Abstract
Dynamin-related protein 1 (DRP1) plays crucial roles in mitochondrial and peroxisome fission. However, the mechanisms underlying the functional regulation of DRP1 in adipose tissue during obesity remain unclear. To elucidate the metabolic and pathological significance of diminished DRP1 in obese adipose tissue, we utilized adipose tissue–specific DRP1 KO mice challenged with a high-fat diet. We observed significant metabolic dysregulations in the KO mice. Mechanistically, DRP1 exerts multifaceted functions in mitochondrial dynamics and endoplasmic reticulum (ER)-lipid droplet crosstalk in normal mice. Loss of function of DRP1 resulted in abnormally giant mitochondrial shapes, distorted mitochondrial membrane structure, and disrupted cristae architecture. Meanwhile, DRP1 deficiency induced the retention of nascent lipid droplets in ER, leading to perturbed overall lipid dynamics in the KO mice. Collectively, dysregulation of the dynamics of mitochondria, ER, and lipid droplets contributes to whole-body metabolic disorders, as evidenced by perturbations in energy metabolites. Our findings demonstrate that DRP1 plays diverse and critical roles in regulating energy metabolism within adipose tissue during the progression of obesity.
Supplementary key Words: lipids, dyslipidemias, lipid droplets, obesity, mitochondria, nutrition
Overnutrition combined with a sedentary lifestyle is a major contributor to the global increase in obesity prevalence and its associated metabolic derangements. Adipose tissue acts as a storage place for excess calories and can expand significantly in response to overnutrition (1). However, unhealthy expansion of adipose tissue can lead to dysfunctional adipose tissue that secretes abnormal hormones, lipids, and adipokines (2). This can cause local inflammation and fibrosis, ultimately resulting in the development of metabolic diseases such as type 2 diabetes, cardiovascular diseases, nonalcoholic fatty liver disease, and certain types of cancer (2, 3, 4).
There are two types of adipose tissue: white (WAT) and brown adipose tissue (BAT) (1). While BAT was once thought to only be present in rodent animal models and infant humans, multiple studies show that normal BAT activity is crucial for systemic metabolisms, such as fatty acid oxidation and diet-induced thermogenesis (5). This is because BAT mitochondria are highly condensed with uncoupling protein 1 (UCP1), a thermogenic factor (5). The term “browning” of adipose tissue describes the dynamic and reversible transformation of WAT into a more BAT-like appearance and function, at the gene, protein, and phenotypic levels. It is worth noting that various strategies, including cold exposure and physical activity, can induce the "browning" of WAT, and can be harnessed as a way to combat obesity (6). Conversely, during the development of diet-induced obesity, nutritional stresses may cause the "whitening" of BAT. This is characterized by larger and more unilocular lipid droplets, as well as impaired mitochondrial function (7).
The structure of mitochondria is tightly associated with the energy balance between demand and supply, and the mitochondrial life cycle plays a crucial role in maintaining the organellar components in a healthy state, including proper bioenergetic adaptation, heat generation, and ATP production (6, 8). Mitochondrial dynamics include fission, fusion, and mitophagy, which are regulated by complex molecular mechanisms involving multiple factors (8, 9). Among all the mitochondrial dynamic factors, dynamin-related protein 1 (DRP1) is a classic mitochondrial fission factor that, depending on the level of fission/fusion and physiological/pathological conditions, can play dual roles in protecting or impairing cellular functions in multiple tissues/organs (10, 11, 12, 13, 14). Importantly, these dynamics have been implicated in obesity, aging, and metabolic diseases (14, 15, 16, 17). Particularly, obesity disrupts mitochondrial dynamics by interfering with the balance between fission and fusion, leading to muscle atrophy and sarcopenia (18, 19).
In addition to mitochondrial fission, DRP1 plays multifaceted functions on dynamic remodeling of other organelles, such as peroxisome fission and neuronal cell endocytosis (20, 21). The peroxisome function is dependent on its constricting and severing ability, and the GTPase activity is key for its fussogenic function (20). Recent studies have shown that DRP1 translocates onto the endoplasmic reticulum (ER) in response to different cell stimuli (22, 23). Specifically, DRP1 is phosphorylated at Ser 616 by PKA under transient β-adrenergic stimulation, and this phosphorylation event facilitates its translocalization onto ER (23). Interestingly, DRP1 enhances mitophagy and is involved in alternative mitophagy during long-term high-fat diet (HFD) conditions in cardiomyocytes. This process also requires phosphorylation of DRP1 at Ser616, which leads to its translocation onto the ER membrane, where it interacts with protein factors involved in alternative mitophagy (14).
Despite extensive studies on the regulation of DRP1's function in other metabolically active tissues like the liver, muscle, and cardiovascular system, its regulation in response to nutritional stress in both WAT and BAT is not yet fully understood. In the current study, we observed the changes in DRP1 and other mitochondria fusion/fission factors during diet-induced obesity. We used an adipose tissue specific DRP1 KO mouse model to study its critical role in regulating the function of mitochondria, ER, and lipid droplets, and the underlying mechanisms. Additionally, we further characterized and evaluated the metabolic consequences that arise in the absence of DRP1 in combination with diet-induced obesity. Our findings emphasize the significant roles of DRP1 in organelle communication and metabolic adaptation under diet-induced nutritional stress conditions.
Materials and Methods
Animal and diet-induced obesity study
The animal experiments conducted in this study were reviewed and approved by the Animal Welfare Committee of the University of Texas Health Science Center at Houston (AWC-21-0019). The mice used in the experiments were housed in the animal facility under a 12-h light-dark cycle at 22 ± 1°C. They had access to water and were fed with a regular diet unless otherwise specified. Two strains of mice, C57BL/6J (Strain # 000664) and Adipoq-Cre (Strain # 010803), were purchased from The Jackson Laboratory. Drp1 flox/flox mouse was generated by Dr Hiromi Sesaki at the University of Johns Hopkins. To generate adipocyte-specific Drp1 knockout (Adipoq-Cre; Drp1flox/flox, referred to as KO) mice, the Adipoq-Cre strain was crossed with Drp1 flox/flox strain. The Drp1flox/flox mice from the littermates were used as controls and from here on referred to as WT. For the diet-induced obesity study, 8-week-old male mice were fed by a HFD (60 kcal% fat, D12492i, Research Diets) for 10 weeks.
Measurement of body weights and body composition
For the diet-induced obesity study, the bodyweights of the HFD-fed mice were meticulously monitored on a weekly basis over the course of 10 weeks. At the initiation and conclusion of the HFD regimen, their body composition—encompassing total body mass, fat mass, and lean mass—was assessed using an Echo 3-in-1 nuclear magnetic resonance minispec MRI instrument manufactured by EchoMRI in Houston, TX.
Glucose tolerance test and insulin tolerance test
The glucose tolerance test and insulin tolerance test (ITT) were performed according to the protocol as previously described (24). For glucose tolerance test, the mice were not fed for 6 h before receiving an injection of glucose (2.5 g/kg of body weight) via intraperitoneal injection. For the ITT, the mice were not fed for 4 h before receiving an injection of insulin (0.75 unit/kg of body weight for lean mice and 1 unit/kg of body weight for obese mice) via intraperitoneal injection. Blood was collected by cutting off 1 mm of the tail tip at the indicated time points and the blood glucose level was measured by glucose m strips.
Cell culture
All cells were cultured at 37°C in a 5% CO2 atmosphere in a Thermo Forma incubator. The stromal vascular fraction (SVF) derived from adipose tissue was obtained and cultured following the procedure outlined in previous study (25). Briefly, adipose tissue was obtained from 8-week-old male mice and minced on ice. The minced tissue was then digested in collagenase solution (0.8 mg/ml collagenase type I, 2.5 mg/ml dispase, 2 mM CaCl2 in Hank’s balanced salt solution) at 37°C with gentle agitation for 30 min. The digestion solution was neutralized using complete growth medium (DMEM supplemented with 10% fetal bovine serum, antibiotic-antimycotic, and 10 mM Hepes) and passed through a 70 μm strainer to remove any undigested tissue debris. The resulting cell solution was centrifuged at 500 g at 4°C for 15 min to pellet the SVF which was resuspended in the complete growth medium and directly plated into the experimental vessels. Once the SVF was cultured to confluence, the growth medium was changed to brown induction medium (growth medium supplemented with 1 mM dexamethasone, 5 g/ml insulin, 0.5 mM 3-isobutyl-1-methylxanthine, 5 μM rosiglitazone, and 1 nM 3,5,3′-triiodothyronine) for 2 days, followed by differentiation medium (growth medium with 5 g/ml insulin and 1 nM 3,5,3′-triiodothyronine) for 5 more days. The DRP1 KO efficiency of the differentiated adipocyte was verified by Western blot.
HeLa cells were obtained from American Tissue Culture Collection and cultured in complete DMEM media. The DRP1 KO HeLa cell line was generated using CRISPR/Cas9 KO plasmid (Santa Cruz, sc-400459) and the single-cell colony was selected by GFP marker that coexpressed in the CRISPR/Cas9 KO plasmid. The DRP1 KO efficiency of the single-cell colony was tested by Western blot.
Isolation of mitochondria
Mitochondria were isolated from the BAT of normal chow diet fed mice. The fresh BAT was minced into pieces and homogenized in cold MSHE buffer (70 mM sucrose, 210 mM mannitol, 5 mM Hepes, and 1 mM EDTA) containing 4% fatty acid–free BSA. The lysate was centrifuged at 800 g at 4°C for 10 min to remove intact cells and cell debris. Then, the supernatant was subjected to a second centrifugation at 8000 g at 4°C for 15 min to pellet the mitochondria. The mitochondria pellet was then resuspended in MAS-1 buffer (70 mM sucrose, 220 mM mannitol, 5 mM K2PO4, 5 mM MgCl2, 2 mM Hepes, 1 mM EGTA, and 0.2% fatty acid–free BSA, at pH 7.2) for further analysis. The protein concentration of the isolated mitochondria was determined using a BCA kit from Thermo Fisher Scientific.
Measurement of mitochondrial respiration
The Seahorse XFe24 Analyzer from Agilent Technologies was used to determine the oxygen consumption rate (OCR) according to the manufacturer's protocols. After a 7-day differentiation period, adipocytes derived from SVF were subjected to the Cell Mito Stress Test (MST). For the Cell MST, XF DMEM supplemented with 10 mM glucose, 2 mM glutamine, and 1 mM pyruvate was used as the assay medium and 2 μM oligomycin, 2 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, and 5 μM rotenone/15 μM antimycin A were subsequently injected into the wells. In addition, 100 μM BSA-conjugated palmitic acid (PA) was added to the assay medium 2 h before the MST analysis and was present during the test. To measure the degree of coupling in isolated mitochondria, 5 μg/well mitochondria were coated into the XF24 cell culture microplate by centrifugation 2,000 g at 4°C for 20 min in MAS-1 buffer. MAS-1 buffer supplemented with 10 mM succinate was used as the assay buffer, and 4 mM ADP, 2 μM oligomycin, 4 μM carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, and 4 μM antimycin A were subsequently injected into the wells. The key parameters of mitochondrial respiration were generated in the Seahorse Wave software.
Metabolic substrate assay in mitochondria
The method used to determine mitochondrial substrate metabolism was followed according to the instructions in the MitoPlate S-1 Assay kit (Cat. #14105, Biolog), which has been described in a previous study (26). To perform the assay, 20 μg of freshly isolated mitochondria were mixed with Redox Dye and then added to the wells of the MitoPlate S-1 that had been precoated with diverse mitochondrial substrates. The addition of electrons from the mitochondrial electron transport chain caused the Redox Dye to turn purple. The plate was then placed in the Cytation 5 Cell Imaging Reader (Agilent) at 37°C and the rate of purple color development was recorded at A 590 for 4 h. The MitoPlateS-1 96-well plate was pre-coated with a set of diverse substrates (supplemental Figs. S4A and S6), including glucose, fatty acids, and amino acids, which can be utilized by mitochondria respiration. The generation of energy-rich NADH or flavin adenine dinucleotide (FADH2) by the mitochondria reduces the redox dye and brings about a color change that can be read. Functional mitochondria were isolated from the BAT of WT and KO mice, and loaded to the MitoPateS-1 plate to incubate at 37°C. By monitoring the real-time color change and comparing the color-changing rate between WT and KO mitochondria, substrate preference was determined.
ER extraction
The ER was isolated from BAT by using the ER Isolation Kit (Cat. # ER0100, Sigma-Aldrich). The process involved homogenizing 0.5 g of the tissue in 2 ml of 1x isotonic extraction buffer and sequentially centrifuging the samples at 1,000 g to remove cell debris, followed by centrifuging at 12,000 g to remove mitochondria. After that, the clear supernatant fraction and the top lipid layer were collected and subjected to ultracentrifugation at 100,000 g for 1 h. The resulting ER pellet was suspended in 600 μl of isotonic extraction buffer for further analysis.
Biochemical tests
Serum nonesterified FFA levels were measured using the FFA assay kit (Cat. # EFFA-100, BioAssay Systems). Triglyceride (TG) levels in serum, tissue, and ER were measured with the TG assay kit (Cat. # ab65336, Abcam). Serum samples were collected from 6-h fasted mice, while tissues and ER samples were prepared as per the manufacturer's instructions. The TG level in tissues and ER was normalized to the sample protein concentration.
Tracking lipid droplet formation and ER labeling
Tracking the nascent lipid droplet dynamics in ER has been previously described (23). Briefly, BODIPY FLC12 (green fluorescent–labeled C12 FFAs) (at a final concentration of 5 μM, Cat. #D3822, Invitrogen) and ER-tracker red (at a final concentration of 1 μM, Cat. #E34250, Invitrogen) were used to label cells and tissues. Fresh adipose tissue explants were stained with BODIPY-C12 for 30 min and then labeled with ER-tracker red in HBSS buffer for 1 h. Images of adipose tissues were acquired using a Leica TCS SP5 Confocal Laser Scanning Microscope. HeLa cells were stained with BODIPY-C12 in complete growth medium for 10 min, washed with PBS twice, and then labeled with ER-tracker red in HBSS buffer for 30 min. After that, the cells were fixed and imaged using Nikon nSIM super-resolution microscopy.
Quantitative-PCR analysis
Quantitative-PCR (qPCR) analysis was previously described (25). To summarize, total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Cat. #15596026) and Illustra RNA spin Mini Kit (Cat. #25050072, GE Healthcare Life Sciences). Then, 1 μg of total RNA was reverse-transcribed into cDNAs with RevertAid Reverse Transcription Kit (Cat. #K1691, Thermo Fisher Scientific). qPCR was performed on a Bio-Rad CFX96 system (Bio-Rad Laboratories). The results were calculated using the 2-ΔΔCt method and normalized by endogenous b-actin. Sequences of the used primers are previously described or listed in supplemental Fig. S7.
Western blot
The preparations of tissue samples have been previously described (25). The protein samples were separated using an SDS gel and transferred onto PVDF membranes. The membranes were then blocked with 5% BSA in 1 × Tris-buffered saline with Tween 20 (TBST). After that, the membranes were incubated with primary antibodies in 1% BSA in 1 × TBST at 4°C overnight. The blots were probed with IR-Dye 800CW or 680RD secondary antibodies (LI-COR) and then visualized with the Odyssey Imaging System (LI-COR) after being washed three times with 1 × TBST. Anti-β-ACTIN antibody was used to control for equal loading. The following antibodies were used: Drp1 (Abcam, ab18274), p-DRP1 (Ser600/637) (#4867, Cell Signaling Technology), HSL (#4107, Cell Signaling Technology), PLIN3 (#ab47639, Abcam), PLIN1 (#ab61682, Abcam,), BIP (#PA1-014A, Thermo Fisher Scientific,), ATGL (#sc-36527, Santa Cruz Biotechnology), CGI58 (#sc-365278, Santa Cruz Biotechnology), β-ACTIN (#61256, BD Biosciences), and UCP1 (#ab10983, Abcam).
Immunofluorescent staining
The process of preparing paraffin slides was previously described in a report by Zhao et al. in 2016 (25). To prepare the sections of subcutaneous WAT (sWAT) and BAT, they were first deparaffinized and permeabilized for 10 min in PBS containing 0.2% Triton ×-100. After three washes with PBS, the sections were incubated in sodium citrate buffer at 95°C for 30 min to retrieve antigens. Following this, the sections were blocked using 5% BSA in 1x PBST (1 × PBS containing 0.1% Tween 20) at room temperature for 1 h and then incubated with primary antibodies overnight at 4°C. After washing three times with 1x PBST, the sections were incubated with fluorescent signal–conjugated secondary antibodies at room temperature for 1 h. The sections were then washed three times with 1 × PBST and rinsed with double distilled water before being mounted with Diamond Antifade Mountant (Cat. #P36970, Thermo Fisher Scientific) for imaging using confocal microscopy.
Transmission electron microscopy
Transmission electron microscopy (TEM) was performed using the BATs from the normal chow diet (NCD)-fed mice. The mice were anesthetized and perfused with 4% paraformaldehyde (PFA) in 1 × PBS, 2% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2). Then, the BAT was dissected, cut into small pieces, and fixed in 4% PFA, 2% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) for 48 h, followed by a secondary fixation in 1% OsO4 for 2 h. Subsequently, the tissue samples were dehydrated in graded ethanol/propylene oxide series and embedded in Embed-812 resin (Electron Microscopy Science, PA) for 72 h. After being cured in the blocks, the sample blocks were sectioned using a Leica UC7 microtome followed by staining with 1% uranyl acetate and 2.5% lead citrate. The images were acquired using a JEOL JEM 1010 transmission electron microscope with an AMT XR-16 mid-mount 16-megapixel camera. For HeLa cells, the cells were fixed in 4% PFA, 2% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) for 48 h, followed by a secondary fixation in 1% OsO4 for 2 h. Then, they were processed with same next steps as above.
Quantification of ER-lipid droplet colocalization
ImageJ software was used to analyze lipid droplets based on particle selection, by setting specific parameters for particle size and circularity. The size of the dots was set to 200–500, and circularity discrimination was set to 0.50–1.00. The outline selection and exclude on edges functions were used to summarize the particle count. Colocalized dots were counted manually, and the colocalization ratio was calculated by dividing the number of colocalized dots by the total number of dots.
Metabolomic analyses of mouse whole blood samples
Polar metabolites were extracted from mouse whole blood samples using ice-cold 80/20 (v/v) methanol/water with 0.1% ammonium hydroxide. Extracts were centrifuged at 17,000 g for 5 min at 4°C, and supernatants were transferred to clean tubes, followed by evaporation to dryness under nitrogen. Dried extracts were reconstituted in deionized water, and 10 μl was injected for analysis by ion chromatography-mass spectrometry. Ion chromatography mobile phase A (MPA; weak) was water, and mobile phase B (MPB; strong) was water containing 100 mM KOH. A Thermo Scientific Dionex ICS-6000+ system included a Thermo IonPac AS11 column (4 μm particle size, 250 × 2 mm) with column heated at 35°C. The autosampler tray was chilled to 4°C. The mobile phase flow rate was 360 μl/min, and the gradient elution program was: 0–5 min, 1% MPB; 5–25 min, 1%–35% MPB; 25–39 min, 35–99% MPB; 39–49 min, 99% MPB; and 49%–50%, 99%–1% MPB. The total run time was 50 min. Methanol was delivered by an external pump and combined with the eluent via a low dead volume mixing tee. Data were acquired using a Thermo Orbitrap IQ-× Tribrid Mass Spectrometer under ESI negative ionization mode.
For carnitine analysis, the same samples were analyzed by hydrophilic interaction liquid chromatography tandem mass spectrometry. MPA was 95/5 (v/v) water/acetonitrile containing 20 mM ammonium acetate and 20 mM ammonium hydroxide and MPB was acetonitrile. A Thermo Vanquish LC system included a Xbridge BEH Amide column (3.5 μm particle size, 100 × 4.6 mm) with column compartment kept at 30°C. The autosampler tray was chilled to 4°C. The mobile phase flow rate was 300 μl/min, and the gradient elution program was 0–3 min, 85% MPB; 3–10 min, 85%–30% MPB; 10–20 min, 30%–2% MPB; 20–25 min, 2% MPB; and 26–30 min, 2%–85% MPB. The total run time was 30 min. Data were acquired using a Thermo Orbitrap Exploris 240 Mass Spectrometer under ESI positive ionization mode at a resolution of 240,000.
For CoA analysis, the same samples were analyzed by C30-MS using a Thermo Vanquish LC system containing a Thermo Accucore C30, 250 × 2.1 mm column with 2.6 μm particle size. MPA was 5 mM ammonium acetate. MPB was methanol. The flow rate was 150 μl/min (at 35°C), and the gradient conditions were initial 5% MPB, increased to 95% MPB at 5.5 min, held at 95% MPB for 9 min, returned to initial conditions, and equilibrated for 5 min. The total run time was 20 min. Data were acquired using a Thermo Orbitrap Exploris 240 mass spectrometer under ESI positive ionization mode at a resolution of 240,000.
All raw data files were imported into Thermo Trace Finder 5.1 software for analysis, and the relative metabolite levels of metabolites were normalized by blood volume or total peak intensity.
Untargeted lipidomics analysis
Ten microliters of serum sample was used for untargeted lipidomics analysis. Serum samples were extracted by adding 100 μl of ice-cold ethanol containing 1% 10 mM butylated hydroxytoluene and 2% Avanti SPLASH® LIPIDOMIX® Mass Spec Standard. The samples were vortexed for 10 min, placed on ice for 10 min, and centrifuged at 17,000 g for 10 min at 4°C. The supernatants were then collected for LC-MS analysis. MPA was 40:60 acetonitrile: water with 0.1% formic acid and 10 mM ammonium formate. MPB was 90:9:1 isopropanol:acetonitrile: water with 0.1% formic acid and 10 mM ammonium formate. The chromatographic method included a Thermo Fisher Scientific Accucore C30 column (2.6 μm, 150 × 2.1 mm) maintained at 40°C, a mobile phase flow rate of 0.200 ml/min, an autosampler tray chilling at 8°C, and a gradient elution program as follows 0–3 min, 30% MPB; 3–13 min, 30–43% MPB; 13.1–33 min, 50%–70% MPB; 33–48 min, 70%–99% MPB; 48–55 min, 99% MPB; and 55.1–60 min, 30% MPB. The injection volume was 10 μl. A Thermo Fisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer with heated electrospray ionization source was operated in data-dependent acquisition mode, in both positive and negative ionization modes, with scan ranges of 150–1500 m/z. An Orbitrap resolution of 240,000 (full width at half maximum) was used for MS1 acquisition and spray voltages of 3,600 and −2900 V were used for positive and negative ionization modes, respectively. Vaporizer and ion transfer tube temperatures were set at 275°C and 300°C, respectively. The sheath, auxiliary, and sweep gas pressures were 35, 10, and 0 (arbitrary units), respectively. For MS2 and MS3 fragmentation, a hybridized higher-energy collisional dissociation/collision-induced dissociation approach was used. Data were analyzed using Thermo Scientific Lipid-Search software (version 5.1) and R scripts written in house.
Statistical analysis
The data is presented as Mean ± SEM. Statistical analyses were performed using GraphPad Prism 9 software (GraphPad Software Inc.). The statistical differences were analyzed using a two-tailed Student’s t test or one-way ANOVA, followed by Tukey's post hoc test for multiple groups. A P value less than 0.05 was considered statistically significant.
Results
DRP1 and other mitochondrial fission/fusion factors are dynamically regulated in adipose tissue during diet-induced obesity development
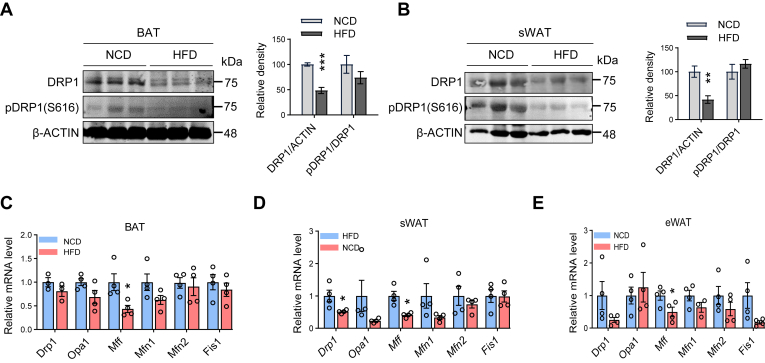
In this study, we investigated the role of DRP1 in metabolic regulation in adipose tissue during obesity. We first examined the dynamic changes in its total and phosphorylated protein levels in adipose tissues. We conducted experiments using WT C57/BL6 mice fed a HFD for 10 weeks and analyzed the protein levels of DRP1 in various adipose tissues. The results showed a significant decrease in both phosphorylated and total levels of DRP1 in sWAT and BAT of mice fed the HFD, in comparison to their littermate controls fed a NCD (Fig. 1A, B). Of note, the decrease in phosphorylation is attributed to a decrease in the total protein levels under both diet conditions. qPCR results further confirmed a significant downregulation of the mRNA level of the Drp1 gene under the HFD feeding condition in WAT, while revealing a downregulation trend in BAT (Fig. 1C–E). Intriguingly, other mitochondrial fission/fusion factors, such as Opa1, Mff, Mfn1/2, and Fis1 also showed divergent regulations on mRNA levels in the HFD-induced obese adipose tissues. Among them, Mff was significantly downregulated, and other factors did not show significant changes (Fig. 1C–E). In summary, these findings highlight the impact of HFD on DRP1 protein levels and gene expression in different adipose tissues, suggesting a potential link between DRP1-induced mitochondrial dysfunction and obesity-related metabolic changes.
Fig. 1.
DRP1 and other mitochondrial dynamic factors are regulated in adipose tissue during diet-induced obesity development. (A, B) Representative western blots of DRP1 and p-DRP1 (p-Ser616) protein levels in the brown adipose tissues (BATs) (A) and subcutaneous white adipose tissues (sWATs) (B) of normal chow diet (NCD) and high-fat diet (HFD) fed mice along with densitometric analysis (n = 3/group). (C–E) qPCR analysis of the mitochondrial dynamic-related genes in BAT, sWAT, and eWAT of NCD and HFD-fed mice (n = 4/group). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, unpaired Student’s t test. DRP1, dynamin-related protein 1; eWAT, epididymal WAT; qPCR, quantitative-PCR.
DRP1 KO in adipose tissue leads to impaired glucose and lipid metabolism
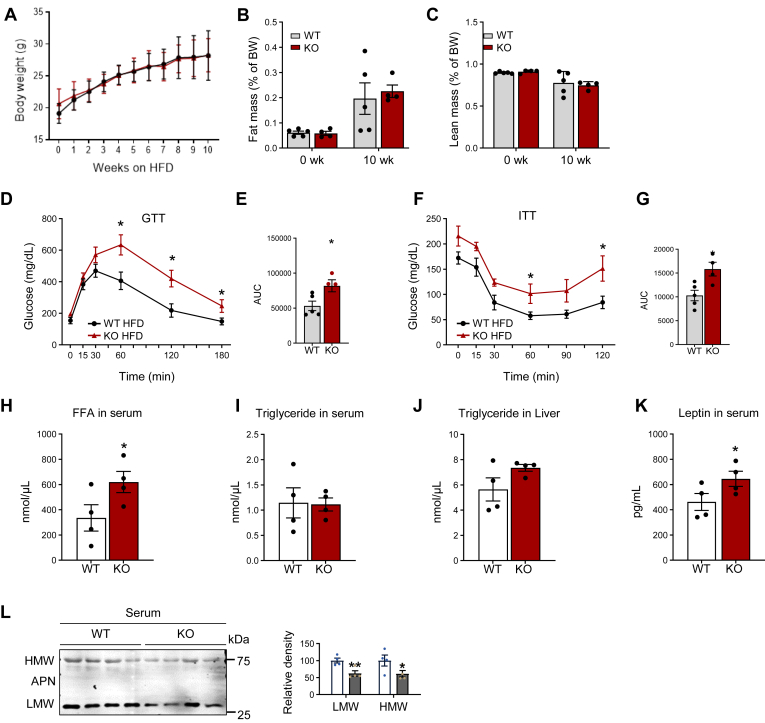
To address the specific role of DRP1 in adipose tissue remodeling under various metabolic conditions, we created an adipose tissue–specific Drp1 KO mouse model (referred to as Drp1 fat tissue specific KO or KO) by crossing the adiponectin-Cre strain with the Drp1fl/fl strain (23). In this study, we subjected both KO mice and their littermate controls (refer to as WT) to a HFD for 10 weeks. Throughout the HFD feeding period, we monitored the changes in body weights and body composition of KO mice. Surprisingly, we observed no significant alterations in either body weight or body composition when compared to the WT controls (Fig. 2A–C). However, the KO mice exhibited severely impaired glucose tolerance and insulin sensitivity, as evidenced by an intraperitoneal glucose tolerance test, and an ITT (Fig. 2D–G). Importantly, these changes in glucose tolerance were not observed in lean KO mice when fed by an NCD (supplemental Fig. S1A–D). This unexpected finding under HFD highlights the crucial involvement of DRP1 in glucose metabolism regulation within adipose tissue, even in the absence of significant changes in body weight or composition.
Fig. 2.
DRP1 knockout in adipose tissue leads to impaired glucose and lipid metabolism. (A) Measurement gains of bodyweight of Adipoq-Cre; Drp1flox/flox (KO); and their Drp1flox/flox WT littermates during HFD feeding for 10 weeks (n = 5/group). (B, C) MRI analysis for fat and lean mass of WT and KO mice before and after HFD feeding for 10 weeks (n = 5/group). (D, E) Glucose tolerance test (GTT) with the area under the curve (AUC) in HFD-fed WT and KO mice (n = 5/group). (F, G) Insulin tolerance test (ITT) with AAC in HFD-fed WT and KO mice (n = 5/group). (H, I) The levels of FFAs and triglyceride (TG) in fasted serum of HFD-fed WT and KO mice (n = 4/group). (J) The levels of triglyceride in the liver of HFD-fed WT and KO mice (n = 4/group). (K) The levels of leptin in the serum of HFD-fed WT and KO mice (n = 4/group). (L) Representative Western blots and densitometric analysis of protein levels of adiponectin (APN) in the serum of HFD-fed WT and KO mice (n = 4/group). ∗P < 0.05 and ∗∗P < 0.01, unpaired Student’s t test. DRP1, dynamin-related protein 1; HFD, high-fat diet.
To further investigate the metabolic consequences resulting from the loss of function of DRP1 in adipose tissue, we evaluated the circulating levels of lipids and metabolism-related cytokines/hormones. Our results revealed a significant increase in the levels of FFAs in serum (Fig. 2H), while the level of TGs remained unchanged (Fig. 2I). Additionally, we observed a trend of elevated TG levels in the livers of HFD-fed KO mice (Fig. 2J). Notably, there was a substantial reduction in adiponectin levels (both high molecular and low molecular adiponectin), and an increase in leptin levels in serum of the HFD-fed KO mice (Fig. 2K, L). Of note, we did not observe the changes outlined above in the chow-diet fed KO mice (supplemental Fig. S2B–F).
Deficiency of DRP1 induces “whitening” of BAT during obesity
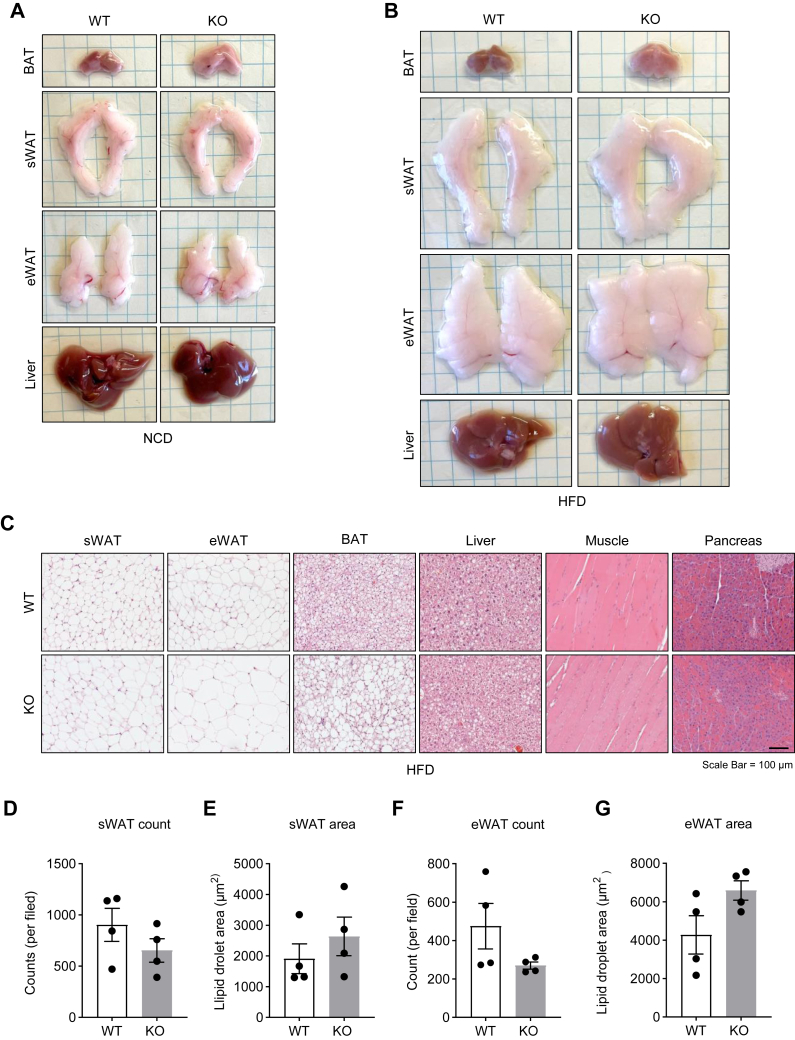
To determine the impact of DRP1 ablation on the morphology of adipose tissue and other metabolic tissues, we conducted histological investigations. We observed that sWAT and epididymal WAT in KO mice exhibited similar fat masses as in the control mice. In contrast, BAT was significantly larger in the KO mice (Fig. 3A, B). Notably, this effect was more pronounced under HFD conditions (Fig. 3B). H&E staining revealed that although the size of adipocytes in WAT did not differ significantly in the KO mice, BAT exhibited a notable "whitening" effect, characterized by much larger unilocular lipid droplets. Remarkably, this effect was more prominent under HFD feeding conditions (Fig. 3C and supplemental Fig. S2A). Moreover, the sizes of lipid droplets did not show significant differences in WAT, even under HFD feeding condition (Fig. 3D–H). Notably, we did not observe more severe lipid accumulation in other metabolic tissues, such as the liver, skeletal muscles, and pancreas in the KO mice (Fig. 3C).
Fig. 3.
DRP1 knockout in adipose tissue induces “whitening” of BAT. (A) Representative images of fresh adipose tissues and liver collected from NCD-fed WT and KO mice. (B) Representative images of fresh adipose tissues and liver collected from HFD-fed WT and KO mice. (C) Representative H&E staining of sWAT, eWAT, BAT, liver, muscle, and pancreas collected from the WT and KO mice fed by HFD. (D–G) Quantification of the adipocyte numbers and sizes in the sWAT and eWAT collected from the WT and KO mice fed by HFD. BAT, brown adipose tissue; DRP1, dynamin-related protein 1; eWAT, epididymal WAT; HFD, high-fat diet; NCD, normal chow diet; sWAT, subcutaneous WAT.
DRP1 KO in adipose tissue causes unique morphological changes of mitochondria in BAT
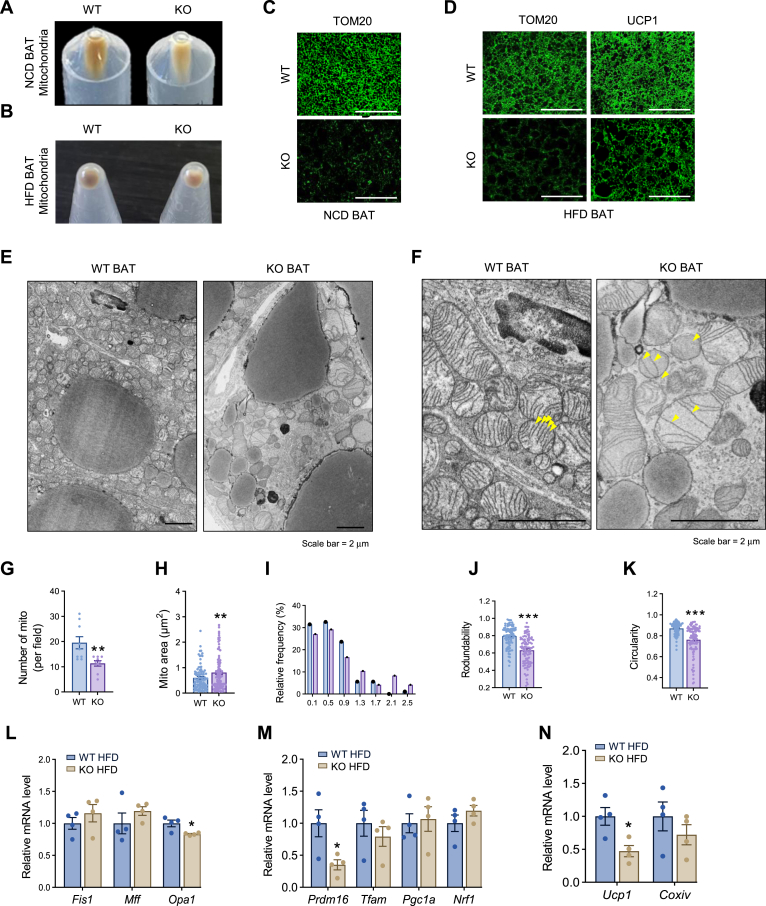
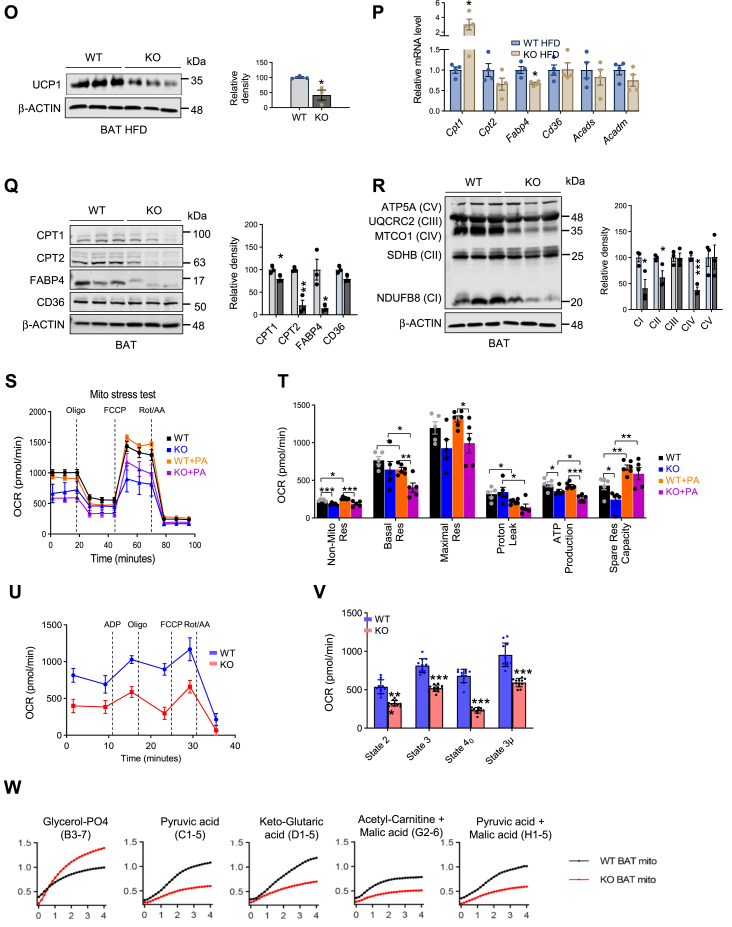
Given the direct role of DRP1 in regulating mitochondrial fission, we purified mitochondria from the KO adipose tissue for functional assays. Upon pooling mitochondria isolated from the BAT of KO and WT mice, we observed that the KO BAT mitochondrial pellet was considerably smaller, indicating a reduced mitochondrial quantity (Fig. 4A). Intriguingly, the color of the WT samples was dark brown, whereas the pellets collected from the KO BAT exhibited a noticeably lighter, paler color (Fig. 4A, B). We further determined the mitochondrial density in the tissue by immunofluorescent staining with anti-Tom20, a specific antibody that recognizes the mitochondrial marker protein Tom20 (27). We found that under both HFD and NCD conditions, the BAT showed significant less density of mitochondria in the KO group (Fig. 4C, D). We conducted a study to examine the morphological changes in brown adipocytes isolated from KO and WT mice using TEM. Our imaging results from TEM showed that adipocytes in the KO BAT had fewer but much larger mitochondria (Fig. 4E, F under higher magnification). Upon further inspection, we found that the mitochondria in the KO cells displayed abnormal shapes. Specifically, some adopted an irregular pod-like structure, and others presented with other irregular shapes, with most displaying compromised ultrastructure, including impaired membrane structure (Fig. 4F). Intriguingly, the architecture of the cristae was also distorted, displaying lower inner membrane density within the mitochondria of the KO cells (Fig. 4F, arrows pointing to the affected area). Quantitative analysis revealed that although the total number of mitochondria decreased (Fig. 4G), the overall area of mitochondria in the cells increased with higher frequency of bigger mitochondria (Fig. 4H, I). Abnormal mitochondrial shapes in the KO cells were further evident from the decreased circularity and roundness (refer to Fig. 4J, K). We next examined the regulation of other mitochondrial dynamic regulators, including Fis1, Mff, and Opa1. Unexpectedly, we did not detect significant changes in the expression levels of these factors except for Opa1 in KO adipocytes (Fig. 4L). We also did not observe significant changes in mitochondrial biogenesis–related genes, such as Tfam, Pgc1α, and Nrf1 (Fig. 4M). However, the thermogenic genes, including Prdm16 and Ucp1, were dramatically downregulated in the KO brown adipocytes (Fig. 4M, N). In agreement, we observed reduced protein levels of UCP1 in the KO brown adipocytes (Fig. 4O), suggesting decreased thermogenesis in BAT.
Fig. 4.
DRP1 knockout in adipose tissue causes unique morphological and pathological changes in BAT. (A) Images of isolated mitochondrial pellets from one entire BAT of WT and KO fed on NCD (representative of 6 mice/group). (B) Images of isolated mitochondrial pellets from one entire BAT of WT and KO fed on HFD (representative of 5 mice/group). (C) IF staining of TOM20 in the BAT of NCD-fed WT and KO mice. Scale bar = 100 μm. (D) IF staining of TOM20 and UCP1 in the BAT of WT and KO mice after HFD feeding. Scale bar = 100 μm. (E) Transmission electron microscope (TEM) images of BAT in the WT and KO mice. (F) High magnification for TEM images of BAT in the WT and KO mice. Scale bar in (E, F) = 2 μm. (G–K) Morphometric quantification of mitochondria for the TEM analysis in (E). (L) qPCR analysis of the mitochondrial dynamic–related genes in the BAT of HFD-fed WT and KO mice (n = 4/group). (M, N) qPCR analysis of the mitochondrial function and biogenesis-related genes in the BAT of HFD-fed WT and KO mice (n = 4/group). (O) Representative Western blots and densitometric analysis of UCP1 in the BAT of HFD-fed WT and KO mice (n = 3/group). (P) qPCR analysis of the fatty acid metabolism–related genes in the BAT of HFD-fed WT and KO mice (n = 4/group). (Q) Representative Western blots and densitometric analysis of CPT1, CPT2, and UCP1 in the BAT of HFD-fed WT and KO mice (n = 3/group). (R) Representative western blots and densitometric analysis of oxidative phosphorylation (OXPHOS) protein complexes in the BAT of HFD-fed WT and KO mice (n = 3/group). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, unpaired Student’s t test. (S) Oxygen consumption rate (OCR) for Mitochondrial Stress Test (MST) of the differentiated adipocytes of WT and KO mice. BSA-conjugated palmitic acid (PA, 100 μM) was added to the assay medium 2 h before the MST (n = 5). (T) Parameters for mitochondrial respiratory based on data for the OCR in (T). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, one-way ANOVA followed by Turkey’s post hoc test. (U, V) OCR for electronic coupling assay of the isolated mitochondria from the BAT of WT and KO mice fed on NCD (n = 11/group). ∗∗∗P < 0.001, unpaired Student’s t test for (W). (W) Kinetics of electron transport of isolated mitochondria in the presence of several mitochondrial substrates. Mitochondria were isolated from the BAT of WT and KO fed on NCD. BAT, brown adipose tissue; DRP1, dynamin-related protein 1; HFD, high-fat diet; IF, immunofluorescent; NCD, normal chow diet; qPCR, quantitative-PCR; UCP1,uncoupling protein 1.
Deletion of DRP1 in adipose tissue results in reduced FFA uptake, β-oxidation, and energy production
A key function of mitochondria is to oxidize FFA to produce energy via β-oxidation and oxidative phosphorylation (28). We thus investigated the key factors involved in the transportation and utilization of FFA in mitochondria in the KO adipocytes. Our results revealed that Cpt1, the key enzyme that control the rate of FFA uptake by mitochondria (29), was significantly upregulated on mRNA level (Fig. 4P). However, Cpt2, along with the FFA binding protein 4 (Fabp4) (29), were significantly downregulated in the BAT of KO mice fed on HFD (Fig. 4P). Interestingly, scavenger receptor, a rate-limiting fatty acid and coenzyme update factor in BAT (30), did not exhibit significant changes in the KO mice (Fig. 4P). Paradoxically, the protein levels of CPT1, together with other factors, including CPT2, FABP4 were dramatically decreased in the KO BAT (Fig. 4Q), reflecting some unknown posttranslational modification might further regulate CPT1 levels. We further examined the changes in oxidative-phosphorylation and found decreased levels of NDUFBS (CI), UQCRC2 (CIII), and MTCO1 (CIV) in the BAT of the KO mice fed on HFD (Fig. 4R). Moreover, we observed that under NCD, only CI was dramatically reduced in the KO mice (supplemental Fig. S3B), suggesting that nutritional stress under HFD further impaired oxidative phosphorylation and energy production in the adipocytes depleted of DRP1.
DRP1 deficiency has a profound impact on metabolic function and dysfunction of mitochondria in adipocytes
To investigate the effect of DRP1 deficiency on mitochondrial function, we conducted Seahorse XF analysis on mature adipocytes that were differentiated from the isolated SVF of WT and KO mice. Seahorse XF assay was performed on day 7 of differentiation conditions. We observed a significant difference in mitochondrial respiration between the WT and KO adipocytes. The KO adipocytes showed dramatically impaired mitochondrial respiration, as reflected by the greatly reduced overall OCR (Fig. 4S). Particularly, the KO adipocytes demonstrated a reduced basal and maximal respiration capacity trend, as well as significantly decreased nonmitochondrial respiration, ATP production, and spare respiration capacity (Fig. 4T). To mimic the HFD effect, we added BSA-conjugated PA to the cell culture media for the Seahorse XF assay. We found that the respiration capacity of KO adipocytes was impaired in the presence of PA (Fig. 4S). However, the amount of reduction of respiration capacity, especially the basal and maximal respiration rate, was much more dramatic in KO adipocytes (Fig. 4T), suggesting lipid stress worsened the impaired mitochondrial function in the DRP1-deficient adipocytes. Of cause, it remains possible that PA might have direct effect on mitochondrial membrane fluidity and potential, which further influence the functional outcomes in vitro. To gain insights into the in vivo mitochondria function, we isolated the mitochondria from the BAT of WT and KO mice and performed Seahorse XF analysis. We observed declined OCR in the isolated BAT mitochondria from the KO mice (Fig. 4U, V), which is in line with the results from the whole-cell analysis.
To investigate the substrate preference of the mitochondria, we added isolated BAT mitochondria of WT and KO mice into the MitoPlateS-1, which was precoated with diverse NADH or FADH2-producing substrates (supplemental Fig. S6) and subsequently monitored the electron-flow visualized by color reaction of redox dye secondary to reduction (supplemental Fig. S4A). We found that the metabolic rate of FADH-producing substrates (glycerol-PO4) was increased, while the metabolic rates of NADH2-producing substrates (pyruvic acids, keto-glutaric acid, acetyl-carnitine + malic acid, and pyruvic acid + malic acid) were decreased in KO mitochondria (Fig. 4W and supplemental Fig. S4B), suggesting an overall impaired mitochondrial function and increased glycolysis function in the KO BAT.
Loss of function of DRP1 leads to retention of nascent lipid droplets on ER
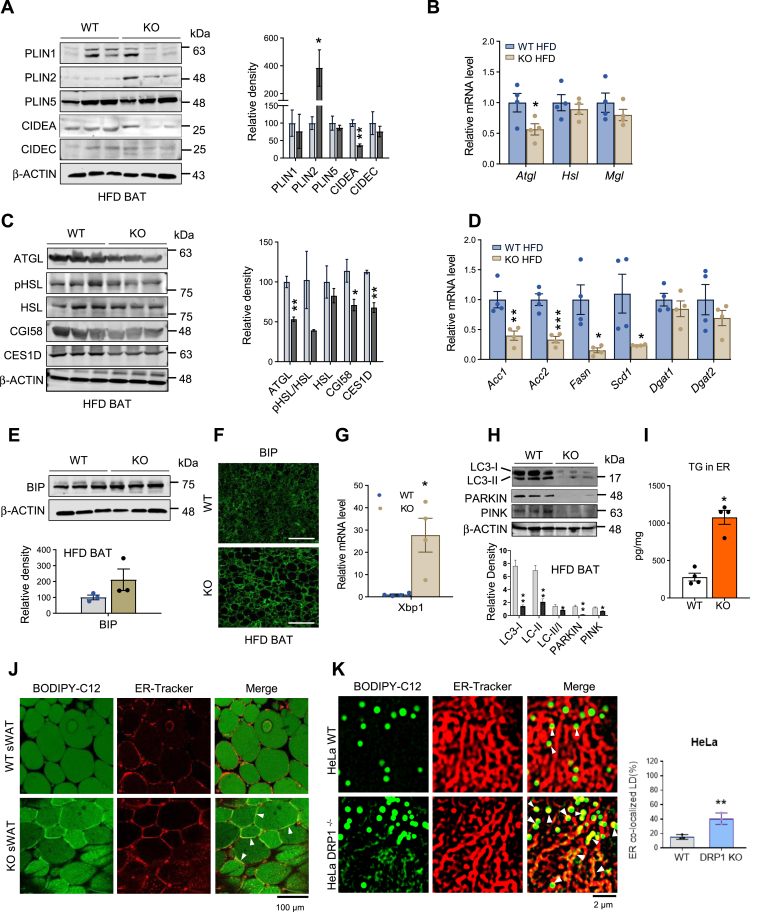
In addition to its established fission function on mitochondria, our recent findings reveal that DRP1 translocates onto the ER, participating in the release of nascent lipid droplets into the cytosol, ultimately forming mature lipid droplets (23). This discovery was made under conditions of β-adrenergic stimulation (23). Here, our study delved into the multifaceted functions of DRP1 in regulating ER-lipid droplet dynamics during HFD feeding. Histological examination (H&E staining) illustrated the presence of larger and unilocular lipid droplets in the cytosol in the BAT (Fig. 3C). Surprisingly, mature lipid droplet surface markers, including PLIN1 and PLIN5, showed no significant differences between WT and KO BAT (Fig. 5A). However, the dynamic regulator of lipid droplets, CIDEA, exhibited reduced levels in the KO BAT (Fig. 5A). Notably, the nascent lipid droplet marker PLIN2 significantly increased in the KO BAT (Fig. 5A). Expression of the lipid droplet-associated lipase, Atgl, was downregulated in the KO BAT (Fig. 5B). Consistently, the protein levels of CGI58-mediated ATGL-HSL lipolysis factors, including ATGL levels, the p-HSL/HSL ratio, and CGI58, exhibited reductions or a trend toward reduction (Fig. 5C). CES1D, a recently identified lipid droplet targeting lipase (31, 32, 33), also displayed reduced levels in the KO BAT (Fig. 5C). Key lipogenesis factors, such as Acc1/2, Fasn, and Scd1, were downregulated in the KO BAT (Fig. 5D), further emphasizing the profound impact of DRP1 on lipid droplet dynamics.
Fig. 5.
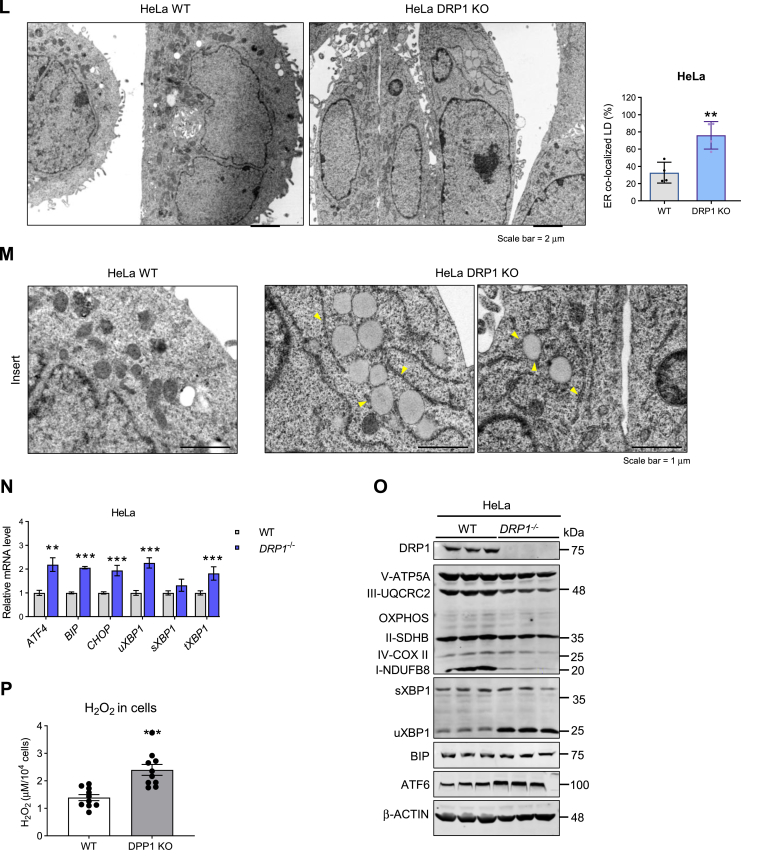
Loss of function of DRP1 leads to retention of nascent lipid droplets on ER. (A) Representative western blots and densitometric analysis of lipid droplet–related proteins in the BAT of HFD-fed WT and KO mice (n = 3/group). (B) qPCR analysis of the lipolysis-related genes in the BAT of HFD-fed WT and KO mice (n = 4/group). (C) Representative Western blots and densitometric analysis of lipases and related proteins in the BAT of WT and KO mice fed on HFD (n = 3/group). (D) qPCR analysis of lipogenesis-related genes in the BAT of WT and KO mice fed on HFD (n = 4/group). (E) Representative Western blots and densitometric analysis of BIP in the BAT of WT and KO mice fed on HFD (n = 3/group). (F) Representative immunofluorescent staining of BIP in the BAT of WT and KO mice after HFD feeding (Scale bars: 100 μm). (G) qPCR analysis of the Xbp1 in the BAT of WT and KO mice fed on HFD (n = 4/group). (H) Representative Western blots and densitometric analysis of LC3-I/II, Parkin, and Pink in the BAT of WT and KO mice fed on HFD (n = 3/group). Bottom panel shows quantitation of the band densities. (I) TG levels in the ER isolated from BAT of the WT and KO mice fed on HFD. The TG levels were normalized to protein concentrations (n = 4/group). (J) Costaining of BODIPY-C12 and ER-tracker in the sWAT explants from WT and KO mice fed on HFD. The yellow colocalization effect was pointed by the white arrows. The right panel indicates the quantification of the colocalized LDs with ER. (K) Costaining of BODIPY-C12 and ER-tracker in the WT and DRP1−/− HeLa cells. (L, M) Transmission electron microscope images of WT and DRP1−/− KO HeLa cells. (N) qPCR analysis of the ER stress–related genes in WT and DRP1−/− HeLa cells (n = 6). (O) Representative Western blots of DRP1, OXPHOS, ER stress–related proteins in WT, and DRP1−/− HeLa cells (n = 3/group). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, unpaired Student’s t test. (P) H2O2 measurement in the WT and DRP1−/− HeLa cells (n = 6/group). BAT, brown adipose tissue; DRP1, dynamin-related protein 1; ER, endoplasmic reticulum; HFD, high-fat diet; H2O2, hydrogen peroxide; OXPHOS, oxidative phosphorylation; qPCR, quantitative-PCR; sWAT, subcutaneous WAT; TG, triglyceride.
Lipid accumulation and aberrant lipid metabolism may lead to ER stress (34, 35, 36). We confirmed an increase in the ER stress marker BIP protein in the KO BAT by immunofluorescence staining, while the Western blotting result only reflected a slight increase (Fig. 5E, F). qPCR results indicated a marked upregulation of Xbp1 mRNA levels in the KO BAT (Fig. 5G). Moreover, autophagy and mitophagy was suppressed in the KO BAT, evidenced by an increased ratio of LC3II/I, as well as decreased levels of Parkin and Pink, the markers of mitophagy (Fig. 5H).
A direct indication of ER lipid retention is the substantial elevation in the total level of TG in the ER of the KO BAT (Fig. 5I). We further designed an experiment to visualize ER retention of nascent lipid droplets in KO BAT. We monitored the formation of Bodipy-C12–labeled nascent lipid droplets within the ER of BAT (23). Consistent with our previous observations during β-adrenergic stimulation, we identified pronounced ER structural changes in the KO sWAT (Fig. 5J). Furthermore, an increase in the yellow merged area between the ER and lipid droplets was observed, suggesting an enhanced ER retention effect in the KO white adipocytes (Fig. 5J, pointed by the white arrows).
We extended our analysis to DRP1 KO HeLa cells to provide a clearer manifestation of these effects. These cells naturally exhibit smaller lipid droplets and higher ER density than adipocytes. In HeLa cells, we confirmed the ER retention of nascent lipid droplets in the absence of DRP1. Notably, we not only observed ER-retained lipid droplets indicated by the yellow area within the ER but also found an increased yellow merged area between the formed lipid droplets (green) and the ER (red) (Fig. 5K, pointed by the white arrows, quantified yellow areas were shown in the right panel). The TEM examination further revealed that the lipid droplets in the KO cells are much larger (Fig. 5L, M, panel M presents the zoomed in effect). More strikingly, we observed significantly more nascent lipid droplets stuck with the ER in the KO cells (Fig. 5M, arrows pointed in the right panel), suggesting the ER retention of the lipid droplets at the absence of DRP1 in the cells. As anticipated, we detected upregulation of ER stress-related factors, including Atf4, Bip, Chop, and Xbp1 in KO HeLa cells (Fig. 5N). At the protein level, we observed increased levels or a trending increase in uXBP1 and ATF6 in KO cells (Fig. 5O). Of note, we detected reduced levels of mitochondrial complex I and III in the KO cells (Fig. 5O). The total hydrogen peroxide level also increased, further corroborating the other evidence of overall ER stress conditions in the KO cells (Fig. 5P).
The loss of function of DRP1 in adipocytes induces a unique alteration in the circulating metabolomic profile
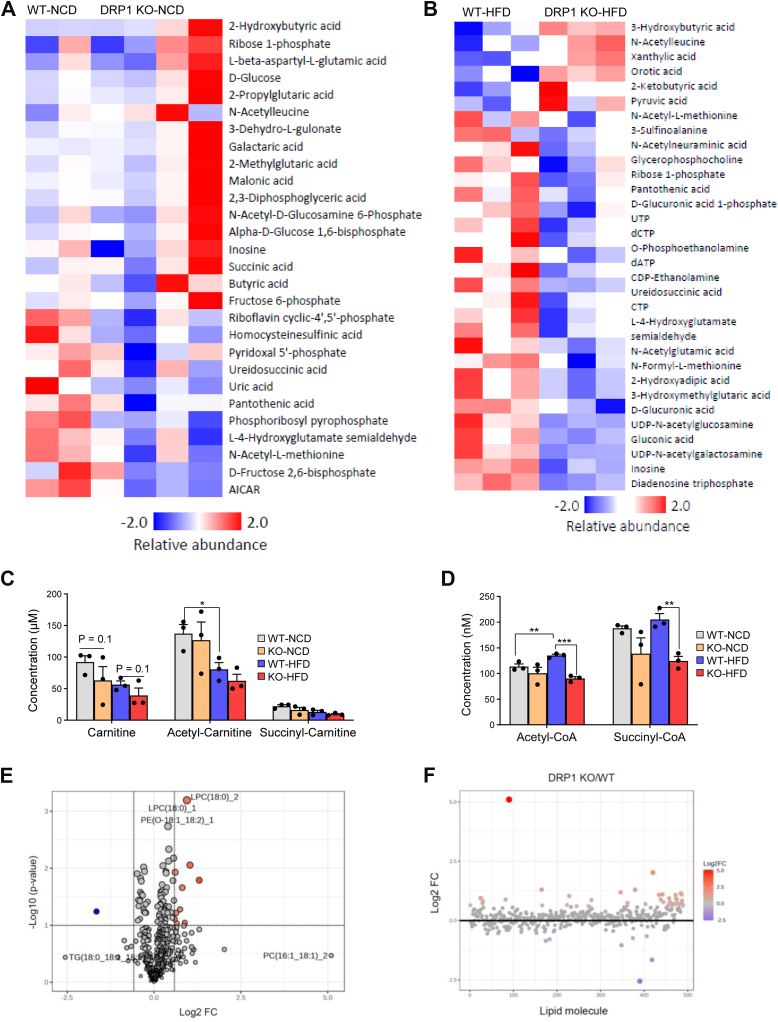
Given the multifaceted role of DRP1 in mitochondrial energy production and lipid dynamics on the ER, we conducted a comprehensive investigation into its combined effects on whole-body energy metabolism. Metabolomic analyses were performed on whole blood samples collected from both KO and WT mice under NCD and HFD feeding conditions. The heat maps illustrate the alterations in circulating metabolites under these dietary conditions (Fig. 6A, B). Notably, various isoforms of ketone bodies (2-hydroxybutyric acid, 3-hydroxybutyric acid, and 2-ketobutyric acid) were elevated in KO mice under both diet conditions, consistent with impaired insulin sensitivity in these mice (Fig. 6A, B).
Fig. 6.
Loss of function of DRP1 in adipocytes results in profound metabolomic and lipidomic profiles in circulation. (A, B) Metabolomics analysis on the whole blood collected from WT and KO mice fed on NCD or HFD. The significantly different metabolites of WT and KO mice fed on NCD (A) or fed on HFD (B). (C) Carnitine, acetyl-carnitine, and succinyl-carnitine levels in the whole blood of WT and KO mice fed on NCD or HFD. (D) Acetyl-CoA and succinyl-CoA levels in the whole blood of WT and KO mice fed on NCD or HFD. (E) Lipidomic analysis of the sera collected from WT and KO mice upon HFD feeding for 10 weeks. The volcano plot analysis visualizes the statistical significance and magnitude of changes in lipid species (a P value of <0.05 and a fold change threshold of 1.5). (F) The fold change ratio between KO and WT indicates a large proportion of lipid species were enriched in the KO serum. (G) Heat map for comparisons of different lipid species between WT and KO groups. (H) Heat map for comparisons of different lipid species, highlighting TG and DG between WT and KO groups. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and #P < 0.05. KO versus. WT (under HFD), one-way ANOVA followed by Turkey’s post hoc test. BAT, brown adipose tissue; DG, diacylglycerols; DRP1, dynamin-related protein 1; HFD, high-fat diet; NCD, normal chow diet.
The most pronounced changes were observed in the overall decrease of metabolites involved in purine and pyrimidine metabolism. Specifically, under normal chow feeding conditions, metabolites such as pyridoxal 5′-phosphate, ureidosuccinic acid, and uric acid were reduced, while under HFD feeding, pyruvic acid, UTP, and inosine exhibited decreased levels (Fig. 6A, B). We found that the levels of free carnitine, acetyl-carnitine and succinyl-carnitine downtrended in the KO under both NCD and HFD conditions (Fig. 6C). Additionally, acetyl-CoA and succinyl-CoA levels were dramatically reduced in KO mice under HFD conditions, with a noticeable downtrend observed under NCD feeding (Fig. 6D).
Pathway analysis further highlighted purine metabolism as the major downregulated pathway in KO mice under NCD, and this downregulation was further enhanced under HFD conditions (Fig. 6E, blue color highlighted). Further detailed analysis of metabolite changes revealed specific molecules (highlighted in blue) that exhibited altered levels in KO mice. Notably, CTP, inosine, dATP, and dCTP showed a decreasing trend in KO mice under chow diet, with significantly reduced levels under HFD (supplemental Fig. S5A). Another crucial category of metabolites related to mitochondrial energy balance is the vitamin B group, known for their involvement in the regulation of energy balance from proteins, sugars, and fats (supplemental Fig. S5A). Metabolomic analysis indicated a decreasing trend in vitamin B2 levels in KO mice under NCD condition, which diminished under HFD feeding (supplemental Fig. S5B). Furthermore, vitamin B5 and B6 exhibited significant decreases in KO mice under NCD feeding, with only a trend of decrease detected in the HFD feeding groups (supplemental Fig. S5C). These divergent effects reflect the complexity of diet regulation in KO mice.
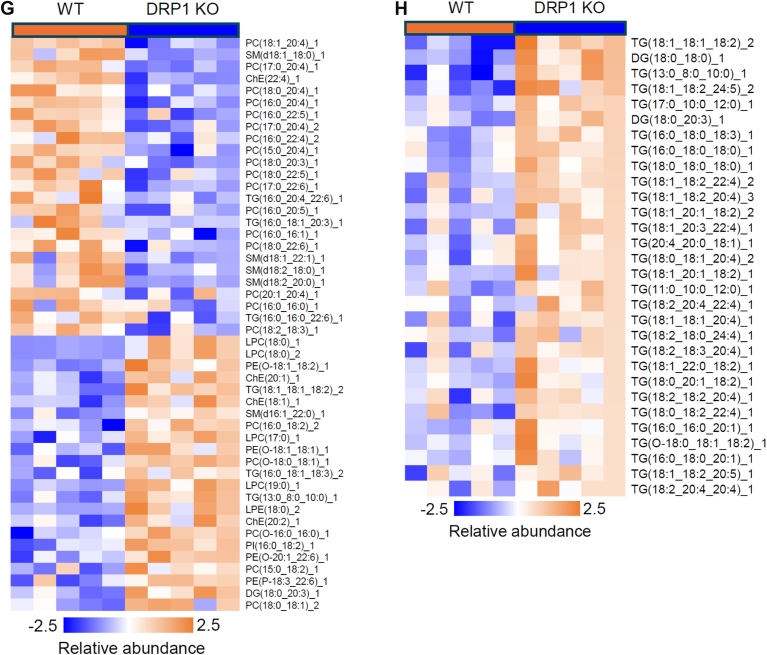
To further characterize the consequence of loss of function of DRP1 in lipid metabolism, we performed lipidomic analysis on the serum of KO and the WT mice upon HFD feeding. The clustering analysis revealed a distinct modulation of the lipid profile in the serum of KO mice. Particularly, the volcano plot, which visualizes the statistical significance and magnitude of changes in lipid species, identified a substantial number of lipids that were significantly altered based on a P value threshold of <0.05 and a fold change threshold of 1.5 (Fig. 6E). Interestingly, the fold change analysis indicates that a large proportion of the significantly altered lipids were enriched in the KO serum, with fold increases exceeding 1.5, suggesting that the DRP1 deficiency in adipose tissue leads to an accumulation of different lipids in circulation (Fig. 6F). Intriguingly, not all the lipid species increased their levels in KO serum. Indeed, some of them, especially phosphatidylcholine with different lengths were reduced (Fig. 6G). However, we do observe dramatic increase of most of TG and some of the DG in the KO group (Fig. 6H). In summary, these findings highlight the importance of DRP1 in regulating lipid homeostasis, and its absence may disrupt the normal balance of lipid synthesis, degradation, and transportation.
Loss of function of DRP1 triggers local inflammation in obese adipose tissue
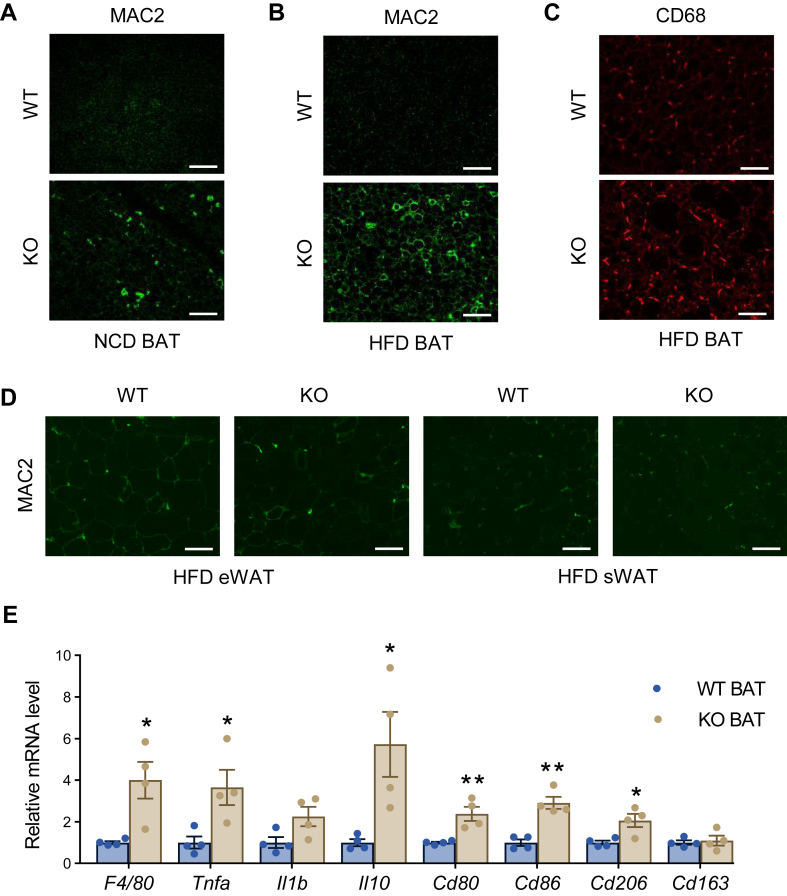
Metabolic dysregulation may further impact adipose tissue remodeling, leading to proinflammatory microenvironment in adipose tissue (37). We thus analyzed the levels of local inflammation and fibrosis in the KO mice. Mac2 staining revealed a significant accumulation of macrophages in the BAT under NCD feeding, while the effect is enhanced under HFD feeding (Fig. 7A, B). The accumulated macrophages in the BAT were of the proinflammatory state, as demonstrated by the immunofluorescent staining by proinflammatory marker CD68 (38) (Fig. 7C). We further detected more macrophage accumulation in the epididymal WAT and sWAT in the KO mice under HFD feeding conditions (Fig. 7D). Consistent with the histological findings, qPCR results demonstrated significant upregulation of proinflammatory genes, including Il-1β, Il-6, and Tnfα, in the BAT of KO mice fed with HFD (Fig. 7E), indicating that the loss of function of DRP1 leads to proinflammatory unhealthy expansion of adipose tissue in the KO mice (37).
Fig. 7.
DRP1 knockout in adipose tissue triggers local inflammation. (A, B) Representative IF staining of MAC2 in the BAT of WT and KO mice fed on NCD (A) or HFD (B) (Scale bars: 100 μm). (C) Representative IF staining of CD68 in the BAT of WT and KO mice after HFD feeding (Scale bars: 100 μm). (D) Representative IF staining of MAC2 in the eWAT and sWAT of WT and KO mice after HFD feeding (Scale bars: 100 μm). (E) qPCR analysis of inflammation-related genes in the BAT of WT and KO mice fed on HFD (n = 4/group). Scale bar = 100 μm. ∗P < 0.05, ∗∗P < 0.01, unpaired Student’s t test. BAT, brown adipose tissue; DRP1, dynamin-related protein 1; eWAT, epididymal WAT; HFD, high-fat diet; IF, immunofluorescent; qPCR, quantitative-PCR; NCD, normal chow diet; sWAT, subcutaneous WAT.
Discussion
DRP1 plays a crucial role in mitochondrial fission across various tissues, proving particularly significant in age-related diseases such as obesity, type-2 diabetes, neuronal degenerative diseases, and cancer. Its impact on mitochondrial biogenesis, mitophagy, and essential cellular processes like proliferation, differentiation, and apoptosis is well established. Nevertheless, the underlying mechanisms regulating these processes warrant further exploration (39, 40). Notably, DRP1's role has been extensively studied in metabolic tissues such as the skeletal muscle, heart, blood vessels, and liver (14, 15, 16, 41, 42, 43). Based on these studies, its impact on mitochondrial dynamics and functional regulations varies across different tissues, displaying tissue-specific effects. In skeletal muscles, DRP1 deficiency induces functionally abnormal mitochondria, reduced muscle mass and even animal death (16). In cardiomyocytes, it has a profound effect on mitochondrial functional regulation and the dynamics of the mitochondria-ER membrane (14). Loss of function of DRP1 in heart has been linked to cardiomyocyte apoptosis, cardiomyopathy, and heart failure (41, 44). Paradoxically, disruption of DRP1 in liver protected mice from obesity induced metabolic deterioration, which is related to ameliorated ER stress in hepatocytes (45). These distinct actions of DRP1 highlight the intricate tissue-specific mechanisms that govern mitochondrial dynamics and function. Despite the wealth of reports about DRP1 in various organs and tissues, the function and regulation of DRP1 in adipose tissue, especially under diet-induced obesity conditions, has not been thoroughly investigated yet. In this study, we discovered that the total levels of DRP1 were decreased in response to nutritional stress in adipose tissues, including WAT and BAT. Importantly, other mitochondrial dynamic factors, such as RalA, Opa1, Mff, Mfn2, and Fis1 also exhibited changes in the whole process, demonstrating the tight regulation of mitochondria biogenesis and dynamics in response to nutritional stresses. Our findings are in line with previous findings about their key functions in mitochondrial energy balance (46, 47, 48, 49, 50, 51). Based on these reports, we believe that these factors or co-factors also affect lipid droplet dynamics. However, given the facts that they do not directly translocate onto ER, we think that their influence on ER-lipid droplet dynamics is mediated through mitochondrial regulation, representing an indirect effect. Since both we and others have observed correlations between changes in these factors under various physiological/pathological conditions, it will be crucial to further elucidate the potential crosstalk among them in the dynamic regulation of mitochondria, the ER, and lipid droplets.
To mimic the loss of function of DRP1 during obesity in adipose tissue, we generated an adipose tissue-specific DRP1 KO mouse model. We observed that under NCD conditions, the KO mice appear to exhibit normal characteristics when compared to the WT mice. This stands in stark contrast to the phenotype observed with the deletion of the Drp1 gene in other tissues and organs, including skeletal muscles, the cardiovascular system, and the liver (14, 15, 16, 41, 42, 43). For instance, the constitutive knockout of DRP1 specifically in muscle tissue decelerates growth and can even lead to animal death (16). In our study, we noted that the adipose tissue specific KO mice are much more sensitive to cold exposure (23). More surprisingly, the adipose tissue specific KO mice did not exhibit significant weight gain compared to their littermate control WT mice when exposed to an HFD. However, the KO mice exhibited impaired glucose tolerance and reduced insulin sensitivity in response to nutritional stress. The most notable pathological alteration observed was the "whitening" of BAT, even preceding HFD feeding. Our investigation thus focused on elucidating the distinctive morphological and pathological changes in the BAT of the KO mice. Remarkably, the mitochondria in the KO BAT displayed abnormal enlargement and structural alterations in the cristae. This morphological aberration not only directly disrupted proper mitochondrial OCR but also led to reduced FFA uptake, impaired β-oxidation, and compromised energy electron chain function and oxidative phosphorylation. Significantly, DRP1 deficiency contributed to the retention of nascent lipid droplets on ER, resulting in abnormal lipid droplet dynamics, as well as perturbed ER structure and stress. The cumulative defects of DRP1 on mitochondria and ER-lipid droplet interfaces ultimately induced profound changes in the circulating metabolite profile. One notable alteration was the diminished purine and pyrimidine metabolism, leading to a lower total level of energy metabolism in the KO mice. These findings underscore the intricate interplay between DRP1 regulation, mitochondrial dynamics, ER function, and lipid metabolism, thus shedding light on the complex molecular mechanisms underlying the observed metabolic disturbances in the context of DRP1 deficiency in adipose tissue. Notably, recent findings suggest that during obesity, other factors, such as RalA, may promote DRP1 function (49). This gain-of-function regulation could further cause mitochondrial fragmentation and dysfunction (49). Intriguingly, in our study, a complete loss of DRP1 function also leads to dysfunctional regulation of mitochondria. These results clearly suggest that proper fission function of DRP1 is required for normal mitochondrial functions, while malfunction of DRP1 may induce pathological overfission of mitochondria, causing organelle fragmentation.
Since DRP1 levels dramatically decreased in both WAT and BAT during obesity, we propose that the adipose tissue–specific DRP1 KO mouse is an appropriate model to investigate the metabolic implications of DRP1 deficiency under such a nutritionally stressed condition. Remarkably, the most notable morphological alteration observed is the "whitening" of BAT, and all the metabolic consequences align with this phenotype. It is widely established that while the primary function of WAT is lipid storage, BAT uniquely plays a pivotal role in energy expenditure by oxidizing lipids to fuel thermogenesis (1, 52, 53). Given that DRP1's function involves mitochondrial fission, the observed pathological changes are reasonably associated with its deficiency. Unexpectedly, the KO mice did not experience significant body weight gain, even when subjected to a HFD, suggesting the compensatory effects of other metabolic tissues/organs in response to the defective mitochondrial function in the adipose tissue. We also noted that HFD exposure led to significantly impaired insulin sensitivity in the KO mice. Additionally, the KO mice displayed abnormal lipid metabolism and dysregulation of adipokines and cytokines, all contributing to an overall proinflammatory state in the KO mice under HFD conditions.
Balance of fission and fusion is a crucial process that regulates mitochondrial function in response to various cellular stimuli (54, 55). Loss of DRP1 during this process induces significant morphological and functional changes in different types of cells (54). Here, we found that in the BAT, crude extracts of mitochondria revealed notable differences. It is well recognized that normal mitochondria extracts exhibit a yellowish or brownish color (56). As expected, the mitochondria isolated from WT BAT were yellowish and brownish in color. Interestingly, KO mitochondria were lower in density and appeared lighter or whiter in color, under both normal chow and HFD conditions. The lighter color in KO mitochondria suggests important abnormalities, indicating a deficiency in essential components such as functional iron within the inner and outer membranes of the mitochondria. Further study is needed to elucidate the molecular meaning behind the color differences between KO and WT mitochondria.
TEM provided additional insights into the abnormal structure and size of KO BAT mitochondria. Unlike the regular shape and normal ultrastructure of WT mitochondria, KO BAT mitochondria appeared abnormally giant with heterogeneous structural changes. Some adopted a pod-like structure, and others exhibited irregular shapes, with most displaying compromised ultrastructure, including the membrane organization. Notably, the cristae in KO mitochondria showed impaired structure, with less folds and lower density, indicative of potential dysregulation of essential enzymes in the inner membrane, including key enzymes involved in lipid metabolism, oxidative phosphorylation (OXPHOS), and energy balance (57, 58, 59). These observations align with findings of lipid dysregulation and impaired energy production in KO BAT. As evidence of these issues, we noted a reduced level of UCP1, indicating compromised thermogenesis in the KO BAT. Additionally, CPT1/2, OXPHOS complexes, and other FFA oxidation enzymes presented at decreased levels, implying impaired lipid oxidation and energy production in the KO BAT.
Our Seahorse XF tests further confirmed impaired mitochondrial functions in both brown adipocytes differentiated from SVF and isolated BAT mitochondria. Notably, in differentiated brown adipocytes, the addition of PA FFA to mimic HFD-induced nutritional stress resulted in more severe mitochondrial functional damage, as evidenced by changes in OCR at different time intervals. This observation aligns with the more pronounced metabolic phenotype observed in HFD-fed KO mice. Intriguingly, in vitro substrate assays on isolated mitochondria revealed that KO mitochondria lost the ability to uptake substrates for NADH production. This corresponds with the significantly reduced OXPHOS complex I in KO BAT, responsible for electron transport and generation of proton gradient across the inner membrane (60). Remarkably, we observed a notable increase in the uptake of glycerol-PO4 by the KO mitochondria; this substrate was the only one which demonstrated this enhanced uptake by the KO mitochondria. It is established that the glycerol-3-phosphate shuttle is a vital pathway for forming FADH2 in the mitochondrial matrix, with glycolysis contributing to the production of glycerol-phosphate. While the effects of DRP1 activation on glycolysis have been reported differently (61, 62, 63), here, our data suggested that that the KO mitochondria may preferentially utilize the by-products of glycolysis for energy production.
In addition to its typical translocation to mitochondria and peroxisomes, DRP1 has been reported to translocate to the ER. In this context, it performs significant functions, both with and without its GTPase activity, across various tissues (22, 23, 44, 64). Specifically, we have recently uncovered that under acute β-adrenergic stimulation conditions, DRP1 translocated to the interface of ER-lipid droplets in adipocytes (both sWAT and BAT). In this context, it plays a pivotal role in the initiation of nascent lipid droplet biogenesis originating from the ER (23). In this study, we further revealed that DRP1 exerts the same function under chronic diet–induced obesity conditions. By knocking out DRP1 in adipocytes, we noted that more nascent lipid droplets get stuck in the ER lumen, and the preexisting lipid droplets become larger and remain tethered to the ER surface. These findings suggest that fission by DRP1 is a fundamental molecular event that regulates the dissociation of lipid droplets from the ER to release into the cytosol under different cell stimuli. As a demonstration, we observed the same effect in DRP1 KO HeLa cells. Since HeLa cells naturally have denser ER and relatively smaller lipid droplets than adipocytes, we were able to achieve higher-resolution imaging with clearer indication that DRP1 is required in the lipid droplet dissociation from the ER. Importantly, we also found that the lipid droplet lipolysis-related factors and enzymes, such as CGI58 and its mediated ATGL-HSL axis (65, 66, 67), were downregulated on mRNA levels, or decreased on protein levels, suggesting abnormal lipid droplet dynamics in response to the loss of function of DRP1. On the other hand, the nascent lipid droplet marker, Plin2, exhibited a significant upregulation, suggesting the accumulation of perpetually immature or younger lipid droplets without undergoing the typical maturation processes. The observed ER-lipid droplet phenotype closely resembles that reported in the context of loss of function of Seipin (68, 69, 70, 71, 72), implying potential shared mechanisms between DRP1 and seipin during the lipid droplet dissociation process. However, a comprehensive understanding of the detailed molecular and cellular mechanisms involved remains to be elucidated.
Given the profound impact of DRP1 on the regulation of both mitochondria and ER-lipid droplets, we proposed that the observed metabolic phenotype, characterized by impaired insulin sensitivity, abnormal lipid metabolism, and disrupted energy expenditure, arises from the combined effects of mitochondrial dysregulation and deranged ER-lipid droplet dynamics. To further explore the multifaceted functions of DRP1 in non–cell autonomous whole-body metabolism, we conducted an unbiased metabolomic analysis of circulating metabolites in whole blood collected from KO and WT mice. Our analysis revealed dysregulation in numerous factors that serve as key nodes of glucose and lipid metabolism, or are involved in the overall metabolic flexibility, including acetyl-carnitine, succinyl-carnitine, free carnitine, acetyl-CoA, and succinyl-CoA (73, 74), aligning with DRP1's roles in both mitochondria and ER-lipid droplets. Notably, we observed a substantial impairment in the purine and pyrimidine metabolism pathways in the KO groups, with the degree of impairment exacerbated under HFD condition. Previous studies have established the critical roles of purine and pyrimidine metabolites, including key molecules like uric acid in energy balance. Disruptions in their levels may compromise overall energy homeostasis, leading to severe cellular pathological changes, including cell apoptosis (75, 76, 77). Metabolites from purine and pyrimidine pathways, including uric acid play a vital role in providing cells with the necessary energy and cofactors for cell survival and proliferation (76, 77). Importantly, recent research has provided deeper understanding on how purine metabolism may contribute to cancer progression, revealing a novel perspective on cellular purine regulation (78). Particularly, under conditions of high purine demand, de novo purine biosynthetic enzymes cluster near mitochondria and microtubules to synthesize the necessary initial and intermediate molecules for continued production. Additionally, purines can be incorporated into complex biomolecules and serve as cofactors, such as NAD and coenzyme A. These fundamental processes utilize glycolytic and citric acid cycle intermediates for NADPH production (79). Our findings align with these reports, as evidenced by the decreased levels of NADPH observed in the KO tissue samples, providing further insight into the intricate interplay between DRP1, purine metabolism, and cellular energetics. Of note, another mitochondrial fission/fusion factor, Opa1, has been reported to promote adipocyte browning via regulation of urea cycle metabolites (46). The mechanisms between mitochondrial fission/fusion and purine/pyrimidine metabolisms need further investigation.
Interestingly, we observed decreased levels of vitamin B family factors in the KO groups. It is widely acknowledged that the vitamin B family plays a crucial role in regulating essential enzymes within mitochondria, influencing various aspects of protein, glucose, and lipid metabolism (80). Thus, maintaining a balanced pool of vitamin B constituents is vital for normal mitochondrial function and energy homeostasis (81). In our study, we noted a reduction in circulating vitamin B in KO mice, indicating potential impairment in mitochondrial energy balance. However, the specific mechanisms underlying the decreased circulating vitamin Bs require further investigation.
The lipidomic analysis of DRP1 KO mice has revealed a distinct lipid profile in the KO serum, suggesting that DRP1, as a key protein involved in mitochondria-ER-lipid droplet dynamics, plays a significant role in whole-body lipid metabolism. Absence of DRP1 in adipose tissue may not only disrupt the normal balance between lipid synthesis and degradation in these organelles, but also affect the nascent lipid droplet release from the ER. Consequently, the whole lipid profile in the body changes significantly. Further investigation into the specific classes and functions of the enriched lipid species may provide valuable insights into the mechanic link between mitochondria, ER, and lipid droplets (82).
Lipid disorders and mitochondrial dysfunction have been associated with localized inflammation in adipose tissue (83, 84, 85). Consistent with this paradigm, we observed an elevated level of macrophage accumulation in the adipose tissues of KO mice. Intriguingly, we identified an even higher level of local inflammation in the BAT than the WATs. The presented qPCR results reflected polarization of macrophages to proinflammatory state in response to nutritional stresses in the KO BAT. Given the crucial role of BAT as a significant energy expenditure organ, the heightened inflammation within BAT may contribute to a systemic reduction in energy balance, thereby exacerbating whole-body metabolic dysregulations. This aligns with our observations in the KO mice, particularly under HFD conditions.
In conclusion, our study delineates the multifaceted functions of DRP1 in regulating the dynamics of mitochondria, ER, and lipid droplets. The dysregulation of these functions may contribute to systemic metabolic disorders. Although detailed mechanistic studies are imperative, our investigation sheds light on a novel aspect of DRP1 in a tissue-specific manner within adipose tissue, opening avenues for innovative strategies in addressing obesity and related metabolic diseases.
Data availability
All the data in this reported study is open to readers. They are available from K. S. at the University of Texas Health Science Center at Houston upon reasonable request.
Supplemental data
This article contains supplemental data.
Conflict of interests
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors are grateful to our colleagues at the Center for Metabolic and Degenerative Diseases in UTHealth for their critical discussions. We thank Dr Zhengmei Mao at the Microscopy Core of the Institute of Molecular Medicine for assistance with imaging and tissue processing.
Author contributions
X. L., H. S., P. L., and K. S. writing–review and editing; X. L., L. J. S., and K. S. visualization; X. L., H. S., P. L., and K. S. validation; X. L., P. L., and K. S. software; X. L. and K. S. project administration; X. L., I. M., L. T., P. L., and K. S. methodology; H. S., P. L., and K. S. resources; X. L., K. P., J. Y., I. M., L. T., B. W., L. J. S., M. E., R. H., F. E., P. L., and K. S. investigation; X. L., K. P., I. M., L. T., B. W., P. L., and K. S. data curation; X. L., J. Y., I. M., B. W., P. L., and K. S. formal analysis; P. L. and K. S. writing–original draft; P. L. and K. S. supervision; K. S. funding acquisition; P. L. and K. S. conceptualization.
Funding and additional information
This work was supported by the National Institutes of Health [grant number: R01DK129815]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplemental data
References
- 1.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown K.A. Scherer PE: update on adipose tissue and cancer. Endocr. Rev. 2023;44:961–974. doi: 10.1210/endrev/bnad015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gliniak C.M., Pedersen L., Scherer P.E. Adipose tissue fibrosis: the unwanted houseguest invited by obesity. J. Endocrinol. 2023;259 doi: 10.1530/JOE-23-0180. [DOI] [PubMed] [Google Scholar]
- 4.Sun K., Li X., Scherer P.E. Extracellular matrix (ECM) and fibrosis in adipose tissue: overview and perspectives. Compr. Physiol. 2023;13:4387–4407. doi: 10.1002/cphy.c220020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajimura S., Spiegelman B.M., Brown S.P., Fat B. Physiological roles beyond heat generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu I., Walsh K. The whitening of Brown fat and its implications for weight management in obesity. Curr. Obes. Rep. 2015;4:224–229. doi: 10.1007/s13679-015-0157-8. [DOI] [PubMed] [Google Scholar]
- 8.Liesa M., Shirihai O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sesaki H., Jensen R.E. Division versus fusion: dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D., Tian L., Hoskin V., Dasgupta A. Editorial: the effects of mitochondrial dysfunction on the cell cycle. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1303834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokuyama T., Yanagi S. Role of mitochondrial dynamics in heart diseases. Genes (Basel) 2023;14 doi: 10.3390/genes14101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao S., Huang H., Ma R.Y., Zeng X., Duan C.Y. Multifaceted functions of Drp1 in hypoxia/ischemia-induced mitochondrial quality imbalance: from regulatory mechanism to targeted therapeutic strategy. Mil. Med. Res. 2023;10:46. doi: 10.1186/s40779-023-00482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Titus A.S., Sung E.A., Zablocki D., Sadoshima J. Mitophagy for cardioprotection. Basic Res. Cardiol. 2023;118:42. doi: 10.1007/s00395-023-01009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong M., Mukai R., Mareedu S., Zhai P., Oka S.I., Huang C.Y., et al. Distinct roles of DRP1 in conventional and alternative mitophagy in obesity cardiomyopathy. Circ. Res. 2023;133:6–21. doi: 10.1161/CIRCRESAHA.123.322512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steffen J., Ngo J., Wang S.P., Williams K., Kramer H.F., Ho G., et al. The mitochondrial fission protein Drp1 in liver is required to mitigate NASH and prevents the activation of the mitochondrial ISR. Mol. Metab. 2022;64 doi: 10.1016/j.molmet.2022.101566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Favaro G., Romanello V., Varanita T., Andrea Desbats M., Morbidoni V., Tezze C., et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 2019;10:2576. doi: 10.1038/s41467-019-10226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rana A., Oliveira M.P., Khamoui A.V., Aparicio R., Rera M., Rossiter H.B., et al. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun. 2017;8:448. doi: 10.1038/s41467-017-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Potes Y., Perez-Martinez Z., Bermejo-Millo J.C., Rubio-Gonzalez A., Fernandez-Fernandez M., Bermudez M., et al. Overweight in the elderly induces a switch in energy metabolism that undermines muscle integrity. Aging Dis. 2019;10:217–230. doi: 10.14336/AD.2018.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tardif N., Salles J., Guillet C., Tordjman J., Reggio S., Landrier J.F., et al. Muscle ectopic fat deposition contributes to anabolic resistance in obese sarcopenic old rats through eIF2alpha activation. Aging Cell. 2014;13:1001–1011. doi: 10.1111/acel.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamerkar S.C., Kraus F., Sharpe A.J., Pucadyil T.J., Ryan M.T. Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat. Commun. 2018;9:5239. doi: 10.1038/s41467-018-07543-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh K., Murata D., Kato T., Yamada T., Araki Y., Saito A., et al. Brain-specific Drp1 regulates postsynaptic endocytosis and dendrite formation independently of mitochondrial division. Elife. 2019;8 doi: 10.7554/eLife.44739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adachi Y., Kato T., Yamada T., Murata D., Arai K., Stahelin R.V., et al. Drp1 tubulates the ER in a GTPase-independent manner. Mol. Cell. 2020;80:621–632. doi: 10.1016/j.molcel.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Yang L., Mao Z., Pan X., Zhao Y., Gu X., et al. Novel role of dynamin-related-protein 1 in dynamics of ER-lipid droplets in adipose tissue. FASEB J. 2020;34:8265–8282. doi: 10.1096/fj.201903100RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Zhao Y., Chen C., Yang L., Lee H.H., Wang Z., et al. Critical role of matrix metalloproteinase 14 in adipose tissue remodeling during obesity. Mol. Cell Biol. 2020;40:e00564–e005619. doi: 10.1128/MCB.00564-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y., Gu X., Zhang N., Kolonin M.G., An Z., Sun K. Divergent functions of endotrophin on different cell populations in adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2016;311:E952–E963. doi: 10.1152/ajpendo.00314.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An Y.A., Chen S., Deng Y., Wang Z.V., Funcke J.B., Shah M., et al. The mitochondrial dicarboxylate carrier prevents hepatic lipotoxicity by inhibiting white adipocyte lipolysis. J. Hepatol. 2021;75:387–399. doi: 10.1016/j.jhep.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalier L., Mignard V., Joalland M.P., Lanoe D., Cartron P.F., Manon S., et al. TOM20-mediated transfer of Bcl2 from ER to MAM and mitochondria upon induction of apoptosis. Cell Death Dis. 2021;12:182. doi: 10.1038/s41419-021-03471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houten S.M., Wanders R.J. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J. Inherit. Metab. Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreurs M., Kuipers F., van der Leij F.R. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes. Rev. 2010;11:380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 30.Anderson C.M., Kazantzis M., Wang J., Venkatraman S., Goncalves R.L., Quinlan C.L., et al. Dependence of brown adipose tissue function on CD36-mediated coenzyme Q uptake. Cell Rep. 2015;10:505–515. doi: 10.1016/j.celrep.2014.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L., Li X., Tang H., Gao Z., Zhang K., Sun K. A unique role of carboxylesterase 3 (Ces3) in beta-adrenergic signaling-stimulated thermogenesis. Diabetes. 2019;68:1178–1196. doi: 10.2337/db18-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G., Li X., Mahmud I., Ysaguirre J., Fekry B., Wang S., et al. Interfering with lipid metabolism through targeting CES1 sensitizes hepatocellular carcinoma for chemotherapy. JCI Insight. 2023;8 doi: 10.1172/jci.insight.163624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G., Li X., Yang L., Wang S., Dai Y., Fekry B., et al. Adipose tissue-specific ablation of Ces1d causes metabolic dysregulation in mice. Life Sci. Alliance. 2022;5 doi: 10.26508/lsa.202101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentile C.L., Frye M.A., Pagliassotti M.J. Fatty acids and the endoplasmic reticulum in nonalcoholic fatty liver disease. Biofactors. 2011;37:8–16. doi: 10.1002/biof.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo W., Wong S., Xie W., Lei T., Luo Z. Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3-L1 and rat primary preadipocytes. Am. J. Physiol. Endocrinol. Metab. 2007;293:E576–E586. doi: 10.1152/ajpendo.00523.2006. [DOI] [PubMed] [Google Scholar]
- 37.Sun K., Kusminski C.M., Scherer P.E. Adipose tissue remodeling and obesity. J. Clin. Invest. 2011;121:2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atkinson S.J., Roghi C., Murphy G. MT1-MMP hemopexin domain exchange with MT4-MMP blocks enzyme maturation and trafficking to the plasma membrane in MCF7 cells. Biochem. J. 2006;398:15–22. doi: 10.1042/BJ20060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kageyama Y., Zhang Z., Sesaki H. Mitochondrial division: molecular machinery and physiological functions. Curr. Opin. Cell Biol. 2011;23:427–434. doi: 10.1016/j.ceb.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji W.K., Hatch A.L., Merrill R.A., Strack S., Higgs H.N. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4 doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong M., Zablocki D., Sadoshima J. The role of Drp1 in mitophagy and cell death in the heart. J. Mol. Cell Cardiol. 2020;142:138–145. doi: 10.1016/j.yjmcc.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Li X., Hanada Y., Hasuzawa N., Moriyama Y., Nomura M., et al. Dynamin-related protein 1 deficiency accelerates lipopolysaccharide-induced acute liver injury and inflammation in mice. Commun. Biol. 2021;4:894. doi: 10.1038/s42003-021-02413-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma X., Chen A., Melo L., Clemente-Sanchez A., Chao X., Ahmadi A.R., et al. Loss of hepatic DRP1 exacerbates alcoholic hepatitis by inducing megamitochondria and mitochondrial maladaptation. Hepatology. 2023;77:159–175. doi: 10.1002/hep.32604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirakabe A., Zhai P., Ikeda Y., Saito T., Maejima Y., Hsu C.P., et al. Response by Shirakabe et al to letter regarding article, "Drp1-Dependent mitochondrial autophagy plays a protective role against pressure overload-induced mitochondrial dysfunction and heart failure". Circulation. 2016;134:e75–e76. doi: 10.1161/CIRCULATIONAHA.116.023667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L., Ishihara T., Ibayashi Y., Tatsushima K., Setoyama D., Hanada Y., et al. Disruption of mitochondrial fission in the liver protects mice from diet-induced obesity and metabolic deterioration. Diabetologia. 2015;58:2371–2380. doi: 10.1007/s00125-015-3704-7. [DOI] [PubMed] [Google Scholar]
- 46.Bean C., Audano M., Varanita T., Favaretto F., Medaglia M., Gerdol M., et al. The mitochondrial protein Opa1 promotes adipocyte browning that is dependent on urea cycle metabolites. Nat. Metab. 2021;3:1633–1647. doi: 10.1038/s42255-021-00497-2. [DOI] [PubMed] [Google Scholar]
- 47.Ejarque M., Ceperuelo-Mallafre V., Serena C., Maymo-Masip E., Duran X., Diaz-Ramos A., et al. Adipose tissue mitochondrial dysfunction in human obesity is linked to a specific DNA methylation signature in adipose-derived stem cells. Int. J. Obes. (Lond) 2019;43:1256–1268. doi: 10.1038/s41366-018-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin X., Lanza I.R., Swain J.M., Sarr M.G., Nair K.S., Jensen M.D. Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. J. Clin. Endocrinol. Metab. 2014;99:E209–E216. doi: 10.1210/jc.2013-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia W., Veeragandham P., Cao Y., Xu Y., Rhyne T.E., Qian J., et al. Obesity causes mitochondrial fragmentation and dysfunction in white adipocytes due to RalA activation. Nat. Metab. 2024;6:273–289. doi: 10.1038/s42255-024-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finocchietto P., Perez H., Blanco G., Miksztowicz V., Marotte C., Morales C., et al. Inhibition of mitochondrial fission by Drp-1 blockade by short-term leptin and Mdivi-1 treatment improves white adipose tissue abnormalities in obesity and diabetes. Pharmacol. Res. 2022;178 doi: 10.1016/j.phrs.2021.106028. [DOI] [PubMed] [Google Scholar]
- 51.Mooli R.G.R., Mukhi D., Chen Z., Buckner N., Ramakrishnan S.K. An indispensable role for dynamin-related protein 1 in beige and brown adipogenesis. J. Cell Sci. 2020;133 doi: 10.1242/jcs.247593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kotzbeck P., Giordano A., Mondini E., Murano I., Severi I., Venema W., et al. Brown adipose tissue whitening leads to brown adipocyte death and adipose tissue inflammation. J. Lipid Res. 2018;59:784–794. doi: 10.1194/jlr.M079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Townsend K., Tseng Y.H. Brown adipose tissue: recent insights into development, metabolic function and therapeutic potential. Adipocyte. 2012;1:13–24. doi: 10.4161/adip.18951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott I., Youle R.J. Mitochondrial fission and fusion. Essays Biochem. 2010;47:85–98. doi: 10.1042/bse0470085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 56.Samali A., Cai J., Zhivotovsky B., Jones D.P., Orrenius S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J. 1999;18:2040–2048. doi: 10.1093/emboj/18.8.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joubert F., Puff N. Mitochondrial cristae architecture and functions: lessons from minimal model systems. Membranes (Basel) 2021;11:465. doi: 10.3390/membranes11070465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cogliati S., Enriquez J.A., Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 61.Hasani S., Young L.E.A., Van Nort W., Banerjee M., Rivas D.R., Kim J., et al. Inhibition of mitochondrial fission activates glycogen synthesis to support cell survival in colon cancer. Cell Death Dis. 2023;14:664. doi: 10.1038/s41419-023-06202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagdas S., Kashatus J.A., Nascimento A., Hussain S.S., Trainor R.E., Pollock S.R., et al. Drp1 promotes KRas-driven metabolic changes to drive pancreatic tumor growth. Cell Rep. 2019;28:1845–1859. doi: 10.1016/j.celrep.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan K., Ding X., Zang Z., Zhang Y., Tang X., Pei X., et al. Drp1-Mediated mitochondrial metabolic dysfunction inhibits the tumor growth of pituitary adenomas. Oxid. Med. Cell Longev. 2022;2022 doi: 10.1155/2022/5652586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ilamathi H.S., Benhammouda S., Lounas A., Al-Naemi K., Desrochers-Goyette J., Lines M.A., et al. Contact sites between endoplasmic reticulum sheets and mitochondria regulate mitochondrial DNA replication and segregation. iScience. 2023;26 doi: 10.1016/j.isci.2023.107180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu L., Li Y., Grise A., Wang H. CGI-58: versatile regulator of intracellular lipid droplet homeostasis. Adv. Exp. Med. Biol. 2020;1276:197–222. doi: 10.1007/978-981-15-6082-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lord C.C., Brown J.M. Distinct roles for alpha-beta hydrolase domain 5 (ABHD5/CGI-58) and adipose triglyceride lipase (ATGL/PNPLA2) in lipid metabolism and signaling. Adipocyte. 2012;1:123–131. doi: 10.4161/adip.20035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin H., Ma Y., Chanturiya T., Cao Q., Wang Y., Kadegowda A.K.G., et al. Lipolysis in Brown adipocytes is not essential for cold-induced thermogenesis in mice. Cell Metab. 2017;26:764–777. doi: 10.1016/j.cmet.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arlt H., Sui X., Folger B., Adams C., Chen X., Remme R., et al. Seipin forms a flexible cage at lipid droplet formation sites. Nat. Struct. Mol. Biol. 2022;29:194–202. doi: 10.1038/s41594-021-00718-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., et al. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim S., Chung J., Arlt H., Pak A.J., Farese R.V.J., Walther T.C., et al. Seipin transmembrane segments critically function in triglyceride nucleation and lipid droplet budding from the membrane. Elife. 2022;11:e75808. doi: 10.7554/eLife.75808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lv X., Liu J., Qin Y., Liu Y., Jin M., Dai J., et al. Identification of gene products that control lipid droplet size in yeast using a high-throughput quantitative image analysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:113–127. doi: 10.1016/j.bbalip.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 72.Liu L., Jiang Q., Wang X., Zhang Y., Lin R.C., Lam S.M., et al. Adipose-specific knockout of SEIPIN/BSCL2 results in progressive lipodystrophy. Diabetes. 2014;63:2320–2331. doi: 10.2337/db13-0729. [DOI] [PubMed] [Google Scholar]
- 73.Virmani M.A., Cirulli M. The role of l-carnitine in mitochondria, prevention of metabolic inflexibility and disease initiation. Int. J. Mol. Sci. 2022;23:2717. doi: 10.3390/ijms23052717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shi L., Tu B.P. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moffatt B.A., Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book. 2002;1 doi: 10.1199/tab.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng Y., Wang Z.V., Gordillo R., Zhu Y., Ali A., Zhang C., et al. Adipocyte Xbp1s overexpression drives uridine production and reduces obesity. Mol. Metab. 2018;11:1–17. doi: 10.1016/j.molmet.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sautin Y.Y., Johnson R.J. Uric acid: the oxidant-antioxidant paradox. Nucleosides Nucleotides Nucleic Acids. 2008;27:608–619. doi: 10.1080/15257770802138558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin J., Ren W., Huang X., Deng J., Li T., Yin Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front Immunol. 2018;9:1697. doi: 10.3389/fimmu.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang R., Yang C., Ma L., Zhao Y., Guo Z., Niu J., et al. Identification of purine biosynthesis as an NADH-sensing pathway to mediate energy stress. Nat. Commun. 2022;13:7031. doi: 10.1038/s41467-022-34850-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Depeint F., Bruce W.R., Shangari N., Mehta R., O'Brien P.J. Mitochondrial function and toxicity: role of the B vitamin family on mitochondrial energy metabolism. Chem. Biol. Interact. 2006;163:94–112. doi: 10.1016/j.cbi.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 81.Janssen J.J.E., Grefte S., Keijer J., de Boer V.C.J. Mito-nuclear communication by mitochondrial metabolites and its regulation by B-vitamins. Front. Physiol. 2019;10:78. doi: 10.3389/fphys.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo J., Yang H., Song B.L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 83.Joffin N., Gliniak C.M., Funcke J.B., Paschoal V.A., Crewe C., Chen S., et al. Adipose tissue macrophages exert systemic metabolic control by manipulating local iron concentrations. Nat. Metab. 2022;4:1474–1494. doi: 10.1038/s42255-022-00664-z. [DOI] [PubMed] [Google Scholar]
- 84.Joffin N., Paschoal V.A., Gliniak C.M., Crewe C., Elnwasany A., Szweda L.I., et al. Mitochondrial metabolism is a key regulator of the fibro-inflammatory and adipogenic stromal subpopulations in white adipose tissue. Cell Stem Cell. 2021;28:702–717. doi: 10.1016/j.stem.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murano I., Rutkowski J.M., Wang Q.A., Cho Y.R., Scherer P.E., Cinti S. Time course of histomorphological changes in adipose tissue upon acute lipoatrophy. Nutr. Metab. Cardiovasc. Dis. 2013;23:723–731. doi: 10.1016/j.numecd.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data in this reported study is open to readers. They are available from K. S. at the University of Texas Health Science Center at Houston upon reasonable request.