This work reports the structure of an enteric adenovirus and its differences with respiratory and ocular adenoviruses.
Abstract
Enteric adenoviruses, one of the main causes of viral gastroenteritis in the world, must withstand the harsh conditions found in the gut. This requirement suggests that capsid stability must be different from that of other adenoviruses. We report the 4-Å-resolution structure of a human enteric adenovirus, HAdV-F41, and compare it with that of other adenoviruses with respiratory (HAdV-C5) and ocular (HAdV-D26) tropisms. While the overall structures of hexon, penton base, and internal minor coat proteins IIIa and VIII are conserved, we observe partially ordered elements reinforcing the vertex region, which suggests their role in enhancing the physicochemical capsid stability of HAdV-F41. Unexpectedly, we find an organization of the external minor coat protein IX different from all previously characterized human and nonhuman mastadenoviruses. Knowledge of the structure of enteric adenoviruses provides a starting point for the design of vectors suitable for oral delivery or intestinal targeting.
INTRODUCTION
Adenoviruses (AdVs) are pathogens that can be engineered to become therapeutic tools (1, 2). More than 100 types of human AdVs (HAdVs) have been described so far (http://hadvwg.gmu.edu/; https://talk.ictvonline.org/) and grouped in seven species (HAdV-A to HAdV-G). HAdVs cause acute infections in the respiratory and gastrointestinal tracts, as well as conjunctivitis. Most AdV infections are subclinical, but they can cause significant morbidity and even mortality in immunocompromised patients (3). There are currently no vaccines for use in the general population (although some are available for U.S. military personnel) or approved antiviral therapies for AdV infections.
High-resolution (3.2 to 3.5 Å) structures are available only for two HAdVs: HAdV-C5 and HAdV-D26 (4–6). The AdV capsid stands out among the nonenveloped viruses because of its large size (~950 Å, 150 MDa), triangulation number (pseudo-T = 25), and complex composition. The facets are formed by 240 trimeric hexons, while pentamers of penton base protein fill the vertices. Receptor-binding trimeric fibers of various lengths, depending on the virus type, project from the penton bases (7). Minor coat proteins IIIa and VIII on the inner capsid surface, and IX on the outer one, contribute to modulate the quasi-equivalent icosahedral interactions. The membrane lytic protein VI competes with genome-condensing protein VII for binding to an internal cavity in hexons (5, 8).
HAdVs in species F are unusual in that they have a narrow enteric tropism. HAdV-F40 and HAdV-F41 are one of the three major causes of viral gastroenteritis in young children, together with rota- and noroviruses (9, 10). They account for more than half of the AdVs identified in stools of immunocompetent symptomatic children (11–13). Differences in the capsid structure of HAdV-F, relative to HAdV-C, may confer stability to gastric conditions en route to the intestine in vivo. Infectivity assays after incubation under acidic conditions have shown that HAdV-F41 is stable, and even possibly activated, at low pH, in contrast to HAdV-C5 (14). The fact that HAdV-F viruses code for two different fibers has been related with their enteric tropism, although the exact relation is unclear (15, 16). The long fiber binds to the Coxsackie and AdV receptor (CAR) protein like the single fibers of many other nonenteric AdV species (17), but no receptor has been identified for the short fiber. However, pseudotyping HAdV-C5 with the short fiber of HAdV-F40 resulted in resistance to inactivation under acidic conditions and the ability to produce infection in the intestine after oral or rectal administration (18).
Here, we use cryo–electron microscopy (cryo-EM) to obtain the structure of the HAdV-F41 virion and compare it with the two other known HAdV structures. We interpret the structural differences in the context of the high physicochemical stability of the enteric AdV capsid.
RESULTS
Physicochemical stability of HAdV-F41 virions
To evaluate the stability of HAdV-F41 virions, we assessed their infectivity after treatment with synthetic gastric, intestinal, or gastric followed by intestinal fluids. Green fluorescent protein (GFP) fluorescence measured by flow cytometry was used to determine the number of infected cells in human embryonic kidney (HEK) 293 cultures. Neither gastric nor intestinal conditions reduced infectivity. Consecutive incubation in synthetic gastric and intestinal fluid resulted in a clear increase in GFP expression, indicating enhanced infectivity (fig. S1A). That is, not only is HAdV-F41 resistant to acidic pH as previously shown (14), but it is also activated by the combined action of acidification, salt, and proteases found in the sequential gastric and intestinal treatment.
We also examined the physical stability of HAdV-F41 particles in comparison to HAdV-C5 using extrinsic fluorescence of the DNA-intercalating agent YOYO-1 to characterize capsid disruption as a function of temperature. The fluorescence emission of YOYO-1 increased with temperature for both specimens, as genomes became exposed to the solvent (fig. S1B). While the half-transition temperature (T0.5) estimated for HAdV-C5 was 47°C (8), for HAdV-F41, the maximum rate of DNA exposure happened at near 51°C. This result indicates that HAdV-F41 particles are thermally more stable than HAdV-C5.
Overall HAdV-F41 structure
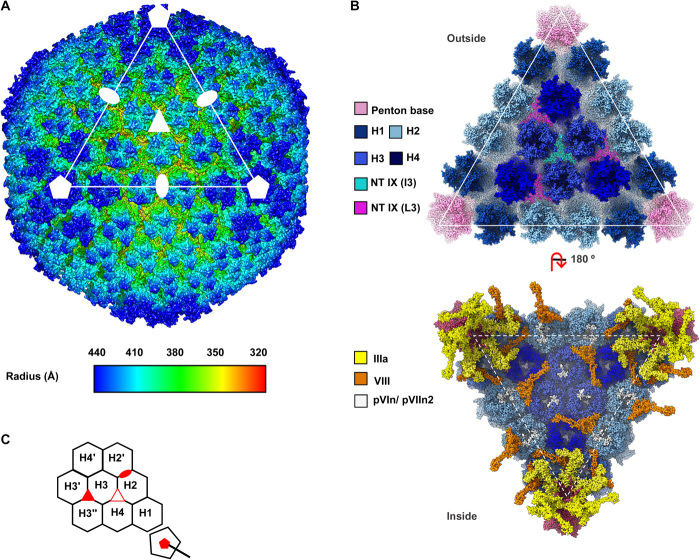
The cryo-EM map of HAdV-F41 at 4.0-Å resolution (Fig. 1, fig. S2, and table S1) shows the general characteristics common to all previous AdV structures: a ~950-Å-diameter pseudo T = 25 icosahedral particle with 12 trimeric hexon capsomers per facet plus the pentameric pentons at the vertices. The N terminus of protein IX lays on the outer capsid surface, in the valleys formed by hexon trimers (Fig. 1B, top). Minor coat proteins IIIa and VIII, plus small fragments of VI and core protein VII, can be observed on the inner surface of the icosahedral shell, oriented toward the virion core (Fig. 1B, bottom). Because of their flexibility and symmetry mismatch with the penton base, fibers cannot be resolved by cryo-EM when imposing icosahedral symmetry as done here.
Fig. 1. The cryo-EM structure of HAdV-F41 at 4.0 Å.
(A) Radially colored surface rendering of the HAdV-F41 map viewed down the twofold icosahedral axis. One facet is identified by a white triangle. Icosahedral symmetry axes are indicated with white symbols: fivefold (pentagon), threefold (triangle), and twofold (oval). A Gaussian filter has been applied to the map to accentuate the main features. (B) The molecular models of the major and minor coat proteins traced in the icosahedral HAdV-F41 facet. Top: view from outside the capsid. Bottom: view inside the capsid. Protein colors as indicated by the legend at the left hand side. The four hexon trimers in the asymmetric unit (AU) are numbered H1 to H4. (C) Cartoon depicting the icosahedral AU and surrounding hexons, as seen from outside the capsid. The icosahedral symmetry axes are indicated with red filled symbols. A hollow red triangle indicates the L3 axis between hexons 2, 3, and 4.
Except for proteins VI and VIII, the HAdV-F41 structural polypeptides are shorter than their HAdV-C5 counterparts (table S2). Sequence identity varies between 52 and 78%. The predicted isoelectric points for hexon and penton base suggest a more basic capsid surface for HAdV-F41 (14), except for the contribution of the external cementing protein IX, which is remarkably more acidic (table S2). On the inner capsid surface, proteins IIIa, VI, and VIII are more acidic in the enteric AdV.
Major coat protein: Hexon
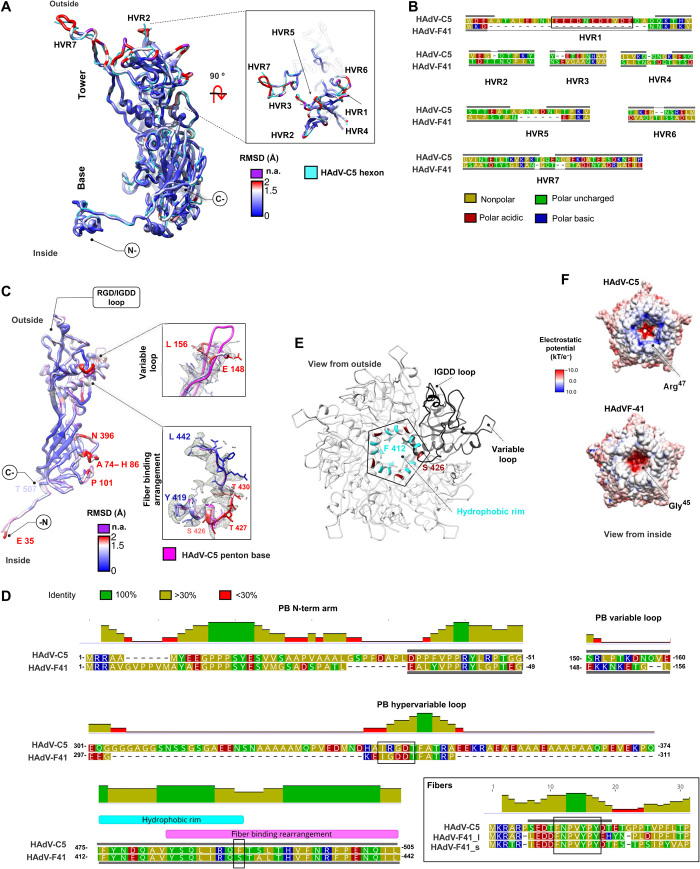
The architecture of the HAdV-F41 hexon is very similar to that of HAdV-C5 (Fig. 2A and table S2) (5). The double jelly roll in each monomer that forms the trimer pseudo-hexagonal base is highly conserved. As expected (19), the main differences between the structures are in the loops located in the hexon towers, the so-called hypervariable regions (HVRs) that constitute serotype-specific epitopes exposed on the virion surface (Fig. 2, A and B, and table S3). As in HAdV-C5, HVRs in HAdV-F41 are flexible (Supplementary Text and fig. S3A), but we were able to fully trace all of them except HVR4 (Fig. 2, A and B, and table S4). The 32-residue acidic loop in HVR1, unique to HAdVs in species C (20), has not been traced in any of the available HAdV-C hexon structures. We were able to completely trace HVR1 in HAdV-F41, probably because its shorter length limits flexibility (Fig. 2B and table S3). Residue Tyr784 also has a root mean square deviation (RMSD) of >2 Å between HAdV-C5 and HAdV-F41 and could be involved in the different interaction with protein IX (see the “Disordered density for protein IX” section below). Besides the HVRs, other regions already described for HAdV-C5 and HAdV-D26 also display variability among the 12 hexon monomers in the icosahedral asymmetric unit (AU) in HAdV-F41 (Supplementary Text and fig. S3A).
Fig. 2. Hexon and penton base structures.
(A) Superposition of the HAdV-C5 [Protein Data Bank (PDB) ID: 6B1T chain A, cyan] and HAdV-F41 hexon monomers, colored by root mean square deviation (RMSD). Residues exceeding the Chimera RMSD cutoff, or where RMSD calculation is not possible because they are not traced in HAdV-C5, are in purple. The orientation with respect to the capsid (outside/inside) is indicated. n.a., not available. (B) Sequence alignment of hexon HVRs from HAdV-C5 and HAdV-F41. Here and in (D), gray bars indicate traced regions. The HAdV-C5 acidic region is highlighted by a black rectangle. Amino acids are colored by polarity. (C) Superposition of the HAdV-C5 penton base monomer (PDB ID: 6B1T chain M, pink) and HAdV-F41 colored by RMSD as in (A). The variable loop and fiber binding rearrangement regions are shown with the map in gray mesh. (D) Sequence alignments focusing on the penton base (PB) N-terminal arm, variable and hypervariable loops, and hydrophobic rim; and on the N-terminal peptides of the HAdV-C5 (PDB ID: 3IZO) and the l (long) and s (short) HAdV-F41 fibers. Black rectangles highlight the RGD/IGDD sequence motifs, the conserved fiber peptide, and the nonconserved Ser426 in the hydrophobic rim. The histogram indicates the mean pairwise identity over all pairs in the column. (E) The penton base pentamer is shown with one monomer in black and the rest in white. Hydrophobic residues conserved with HAdV-C5 in the fiber binding ring are colored cyan (Phe412, Tyr413, Tyr419, and Leu422), and those nonconserved (Ser426) are in maroon. (F) HAdV-C5 and HAdV-F41 penton base pentamers colored by surface electrostatic potential.
Vertex capsomer: Penton base
The architecture of the HAdV-F41 penton base protein is very similar to that of HAdV-C5 (Fig. 2C and table S2) (5). As in previously solved structures (4, 6), the first 34 amino acids could not be traced (Fig. 2D and table S4). This region is either flexible or buried within the noisy, nonicosahedral core density. However, weak density suggests its location in HAdV-F41 (see section on “Proteins VI and VII and additional internal elements” below). The protein is 63 amino acids shorter than in HAdV-C5 (table S2). This length difference corresponds mainly to a hypervariable loop at the periphery of the pentamer, which in HAdVC-5 bears the integrin-binding RGD (Arg-Gly-Asp) sequence motif (Fig. 2, C and D). All HAdVs bear this RGD motif, except HAdV-F40 and HAdV-F41, which instead have RGAD (Arg-Gly-Ala-Asp) and IGDD (Ile-Gly-Asp-Asp) (21), and HAdV-D60, which has a deletion of the loop (22). The RGD loop is highly flexible, precluding its tracing in any of the available structures. In HAdV-F41, the IGDD-containing region is 57 residues shorter than the HAdV-C5 RGD loop but could not be traced either (Fig. 2, C and D, and table S4), indicating that despite being much shorter, it is also flexible. The variable loop (Glu148 to Leu156), one of the least conserved regions in the penton base sequences and likewise exposed at the periphery of the pentamer, has also a different conformation in HAdV-F41 (Fig. 2, C and D). The role of this loop is unknown, but it has been proposed as a site to be engineered for gene therapy (23).
The N-terminal peptides of AdV fibers (Fig. 2D) bind to the groove formed at the interface between monomers in the penton base pentamer. Both penton base and fiber residues involved in this interaction are conserved (23), and we do not observe structural differences in this region. However, we notice a possible difference in the interaction between the start of the fiber shaft and a ring of hydrophobic residues at the center of the pentamer (23, 24). In this region, fiber sequences are not conserved (Fig. 2D). In the penton base of HAdV-F41, the hydrophobic residues forming the ring are conserved, except for HAdV-C5 Phe489, which is instead a polar residue (Ser426) in HAdV-F41 (Fig. 2, D and E). Next to Ser426, Thr427 to Thr430 have a large RMSD from the equivalent residues in HAdV-C5 (Fig. 2C). This region forms part of a larger stretch (Tyr419 to Leu442) that undergoes a conformational rearrangement upon fiber binding (23). These observations suggest a different fiber-penton binding mode in HAdV-F41, with less hydrophobic and more hydrogen-bridging interactions than in HAdV-C5. We also find some differences between HAdV-C5 and HAdV-F41 in the interactions between penton base monomers and between penton base and the surrounding peripentonal hexons, which are described in Supplementary Text and fig. S3 (B and C). Last, at the pentamer cavity oriented toward the viral core, Arg47 residues in HAdV-C5 form a positively charged ring absent in HAdV-F41, which has Gly45 instead (Fig. 2F). This charge variation suggests a different interaction between penton base and the viral genome or its packaging machinery.
External cementing network: Protein IX
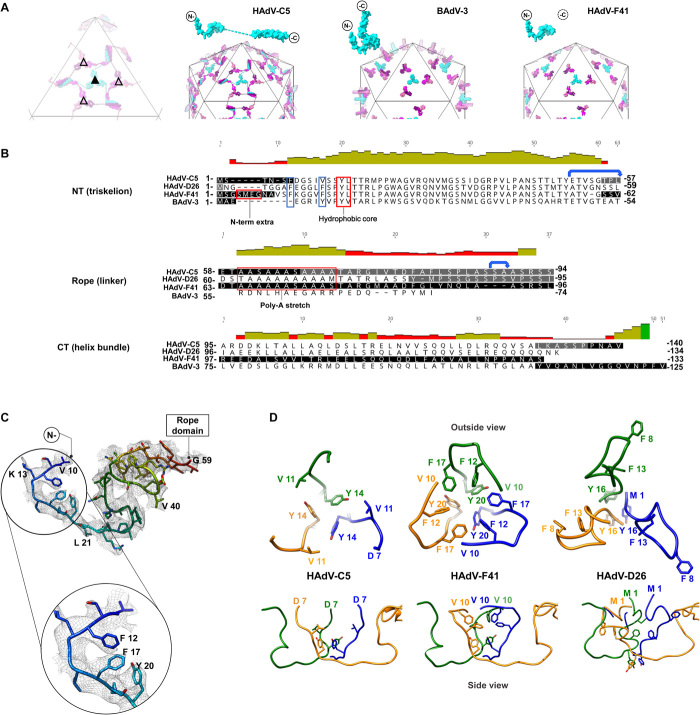
Protein IX is the only cementing protein located on the outer capsid surface of mastadenoviruses. Previous studies (4, 6) have shown that each of the 12 monomers of protein IX in a facet of the AdV capsid presents an extended conformation and is composed of three domains. The N-terminal domains of three IX molecules associate in a triskelion-shaped feature. One of the four triskelions in each facet occupies the valley between hexons at the icosahedral threefold symmetry axis (I3), while the other three lay at the local threefold axes (L3) formed by hexons 2, 3, and 4 in the AU (Figs. 1B and 3A). The central domain of protein IX is highly flexible and has also been termed “rope domain.” In HAdV-C5 and HAdV-D26, the rope domains of three conformationally unique IX molecules, each one originating in a different triskelion, crawl around the hexons on the surface of the icosahedral facet until they reach the edge. There, the C-terminal domains of these three molecules join a fourth one coming from the neighboring facet to form a coiled coil with three parallel and one antiparallel α-helices. There are three of these four-helix bundles per facet (Fig. 3A). In nonhuman mastadenoviruses, the rope domain is shorter (Fig. 3B), and the C-terminal domains of IX form coiled coils with only three parallel α-helices are located directly on top of their N-terminal triskelions (four such bundles per facet), as exemplified in the bovine AdV BAdV-3 structure (Fig. 3A) (25–28). Of all HAdV-F41 structural proteins, protein IX is the least similar to its HAdV-C5 counterpart (table S2).
Fig. 3. Protein IX triskelion structure.
(A) Schematics showing the organization of protein IX in HAdV-C5, BAdV-3, and HAdV-F41, where only the N-terminal domain is ordered. Triskelions located at the I3 symmetry axis (filled triangle) are in cyan, and those located at the L3 axis (empty triangle) are in several shades of pink. Dashed lines: rope domains in HAdV-C5 One IX monomer is depicted in cyan on top of each schematic. (B) Structure-based sequence alignment of protein IX in HAdV-C5, HAdV-D26, HAdV-F41, and BAdV-3. Black text: residues modeled in the different structures. White text on black background: regions not modeled. Gray background: regions modeled, but not in all independent copies of IX. Red boxes: N-terminal extra residues in HAdV-F41, triskelion hydrophobic core, and poly-Ala stretch (absent in BAdV-3). Blue boxes highlight the two Phe residues discussed in the text. Blue arrows: the two flexible bends. (C) N-terminal domain of HAdV-F41 protein IX rainbow colored from blue (N terminus) to red (last traced residue), with the density map in gray mesh and a zoom into the hydrophobic residues discussed in the text. (D) Comparison of the triskelions in HAdV-C5 (PDB ID: 6B1T), HAdV-F41, and HAdV-D26 (PDB ID: 5TX1). Top row: triskelions as seen from outside the capsid. Bottom: a view rotated by 90°, so that the hexon shell surface would be located at the bottom.
We have traced residues Val10 to Gly59 (Fig. 3C), which form the triskelion in a manner very similar to that of HAdV-C5. As previously reported (4), the triskelion is underpinned by a core of hydrophobic residues, Tyr20 to Leu21 in HAdV-F41 (Fig. 3, B and D). The first residues traced fold over the triskelion center. Notably, this arrangement implies that Phe12 and Phe17 add two tiers of hydrophobic interactions to the triskelion core (Fig. 3D). In HAdV-C5, one of the two phenylalanines is absent (Val11; Fig. 3B), and the region containing the other (Phe6) could not be traced, implying lack of icosahedral order (Fig. 3D). Both phenylalanine residues (Phe8 and Phe13; Fig. 3B) are modeled in HAdV-D26 protein IX but in a different conformation, oriented outward from the triskelion core (Fig. 3D).
Protein IX in HAdV-F41 has a five-residue insertion at the N terminus when compared to HAdV-C5 (four-residue insertion compared to HAdV-D26). The N-terminal domain is even shorter in BAdV-3 (Fig. 3B). We hypothesize that, in HAdV-F41, the region containing the two phenylalanine residues is more ordered than in HAdV-C5, and in a different conformation from HAdV-D26, because of the presence of these extra residues. Weak density capping the triskelion at the I3 axis could account for the presence of the longer N terminus (Fig. 4, A and B). Both the dense network of hydrophobic residues at the triskelion core and the presence of extra residues at the N terminus of IX would reinforce the intermolecular interactions within the triskelions, likely contributing to stabilize the protein IX trimer. It has previously been shown that the triskelion is sufficient to provide capsid thermostability in HAdV-C5 (29). More stable triskelions may enhance the physicochemical stability of HAdV-F41.
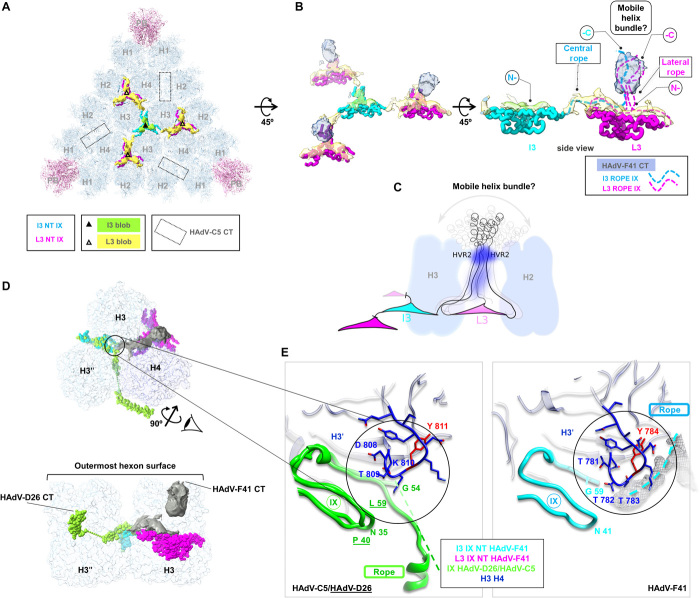
Fig. 4. Organization of protein IX.
(A) External remnant density (green and yellow, unsharpened, 0.2σ). Modelled triskelions are in cyan (I3 axis, I3 NT) and magenta (L3 axes, L3 NT). Dotted rectangles: location of helix bundles in HAdV-C5. (B) Interpretation of the remnant map. Apart from elements shown in (A), density protruding at the L3 axes is in blue, and the proposed path for the rope domains corresponding to each triskelion in dashed lines. (C) Cartoon representing the hypothesis that the C-terminal domains associate forming a mobile, spike-like structure whose density is averaged away. Hexon HVR2 loops would constrain the flexibility of IX, producing the most evident blurry density (dark blue splotch). Less intense dark blue represents weaker density observed in some localized reconstruction classes (fig. S5B). (D) The rope domain of the IX molecules forming the I3 triskelion follows different paths in HAdV-C5/D26 and HAdV-F41. One of the IX monomers forming the HAdV-D26 I3 triskelion is in green; the equivalent monomer in HAdV-F41 in cyan; and the HAdV-F41 L3 triskelion in magenta. Grey surface: HAdV-F41 remnant map. Hexons (H3, H3′, H4) in semi-transparent surface. Top: view from outside the capsid as in (A). Bottom: side view after rotating as indicated. Notice that the blurry density at L3 protrudes above the hexon towers. (E) Zoom in on the region where the triskelion ends and the rope domain turns. Green, overlapped structures of IX in HAdV-C5 and HAdV-D26. HAdV-D26 residue labels are underlined. Dashed lines: untraced rope domains in HAdV-C5 and HAdV-F41. Grey mesh: HAdV-F41 remnant density corresponding to the rope domain. Hexon amino acids with RMSD above 2 Å are in red.
Disordered density for protein IX
Unexpectedly, in the HAdV-F41 map, there is no density corresponding to the C-terminal four-helix bundle at the icosahedron edges. Instead, we observe blurry density protruding radially between the towers of hexons 2, 3, and 4, on top of the triskelions at the L3 axis (Fig. 4B and fig. S4). The presence of this protrusion would suggest that in HAdV-F41, the protein IX C-terminal domains could be arranged in a similar way to the shorter protein IX in BAdV-3, forming a three-helix bundle directly adjacent to each triskelion (Fig. 3A). However, in our HAdV-F41 map, there is no equivalent weak density near the triskelion at the threefold icosahedral axis (Fig. 4B and fig. S4), indicating that the organization of IX is not the same as in BAdV-3. Localized reconstruction and classification without symmetry enforcement did not yield any subset of particles with a protrusion at the I3 axis or with density that could indicate the presence of a four-helix bundle at the capsid edges (fig. S5). Therefore, the arrangement of the C-terminal domain of protein IX in HAdV-F41 does not seem to follow either the typical human or the nonhuman AdV architectures previously observed.
A remnant map (showing weak density on the capsid surface unoccupied by the molecules traced so far) indicates a connection between the triskelion at the L3 axis and the triskelion at the center of each facet (Fig. 4, A and B). We interpret that this connection corresponds to the rope domain of one of the monomers in the central triskelion, which, as in HAdV-C5, runs on the capsid surface toward the facet periphery. If this is the case, it is possible that in HAdV-F41, the C-terminal domains of four copies of protein IX (three from the L3 triskelion and one from I3) interact not at the capsid edges but at the L3 axis (Fig. 4B). Why, then, is there no helix cluster resolved in our map? Secondary structure predictions indicate that the C-terminal helix in HAdV-F41 is shorter than their counterparts in HAdV-C5, HAdV-D26, and BAdV-3 (fig. S6), so one possibility is that the helix bundle is not formed or is unstable. However, in that case, one would expect similar blurry (or absent) densities generated by the disordered C-terminal domains at both the I3 and L3 axes, which is not the case. In addition, coiled-coil formation is still predicted for HAdV-F41, even if the helices are shorter (fig. S6, Lupas). It seems likely, therefore, that there is some kind of association between the C termini of the I3 molecules and those at the L3. Another possibility to explain the lack of well-defined density is that the four-helix bundle is formed directly on top of the L3 triskelion, but since the flexible rope domain is longer in HAdV-F41 than in BAdV-3 (Fig. 3B), the coiled coil is not constrained by interactions with the hexon towers and forms a mobile spike-like structure that is averaged away (Fig. 4C). This would be similar to the averaging out of the distal part of the fibers, linked to the penton base by a flexible shaft.
A remaining question is why the four-helix bundle is not formed at the facet edges as in HAdV-C5 and HAdV-D26, if the length of the rope domain is similar in all three viruses (Fig. 3B). It has been proposed that protein IX in HAdV has two flexible bends that facilitate its coupling to the hexon contours (Fig. 3B) (28). In HAdV-C5 and HAdV-D26, the first flexible bend causes a clockwise turn of the rope domain at the exit of the central triskelion. In HAdV-F41, the remnant map indicates that the turn would be counterclockwise instead (Fig. 4D). This different path may be determined by differences in the hexon-IX interactions near the first bend. Sequence alignment indicates that the rope domain in HAdV-F41 is three residues shorter than in HAdV-C5 and HAdV-D26, with deletions located precisely at both bends (Fig. 3B). We also observe that hexon Tyr784, located near the end of the triskelion (residues 56 to 59), is one of the few residues presenting high RMSD when comparing the HAdV-F41 and HAdV-C5 hexon structures (Figs. 2A and 4E). HAdV-C5 also has a tyrosine in this position, but the upstream sequence is very different in both viruses, with a conspicuous threonine triplet in HAdV-F41 (residues 781 to 783). All these changes may result in different interactions causing protein IX to bend in a different direction in the enteric virus. As a consequence, the C-terminal helix would never reach the location where the coiled coil is formed in the other solved HAdV.
Proteins IIIa and VIII
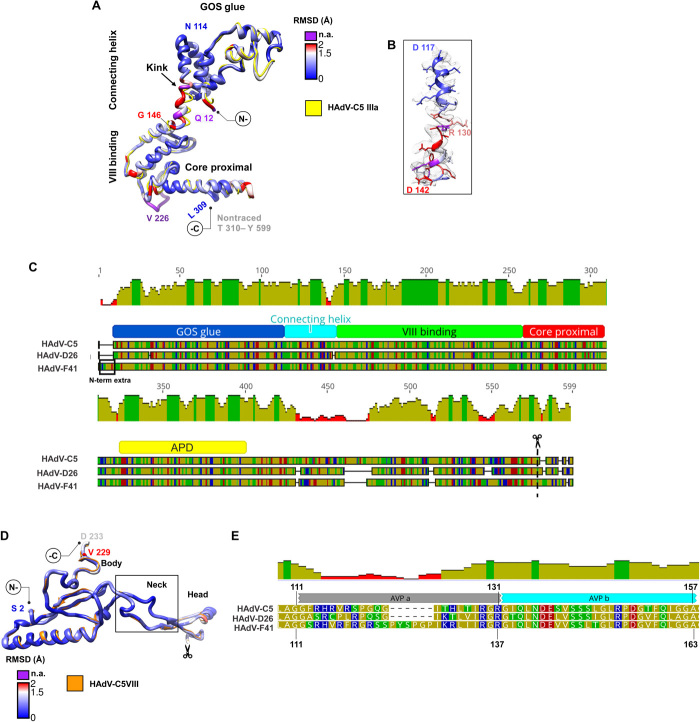
Minor coat proteins IIIa and VIII line the HAdV-F41 internal capsid surface. Five copies of protein IIIa are located beneath each vertex, bridging the penton and the peripentonal hexons (Fig. 1B), and presumably interacting with packaging proteins during assembly (30). The extent of protein IIIa traced and its architecture are very similar to those of HAdV-C5 (tables S2 and S4), with the domain organization previously described (Fig. 5, A and B) (4). HAdV-F41 protein IIIa has an eight–amino acid insertion at the N terminus with predicted α-helical structure (residues 2 to 9; Fig. 5C), which could form extra intramolecular interactions in the peripentonal region and contribute to make the vertex region more stable. However, we did not observe interpretable density for this insertion or for the appendage domain (APD) traced in HAdV-D26 (Fig. 5C) (6). The largest difference with HAdV-C5 corresponds to a kink between the fourth and fifth turns in the helix connecting the GOS-glue domain to the VIII-binding domain (Fig. 5, A and B) (4). This kink may cause differences in the interactions between neighboring molecules beneath the vertex (see next section).
Fig. 5. Proteins IIIa and VIII.
(A) Superposition of HAdV-C5 IIIa (PDB ID: 6B1T, chain N) in yellow and HAdV-F41 IIIa colored by RMSD as in Fig. 2. The GOS glue, connecting helix, VIII-binding, and core proximal domains are indicated. (B) Detail of the connecting helix to show its fit into the cryo-EM map. (C) Schematics showing the alignment of protein IIIa sequences in HAdV-C5, HAdV-D26, and HAdV-F41. Traced domains are indicated below, in different colors. APD is the appendage domain traced in HAdV-D26, but not in the other two viruses. The N-terminal extension in HAdV-F41 (N-term extra) and maturation cleavage site (scissors) are also indicated. (D) Superposition of HAdV-C5 protein VIII (PDB ID: 6B1T Chain O) in orange and HAdV-F41 VIII colored by RMSD. The body, neck, and head domains are indicated, as well as the gap corresponding to the peptide cleaved during maturation (scissors). (E) Sequence alignment showing the two central peptides of protein VIII cleaved by AVP (AVPa and AVPb) in HAdV-C5, HAdV-D26, and HAdV-F41. Amino acid and mean pairwise identity histogram color schemes are the same as in Fig. 2.
Two copies of protein VIII are present in each AU of the AdV icosahedral shell. One of them interacts with protein IIIa and contributes to stabilize the vertex, while the second one helps keep the nonperipentonal hexons together in the central plate of the facet (Fig. 1B). HAdV-F41 protein VIII is highly conserved compared to HAdV-C5, both in sequence and structure (Fig. 5D and table S2). Only the central region of the protein, cleaved by the adenovirus maturation protease (AVP) (31), presents sequence divergence (Fig. 5E and see next section).
Proteins VI and VII and additional internal elements
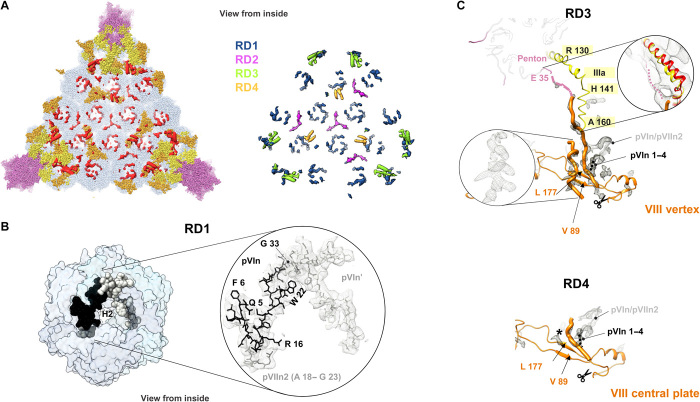
Analysis of density unoccupied by the molecules traced so far in the HAdV-F41 cryo-EM map [remnant density (RD)] revealed several features of interest (Fig. 6A). At the rim of the central cavity of the hexon trimers, we observe discontinuous weak density (RD1) that, by similarity with HAdV-C5, corresponds to peptides pVIn and pVIIn2, cleaved from precursor proteins pVI and pVII during maturation (5, 8). Since side chains are poorly defined in these regions due to lack of order and low occupancy, we have only tentatively traced four copies of pVIn and two of pVIIn2, out of the 12 possible equivalent sites in the AU (Figs. 1B and 6B and table S4) (8). Protein VI is longer in HAdV-F41 than in HAdV-C5 (table S2) and bears a particular epitope common to enteric AdVs (species A and F) in its central domain (residues 114 to 125) (32). However, we do not observe any interpretable density that could inform about either extra interactions between the longer VI chain and hexon or the central region traced in HAdV-C5 (5).
Fig. 6. Additional internal components of the HAdV-F41 capsid.
(A) Left: remnant densities in one facet (red), not occupied by traced proteins hexon (light blue), penton (light pink), IIIa (yellow), or VIII (orange). Right: remnant map components color-coded as RD1 to RD4 (see text). Densities correspond to the unsharpened map contoured at 1.5σ. (B) N-terminal peptides of proteins VI (pVIn, two copies) and VII (pVIIn2) tentatively traced in RD1, inside the hexon cavity. (C) Proposed interpretation of remnant densities RD3 and RD4. Except for the inset at the right hand side, RD in the sharpened map is shown, contoured at 2.5σ. Orange thin ribbon: traced part of the two copies of protein VIII (VIII vertex and VIII central plate). Orange thick worms: poly-Ala peptides modeled in the unassigned densities proposed to correspond to the central peptide of VIII. Left inset: possibly α-helical shape of one of the remnant densities in RD3. This density is much weaker in RD4, where a black star indicates its position. Scissors: AVP cleavage site in protein VIII. Peptide pVIn is represented as a gray ribbon, with the possible location of its four untraced N-terminal residues as a black dotted line. In the RD3 panel, the IIIa connecting helix is in yellow, the first residue traced in the penton base protein (E 35) is colored pink, and a dotted pink line indicates the possible trajectory of the untraced 34 residues. Right framed zoom: the IIIa connecting helix is shown overlapped with the HAdV-C5 helix in red, and the unsharpened map contoured at 1σ is shown as a gray mesh, to emphasize low-resolution density proposed to correspond to the penton base N-terminal arm.
L-shaped densities at the I3 axis and the L3 axis between hexons 2, 3, and 4 (RD2; Fig. 6A) were also observed in HAdV-D26, but their assignment is uncertain (6). In HAdV-F41, we observe two additional RDs that were not reported in previous studies (5, 6). RD3 and RD4 are located near the two independent copies of protein VIII and have similar shapes, but RD3 (located beneath the penton region) has stronger density. Both RD3 and RD4 are near the gap in protein VIII left by AVP cleavage during maturation (Figs. 5, D and E, and 6, A and C). In RD3, we could model two poly-Ala peptides. One of them could correspond to a 21-residue α-helix, while the other one is an extended 23-residue peptide (Fig. 6C, top). These lengths correlate well with those of the pVIII peptides cleaved by AVP (Fig. 5E). In HAdV-F41, one of these peptides is six residues longer than in HAdV-C5 and HAdV-D26 and has low sequence conservation (Fig. 5E). In RD4, the density is weaker, and only a 10-residue extended peptide could be modeled (Fig. 6C, bottom). We hypothesize that at least part of RD3 and RD4 correspond to these excised peptides of protein VIII, which, in HAdV-F41, would be ordered and reinforcing the network of contacts on the inner capsid surface (particularly beneath the vertex), therefore contributing to a higher capsid stability. RD3 and RD4 also connect to density corresponding to pVIn and could account for its untraced first four residues (Fig. 6C, black dotted lines, and table S4). Fragmented density in RD3, which is not present in RD4, seems to connect to the first traced residue in penton base, Glu35, and runs parallel to the IIIa connecting helix. We propose that this density corresponds to the untraced N terminus of penton base (Fig. 6C, pink dotted line) that would interact with the kinked connecting helix in IIIa, a contact which would not happen in HAdV-C5, where the helix is not kinked (Fig. 5A) and the sequence of the penton base N-terminal sequence differs (Fig. 2D).
Determining the identity of weak, discontinuous density patches in AdV is problematic, as there are many virion components that do not follow icosahedral symmetry (packaging proteins, protein VI, half of protein IIIa, cleaved peptides, and core proteins), making the combinatorial problem of sequence assignment highly challenging and error-prone (8, 30, 33). Nevertheless, our interpretation suggests that the central peptides of pVIII, the N terminus of penton base, and the kinked IIIa helix would collaborate to strengthen the HAdV-F41 vertex region in a manner different to that previously observed in other HAdVs.
DISCUSSION
The narrow enteric tropism of HAdV species F is not well understood. Infectivity analyses have shown that unlike HAdV-C5, these viruses are not inactivated by acidic environments (14), and a main player conferring this resistance to low pH seems to be the short fiber, one of the differentiating structural features of HAdV-F (18). Here, we show that HAdV-F41 capsids are more thermostable than HAdV-C5, requiring higher temperatures to open up and expose its genome to the solvent, and that infectivity is stable, even activated, under simulated gastric and intestinal conditions. We have solved the structure of the HAdV-F41 capsid at near-atomic resolution and analyzed it, looking for possible determinants of its tropism and enhanced physicochemical stability.
While the overall structures of hexon and penton base are conserved, there are differences in external loop regions involved in interaction with host factors that could play a role in the enteric tropism. The hexon HVR1 loop is much shorter and rigid in HAdV-F41 than in HAdV-C5. Lack of the long acidic region reflects on the absence of a large negatively charged patch in the hexon towers of HAdV-F41 and could have an influence in the interplay with host factors under the extreme pH conditions in the gastrointestinal tract. However, the acidic stretch is also absent in HAdV-D26, which has ocular tropism and where HVR1 is also wholly traced (6). In penton base, the RGD motif is substituted by IGDD and the loop bearing it is much shorter. Lack of the RGD sequence is not a determinant for enteric tropism either, as this motif is present in other enteric AdVs such as HAdV-A31 (UniProtKB entry: D0Z5S7_ADE31) and HAdV-G52 (A0MK51_9ADEN). Despite lacking the RGD motif, it has recently been shown that HAdV-F41 can bind to laminin-binding integrins (34).
AdV in species F are resistant to neutralization by human α-defensins, small cationic antimicrobial peptides abundantly expressed in the small intestine that inhibit HAdV-C5 infection (35). Defensins bind to the capsid surface, preventing uncoating, and the binding is dependent on salt concentration, suggesting an electrostatic interaction (36). It is then tempting to consider that HAdV-F resistance to defensins is related to the lack of negative charges in the hexon surface. In support of this hypothesis, it has recently been shown that mutations introducing positive charge in HAdV-C5 hexon HVR1 confer resistance to defensin neutralization (37). It remains to be seen whether this is also the case for other respiratory AdVs. AdVs in species A also have enteric tropism and lack the acidic HVR1. The RGD loop is also necessary for defensin neutralization (37). Viruses in HAdV-A carry this loop, while those in HAdV-F do not, and defensins cause a modest inhibition of HAdV-A12 (35). We speculate that the combination of a shorter, basic HVR1 loop in hexon with the lack of the long RGD loop may play a role in determining the enteric tropism of HAdVs in species F by helping them resist neutralization by enteric defensins.
The exact molecular basis of virus capsid stability is difficult to unravel and is still a subject of intense investigation even for so-called simple viruses (38). We describe some intermolecular contacts that could have a different nature in HAdV-F41 and HAdV-C5, but it is difficult to conclude whether these differences will have an effect in capsid stability, due to the large complexity and sheer number of interactions present in AdV virions. However, we do find partially ordered features that suggest collaboration between the N terminus of penton base, protein IIIa, pVIn, and the cleaved peptides of VIII in stabilizing the vertex region. On the outer capsid surface, the protein IX triskelion has a stronger hydrophobic core than in HAdV-C5, while the rest of the protein is disordered, notably lacking the four-helix bundle at its C terminus. It has previously been observed that the four-helix bundle in HAdV-C5 is easily disturbed, becoming disordered when protein IX was modified by a C-terminal fusion (39) or antibody labeling (40). It is possible that the lack of well-defined density for the C-terminal region of IX in our HAdV-F41 map is simply caused by partial disruption accidentally occurring during sample preparation. However, in studies where the HAdV-C5 helix bundle was not clearly visible, no weak density was observed in other capsid locations. Differences in sequence and weak densities in the map support the presence of an organization for protein IX in HAdV-F41, different from all described for other AdVs. Our observations also imply that the formation of a well-ordered helix bundle is not required to enhance capsid stability, reinforcing the idea that the triskelion is the crucial part for the cementing action of the protein (29). Further, our model suggests that seemingly minor alterations in the rope domain may be responsible for radically changing the organization of protein IX in the capsid. Last, in our model, the C terminus of protein IX protrudes from the outermost capsid surface (Fig. 4, C and D), which would make it an interesting locale for exogenous peptide insertion for retargeting, epitope display, or other biotechnological purposes, as previously suggested for protein IX in BAdV-3 (41). Variations in protein IX length, sequence, and organization across AdV types indicate that this protein undergoes evolutionary pressure induced by interaction with the host and highlight its potential role as a determinant of virus-host specificity.
MATERIALS AND METHODS
Virus production
All experiments reported in this work were carried out using EGFP-expressing, E1-deleted HAdV-F41–EGFP virus (provided by D. Brough, GenVec/Precigen), propagated in 2V6.11 cells (American Type Culture Collection, JHU67) with E4 orf6 protein induced with ponasterone A (1 μg/ml) 24 hours before infection. Cells were infected at an input multiplicity of infection of <0.1 fluorescent units (FUs) per cell, and cultures were incubated until cytopathic effect (CPE) was complete or almost complete but before cells showed signs of disintegration. Flasks were hit sharply to dislodge infected cells from the surface, and cells were harvested by centrifugation at 1600g for 10 min, washed with PBS, and then resuspended in a small volume of serum-free medium. Any cells remaining attached to the flask were refed with a 1:1 mixture of fresh medium and clarified medium from the infected flasks. If uninfected cells had become confluent, they were then subcultured to allow for virus spread. The process was continued until CPE was complete, typically about 1 to 2 weeks after infection. Progeny virus was released by five cycles of freezing-thawing, and the cell lysate was clarified by centrifugation at 1600g for 10 min. Viruses were used in experiments at passage level 3.
Virus was purified by two cycles of caesium chloride (CsCl) density gradient centrifugation. Clarified cell lysate (~4.5 ml) was underlaid with 4 ml of CsCl [1.2 g/ml in 50 mM tris-HCl (pH 8.1)] and 2 ml of CsCl [1.4 g/ml in 50 mM tris-HCl (pH 8.1)]. The gradient was centrifuged in a SW41 rotor at 120,000g at 4°C for 1 hour. Virions were collected from the interface of each step gradient and diluted in 50 mM tris-HCl to a density <1.2 g/ml, as determined by measuring refractive index, and layered on top of a preformed gradient (1.2 to 1.4 g/ml) that was prepared using the Gradient Master (BioComp). The gradient was centrifuged in a SW41 rotor at 120,000g at 4°C for 2 hours. The viral band of mature virus particles was recovered and dialyzed at 4°C against three changes of storage buffer [50 mM tris-HCl (pH 8.1), 150 mM NaCl, 10 mM MgCl2, and 10% glycerol] with a buffer change every 45 min. Optical density was measured with a NanoDrop ND-1000 spectrophotometer (v3.3), and virion concentration was calculated using the equation: 1A260 = 1.1 × 1012 particles/ml (42). Infectivity was determined by endpoint dilution assays with HEK293 cells, in duplicate, in 60-well Terasaki plates as previously described (43). Assays were scored for the presence of green cells (expressing EGFP), and titers, calculated by the Reed and Muench method (44), were expressed as FU/ml. Purified HAdV-F41 particles from two different preparations were pooled and concentrated by centrifugation in a CsCl gradient and stored in 20 mM Hepes (pH 7.8), 150 mM NaCl, and 10% glycerol at −80°C.
HAdV-C5 used as a control for extrinsic fluorescence experiments was the E1-E3–deleted, GFP-expressing Ad5/attP vector, which is structurally wild type (45), propagated at 37°C in HEK293 cells, harvested at 36 hours after infection, and purified as described (8).
Infectivity assays under simulated gastrointestinal conditions
HEK293 cells were seeded in six-well plates at a density of 5 × 105 cells per well 1 day before infection. HAdV-41–EGFP preparations (~2 × 1011 particles/ml, 5 × 106 FU/ml) were dialyzed against serum-free medium and frozen for a short period. Virus (50 μl) was then mixed 1:1 with 2× synthetic gastric fluid according to the U.S. Pharmacopoeia at 0.01 N HCl [4.0 g of NaCl and 6.4 g of pepsin (Sigma-Aldrich) dissolved in 14 ml of 1 N HCl, made up to 1 liter with ddH2O (pH 1.2)] and/or 2× synthetic intestinal fluid [13.6 g of monobasic potassium phosphate in 250 ml of ddH2O, 77 ml of 0.4 N NaOH, 500 ml of ddH2O, and then 20.0 g of pancreatin (Sigma-Aldrich), adjusted to pH 6.8 and then diluted with water to 1 liter]. Virus suspension was incubated for 1 hour at 37°C. For sequential treatments, virus was treated with synthetic gastric fluid for 1 hour at 37°C, then mixed 1:1 with synthetic intestinal fluid, and incubated for an additional hour. To neutralize pH and proteases, complete minimum essential medium containing 10% fetal bovine serum was added to final volume of 0.3 ml. HEK293 cells were infected with treated or mock-treated virus for 1 hour at 37°C. Inoculum was removed, cells were harvested 19 to 21 hours after infection, and fixed with 2% paraformaldehyde for flow cytometry. GFP expression was analyzed with an LSR Fortessa (BD Biosciences), and data were analyzed using FACSDiva (BD Biosciences) gating out doublets. A total of 10,000 cells were counted per well. Each experiment was done in duplicate.
Extrinsic fluorescence thermostability assays
The data for HAdV-C5 have been reported previously (8). The data for HAdV-F41 were collected in the same time period, as part of a large study comparing the thermostability of different AdV specimens. HAdV-C5 and HAdV-F41 samples (5 × 109 particles/ml in a final volume of 800 μl of buffer) were incubated overnight at 4°C in 8 mM Na2HPO4, 2 mM KH2PO4, 150 mM NaCl, and 0.1 mM EDTA (pH 7.4). Fluorescence emission spectra from virus samples mixed with YOYO-1 were obtained every 2°C along a temperature range from 20 to 70°C in a Horiba FluoroLog spectrophotometer. The dye was excited at 490 nm and maximal emission intensity was achieved at 509 nm. Raw spectra were corrected by subtraction of the YOYO-1 spectrum in buffer at each tested temperature. A total of six (HAdV-C5) and four (HAdV-F41) independent experiments were considered for each averaged curve. The half-transition temperatures (T0.5) were calculated from the fitting of the averaged fluorescence intensity curve as a function of temperature to a Boltzmann sigmoid equation as described (8).
Cryo-EM sample preparation and data collection
Pooled and concentrated HAdV-F41 samples (as described above) were dialyzed against 50 mM tris-HCl (pH 7.8), 150 mM NaCl, and 10 mM MgCl2 for 1 hour at 4°C and further concentrated by spinning in a Microcon YM-100 device for 15 min at 4°C, for a final estimated concentration of 3.1 × 1012 viral particles/ml. Samples were deposited in glow-discharged, Quantifoil R2/4 300 mesh Cu/Rh grids and vitrified in liquid ethane after manual blotting in a Leica CPC device. Cryo-EM images (table S1) were recorded at the Central European Institute of Technology (CEITEC) facility (Brno, Czech Republic) using a 300-kV Titan Krios microscope equipped with a Falcon II detector, with a total dose of 42 e−/Å2 distributed over 25 frames, at a nominal pixel size of 1.38 Å and a defocus range between −1 and −2.5 μm.
Cryo-EM data analysis
All image processing and three-dimensional (3D) reconstruction tasks were performed within the Scipion framework (46). Frames 2 to 24 of each movie were aligned using whole-image motion correction implemented in Xmipp, followed by correction of local movements using Optical Flow (47). The contrast transfer function (CTF) was estimated using CTFFIND4 (48). Particles were semi-automatically picked from micrographs corrected for the phase oscillations of the CTF (phase-flipped), extracted into 780 × 780 pixel boxes, normalized, and downsampled by a factor of 2, using Xmipp (49). All 2D and 3D classifications and refinements were performed using RELION (50). 2D classification was used to discard low-quality particles and run for 25 iterations, with 50 classes, angular sampling of 5°, and regularization parameter T = 2. Classification in 3D was run for 40 iterations, with three classes, starting with an angular sampling of 3.7° and sequentially decreasing to 0.5°, and regularization parameter T = 4. Icosahedral symmetry was imposed throughout the refinement process. The initial reference for 3D classification was a nonhuman AdV cryo-EM map (51), low-pass–filtered to 60-Å resolution. The class yielding the best resolution was individually refined using the original 780-px boxed particles and the map obtained during the 3D classification as a reference, producing a final map at 4.0-Å resolution, as estimated according to the gold-standard Fourier Shell Correlation (FSC) = 0.143 criterion implemented in RELION autorefine and postprocess routines (52). Local resolution was estimated with ResMap (53). A global B factor was estimated after dividing the map Fourier transform by the modulation transfer function of the Falcon II detector. The actual sampling for the map was estimated by comparison with a HAdV-C5 model [Protein Data Bank (PDB) ID: 6B1T] (5) in UCSF Chimera (54), yielding a value of 1.36 Å/px.
Localized reconstruction and classification of selected regions of interest were carried out following the general methodology described in (55), using RELION as implemented in Scipion (56). The procedure consists of the following steps. (i) A mask excluding the region of interest was applied to the 3D map obtained from the refinement with icosahedral symmetry enforced. (ii) The masked map was projected in the directions determined by the icosahedral refinement, and the projections were subtracted from the corresponding experimental images. (iii) The position of the feature of interest in the 3D map was determined and projected along the projection direction assigned to each experimental image and its 59 symmetry-related directions. This process produced a set of 595,560 coordinates (60 × 9926; table S1). (iv) The set of coordinates was pruned, so that those coordinates closer than 180 px to the projection of the virus center were discarded to minimize interference from the capsid and core residual signal. (v) Using the resulting coordinates, approximately 490,000 subparticles were extracted from the original experimental images in 100 × 100 px boxes. (vi) The projection direction assigned to each subparticle was the one used to create its corresponding coordinate. (vii) These images were then subjected to 3D classification in four classes, without orientation change or symmetry enforcement (C1 symmetry).
Once the classification process was carried out, the resolution of each class was estimated following two approaches. In the first one, the set of images was divided in two random groups and the FSC at 0.143 threshold criterion was used. In the second approach, the local resolution was computed using ResMap (53). The results of the first approach seemed unrealistically high (fig. S5), as they were close to Nyquist, were not corroborated by the map aspect, and the FSC curve did not present a sharp drop. This observation points out that random division of the subparticles after classification does not conserve the independent subsets used by the icosahedral refinement and required for meaningful FSC calculation. On the other hand, ResMap, which compares locally signal and noise, seems to provide a more realistic estimation.
Model building and analysis
Interpretation of the HAdV-F41 3D map was performed using the molecular modeling workflow based on sequence homology in Scipion (57). To facilitate chain tracing, we used MonoRes (58) and Localdeblur (59) to sharpen the map according to the local resolution. The initial model for each polypeptide chain was predicted with Modeller (60), using as input template the structure of the respective HAdV-C5 homolog chain (PDB ID: 6B1T). UCSF Chimera (54) was used to perform a rigid fitting of each chain initial model into the sharpened map. Next, the fitted model of each chain was refined using Coot (61) and Phenix real-space refine (62). Validation metrics to assess the quality of the atomic structure were computed with the Phenix comprehensive validation (cryo-EM) algorithm. Once we generated the whole structure of the AU, Chimera findclash (integrated in the Scipion protocol chimera-contacts) was executed to identify possible contacts between pairs of chains contained in the AU or between a chain in the AU and a chain from a neighboring AU (file S1). The default parameters, cutoff (−0.4) and allowance (0.0), were used to report as possible bonds all pair of atoms whose distance is no higher than the sum of the corresponding van der Waals radii plus 0.4 Å. To identify additional unmodeled densities, we generated remnant maps with Chimera in two alternative manners: (i) subtracting from the cryo-EM map a map generated with molmap around the modeled atoms (resolution of 2.0 and gridSpacing of 1.36) or (ii) masking off from the initial map a region within a 3-, 4-, or 8-Å radius from the modeled atoms (vol zone invert true).
Protein charge at pH 7.0 was calculated as described in http://isoelectric.org/index.html, with amino acid pKa (where Ka is the acid dissociation constant) values taken from (63) and general pKa values for terminal amino and carboxyl groups taken from (64). Protein sequence alignments were performed with Clustal W2 (www.clustal.org/clustal2/) integrated in Geneious version 2019.0 (www.geneious.com). For secondary structure prediction, we used Jpred4 Server Jnet version 2.3.1 (www.compbio.dundee.ac.uk/jpred) (65) or PSIPRED (66). Molecular graphics and analyses including molecule superposition, RMSD calculation, map rendering, map normalization, coloring and measuring surface blobs, and surface electrostatic potential rendering were performed with UCSF Chimera and ChimeraX (54, 67). UCSF Chimera Hide dust was used for clarity when composing figures representing density maps.
Acknowledgments
We acknowledge J. Novacek and M. Peterek at the CEITEC facility (Brno, Czech Republic) for the cryo-EM data collection. The initial seed for HAdV-F41-EGFP was a gift from D. Brough (GenVec/Precigen). We thank M. J. van Raaij (CNB-CSIC) for careful reading and insightful comments on the manuscript; M. Menéndez (IQFR-CSIC), M. Castellanos, and L. A. Campos (CNB-CSIC) for the advice with fluorescence measurements and analyses; M. I. Laguna (CNB-CSIC) for the expert technical help; M. Chagoyen (CNB-CSIC) for the advice on bioinformatics tools; and J. M. Carazo (CNB-CSIC) for the constant support. Funding: This work was supported by grants PID2019-104098GB-I00/AEI/10.13039/501100011033 and BFU2016-74868-P, cofunded by the Spanish State Research Agency and the European Regional Development Fund; BFU2013-41249-P and BIO2015-68990-REDT (the Spanish Adenovirus Network, AdenoNet) from the Spanish Ministry of Economy, Industry, and Competitiveness; and the Agencia Estatal CSIC (2019AEP045) to C.S.M. The CNB-CSIC is further supported by a Severo Ochoa Excellence grant (SEV 2017-0712). Work in M.B’s. lab was supported by grant 194562-08 from the Natural Sciences and Engineering Research Council of Canada. M.H.-P. is a recipient of a Juan de la Cierva postdoctoral contract funded by the Spanish State Research Agency. M.P.-I. holds a predoctoral contract from La Caixa Foundation (ID 100010434), under agreement LCF/BQ/SO16/52270032. Access to CEITEC was supported by iNEXT, project number 653706, funded by the Horizon 2020 Programme of the European Union. The CEITEC Cryo-electron Microscopy and Tomography core facility is supported by MEYS CR (LM2018127). Author contributions: M.B., R.M., and C.S.M. designed the research. M.P.-I., M.M., G.N.C., M.H.-P., C.M., M.B., R.M., and C.S.M. performed the research. M.P.-I., M.M., M.H.-P., C.M., M.B., R.M., and C.S.M. analyzed data. M.M. and R.M. contributed analysis tools. M.P.-I. and C.S.M. wrote the paper, with input from all other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The HAdV-F41 cryo-EM map and model are deposited at the Electron Microscopy Data Bank (EMDB; www.ebi.ac.uk/pdbe/emdb) and PDB (www.ebi.ac.uk/pdbe/) with accession numbers EMD-10768 and 6YBA, respectively. The validation report is included as data file S2.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/9/eabd9421/DC1
REFERENCES AND NOTES
- 1.Appaiahgari M. B., Vrati S., Adenoviruses as gene/vaccine delivery vectors: Promises and pitfalls. Expert Opin. Biol. Ther. 15, 337–351 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Baker A. T., Aguirre-Hernandez C., Hallden G., Parker A. L., Designer oncolytic adenovirus: Coming of age. Cancers 10, 201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lion T., Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 27, 441–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H., Jin L., Koh S. B. S., Atanasov I., Schein S., Wu L., Zhou Z. H., Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329, 1038–1043 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai X., Wu L., Sun R., Zhou Z. H., Atomic structures of minor proteins VI and VII in human adenovirus. J. Virol. 91, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X., Veesler D., Campbell M. G., Barry M. E., Asturias F. J., Barry M. A., Reddy V. S., Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci. Adv. 3, e1602670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicklin S. A., Wu E., Nemerow G. R., Baker A. H., The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol. Ther. 12, 384–393 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Hernando-Pérez M., Martín-González N., Pérez-Illana M., Suomalainen M., Condezo G. N., Ostapchuk P., Gallardo J., Menéndez M., Greber U. F., Hearing P., de Pablo P. J., San Martín C., Dynamic competition for hexon binding between core protein VII and lytic protein VI promotes adenovirus maturation and entry. Proc. Natl. Acad. Sci. U.S.A. 117, 13699–13707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chhabra P., Payne D. C., Szilagyi P. G., Edwards K. M., Staat M. A., Shirley S. H., Wikswo M., Nix W. A., Lu X., Parashar U. D., Vinjé J., Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008-2009. J Infect Dis 208, 790–800 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Corcoran M. S., van Well G. T., van Loo I. H., Diagnosis of viral gastroenteritis in children: Interpretation of real-time PCR results and relation to clinical symptoms. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1663–1673 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Uhnoo I., Wadell G., Svensson L., Johansson M. E., Importance of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children. J. Clin. Microbiol. 20, 365–372 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt C. D., Kim H. W., Rodriguez W. J., Arrobio J. O., Jeffries B. C., Stallings E. P., Lewis C., Miles A. J., Gardner M. K., Parrott R. H., Adenoviruses and pediatric gastroenteritis. J Infect Dis 151, 437–443 (1985). [DOI] [PubMed] [Google Scholar]
- 13.Brown M., Laboratory identification of adenoviruses associated with gastroenteritis in Canada from 1983 to 1986. J. Clin. Microbiol. 28, 1525–1529 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favier A. L., Burmeister W. P., Chroboczek J., Unique physicochemical properties of human enteric Ad41 responsible for its survival and replication in the gastrointestinal tract. Virology 322, 93–104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidd A. H., Chroboczek J., Cusack S., Ruigrok R. W. H., Adenovirus type 40 virions contain two distinct fibers. Virology 192, 73–84 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Favier A.-L., Schoehn G., Jaquinod M., Harsi C., Chroboczek J., Structural studies of human enteric adenovirus type 41. Virology 293, 75–85 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Roelvink P. W., Lizonova A., Lee J. G. M., Li Y., Bergelson J. M., Finberg R. W., Brough D. E., Kovesdi I., Wickham T. J., The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72, 7909–7915 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez E., Romero C., Río A., Miralles M., Raventós A., Planells L., Burgueño J. F., Hamada H., Perales J. C., Bosch A., Gassull M. A., Fernández E., Chillon M., Short-fiber protein of ad40 confers enteric tropism and protection against acidic gastrointestinal conditions. Hum. Gene Ther. Methods 24, 195–204 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Crawford-Miksza L., Schnurr D. P., Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70, 1836–1844 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebner K., Pinsker W., Lion T., Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: Phylogenetic, taxonomic, and clinical implications. J. Virol. 79, 12635–12642 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albinsson B., Kidd A. H., Adenovirus type 41 lacks an RGD alpha(v)-integrin binding motif on the penton base and undergoes delayed uptake in A549 cells. Virus Res. 64, 125–136 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Robinson C. M., Zhou X., Rajaiya J., Yousuf M. A., Singh G., DeSerres J. J., Walsh M. P., Wong S., Seto D., Dyer D. W., Chodosh J., Jones M. S., Predicting the next eye pathogen: Analysis of a novel adenovirus. MBio 4, e00595-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zubieta C., Schoehn G., Chroboczek J., Cusack S., The structure of the human adenovirus 2 penton. Mol. Cell 17, 121–135 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Liu H., Wu L., Zhou Z. H., Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy. J. Mol. Biol. 406, 764–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng L., Huang X., Li X., Xiong W., Sun W., Yang C., Zhang K., Wang Y., Liu H., Huang X., Ji G., Sun F., Zheng C., Zhu P., Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology 450-451, 174–181 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Schoehn G., el Bakkouri M., Fabry C.´. M. S., Billet O., Estrozi L. F., le L., Curiel D. T., Kajava A. V., Ruigrok R. W. H., Kremer E. J., Three-dimensional structure of canine adenovirus serotype 2 capsid. J. Virol. 82, 3192–3203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackenbrack N., Rogers M. B., Ashley R. E., Keel M. K., Kubiski S. V., Bryan J. A., Ghedin E., Holmes E. C., Hafenstein S. L., Allison A. B., Evolution and cryo-EM capsid structure of a North American bat adenovirus and its relationship to other mastadenoviruses. J. Virol. 91, e01504-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy V. S., The role of hexon protein as a molecular mold in patterning the protein IX organization in human adenoviruses. J. Mol. Biol. 429, 2747–2751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vellinga J., van den Wollenberg D. J., van der Heijdt S., Rabelink M. J., Hoeben R. C., The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J. Virol. 79, 3206–3210 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Condezo G. N., Marabini R., Ayora S., Carazo J. M., Alba R., Chillón M., San Martín C., Structures of adenovirus incomplete particles clarify capsid architecture and show maturation changes of packaging protein L1 52/55k. J. Virol. 89, 9653–9664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangel W. F., San Martín C., Structure, function and dynamics in adenovirus maturation. Viruses 6, 4536–4570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grydsuk J. D., Fortsas E., Petric M., Brown M., Common epitope on protein VI of enteric adenoviruses from subgenera A and F. J. Gen. Virol. 77 (Pt 8), 1811–1819 (1996). [DOI] [PubMed] [Google Scholar]
- 33.San Martín C., Latest insights on adenovirus structure and assembly. Viruses 4, 847–877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajan A., Persson B. D., Frängsmyr L., Olofsson A., Sandblad L., Heino J., Takada Y., Mould A. P., Schnapp L. M., Gall J., Arnberg N., Enteric species F human adenoviruses use laminin-binding integrins as co-receptors for infection of Ht-29 cells. Sci. Rep. 8, 10019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith J. G., Silvestry M., Lindert S., Lu W., Nemerow G. R., Stewart P. L., Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLOS Pathog. 6, e1000959 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith J. G., Nemerow G. R., Mechanism of adenovirus neutralization by human α-defensins. Cell Host Microbe 3, 11–19 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Diaz K., Hu C. T., Sul Y., Bromme B. A., Myers N. D., Skorohodova K. V., Gounder A. P., Smith J. G., Defensin-driven viral evolution. PLOS Pathog. 16, e1009018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mateu M. G., Assembly, stability and dynamics of virus capsids. Arch. Biochem. Biophys. 531, 65–79 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Marsh M. P., Campos S. K., Baker M. L., Chen C. Y., Chiu W., Barry M. A., Cryoelectron microscopy of protein IX-modified adenoviruses suggests a new position for the C terminus of protein IX. J. Virol. 80, 11881–11886 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabry C. M., Rosa-Calatrava M., Moriscot C., Ruigrok R. W. H., Boulanger P., Schoehn G., The C-terminal domains of adenovirus serotype 5 protein IX assemble into an antiparallel structure on the facets of the capsid. J. Virol. 83, 1135–1139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matteson N. L., Barry M. A., Reddy V. S., Structure-based assessment of protein-protein interactions and accessibility of protein IX in adenoviruses with implications for antigen display. Virology 516, 102–107 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Maizel J. V. Jr., White D. O., Scharff M. D., The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36, 115–125 (1968). [DOI] [PubMed] [Google Scholar]
- 43.Brown M., Selection of nonfastidious adenovirus species in 293 cells inoculated with stool specimens containing adenovirus 40. J. Clin. Microbiol. 22, 205–209 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed L. J., Muench H., A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
- 45.Alba R., Hearing P., Bosch A., Chillon M., Differential amplification of adenovirus vectors by flanking the packaging signal with attB/attP-ΦC31 sequences: Implications for helper-dependent adenovirus production. Virology 367, 51–58 (2007). [DOI] [PubMed] [Google Scholar]
- 46.de la Rosa-Trevin J. M., Quintana A., Del Cano L., Zaldívar A., Foche I., Gutiérrez J., Gómez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V., Vargas J., Otón J., Sharov G., Vilas J. L., Navas J., Conesa P., Kazemi M., Marabini R., Sorzano C. O. S., Carazo J. M., Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Abrishami V., Vargas J., Li X., Cheng Y., Marabini R., Sorzano C. Ó. S., Carazo J. M., Alignment of direct detection device micrographs using a robust Optical Flow approach. J. Struct. Biol. 189, 163–176 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Rosa-Trevin J. M., Otón J., Marabini R., Zaldívar A., Vargas J., Carazo J. M., Sorzano C. O. S., Xmipp 3.0: An improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marabini R., Condezo G. N., Gómez-Blanco J., San Martín C., Near atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins. bioRxiv 10.1101/2020.07.24.220046, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S., McMullan G., Faruqi A. R., Murshudov G. N., Short J. M., Scheres S. H. W., Henderson R., High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Ilca S. L., Kotecha A., Sun X., Poranen M. M., Stuart D. I., Huiskonen J. T., Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abrishami V., Ilca S. L., Gomez-Blanco J., Rissanen I., de la Rosa-Trevín J. M., Reddy V. S., Carazo J. M., Huiskonen J. T., Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in Cryo-EM data. Prog. Biophys. Mol. Biol., (2020). [DOI] [PubMed] [Google Scholar]
- 57.Martínez M., Jiménez-Moreno A., Maluenda D., Ramírez-Aportela E., Melero R., Cuervo A., Conesa P., Del Caño L., Fonseca Y. C., Sánchez-García R., Strelak D., Conesa J. J., Fernández-Giménez E., de Isidro F., Sorzano C. O. S., Carazo J. M., Marabini R., Integration of cryo-EM model building software in Scipion. J. Chem. Inf. Model. 60, 2533–2540 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Vilas J. L., Gómez-Blanco J., Conesa P., Melero R., de la Rosa-Trevín J. M., Otón J., Cuenca J., Marabini R., Carazo J. M., Vargas J., Sorzano C. O. S., MonoRes: Automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Ramirez-Aportela E., Vilas J. L., Glukhova A., Melero R., Conesa P., Martínez M., Maluenda D., Mota J., Jiménez A., Vargas J., Marabini R., Sexton P. M., Carazo J. M., Sorzano C. O. S., Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 5.6.1–5.6.37 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallograph. Sec. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallograph. Sec. D 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D. R. Lide, W. M. M. Haynes, CRC Handbook of Chemistry and Physics: A Ready-Reference Book of Chemical and Physical Data: 2006–2007 (CRC, Boca Ratón., ed. 90, 2009). [Google Scholar]
- 64.J. M. Berg, J. L. Tymoczko, L. Stryer, Biochemistry (WH Freeman & Company, New York, ed. 8, 2015), pp. XXXII, 1056, 1048, 1043, 1043.
- 65.Drozdetskiy A., Cole C., Procter J., Barton G. J., JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchan D. W. A., Jones D. T., The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/9/eabd9421/DC1