Abstract
The plant hormone auxin elicits many specific context-dependent developmental responses. Auxin promotes degradation of Aux/IAA proteins that prevent transcription factors of the auxin response factor (ARF) family from regulating auxin-responsive target genes. Aux/IAAs and ARFs are represented by large gene families in Arabidopsis. Here we show that stabilization of BDL/IAA12 or its sister protein IAA13 prevents MP/ARF5-dependent embryonic root formation whereas stabilized SHY2/IAA3 interferes with seedling growth. Although both bdl and shy2-2 proteins inhibited MP/ARF5-dependent reporter gene activation, shy2-2 was much less efficient than bdl to interfere with embryonic root initiation when expressed from the BDL promoter. Similarly, MP was much more efficient than ARF16 in this process. When expressed from the SHY2 promoter, both shy2-2 and bdl inhibited cell elongation and auxin-induced gene expression in the seedling hypocotyl. By contrast, gravitropism and auxin-induced gene expression in the root, which were promoted by functionally redundant NPH4/ARF7 and ARF19 proteins, were inhibited by shy2-2, but not by bdl protein. Our results suggest that auxin signals are converted into specific responses by matching pairs of coexpressed ARF and Aux/IAA proteins.
Keywords: Arabidopsis, ARF transcription factor, Aux/IAA protein, embryo, root gravitropism
Introduction
The small signaling molecule auxin elicits a multitude of developmental and physiological responses, such as patterning in embryogenesis, apical dominance, cell elongation and gravitropism (Berleth and Sachs, 2001). The cellular response to auxin involves changes in gene regulation. Genes upregulated by auxin contain in their promoters auxin-response elements (AuxRE), which bind transcription factors of the auxin response factor (ARF) family (Ulmasov et al, 1997a, 1999). At low auxin concentrations, ARFs are thought to be inhibited by interacting proteins of the Aux/IAA family via domains III and IV that are conserved between the two protein families (Ulmasov et al, 1997b). Aux/IAA genes were originally identified as genes that are rapidly upregulated in response to auxin (Abel et al, 1994). High auxin concentrations promote degradation of Aux/IAA proteins, which would release interacting ARFs from inhibition (Tiwari et al, 2001, 2003). Degradation of Aux/IAA proteins involves their conserved domain II, which mediates interaction with the SCFTIR1 ubiquitin-ligase complex for targeting of Aux/IAAs to the proteasome (Gray et al, 2001). Amino-acid exchanges in conserved residues of domain II affect the interaction with the SCFTIR1 ubiquitin-ligase complex, stabilizing mutant Aux/IAA proteins (Ramos et al, 2001). Such stabilizing mutations have been reported for 10 Aux/IAA genes (Reed, 2001; Hellmann and Estelle, 2002; Tatematsu et al, 2004; Yang et al, 2004).
How is a generic signal such as auxin converted into specific context-dependent developmental responses? Auxin can increase the affinity between the SCFTIR1 ubiquitin-ligase complex and Aux/IAA proteins in a cell-free system without modifying the latter (Dharmasiri et al, 2003; Tian et al, 2003; Kepinski and Leyser, 2004). This observation suggests that the specificity of response to auxin is generated by interacting Aux/IAA and ARF proteins present in the auxin-responsive cell. The Arabidopsis genome encodes 22 ARF and 29 Aux/IAA proteins (Remington et al, 2004). Several ARFs have been assigned roles in specific developmental processes on the basis of their loss-of-function mutant phenotypes (Berleth and Jürgens, 1993; Przemeck et al, 1996; Sessions et al, 1997; Hardtke and Berleth, 1998; Harper et al, 2000; Nemhauser et al, 2000; Li et al, 2004; Tian et al, 2004). Although ARFs appear to have unique functions in some contexts, they display overlapping functions in others (Hardtke et al, 2004; Li et al, 2004). For example, MP/ARF5 is required for embryonic root initiation whereas both MP and NPH4/ARF7 contribute to cotyledon development (Hardtke et al, 2004). A larger number of Aux/IAA proteins have been implicated in diverse processes on the basis of their gain-of-function mutant phenotypes (Reed, 2001; Tatematsu et al, 2004; Yang et al, 2004). The mutant phenotypes are distinct for some Aux/IAA proteins but related for others, suggesting both distinct and overlapping roles in development. For example, stabilized BDL/IAA12 protein interferes with embryonic root initiation as does the loss of MP/ARF5 protein, suggesting that these two proteins generate a specific developmental response (Hamann et al, 2002). In contrast to ARF genes, no loss-of-function phenotypes have been reported for Aux/IAA genes except SHY2/IAA3, which is involved in seedling development (Tian and Reed, 1999). Most of the Aux/IAA genes exist as sister genes that appear to have originated by segmental duplications of the genome whereas ARF genes are not located in duplicated segments (Remington et al, 2004). For example, one pair of sister genes consists of BDL/IAA12, which is involved in embryonic root initiation, and IAA13 (Hamann et al, 1999, 2002). It is not known whether IAA13 performs a comparable role to BDL or rather acts in a different process. Furthermore, although mutations in different ARF and Aux/IAA genes cause distinct phenotypes, it is unclear how these proteins contribute to specificity of action.
Here we address how Aux/IAA and ARF proteins generate specific responses to auxin. The effects of stabilized BDL and SHY2 proteins on embryonic root formation and seedling development were analyzed by swapping their gene promoters. These proteins were also assayed for their ability to inhibit MP-dependent gene activation in the absence of plant-specific accessory factors. Finally, candidate ARF proteins for interaction with BDL or SHY2 were examined for roles in BDL- and SHY2-dependent processes. Our results suggest that transcriptionally regulated optimized pairs of interacting Aux/IAA–ARF proteins generate developmental specificity of auxin response.
Results
IAA13 is a functional paralog of BDL/IAA12
Many Aux/IAA genes, including BDL/IAA12 and its closest homolog IAA13 (At2g33310), appear in regions of segmental genome duplications (Remington et al, 2004). To examine whether IAA13 is functionally related to BDL/IAA12, we first introduced a proline to serine mutation (iaa13P80S; Figure 1A) in the conserved domain II of a Myc-epitope-tagged transgenic copy of the IAA13 gene. The homologous mutation in the BDL gene causes semidominant gain-of-function phenotypes, both in the bdl mutant and when provided as a transgene (Hamann et al, 2002). Plants carrying the iaa13P80S transgene resembled bdl mutants in all respects. A single transgene copy caused stunted growth (not shown), whereas two copies caused embryonic phenotypes (Figure 1B). Homozygous iaa13P80S seedlings had no root, and the origin of this defect could be traced to a failure in the specification of the hypophysis-, the embryonic root meristem precursor- and subsequent abnormal cell division patterns (Figure 1B). Western blot analysis showed that the engineered iaa13P80S mutation led to the stabilization of the IAA13 protein (Figure 1C). This effect was quantitatively similar to the stabilization of the BDL protein in bdl mutants (Figure 1C), and caused comparable frequencies of embryo phenotypes (Table I).
Figure 1.
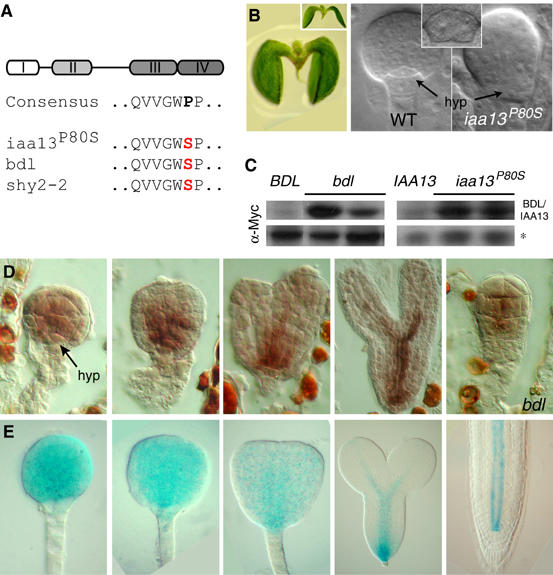
Expression of IAA13, and analysis of an iaa13 stabilizing mutation. (A) Domain structure of Aux/IAA proteins. The four conserved domains are depicted. Below is the consensus amino-acid sequence in conserved domain II of the engineered iaa13P80S mutation, of bdl and of shy2-2. (B) Rootless seedling homozygous for the iaa13P80S mutation (left); inset: bdl seedling. Comparison of a wild-type (middle) and homozygous iaa13P80S (right) globular stage embryo shows defects in hypophysis (hyp) specification; inset: abnormal division of hypophysis in bdl embryo. (C) Western blots (Myc antibody) of protein extracts from pBDL∷BDL, pBDL∷bdl, pIAA13∷IAA13 and pIAA13∷iaa13P80S seedlings. Individual lanes represent independent transgenics. Asterisk: unspecific crossreacting band demonstrating equal loading. (D) mRNA in situ hybridization with an IAA13 antisense probe in wild-type and bdl (right) embryos. RNA signals are in red-brown. (E) GUS activity in pIAA13∷GUS embryos and seedling root tip (right).
Table 1. Frequencies of rootless phenotypes in genotypes used in this study.
| Genotype | Line # | Rootless seedlings (% (N)) | Defective embryos (% (N)) |
|---|---|---|---|
| Columbia WT | 1.4 (141) | ||
| pBDL∷SHY2 | 2 | 1.1 (72) | |
| 6 | 0.7 (124) | 0.5 (187) | |
| pBDL∷shy2-2 | 5 | 0 (430) | |
| 19 | 4.4 (280) | ||
| 20 | 9.8 (283) | 7.1 (70) | |
| 22 | 8.0 (63) | 9.7 (124) | |
| 24 | 9.1 (212) | ||
| 29 | 27.6 (134) | ||
| 38 | 9.4 (112) | ||
| pBDL∷BDL | 38 | 0 (148) | 0 (81) |
| pBDL∷bdl | 27 | 19.4 (66) | 21.1 (19) |
| 41 | 24.6 (61) | 32.2 (59) | |
| 73 | 60.8 (51) | ||
| pBDL∷IAA13 | 11 | 2.5 (115) | 2.2 (138) |
| pBDL∷iaa13P80S | 5 | 25.2 (283) | 25.5 (108) |
| 28 | 29.6 (68) | ||
| 38 | 12.2 (317) | 28.3 (53) | |
| pIAA13∷IAA13 | 12 | 0.9 (94) | |
| 17 | 0.8 (129) | ||
| pIAA13∷iaa13P80S | 4 | 32.6 (177) | |
| 11 | 23.9 (102) | ||
| 23 | 29.0 (62) | ||
| pIAA13∷BDL | 5 | 0 (131) | |
| 8 | 1.3 (130) | ||
| pIAA13∷bdl | 1 | 26.2 (166) | |
| 7 | 28.5 (35) | 29.4 (68) | |
| 30 | 23.1 (337) | 22.8 (206) | |
| pSHY2∷shy2-2 | 11 | 0 (55) | |
| pSHY2∷bdl | 4 | 0 (48) | 0.7 (150) |
| 12 | 0 (30) | ||
| pSHY2∷iaa13P80S | 7 | 2.5 (38) | 1.9 (161) |
| 11 | 5.0 (20) | ||
| bdl | 22.3 (264) |
The phenotypic equivalence of bdl and iaa13P80S gain-of-function mutations suggests that the two genes are expressed in a similar way. mRNA in situ hybridization revealed that the IAA13 gene is first transcribed specifically in the globular proembryo, but not the hypophysis (Figure 1D). Later, expression extends basally to the lens-shaped apical daughter cell of the hypophysis (Figure 1D). Finally, IAA13 mRNA accumulation is restricted to the future vascular tissue (Figure 1D). The identical expression pattern was previously detected for BDL (Hamann et al, 2002). IAA13 mRNA expression was unchanged in bdl mutants (Figure 1D), excluding the possibility that IAA13 acts downstream of BDL. Furthermore, IAA13 promoter-GUS fusions revealed that the IAA13 expression pattern is regulated at the level of gene transcription (Figure 1E). In conclusion, the IAA13 gene is a functional paralog of BDL.
Aux/IAA specificity in embryogenesis is transcriptionally regulated
To assess the relative contributions of transcriptional regulation of Aux/IAA genes and Aux/IAA protein determinants in specificity of action in the embryo, a promoter-swap strategy was adopted. The promoters of BDL or IAA13 were fused to Myc-epitope-tagged genomic coding regions of the SHY2/IAA3, BDL or IAA13 genes. Homologous stabilizing proline to serine domain II mutations (SHY2P69S—shy2-2 (Tian and Reed, 1999); BDLP72S—bdl (Hamann et al, 2002); IAA13P80S) were introduced into each construct, and wild-type versions of the transgenes were analyzed as controls.
In accordance with the similar BDL and IAA13 gene activities, pBDL∷iaa13P80S and pIAA13∷bdl plants showed embryonic and postembryonic phenotypes comparable to the bdl and iaa13P80S mutants (Figure 2A; Table I; not shown). Notably, however, whereas shy2-2 mutants have no embryonic phenotypes (Tian and Reed, 1999; Table I), pBDL∷shy2-2 plants showed a rootless phenotype similar to that of bdl (Figure 2A). In addition, pBDL∷shy2-2 plants showed bdl-like postembryonic growth abnormalities (Figure 2B). Although phenotypes were qualitatively similar in all genotypes, the frequency of embryonic phenotypes was significantly lower in pBDL∷shy2-2 plants (Table I). However, Western blot analysis showed that shy2-2, bdl and iaa13P80S proteins accumulated to comparable levels (Figure 2C). These results suggest that the specificity of BDL and IAA13 action in embryogenesis is mainly regulated at the level of gene transcription, but other Aux/IAA proteins may also affect root formation when expressed in the embryo.
Figure 2.

BDL promoter-swap experiments. (A) Rootless pBDL∷shy2-2 and pBDL∷iaa13P80S homozygous seedlings; inset: pBDL∷bdl seedling. (B) Flowering plants (4 weeks old) are bushy and short; inset: heterozygous bdl plant. (C) Western blots of protein extracts from pBDL∷SHY2, pBDL∷shy2-2, pBDL∷IAA13 and pBDL∷iaa13P80S seedlings. Asterisk: unspecific crossreacting band demonstrating approximately equal loading. Percentage of rootless seedlings (%RL) is indicated for each line.
The ARF transcription factor that likely acts in concert with BDL and IAA13 in the embryo is MONOPTEROS (MP)/ARF5 (Hardtke and Berleth, 1998; Hamann et al, 2002). To examine whether the ability to inhibit MP activity reflects the activity of bdl and shy2-2 in the embryo, we developed a heterologous assay for ARF and Aux/IAA activity. A direct repeat of eight ARF-binding DR5(rev) repeat sequences (Ulmasov et al, 1997b) was placed upstream of a minimal promoter for expression in yeast, and this yDR5 (yeast-DR5) promoter was fused to the lacZ gene for β-galactosidase. When HA-epitope-tagged MP cDNA was expressed in yDR5∷lacZ yeast cells, the activity of the reporter was induced several-fold (Figure 3A). Next, HA-epitope-tagged cDNAs of SHY2 or BDL were introduced on the same plasmid as MP:HA. Coexpression of SHY2:HA or BDL:HA with MP:HA nearly completely repressed MP:HA activity (Figure 3A), showing that both proteins can inhibit MP activity. Western blot analysis consistently showed that MP:HA, SHY2:HA and BDL:HA were expressed in yeast, indicating that SHY2 and BDL did not interfere with MP expression. However, SHY2:HA consistently accumulated to higher levels than BDL:HA (Figure 3B), presumably because of cleavage of the BDL:HA protein (not shown). This assay shows that both SHY2 and BDL can directly inhibit MP transcription factor activity in the absence of plant-specific accessory factors, and BDL may be more potent than SHY2.
Figure 3.
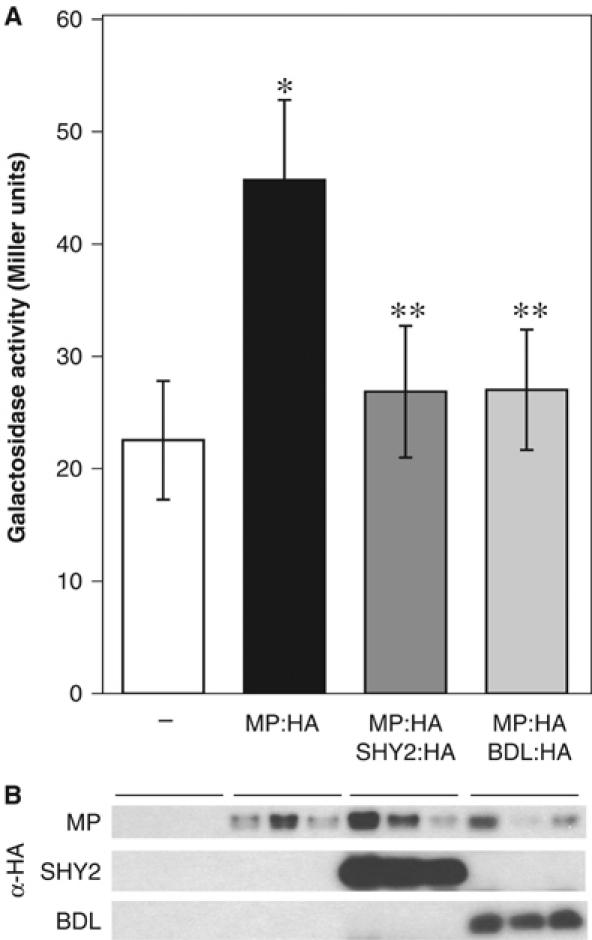
Repression of MP activity by SHY2 and BDL in yeast. (A) Galactosidase activity in yDR5 yeast cells expressing the empty vector (−), MP:HA (MP), MP:HA and SHY2:HA (MP+SHY2) or MP:HA and BDL:HA (MP+BDL). Values (±s.d.) are the average of 12 independent transformants. Asterisks represent statistically significant difference between MP and – (*P<0.001, two-tailed Student's t-test), MP+SHY2 and MP (**P<0.001, two-tailed Student's t-test), and MP+BDL and MP (**P<0.001, two-tailed Student's t-test). (B) Western blot (HA antibody) with equal amounts of protein extracts from three independent yeast transformants for each plasmid. The regions from the same blot that represent MP:HA (105 kDa), SHY2:HA (25 kDa) and BDL:HA (29 kDa) are depicted. Note that expression levels of MP:HA protein vary between different colonies of the same genotype.
Aux/IAA specificity in hypocotyl and shoot is transcriptionally regulated
Elongation of the seedling hypocotyl involves SHY2-dependent auxin responses (Tian and Reed, 1999; Tian et al, 2002). In contrast, bdl mutants show normal hypocotyl elongation (Hamann et al, 1999). SHY2 is predominantly expressed in cotyledons and hypocotyl including peripheral cell layers whereas BDL expression is largely confined to the central vascular strands (Hamann et al, 2002; Tian et al, 2002). To assess if the specificity of SHY2 action in the hypocotyl is also subject to transcriptional regulation, we expressed Myc-epitope-tagged bdl or iaa13P80S proteins from the SHY2 promoter, and compared their phenotypes with pSHY2∷shy2-2 seedlings.
The pSHY2∷shy2-2 construct induced hypocotyl elongation defects in both light- and dark-grown seedlings (Figure 4A), and the severity of defects correlated well with the level of mutant protein accumulation (Figure 4B). By comparison, pSHY2∷bdl and pSHY2∷iaa13P80S seedlings showed a slightly stronger inhibition of hypocotyl elongation (Figure 4A) although mutant proteins accumulated to lower levels than in pSHY2∷shy2-2 seedlings (Figure 4B). These results suggest that, as in embryonic root formation, bdl and iaa13 mutant proteins are more effective than the shy2-2 protein in inhibiting auxin responses. Subsequent shoot development in pSHY2∷shy2-2, pSHY2∷bdl and pSHY2∷ iaa13P80S plants resembled that of shy2-2 mutants (Figure 4C; Tian and Reed, 1999). However, the phenotypes were again quantitatively different between the genotypes. Plants from different pSHY2∷shy2-2 lines showed different phenotypic strengths, ranging from those seen in shy2-2 heterozygotes to those of shy2-2 homozygotes, whereas the phenotypes of the other two transgenic genotypes strongly resembled shy2-2 homozygotes (Figure 4C).
Figure 4.
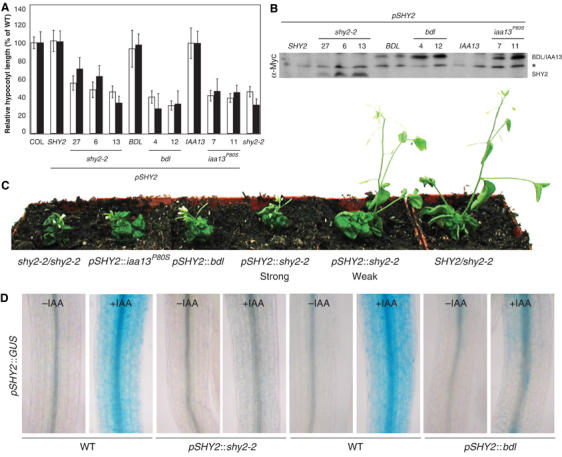
Inhibition of auxin responses in the shoot by stabilized Aux/IAA proteins. (A) Hypocotyl length in light-grown (white bars) or dark-grown (black bars) seedlings of wild type (COL), pSHY2∷SHY2, pSHY2∷shy2-2 (lines #27, #6 and #13), pSHY2∷BDL, pSHY2∷bdl (lines #4 and #12), pSHY2∷IAA13, pSHY2∷iaa13P80S (lines #7 and #11) and shy2-2. Hypocotyl length (±s.d.) is represented as percentage of COL. (B) Western blot of protein extracts from light-grown seedlings in (A). Blot was probed with anti-Myc antibodies; asterisk: unspecific crossreacting band demonstrating equal loading. (C) Phenotypes of flowering plants. (D) GUS activity in hypocotyl of seedlings hemizygous for pSHY2∷GUS and pSHY2∷shy2-2, pSHY2∷bdl or wild-type controls from same cross. Seedlings were treated with IAA for 5 h and stained for GUS activity.
Aux/IAA proteins act primarily at the level of auxin-dependent gene expression (Tiwari et al, 2001). To test whether this process is similarly affected in pSHY2∷shy2-2 and pSHY2∷bdl seedlings, we analyzed auxin-dependent pSHY2∷GUS activity. In wild-type hypocotyls, pSHY2∷GUS was induced by auxin in the outer cell layers (Figure 4D). This auxin-induced expression was almost completely lost in the hypocotyl of both pSHY2∷shy2-2 and pSHY2∷bdl seedlings (Figure 4D). Thus, SHY2 activity feeds back on SHY2 gene expression, and this function can be taken over by BDL protein, indicating functional equivalence of the two Aux/IAA proteins in this specific auxin response.
At similar protein concentrations, bdl appeared to have a stronger effect than did shy2 on the hypocotyl and shoot phenotypes, just as it did for embryo phenotypes. Taken together, these results suggest that specificity of SHY2 action in hypocotyl and shoot is determined by the activity of its promoter, with protein determinants affecting only the extent to which auxin responses are inhibited.
Aux/IAA protein specificity in auxin-mediated root development
The shy2-2 mutation not only affects auxin responses in hypocotyl and shoot, but also in the root (Tian and Reed, 1999). To examine whether the specificity of SHY2 action in the root is also transcriptionally regulated, we studied root-specific auxin responses in pSHY2∷shy2-2 and pSHY2∷bdl seedlings. Alignment of the root tip with a changing gravity vector requires auxin response, and this response is strongly diminished in shy2-2 as well as in pSHY2∷shy2-2 roots (Tian and Reed, 1999; Figure 5A). Surprisingly, roots of pSHY2∷bdl seedlings responded almost normally to gravity although their hypocotyls displayed strong inhibition of elongation (Figure 5A, compare with Figure 4A). Similarly, although root growth was comparably reduced in both pSHY2∷shy2-2 and pSHY2∷bdl seedlings under normal conditions, the two genotypes differed in their response to the growth-inhibiting effects of auxin (Figure 5B). Whereas pSHY2∷bdl root growth was sensitive, shy2-2 and pSHY2∷shy2-2 root growth was partially resistant (Figure 5B). This difference in auxin response between pSHY2∷shy2-2 and pSHY2∷bdl roots must lie in the shy2-2 and bdl proteins themselves because both are expressed in the root (Figure 5C).
Figure 5.
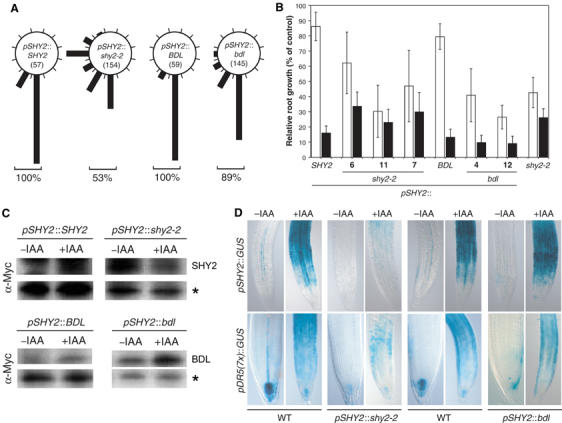
Inhibition of auxin responses in the root by shy2-2, but not by bdl. (A) Gravitropic response of pSHY2∷SHY2, pSHY2∷shy2-2, pSHY2∷BDL and pSHY2∷bdl seedlings after reorientation by 90° (data from two to five independent transgenic lines for each genotype). The percentages represent the fraction of seedlings (numbers analyzed in parentheses) with normal gravitropic response. (B) Inhibition of root growth by 2,4-D. Root length (±s.d.) was measured after 3 days of vertical growth on medium with (black bars) or without (white bars) 0.1 μM 2,4-D. Growth is represented as percentage of root length in wild type (COL) on control media. (C) Western blots of protein extracts from IAA-treated (10 μM for 5 h) or untreated roots of pSHY2∷SHY2, pSHY2∷shy2-2, pSHY2∷BDL and pSHY2∷bdl seedlings. Asterisks: unspecific crossreaction as loading control. Note that SHY2 or BDL accumulation is induced by auxin in pSHY2∷SHY2, pSHY2∷BDL and pSHY2∷bdl, but not in pSHY2∷shy2-2. (D) GUS activity in root tips of F1 seedlings from crosses between hemizygous pSHY2∷shy2-2 or pSHY2∷bdl lines and homozygous pSHY2∷GUS or pDR5(7x)∷GUS lines. Controls (WT) are wild-type segregants from the same cross. −IAA, untreated; +IAA, 10 μM IAA for 5 h. GUS staining time was 3 h for untreated and 1.5 h for IAA-treated roots.
As in the hypocotyl, auxin responses in the root involve changes in gene expression. The shy2-2 mutation and pSHY2∷shy2-2 prevented the auxin-induced expression of pSHY2∷GUS and pDR5(7x)∷GUS (Figure 5D; Tian et al, 2002). In contrast, pSHY2∷bdl roots showed normal auxin-induced expression of both reporters (Figure 5D). The noninduced expression in the root vascular tissues and the distal tip was similarly affected in both pSHY2∷shy2-2 and pSHY2∷bdl (Figure 5D). In summary, bdl did not regulate auxin-induced gene expression or gravitropism in roots, even when present at similar protein levels to levels of shy2-2 that have a strong effect on these phenotypes. These results indicate that shy2-2 is effective in inhibiting auxin-mediated root development whereas bdl is not, which is in contrast to the stronger activity of bdl in embryo development, auxin response in the hypocotyl, and shoot and root growth.
ARF7 and ARF19 as targets of Aux/IAA inhibition in auxin-mediated root development
Aux/IAA proteins inhibit auxin responses through interactions with ARF transcription factors (Tiwari et al, 2003). Hence, a plausible explanation for the differential effects of pSHY2∷shy2-2 and pSHY2∷bdl on auxin-dependent root development would be that shy2-2, but not bdl, interacts with a yet unidentified ARF that regulates these auxin responses. To date, no arf mutant has been reported to have root phenotypes that resemble shy2-2. We took a candidate approach to identify ARF protein(s) involved in auxin-dependent root development. Initially, we analyzed double mutants for ARF10 and ARF16, two closely related genes (Remington et al, 2004) that are highly expressed in elongating root epidermis cells (Birnbaum et al, 2003). These double mutants showed normal gravitropism and auxin response in the primary root (not shown). Thus, ARF10 and ARF16 are unlikely to be targets of shy2-2 inhibition.
The NPH4/ARF7 gene is required for shoot tropisms, but the nph4 mutant has no root gravitropism defect (Liscum and Briggs, 1996; Watahiki and Yamamoto, 1997; Tatematsu et al, 2004; Figure 6A). However, the ARF19 gene is highly related to NPH4 (Remington et al, 2004), and it has recently been shown that ARF19 acts redundantly with NPH4 in plant growth, including gravi- and phototropism in seedlings (Okushima et al, 2005). To test whether ARF19 regulates the same responses that are disturbed in shy2-2 mutants, two mutant alleles of ARF19 were tested. One of these, arf19-4, has a weak but significant phenotype in auxin-mediated root development. Gravitropic response as well as growth sensitivity to 2,4-D is impaired (Figure 6A and B). An nph4-1 arf19-4 double mutant, however, was severely impaired in gravitropism (Figure 6A) and also showed nearly complete auxin-resistant root growth (Figure 6B). Correspondingly, auxin-induced SHY2 gene expression was partially impaired in each single mutant, and nearly completely lost in the nph4-1 arf19-4 double mutant (Figure 6C). These results suggest that NPH4 and ARF19 act redundantly in auxin responses in the primary root tip, and are therefore good candidates for targets of inhibition by shy2-2.
Figure 6.
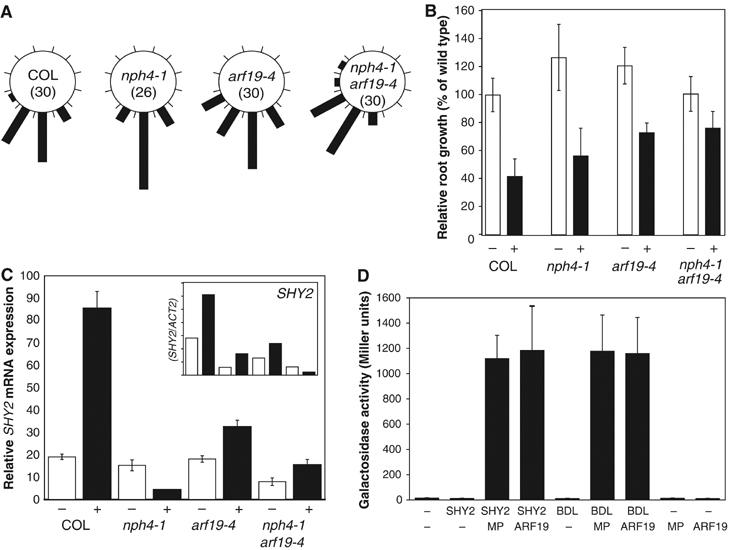
NPH4 and ARF19 as targets for shy2 inhibition in the root. (A) Gravitropic response of wild-type (COL), nph4-1, arf19-4 and nph4-1 arf19-4 seedling roots (numbers analyzed in parentheses) upon reorientation by 90°. (B) Inhibition of root growth by 2,4-D. Root length (±s.d.) was measured after 3 days of vertical growth on medium with (black bars) or without (white bars) 0.1 μM 2,4-D. Growth is represented as percentage of root length in wild type (COL) on control media. (C) SHY2 mRNA expression in COL, nph4-1, arf19-4 and nph4-1 arf19-4 seedlings treated with unsupplemented medium (white bars) or with medium containing 50 μM IAA (black bars) for 3 h. Average values (±s.d.) are taken from five to six replicate real-time PCR reactions (quantitative RT–PCR); inset: semiquantitative RT–PCR experiment on dissected roots from three replicates. Expression is relative to ACT2 expression in the same cDNA samples. (D) Interaction of SHY2 or BDL with MP and ARF19 in yeast two-hybrid assays. Galactosidase activity (±s.d.) was measured in at least 12 independent colonies expressing each of the depicted plasmids (−, empty vector control).
Yeast two-hybrid assays were performed to test whether ARF7 and ARF19 can be targets for SHY2 action in the root, and whether these could be the effectors that discriminate between shy2-2 and bdl. Because ARF19 seems to contribute most strongly to the physiological responses that are inhibited by shy2-2, we tested the interaction between full-length SHY2 or BDL proteins and C-terminal regions corresponding to domains III and IV of ARF19. As a control, interactions between SHY2 or BDL and domains III and IV of MP were tested.
Both SHY2 and BDL interacted with MP as well as with ARF19 (Figure 6D). Unfortunately, although preferential interactions between MP and BDL, on the one hand, and SHY2 and ARF19, on the other, were seen in several independent experiments, the resolution of yeast two-hybrid assays was too coarse to substantiate differential Aux/IAA–ARF affinities. Nonetheless, the ability of SHY2 to interact with ARF19, combined with the fact that mutant phenotypes of shy2-2, arf19-4 and nph4-1 arf19-4 roots are very similar, suggests that NPH4 and ARF19 are targets for SHY2 activity. Whether differential Aux/IAA–ARF interactions underlie specificity of SHY2 in the root tip remains unresolved.
Specific ARF activity in embryogenesis involves protein determinants
To address the relative contributions of transcriptional control and protein determinants in specificity of embryonic ARF action, we performed similar promoter-swap experiments to those described above for Aux/IAAs. We fused the cDNAs of two distantly related ARFs, MP/ARF5 and ARF16, to the BDL cis-regulatory sequences, and assessed to what extent these transgenes can complement the rootless phenotype of mp mutant seedlings. The mp mutation prevents the formation of the embryonic root in approximately 25% of the progeny of heterozygous plants (Berleth and Jürgens, 1993). When pBDL∷MP was transformed into mp heterozygotes, 11 out of 20 plants that carried the mp mutation had a strongly reduced frequency of rootless seedlings (Figure 7A), indicating complementation of the embryonic phenotype. In contrast, the pBDL∷ARF16 transgene reduced the frequency of rootless seedlings in only two out of 11 lines (Figure 7B). Thus, ARF16 protein can potentially complement for the absence of MP. However, MP is much more effective than ARF16, suggesting that features specific to MP protein are required for effectiveness, although not strictly for specificity of action in embryogenesis.
Figure 7.

Specificity of MP action during embryogenesis. Percentage of rootless seedlings in (A) 20 independent pBDL∷MP transgenic lines and (B) 11 independent pBDL∷ARF16 transgenic lines, all carrying the mp mutation. The line at 25% marks the frequency of rootless seedlings in the untransformed mp mutant.
Discussion
Auxin response is mediated by ARF transcription factors and their Aux/IAA inhibitors. Here, we examined Aux/IAA protein functions in their physiological contexts, by using promoter-swapping experiments with stabilized variants. As controls we used wild-type versions of the same proteins. These accumulated to barely detectable levels and did not cause physiological changes, unlike the stabilized forms, which is in line with previous studies on ARX3/IAA17 (Worley et al, 2000). For this reason, we focused our study on qualitative and quantitative differences between stabilized proteins expressed in the same developmental context. In different transgenic lines expressing either bdl or shy2-2 protein, the amount of detectable Aux/IAA protein correlated quantitatively with the strength of the resulting phenotypes. However, bdl had a stronger effect on embryo and hypocotyl phenotypes, whereas shy2-2 had a stronger effect on root phenotypes. Taken together, the results indicate that the specificity of auxin response is generated by both transcriptional regulation and protein function.
Regulation of auxin response in embryogenesis by the MP and BDL or IAA13 pairs
Embryonic root initiation is promoted by MP/ARF5 and inhibited by stabilized BDL/IAA12 (Berleth and Jürgens, 1993; Hardtke and Berleth, 1998; Hamann et al, 1999, 2002; this study). MP and BDL mRNAs accumulate in the same cells of the young embryo, and the two proteins interact in the yeast two-hybrid assay (Hamann et al, 2002). In addition, MP-mediated activation of DR5∷LacZ expression in yeast is inhibited by BDL in the absence of accessory plant-specific factors, as shown here. Overexpression of MP rescues bdl mutant plants and conversely, bdl suppresses the floral defects caused by overexpression of MP (Hardtke et al, 2004). These results suggest that BDL counteracts MP in auxin response. Here we identified IAA13 as the functionally equivalent sister gene of BDL. IAA13 displayed the same expression pattern as BDL, and a genomic fragment of IAA13 carrying a bdl homologous mutation gave rootless seedlings at comparable frequencies. It is thus likely that both BDL and IAA13 need to be degraded in early embryogenesis for MP to promote root initiation (Figure 8).
Figure 8.
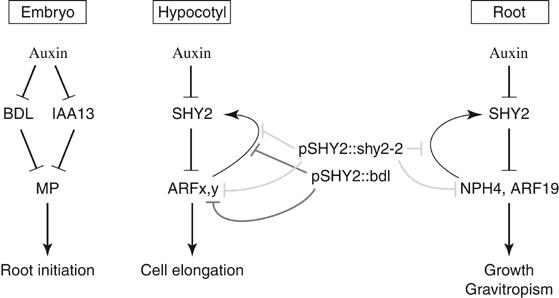
Model for Aux/IAA- and ARF-mediated auxin responses. In all tissues, auxin promotes the degradation of Aux/IAA proteins. In the embryo, BDL and IAA13 likely inhibit MP. When released from inhibition, MP promotes root initiation. In hypocotyl, SHY2 inhibits yet unknown ARF(s) (ARFx,y). When released from inhibition, ARFx,y promote cell elongation and enhance the transcription of SHY2. In root tips, SHY2 inhibits NPH4 and ARF19, which, when released, promote root growth and gravitropic response. NPH4 and ARF19 activity also stimulates SHY2 transcription. When expressed from the SHY2 promoter, shy2-2 mutant protein (pSHY2∷shy2-2) constitutively inhibits ARFx,y activity in the hypocotyl and NPH4 and ARF19 activity in the root, including feedback regulation of SHY2 transcription. Mutant bdl protein, expressed from the SHY2 promoter (pSHY2∷bdl), inhibits ARFx,y activity in the hypocotyl, but not NPH4 and ARF19 activity in the root.
To examine whether transcriptional regulation determines the specificity of auxin response in embryogenesis, we replaced BDL and MP by their distant relatives, SHY2 and ARF16, respectively. When expressed from the BDL promoter, the stabilized shy2-2 protein also caused rootless seedlings. However, syh2-2 was much less efficient than bdl. Similarly, ARF16 was much less effective than MP to rescue mp mutant embryos when expressed from the BDL promoter. Thus, specificity of auxin response in embryogenesis not only requires transcriptional regulation but also features of both ARF and Aux/IAA proteins, suggesting that MP and BDL or IAA13 are optimized ARF–Aux/IAA pairs.
Regulation of auxin response in the root by the SHY2 and NPH4 or ARF19 pairs
SHY2 plays a role in auxin response in the seedling root (Tian and Reed, 1999). However, no corresponding ARF has been identified. Here, we showed that SHY2 acts through inhibiting the redundant action of the previously known NPH4/ARF7 and the newly identified ARF19. Like shy2-2, double mutants lacking NPH4/ARF7 and ARF19 were defective in auxin-dependent root growth and root gravitropism, and also failed to activate the auxin-inducible SHY2 gene. To address whether, like BDL and MP in the embryo, SHY2 forms pairs with NPH4 and ARF19 for auxin responses in the root, we expressed stabilized bdl protein from the SHY2 promoter. In contrast to shy2-2, bdl did not interfere with root gravitropism, auxin responses and auxin-induced gene expression (Figure 8). Thus, BDL cannot replace SHY2 in the root at comparable expression levels. Hence, the contribution of protein determinants to auxin response specificity in the root is even more pronounced than in the embryo. This is also supported by the finding that heat shock promoter-driven expression of two stabilized Aux/IAA proteins, shy2-2 and axr3-1, in the root tip caused different responses of the same cell (Knox et al, 2003). Our findings imply that SHY2 forms optimized pairs with NPH4/ARF7 and ARF19 in the root. Furthermore, different optimized ARF–Aux/IAA pairs appear to regulate different developmental auxin responses.
Possible mechanisms of specificity
Auxin directly stimulates the interaction between Aux/IAA proteins and SCFTIR1 E3 ubiquitin-ligase complexes, resulting in the degradation of Aux/IAA proteins (Dharmasiri et al, 2003; Tian et al, 2002; Kepinski and Leyser, 2004). In this way, ARF transcription factors are released from inhibition and can regulate the expression of auxin-responsive target genes (Weijers and Jürgens, 2004). Thus, ARFs and interacting Aux/IAA inhibitors are prime candidates for converting the generic signal into a specific response. If any ARF could interact with any Aux/IAA protein, there would be more than 600 possible pairwise combinations. One way to reduce this complexity is to regulate ARFs and Aux/IAA gene transcription. Our results suggest that transcriptional regulation of ARF and Aux/IAA genes indeed contributes to specifying developmental responses to auxin. However, when expressed in the same developmental context, different members of the ARF and Aux/IAA protein families are functionally diverse and thus protein determinants also contribute to the specificity of response to auxin.
Auxin-dependent regulation of a target gene is the outcome of the activity of a DNA-bound ARF transcription factor, the activity of the associated Aux/IAA inhibitor and the affinity between the two. Transcriptional regulation by ARFs has been studied in cell cultures, using artificial reporter genes, but nothing is known about their natural targets in plant development. Their DNA-binding specificities for these target genes may differ, but this has not been addressed experimentally. The cell-culture studies suggest that ARFs differ in their activation potential, which has been related to sequences in the middle region (Tiwari et al, 2003). Similar to ARFs, Aux/IAA proteins could have differential effects. For example, their domain I has an LxLxL motif that, when fused to a DNA-binding domain, confers transcriptional repression of a reporter gene, and this motif is conserved between BDL and IAA13 but different in SHY2 (Tiwari et al, 2004).
Regarding interactions of ARFs with Aux/IAAs, all currently available data are from yeast studies, in particular two-hybrid studies. ARFs can activate auxin-responsive genes in yeast and their activity can be counteracted by Aux/IAAs, as shown here. Yet, none of these two-hybrid studies revealed differential interactions (Ouellet et al, 2001; Hamann et al, 2002; Hardtke et al, 2004; Tatematsu et al, 2004; this study). Although this might be taken to indicate that there are no differential interactions between ARFs and Aux/IAA proteins in the plant, there are caveats to yeast two-hybrid experiments that make this possibility unlikely. For example, the high levels of protein expression used in the yeast assays may allow for indiscriminate interactions between any protein of one family and any protein of the other family. Furthermore, yeast may lack factors that increase or prevent particular ARF–Aux/IAA interactions in the plant. Thus, direct ARF–Aux/IAA interaction needs to be analyzed in planta, which cannot be carried out at present. Considering the genetic evidence for optimized ARF–Aux/IAA pairs requiring both a specific ARF and a specific Aux/IAA, differential interactions are plausible.
In summary, we propose that the specificity of response to the generic signal auxin in different developmental and physiological contexts is generated at the level of interacting ARF–Aux/IAA proteins by two layers of control. First, transcriptional regulation of ARF and Aux/IAA genes limits the options of the responsive cell. Second, optimized pairs of interacting ARF and Aux/IAA proteins increase the specificity of response. The same ARF may be inhibited by one or more Aux/IAA proteins, depending on functional redundancy of duplicated Aux/IAA genes. It remains to be determined whether a single cell is capable of responding to auxin in several specific ways by expressing different optimized pairs of interacting ARF and Aux/IAA proteins.
Materials and methods
Plant material
bdl, shy2-2 and nph4-1 mutants have been described (Hamann et al, 1999; Tian and Reed, 1999; Harper et al, 2000). T-DNA insertion lines arf10-1 (SALK_143232), arf10-2 (SALK_087247), arf16-1 (SALK_021448), arf16-2 (SALK_021432), arf19-3 (SALK_021481) and arf19-4 (SALK_009879) were obtained from the Arabidopsis Biological Resource Center (Alonso et al, 2003). Double homozygotes for both alleles of arf10 and arf16 were identified in the F2 generation. Construction of nph4-1 arf19-3 and nph4-1 arf19-4 double mutants will be described elsewhere (JC Wilmoth and JW Reed, unpublished results).
Cloning of promoter∷GUS fusions and promoter swapping
pSHY2∷GUS was described previously (Hamann et al, 2002). Similarly, the IAA13 promoter was amplified by PCR as a 2.11 kb fragment upstream of the translational start (sense primer 5′CGCGAGCTCCTCCATCATTTATCTTCAACCA3′, antisense primer 5′CGCGGATCCCAGAGAGACCACAACAACAACA3′) and cloned in the plant transformation vector pVKH35sGUSpA (Hamann et al, 2002) using SacI/BamHI to result in pVKHIAA13GUSpA. The constructs/vectors pVKHSHY2GUSpA, pVKHBDLGUSpA (Hamann et al, 2002; contain 1.76 kb and 1.96 kb of sequence upstream of the ATG of SHY2 and BDL, respectively) or pVKHIAA13GUSpA were used to generate promoter fusions with genomic fragments spanning the complete coding sequence for SHY2, BDL or IAA13 genes, respectively, with 1.4 kb (SHY2), 1.95 kb (BDL) or 1.99 kb (IAA13) fragments downstream of ATG obtained by PCR amplification. SHY2-2, BDL and IAA13 coding sequences were amplified from genomic DNA of bdl, shy2-2 and Col-0, respectively. A sequence coding for an N-terminal c-Myc tag was added to each forward primer. A P80S mutation was introduced into IAA13 by PCR (primers: 5′ATAGGAGACCATCCAACAACTTG3′, 5′CAAGTTGTTGGATGGTCTCCTAT3′). Coding sequences were cloned in the plant transformation vectors pVKHIAA3GUSpA, pVKHIAA12GUSpA or pVKHIAA13GUSpA, respectively, by replacing the GUS coding sequence using BamHI/SalI restriction sites. Agrobacterium and Arabidopsis Col-0 ecotype plant transformations were performed as described (Hamann et al, 2002). Transgenic plants were selected using 15 mg/l hygromycin.
Growth conditions and phenotypic analysis
Plants were grown on a 16 h light/8 h dark cycle at 18 or 23°C. Exogenous drug application was performed by incubation of 7-day-old seedlings in liquid minimal salt media (2.1 g/l MS salts, 1% sucrose) supplemented with 10 μM IAA for 5 h.
For hypocotyl measurements, seedlings were grown on minimal salt media (0.7% agar) along the surface of vertical agar plates on a 16 h light/8 h dark cycle or in constant darkness. Hypocotyl lengths were measured from digital images taken after 7 or 5 days of growth.
For auxin resistance assays, seedlings grown for 5 days as described above were transferred to MS medium or MS medium supplemented with 0.1 μM 2,4-D. Root elongation was measured 3 days after transfer.
For gravitropism assays, seedlings were vertically grown for 7–9 days in a light/dark cycle, and then transferred to darkness and reoriented by 90°. The angle between the root tip and gravity vector was determined from digital images taken 24 h after reorientation.
Analysis of GUS expression and in situ hybridization
GUS expression and RNA in situ hybridization were performed as described (Hamann et al, 2002). The fragment for the IAA13 probe was amplified from cDNA by PCR using primers IAA13S (5′TCTGATCGATATGCTGGTTCATCTCCTCCTCG3′) and IAA13AS (5′GTCTCTCTAGAGGTTCTTGATTTCGAGCAGCGA3′) and subcloned in pBluescript SK+.
RT–PCR
For analysis of SHY2 expression, seedlings were incubated in control medium or medium containing 50 μM IAA at room temperature for 2 h. In one experiment, entire 3-day-old seedlings were used for RNA isolation using an RNeasy kit (QIAgen) according to the manufacturer's instructions. Poly(A)+-RNA was isolated using a Dynal mRNA DIRECT kit (Dynal, Oslo, Norway). cDNAs were synthesized using MMuLV-RT (Invitrogen), and real-time quantitative RT–PCR reactions were performed on an icycler machine (BIO-RAD). ACT2 (sense: 5′ATTCAGATGCCCAGAAGTCTTGTTC3′; antisense: 5′GCAAGTGCTGTGATTTCTTTGCTCA3′) and SHY2 (sense: 5′GTCGACGAATTCATGGGGGAGCAAAAGCTTATTTCTGAGGAGGATGATGAGTTTGTTAACC; antisense: 5′GAATTCGGATCCTCATACACCACAGCC3′) PCRs were performed on three different serial dilutions of each cDNA and each reaction was performed at least five times. In a separate experiment, cDNAs were synthesized from poly(A)+-RNA isolated from roots of 5- to 10-day-old seedlings as above, and PCR reactions for ACT2 and SHY2 were performed using different dilutions of cDNA. Intensities of bands on ethidium bromide-stained gels were quantified from scans using the ImageJ program, and normalized SHY2/ACT2 values are represented. RT–PCRs were performed five times on two independent RNA batches, and results were comparable in all experiments.
Analysis of protein expression
Protein extracts from 7-day-old seedlings or roots of 12-day-old seedlings were prepared by grinding and boiling in Laemmli buffer (Laemmli, 1970). Proteins were separated on polyacrylamide gels and transferred to PVDF membranes by Western blotting. Membranes were incubated with monoclonal anti-Myc antibody (9E10) at 1:600 dilution and then with alkaline phosphatase-conjugated anti-mouse secondary antibody at dilution 1:5000, and signals were detected with the Western Star detection kit (Tropix). For Western blot analysis of yeast cells, crude protein extracts were prepared by boiling pelleted yeast cells in 4 volumes of Laemmli buffer. Proteins were separated, blotted and incubated with 1:1000 diluted horseradish peroxidase (POD)-coupled anti-HA monoclonal antibody (Sigma), and washed and detected according to the manufacturer's recommendations.
Yeast assays
Yeast (Saccharomyces cerevisiae) strain YPH500 was transformed with plasmid yDR5∷LacZ containing a direct (8 ×) repeat of the DR5-rev sequence (Ulmasov et al, 1997b), fused to a minimal CYC1 promoter and the lacZ coding sequence in pLacZi (Clontech). Two to four independent yDR5∷LacZ transformants were used in all experiments. yDR5∷LacZ strains were transformed with pESC-TRP (Stratagene) plasmids containing either C-terminal HA-epitope-tagged cDNAs of ARF5/MP alone, or in combination with C-terminal HA-epitope-tagged cDNAs of IAA3/SHY2 or IAA12/BDL. All cDNAs were amplified by PCR and sequenced. Approximately 12 yeast colonies per plasmid were grown overnight in minimal medium lacking tryptophan and containing 2% glucose. Cultures were then diluted to a final OD600 of 0.2 in the same medium now containing 2% raffinose and 2% galactose for GAL1/10 promoter induction. Galactosidase activity was measured after 16 h induction according to Meijer et al (2000). All experiments were repeated at least three times and gave comparable results. For yeast two-hybrid experiments, entire SHY2 or BDL open reading frames were amplified from wild-type cDNA. C-terminal regions of MP (aa 777–902) and ARF19 (aa 948–1086) were PCR-amplified from cDNA libraries (Grebe et al, 2000). PCR products were subcloned into pGEM-T and subsequently cloned in-frame with LexA in pEG202 and with B42 in pJG4-5. Yeast strain EGY48 was first transformed with plasmid pSH18-34, and subsequently cotransformed with all possible combinations of pEG and pJG plasmids (in both directions). Experiments with Aux/IAA proteins as LexA fusions gave more consistent results, and are presented here. Galactosidase activity was quantified in cultures from at least 12 colonies for each plasmid combination in each of five experiments.
Complementation of monopteros
MP was C-terminally HA epitope tagged by exchanging a 3′ region of the cDNA (MunI–AflII) with a corresponding PCR fragment containing a C-terminal HA tag (primers: MpcMunI-S, 5′GATCAATTGATGTCACAAGCTTTAAAGAC3′ and MpcHAAflII-AS, 5′GCTTAAGAGCATAATCAGGAACATCATAAGGATAATCGTTAATGCCTGC3′). The HA-tagged cDNA was then used to replace the BDL coding region in a 4.5 kb genomic BDL fragment in pGreenII0229 (Roger et al, 2000) with BamHI and SpeI sites generated at start and stop codons, respectively. The ARF16 cDNA was amplified from a cDNA library with primers ARF16-CDS-S (5′CCGGATCCGAATTCATGATAAATGTGATGAATCC3′) and ARF16-CDS-HA-AS (5′CCACTAGTTTAAGCATAATCAGGAACATCATAAGGATATACTACAACGCTCTCAC3′), the latter containing a C-terminal HA tag. The PCR fragment was used to replace the BDL coding region in pGreenII BDL as described above. Constructs were transformed into mpB4149 (Columbia ecotype; a gift from B Scheres) heterozygotes, and frequencies of rootless seedlings were determined in T2 seedlings from transgenics carrying the mpB4149 allele.
Acknowledgments
We thank Claus Schwechheimer and Jarmo Schrader for helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (GJ; DFG 179/9-1, SFB 446/B8), the Margarete von Wrangell-Habilitationsprogramm (EB) and the European Molecular Biology Organization (DW; ALTF 582-2001).
References
- Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587 [Google Scholar]
- Berleth T, Sachs T (2001) Plant morphogenesis: long-distance coordination and local patterning. Curr Opin Plant Biol 4: 57–62 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M (2003) Auxin action in a cell-free system. Curr Biol 13: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUU/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jürgens G (2000) A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12: 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2004) Auxin-induced SCFTIR1–Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA 101: 12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7: 193–204 [DOI] [PubMed] [Google Scholar]
- Liscum E, Briggs WR (1996) Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol 112: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AH, Schouten J, Ouwerkerk PBF, Hoge JHC (2000) In Plant Molecular Biology Manual, SB Gelvin, RA Schilperoort (eds) E3 pp 1–28. Dordrecht, the Netherlands: Kluwer Academic Publishers [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC (2000) Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, Onodera C, Quach H, Smith A, Yu G, Theologis A (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3. Biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200: 229–237 [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger P, Hellens E, Edwards A, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC (1997) ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian CE, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT (2004) Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J 40: 333–343 [DOI] [PubMed] [Google Scholar]
- Tian Q, Nagpal P, Reed JW (2003) Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J 36: 643–651 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997a) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki MK, Yamamoto KT (1997) The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol 115: 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Jürgens G (2004) Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol 7: 687–693 [DOI] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21: 553–562 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCF. Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]


