Abstract
Remodelling of the nuclear membrane is essential for the dynamic changes of nuclear architecture at different stages of the cell cycle and during cell differentiation. The molecular mechanism underlying the regulation of nuclear membrane biogenesis is not known. Here we show that Smp2, the yeast homologue of mammalian lipin, is a key regulator of nuclear membrane growth during the cell cycle. Smp2 is phosphorylated by Cdc28/Cdk1 and dephosphorylated by a nuclear/endoplasmic reticulum (ER) membrane–localized CPD phosphatase complex consisting of Nem1 and Spo7. Loss of either SMP2 or its dephosphorylated form causes transcriptional upregulation of key enzymes involved in lipid biosynthesis concurrent with a massive expansion of the nucleus. Conversely, constitutive dephosphorylation of Smp2 inhibits cell division. We show that Smp2 associates with the promoters of phospholipid biosynthetic enzymes in a Nem1–Spo7-dependent manner. Our data suggest that Smp2 is a critical factor in coordinating phospholipid biosynthesis at the nuclear/ER membrane with nuclear growth during the cell cycle.
Keywords: lipin, nuclear membrane, phospholipid biosynthesis, Smp2
Introduction
The defining characteristic of eukaryotic cells is the compartmentalization of chromatin inside the nucleus. The nucleoplasm and the cytoplasm are separated by the nuclear envelope that consists of two closely opposed lipid bilayers, the inner and outer nuclear membranes (Gant and Wilson, 1997). The outer nuclear membrane is continuous with the endoplasmic reticulum (ER) and functions in lipid biosynthesis and protein secretion whereas the inner nuclear membrane faces the nucleoplasm and provides an anchoring site for chromatin at the nuclear periphery. In metazoans, the inner nuclear membrane is lined with a filamentous network of intermediate filaments, the nuclear lamina (Gruenbaum et al, 2003). The two membranes are connected at the nuclear pore complexes (NPCs), evolutionarily conserved multiprotein assemblies that mediate nucleocytoplasmic transport (Suntharalingam and Wente, 2003).
Nuclear assembly is a highly complex process that requires the coordinated biosynthesis, targeting and interaction of nuclear membrane with nuclear pores, inner nuclear membrane proteins and chromatin (reviewed in Mattaj, 2004). In animal cells, the nuclear envelope breaks down at the onset of mitosis so that the spindle can access and attach to the chromosomes. In vitro experiments using frog oocyte extracts (reviewed in Burke and Ellenberg, 2002) have demonstrated that following the postmitotic nuclear envelope assembly, the closed nuclear membrane undergoes expansion, presumably to accommodate further chromatin decondensation and later on DNA replication. In contrast to animal cells, budding yeast, like other unicellular fungi, separate its chromosomes within a single intact nucleus that partitions between mother and daughter cell (Byers and Goetsch, 1975). The hallmark of the so-called ‘closed mitosis' of yeast cells is the rapid expansion of the nucleus along the mother–daughter axis that takes place during anaphase and is driven by the elongation of the intranuclear spindle (Yeh et al, 1995). Although the cytology of nuclear division in yeast and animal cells is different, in both cases the nucleus expands during the cell cycle. The molecular mechanism underlying nuclear membrane growth is poorly understood: nuclear growth in cell-free systems takes place via homotypic fusion of vesicles with the outer nuclear membrane and depends on the triple AAA ATPase p97 in complex with p47, Ran-dependent nuclear import and lamins (Newport et al, 1990; Zhang and Clarke, 2000; Hetzer et al, 2001), but how exactly these factors regulate incorporation of a new membrane into the nuclear envelope in vitro and whether they also function in vivo is not known.
The ER is a complex network of membrane tubules and cisternae that is continuous with the outer nuclear membrane, or ‘nuclear ER' and extends throughout the cytoplasm to form the ‘peripheral ER'. In budding yeast, most of the peripheral ER forms a continuous tubular network underlying the plasma membrane (Novick et al, 1980; Preuss et al, 1991). The ER membrane is the major site of lipid biosynthesis and in yeast consists largely of phosphatidylinositol and phosphatidylcholine (Paltauf et al, 1992). Membrane biogenesis at the ER in yeast is regulated primarily by the intracellular concentration of the essential phospholipid precursors inositol and choline (Henry and Patton-Vogt, 1998). When inositol levels are low, a transcription factor complex composed of the basic helix–loop–helix proteins Ino2p and Ino4p activates the expression of many genes encoding phospholipid, fatty acid and sterol biosynthetic enzymes. Conversely, high levels of inositol induce nuclear translocation of Opi1, an ER-localized transcription factor, to repress transcription of phospholipid biosynthetic genes (Loewen et al, 2004). Interestingly, phospholipid biosynthesis is also transcriptionally induced in response to the need for more ER membrane during the unfolded protein response (Cox et al, 1997), suggesting that lipid metabolism is coordinated with ER growth.
The Nup84 complex is an evolutionarily conserved component of the nuclear pore required for nuclear pore biogenesis in yeast (Siniossoglou et al, 1996) and animal cells (Walther et al, 2003). Through a synthetic lethality screen with a nup84 mutant, we have previously identified a complex of two integral nuclear/ER membrane proteins, Nem1 and Spo7, essential for correct nuclear morphology (Siniossoglou et al, 1998). As of yet the function of these proteins have remained elusive. In this study, we show that the Nem1–Spo7 complex is a novel phosphatase, which regulates nuclear growth by controlling recruitment of Smp2, the yeast homologue of the adipogenic factor Lipin 1, onto promoters of phospholipid biosynthetic genes. Our data provide a first evidence for a link between membrane biosynthesis and nuclear envelope growth.
Results
Overproduction of SMP2 restores nuclear membrane structure in nem1 and spo7 cells
In a screen looking for mutations that are synthetically lethal when combined with a knockout of the nucleoporin NUP84, we have previously identified a complex of two integral nuclear and ER membrane proteins, Nem1 and Spo7 (Siniossoglou et al, 1998). Nem1Δ and spo7Δ mutants exhibit a striking proliferation of the nuclear membrane that leads to a substantial expansion of the nucleus and the presence of long nuclear membrane extensions that penetrate into the cytoplasm. These elongations contain nuclear pores and intranuclear soluble proteins but, strikingly, no DNA. Interestingly, despite these defects in nuclear structure, nem1Δ and spo7Δ mutants grow normally at 30°C and do not display defects in nucleocytoplasmic transport (Siniossoglou et al, 1998). Although overexpression of certain ER membrane proteins can induce proliferation of ER membrane stacks, often referred to as ‘karmellae' in yeast and ‘crystalloid ER' in animal cells (Wright et al, 1988; Wanker et al, 1995; Koning et al, 1996), these do not appear to affect the overall size and structure of the nucleus. Thus, Nem1–Spo7 could have a specific role in controlling ER membrane flow and nuclear membrane production.
To elucidate the function of Nem1–Spo7, we decided to search for genes that could suppress the nuclear membrane proliferation of spo7Δ knockout cells. We reasoned that rescuing the nuclear defects of the spo7Δ mutant could also suppress its synthetic lethality with the nup84Δ. Therefore, we conducted a screen for genes that, in high copy, can suppress the synthetic lethality of the nup84Δspo7Δ double knockout strain. This genetic approach identified SMP2 (Figure 1A), a gene originally isolated in a screen for mutants showing increased stability of heterologous plasmids (Irie et al, 1993). Smp2Δ deletion mutants are viable but grow slowly at 30°C and exhibit temperature-sensitive growth at 37°C (data not shown). We found that overexpression of SMP2 in spo7Δ, nem1Δ and nem1Δspo7Δ mutants can efficiently (a) suppress their slow growth phenotypes at 37°C (Figure 1B and data not shown) and (b) restore a normal nuclear envelope and nuclear structure in all three mutants, as judged by a nuclear/ER membrane (Sec63-GFP) and nucleoplasmic (GFP-Pus1) reporters, respectively (Figure 1C and data not shown). In contrast, overproduction of SMP2 does not suppress the temperature sensitivity of the nup84Δ mutant (data not shown). We conclude that overexpression of SMP2 can overcome the requirement for the Nem1–Spo7 complex in nuclear membrane organization.
Figure 1.
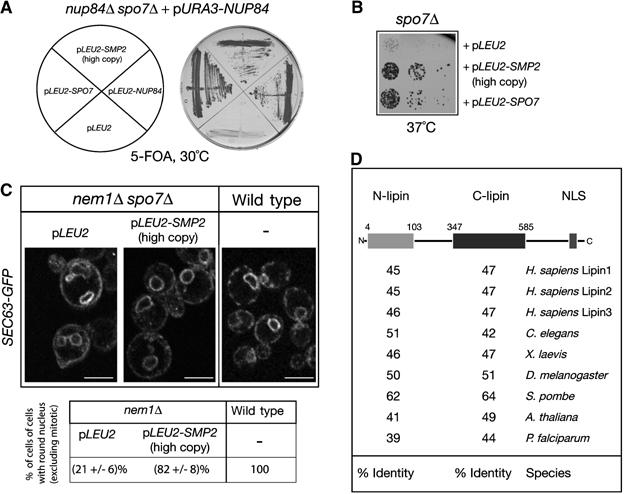
Functional interaction between NEM1–SPO7 and SMP2. (A) Identification of SMP2 as a high-copy number suppressor of the nup84Δ spo7Δ synthetic lethal mutant. The nup84Δ spo7Δ double deletion strain carrying a centromeric vector expressing NUP84 was transformed with the indicated plasmids. Transformants were grown on plates containing 5-FOA for 3 days. (B) SMP2 rescues the temperature-sensitive growth defect of spo7Δ cells. Spo7Δ cells transformed with the indicated plasmids were diluted in YEPD, spotted onto selective (-Leu) plates and grown at 37°C for 2 days. (C) Overexpression of SMP2 suppresses the nuclear membrane proliferation of nem1Δ spo7Δ cells. Upper panel: the nem1Δ spo7Δ mutant or the isogenic wild-type strain, expressing the ER marker Sec63-GFP were transformed with the indicated plasmids. Transformants were visualized by confocal microscopy. Bars, 5 μm. Lower panel: percentage of cells with no or small buds containing a round nucleus in wild-type and nem1Δ strains expressing GFP-Pus1. Three different transformants per strain were analyzed and for each one the number of cells counted was n=200. (D) Smp2 is evolutionarily conserved. Schematic representation of the primary structure of Smp2. The gray and black boxes indicate the highly conserved amino-terminal (N-lipin) and C-terminal (C-lipin) domains within Smp2. The percent sequence identity between the yeast domains and putative orthologues in various species is given.
Smp2 belongs to a large family of evolutionarily conserved proteins present in eukaryotic species. The founding member of this family is Lipin1, a recently identified nuclear protein required for adipocyte differentiation whose mutation results in the fatty liver dystrophy (fld) phenotype in mice (Peterfy et al, 2001). All members of the Lipin family share a high degree of similarity in two domains of unknown function, N-lipin located next to their N-termini, and C-lipin found close to their C-termini (Figure 1D). PSI-BLAST searches show that the C-lipin domain is distantly related to the conserved C-terminal domain of the rdg/Nir family of proteins involved in lipid trafficking and signalling (Lev, 2004; our unpublished observations). Mammalian species express three Lipin orthologues that share no significant homology outside the two conserved domains. Smp2, like most members of the Lipin family, contain a predicted C-terminal nuclear localization signal.
Thin section electron microscopy (Figure 2A) and double labelling using an intranuclear GFP fusion that depicts nuclear structure (Siniossoglou et al, 1998) and DNA staining revealed that cells lacking SMP2, like the spo7Δ, and nem1Δ mutants, have enlarged and irregularly shaped nuclei, often consisting of two or more interconnected lobes within a single cell (Figure 2B). The most remarkable feature of the smp2Δ cells is the presence of long nuclear membrane projections that appear to be directly connected to the main body of the nucleus. We noticed often that these projections appear to associate with the peripheral ER at the cell cortex. Although we can occasionally see short arrays of stacked ER membranes tightly associated with the nuclear envelope, these do not have the ordered arrangement of karmellae membranes around the nucleus.
Figure 2.
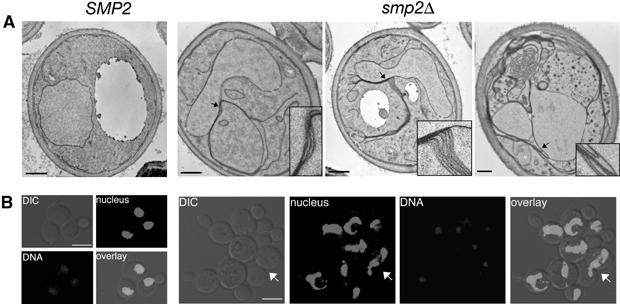
Deletion of SMP2 induces nuclear membrane proliferation and nuclear expansion. (A) Thin section electron microscopy of wild-type (SMP2) or smp2Δ cells grown at 30°C and stained with potassium permanganate. Detail panels show enlargements of areas of the nuclear envelope in smp2Δ cells (highlighted by arrows) that display membrane proliferation. Bars, 0.5 μm. (B) DNA staining of wild-type (SMP2) or smp2Δ knockout cells expressing an intranuclear GFP-reporter (GFP-Pus1) used to depict nuclear structure (‘nucleus'). Cells were grown in selective medium at 30°C, fixed for 30 min and inspected by confocal microscopy. The white arrow points to a dividing yeast cell. Bars, 5 μm.
The Nem1–Spo7 complex dephosphorylates Smp2
In order to understand the role of Smp2 in nuclear membrane biogenesis, we set out to define its functional link with Nem1–Spo7. Multiple sequence alignments reveal that the conserved carboxy-terminal domain (CTD) of Nem1 contains a predicted Ser/Thr phosphatase CPD (CTD-like phosphatase) domain. The hallmark of the CPD phosphatases is a stretch of 11 residues, different from the phosphatase motifs of the other known protein phosphatase families, the DXDX (T/V) motif, with the first aspartate acting as the phosphoryl acceptor residue (Kobor et al, 1999). To test directly whether Nem1–Spo7 has phosphatase activity, we affinity purified the native complex from detergent-solubilized extracts derived from yeast cells expressing a functional Nem1-PtA using IgG-Sepharose chromatography (Figure 3A) and assayed it against an artificial substrate (p-nitrophenylphosphate ‘p-NPP'). As seen in Figure 3B, the Nem1p–Spo7p complex exhibits phosphatase activity. This effect is specific since no p-NPP hydrolysis was observed when the Nem1[D257A]–Spo7 complex, carrying a point mutation in the Nem1 phosphoacceptor site, was used in the assay. Moreover, in the absence of Spo7, Nem1 did not provide any detectable activity. This finding explains why the individual nem1Δ and spo7Δ cells display very similar, if not identical, phenotypes and genetic interactions with the nup84Δ mutant (Siniossoglou et al, 1998). Taken together, these data strongly suggest that within the Nem1–Spo7 holoenzyme, Nem1p is the catalytic subunit and Spo7 the regulatory subunit necessary for catalytic activity in vitro.
Figure 3.
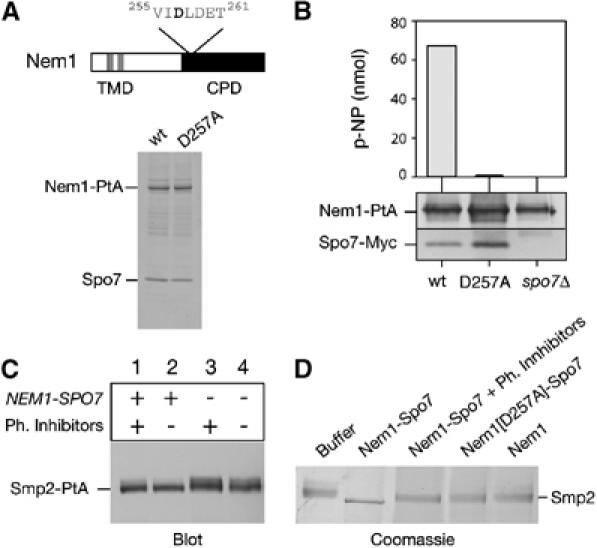
Dephosphorylation of Smp2 by the Nem1–Spo7 complex. (A) Upper panel: domain organization of Nem1. TMD, transmembrane domain. CPD, CTD Phosphatase domain. The DLD phosphoacceptor site is indicated. Lower panel: affinity purification of Nem1-PtA (‘wt') and Nem1[D257A]-PtA (‘D257A'). Purified proteins were analyzed by SDS–PAGE and Coomassie staining. The positions of the PtA-fusions and copurifying Spo7 are indicated. (B) The Nem1–Spo7 complex exhibits phosphatase activity in vitro. In vitro dephosphorylation of p-nitrophenylphosphate (p-NPP) by the Nem1–Spo7 complex. IgG-Sepharose beads loaded with (Nem1-PtA)–(Spo7-Myc), (Nem1[D257A]-PtA)–(Spo7-Myc) or Nem1-PtA fusions, were tested for the ability to hydrolyze p-NPP as described under Materials and methods. Absorbance of the generated p-nitrophenol (p-NP) was measured at 410 nm. The amount of Nem1 and Spo7 in each reaction was followed by Western blot with anti-PtA and anti-Myc antibodies respectively. (C) Nem1–Spo7 is a phosphatase for Smp2. Protein extracts from smp2Δ (lanes 1 and 2) or nem1Δ spo7Δ smp2Δ (lanes 3 and 4) strains expressing a Smp2-PtA fusion were prepared in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of phosphatase inhibitors. Smp2-PtA was detected by western blot using anti-PtA antibody. (D) In vitro dephosphorylation of Smp2 by the Nem1–Spo7 complex. Native Smp2 (2 μg) was incubated with IgG-Sepharose beads alone (‘buffer') or beads containing the indicated protein A fusions for 30 min at 30°C. Reactions were resolved by 7% SDS–PAGE and Coomassie stained.
This result prompted us to test whether Smp2 could be the physiological substrate of Nem1–Spo7 in vivo. To this end, we tried to resolve Smp2 mobility shifts by SDS–PAGE, which are often indicative of phosphorylation. A functional Protein A tagged Smp2 fusion (Smp2-PtA) expressed from the SMP2 promoter in smp2Δ cells migrates predominantly as a single band in wild-type cells (Figure 3C, lane 2). A second band of reduced electrophoretic mobility could also be detected in the wild-type extracts prepared in the presence of phosphatase inhibitors (Figure 3C, lane 1). Furthermore, the reduced electrophoretic mobility band was stronger in extracts from the nem1Δspo7Δ strain (Figure 3C, lanes 3 and 4). These results show that Smp2 is phosphorylated in vivo and that Nem1–Spo7 is required for Smp2 dephosphorylation.
To test whether Nem1–Spo7 directly catalyses Smp2 dephosphorylation, we purified Smp2-PtA from a nem1Δspo7Δ smp2Δ strain and eluted the native Smp2 off the IgG-Sepharose beads by TEV protease cleavage. Purified Smp2 was then incubated with IgG-Sepharose beads containing Nem1-PtA–Spo7 complex in vitro. The Smp2 doublet collapses down to a faster migrating form in a phosphatase inhibitor-dependent manner (Figure 3D). Importantly, the Nem1[D257A]–Spo7 complex containing the phosphoacceptor mutant was inactive in this assay showing that this mobility shift depends on a catalytically active Nem1–Spo7 complex. Overall, these data show that Nem1–Spo7 complex is a phosphatase for Smp2. The fact that deletion of NEM1-SPO7, like that of SMP2, induces nuclear membrane proliferation suggests that phosphorylation of Smp2 inactivates SMP2 and drives nuclear membrane production.
Smp2 is phosphorylated by Cdc28 at the onset of mitosis
Smp2 was recently identified in a large-scale screen for in vitro substrates of Cdc28/Cdk1 (Ubersax et al, 2003). In budding yeast, Cdc28 is the only cyclin-dependent kinase that, in complex with any of the three G1 and six B-type cyclins, drives cell cycle progression (Mendenhall and Hodge, 1998). Several lines of evidence imply that Cdc28/Cdk1 phosphorylates in vivo Smp2: first, we find that the phosphorylation of an Smp2-PtA fusion is cell cycle regulated, with the modification appearing at the onset of mitosis (Figure 4A) and causing a mobility shift similar to that seen in the nem1Δspo7Δ mutant. Second, Smp2 phosphorylation was abolished in the cdc28-4 mutant after shifting the cells to the restrictive temperature (Figure 4B, left panel). Third, although phosphorylation of Smp2 is mostly unaffected in the six single B-type yeast cyclin deletion mutants (CLB1 to 6, data not shown), we found that it decreased in cells lacking the pair of the closely homologous mitotic cyclins CLB3 and CLB4 but not in cells lacking the S-phase cyclins CLB5 and CLB6 (Figure 4B, right panel). Fourth, Smp2 is recognized in vivo by an antibody specific for phosphorylated Cdc28/Cdk1 sites (anti-MPM2, Davis et al, 1983), in a Nem1–Spo7- dependent manner (Figure 4C). Taken together, these data are consistent with Smp2 being phosphorylated in a cell-cycle-dependent manner by mitotic cyclin-Cdc28/Cdk1 complexes.
Figure 4.
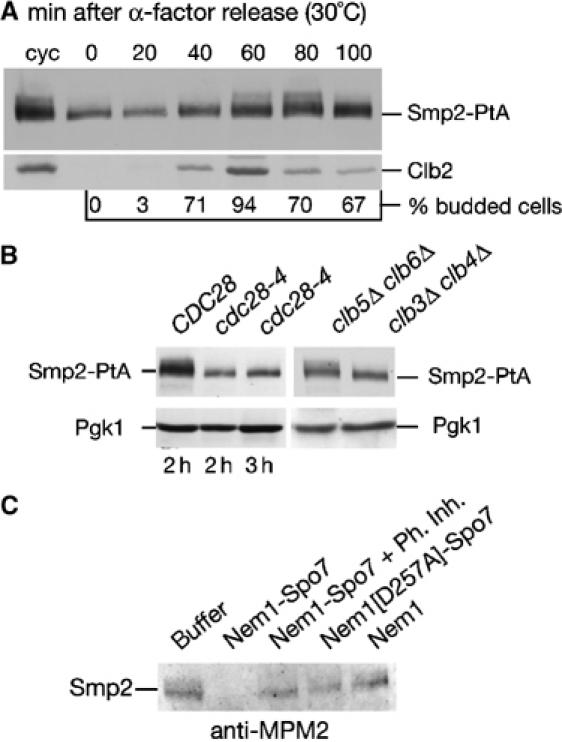
Smp2 is phosphorylated in a cell-cycle-dependent manner by Cdc28. (A) Phosphorylation of Smp2 takes place during mitosis. Cells expressing Smp2-PtA were synchronized by alpha-factor arrest/release. Protein extracts were prepared every 20 min and analyzed by Western blot using the indicated antibodies. Cell cycle stages were monitored by Clb2 cyclin levels and budding index. (B) Left panel: protein extracts from wild-type (‘CDC28') or cdc28-4 cells expressing Smp2-PtA grown for the indicated times at 37°C were analyzed by Western blot using anti-PtA antibodies. Right panel: protein extracts from clb3Δclb4Δ and clb5Δclb6Δ cells expressing Smp2-PtA were analyzed as above. (C) Native Smp2 is recognized by the MPM2 antibody in a Nem1–Spo7-dependent manner. Samples from the experiment in Figure 3D were transferred onto nitrocellulose membrane and incubated with the MPM2 antibody.
Deletion of phosphorylated Smp2 inhibits cell division
Deletion of the Nem1–Spo7 induces nuclear expansion that can be suppressed by overexpression of its substrate Smp2 (Figure 1C). This result supports the idea that Nem1–Spo7 mediated dephosphorylation of Smp2 represses nuclear growth during interphase. What could be the consequence of constitutive dephosphorylation of Smp2? To address this question, we coexpressed NEM1 and SPO7 under the control of the strong inducible GAL1/10 promoter. As seen in Figure 5A, overexpression of the Nem1–Spo7 complex is lethal, but importantly, only in the presence of its substrate Smp2. Thus, the toxic effect of Nem1–Spo7 overexpression in wild-type cells is mediated by Smp2. Western blot analysis of lysates prepared from galactose-induced cells shows that Nem1-PtA levels reach a maximum by 8 h (data not shown) while an Smp2-PtA fusion collapses to a faster migrating form, consistent with its in vivo dephosphorylation by Nem1–Spo7 (Figure 5B). An examination of the morphology of cells overexpressing Nem1–Spo7 for 9 h showed that they contained a significantly higher percentage of large-budded cells with GFP-Pus1 labeled nuclei at the bud neck (data not shown). In agreement with an increase on the proportion of mitotic cells, spindle length measurements using a tubulin-GFP fusion revealed that 50% of the cells at 9 h induction contain short spindles positioned at the bud neck (Figure 5C). Cells overexpressing Nem1–Spo7 continued to grow in size until 21 h of induction (Figure 5D, compare size of Nem1–Spo7 and vector control) after which they lysed. The fact that deletion of SMP2 rescues these defects (Figure 5C and D), suggests that the accumulation of dephosphorylated Smp2 inhibits mitotic division.
Figure 5.
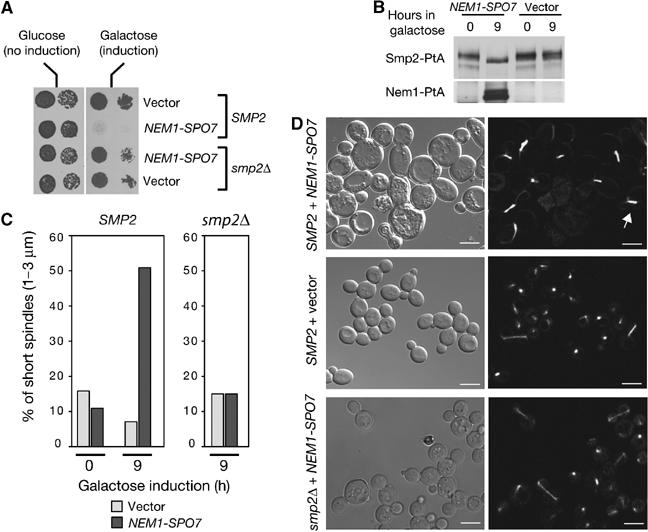
Accumulation of dephosphorylated Smp2 inhibits cell division. (A) Overexpression of the Nem1–Spo7 complex is lethal only in the presence of Smp2. Wild-type (‘SMP2') or smp2Δ cells transformed with centromeric vectors expressing NEM1 and SPO7 under the control of the GAL1/10 promoter, or with the corresponding empty vectors, were spotted onto selective plates supplemented with glucose or galactose and grown at 30°C. (B) Cells overexpressing NEM1–SPO7 for the indicated times were analyzed for Smp2-PtA mobility shifts by Western blot using anti-PtA antibodies. (C) Wild-type or smp2Δ cells overexpressing NEM1–SPO7 or empty vectors were scored for short spindles (1–3 μm) at the indicated times by using a tubulin-GFP reporter. (D) Cell (left panels) and spindle (right panels) morphologies using a tubulin-GFP fusion of wild-type or smp2Δ strains overexpressing NEM1–SPO7 or empty vector for 21 h. The white arrow points to a typical short spindle. Bars, 5 μm.
Smp2-mediated nuclear growth depends on phopholipid biosynthesis
How does Smp2 control nuclear membrane growth? Since the outer nuclear membrane is continuous with the ER, we hypothesized that factors controlling ER membrane production could also be responsible for the nuclear membrane expansion observed in smp2Δ, spo7Δ and nem1Δ mutants. The major lipid components of the ER membrane in yeast are phosphatidylinositol and phosphatidylcholine. Production of these lipids is regulated primarily by the availability of the soluble phospholipid precursor inositol: repression is mediated by the negative regulator Opi1 (White et al, 1991) and activation by the global phospholipid transcription complex Ino2/Ino4 (Hoshizaki et al, 1990; Nikoloff et al, 1992). Two experiments show that de novo phospholipid biosynthesis is actually required for the nuclear membrane growth in smp2Δ and nem1Δ spo7Δ cells. First, disruption of phospholipid biosynthesis, by deleting the activator INO2 represses nuclear expansion and restores a spherical nucleus in 90% of the nem1Δ spo7Δ and smp2Δ cells as seen by labelling the nuclear membrane with Sec63-GFP (Figure 6, compare panel B with C and panel E with F) or the nucleus with Pus1-GFP (Supplementary Figure 1). Similarly, overexpression of the repressor OPI1 (which renders cells inositol auxotrophs) in the nem1Δ spo7Δ mutant results in an efficient suppression of the nuclear membrane proliferation (Figure 6, panel D). Second, the mRNA levels of INO1, the rate-limiting enzyme for phosphatidylinositol synthesis and its transcriptional activator INO2 showed a significant upregulation in cells lacking SMP2 or NEM1–SPO7 with respect to the wild type, as monitored by standard and quantitative RT–PCR experiments (Figure 7A and B). Moreover, transcription of OPI3, the enzyme catalyzing the final steps in the production of phosphatidylcholine (McGraw and Henry, 1989) is also upregulated. The increase in the mRNA levels is higher in smp2Δ than nem1Δ spo7Δ mutants, consistent with the fact that smp2Δ cells exhibit a more severe nuclear membrane proliferation than its phophatase mutants. Interestingly, mRNA levels of the resident ER membrane protein Sec63, or nuclear envelope components like the nucleoporins Nup49 and Nup84 are not altered (Figure 7A and B). Thus, deletion of SMP2 or its phosphatase complex NEM1–SPO7 induce the expression of enzymes involved in lipid synthesis of the ER/nuclear membrane but not the protein components that assemble on them.
Figure 6.
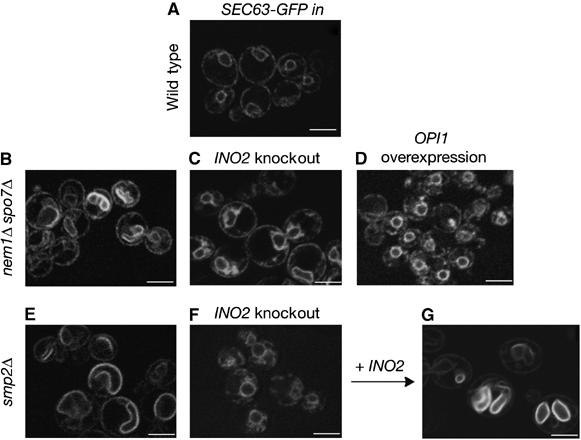
Inhibition of the phospholipid biosynthetic pathway restores normal nuclear membrane structure in smp2Δ and nem1Δ spo7Δ cells. SEC63-GFP was used to visualize nuclear membrane structure in wild type (A), nem1Δ spo7Δ (B), nem1Δ spo7Δ ino2Δ (C), nem1Δ spo7Δ overexpressing OPI1 (D), smp2Δ (E), smp2Δ ino2Δ (F) and smp2Δ ino2Δ complemented by a plasmid expressing INO2 (G) strains. Transformants in early logarithmic phase were visualized by confocal microscopy. Bars, 5 μm.
Figure 7.
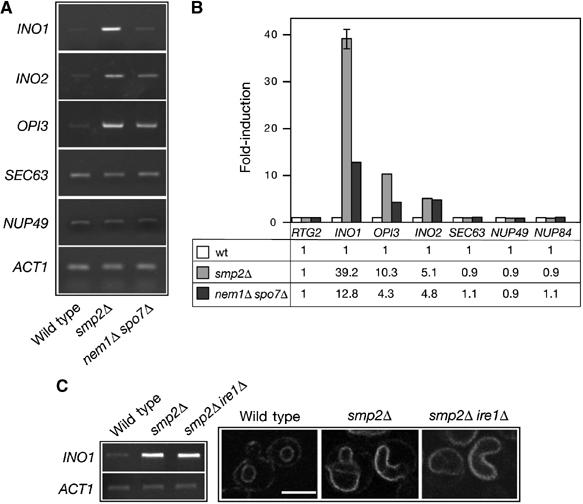
Smp2 regulates expression of phospholipid biosynthetic genes. (A) Transcription of key genes involved in phospholipid biosynthesis is upregulated in smp2Δ and nem1Δ spo7Δ mutants. The mRNA levels of INO1, INO2, OPI3, SEC63, NUP49 and ACT1 were analyzed in the smp2Δ, nem1Δ spo7Δ and isogenic wild-type strains by semiquantitative RT–PCR. (B) As in (A), but the mRNA levels of INO1, INO2, OPI3, SEC63, NUP49 and NUP84 were analyzed by quantitative RT–PCR. Amplification of each sample was performed in triplicate and normalized to a control gene, RTG2, which is expressed at similar level to those analyzed and is unaffected by smp2Δ or nem1Δ spo7Δ mutations. Errors were less than 5% except for the smp2Δ INO1 sample. The fold-difference for the three strains is given below. (C) Upregulation of phospholipid biosynthesis in smp2Δ is independent of the unfolded protein response (UPR) pathway. The mRNA levels of INO1 and ACT1 were analyzed in the smp2Δ, smp2Δ ire1Δ and isogenic wild-type strains by RT–PCR (left panel). The nuclear morphology of smp2Δ, smp2Δ ire1Δ and isogenic wild-type cells expressing Sec63-GFP reporter was visualized by confocal microscopy (right panel). Bar, 5 μm.
Since phospholipid biosynthesic genes can be induced by the unfolded protein response (UPR) pathway (Cox et al, 1997; Travers et al, 2000), we tested whether the upregulation of INO1 expression observed in smp2Δ mutants depends on Ire1. Ire1 initiates the UPR and mediates the splicing of a precursor mRNA encoding the transcription factor Hac1 that upregulates expression of many genes, including INO1 (Cox et al, 1997). As shown in Figure 7C, Ire1 is dispensable both for the upregulation of INO1 mRNA levels and the nuclear membrane expansion in smp2Δ cells. The same results were obtained with the nem1Δ spo7Δ ire1Δ mutant (data not shown). These data suggest that the regulation of nuclear membrane growth by Smp2 takes place by an UPR-independent mechanism.
Phosphorylation of Smp2 regulates its recruitment on promoters of lipid biosynthetic enzymes
The above results prompted us to test whether Smp2 could play a more direct role in the regulation of lipid biosynthetic genes. Although Smp2 does not contain any sequence that might resemble a DNA-binding motif, it could be indirectly recruited to specific promoters by interaction with other factors. To test this possibility, we performed chromatin inmunoprecipitation of an Smp2-PtA fusion, followed by real-time PCR analysis. We found that a pool of Smp2 associates in fact with the INO1, INO2 and OPI3 promoters (Figure 8A, black bars). The amounts of Smp2 in these promoters oscillate between 3.4- and 4-fold over the background levels detected at an intergenic region in chromosome V. This interaction is specific since no significant amount of Smp2 could be detected at the promoters of unrelated genes such as MPP10 (Figure 8A). Moreover, chromatin immunoprecipitation (ChIP) from an Smp2 untagged strain or strain expressing a control Protein-A fusion (Psr1-PtA) yielded no product (data not shown). Interestingly, no Smp2-PtA could be crosslinked to the coding region of INO1, INO2 or OPI3. The absence of Smp2 from the open reading frames of the lipid biosynthetic genes is consistent with the role of Smp2 as a transcriptional repressor and the increase in the mRNA levels observed in an smp2Δ strain (Figure 7A). Importantly, the association of Smp2 with the promoters of INO1, INO2 and OPI3 was disrupted in a strain harboring a deletion of the Smp2 phosphatase, NEM1–SPO7 (Figure 8A, gray bars). The lack of association of Smp2 with chromatin was not due to a decrease in the intracellular pool of Smp2 in the nem1Δ spo7Δ strain (our unpublished data, see also Figure 3C). Hence, recruitment of Smp2 on the promoters of the lipid biosynthetic genes is regulated by phosphorylation: loss of the dephosphorylated form of Smp2 due to deletion of its phosphatase abrogates its association with chromatin.
Figure 8.
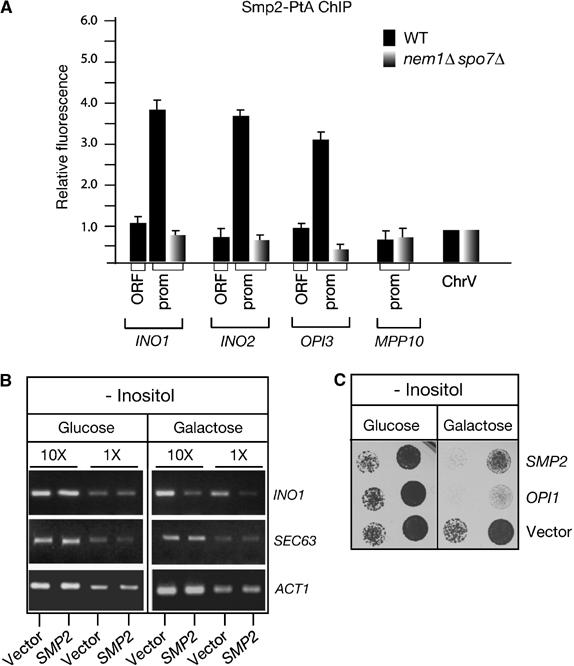
Recruitment of Smp2 on the promoters of phospholipid biosynthetic genes. (A) Smp2 associates with the promoters of lipid biosynthetic genes. Chromatin inmunoprecipitation of Smp2-PtA was performed in wild-type and isogenic nem1Δ spo7Δ strains. The chromatin associated Smp2 was quantified by real-time PCR. Histogram bars represent relative fluorescence units, calculated as described under Materials and methods. An intergenic region in chromosome V was used as background control and arbitrarily given the value 1. (B) Transcription of INO1 is downregulated in cells overexpressing SMP2. The mRNA levels of INO1, SEC63 and ACT1 were analyzed by RT–PCR in cells overexpressing SMP2. Two different dilutions of the mRNA are shown. (C) Overexpression of Smp2 inhibits growth in media lacking inositol. Wild-type cells transformed with plasmids expressing SMP2 or OPI1 under the control of the GAL1/10 promoter or the respective empty vector, were spotted onto selective plates lacking inositol and supplemented with either glucose or galactose as carbon source and grown at 25°C.
Wild-type cells respond to inositol starvation by a strong induction of phospholipid biosynthetic genes (Henry and Patton-Vogt, 1998). If Smp2 can repress transcription of inositol-responsive genes, increasing the Smp2 levels could inhibit their induction and cause defects similar to those described in cells overexpressing the corepressor Opi1 (White et al, 1991; Wagner et al, 1999). To test this hypothesis, the Smp2 gene was expressed under the control of the GAL1/10 promoter in media lacking inositol. Indeed, overexpression of SMP2 results in a reduction of INO1 (Figure 8B) and OPI3 (not shown) mRNA expression and severely impairs growth of wild-type cells in the absence of inositol (Figure 8C). These data, together with the fact that Smp2 is recruited onto the INO1, INO2 and OPI3 promoters, support the role of Smp2 as a transcriptional regulator of lipid biosynthetic enzymes.
Discussion
During the cell cycle, membrane-bound organelles must increase in size before they can divide and be delivered to the daughter cell. The nuclear membrane undergoes expansion, following its postmitotic reassembly in metazoan cells or during anaphase, in fungi. The molecular mechanism underlying nuclear expansion is not understood but certainly involves a coordinated increase on the lipid (nuclear membrane) and protein components (nuclear pores and, in metazoans, lamina) of the nuclear envelope. In this study, we show that the yeast lipin Smp2 provides a novel link between nuclear growth and membrane biogenesis. Lack of SMP2 or its dephosphorylated form generated by the phosphatase Nem1–Spo7 induces transcriptional upregulation of key genes involved in phospholipid biosynthesis concomitant with a massive expansion of the nucleus. Importantly, Smp2 is recruited onto the promoters of phospholipid biosynthetic enzymes in a Nem1–Spo7 dependent manner. These data have important implications on the mechanisms by which cells remodel nuclear envelope structure at different stages of the cell cycle.
Nem1 and Spo7 were originally identified as essential for the maintenance of a spherical nucleus in yeast (Siniossoglou et al, 1998). The two proteins form a complex that is anchored to the nuclear membrane and ER. Here we show that the Nem1–Spo7 complex is a phosphatase and that both subunits are required for the catalytic activity against its in vivo substrate, Smp2. Nem1, the catalytic subunit of the complex, belongs to a new phosphatase family, typified by Fcp1, the major phosphatase of the CTD of RNA Polymerase II (Kobor et al, 1999) and contains an essential DXDX(T/V) phosphoacceptor motif. The role of Spo7 is less clear. The fact that it binds tightly to the Nem1 C-terminal catalytic domain (Siniossoglou et al, 1998) and that it is necessary for Nem1 activity in vitro (Figure 3B and C), advocate its function as a cofactor required for the phosphatase activity and/or substrate specificity in vivo.
Several lines of evidence indicate that phosphorylated Smp2 is a Nem1–Spo7 substrate. First, Smp2 undergoes phosphatase inhibitor-dependent mobility shifts in vivo as a result of NEM1 and SPO7 deletion. Second, overexpression of the Nem1–Spo7 complex in wild-type cells induces the collapse of Smp2 to a faster migrating band. Third, purified Nem1–Spo7 complex catalyze the dephosphorylation of Smp2 in vitro in a DXDX(T/V)-dependent manner. These evidences are supported by the suppression of nem1Δ spo7Δ mutants by SMP2 and the fact that Smp2 can be co-precipitated with Nem1–Spo7 from cell lysates (S Siniossoglou, unpublished data). The fact that deletion of SMP2 causes the same phenotype as deletion of its phosphatase, indicates that it is the dephosphorylated Smp2 that represses phospholipid biosynthesis and nuclear growth. Consistent with this model, accumulation of dephosphorylated Smp2 is toxic and results in a significant increase of cells with short metaphase spindles. Importantly, removal of NEM1 or SPO7 from an smp2Δ strain does not result in additional growth impairment, nuclear membrane proliferation or transcriptional activation of phospholipid biosynthetic genes (our unpublished data). Moreover, the growth defects and phenotypes of nem1Δ, spo7Δ and nem1Δspo7Δ can be efficiently rescued by overexpression of SMP2. These observations argue that the major, if not the only, role of the Nem1–Spo7 complex is to repress nuclear growth by dephosphorylating Smp2.
Different environmental or developmental signals can regulate membrane biogenesis in the ER according to various cellular needs. Transcription of phospholipid biosynthetic enzymes is induced when yeast cells need to produce more ER-membrane during the unfolded protein response to accommodate increased levels of chaperones (Cox et al, 1997). The transcription factor XBP1 has been shown recently to mediate the massive expansion of the ER membrane network during the differentiation of B cells into antibody-secreting plasma cells (Shaffer et al, 2004; Sriburi et al, 2004). Our data indicate that Smp2 plays a critical role in remodelling nuclear structure by controlling expression of phospholipid biosynthetic genes. What could be the function of a pathway that controls nuclear growth in yeast? Fungi undergo a closed mitosis where the nuclear envelope remains intact throughout nuclear division (Byers and Goetsch, 1975). Spindle elongation during the metaphase to anaphase transition is accompanied by a rapid extension of the nucleus through the bud neck. The spindle elongates from around 2 μm length in the round metaphase nucleus to 12 μm in the late anaphase, when the nucleus adopts a characteristic ‘hour-glass' shape with two lobes interconnected by a narrow stem (Yeh et al, 1995). The presence of an intact nuclear envelope throughout the spindle polymerization process requires the addition of new membrane components to the envelope, so that the elongating nucleus can partition between mother and daughter cells. Phosphorylation of Smp2 by Cdc28/Cdk1 at the onset of mitosis could play a critical function in this process by derepressing transcription of phospholipid enzymes and inducing a short pulse of membrane synthesis. This could provide a mechanism by which yeast cells coordinate lipid biosynthesis in the nuclear/ER membrane with nuclear growth during mitosis.
The nuclear membrane is generally considered as a specialized domain of the ER and several studies demonstrated that small molecules freely diffuse between the two compartments (Baumann and Walz, 2001). A key question is what limits diffusion of bulk ER membrane at the outer nuclear membrane–ER junctions. Cells that overproduce cytosolic ER stacks originating from the outer nuclear membrane known as karmellae (Wright et al, 1988; Wanker et al, 1995; Koning et al, 1996) or intranuclear membrane arrays (Bastos et al, 1996; Marelli et al, 2001) appear to have a normal nucleus suggesting the existence of a barrier at the nuclear envelope that controls nuclear expansion. It is possible that chromatin tethering to the inner nuclear membrane restricts the expansion of the nucleus during interphase. Temporal loss of tethering during mitosis (Hediger et al, 2002) combined with a short pulse of membrane synthesis at the ER might drive nuclear growth. Alternatively, phosphorylated Smp2 might, directly or indirectly, mark sites onto the nuclear membrane where phospholipid biosynthesis could take place.
The function of Smp2 seems to be conserved since mutations in the Smp2 homologue of the distantly related yeast Saccharomyces pombe, cause similar defects in nuclear structure (Tange et al, 2002). Recent studies have shown that a mouse homologue of Smp2, Lipin1, is involved in the regulation of key transcription factors controlling adipogenesis including the peroxisome proliferator activated receptor (PPAR)γ and CCAAT enhancer binding protein (C/EBP)α (Phan et al, 2004). Importantly, BLAST searches indicate that Nem1 is also conserved in higher eukaryotes. We have identified a human gene encoding a membrane bound CPD phosphatase closely related to Nem1p that localizes to the nuclear envelope and ER (our unpublished data). A recent proteomic screen for novel nuclear envelope proteins has identified the same protein as an integral nuclear envelope component (Schirmer et al, 2003). Although the open mitosis of animal cells places different constraints in the biogenesis of the nuclear envelope, a number of studies have shown that a significant increase in nuclear size occurs following the postmitotic nuclear assembly in cell-free systems (Hetzer et al, 2001) and during G1 phase in mammalian cells (Maul et al, 1972; Yen and Pardee, 1979; Yang et al, 1997). It will be interesting to determine whether Lipin homologues could have an analogous function in nuclear growth in mammalian cells.
Materials and methods
Yeast strains, media and plasmids; high copy number suppressor screen with a nup84Δspo7Δ mutant; affinity purifications of Protein A fusions and phosphatase assays; antibodies are available as Supplementary data at The EMBO Journal Online.
RT–PCR analysis
Yeast strains were grown on rich media (YPD) to an early logarithmic phase. Total mRNA was isolated using the Rneasy plus Oligotex mRNA Kit (Qiagen). RT–PCR was performed following the instructions of the OneStep RT–PCR (Qiagen). A total of 26 cycles of PCR reactions were performed on 40, 4 and 0.4 ng of mRNA to ensure the linear range of amplification. The annealing temperature was 62°C. Controls carried out in the absence of RT yielded no PCR products, thus indicating that amplification was not caused by DNA contaminants. Primer sequences are available upon request.
For quantitative RT–PCR analysis, the total mRNAs prepared for the standard RT–PCR were used to synthesize cDNA with Superscript IITM Rnase H-reverse transcriptase (Invitrogen) and 18 mer oligo dT primers (Ambion). The resulting cDNAs were analyzed by real-time PCR on an ABI Prism 7000 (ABI) using Sybr Green following the same procedure described for the ChIP analysis. The cDNAs were quantified by reference to a standard curve of wild-type yeast genomic DNA (of known concentration) diluted 1:10, 1:100, 1:1000, 1:10 000 and 1:100 000. The results from each strain were standardized with respect to an average expressed mRNA (RTG2).
Chromatin immunoprecipitation
ChIP analysis was performed as described (Morillon et al, 2003). Chromatin was prepared from cells expressing a functional Smp2-PtA fusion, a nontagged Smp2 and a control Protein A tagged plasma membrane fusion Psr1-PtA. Chromatin was sonicated to produce fragments with an average size of 400–500 bp (size range determined for each experiment) and immunoprecipitated with 20 μl of IgG Sepharose (Amersham Bioscience) or 20 μl of Sepharose (non-IgG control) per ChIP. PtA-Smp2p tagged, Smp2p nontagged (negative control) and PtA-Psr1 (negative control) strains were processed in parallel.
Real-time PCR analysis was performed on an ABI Prism 7000 (ABI) using Sybr Green. Standard curve of wild-type yeast genomic DNA serially diluted was done for each measurement. To get amplification within the linear range, the input chromatin was diluted 1/30 and IPs were nondiluted. Each measurement was triplicated and the experiments repeated in two different genetic backgrounds. After each run, a melting curve was performed to ensure that no primer dimmers contaminate the quantification and that the product has the expected melting temperature. Samples were also subject to agarose gel electrophoresis to confirm the integrity of the product. Relative fluorescent intensities are calculated from the following formula:
[(∑IP signals/∑IP samples)−(∑No IgG signals/∑No Ab samples)]/(∑input signals/∑input samples).
An intergenic region in chromosome V was used as background control and arbitrary given value 1. The primers used for PCR analysis are available on request.
Microscopy
To view live cells expressing the various GFP fusions, cells were grown in the appropriate selective medium at 25°C to early logarithmic phase and examined with an LSM 510 Zeiss confocal microscope. Spindle length measurements were performed in living cells expressing TUB1-GFP at 30°C grown in selective medium lacking uracil and leucine and supplemented with 2% galactose. Thin section electron microscopy was performed as previously described (Kaiser and Schekman, 1990).
Supplementary Material
Supplementary Information
Supplementary Figure 1
Supplementary Table I
Acknowledgments
We are particularly grateful to Hugh Pelham for stimulating discussions and generous support during the initial phase of this project in the MRC-LMB. We thank Drs E Hettema, HJ Schueller and M Segal for reagents and strains, Dr T Kouzarides for sharing the ABI Prism 7000 cycler, Dr N Bright for help with the electron microscopy and Drs E Hurt, P Luzio and H Pelham for comments on the manuscript. This work was supported by a Wellcome Trust Career Development Fellowship in Basic Biomedical Science to SS.
References
- Basco RD, Segal MD, Reed SI (1995) Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol Cell Biol 15: 5030–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B (1996) Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol 134: 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Walz B (2001) Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205: 149–214 [DOI] [PubMed] [Google Scholar]
- Burke B, Ellenberg J (2002) Remodelling the walls of the nucleus. Nat Rev Mol Cell Biol 3: 487–497 [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol 124: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FM, Tsao TY, Fowler SK, Rao PN (1983) Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA 80: 2926–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant TM, Wilson KL (1997) Nuclear assembly. Annu Rev Cell Dev Biol 13: 669–695 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Goldman RD, Meyuhas R, Mills E, Margalit A, Fridkin A, Dayani Y, Prokocimer M, Enosh A (2003) The nuclear lamina and its functions in the nucleus. Int Rev Cytol 226: 1–62 [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM (2002) Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Henry SA, Patton-Vogt JL (1998) Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol 61: 133–179 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol 3: 1086–1091 [DOI] [PubMed] [Google Scholar]
- Hoshizaki DK, Hill JE, Henry SA (1990) The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem 265: 4736–4745 [PubMed] [Google Scholar]
- Irie K, Takase M, Araki H, Oshima Y (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol Gen Genet 236: 283–288 [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R (1990) Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61: 723–733 [DOI] [PubMed] [Google Scholar]
- Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Youn RA, Greenblatt J (1999) An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol Cell 4: 55–62 [DOI] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL (1996) Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell 7: 769–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S (2004) The role of the Nir/rdgB protein family in membrane trafficking and cytoskeleton remodeling. Exp Cell Res 297: 1–10 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW (2001) A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J Cell Biol 153: 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW (2004) Sorting out the nuclear envelope from the endoplasmic reticulum. Nat Rev Mol Cell Biol 5: 65–69 [DOI] [PubMed] [Google Scholar]
- Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY-L, Borun TW (1972) Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol 55: 433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P, Henry SA (1989) Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics 122: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, O'Sullivan J, Azad A, Proudfoot N, Mellor J (2003) Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science 300: 492–495 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Reed SI (1980) Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci USA 77: 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport JW, Wilson KL, Dunphy WG (1990) A lamin-independent pathway for nuclear envelope assembly. J Cell Biol 111: 2247–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloff DM, McGraw P, Henry SA (1992) The INO2 gene of Saccharomyces cerevisiae encodes a helix–loop–helix protein that is required for activation of phospholipid synthesis. Nucleic Acids Res 20: 3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R (1980) Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21: 205–215 [DOI] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K (2001) Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet 27: 121–124 [DOI] [PubMed] [Google Scholar]
- Phan J, Peterfy M, Reue K (2004) Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J Biol Chem 279: 29558–29564 [DOI] [PubMed] [Google Scholar]
- Paltauf F, Kohlwein S, Henry S (1992) Regulation and compartmentalization of lipid synthesis in yeast. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, Jones E, Pringle J, Broach J (eds), pp 415–500. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D (1991) Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast 7: 891–911 [DOI] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI (1992) Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev 6: 2021–2034 [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR III, Gerace L (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301: 380–1382 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21: 81–93 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Hurt EC, Pelham HR (2000) Psr1p/Psr2p, two plasma membrane phosphatases with an essential DXDX(T/V) motif required for sodium stress response in yeast. J Biol Chem 275: 19352–19360 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E (1998) A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J 17: 6449–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW (2004) XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167: 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M, Wente SR (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev Cell 4: 775–789 [DOI] [PubMed] [Google Scholar]
- Tange Y, Hirata A, Niwa O (2002) An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci 115: 4375–4385 [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Wagner C, Blank M, Strohmann B, Schuller HJ (1999) Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast 15: 843–854 [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Wanker EE, Sun Y, Savitz AJ, Meyer DI (1995) Functional characterization of the 180-kDa ribosome receptor in vivo. J Cell Biol 130: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Hirsch JP, Henry SA (1991) The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem 266: 863–872 [PubMed] [Google Scholar]
- Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt EC (1992) A new subclass of nucleoporins that functionally interact with nuclear pore protein NSP1. EMBO J 11: 5051–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Basson M, D'Ari L, Rine J (1988) Increased amounts of HMG-CoA reductase induce ‘karmellae': a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol 107: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L (1997) Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J Cell Biol 139: 1077–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K (1995) Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol 130: 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen A, Pardee AB (1979) Role of nuclear size in cell growth initiation. Science 204: 1315–1317 [DOI] [PubMed] [Google Scholar]
- Zhang C, Clarke PR (2000) Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 288: 1429–1432 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figure 1
Supplementary Table I


