Abstract
The molecular mechanisms governing early cardiogenesis are still largely unknown. Interestingly, the retinoblastoma protein (Rb), a regulator of cell cycle, has recently emerged as a new candidate regulating cell differentiation. Rb−/− mice die at midgestation and mice lacking E2f1/E2f3, downstream components of the Rb-dependent transcriptional pathway, die of heart failure. To gain insight into the function of Rb pathway in early cardiogenesis, we used Rb−/− embryonic stem (ES) cells differentiating into cardiomyocytes. Rb−/− cells displayed a dramatic delay in expression of cardiac-specific transcription factors and in turn in the whole process of cardiac differentiation. The phenotype of Rb−/− ES cell-derived cardiomyocytes was rescued by reintroducing Rb in cardiac progenitors, by stimulating the BMP-dependent cardiogenic pathway or by overexpression of Nkx2.5. ES cells deficient in the recently identified factor LEK1, a murine homolog of the cardiomyogenic factor 1, or specific disruption of Rb–LEK1 interaction into the nucleus of differentiating ES cells recapitulated the delay in cardiac differentiation of Rb−/− ES cells. Thus, we provide evidence for a novel Rb/LEK1-dependent and BMP-independent transcriptional program, which plays a pivotal role in priming ES cells toward a cardiac fate.
Keywords: cardiogenesis, cell differentiation, stem cell
Introduction
The retinoblastoma protein (Rb) is the archetype for the family of pocket proteins comprising Rb, p107 and p130. Inactivation of Rb in many cancer types has highlighted its role in cell cycle progression (Weinberg, 1995). In G1 phase of the cell cycle, hypophosphorylated Rb sequesters the transcription factors E2F bound to their DP partners. Phosphorylation of the pocket protein by cyclin D- and cyclin E-dependent kinases 4/6 releases E2F–DP complex allowing for transcriptional activity of E2F and progression of the cell through the S phase of the cell cycle (Sears and Nevins, 2002; Sherr and McCormick, 2002). The pivotal role of Rb in the G1/S transition of the cell cycle has been extensively investigated. Rb has also recently been involved in cell differentiation, senescence and apoptosis (Lipinski and Jacks, 1999). A function of Rb in cell lineage specification might also be surmised since the retinoblastoma gene family is expressed early during embryogenesis (Jiang et al, 1997). One of the earliest evidence for the role of Rb in the process of cell differentiation was evoked by the phenotype of Rb−/− mice generated by gene targeting (Clarke et al, 1992; Jacks et al, 1992; Lee et al, 1992). Embryos carrying inactivation of both Rb alleles die in midgestation because of several defects in neurogenesis, hematopoiesis and myogenesis potentially as a consequence of a disrupted placenta (Wu et al, 2003) while p107−/− or p130−/− mice are viable (Cobrinik et al, 1996; Lee et al, 1996). A key role of pocket proteins in mesenchymal lineage differentiation has further emerged soon after generation of the knockout mice. Indeed, cell studies revealed a crucial role of the family of pocket proteins in myogenesis, adipogenesis and chondrogenesis (Schneider et al, 1994; Chen et al, 1996; Cobrinik et al, 1996). In myoblasts, Rb works together with MyoD to induce the transcriptional activity of Mef2c. The protein is required for cell cycle arrest and in turn fusion of quiescent myoblasts (Novitch et al, 1996). In osteoclasts, Rb also interacts with the specific CBFA1 transcription factor to promote osteogenic differentiation (Thomas et al, 2003). There is thus a developing concept that Rb may act as a partner that cooperates with tissue-specific factors in a complex transcriptional network turned on during cell differentiation.
In contrast to skeletal muscle cells, the cardiomyocytes proliferate while differentiating during embryogenesis. Soon after birth, the cardiomyocytes withdraw from the cell cycle in the perinatal period and are locked in the G0/G1 phase of the cell cycle (McGill and Brooks, 1995; McBride and Nemer, 1998). Rb is expressed in blastocysts (Xie et al, 2005) and low level of Rb was still observed in embryonic hearts while its expression drops in neonates and adults (Kim et al, 1994). Although Rb was taken as the basis for irreversible cell cycle exit in postmitotic myotubes, whether this may apply to cardiac myocytes is still questionable (Kirshenbaum and Schneider, 1995), especially since p130 seems to play already such a function in the heart (LeCouter et al, 1998). The Rb−/− mice are not viable and thus did not allow to investigate specifically the role of Rb in cardiac cell differentiation. Although in Mox2+/Cre Rb−/loxP mice with a reconstituted placenta, no apparent histological cardiac defect was observed (de Bruin et al, 2003), a role of the pocket protein in the transcriptional program of cardiogenesis is still presumed, since disruption of downstream targets of Rb, E2F1 and E2F3 leads to embryonic congestive heart failure attributed to a myocardial defect (Cloud et al, 2002). Interestingly, disruption of E2F3 gene in Rb−/− mice dramatically rescued the phenotype of Rb−/− mice, which however die of heart failure due to a thinning of the ventricular walls (Ziebold et al, 2001) suggesting growth retardation. Furthermore, E2Fs are required in cardiac cells to undergo hypertrophy in response to growth factors, an event that partially recapitulates the genetic program of cardiogenesis (Vara et al, 2003).
Cardiac cell specification and differentiation occur under the control of the endoderm secreting morphogens belonging to the TGFβ superfamily. These comprise nodal, activin, TGFβ and bone morphogenetic proteins (BMPs). More specifically, a BMP-dependent pathway plays a major role in this process activating a transcriptional cascade including the factors Nkx2.5, Mef2c and GATA4 (Srivastava and Olson, 2000; Harvey, 2002; Olson and Schneider, 2003). A BMP-independent pathway has been more recently described and implicates an Rb binding cardiomyogenic factor (cardiomyogenic factor 1 (CMF1)) in avian heart (Pabon-Pena et al, 2000; Redkar et al, 2002). A member of the same gene family as CMF1, namely LEK1, was found in murine embryonic heart (Goodwin et al, 1999). Thereby, we hypothesized that an Rb-dependent pathway might play a role in the process of early cardiogenesis.
To overcome the embryonic lethality of Rb−/− mice and to address specifically the role of Rb in early stages of cardiac specification and differentiation, before cardiac morphogenesis, we used Rb−/− embryonic stem (ES) cells and differentiated them toward a cardiac lineage. We found a novel function of the cell cycle regulator Rb. We show that Rb by interacting with the recently identified factor LEK1 plays a pivotal role in determining stem cells toward a cardiac lineage in a BMP-independent manner.
Results
Beating activity of Rb−/− ES cell-derived cardiomyocytes is delayed
ES4Rb−/− ES cells divided in the serum- and leukemia inhibitory factor (LIF)-containing medium with the same rate as wild-type (WT) BS1 ES cells (data not shown). Using the hanging drop method (Meyer et al, 2000), ES cells aggregated within an embryoid body (EB), a three-dimensional cell structure comprising the three embryonic layers (i.e., endoderm, mesoderm and ectoderm). After 7 days, spontaneous beating areas appeared within the mesoderm revealing differentiation of ES cells in cardiac myocytes (Doetschman et al, 1985; Maltsev et al, 1994). At day 10, maximal beating activity was observed in WT EBs generated from BS1 ES cells, while only 20% of Rb−/− EBs generated from ES4 ES cells featured contractile areas. Maximal beating activity was only reached at day 15 in Rb−/− EBs (Figure 1A). The other pocket proteins did not compensate the loss of Rb function in ES4 ES cells although p107 or p130 was expressed to the same extent in WT as in Rb−/− EBs at differentiating days 5, 7 or 9 (Figure 1B).
Figure 1.
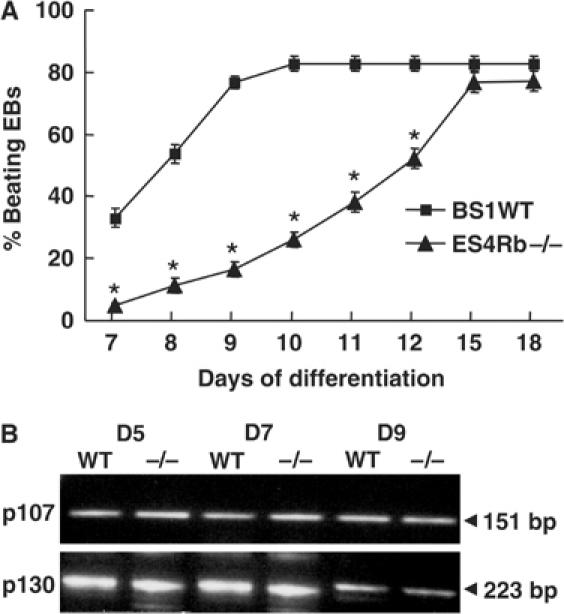
Impaired beating activity in Rb−/− ES cell-derived EBs. (A) Beating activity of EBs generated from BS1WT (▪) and ES4Rb−/− (▴) ES cells. An EB was considered as beating if at least three independent contractile areas were observed in the mesoderm (median layer of the EB) (n=10 experiments, 100 EBs counted/experiment). (B) p107 and p130 expression in BS1WT (WT) and ES4Rb−/− (−/−) EBs at days 5, 7 and 9. The gel is representative of three RT–PCR experiments. *Significantly different from WT (P⩽0.01).
Cardiac differentiation of Rb−/− ES cells is delayed
We next asked whether the delayed appearance of beating activity of Rb−/− EBs at early stages of cardiac differentiation was due to a delay in activation of the cardiac transcriptional network including Nkx2.5 and Mef2c governing the fate of cardiac progenitors.
Real-time quantitative PCR revealed that expression of the earliest cardiac-specific transcription factor Nkx2.5 was two-fold decreased in ES4Rb−/− EBs at days 5 and 7 when compared to BS1 WT EBs (Figure 2A). Expression of Mef2c, another cardiac marker, was even more dramatically decreased (three-fold) at days 5 and 7 in ES4Rb−/− EBs (Figure 2B). At day 9, both Nkx2.5 and Mef2c mRNA levels in ES4Rb−/− EBs were close to the ones measured in BS1 WT EBs (Figure 2A and B). RT–PCR showed that MLC2v expression was also dramatically impaired in Rb−/− EBs (data not shown).
Figure 2.
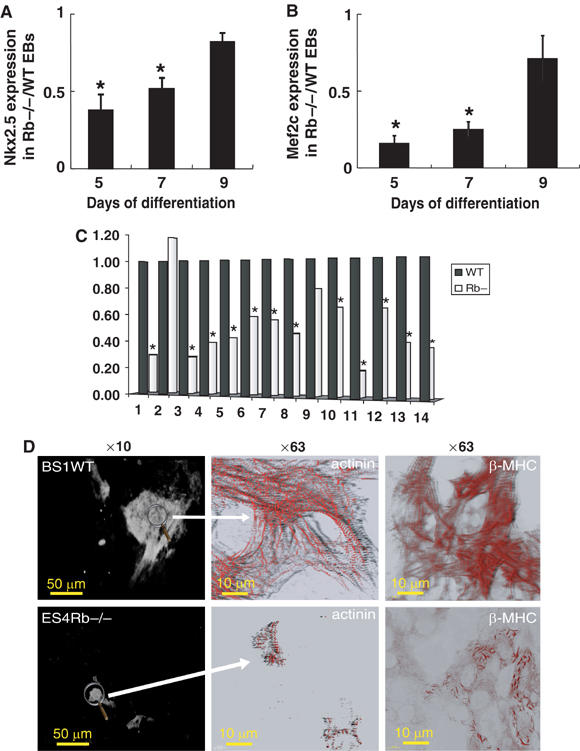
Cardiac differentiation is delayed in Rb−/− EBs. Expression of early cardiac markers in BS1WT or ES4Rb−/− EBs was measured by real-time quantitative PCR. Results are expressed as the ratio of Nkx2.5 (A) and Mef2c (B) expression in Rb−/− to WT EBs at days 5, 7 and 9 of differentiation. Results are normalized to β-tubulin expression. (C) Gene expression in Rb−/− compared to WT EBs at day 2. Results are normalized to tubulin expression (n=2 separate experiments). 1, Brachyury; 2, SRF; 3, Mesp1; 4, Mesp2; 5, myocardin; 6, Tbx6; 7, Tbx5; 8, Tbx2; 9, Isl1; 10, Mef2c, 11, Nkx2.5; 12, GATA6; 13, GATA4; 14, α-actin. *Levels of expression of genes are significantly different in Rb−/− EBs than in WT EBs by paired t-test (P⩽0.05). (D) Immunostaining of day 10 BS1WT (upper panels) and ES4Rb−/− (lower panels) EBs with an anti-sarcomeric actinin or β-MHC antibody. Images were acquired at × 10 (left panels) and × 63 (right panels) magnification. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01).
We further investigated in early EBs (day 2) generated from WT or Rb −/− EBs, expression of a panel of mesodermal and cardiac-specific genes involved in early cardiogenesis. Real-time quantitative PCR showed that expressions of brachyury, Mesp1 and 2, myocardin, Tbx6, Tbx5, Tbx2, Mef2c, Nkx2.5 GATA4 and 6 and α-actin were downregulated while expression of SRF and Isl1 (Cai et al, 2003) remained unchanged in Rb−/− EBs when compared to WT EBs (Figure 2C). Expression of some of the downregulated genes was still low at day 5 in Rb−/− EBs and slowly recovered to a normal level at day 8 (Supplementary Figure 1).
Visualization of cardiomyocytes within EBs using immunolabelling of sarcomeric α-actinin showed much less cardiomyocytes in day 10 ES4Rb−/− EBs than in WT BS1 EBs (Figure 2D, left panels). Observation of the sarcomeric network labelled with either an anti-α-actinin or anti-β-MHC antibody at high magnification further revealed the presence of cytosolic myosin fibrils and an incomplete organization of the sarcomeric units in Rb−/− EBs, suggesting an early stage of cardiac cell differentiation (Figure 2D, central and right panels).
Targeted expression of Rb in ES cell-derived cardiac progenitors rescued cardiac cell differentiation of Rb−/− ES cells
To investigate whether Rb played a role restricted to the cardiac lineage, we reintroduced Rb in the ES cell-derived cardiac progenitors using the Nkx2.5 promoter (Reecy et al, 1999) to drive expression of the Rb cDNA. A stable Rb−/− ES cell line was generated using this construct and allowed to differentiate within EBs. The kinetic of beating activity of EBs with cardiac-restricted expression of Rb (Figure 3A) was significantly improved when compared to parental Rb−/− EBs (Figure 3A) and not different from WT EBs (Figure 1). In support of these data, the time course of expression of both Nkx2.5 and Mef2c in the EBs was indistinguishable from that of WT cells (Figure 3B and C). Furthermore, numerous cardiomyocytes were observed in the mesodermal area of EBs with cardiac-restricted expression of Rb. Microscopic observation at high magnification of the actinin-stained cardiomyocytes showed well-formed sarcomeric units in aligned myofibrils (Figure 3D).
Figure 3.
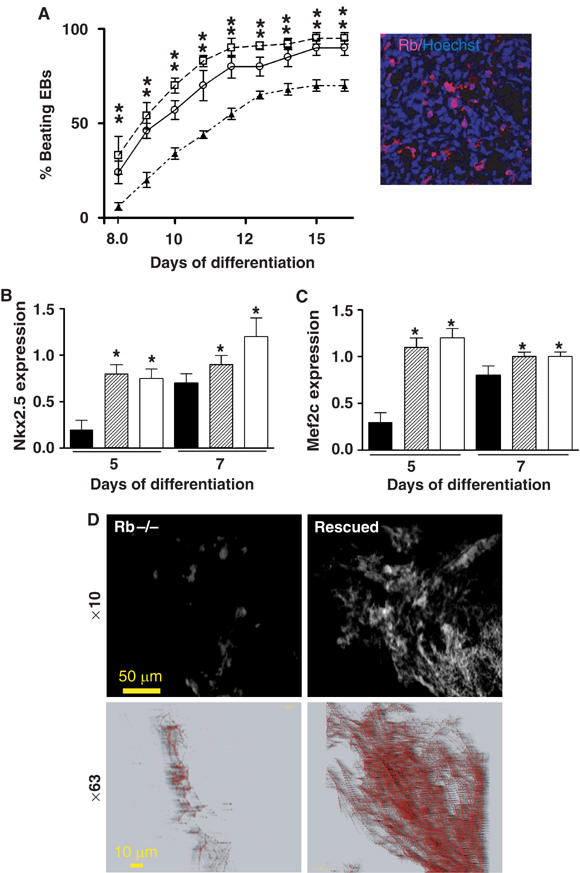
Targeted expression of Rb in cardiac progenitors rescues cardiac differentiation of Rb−/− ES cells. (A) Beating activity of EBs generated from ES4Rb−/− cells (▴) or ES4Rb−/− cells expressing Rb under the transcriptional control of the Nkx2.5 promoter (clones 7 ○ and 9 □). The inset shows expression of Rb in the mesodermal area of an EB generated from the rescued clone 7. Nuclei were visualized by Hoechst staining. Expression of early cardiac markers in ES4Rb−/− EBs (▪) or Rb rescued EBs (clones 7  and 9 □) was measured by real-time quantitative PCR. Results are expressed as the ratio of Nkx2.5 (B) and Mef2c (C) expression in Rb−/− or rescued to WT EBs at days 5 and 7 of differentiation. Results are normalized to β-tubulin expression. (D) Immunostaining of day 10 ES4Rb−/− or Rb-rescued EBs with an anti-sarcomeric actinin antibody. Images were acquired at × 10 (top panels) and × 63 (bottom panels) magnification. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01).
and 9 □) was measured by real-time quantitative PCR. Results are expressed as the ratio of Nkx2.5 (B) and Mef2c (C) expression in Rb−/− or rescued to WT EBs at days 5 and 7 of differentiation. Results are normalized to β-tubulin expression. (D) Immunostaining of day 10 ES4Rb−/− or Rb-rescued EBs with an anti-sarcomeric actinin antibody. Images were acquired at × 10 (top panels) and × 63 (bottom panels) magnification. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01).
Activation of the cardiogenic BMP-dependent pathway rescues cardiac cell differentiation of Rb−/− ES cells
Cardiac specification results from activation of a major BMP-dependent transcriptional pathway including expression of both Nkx2.5 and Mef2c (Srivastava and Olson, 2000; Olson and Schneider, 2003). In an attempt to rescue the genetic program of cardiac cell differentiation in Rb−/− EBs, ES4 ES cells were pretreated with BMP2 to induce expression of Nkx2.5 and Mef2c, a treatment that commits stem cells toward a cardiac lineage (Behfar et al, 2002). Stimulation of ES4Rb−/− ES cells with BMP2 (2.5 ng/ml) for 24 h prior to differentiation restored the beating activity of EBs at early days 7–8 and the percentage of contractile ES4 EBs at day 9 was maximal and indistinguishable from that of WT BS1 EBs (Figure 4A). Nkx2.5 and Mef2c genes were expressed to the same extent in days 5 and 9 Rb−/− EBs generated from ES4Rb−/− cells pretreated with BMP2 as in WT EBs pretreated with BMP2 (Figure 4B and C). Myofibrillogenesis was restored in day 10 Rb−/− EBs generated from ES4 cells pretreated with BMP2 (Figure 4D). Another member of the TGFβ superfamily, namely TGFβ, applied for 24 h to Rb−/− ES cells prior to cell differentiation restored as early as day 2 expression of mesodermal and cardiac genes as well as myofibrillogenesis and in turn beating activity of EBs (Supplementary Figure 2).
Figure 4.
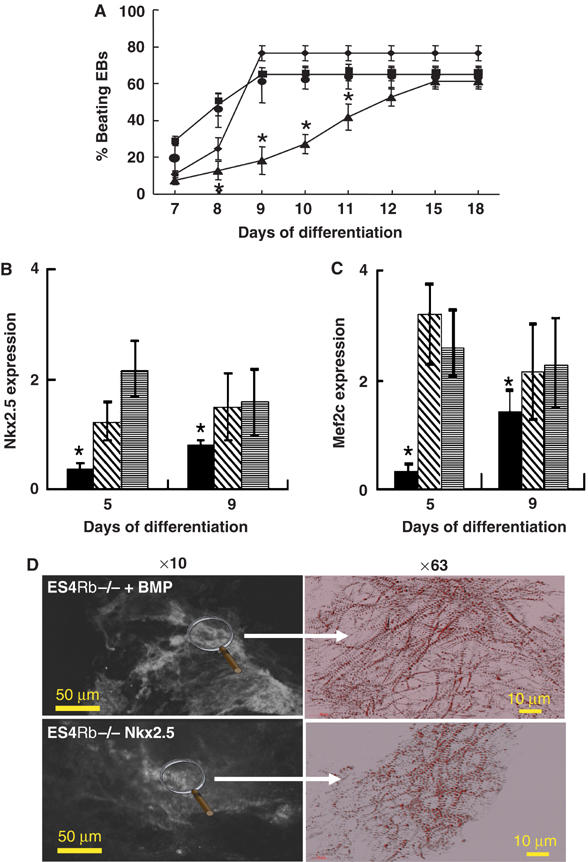
Stimulation of the BMP-dependent cardiogenic pathway rescues cardiac differentiation of ES4Rb−/− ES cells. (A) Beating activity of EBs generated from BS1WT (▪), ES4Rb−/−(▴), BMP-treated ES4Rb−/− ES cells (•) and ES4Rb−/− cells overexpressing Nkx2.5 under control of the α-actin promoter (⧫). Expression of Nkx2.5 (B) and Mef2c (C) in EBs generated from ES4Rb−/− (▪), BMP2-treated ES4Rb−/− cells ( ) and ES4Rb−/− ES cells overexpressing Nkx2.5 under control of α-actin promoter (
) and ES4Rb−/− ES cells overexpressing Nkx2.5 under control of α-actin promoter ( ). Results are expressed as the ratio of gene expression in BMP2-treated ES4Rb−/− or genetically modified Nkx2.5-overexpressing ES4Rb−/− EBs to BS1WT EBs treated with BMP2 or BS1WT EBs overexpressing Nkx2.5, respectively, and normalized to β-tubulin expression. (D) Immunostaining of day 10 Rb−/− EBs generated from BMP-treated ES4Rb−/− ES cells (+BMP) or from ES4Rb−/− cells overexpressing Nkx2.5 under control of α-actin promoter (+Nkx2.5) with an anti-sarcomeric α-actinin antibody. The figure is representative of at least three separate experiments. *Significantly different from WT (P⩽0.01).
). Results are expressed as the ratio of gene expression in BMP2-treated ES4Rb−/− or genetically modified Nkx2.5-overexpressing ES4Rb−/− EBs to BS1WT EBs treated with BMP2 or BS1WT EBs overexpressing Nkx2.5, respectively, and normalized to β-tubulin expression. (D) Immunostaining of day 10 Rb−/− EBs generated from BMP-treated ES4Rb−/− ES cells (+BMP) or from ES4Rb−/− cells overexpressing Nkx2.5 under control of α-actin promoter (+Nkx2.5) with an anti-sarcomeric α-actinin antibody. The figure is representative of at least three separate experiments. *Significantly different from WT (P⩽0.01).
To further delineate the BMP-dependent rescue pathway, we genetically modified both WT BS1 and ES4Rb−/− ES cells to overexpress Nkx2.5 under the control of the α-actin promoter, which allows expression of the transcriptional factor as early as days 3–5. Beating activity of Rb−/− EBs overexpressing Nkx2.5 was significantly improved over that of Rb−/− EBs at day 7 and reached the same percentage as the WT EBs at day 9 (Figure 4A). In BS1 ES cell-derived EBs overexpressing Nkx2.5, beating activity did not occur earlier than in WT Ebs, suggesting that expression of the endogenous factor is not limiting. Expression of Nkx2.5 gene was two times increased in day 5 Rb−/− EBs when compared to WT EBs (data not shown), thus confirming overexpression of the transcription factor (Figure 4B). Expression of Mef2c was comparable in Rb−/− EBs overexpressing Nkx2.5 and in parental BS1 ES cell-derived EBs overexpressing Nkx2.5 (Figure 4C). In line with a restored cardiac transcriptional program, α-actinin immunostaining showed a normal myofibrillogenesis in Rb−/− EBs overexpressing Nkx2.5 (Figure 4D).
Knockdown of LEK1 mimics Rb−/− phenotype
A Rb binding cardiomyogenic factor was recently discovered in avian heart (Pabon-Pena et al, 2000; Redkar et al, 2002) and a member of the same gene family, namely LEK1, was found in murine embryonic heart (Goodwin et al, 1999). LEK1 was expressed as early as in undifferentiated ES cells and its level of expression did not significantly change in the course of cardiac differentiation. LEK1 was still expressed in proliferating myoblasts but no longer in postnatal murine heart (Figure 5A). Western blot analysis further showed that LEK1 protein migrating with an apparent molecular weight of 310 kDa was expressed at early stages of cardiogenesis (heart at E9) (Goodwin et al, 1999) as well as in EBs (day 5) (Figure 5B).
Figure 5.
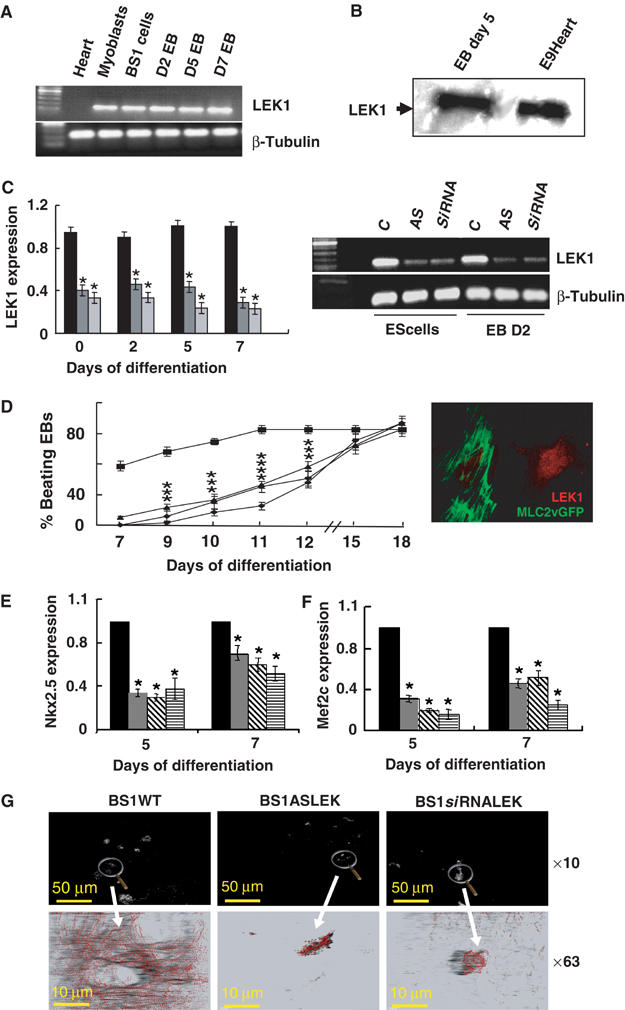
Knockdown of LEK1 in differentiating ES cells mimics Rb−/− phenotype. (A) RT–PCR analysis of expression of LEK1 in undifferentiated BS1WT, D2, D5 and D7 EBs, adult heart (heart) and in C2C12 myoblastic cells (myoblasts). (B) Western blot of LEK1 from protein extracted from EB day 5 and heart from E9 embryos (E9Heart). (C) Expression of LEK1 in BS1WT, BS1ASLEK (AS) and BS1siRNALEK (siRNA) ES cells and D2 EBs and quantification by semiquantitative PCR expressed as the ratio of expression in BS1 (▪), BS1ASLEK ( ) or BS1siRNALEK (
) or BS1siRNALEK ( ) to WT BS1 ES cells and D2, D5 or D7 EBs normalized to β-tubulin expression. The inset shows cells cotransfected with siRNALEK and MLC2vGFP cDNA and immunostained 72 h later with the anti-LEK1 antibody. *Significantly different from WT (P⩽0.01). (D) Beating activity of EBs generated from WT BS1 (▪), BS1ASLEK1 (⧫), BS1siRNALEK1 (•) and ES4Rb−/− cells (▴). Expression of Nkx2.5 (E) and Mef2c (F) in EBs generated from BS1WT (▪), BS1ASLEK1 (
) to WT BS1 ES cells and D2, D5 or D7 EBs normalized to β-tubulin expression. The inset shows cells cotransfected with siRNALEK and MLC2vGFP cDNA and immunostained 72 h later with the anti-LEK1 antibody. *Significantly different from WT (P⩽0.01). (D) Beating activity of EBs generated from WT BS1 (▪), BS1ASLEK1 (⧫), BS1siRNALEK1 (•) and ES4Rb−/− cells (▴). Expression of Nkx2.5 (E) and Mef2c (F) in EBs generated from BS1WT (▪), BS1ASLEK1 ( ), BS1siRNALEK1 (
), BS1siRNALEK1 ( ) and ES4Rb−/− (
) and ES4Rb−/− ( ) ES cells, measured by real-time quantitative PCR. Results are expressed as the ratio of gene expression in BS1ASLEK, BS1siRNALEK1 or ES4Rb−/− to WT BS1 at days 5 and 7 of differentiation and were normalized to β-tubulin expression. (G) Immunostaining of day 10 BS1WT, BS1ASLEK1 and BS1siRNALEK1 EBs with an anti-sarcomeric actinin antibody. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01).
) ES cells, measured by real-time quantitative PCR. Results are expressed as the ratio of gene expression in BS1ASLEK, BS1siRNALEK1 or ES4Rb−/− to WT BS1 at days 5 and 7 of differentiation and were normalized to β-tubulin expression. (G) Immunostaining of day 10 BS1WT, BS1ASLEK1 and BS1siRNALEK1 EBs with an anti-sarcomeric actinin antibody. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01).
To address the question of whether the loss of LEK1 could mimic the phenotype of EBs lacking Rb, we knocked down expression of the gene using two separate approaches. First, a 200 bp LEK1-specific PCR fragment was amplified from total RNA extracted from BS1 ES cells and subcloned in the antisense orientation in a pcDNA expression vector. Second, a specific LEK1 oligonucleotide was selected to design an siRNA. The siRNA was subcloned in the pSuper expression vector. Three BS1 stable cell lines were then generated to express the LEK1 antisense or the LEK1 siRNA. Cells were allowed to differentiate within EBs. Semiquantitative RT–PCR revealed that in ES cells or in differentiating EBs expressing the antisense or the siRNA, LEK1 mRNA was significantly decreased when compared to WT BS1 ES cells or EBs. On average, only 30–40% of LEK1 mRNA remained in ES cells or EBs at days 2, 5 and 7 expressing the antisense or the siRNA (Figure 5C). This resulted in a low expression of a protein, barely detectable in cells expressing the antisense or the siRNA while LEK1 was mainly located in the nucleus of the neighbor nontransfected cell (Figure 5C, inset). Beating activity of EBs expressing either the antisense or the siRNA was severely compromised. The time course of appearance of contractile activity tightly followed the one observed in Rb−/− EBs (Figure 5D). Expression of both Nkx2.5 and Mef2C was also dramatically reduced in days 5 and 7 EBs in which LEK1 was knocked down (Figure 5E and F). The number of cardiomyocytes within these EBs was much less than in WT EBs, and anti-α-actinin immunostaining showed an incomplete sarcomerogenesis in EBs expressing LEK1 antisense or siRNA (Figure 5G).
LEK1–Rb interaction is specifically required for early cardiogenesis
We next hypothesized that a structural interaction of Rb with LEK1 was required to drive ES cells along the cardiogenic pathway. To disrupt such an interaction, we designed a DNA vector including a short sequence to express the atypical Rb binding domain of LEK1 fused to GFP, and a nuclear localization sequence (NLS) to target the domain into the nucleus. Expression of this short peptide fused to GFP within the nucleus of cells should act as a dominant-negative protein and prevent Rb from binding to LEK1. Indeed, expression of the peptide in C2C12 myoblasts, which express both Rb and LEK1 (Ashe et al, 2003), significantly decreased the interaction of Rb with LEK1, as shown by co-immunoprecipitation experiments (Figure 6A). Next, we investigated whether LEK1Rb binding domain-GFP affected Rb function in regulation of cell cycle. The cDNA encoding the LEK1Rb binding domain-GFP was transfected in proliferating embryonic cardiomyocytes. The proliferative activity of LEK1Rb binding domain-transfected cardiomyocytes assessed by Ki67 immunostaining was not different from the control (i.e., mock cells or NLSYFP-transfected cells) (Figure 6B and C). Similarly, LEK1Rb domain-GFP did not affect the proliferative activity of nondifferentiated cells, the NIH3T3 cells (Figure 6C). In contrast, expression of LEK siRNA dramatically decreased the number of NIH3T3 undergoing G1/S transition, as indicated by histone H3 labelling (Supplementary Figure 3).
Figure 6.
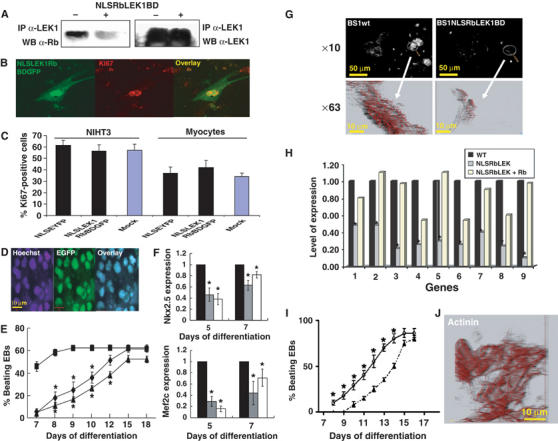
Disruption of LEK1–Rb interaction in differentiating ES cells mimics Rb−/− phenotype and does not affect cell proliferation. (A) LEK1 was immunoprecipitated from a whole lysate of C2C12 mock cells or cells transfected with NLSRbLEK1BD cDNA. The immunoprecipitated complex was subjected to 5% SDS–PAGE and Western blot analysis performed using an anti-Rb antibody and then anti-LEK1 antibody. A LEK1Rb-GFP binding domain (NLSRbLEK1BD) was transfected in E9 proliferating embryonic cardiomyocytes (B) or NHT3 cells (C). After 2 days, cells were immunostained with an anti-Ki67 antibody and a secondary alexa540-conjugated antibody. LEK1Rb-GFP binding domain was expressed and targeted in the nuclei of differentiating ES cells to disrupt LEK1–Rb interaction. (D) Intranuclear localization of LEK1Rb-GFP binding domain in EBs day 10 with nuclear DNA stained by Hoechst. Images (Hoechst), EGFP and overlaid images were simultaneously acquired by confocal microscopy. (E) Beating activity of EBs generated from WT BS1 (▪), BS1ES cells expressing the LEK1Rb-GFP binding domain (⧫) and Rb−/− ES4 (▴) ES cells. (F) Expression of Nkx2.5 and Mef2c in EBs generated from BS1WT ES cells (▪), BS1ES cells expressing the LEK1Rb-GFP binding domain ( ) and Rb−/− ES4 (□) ES cells measured by real-time quantitative PCR. Results are expressed as the ratio of gene expression in BS1WT ES cells, GFP-LEK1Rb binding domain-expressing ES cells and Rb−/− ES4 ES cells to WT BS1 ES cells, and were normalized to β-tubulin expression. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01). (G) Immunofluorescence of anti-sarcomeric actinin of EBs day 10 generated from WT BS1 and of BS1 expressing NLSRbLEK1GFP binding domain. (H) Quantitative PCR of 1, brachyury; 2, Mesp1; 3, Mesp2; 4, myocardin; 5, Tbx6; 6, Mef2c; 7, Nkx2.5; 8, GATA4; 9, GATA6 mRNA extracted from EBs day 5 generated from WT BS1, and from BS1 expressing NLSRbLEK1GFP binding domain and overexpressing or not Rb (n=2). *Levels of expression of genes are significantly different in EBs expressing NLSRbLEK1GFP binding domain when compared to those in WT EBs (WT) (paired t-test, P⩽0.05). (I) Beating activity of EBs generated from BS1 expressing NLSRbLEK1GFP binding domain overexpressing (○) or not (▴) Rb (average of at least 50 EBs; *significantly different P⩽0.05). (J) Immunofluorescence of sarcomeric actinin in EBs generated from BS1 expressing NLSRbLEK1GFP binding domain and overexpressing Rb.
) and Rb−/− ES4 (□) ES cells measured by real-time quantitative PCR. Results are expressed as the ratio of gene expression in BS1WT ES cells, GFP-LEK1Rb binding domain-expressing ES cells and Rb−/− ES4 ES cells to WT BS1 ES cells, and were normalized to β-tubulin expression. The figure is representative of three separate experiments. *Significantly different from WT (P⩽0.01). (G) Immunofluorescence of anti-sarcomeric actinin of EBs day 10 generated from WT BS1 and of BS1 expressing NLSRbLEK1GFP binding domain. (H) Quantitative PCR of 1, brachyury; 2, Mesp1; 3, Mesp2; 4, myocardin; 5, Tbx6; 6, Mef2c; 7, Nkx2.5; 8, GATA4; 9, GATA6 mRNA extracted from EBs day 5 generated from WT BS1, and from BS1 expressing NLSRbLEK1GFP binding domain and overexpressing or not Rb (n=2). *Levels of expression of genes are significantly different in EBs expressing NLSRbLEK1GFP binding domain when compared to those in WT EBs (WT) (paired t-test, P⩽0.05). (I) Beating activity of EBs generated from BS1 expressing NLSRbLEK1GFP binding domain overexpressing (○) or not (▴) Rb (average of at least 50 EBs; *significantly different P⩽0.05). (J) Immunofluorescence of sarcomeric actinin in EBs generated from BS1 expressing NLSRbLEK1GFP binding domain and overexpressing Rb.
In differentiating EBs, the LEK1Rb domain-GFP was targeted into the nucleus, as revealed by a colocalization of GFP fluorescence with the one of nuclear DNA stained by Hoechst (Figure 6D). The time course of beating activity of EBs expressing the LEK1Rb domain-GFP tightly followed that of Rb−/− EBs and was thus significantly slower than that of WT EBs (Figure 6E). Expression of Nkx2.5 and Mef2c was also impaired in days 5 and 7 EBs and was similar to that measured in Rb−/− EBs (Figure 6F). As a functional consequence, EBs featured smaller areas stained by the anti-α-sarcomeric actinin antibody at day 10 and observation of these later at high magnification showed incomplete myofibrillogenesis (Figure 6G).
To further check the specificity of LEK1Rb domain-GFP peptide, the ES cell line expressing the LEK1Rb domain-GFP was transfected with Rb cDNA to overexpress the pocket protein in order to displace the interaction of the peptide with Rb. Cells were then allowed to differentiate within EBs. Figure 6H reveals that expression of mesodermal and cardiac-specific genes (i.e., brachyury, Mesp1, Mesp2, myocardin, Tbx6, Mef2c, Nkx2.5 GATA4, GATA6) impaired in EBs day 5 generated from the LEK1Rb domain-GFP cells as found in Rb−/− EBs was partially or fully restored after overexpression of Rb. This was translated into early appearance of beating activity of EBs when Rb was overexpressed in cells expressing the LEK1Rb domain-GFP (Figure 6I), as expected from the presence of cardiomyocytes with well-developed contractile apparatus in these EBs stained with an anti-actinin antibody (Figure 6J).
In contrast, p107 and/or p130 expression in LEK1Rb domain-GFP ES cells did not restore expression of genes or beating activity of LEK1Rb domain-GFP-expressing ES cell-derived cardiomyocytes (data not shown).
Discussion
The role of Rb in cell cycle G1/S progression has been extensively studied. A potential participation of the protein in cell differentiation is emerging. Several observations, arising from Rb/E2f3- (Ziebold et al, 2001) or E2f1/E2f3-deficient mice (Cloud et al, 2002) featuring cardiac growth retardation, pointed to a role of Rb in cardiogenesis. However, the molecular mechanisms underlying this cardiac defect have not been elucidated. We hypothesized that myocardial growth retardation might result from an impaired specification or differentiation of stem cells or mesodermal progenitors.
To gain insight into the potential role of Rb in early stages of cardiogenesis, we used ES cells differentiating toward all cardiac cell types (Maltsev et al, 1994), a model that faithfully recapitulates the first week of embryogenesis (Leahy et al, 1999). We have found that the cell cycle regulator Rb is a key molecular component in cardiac cell differentiation in the first stages of the process but is dispensable at later stages. We have also engineered ES cell clones knocked down for a key partner of Rb, namely LEK1, or in which we disrupted Rb–LEK1 interaction. These approaches provide direct evidence for a participation of this cardiogenic factor also in the early steps of the cardiac differentiation process. Thus, the presence of both Rb and LEK1 in mesodermal cardiac progenitors allows them to function in concert and to regulate the early cardiac transcriptional program of differentiation.
Cardioblasts undergo the process of differentiation while still proliferating. This thus excludes a central role of Rb in cell cycle exit in that model. The recent finding that Rb binds CMF1 (Redkar et al, 2002), a cardiomyogenic factor (Wei et al, 1996; Pabon-Pena et al, 2000), rather suggests its participation in cardiogenesis, extending its function beyond its ability to regulate cell cycle progression. Indeed, ES cells lacking Rb feature a dramatic delay in their process of differentiation that was not compensated by the other pocket proteins p107 and p130, the expression of which was not affected in Rb−/− cells. Expression of the earliest cardiac transcription factor Nkx2.5 is severely compromised in the first days of the differentiation process. Similarly, the downstream component of the transcriptional cascade, Mef2c, and one of its direct target gene MLC2v (data not shown) as well as GATA4,6 or earlier cardiogenic markers Mesp1, Mesp2 and Tbxs are poorly expressed in Rb−/− EBs. The mesodermal gene brachyury was also downregulated in Rb−/− EBs, suggesting a partial impairment in mesodermal commitment of ES cells. The first striking consequence of the deficiency in the cardiac genetic program is the severe delay in the contractile activity of EBs. This may be attributed to a decrease in the number of cardioblasts within the EBs and/or an impaired cell differentiation including myofibrillogenesis, as visualized by actinin immunostaining of sarcomeric units in EBs (Figure 2D). The phenotype of Rb−/− cardiac progenitors is fully rescued by reintroducing Rb in cardiac progenitors. This strongly argues against the requirement of Rb in endodermal or other mesodermal cell lineage to induce the cardiac lineage. Rather, the function of Rb is specifically required in cardiac precursors to timely differentiate toward a cardiac phenotype. Thus, in contrast to the scenario observed in skeletal muscle (Novitch et al, 1996; Huh et al, 2004), the lack of Rb in cardioblasts does not lead to cell hyperproliferation. Rather, it affects the specification of cardiac myocytes.
To further address this question, we took advantage of the BMP-dependent canonical cardiogenic pathway (Srivastava and Olson, 2000; Harvey, 2002) still functional in Rb−/− ES cells. This pathway is a key pathway since treatment of EBs by the BMP antagonist noggin does not delay but severely disrupts cardiac differentiation resulting from a complete loss in Nkx2.5 expression (data not shown) as it does in vivo (Schlange et al, 2000). Direct stimulation of this pathway by BMP or TGFβ, or restoration of the level of Nkx2.5 by overexpression of the factor in EBs, rescues the phenotype of Rb−/− cardiomyocytes. This finding further points to a requirement of Rb in the process of cardiac cell specification and differentiation rather than in cardioblast proliferation. Altogether, these data strongly suggest that the BMP pathway is not deficient in these cells and that Rb is a component of a BMP-independent cardiogenic road. This also implies that both the BMP-dependent and -independent pathways work in concert to determine the cardiac lineage.
Rb binds the Id protein 2, an inhibitor of differentiation, and loss of Id2 rescues part of the Rb−/− phenotype (Lasorella et al, 2000, 2001). Since Id proteins (Id1, Id2 and Id3) are targets of the BMP-dependent pathway including in ES cells (Hollnagel et al, 1999; Kowanetz et al, 2004), they could in principle play a role in the rescue of Rb−/− cardiomyocytes in BMP2-treated EBs. However, the role of Id in cardiac cell differentiation is unclear (Evans et al, 1993) and loss of Rb is expected to relieve its block on Id2. Although Id may favor the proliferation of BMP2-induced Rb−/− cardiac progenitors, it is expected to prevent their differentiation (Kowanetz et al, 2004), which is not the case of rescued cardiomyocytes, which fully differentiated (Figure 4). Thus, Id proteins are unlikely to play a role in the Rb-dependent cardiac specification and differentiation of ES cells.
A BMP-independent pathway has been first described in avians (Wei et al, 1996). This transcriptional pathway now described in mouse involves CMF1, a member of an emerging family of developmental genes including LEK1. CMF1 and LEK1 have the ability to bind Rb (Redkar et al, 2002; Ashe et al, 2003). We knocked down expression of LEK1 in ES cell clones using an antisense or an siRNA directed to a very specific LEK1 domain. Both approaches were successful and significantly decreased LEK1 expression in stable ES cell clones and differentiated myogenic cells in which LEK1 is also expressed. LEK1 knockdown ES cells feature the same delay in cardiac cell differentiation as that observed in Rb−/− cells. All characteristics of the phenotype (i.e., decreased expression of Nkx2.5, Mef2c and MLC2v, misassembly of sarcomeric units and delay in beating activity) are shared by both Rb−/− and LEK1 knockdown differentiating cardiomyocytes. Furthermore, inhibition of Rb–LEK1 binding by expression of the peptide RbLEK1 binding domain recapitulated the phenotype of Rb−/− ES cell-derived cardiac progenitors and cardiomyocytes characterized by downregulation of a broad panel of mesodermal and cardiac genes and disruption of myofibrils. Although it is difficult to ascertain that no difference exists in both phenotypes, it is unlikely to occur at the level of gene expression.
The LEK knockout phenotype may be partly attributed to a delay or arrest in proliferation of cardiac cell progenitors since LEK1 siRNA (our data) or morpholino (Ashe et al, 2003) slows down mitotic activity of NIHT3 cells and C2C12 myoblastic cell line. However, overexpression and nuclear targeting of the Rb binding domain of LEK1 in ES cell-derived cardiomyocytes fully mimics the phenotype of Rb−/− or LEK1 knockdown while not affecting the proliferation of embryonic cardiomyocytes or NIH3T3 cells. This demonstrates that interaction of Rb with LEK1 is not necessary for cell proliferation but is required to secure a timely differentiation of cardiomyocytes including a complete sarcomerogenesis, although it does not exclude an Rb-independent role of LEK1 in proliferation of cardiac progenitors. Binding of LEK1 to Rb was suggested to retain cells in an active proliferative state preventing differentiation during development (Ashe et al, 2003). This might explain that LEK1 as well as Rb depletion prevents skeletal muscle terminal differentiation (Ashe et al, 2003). However, this mechanism cannot account for the delay in cardiac differentiation since contrary to myoblasts, which differentiate after exit of the cell cycle, proliferation and differentiation occur together and are not mutually exclusive in cardioblasts. Rather, Rb–LEK1 interaction participates in a BMP-independent cardiogenic pathway. The striking delay but not a block in cardiac differentiation of Rb−/− and LEK1 knockdown ES cells, or in cells in which Rb–LEK1 interaction was disrupted, rather places Rb–LEK1 in an early cardiogenic transcriptional program upstream of Nkx2.5 and which adds to the BMP pathway. How Rb–LEK1 interaction primes stem cells toward a cardiac lineage requires further investigation. Nonetheless, this study shows that such a protein–protein interaction is required at a very early stage, likely to specify cardiogenic mesodermal cells toward a definite cardiac fate (Figure 7). Our data suggest that the interaction of Rb with LEK might play a role as a complex of transcriptional coactivators. We speculate that Rb–LEK complex might recruit enzymes modifying chromatin and/or transcriptional coactivators (Taubert et al, 2004) to coordinate and to integrate multiple transcriptional networks.
Figure 7.
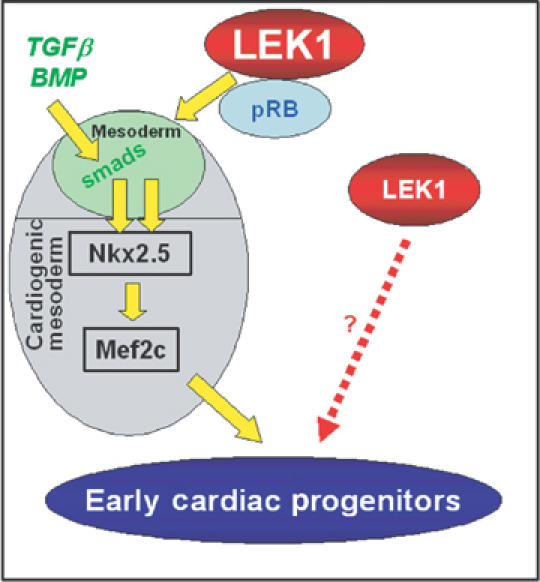
The Rb–LEK1 interaction plays a key role in early cardiogenesis: working model. Rb–LEK1 interaction is required to turn on a BMP-independent cardiac transcriptional pathway, which together with the BMP-dependent pathway leads to cardiac determination of ES cells. The differentiation factor responsible for stimulating the Rb–LEK pathway remains to be identified. This model does not exclude an Rb-independent role of LEK1 in proliferation of early cardioblasts.
Altogether, our data reveal that both Rb and LEK1 are required to prime stem cells toward a cardiac fate. Furthermore, our findings open new insight into the understanding of the molecular mechanisms of early cardiogenesis, which might be helpful to better comprehend the etiology of cardiac congenital diseases.
Materials and methods
Cell culture and differentiation of EBs
The ES cell lines BS1WT and ES4Rb−/− (Jacks et al, 1992) were used throughout this study. Cells were cultured on mouse embryonic fibroblasts (MEFs) treated for 3 h with 10 μg mitomycin C in DMEM and 10% fetal calf serum (FCS; Hyclone). The propagation medium was DMEM supplemented with sodium pyruvate, nonessential amino acids, L-glutamine, penicillin–streptomycin, β-mercaptoethanol (10−7 M), 7.5% FCS (Biomedia, France), and the LIF obtained from LIF-D cells (Meyer et al, 2000). ES cells were passed twice on gelatin-coated (0.1%) tissue culture dishes without MEFs, before starting the differentiation experiments. Differentiation was carried out in hanging drops of differentiation medium (20% FCS without LIF) in which EBs were formed. EBs were cultured in hanging drops for 2 days (D0–2) and subsequently in suspension for 3 days (D2–5) in bacterial Petri dishes (Falcon). The 5-day-old EBs were plated onto gelatin-coated (0.1% gelatin) tissue culture dishes for at least 13 days (D18). Where needed, ES cells were treated with BMP2 (2.5 ng/ml) 24 h before starting the differentiation experiments.
NIHT3 experiments
NIHT3 cells were cultured in DMEM with 10% FCS. Cells were transfected with the pSuper vector to express LEK1 siRNA, a vector encoding EYFP using the NLS nuclear targeting sequence or the pNLSEGFP-RbLEK1 binding domain vector using lipofectamine.
RT–PCR, quantitative PCR
Total RNA was prepared from ES cells and EBs using an RNA extraction kit (Zymo Research, CA, USA). After reverse transcription, 10 ng of cDNA was used for real-time quantitative PCR, performed with a light cycler and the SYBR green fast start kit (Roche, Germany). Primers used in real-time quantitative PCR were as follows:
β-tubulin, forward 5′-CCGGACAGTGTGGCAACCAGATCGG-3′ and
reverse 5′-TGGCCAAAAGGACCTGAGCGAACGG-3;
Nkx2.5, forward 5′-CATTTACCCGGGAGCCTACG-3′ and
reverse 5′-GCTTTCCGTCGCCGCCGTGCGCGTG-3′;
Mef2c, forward 5′-AGATACCCACAACACACCACGCGCC-3′ and
reverse 5′-ATCCTTCAGAGAGTCGCATGC-3′;
SRF, forward 5′-CTCCGCCCCGCTCAGACCCCACCACAGA-3′ and
reverse 5′-AGGTAGTTGGTGATGGGGAAGGA-3′;
Mesp1, forward 5′-GCGACATGCTGGCTCTTCTA-3′ and
reverse 5′-TGGTATCACTGCCGCCTCTTCC-3′;
Mesp2, forward 5′-GTGCCTTTATCTGCCTCTTCTG-3′ and
reverse 5′-AGCGGGGGTGTCTTGTCTC-3′;
α-actin, forward 5′-CACTGAAGCCCCGCTGAACG-3′ and
reverse 5′-TCGCCAGAATCCAGAACAATGC-3′;
Tbx6, forward 5′-AGGCCCGCTACTTGTTTCTTCTGG-3′ and
reverse 5′-TGGCTGCATAGTTGGGTGGCTCTC-3′;
Tbx5, forward 5′-CCAGCTCGGCGAAGGGATGTTT-3′ and
reverse 5′-CCGACGCCGTGTACCGAGTGAT-3′;
Tbx2, forward 5′-GGTGCAGACAGACAGTGCGT-3′ and
reverse 5′-AGGCCAGTAGGTGACCCATG-3′;
Isl1, forward 5′-CATCGAGTGTTTCCGCTGTGTAG-3′ and
reverse 5′-GTGGTCTTCTCCGGCTGCTTGTGG-3′;
GATA4, forward 5′- GGTTCCCAGGCCTCTTGCAATGCGG-3′ and
reverse 5′-AGTGGCATTGCTGGAGTTACCGCTG-3′;
Brachyury, forward 5′-GACTTCGTGACGGCTGACAA-3′ and
reverse 5′-CGAGTCTGGGTGGATGTAG-3′;
GATA6, forward 5′-CCGCGAGTGCGTGAACT-3′ and
reverse 5′-CGCTTCTGTGGCTTGATGAG-3′;
myocardin, forward 5′-CATTGTTTCCCAAGGAGATTC-3′ and
reverse 5′-GCGATGTTACCCTCCTCAAAA-3′.
The reaction contained 1 μl of Master SYBR green I mix (Taq DNA polymerase, buffer, deoxynucleoside trisphosphate mix and SYBR green I dye), 3 mM MgCl2 and 0.5 μM of each primer to which 2 μl of diluted cDNA was added. Amplification included initial denaturation at 95°C for 8 min, 45 cycles of denaturation at 95°C for 3 s, annealing at 60–65°C for 8–10 s and extension at 72°C for 7–10 s performed at a temperature transition rate of 20°C/s. Fluorescence was measured at the end of each extension step. After amplification, a melting curve acquired by heating the product to 95°C, cooling to and maintaining at 70°C for 20 s, then slowly (0.3°C/s) heating to 95°C was used to determine the specificity of PCR products, confirmed by gel electrophoresis. Quantification of data was performed according to the mathematical approach (Pfaffl, 2001). Data were normalized to expression of the housekeeping gene β-tubulin. A 300 ng portion of cDNA was used in the amplification of genes using semiquantitative PCR and 100 ng of cDNA for β-tubulin. Primers used were as follows:
p107, forward 5′-ATATGTTGCTTGCCGCAAGA-3′ and
reverse 5′-TTTCTAGCCTTTCTATCCGC-3′;
p130, forward 5′-AGGCCTGGAGCAGTTACCGC-3′ and
reverse 5′-TAATCTTTCAGTACGTTCTC-3′;
Lek1, forward 5′-GCTTAATTCTTCCATTCCAGG-3′ and
reverse 5′-TCGGTTTCCTGCTCGTGCTCAG-3′.
PCR reaction was performed in a thermocycler. The cycle sequence consisted of an initial denaturation step for 5 min at 95°C, followed by 35 cycles (25 cycles for β-tubulin) of 1 min at 95°C, 45 s at 50–60°C and 1 min at 72°C. A final 10 min extension at 72°C was also performed. PCR products were analyzed in 1% agarose gel. DNA bands on gels were quantified using NIH image software.
Transfection of C2C12 cells and LEK1 immunoprecipitation
C2C12 cells were cultured in DMEM supplemented with 10% FCS. Cells were transfected with lipofectamine as described by the manufacturer. The antibody was raised against the LEK1-specific sequence (KVHIDADEKKHQNILEAA 2128–2142) in the C-terminal domain of LEK1. The peptide was coupled to KLH before immunization of the rabbits. Immunoprecipitation of LEK1 and Western blotting using an Rb or LEK1 antibody were performed as previously described (Puceat et al, 1998).
Generation of ES cell clones expressing Nkx2.5 and Rb
The cDNA of Nkx2.5 was subcloned into the EcoRI restriction site of the α-actinIRESEGFP vector carrying a neomycin resistance cassette (Clontech). The BS1WT/ES4Rb−/− ES cell lines were transfected with the α-actinNkx2.5 vector using lipofectamine (Invitrogen, France) and selected for 15 days with 250 μg/ml G418 (Invitrogen).
WT Rb cDNA was subcloned downstream of the Nkx2.5 promoter using SmaI restriction site and then linearized before electroporation into ES4Rb−/− ES cells together with a pcDNA hygromycin vector according to standard protocol (Meyer et al, 2000). The ES cells were selected for 10 days with 250 μg/ml hygromycin (Invitrogen) and the individual colonies screened by PCR.
Stable expression of siRNA and antisense RNA targeting LEK1
siRNA. Two oligonucleotides were designed and hybridized to specifically target LEK1 (sense 5′-AGACAGCGAAACTGTTCTCTTCAAGAGAGAGA ACAGTTTCGCTGTCTTTTTTT-3′ and antisense 5′-AATTAAAAAAAGACAGCGAAACTGTTCTCTCT CTTGAAGAGAACAGTTTCGCTGTC-3′) and were inserted in the ApaI/EcoRI site of a polylinker subcloned into the pSuper vector (Brummelkamp et al, 2002). The BS1WT cell line was cotransfected with the pSuperLEK together with a pcDNA vector including a neomycin resistance cassette using lipofectamine and selected for 15 days with 250 μg/ml G418. Individual ES cell clones were screened by PCR.
Antisense RNA. A 200 bp LEK1-specific fragment was amplified by PCR using the following primers: forward 5-ATTTGCTCGAGTCCAGAGGATCTTC-3′ and reverse 5′-GCTATATGTTCTGGAATTCGGCCC-3′. This fragment was subcloned in the pcDNA3.1(+) (Clonetics) in the XhoI/EcoRI site. The BS1WT cell line was transfected with the antisense LEK (ASLEK) vector, which includes the neomycin resistance cassette, using lipofectamine and selected for 15 days with 250 μg/ml G418.
Generation of the ES cell clone expressing the atypical Rb domain of LEK fused to GFP
The atypical Rb domain of LEK1 downstream of an NLS sequence was amplified by PCR from RNA extracted from BS1 ES cell line using the forward primer 5′-GGCCAAAATAAAGCTTCAGG-3′ and the reverse primer 5′-ATGGGGAGGATCCCACTAAC-3′ including a HindIII and a BamH1 site, respectively. The PCR fragment was then subcloned in-frame in pEGFP-N2 vector (Clontech) using the HindIII and BamH1 sites to fuse the NLSRbLEK1 binding domain to GFP. The ES cell line was generated as described previously.
Immunostaining and cell imaging
EBs (10-day-old) were fixed in 3% paraformaldehyde for 30 min, permeabilized for 30 min with 1% Triton X-100 and immunostained as described previously (Meyer et al, 2000). In situ immunostained sarcomere-specific α-actinin was visualized in 0.2 μm optically z-sectioned EBs. Fluorescent images of EBs were acquired on a LEICA microscope with objectives mounted on a piezo-electric device, digitized on-line with a Micromax 1300YHS CCD camera (Princeton, NJ), and stored as volume files (‘stack' of z-section images) using the Metamorph software (Universal Imaging, Downington, PA). To improve resolution and signal-to-noise ratio, images were restored using Huygens software (Huygens 2.2.1, Scientific Volume Imaging, Hilversum, The Netherlands) and visualized using Imaris (Bitplane, Switzerland). Calculations were performed on DELL Precision 450 workstations.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
We acknowledge Drs T Jacks and J Sage (Boston) for providing the Rb−/− ES cell line, Dr Pierre Travo (CRBM) for help in image analysis and Claude Sardet and Laurent Le Cam (IGMM, CNRS Montpellier) for providing Rb cDNA and Rb and histone H3 antibodies. EP was a Marie Curie fellow (Mobility Programme of European Community). CM was a fellow from the Groupement de Réflexion sur la Recherche Cardiovasculaire and is a fellow from the Fondation Lefoulon-Delalande. MP is an INSERM established investigator. This study was funded by the Marie Curie EC Contract QLKCT2001 50978 (to MP) and by a grant (no. 8849 to MP) from Association française contre les myopathies (AFM).
References
- Ashe MD, Pabon-Pena LM, Dees E, Price KL, Bader D (2003) LEK1 is a potential inhibitor of pocket protein-mediated cellular processes. J Biol Chem 279: 664–676 [DOI] [PubMed] [Google Scholar]
- Behfar A, Zingman L, Hodgson D, Rauzier J, Kane G, Terzic A, Pucéat M (2002) Stem cell differentiation requires a paracrine pathway in the heart. FASEB J 16: 1558–1566 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S (2003) Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 5: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Riley DJ, Chen Y, Lee WH (1996) Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes dev 10: 2794–2804 [DOI] [PubMed] [Google Scholar]
- Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H (1992) Requirement for a functional Rb-1 gene in murine development. Nature 359: 328–330 [DOI] [PubMed] [Google Scholar]
- Cloud JE, Rogers C, Reza TL, Ziebold U, Stone JR, Picard MH, Caron AM, Bronson RT, Lees JA (2002) Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol Cell Biol 22: 2663–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T (1996) Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev 10: 1633–1644 [DOI] [PubMed] [Google Scholar]
- De Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G (2003) Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci USA 100: 6546–6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R (1985) The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol 87: 27–45 [PubMed] [Google Scholar]
- Evans SM, Walsh BA, Newton CB, Thorburn JS, Gardner PD, van Bilsen M (1993) Potential role of helix–loop–helix proteins in cardiac gene expression. Circ Res 73: 569–578 [DOI] [PubMed] [Google Scholar]
- Goodwin RL, Pabon-Pena LM, Foster GC, Bader D (1999) The cloning and analysis of LEK1 identifies variations in the LEK/centromere protein F/mitosin gene family. J Biol Chem 274: 18597–18604 [DOI] [PubMed] [Google Scholar]
- Harvey RP (2002) Patterning the vertebrate heart. Nat Rev Genet 3: 544–556 [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A (1999) Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem 274: 19838–19845 [DOI] [PubMed] [Google Scholar]
- Huh MS, Parker MH, Scime A, Parks R, Rudnicki MA (2004) Rb is required for progression through myogenic differentiation but not maintenance of terminal differentiation. J Cell Biol 166: 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA (1992) Effects of an Rb mutation in the mouse. Nature 359: 295–300 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Zacksenhaus E, Gallie BL, Phillips RA (1997) The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene 14: 1789–1797 [DOI] [PubMed] [Google Scholar]
- Kim KK, Soonpaa MH, Daud AI, Koh GY, Kim JS, Field LJ (1994) Tumor suppressor gene expression during normal and pathologic myocardial growth. J Biol Chem 269: 22607–22613 [PubMed] [Google Scholar]
- Kirshenbaum LA, Schneider MD (1995) Cardiac cell cycle, pocket proteins and p300. Trends Cardiovasc Med 5: 230–235 [DOI] [PubMed] [Google Scholar]
- Kowanetz M, Valcourt U, Bergstrom R, Heldin CH, Moustakas A (2004) Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol 24: 4241–4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A (2000) Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407: 592–598 [DOI] [PubMed] [Google Scholar]
- Lasorella A, Uo T, Iavarone A (2001) Id proteins at the cross-road of development and cancer. Oncogene 20: 8326–8333 [DOI] [PubMed] [Google Scholar]
- Leahy A, Xiong JW, Kuhnert F, Stuhlmann H (1999) Use of developmental marker genes to define temporal and spatial patterns of differentiation during embryoid body formation. J Exp Zool 284: 67–81 [DOI] [PubMed] [Google Scholar]
- LeCouter JE, Kablar B, Whyte PF, Ying C, Rudnicki MA (1998) Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125: 4669–4679 [DOI] [PubMed] [Google Scholar]
- Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A (1992) Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359: 288–294 [DOI] [PubMed] [Google Scholar]
- Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T (1996) Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev 10: 1621–1632 [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T (1999) The retinoblastoma gene family in differentiation and development. Oncogene 18: 7873–7882 [DOI] [PubMed] [Google Scholar]
- Maltsev VA, Wobus AM, Rohwedel J, Bader M, Hescheler J (1994) Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ Res 75: 233–244 [DOI] [PubMed] [Google Scholar]
- McBride K, Nemer M (1998) The C-terminal domain of c-fos is required for activation of an AP-1 site specific for jun–fos heterodimers. Mol Cell Biol 18: 5073–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill CJ, Brooks G (1995) Cell cycle control mechanisms and their role in cardiac growth. Cardiovasc Res 30: 557–569 [PubMed] [Google Scholar]
- Meyer N, Jaconi M, Ladopoulou A, Fort P, Puceat M (2000) A fluorescent reporter gene as a marker for ventricular specification in ES-derived cardiac cells. FEBS Lett 478: 151–158 [DOI] [PubMed] [Google Scholar]
- Novitch BG, Mulligan GJ, Jacks T, Lassar AB (1996) Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol 135: 441–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Schneider MD (2003) Sizing up the heart: development redux in disease. Genes Dev 17: 1937–1956 [DOI] [PubMed] [Google Scholar]
- Pabon-Pena LM, Goodwin RL, Cise LJ, Bader D (2000) Analysis of CMF1 reveals a bone morphogenetic protein-independent component of the cardiomyogenic pathway. J Biol Chem 275: 21453–21459 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puceat M, Roche S, Vassort G (1998) Src family tyrosine kinase regulates intracellular pH in cardiomyocytes. J Cell Biol 141: 1637–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A, deRiel JK, Xu YS, Montgomery M, Patwardhan V, Litvin J (2002) Characterization of cardiac muscle factor 1 sequence motifs: retinoblastoma protein binding and nuclear localization. Gene 282: 53–64 [DOI] [PubMed] [Google Scholar]
- Reecy JM, Li X, Yamada M, DeMayo FJ, Newman CS, Harvey RP, Schwartz RJ (1999) Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development 126: 839–849 [DOI] [PubMed] [Google Scholar]
- Schlange T, Andree B, Arnold HH, Brand T (2000) BMP2 is required for early heart development during a distinct time period. Mech Dev 91: 259–270 [DOI] [PubMed] [Google Scholar]
- Schneider JW, Gu W, Zhu L, Mahdavi V, Nadal-Ginard B (1994) Reversal of terminal differentiation mediated by p107 in Rb−/− muscle cells. Science 264: 1467–1471 [DOI] [PubMed] [Google Scholar]
- Sears RC, Nevins JR (2002) Signaling networks that link cell proliferation and cell fate. J Biol Chem 277: 11617–11620 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F (2002) The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN (2000) A genetic blueprint for cardiac development. Nature 407: 221–226 [DOI] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B (2004) E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol 24: 4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Yang HS, Alexander K, Hinds PW (2003) Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol Ther 2: 124–130 [PubMed] [Google Scholar]
- Vara D, Bicknell KA, Coxon CH, Brooks G (2003) Inhibition of E2F abrogates the development of cardiac myocyte hypertrophy. J Biol Chem 278: 21388–21394 [DOI] [PubMed] [Google Scholar]
- Wei Y, Bader D, Litvin J (1996) Identification of a novel cardiac-specific transcript critical for cardiac myocyte differentiation. Development 122: 2779–2789 [DOI] [PubMed] [Google Scholar]
- Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett LA, Weinstein M, Cross JC, Robinson ML, Leone G (2003) Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421: 942–947 [DOI] [PubMed] [Google Scholar]
- Xie Y, Sun T, Wandg QT, Wang F, Puschek E, Rappolee DA (2005) Acquisition of essential somatic cell cycle regulatory protein expression and implied activity occurs at the second to third cell division in mouse preimplantation embryos. FEBS Lett 579: 398–408 [DOI] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA (2001) E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev 15: 386–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3


