC, Heller-Harrison R, Lambright DG (2001) Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition. J Biol Chem 276: 13982–13988 [DOI] [PubMed] [Google Scholar]
- Merithew E, Stone C, Eathiraj S, Lambright DG (2003) Determinants of Rab5 interaction with the N terminus of early endosome antigen 1. J Biol Chem 278: 8494–8500 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kitamura A, Sasaki T (2003) Rabring7, a novel Rab7 target protein with a RING finger motif. Mol Biol Cell 14: 3741–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Navaza J, Saludjian P (1997) Amore: an automated molecular replacement program package. Methods Enzymol 276: 581–594 [DOI] [PubMed] [Google Scholar]
- Neu M, Brachvogel V, Oschkinat H, Zerial M, Metcalf P (1997) Rab7: NMR and kinetics analysis of intact and C-terminal truncated constructs. Proteins 27: 204–209 [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Klemm JD, Kim PS, Alber T (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254: 539–544 [DOI] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT (1999) Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 96: 363–374 [DOI] [PubMed] [Google Scholar]
- Panic B, Perisic O, Veprintsev DB, Williams RL, Munro S (2003) Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol Cell 12: 863–874 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC (2000) The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol 301: 1077–1087 [DOI] [PubMed] [Google Scholar]
- Pfeffer SR (2001) Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol 11: 487–491 [DOI] [PubMed] [Google Scholar]
- Press B, Feng Y, Hoflack B, Wandinger-Ness A (1998) Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J Cell Biol 140: 1075–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak A, Pylypenko O, Niculae A, Pyatkov K, Goody RS, Alexandrov K (2004) Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease. Cell 117: 749–760 [DOI] [PubMed] [Google Scholar]
- Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S (2003) Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol 10: 386–393 [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Clague MJ, Huber LA, Chavrier P, Gruenberg J, Gorvel JP (1994) The N-terminal domain of a rab protein is involved in membrane–membrane recognition and/or fusion. EMBO J 13: 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Valencia A, Martinez O, Ullrich O, Goud B, Zerial M (1994) Distinct structural elements of rab5 define its functional specificity. EMBO J 13: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia A, Chardin P, Wittinghofer A, Sanderm C (1991) The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry 30: 4637–4648 [DOI] [PubMed] [Google Scholar]
- Wang T, Hong W (2002) Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell 13: 4317–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wong KK, Hong W (2004) A unique region of RILP distinguishes it from its related proteins in its regulation of lysosomal morphology and interaction with Rab7 and Rab34. Mol Biol Cell 15: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Lu L, Hong W, Song H (2004) Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat Struct Mol Biol 11: 86–94 [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zhu G, Zhai P, Liu J, Terzyan S, Li G, Zhang XC (2004) Structural basis of Rab5–Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol 11: 975–983 [DOI] [PubMed] [Google Scholar]


