Abstract
Primary immunodeficiency diseases encompass a variety of genetic conditions characterized by a compromised immune system and typically results in increased susceptibility to infection. In fact, they also manifest as autoimmunity, autoinflammation, atopic diseases, and malignancy. Currently, the number of recognized monogenic primary immunodeficiency disorders is set at ∼500 different entities, owing to the exponential use of unbiased genetic testing for disease discovery. In addition, the prevalence of secondary immunodeficiency has also been on the rise due to the increased use of immunosuppressive drugs to treat diseases based on immune dysregulation, an increase in the number of individuals undergoing hematopoietic stem cell transplantation, and other chronic medical conditions, including autoimmunity. Although the clinical symptoms of immunodeficiency disorders are broad, an early diagnosis and tailored management strategies are essential to mitigate the risk of infections and prevent disease-associated morbidity. Generally, the medical history and physical examination can provide useful information that can help delineate the possibility of immune defects. In turn, this makes it feasible to select focused laboratory tests that identify immunodeficiency disorders based on the specific immune cells and their functions or products that are affected. Laboratory evaluation involves quantitative and functional classic testing (e.g., leukocyte counts, serum immunoglobulin levels, specific antibody titers in response to vaccines, and enumeration of lymphocyte subsets) as well as genetic testing (e.g., individual gene evaluation via Sanger sequencing or unbiased evaluation based on next-generation sequencing). However, in many cases, a diagnosis also requires additional advanced research techniques to validate genetic or other findings. This article updates clinicians about available laboratory tests for evaluating the immune system in patients with primary immunodeficiency disorders. It also provides a comprehensive list of testing options, organized based on different components of host defense.
Keywords: Diagnosis, Immunodeficiency, Inborn Errors of Immunity, Laboratory test, Immunological evaluation
Primary immunodeficiency disorders (PID) include a broad range of conditions with a broad clinical spectrum of manifestations and established genetic backgrounds.1 Getting a precise and prompt diagnosis of immunodeficiency enables the initiation of the appropriate treatment with the aim of reducing the risk of complications and of improving quality of life and survival. After a comprehensive clinical assessment, the laboratory plays a key role in identifying and classifying immunologic abnormalities. This article focuses on the strategies for optimally using laboratory tests, both classic and genetic, in the diagnosis of PID.
EVALUATING SUSPECTED DEFECTS IN ANTIBODY RESPONSE
Clinical Indication
Primary immunodeficiency that affects the humoral compartment of the immune system represents the most common defect. The clinical scenarios that would typically lead to an evaluation of antibody production are recurrent and/or severe upper and lower respiratory tract infections that typically involve encapsulated bacteria and viruses. Patients also have a higher risk of diarrhea, autoimmune diseases, enteropathy, and lung infiltration.2,3
Methodology
Initial screening for a suspected humoral immunodeficiency involves quantitation of circulating serum immunoglobulins (immunoglobulin G [IgG], IgA, IgM, and IgE) (Table 1). The results must be compared with age-matched controls that typically are provided as 95% confidence intervals. Hypogammaglobulinemia due to causes other than PID should also be considered in any patient with low immunoglobulin.4 Ruling this out may be accomplished by evaluating the serum albumin level to exclude protein loss as a cause of low immunoglobulin levels.
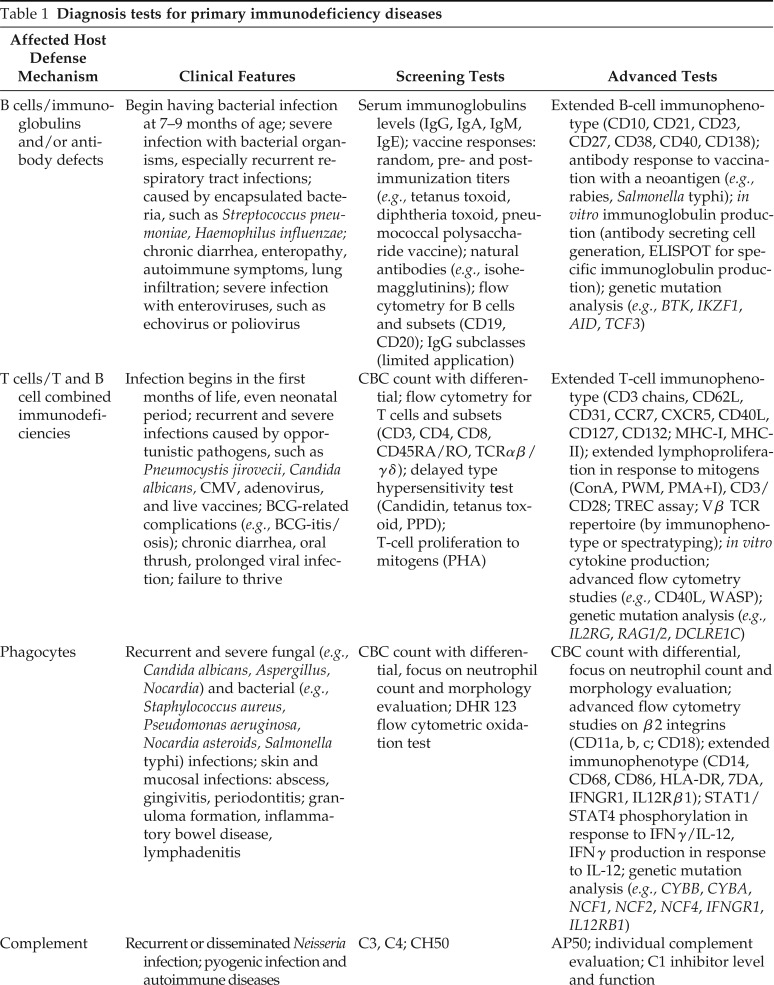
Table 1.
Diagnosis tests for primary immunodeficiency diseases
| Affected Host Defense Mechanism | Clinical Features | Screening Tests | Advanced Tests |
|---|---|---|---|
| B cells/immunoglobulins and/or antibody defects | Begin having bacterial infection at 7–9 months of age; severe infection with bacterial organisms, especially recurrent respiratory tract infections; caused by encapsulated bacteria, such as Streptococcus pneumoniae, Haemophilus influenzae; chronic diarrhea, enteropathy, autoimmune symptoms, lung infiltration; severe infection with enteroviruses, such as echovirus or poliovirus | Serum immunoglobulins levels (IgG, IgA, IgM, IgE); vaccine responses: random, pre- and postimmunization titers (e.g., tetanus toxoid, diphtheria toxoid, pneumococcal polysaccharide vaccine); natural antibodies (e.g., isohemagglutinins); flow cytometry for B cells and subsets (CD19, CD20); IgG subclasses (limited application) | Extended B-cell immunophenotype (CD10, CD21, CD23, CD27, CD38, CD40, CD138); antibody response to vaccination with a neoantigen (e.g., rabies, Salmonella typhi); in vitro immunoglobulin production (antibody secreting cell generation, ELISPOT for specific immunoglobulin production); genetic mutation analysis (e.g., BTK, IKZF1, AID, TCF3) |
| T cells/T and B cell combined immunodeficiencies | Infection begins in the first months of life, even neonatal period; recurrent and severe infections caused by opportunistic pathogens, such as Pneumocystis jirovecii, Candida albicans, CMV, adenovirus, and live vaccines; BCG-related complications (e.g., BCG-itis/osis); chronic diarrhea, oral thrush, prolonged viral infection; failure to thrive | CBC count with differential; flow cytometry for T cells and subsets (CD3, CD4, CD8, CD45RA/RO, TCRαβ/γδ); delayed type hypersensitivity test (Candidin, tetanus toxoid, PPD); T-cell proliferation to mitogens (PHA) | Extended T-cell immunophenotype (CD3 chains, CD62L, CD31, CCR7, CXCR5, CD40L, CD127, CD132; MHC-I, MHC-II); extended lymphoproliferation in response to mitogens (ConA, PWM, PMA+I), CD3/CD28; TREC assay; Vβ TCR repertoire (by immunophenotype or spectratyping); in vitro cytokine production; advanced flow cytometry studies (e.g., CD40L, WASP); genetic mutation analysis (e.g., IL2RG, RAG1/2, DCLRE1C) |
| Phagocytes | Recurrent and severe fungal (e.g., Candida albicans, Aspergillus, Nocardia) and bacterial (e.g., Staphylococcus aureus, Pseudomonas aeruginosa, Nocardia asteroids, Salmonella typhi) infections; skin and mucosal infections: abscess, gingivitis, periodontitis; granuloma formation, inflammatory bowel disease, lymphadenitis | CBC count with differential, focus on neutrophil count and morphology evaluation; DHR 123 flow cytometric oxidation test | CBC count with differential, focus on neutrophil count and morphology evaluation; advanced flow cytometry studies on β2 integrins (CD11a, b, c; CD18); extended immunophenotype (CD14, CD68, CD86, HLA-DR, 7DA, IFNGR1, IL12Rβ1); STAT1/STAT4 phosphorylation in response to IFNγ/IL-12, IFNγ production in response to IL-12; genetic mutation analysis (e.g., CYBB, CYBA, NCF1, NCF2, NCF4, IFNGR1, IL12RB1) |
| Complement | Recurrent or disseminated Neisseria infection; pyogenic infection and autoimmune diseases | C3, C4; CH50 | AP50; individual complement evaluation; C1 inhibitor level and function |
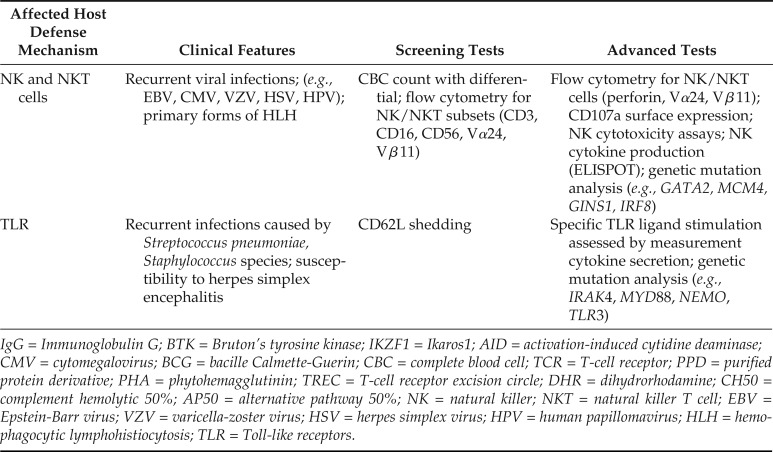
| NK and NKT cells | Recurrent viral infections; (e.g., EBV, CMV, VZV, HSV, HPV); primary forms of HLH | CBC count with differential; flow cytometry for NK/NKT subsets (CD3, CD16, CD56, Vα24, Vβ11) | Flow cytometry for NK/NKT cells (perforin, Vα24, Vβ11); CD107a surface expression; NK cytotoxicity assays; NK cytokine production (ELISPOT); genetic mutation analysis (e.g., GATA2, MCM4, GINS1, IRF8) |
| TLR | Recurrent infections caused by Streptococcus pneumoniae, Staphylococcus species; susceptibility to herpes simplex encephalitis | CD62L shedding | Specific TLR ligand stimulation assessed by measurement cytokine secretion; genetic mutation analysis (e.g., IRAK4, MYD88, NEMO, TLR3) |
IgG = Immunoglobulin G; BTK = Bruton’s tyrosine kinase; IKZF1 = Ikaros1; AID = activation-induced cytidine deaminase; CMV = cytomegalovirus; BCG = bacille Calmette-Guerin; CBC = complete blood cell; TCR = T-cell receptor; PPD = purified protein derivative; PHA = phytohemagglutinin; TREC = T-cell receptor excision circle; DHR = dihydrorhodamine; CH50 = complement hemolytic 50%; AP50 = alternative pathway 50%; NK = natural killer; NKT = natural killer T cell; EBV = Epstein-Barr virus; VZV = varicella-zoster virus; HSV = herpes simplex virus; HPV = human papillomavirus; HLH = hemophagocytic lymphohistiocytosis; TLR = Toll-like receptors.
Measuring IgG subclasses levels has more limited utility. These are often evaluated in patients with clinical manifestations associated with antibody deficiencies but normal total IgG levels and/or selective IgA deficiency. IgG subclass levels also may be tested based on the pattern of vaccination responses. Although not exclusively, IgG1 and IgG3 are primarily associated with responses to protein antigens, whereas IgG2 and IgG4 represent the typical subclass response to polysaccharide antigens. Importantly, IgG subclasses determinations are neither sensitive nor specific by themselves for diagnosing increased susceptibility to recurrent infections. Besides, IgG subclass values can widely vary in infants and young children, which makes interpretation more difficult, and abnormalities in IgG subclasses can be observed in various conditions other than recurrent infections.
To accurately evaluate B-cell function, specific antibody production must be measured. Isohemagglutinins, a natural and predominantly IgM antibody directed at the nonexpressed ABO polysaccharide blood antigens (e.g., individuals who are A+ have anti-B isohemagglutinins), is a simple and readily accessible test. Isohemagglutinin levels are reliable in adults and children ages of >1 year; however, they are absent in individuals with blood type AB+. Patients with impaired antibody production may lack these antibodies or have titers < 1:10.5,6
For specific antibody evaluation, antibody titers to specific antigens from vaccines or natural infections represent the most reliable approach. Both protein- and polysaccharide-based vaccines should be evaluated.7 The response to protein antigens can be assessed by using tetanus toxoid, diphtheria toxoid, conjugated Haemophilus influenzae type B, and conjugated pneumococcal vaccines (e.g., Prevnar 13, Pfizer, Philadelphia, PA). Prevaccine and 4–6 weeks postvaccine titers should be determined; normal responses should raise antigen-specific antibody titers by at least fourfold and/or by reaching specific protective titers. Protective anti-tetanus toxoid antibody titers are typically defined as ≥0.1 IU/mL, although higher levels are expected after recall vaccine challenges. Pneumococcal polysaccharide vaccination (e.g., Pneumovax 23 (Merck & Co., Rahway, NJ) a pnenumococcal polysacharide vaccine 23-valent (PPSV23), and Salmonella capsular polysaccharide vaccine) allows testing of adults and children ages >2 years for polysaccharide responses. Nevertheless, the interpretation of this polysaccharide vaccine responses could be complex because most children are regularly vaccinated with conjugated pneumococcal vaccines during infancy. The PID practice parameter for PPSV23 recognizes a protective titer of ≥1.3 µg/mL, or a two- to fourfold increase in postvaccination titers compared with prevaccination levels. These recommendations use the expected increase to be observed in 50% of tested serotypes for children ages ≤ 5 years and 70% of serotypes for children >5 years of age as well as adults.8,9 Notably, in the context of evaluating patients on immunoglobulin replacement therapy, immunization with a neoantigen (an antigen not previously administered or recognized by the patient) is required. The Salmonella capsular polysaccharide vaccine has been validated as a useful neoantigen to measure a humoral response for most patients on immunoglobulin replacement.10,11 The rabies vaccine is another helpful neoantigen.8
Flow cytometry is used to quantitatively evaluate B lymphocyte counts and their maturation status. CD19 and CD20 are the most widely used pan–B-cell markers. A severely reduced or absent number of B cells (<2% of total lymphocytes) is typically associated with agammaglobulinemia. In addition, immunophenotyping of B cells, including memory (CD27+), naive B cells (CD27–), non–class-switched (IgM+), and class-switched (IgM–) memory B cells help to identify and stratify B-cell defects, not necessarily affecting their total number but their subsets. This more extensive characterization of B cells has proven to be useful in patients with common variable immunodeficiency (CVID).12
EVALUATING SUSPECTED T-CELL DEFECTS
Clinical Indication
T-cell defects result in some of the most severe forms of PID, including life-threatening and lethal (if untreated) forms of severe combined immunodeficiency (SCID). The manifestations commonly experienced by patients with a T-cell defect include neonatal onset recurrent and severe infections caused by opportunistic pathogens (including live vaccines), chronic diarrhea, oral thrush, Pneumocystis jirovecii pneumonia, prolonged viral infections, autoimmunity, and failure to thrive. Noteworthy, T-cell defects can arise later in life, including adulthood.
Methodology
Initial evaluation includes a complete blood cell count, with the differential focusing on the absolute lymphocyte count compared with age-matched control ranges for proper interpretation. As a rule of thumb, lymphopenia in infants is determined when the absolute lymphocyte count is <3000/mm3, and in adults when it is <1000/mm3. Lymphopenia in infants should always be taken as a potentially serious and life-threatening finding. Repeating the lymphocyte count when lymphopenia is detected is a good practice because transient lymphopenia can be found in a variety of common infectious illnesses. Human immunodeficiency virus (HIV) viral load assay also should be evaluated to exclude HIV infection when this disease is part of the differential diagnosis. Mechanical lymphocyte loss can also cause lymphopenia and intact T-cell function, especially in cases after neonatal cardiac surgery or in patients with diarrhea due to lymphangiectasia.
Flow cytometry to evaluate the T cells represents the next step in assessing suspected T-cell or combined T- and B-cell defects. Standard clinical flow cytometry involves immunophenotyping of T cells (CD3+ cells co-expressing either CD4 or CD8), B cells (CD3–, CD19+/20+ cells), and natural killer (NK) cells (CD3–, CD16+/56+ cells). When focusing on T cells, analyzing the expression of markers, including CD45RA, CD62L (or CCR7), and CD45RO, help identify maturation stages (e.g., CD45RA+/CD62L+ naive cells, CD45RA–/CD62L+ central memory cells, CD45–/CD62L– effector memory cells, and CD45RA+/CD62L– effector memory re-expressing CD45RA (T-EMRA) cells; the CD45 isoforms -RA and -RO represent naive and memory populations, respectively. Some of the T-cell subsets could be preferentially affected in different combined immunodeficiency syndromes. Oligoclonal expansion of T cells and maternal T-cell engraftment (as could be found in babies with SCID) result primarily in memory CD45RO+ T cells over-representation, in contrast to the presence of predominantly naive CD45RA+ T cells in healthy infants. Quantitative evaluation of the T-cell compartment should be complemented with in vitro functional evaluation by means of cell proliferation, cytokine production, intracellular signaling, and cytotoxicity assays.
Advanced flow cytometry by using monoclonal antibodies specific for activated cells (CD25+, CD69+), regulatory T cells (CD3+/CD4+/CD25+/FOXP3+), or other subsets of T cells as T helper (Th) 1, Th2, Th17, or T follicular helper can help with the characterization of several PIDs.13 In addition, specific protein expression is another screening test for selected proteins affected in PIDs (e.g., CD40L, WASp), although the presence does not necessarily imply that a protein is functional, a situation that can be seen with missense genetic changes. Thus, functional studies are required to confirm or rule out the activity of expressed proteins. In that regard, and depending on the protein and/or pathway of interest, specific research laboratories might have to be involved in such investigations.
Cutaneous delayed-type hypersensitivity (DTH) testing can be performed to measure the memory response to previously exposed (recall) antigens such as those present in vaccines (purified protein derivative [PPD] after Bacille Calmette-Guerin [BCG] vaccination or tetanus toxoid after tetanus, diphtheria, acellular pertussis [Tdap] vaccination) or environmental and/or common microbes (e.g., Candida). This method has its own pros and cons; although a negative DTH reaction as determined by an experienced reader 48 hours after inoculation does not necessarily imply a T-cell deficiency, a positive reaction does rule out most forms of the SCID.14 Although DTH remains a traditional, relatively accessible, and useful method for evaluating cell-mediated immunity to specific antigens in vivo, the more standardized in vitro cell proliferation tests have mostly replaced it in clinical practice.
T-cell proliferation assays directly assess T-cell capability to replicate. After several days of incubation with external stimuli (3–4 days for mitogens; 5–7 days for recall antigens and allogeneic cells), tritiated thymidine is added for incorporation into the DNA of proliferating cells. After quantitation, the results are reported as counts per minute (cpm), which can be converted into a stimulation index (the ratio of cpm after stimulation divided by cpm of unstimulated/background proliferation).15 The culture supernatant may also be used to detect cytokine production under the same conditions.16 Nonradioactive markers of proliferation include EdU (e.g., 50-ethynyl-20-deoxyuridine) and cell tracking dyes (e.g., carboxyfluorescein diacetate succinimidyl ester, CFSE). These markers are, nowadays, used more routinely because smaller amounts of cells are required and these markers are evaluated via flow cytometry, which also allows for the detection of proliferation by specific cell subsets (e.g., CD4+ versus CD8+ T cells).
T-cell repertoire analysis is valuable for examining T-cell diversity, including the identification of oligoclonal T-cell populations. Methods include evaluation of T-cell receptor variable β (Vβ) chain families by flow cytometry, and T-cell receptor Vβ complementarity determining region 3 detection by polymerase chain reaction, also known as spectratyping. T-cell repertoire studies could be of added value when studying cases in which T-cell rearrangement or thymic output are affected (as in RAG1/RAG2 defects and DiGeorge syndrome, respectively), and when evaluating maternal engraftment or T-cell clonal expansions.
Newborn screening for SCID has been implemented to identify severe T-cell quantitative defects in the newborn period. The determination of T-cell receptor excision circles in peripheral blood is used to evaluate for the presence of recent thymic emigrants on the blood spot of a Guthrie card.17
EVALUATING SUSPECTED PHAGOCYTE DYSFUNCTION SYNDROMES
Clinical Indication
The clinical features of neutrophil defects include recurrent or severe bacterial and fungal infections that typically involve the skin and internal organs. Identifying common pathogens and clinical patterns of infection is crucial for narrowing down the underlying phagocyte dysfunction abnormalities. For example, invasive Serratia marcescens, Chromobacterium violaceum, and Aspergillus nidulans infections are highly suggestive of chronic granulomatous disease (CGD). Screening studies should start with a leukocyte count and differential, and morphologic review. Chronic and cyclic neutropenia as well as morphologic abnormalities should be ruled out. The evaluation should then focus on qualitative defects, which mainly involve two types of disorders: defects in neutrophil migration to sites of infection and defects that impact neutrophil killing of certain bacteria and fungi associated with impaired production of reactive oxygen species.
Methodology
Different forms of leukocyte adhesion deficiencies (LAD), each of them displaying some common and other distinctive features, have been described. LAD1, clinically characterized by recurrent and severe infections with neutrophilia but absent neutrophilic tissue response (e.g., no pus formation) can be evaluated by flow cytometry through the surface expression of the CD18 component of the β2 integrin that dimerizes with three other molecules (CD11a to form LFA1, CD11b to form Mac-1 [CR3], and CD11c to form p150/95 [CR4]). LAD2 is characterized by the absence of fucosylated CD15 (sialyl-Lewis-X) and intellectual disability. In contrast, a diagnosis of LAD3 requires specialized testing of integrin function on leukocytes and platelets, which leads to immune dysfunction and hemorrhagic diathesis, respectively. In CGD, the neutrophil oxidative burst pathway can be screened by using the dihydrorhodamine (DHR) 123 flow cytometry assay or its predecessor, the nitroblue tetrazolium test. The DHR assay depends on the oxidation of the DHR 123 dye and on producing a bright fluorescent signal measured by flow cytometry after cell stimulation with phytohemagglutinin (PHA). This assay can generally distinguish between X-linked CGD and autosomal recessive forms as well as detect female X-linked recessive carriers.18 In research settings, chemotaxis abnormalities can be identified by using the Rebuck skin window technique and, in vitro, by using cell movement to a chemoattractant in a Boyden chamber or a soft agar system.
EVALUATING SUSPECTED COMPLEMENT DISORDERS
Clinical Indication
Different complement disorders result in infectious susceptibility and inappropriate inflammation. Defects in the early components of the classic complement components (C1, C4, and C2) increase the risk of pyogenic infections and autoimmune diseases, including systemic lupus erythematosus. Defects in the terminal components involved in both the classic and the alternative activation pathways are associated with increased susceptibility to invasive Neisseria species infections. C1 esterase inhibitor deficiency causes hereditary angioedema, whereas patients with paroxysmal nocturnal hemoglobinuria often demonstrate decay-accelerating factor or CD59 defects.
Methodology
The total hemolytic complement (CH50) test measures the function of the classic complement cascade, whereas the alternative pathway (AH50) test measures the function of the alternative complement pathway. Patients with C1, C2, or C4 deficiency will have a markedly decreased CH50 with a normal AH50. Finding a decreased AH50, but normal CH50, is consistent with a deficiency in factor B, factor D, or properdin. A decrease in both CH50 and AH50 suggests a deficiency in C3 or the shared terminal complement components C5, C6, C7, C8, or C9. Individual component levels can be examined by using immunoassays (e.g., enzyme-linked immunosorbent assay [ELISA]), and nephelometric techniques to confirm a suspected complement deficiency; however, only a few laboratories conduct functional tests.19 Generally, C1 inhibitor (C1INH) immunoassays are capable of determining C1q, C1INH levels but certain circumstances require component function testing. The evaluation of the mannose binding lectin activation pathway, which involves measuring mannose binding lectin serum levels and performing functional assays to determine its ability to activate the complement pathway, has limited clinical application.
EVALUATING SUSPECTED NK DEFECTS
Clinical Indication
Testing for NK-cell function is indicated in patients with recurrent herpesvirus family infections as well as primary forms of hemophagocytic lymphohistiocytosis (HLH) syndromes.
Methodology
Flow cytometry can be used to assess NK cells (CD3–/CD16+/CD56+). A more in-depth evaluation can include testing of NK/NK T-cell markers as perforin, killer-cell immunoglobulin-like receptors (KIRs), CDG2/CD94, NKp46, CD117, or CD3+Vα24+Vβ11+.20 NK functional evaluation can include flow cytometric CD107a degranulation test and in vitro NK cytotoxic activity against particular cell lines (e.g., the K562 erythroleukemia cell line).
EVALUATING IMMUNE DEFECTS THAT INVOLVES MACROPHAGE ACTIVATION
Clinical Indication
Defects in interferon (IFN)-γ–mediated immunity results in increased susceptibility to mycobacteria and Salmonella species. The two most prevalent genetic defects that result in Mendelian susceptibility of mycobacteria disease are IFNGR1 and IL12RB1 gene mutations.21
Methodology
Flow cytometry by using specific monoclonal reagents is commonly used to assess surface protein expression of IFNγR1 and IFNγR2 (which makes the IFNγ receptor complex, IFNγRC), IL-12Rβ1 and IL-12Rβ2 (makes the IL-12 receptor complex, IL12RC).22 Ex vivo evaluation of signal transducer and activator of transcription 1 (STAT1) phosphorylation in response to IFN-γ in monocytes by means of flow cytometry or Western blotting evaluation can detect abnormalities in the IFNγRC and the downstream signaling pathway that involves Janus kinase 1 (JAK1), JAK2, and STAT1. Evaluation of STAT4 phosphorylation in prestimulated T cells in response to IL-12 by using the same methods described above can detect abnormalities in the IL12RC and downstream signaling pathway. Defects in IL-12 production itself can be detected by evaluating IL-12 production in response to ex vivo stimulation of mononuclear cells with lipopolysaccharide and IFN-γ.
EVALUATING SUSPECTED TOLL-LIKE RECEPTOR DEFECTS
Clinical Indication
Patients with Toll-like receptor (TLR) deficiency exhibit distinctive features due to TLR pathway molecular abnormalities. Interleukin 1 receptor associated kinase 4 (IRAK4), myeloid differentiation primary response protein 88 (MYD88), and NFkB essential modulator (NEMO) defects are associated with recurrent Streptococcus pneumoniae and Staphylococcus species infections. TLR3, UNC93B, TBK1, and IRF3 gene mutations can result in an increased incidence of herpes simplex encephalitis.
Methodology
TLR function screening includes stimulating leukocytes with specific TLR ligands and assessing CD62L shedding from granulocytes by using flow cytometry. Cells with intact TLR signaling should shed CD62L rapidly.23 Laboratory evaluations of TLR activity involve activating mononuclear cells with TLR ligands and measuring cytokine production. In contrast, functional evaluation for TLR defects associated with herpes simplex encephalitis usually requires research tests on samples other than blood. Direct sequencing of the suspected mutant gene involved in the specific TLR signaling pathway would be a further step.
GENETIC DIAGNOSTIC TESTING FOR PID
Because PIDs are defined as genetic diseases with increased susceptibility to infections, immune dysregulation (including autoimmunity, allergy, and autoinflammation), and cancer, molecular testing plays an essential role in establishing these diagnoses. Whereas the timing for genetic testing can be flexible (e.g., “genetics first” versus “genetics last”), depending on the preference of the physician and access at the patient’s medical center, no PID diagnostic evaluation is done until proper genetic testing is completed. Multiple genetic testing modalities exist, ranging from single variant/single gene Sanger sequencing to massively parallel sequencing/next-generation sequencing. The latter, in its multiple testing varieties, has demonstrated to provide the most cost-effective diagnostic rates for patients in the clinical and research settings.
Individual gene or genomic variant testing by using the Sanger method is typically performed when there is a high level of confidence that a specific molecular defect is present in a patient, particularly when there is a family history of an identified genetic diseases or pathogenic variant. Sanger sequencing is also used for confirmation of NGS findings.
NGS tests, including PID genetic panels, whole exome sequencing (WES), and whole genome sequencing (WGS), are now widely used for PID diagnosis. High-throughput NGS methods enable for the simultaneous examination of multiple genes, which results in a rapid, cost-effective, genotype-based approach to a molecular diagnosis. This is the preferred genetic approach for patients with typical or atypical clinical presentations, PID with large genotype-phenotype variability (i.e., clinical heterogeneity) or variable penetrance, and PID with multiple candidate genes that cause the same phenotype (i.e., genetic heterogeneity). Whereas NGS panels and WES and WGS tests mostly rely on the same technology, the genetic information covered in panels (i.e., the coding sequences of up to ∼500 PID-related genes), WES (i.e., the coding sequences of ∼22,000 genes), and WGS (i.e., the coding and noncoding information of the whole genome, which represents ∼100 times more genetic data than WES) widely varies and, with it, the chances of making expected and unexpected genetic diagnoses. However, the larger genetic information tested with WGS carries an increase in analysis complexity and cost.
When focused on NGS testing, the diagnostic yield is higher for patients with more severe phenotypes, in families with more than one affected individual, under conditions of consanguinity, or when “trios” (index and parents) are tested. However, testing of “singletons”/index cases alone is a common practice and should not be discouraged.24 It is important to note that NGS panels and WES analysis are limited to gene coding exons (and close by intronic/splicing areas), and thus will miss most variants in noncoding regions. Most commercially available NGS panels and WES also perform quantitative gene amplification analysis that can detect copy number variations that are responsible for ∼5% of PID due to haploinsufficiency. As a result, targeted NGS panels can be used as a cost-effective first-line genetic approach for evaluating new patients with PID, with second-line testing to exclude disease-causing mutations if needed.24–26 In other words, a nondiagnostic NGS PID panel does not exclude a genetic diagnosis for a PID and is not an uncommon situation.
When requesting a NGS panel for the diagnosis of PID, it is recommended to go broader and assess all known PID-associated genes rather than to restrict the testing to a single clinical phenotype, such as SCID or familial HLH panels. The “think big” approach has been found to be more sensitive and efficient in genetically diagnosing patients who present with HLH, not infrequently identifying defects beyond the classic familial HLH-associated genes.27
WES is routinely used and is being refined to detect known but also new PIDs, as has been the experience with the exponential growth of new PID diagnoses reported in the past two decades.1 Alternatively, WGS, which shares the same aim as WES of providing an unbiased evaluation but on a larger scale, is becoming more accessible. WGS provides broader coverage and can detect a higher number of high-quality variants compared with the WES. It is also more effective in detecting potential disease-causing variants within the targeted regions, particularly single-nucleotide variants. WGS can also capture copy number variations with a higher sensitivity than other NGS methods. Moreover, WGS is also considered to be more powerful and reliable than WES for variant detection within the coding regions of the genome.24–26,28,29
Clinicians should be aware that each of the NGS tests discussed has its own pros and cons in terms of variant detection sensitivity and specificity, and this should be balanced with the needs of their patients, e.g., cost and turn-around time usually sides with NGS PID panel testing, whereas breadth and depth of information usually sides with WES and WGS testing. Furthermore, although there are no “good” or “bad” genetic tests among those herein described, users should be mindful of their strengths and limitations, and manage their expectations; no single test will provide all the diagnoses and, most likely, a combination of genetic and classic laboratory tests not infrequently, including re-analysis of genetic data, will be needed to reach an accurate diagnosis. Importantly, and, as stated above, the genetic diagnostic success rate varies, depending on the patient’s phenotype, e.g., for patients with a CVID phenotype, it has been found to be ∼40% and as low as ∼15% for autoinflammatory syndromes. Thus, not finding a genetic diagnosis for patients with CVID or autoinflammatory diseases (∼60% and ∼85% diagnostic failure rate, respectively), is a frustrating but also, presently, a common situation.
In addition to NGS, chromosome microarray analysis (CMA) remains an effective approach for detecting large-scale chromosomal abnormalities associated with PID, such as deletions, duplications, and other structural variations. CMA is commonly used to confirm the diagnosis of 22q11 deletion syndrome and other chromosomal deletion syndromes. Integrating NGS findings with CMA by targeting specific genes associated with PID enhances diagnostic yield and enables the identification of genetic alterations.30
Dealing with variants of uncertain significance (VUS) is another common issue when interpreting reported variants. Understanding the phenotype associated with the different PID can help in determining the significance of the variant. Genetic counselors can provide further insights based on existing evidence, including population frequency data, family segregation, de novo occurrence, and in silico predictions among other variables. However, functional studies are absolutely required to assess the effect of a VUS on protein expression, function, and the cellular pathway relevant to each PID. These studies also generate information to support variant classification, although they require highly specialized laboratories. In this setting, when facing a VUS as a potential cause for an individual patient, it is prudent and also recommended to contact colleagues with hands-on experience in that particular gene to inquire if they have seen and/or tested a particular VUS.
CONCLUSION
Classic and genetic laboratory testing in PID has shed light on critical defects of the immune system. The identification of specific mutations and altered pathways provides valuable insights for early diagnosis, which, in turn, should result in targeted therapeutic interventions, which emphasizes the need for tailored treatments to reduce the clinical burden of the disease and improve patient outcomes. When focused on laboratory testing for patients suspected of having a PID, providers should be mindful that quantitative and functional evaluation of immune cells (e.g., B, T, NK, and phagocytic cells) and their products (e.g., immunoglobulins, specific antibodies, and cytokines), as well as genetic testing, will be necessary to complete a thorough evaluation of a patient.
CLINICAL PEARLS AND PITFALLS
The clinical manifestation of PID varies broadly; however, on suspicion, a consultation with a clinical immunologist is essential to tailor the testing of the immune system aimed at an early diagnosis and developing a suitable treatment plan.
Optimized laboratory tests based on patients' clinical characteristics are required to provide an accurate diagnosis without performing unnecessary tests, e.g. testing for complement deficiencies via CH50 and AH50 evaluation in patients with disseminated mycobacterial disease is unlikely to be informative, whereas it would be absolutely necessary in patients with invasive or rare neisserial infections.
Some patients with PID may present with normal initial laboratory results, which leads to incorrect reassurance. Repeated testing or a broader and/or more specific panel of tests may be required to detect abnormalities of immune functions.
Certain medications, such as immunosuppressive drugs, corticosteroids, or even acetaminophen, may have an effect on immune tests. Thus, it is essential to consider a patient's medication history when interpreting laboratory results.
Advanced genetic sequencing technologies, such as different NGS modalities, have greatly contributed to the expanded understanding of PID, and each of these tests has its own pros and cons. Consider patient characteristics, clinical phenotype, cost-effectiveness, and data analysis complexity when choosing genetic testing methods.
Footnotes
The authors have no conflicts of interest to declare pertaining to this article
Funding to support this work was provided by Elevare Consulting Group LLC through an unrestricted educational grant to the American Association of Certified Allergists (AACA). The AACA had full responsibility for selecting topics and authors and provided publisher oversight. The contents of this work reflect the opinion(s) of the author(s) and are not intended to replace published guidelines or the clinician’s medical advice in the doctor-patient relationship. Additional support by the Intramural Research Program, National Institutes of Health Clinical Center. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government
REFERENCES
- 1.Tangye SG, Al-Herz W, Bousfiha A, et al. Human inborn errors of immunity: 2022 update on the classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol. 2022; 42:1473–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith T, Cunningham-Rundles C. Primary B-cell immunodeficiencies. Hum Immunol. 2019; 80:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed A, Lippner E, Khanolkar A. Clinical aspects of B cell immunodeficiencies: the past, the present and the future. Cells. 2022; 11:3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otani IM, Lehman HK, Jongco AM, et al. Practical guidance for the diagnosis and management of secondary hypogammaglobulinemia: a work group report of the AAAAI Primary Immunodeficiency and Altered Immune Response Committees. J Allergy Clin Immunol. 2022; 149:1525–1560. [DOI] [PubMed] [Google Scholar]
- 5.Alkan G, Keles S, Reisli İ. Evaluation of clinical and immunological characteristics of children with common variable immunodeficiency. Int J Pediatr. 2018; 2018:3527480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seidel MG, Kindle G, Gathmann B, et al. The European Society for Immunodeficiencies (ESID) Registry Working Definitions for the Clinical Diagnosis of Inborn Errors of Immunity. J Allergy Clin Immunol Pract. 2019; 7:1763–1770. [DOI] [PubMed] [Google Scholar]
- 7.Oliveira JB, Fleisher TA. Laboratory evaluation of primary immunodeficiencies. J Allergy Clin Immunol. 2010; 125(suppl 2):S297–S305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orange JS, Ballow M, Stiehm ER, et al. Use and interpretation of diagnostic vaccination in primary immunodeficiency: a working group report of the Basic and Clinical Immunology Interest Section of the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2012; 130(suppl 2):S1–S24. [DOI] [PubMed] [Google Scholar]
- 9.McNulty CMG, Li JT. Interpretation of post-pneumococcal vaccine antibody levels: concerns and pitfalls. J Allergy Clin Immunol Pract. 2019; 7:1061–1062. [DOI] [PubMed] [Google Scholar]
- 10.Evans C, Bateman E, Steven R, et al. Measurement of Typhi Vi antibodies can be used to assess adaptive immunity in patients with immunodeficiency. Clin Exp Immunol. 2018; 192:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bausch-Jurken MT, Verbsky JW, Gonzaga KA, et al. The use of Salmonella Typhim vaccine to diagnose antibody deficiency. J Clin Immunol. 2017; 37:427–433. [DOI] [PubMed] [Google Scholar]
- 12.Wehr C, Kivioja T, Schmitt C, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008; 111:77–85. [DOI] [PubMed] [Google Scholar]
- 13.Ma CS, Freeman AF, Fleisher TA. Inborn errors of immunity: a role for functional testing and flow cytometry in aiding clinical diagnosis. J Allergy Clin Immunol Pract. 2023; 11:1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yates AB, deShazo RD. Delayed hypersensitivity skin testing. Immunol Allergy Clin North Am. 2001; 21:383–397. [Google Scholar]
- 15.Bonilla FA. Interpretation of lymphocyte proliferation tests. Ann Allergy Asthma Immunol. 2008; 101:101–104. [DOI] [PubMed] [Google Scholar]
- 16.Prussin C, Metcalfe DD. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J Immunol Methods. 1995; 188:117–128. [DOI] [PubMed] [Google Scholar]
- 17.Rosenzweig SD, Kobrynski L, Fleisher TA. Laboratory evaluation of primary immunodeficiency disorders. In Stiehm's Immune Deficiencies, 2nd ed. Sullivan KE, Stiehm ER. (Eds). Cambridge, MA: Academic Press, 115–131, 2020. [Google Scholar]
- 18.Jirapongsananuruk O, Malech HL, Kuhns DB, et al. Diagnostic paradigm for evaluation of male patients with chronic granulomatous disease, based on the dihydrorhodamine 123 assay. J Allergy Clin Immunol. 2003; 111:374–379. [DOI] [PubMed] [Google Scholar]
- 19.Wen L, Atkinson JP, Giclas PC. Clinical and laboratory evaluation of complement deficiency. J Allergy Clin Immunol. 2004; 113:585–593; quiz 594. [DOI] [PubMed] [Google Scholar]
- 20.Nichols KE, Ma CS, Cannons JL, et al. Molecular and cellular pathogenesis of X-linked lymphoproliferative disease. Immunol Rev. 2005; 203:180–199. [DOI] [PubMed] [Google Scholar]
- 21.Bustamante J, Boisson-Dupuis S, Abel L, et al. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol. 2014; 26:454–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delmonte OM, Fleisher TA. Flow cytometry: surface markers and beyond. J Allergy Clin Immunol. 2019; 143:528–537. [DOI] [PubMed] [Google Scholar]
- 23.von Bernuth H, Ku C-L, Rodriguez-Gallego C, et al. A fast procedure for the detection of defects in Toll-like receptor signaling. Pediatrics. 2006; 118:2498–2503. [DOI] [PubMed] [Google Scholar]
- 24.Stray-Pedersen A, Sorte HS, Samarakoon P, et al. Primary immunodeficiency diseases: genomic approaches delineate heterogeneous Mendelian disorders. J Allergy Clin Immunol. 2017; 139:232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platt CD, Zaman F, Bainter W, et al. Efficacy and economics of targeted panel versus whole-exome sequencing in 878 patients with suspected primary immunodeficiency. J Allergy Clin Immunol. 2021; 147:723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoddard JL, Niemela JE, Fleisher TA, et al. Targeted NGS: a cost-effective approach to molecular diagnosis of PIDs. Front Immunol. 2014; 5:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chinn IK, Eckstein OS, Peckham-Gregory EC, et al. Genetic and mechanistic diversity in pediatric hemophagocytic lymphohistiocytosis. Blood. 2018; 132:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belkadi A, Bolze A, Itan Y, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015; 112:5473–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H, Zhang VW, Stray-Pedersen A, et al. Rapid molecular diagnostics of severe primary immunodeficiency determined by using targeted next-generation sequencing. J Allergy Clin Immunol. 2016; 138:1142–1151.e2. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh S, Krux F, Binder V, et al. Array-based sequence capture and next-generation sequencing for the identification of primary immunodeficiencies. Scand J Immunol. 2012; 75:350–354. [DOI] [PubMed] [Google Scholar]