Abstract
Congenital central hypoventilation syndrome (CCHS), a rare genetic disease caused by heterozygous PHOX2B mutations, is characterized by life-threatening breathing deficiencies. PHOX2B is a transcription factor required for the specification of the autonomic nervous system, which contains, in particular, brainstem respiratory centers. In CCHS, PHOX2B mutations lead to cytoplasmic PHOX2B protein aggregations, thus compromising its transcriptional capability. Currently, the only available treatment for CCHS is assisted mechanical ventilation. Therefore, identifying molecules with alleviating effects on CCHS-related breathing impairments is of primary importance. A transcriptomic analysis of cells transfected with different PHOX2B constructs was used to identify compounds of interest with the CMap tool. Using fluorescence microscopy and luciferase assay, the selected molecules were further tested in vitro for their ability to restore the nuclear location and function of PHOX2B. Finally, an electrophysiological approach was used to investigate ex vivo the effects of the most promising molecule on respiratory activities of PHOX2B-mutant mouse isolated brainstem. The histone deacetylase inhibitor SAHA was found to have low toxicity in vitro, to restore the proper location and function of PHOX2B protein, and to improve respiratory rhythm-related parameters ex vivo. Thus, our results identify SAHA as a promising agent to treat CCHS-associated breathing deficiencies.
Keywords: MT: Bioinformatics, PHOX2B, aggregates, breathing, pharmacological treatment, CCHS
Graphical abstract
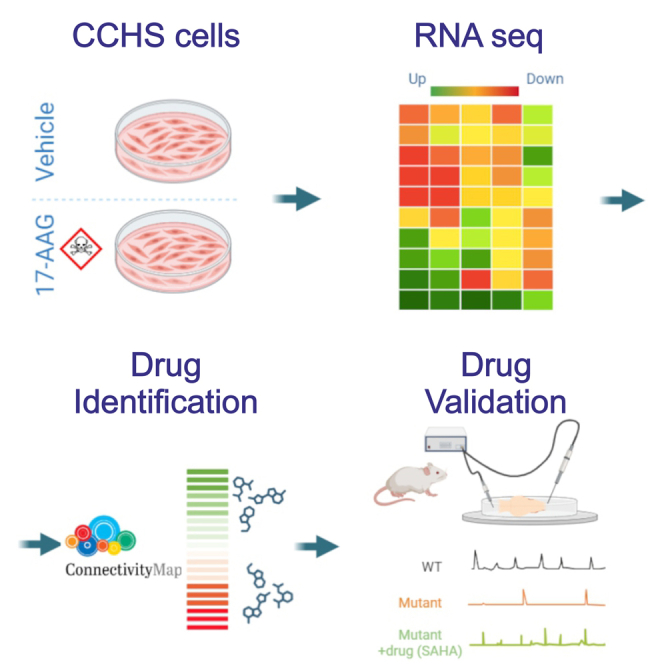
Ceccherini and colleagues demonstrate, using a multidisciplinary approach, that the cellular defects underlying the effects of causative PHOX2B variants can be reversed by pharmacological treatments. These findings represent a first step toward the identification of a molecule potentially effective in alleviating breathing deficiencies associated with CCHS.
Introduction
Congenital central hypoventilation syndrome (CCHS) is a rare genetic disease whose defining symptoms consist of a reduced ventilatory response to hypoxia and hypercapnia, particularly during sleep.1,2 Since no effective drug is available to relieve the potentially lethal outcome of CCHS, current treatments are based on supportive measures such as mechanical ventilation, with continuous monitoring of the patient’s respiratory parameters.3
Heterozygous variants of the PHOX2B gene, encoding a transcription factor crucial for the specification of neurons involved in cardiovascular, digestive, and respiratory reflex circuits,4 have been identified as the primary cause of CCHS. In-frame triplet duplications, leading to an expansion of +4 to +13 additional residues of a 20-polyalanine (polyAla) stretch, account for approximately 90% of causative PHOX2B variants. Furthermore, rare missense, nonsense, and frameshift mutations of PHOX2B may also occur in CCHS patients.5,6,7,8,9
In vitro data have revealed a correlation between the length of the polyAla expansion and the aberrant retention of PHOX2B protein in the cytoplasm in diffuse or aggregated forms,10 thus leading to impaired transactivation of the PHOX2B gene itself and of PHOX2B target genes such as DBH, TH, RET, TLX2, PHOX2A, and TFAP2A.10,11,12,13,14 Furthermore, stimulation of the cellular stress response and proteasome and autophagic pathways by the antibiotic geldanamycin (GA) or, more specifically, its analog 17-allylaminogeldanamycin (17AAG) can (1) reverse the formation of mutant PHOX2B aggregates, (2) induce the removal of multiprotein aggregates (inclusions) by promoting protein folding and/or eliminating mutant PHOX2B, and (3) rescue the nuclear localization of PHOX2B and thereby the transactivation of PHOX2B target genes.15,16 However, the toxicity of both GA and 17AAG has precluded their clinical development for CCHS treatment.
Homozygous Phox2b−/− mice die in utero, showing a lack of sympathetic, parasympathetic, and enteric components of the autonomic nervous system at embryonic day (E) 13.5, whereas heterozygous Phox2b+/− animals express no obvious abnormal phenotype.17 Interestingly, mutant mice carrying a heterozygous PHOX2B+7Ala expansion of the 20 polyAla tract (Phox2b27Ala/+) recapitulate the hypoventilatory features of CCHS.18,19 These knockin animal models offer potentially relevant tools for evaluating the abnormal respiratory parameters associated with CCHS and searching for treatments.
Here, to identify molecules with potential therapeutic effects on CCHS symptoms, RNA-sequencing (RNA-seq) data from cell lines transiently transfected with different PHOX2B constructs were used to perform connectivity mapping (CMap).20,21 Selected compounds were subsequently tested for their ability, (1) in vitro, to restore the correct subcellular localization of PHOX2B and the effective regulation of the DBH target gene and, (2) ex vivo, to improve the autonomously produced central respiratory command of isolated brainstem preparations obtained from Phox2b27Ala/+ mice.
Results
Effect of PHOX2B mutations and 17AAG treatment on gene expression profiles
To identify potential curative molecules for CCHS, our initial goal was to search for an agent with a gene expression profile signature and molecular action similar to those of 17AAG, known for its actions on PHOX2B protein aggregates (see introduction), but less toxic and therefore potentially usable in vivo. To this end, we generated transcription datasets from SK-N-BE cells expressing wild-type (WT) or mutant (+13Ala) PHOX2B proteins, enabling comparison with or without 17AAG treatment.
Gene expression in WT and +13Ala PHOX2B samples was first analyzed to seek differences in genes and associated molecular pathways that are possibly involved in PHOX2B-mediated CCHS pathogenesis. Forty-six genes were found to be significantly differentially expressed (DE) in +13Ala vs. WT, of which 28 were overexpressed and 18 downregulated, with 25 of these (13 up- and 12 downregulated) showing >4-fold changes in expression (Table S1A).
The effects of 17AAG treatment on these transcriptional profiles were then investigated. In WT transfected cells, 16 genes were overexpressed and 7 genes were downregulated by 17AAG, of which 6 were up- and 3 downregulated with a ±4-fold change (Table S1B).
In treated mutant samples (+13Ala(17AAG)), 42 genes were found to be DE, compared to the +13Ala samples, of which 28 genes were overexpressed and 14 downregulated, with 16 (13 up- and 3 downregulated) of these displaying >4-fold changes (Table S1C). Interestingly, most of the upregulated genes in WT(17AAG) vs. WT were also upregulated in +13Ala(17AAG) vs. +13Ala, presumably reflecting non-specific effects of 17AAG.
Finally, we examined whether 17AAG could modify the molecular pathways that were found to be dysregulated by the polyAla mutation, possibly counteracting its effects and restoring near-normal levels of gene expression. Consistent with this idea, comparison of the expression profiles of +13Ala(17AAG) vs. WT revealed that none of the 115 reported genes showed differential expression greater than 2-fold after 17AAG treatment (Table S1D), thus indicating that +13Ala(17AAG) samples had indeed recovered expression profiles comparable to that of WT.
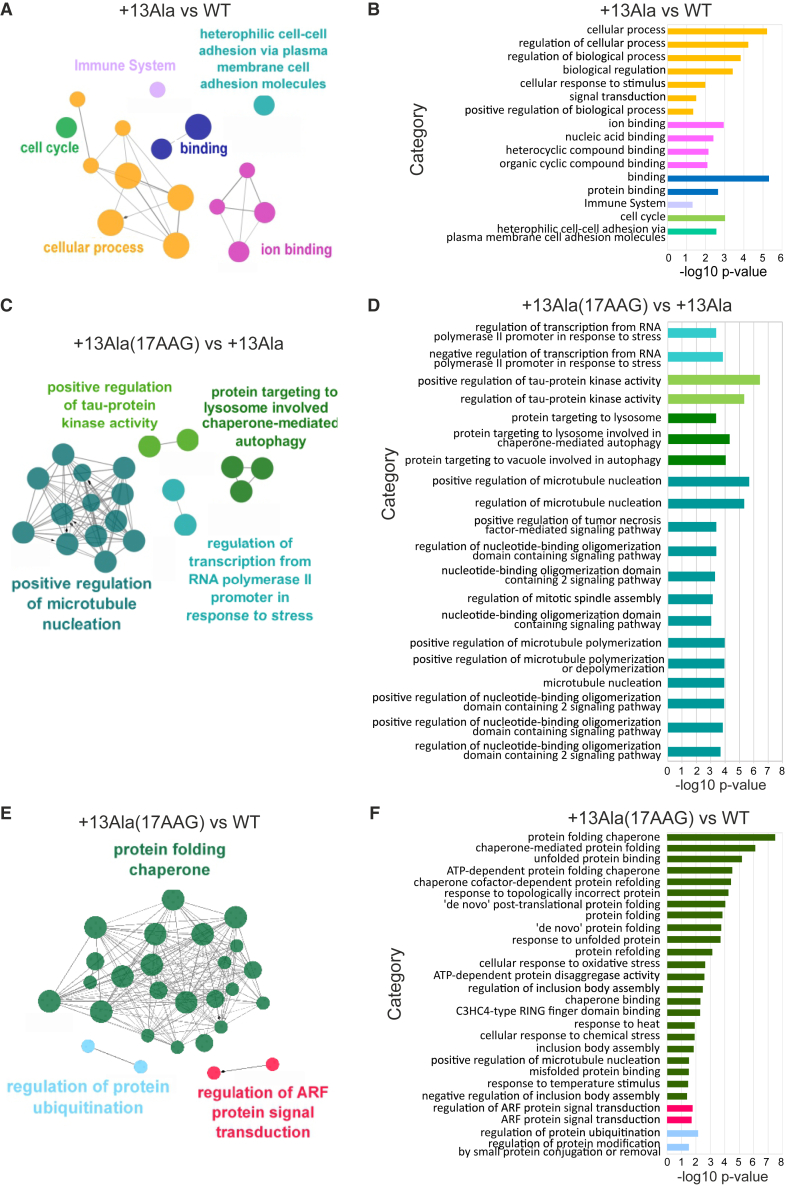
Genes subjected to changes in their expression profiles in the different conditions underwent further validation through Cytoscape analysis (Figure 1). In particular, the ClueGO plug-in was used to find enriched pathways via the same pairwise comparisons as above.
Figure 1.
Cytoscape ClueGO pairwise analysis
Differentially expressed genes (DEGs) were analyzed for their enrichment in specific pathways by using the Cytoscape app ClueGO v.2.5.9. Images show the representation of nodes connecting the terms of enriched pathways (A, C, and E). The corresponding bar diagrams of the detailed gene ontology (GO) terms are ranked according to their p values (B, D, and F) in the following pairwise comparisons: WT vs. +13Ala, +13Ala(17AAG) vs. +13Ala, and +13Ala(17AAG) vs. WT. Functionally grouped networks are shown, with terms as nodes linked based on their kappa score level (≥0.3), where only the label of the most significant term per group is shown. GO levels 1–20 were considered for analysis. Each pathway is represented by a different color ascribed to each GO regulatory action in both the schematics (A, C, and E) and the bar diagrams (B, D, and F). The node dimension represents the enrichment expressed as the ratio between the number of DEGs and the total genes in the corresponding GO term.
In +13Ala vs. WT samples, 28 (75.68%) genes associated with 16 representative terms and pathways related to cell-cell interactions, immune system, cell cycle, and ion-transport processes were found to be enriched, although not interconnected (Figures 1A and 1B). The finding of a dysregulation of the cell cycle due to the +13Ala mutation was not surprising, as PHOX2B is crucial for neuronal differentiation and cell-cycle exit.22
In +13Ala(17AAG) vs. +13Ala, 27 (75%) enriched genes were associated with 55 representative gene ontology (GO) terms and pathways (Figures 1C and 1D). A marked representation of pathways related to the cellular response to protein aggregation, such as protein folding, macro- and chaperone-mediated autophagy, inclusion body assembly, and microtubule polymerization with Tau protein kinetics, was also evident. This finding is in accordance with the known effects of 17AAG in inducing protein quality control by acting on heat shock protein (HSP) expression and mechanisms involved in protein elimination.23
In +13Ala(17AAG) vs. WT, 30 genes were enriched in 27 significant terms and pathways (Figures 1E and 1F), including the ARF gene family known to be involved in protein aggregation and in protein ubiquitination and chaperone folding pathways. No differences were detected in the pathways previously found to be altered by +13Ala mutation compared to WT. This further indicates that 17AAG is able, at least to some extent, to revert the dysregulation resulting from the +13Ala mutation.
Identification of compounds with a gene expression signature similar to that of 17AAG
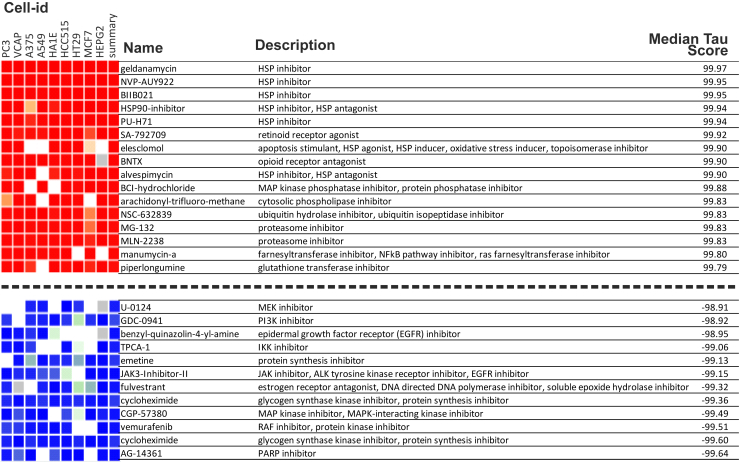
We next used the CMap database and query tool to identify bioactive compounds that could have restorative effects similar to those of 17AAG in the context of PHOX2B+13Ala, but without the latter agent’s toxicity. Of the 30 genes that were significantly upregulated in +13Ala(17AAG)-expressing cells, 12 were components of the L1000 gene expression assay used in CMap (1,000 selected landmark transcripts of the CMap algorithm) and were therefore analyzed further. In particular, we focused on the class of compounds with a median connectivity score (tau) attaining >90, considered a high-significance threshold in CMap. Seventy-four compounds were identified as producing a gene expression profile comparable to that of +13Ala(17AAG) cells (Figure 2). The top-ranked agents were HSP90 inhibitors, including GA and alvespimycin, which are closely related to 17AAG. Of the 74 pre-selected drugs, we focused on those that already have established clinical relevance (Table 1). Accordingly, four agents with different targets and mechanisms of action were chosen to assess whether they have effects similar to those of 17AAG in vitro and, ultimately, if they could be considered as alternative options for CCHS treatment. Those selected were parthenolide, trichostatin A (TSA), guggulsterone, and vorinostat (SAHA). Details on the chemical nature and functional effects of these molecules are provided in the discussion.
Figure 2.
Connectivity Map data for compounds with strongest to weakest connections with the DEG in PHOX2B+13Ala cells treated with 17AAG compared to untreated control cells
The name, action, and median tau score for each compound are indicated. The cell-line identifier is also shown for each column. The median tau score represents the strength of the connection between a query signature and a drug across multiple cell types. Compounds were considered significantly connected with the input signature when the median tau score was above 90.
Table 1.
Compounds with significant connections in CMap analysis that either are clinically available or have already reached the market
| Name | Description | Median tau score | Development |
|---|---|---|---|
| NVP-AUY922 | HSP inhibitor | 99.95 | phase 2 |
| BIIB021 | HSP inhibitor | 99.95 | phase 2 |
| PU-H71 | HSP inhibitor | 99.94 | phase 1 |
| Elesclomol | apoptosis stimulant, HSP agonist, HSP inducer, oxidative stress inducer, topoisomerase inhibitor | 99.90 | phase 3 |
| Alvespimycin | HSP inhibitor, HSP antagonist | 99.90 | phase 2 |
| Thiostrepton | downregulates FOXM1 expression, FOXM1 expression inhibitor, protein synthesis inhibitor | 99.71 | launched |
| Diphencyprone | immunostimulant | 99.67 | phase 2 |
| Parthenolide | NF-κB pathway inhibitor, adiponectin receptor agonist | 99.58 | phase 1 |
| Parthenolide | NF-κB pathway inhibitor, adiponectin receptor agonist | 99.19 | phase 1 |
| Trichostatin A | HDAC inhibitor, CDK expression enhancer, ID1 expression inhibitor | 98.72 | phase 1 |
| Trichostatin A | HDAC inhibitor, CDK expression enhancer, ID1 expression inhibitor | 98.69 | phase 1 |
| Disulfiram | aldehyde dehydrogenase inhibitor, DNA methyltransferase inhibitor, TRPA1 agonist | 98.47 | launched |
| Guggulsterone | estrogen receptor agonist, FXR antagonist, progesterone receptor agonist, cholesterol inhibitor, IKK inhibitor, PXR agonist | 98.06 | launched |
| Sulforaphane | anticancer agent, aryl hydrocarbon receptor antagonist, nuclear factor (erythroid-derived 2)-like (NRF2) activator | 97.15 | phase 2 |
| Vorinostat | HDAC inhibitor, cell-cycle inhibitor | 96.30 | launched |
| BI-2536 | PLK inhibitor, apoptosis stimulant, cell-cycle inhibitor, protein kinase inhibitor | 96.19 | phase 2 |
| Oxibendazole | DNA polymerase inhibitor, tubulin inhibitor | 96.01 | launched |
| Phenethyl-isothiocyanate | cancer cell growth inhibitor, unidentified pharmacological activity | 94.53 | phase 2 |
| Atracurium | acetylcholine receptor antagonist | 92.66 | launched |
| Rifapentine | DNA-directed RNA polymerase inhibitor, DNA-directed DNA polymerase inhibitor | 91.13 | launched |
| Belinostat | HDAC inhibitor, cell-cycle inhibitor | 90.40 | launched |
The selected agents mimic 17AAG’s effects in vitro
As a transcription factor, PHOX2B requires a nuclear localization to exercise function. PolyAla expansions are known to cause abnormal cytoplasmic retention of the PHOX2B protein, often inducing the aggregate formation. Treatments with GA or its analog 17AAG can promote nuclear relocalization of polyAla PHOX2B proteins,10,15,16 but both are highly toxic, thereby preventing their clinical usage. In search of an employable alternative treatment, therefore, the toxicity of the above four compounds identified by the CMap tool was first evaluated in vitro, and the most suitable dose application was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay protocol.24
A low toxicity of all four agents was observed at the lowest doses used in almost all the conditions tested (Figure S1). We then assessed their capacity to promote relocalization of expanded polyAla PHOX2B proteins into cell nuclei using fluorescence microscopy on transfected COS-7 cells. In contrast to drugs with a low tau score in the CMap analysis (e.g., TPCA-1 and CGP-57380 used as negative controls), the four selected drugs increased nuclear relocalization of the mutated proteins (Figure 3A). Parthenolide showed the best relocating effect at high doses, but also a high in vitro toxicity (Figure S1). In contrast, drugs such as SAHA, TSA, and guggulsterone induced a significant nuclear rescue, reaching a protein distribution similar to that in WT, while displaying a lower toxicity than parthenolide (Figure S1).
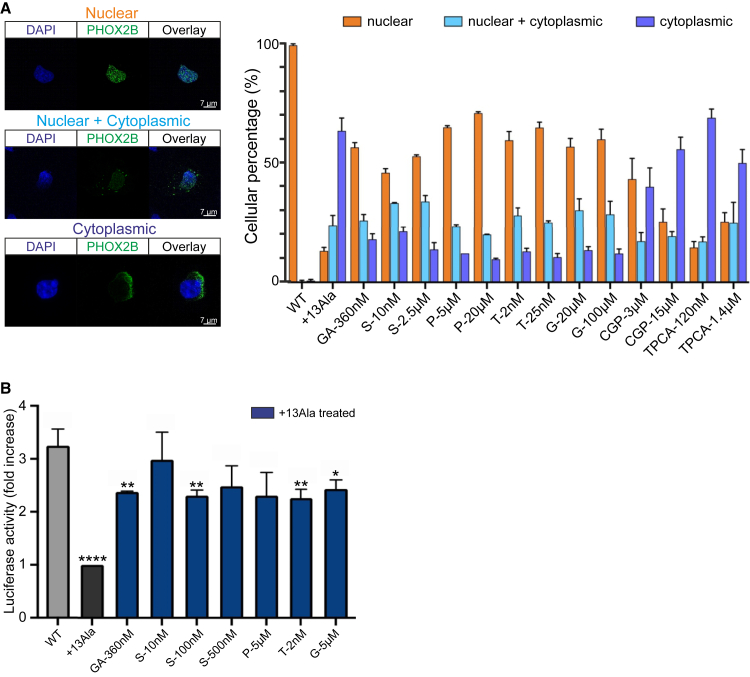
Figure 3.
Drug effects on WT and PHOX2B+13Ala cellular localization and activity
(A) The subcellular localization of WT and PHOX2B+13Ala proteins after pharmacological treatments was analyzed using fluorescence microscopy. Forty thousand COS-7 cells were plated and transfected with plasmids (pcDNA 3.1 CT-GFP PHOX2B WT and pcDNA 3.1 CT-GFP PHOX2B+13Ala). Molecules tested were geldanamycin (GA), SAHA (S), parthenolide (P), trichostatin A (T), guggulsterone (G), CGP-57380 (CGP), and TPCA-1. Fifty COS-7 (PHOX2B-GFP+) transfected cells were counted and classified for each condition in terms of nuclear, cytoplasmic, or nuclear + cytoplasmic PHOX2B localization. Representative images of cells in the three subcellular localization groups are shown on the left, where the blue DAPI is for nuclear staining and the green GFP for PHOX2B protein, and their overlays are also shown. Pictures were taken using a 63× objective microscope. Scale bars, 7 μm. On the right, percentage values are presented in histograms as means ± SEM from three independent experiments. (B) Analysis of luciferase activity on HeLa cells induced by pharmacological treatments and obtained after cell co-transfection of WT or PHOX2B+13Ala with a reporter construct containing the DBH promoter upstream of the luciferase gene. The Renilla luciferase gene was used as an internal control. Raw data were first normalized against the empty vector. Bars represent the increase in luciferase activity under different conditions, normalized to untreated PHOX2B+13Ala. The WT construct (gray bar) shows greater activity on the DBH promoter than the luciferase value obtained in the presence of the expanded polyAla proteins. GA was used as a positive treatment control. Values are represented as means ± SEM of six to eight independent replicate experiments. P at 5 μM, S at 500 nM, and, in particular, S at 10 nM show similar levels of transcriptional activity compared to the WT condition. All the other treatments were less effective, as indicated by statistical comparison with PHOX2B WT (Welch’s t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
Next, using a luciferase-based assay, we tested the ability of the selected compounds to restore PHOX2B+13Ala transcriptional function. All four drugs triggered a significant recovery of mutant PHOX2B transcriptional activity (Figure 3B). In particular, SAHA treatments led to a 2-fold recovery of PHOX2B protein function, which was sufficient to restore transcriptional levels comparable to those of WT.
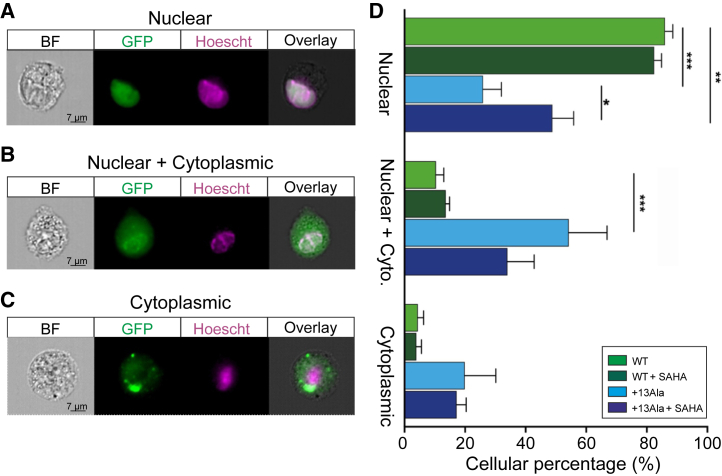
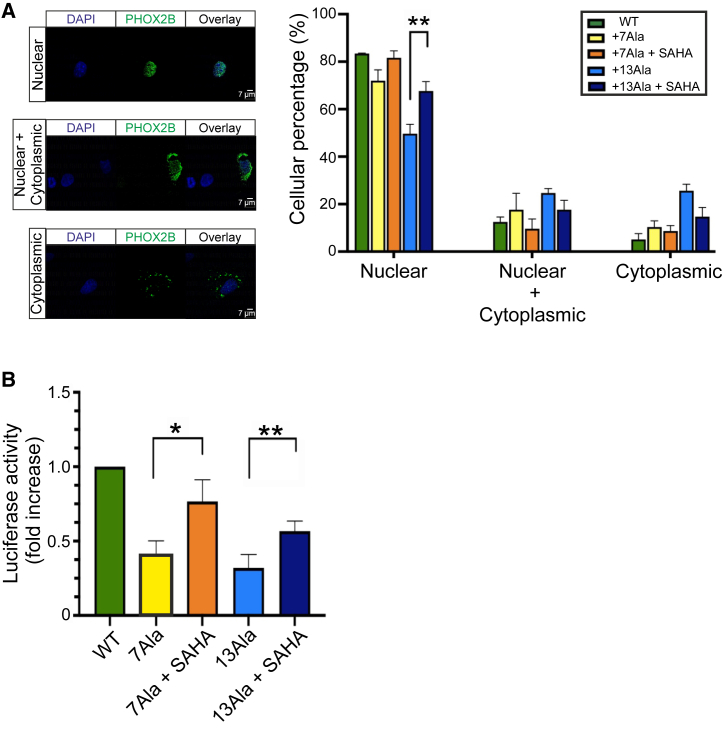
Since, at the lowest dose, SAHA was found to be more effective than the other three agents in causing recovery of mutant PHOX2B protein function, we further characterized its ability to relocate the PHOX2B protein to the nucleus using imaging flow cytometry. Consistent with the findings reported in Figure 3A, a nuclear localization was again observed in the WT condition, whereas the mutant form exhibited cytoplasmic or both nuclear and cytoplasmic retention (Figures 4A, 4B, and 4C). Strikingly, a nuclear relocalization was evident after SAHA treatment (Figure 4D) to a degree that any significant differences in PHOX2B protein localization between WT and +13Ala were abolished. This finding therefore further confirms SAHA’s ability to rescue the nuclear relocation of mutant PHOX2B protein, which in turn would be consistent with an ensuing restoration of transactivation function.
Figure 4.
Subcellular localization analysis of WT and PHOX2B+13Ala proteins after SAHA treatment
COS-7 cells were transfected with pcDNA3.1-CT/GFP PHOX2B WT and pcDNA3.1-CT/GFP PHOX2B+13Ala plasmids and treated with 10 nM SAHA. ImageStream X Mark II was used to analyze PHOX2B nuclear relocalization after the drug treatment. On the left three types of PHOX2B subcellular localization are shown: (A) nuclear localization, (B) nuclear localization with cytoplasmic retention, and (C) cytoplasmic localization alone. PHOX2B was fused to GFP (green), nuclei were stained with Hoechst 33342 (purple), and their co-localization is shown at right (overlay). Approximately 5,000 PHOX2B-GFP-positive cells were acquired for each condition (i.e., WT, WT + SAHA, +13Ala, and +13Ala + SAHA) in four independent experiments. All images in (A)–(C) were taken at 60× magnification. (D) Bar graph representing the percentage of cells (means ± SD) exhibiting the different PHOX2B protein localizations. A paired t test was performed in each group of PHOX2B subcellular compartment localization (nuclear, cytoplasmic, nuclear + cyto) comparing WT vs. +13Ala, WT vs. +13Ala + SAHA, and +13Ala vs. +13Ala + SAHA (paired t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001). Scale bars, 7 μm.
The effect of SAHA also extends to PHOX2B polyAla expansions of different lengths
Previous studies have shown that there is a direct correlation between the severity of symptoms and the length of polyAla expansion.7 Before testing SAHA in an ex vivo model, using tissue samples derived from a knockin mouse Phox2b27Ala/+ (see below), we sought to confirm in vitro the effects of PHOX2B+7Ala, the most common polyAla expansion among CCHS patients, with and without SAHA treatments. To this end, we first assessed the localization of the PHOX2B+7Ala protein within the nucleus in COS-7. As shown in Figure 5A, the WT condition was confirmed to induce correct nuclear localization and, in line with expectations, the PHOX2B+7Ala protein was retained in the cytosol, though to a lesser extent than the PHOX2B+13Ala protein. Treatment with 10 nM SAHA demonstrated a statistically significant proportion of mutated proteins entering the nucleus, including the PHOX2B+7Ala protein, which showed a recovery close to the significance threshold (p = 0.06).
Figure 5.
Subcellular localization and activity of different expanded PHOX2B proteins and SAHA-mediated recovery
(A) Subcellular localization of PHOX2B proteins in COS-7 cells expressing PHOX2B WT, +7Ala, and +13Ala before and after treatment with 10 nM SAHA. The immunofluorescence assay was performed with an antibody against the PHOX2B protein. Representative images of cells in the three subcellular localization groups (cells with PHOX2B in the nucleus, nucleus and cytoplasm, or cytoplasm alone) are shown on the left, where the blue DAPI is for nuclear staining and the green GFP for PHOX2B protein, and their overlays are also shown. Pictures were taken using a Leica SP5 confocal microscope (63× magnification). Scale bars, 7 μm. The histogram on the right represents the mean ± SEM of three independent experiments and shows the percentage values of cells for the different conditions. The nuclear relocalization of the PHOX2B+13Ala protein induced by drug treatment is statistically significant, but the positive effect of SAHA on the subcellular localization of PHOX2B+7Ala is also evident (p = 0.06). (B) Effect of 10 nM SAHA on the transactivation activity of the DBH promoter by expanded polyalanine PHOX2B. HeLa cells were co-transfected with PHOX2B WT, +7Ala, and +13Ala and the DBH promoter carrying a luciferase gene. A Renilla luciferase genetic construct was used as an internal control. The histogram shows the luciferase activity of the different conditions with or without SAHA treatment, normalized compared to PHOX2B WT. The mutated forms appear to lose the ability to activate the DBH promoter, which can be partially rescued by SAHA treatment. Values are expressed as the mean ± SEM of three independent experiments (paired t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
We then investigated the effect of PHOX2B+7Ala protein on restoring DBH promoter transactivation activity on HeLa cells with and without 10 nM SAHA treatment. Once again, we tested the PHOX2B WT protein and the polyAla-expanded PHOX2B protein (+7Ala and +13Ala) and confirmed that the PHOX2B+7Ala protein induces the expected effect compared to WT and +13Ala and that the drug can restore transcriptional activation of the DBH promoter whatever the PHOX2B polyAla expansion tested (Figure 5B).
We finally took into account an additional cellular outcome known to be related to the presence of expanded PHOX2B polyAla proteins, namely the formation of cytoplasmic aggregates. Specifically, we investigated the proportion of cells showing aggregates in the cytosol in the PHOX2B WT, +7Ala, and +13Ala conditions, finding, as expected, an aggregation increase correlated with the size of the expansion (Figure S2). SAHA induces a decrease in the average number of cytoplasmic aggregates per cell in the presence of the more severe mutation, but appears to have no significant effect on the cytosolic aggregates that are also induced by PHOX2B+7Ala but at a lower extent compared to +13Ala.
SAHA improves functional breathing parameters in isolated brainstem preparations of CCHS mutant animals
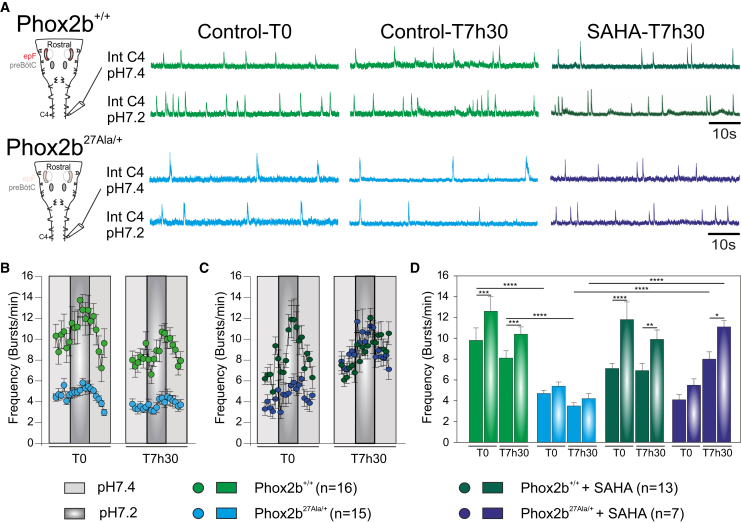
In a final set of experiments, we tested SAHA’s ability to improve actual breathing-related activity in a mouse model of CCHS. Because a +13Ala mouse line does not currently exist, we used the Phox2b27Ala/+ line that bears a +7Ala expansion of the PHOX2B gene, corresponding to the most common mutation found in human CCHS patients.18 However, such mutant mice survive only 1 to 2 h after birth, making in vivo testing nearly impossible. Consequently, to assess the effectiveness of SAHA in restoring respiration-related parameters, we used ex vivo isolated brainstem preparations that contain the two main respiratory neural networks, obtained from embryos harvested at E18.5 (the ultimate prenatal stage) (Figure 6A). Electrophysiological recordings of spontaneous burst activity in phrenic rootlets, which convey the central respiratory motor command to the diaphragm in vivo, were utilized to measure the basal respiratory rhythm frequency and the response to applied acidosis (used to mimic hypercapnia ex vivo), two parameters known to be deficient in CCHS (see details in materials and methods). As previously reported,18 Phox2b27Ala/+ preparations generated so-called “fictive breathing” activity at a lower frequency compared to Phox2b+/+ (WT) preparations and lacked the capability to respond to acidosis that normally leads to a significant rhythm frequency increase (Figures 6A, 6B, and 6D). One group of preparations (n = 13 Phox2b+/+ and n = 7 Phox2b27Ala/+) was then exposed to SAHA (1 μM) during 7 h 30 min, whereas a second group (n = 16 Phox2b+/+ and n = 15 Phox2b27Ala/+) remained under control bathing oxygenated artificial cerebrospinal fluid (aCSF) conditions. After 7 h 30 min, all untreated preparations remained rhythmically active, with the mutant subgroup still exhibiting abnormal respiratory parameters compared to Phox2b+/+ (Figures 6A, 6B, and 6D). In contrast, after the same long-term exposure to SAHA, Phox2b27Ala/+ preparations exhibited an increased spontaneous rhythm frequency and a sensitivity to acidosis that had become comparable to WT preparations (Figures 6A, 6C, and 6D). These results therefore indicate that, at least in the isolated brainstem ex vivo, the main respiratory deficits characterizing CCHS can be significantly compensated for by exposure to SAHA.
Figure 6.
Effects of SAHA on fictive respiratory activity in isolated embryonic mouse brainstem preparations
(A) Left: schematic representation of a brainstem preparation obtained at E18.5 from WT (Phox2b+/+, top) and mutant (Phox2b27Ala/+, bottom) embryos. Such a preparation hosts the main respiratory networks: the embryonic parafacial respiratory group (epF) and the pre-Bötzinger complex (preBötC). Spontaneous respiratory-related motor activity was monitored by an extracellular electrode placed on the phrenic (C4) motor root. Right: integrated phrenic nerve discharge (Int C4) obtained from Phox2b+/+ (two top traces) and Phox2b27Ala/+ (two bottom traces) preparations under normal aCSF (pH 7.4, traces above) and after acidification (pH 7.2, traces below). (B) Quantification of the mean fictive breathing burst frequency over time (measured at 2 min intervals) for 16 Phox2b+/+ (green dots) and 15 Phox2b27Ala/+ (blue dots) preparations at time 0 (T0) and after 7 h 30 (T7h30) in control aCSF conditions (pH 7.4, light gray) and after acidification (pH 7.2, dark gray). Note that mutant preparations exhibited a lower mean respiratory frequency compared to Phox2b+/+ preparations and that acidification induced a significant increase in respiratory frequency solely in the Phox2b+/+ preparations. (C) Same layout as in (B) for preparations (13 Phox2b+/+ and 7 Phox2b27Ala/+) exposed to 1 μM SAHA. Note the increased burst frequency in mutant preparations and a partial restoration of the response to acidosis at T7h30. (D) Bar graphs representing the mean respiratory frequency values measured over 4 min at steady state in pH 7.4 and during the maximal effect at pH 7.2 for all preparations in each condition, at T0 and T7h30. Graphs in B, C, and D are frequency mean values ± SEM. Two-way ANOVA, followed by a post hoc Sidak’s comparison test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Discussion
In this study, transcriptomic and CMap analysis allowed us to identify four compounds with different targets and mechanisms of action. These molecules were also selected because they are clinically available, albeit for the treatment of pathologies other than CCHS, as their toxicity, bioavailability, stability, and therefore applicability to patients have already been tested. In particular, parthenolide shows potent antiinflammatory, antiapoptosis, and antioxidative stress effects;25,26,27 guggulsterone is involved in antiapoptotic signaling, cell survival, cell proliferation, angiogenesis, and chemoresistant activity in tumor cells;28 TSA acts as antitumor, antiinflammatory, and antioxidant agent;29,30 and SAHA is capable of modulating cell signaling pathways and exerting potent apoptotic and antiproliferative effects.31,32,33 SAHA and TSA are already known to downregulate PHOX2B gene expression, resulting in decreased protein and mRNA levels in the IMR32 neuroblastoma cell line stably expressing the luciferase gene under the control of the PHOX2B promoter.34 Moreover, SAHA was recently shown to positively regulate neuronal gene expression and microtubule dynamics, inducing neurite outgrowth and spine density and enhancing synaptic transmission,35,36 thus further pointing to its potential use in the treatment of CNS disorders.
In vitro testing of the ability of these four molecules to relocalize PHOX2B+13Ala and thereby improve the regulation of PHOX2B target genes revealed SAHA to be the most effective candidate. In addition, SAHA was further validated in vitro on a PHOX2B+7Ala cellular model and ex vivo by its impact on fictive breathing parameters in isolated brainstem preparations of CCHS-replicating (Phox2b27Ala/+) mutant mice, including a restoration of respiratory rhythm sensitivity to metabolic acidosis and a return of the basal rhythm frequency to near control values. Furthermore, no significant effect of SAHA was observed in WT, either in vitro or ex vivo, thus further underlining this molecule’s therapeutic potential. This finding appears to be particularly significant in light of the prospective use of SAHA in brain disorders and the strategies that are currently being employed to improve drug efficacy and reduce side effects.35
Our in vitro system, based on transiently transfected neuroblastoma cells, proved suitable for the RNA-seq-based transcriptomic approach in cells expressing PHOX2B WT or the more severe polyAla expansion (PHOX2B+13Ala), either in the presence or in the absence of 17AAG. The bioinformatic analysis of our data allowed us to confirm the involvement of pathways already identified in the pathogenesis of polyAla expansions both in vitro and in patients, such as the cellular response to protein aggregation with induction of protein quality control, subsequently restored by treatment with 17AAG.16,34 Therefore, the RNA-seq approach, applied to in vitro-transfected cell lines, appears to recapitulate well the molecular pathogenesis of PHOX2B polyAla expansion mutations in CCHS, while our in vitro cell system proved suitable as a simple recipient for preliminary drug screening.
However, several limitations must be considered. One is the severe effect exerted by the variant (PHOX2B+13Ala) we selected for most of our in vitro testing, which affects the transactivation of the PHOX2B target promoters mainly by limiting its presence in the cell nucleus. In fact, massive protein aggregation may slow down and even cease a complete drug-mediated restoration of PHOX2B nuclear localization. This was evident from the finding that in no case were the tested drugs able to induce the complete nuclear translocation of PHOX2B+13Ala, a variable proportion of which, depending on the molecule and dose applied, remained sequestered in the cytoplasmic compartment. The massive formation of aggregates in the presence of the PHOX2B+13Ala protein could ultimately lead to underestimation of the drug effect in our in vitro context. However, we have already demonstrated that expansions shorter than +13Ala can recover full nuclear localization with sufficient time, confirming a correlation between the length of the expanded tract, the amount of aggregate formation, and the drug-mediated translocation of the PHOX2B protein into the nucleus.15 Accordingly, here, we have observed that the +7Ala expansion induces a less important effect on PHOX2B protein localization that can nevertheless be reverted by SAHA, at least partially.
Whether PHOX2B-polyAla-associated protein aggregates are observed only in vitro or whether they also occur in vivo in CCHS-affected tissues and organs remains unknown. Such aggregates could be due to the in vitro experimental system, which relies on PHOX2B being overexpressed, a condition favorable for the formation of aggregates. Nonetheless, these remain very useful in vitro markers, capable of revealing the cellular consequences of PHOX2B expanded variants. PHOX2B cellular localization as well as downstream target gene regulation is correlated to PHOX2B misfolding, which is consistent with the fact that the expansion length of polyAla, from +4Ala to +13Ala, has already been correlated with respiratory phenotype severity.7,37
Finally, even if aggregation has so far never been observed in CCHS animal models, SAHA still appears capable of compensating for the abnormal respiratory phenotype observed in the isolated brainstem preparation obtained from Phox2b27Ala/+ embryos. Therefore, although we cannot precisely interpret the nature and significance of our in vitro observations concerning aggregates formation, cytoplasmic retention, and protein quality control involvement, their predictiveness in terms of expected associated cellular and phenotypic outcomes appears compelling.
It is important to note that our in vitro and ex vivo experiments were conducted using several PHOX2B variants (+7Ala and +13Ala in vitro and +7Ala ex vivo), and the results obtained are consistent with one another: all sets of findings point to SAHA as an effective tool in counteracting cellular damage induced by PHOX2B elongations, ensuring recovery of its transactivation capacity, and restoring near-normal functionality of the central respiratory networks of Phox2b27Ala/+ mice. We also chose to mainly develop an in vitro model that would enable compound screening under the most extreme conditions, i.e., the most common expansion of polyAla PHOX2B in humans (+7Ala) and the largest expansion of polyAla PHOX2B (+13Ala), while using the only existing +7Ala mouse line for ex vivo testing.18,19,38,39,40,41,42 Moreover, because of the restricted postnatal survival of such CCHS mutant animals, we were constrained to perform these last experiments on still-viable late embryonic stages.
In terms of how SAHA treatment is able to restore proper PHOX2B localization and target gene regulation, as well as the restoration of proper breathing-related parameters ex vivo, two different but non-exclusive mechanisms of action may occur. First, SAHA might influence PHOX2B through its effect on HSP90, as suggested by the fact that hyperacetylation of HSP90 leads to the inhibition of its chaperone function.43,44,45 Indeed, the regulation of HSP90α and HSP90β by pan-HDAC inhibitors leads to HSP90 activity inhibition by inhibiting HDAC6 and eventually to the degradation of HSP90 client proteins.43,46 Interestingly, inhibiting HSP90 has been shown to be able to trigger HSP90 client protein degradation very rapidly, within minutes to a few hours, depending on the client protein.44,47,48,49 Thus, SAHA could lead to the degradation of PHOX2B aggregates through indirect inhibition of HSP90 in a time frame compatible with our ex vivo observations. Second, in addition to this chaperone effect, SAHA might also act by downregulating PHOX2B transcription, an effect already observed in SK-N-BE cells transfected with a luciferase reporter plasmid carrying 1 kb of the PHOX2B regulatory region.34 Indeed, we cannot exclude the possibility that the beneficial effects of SAHA demonstrated ex vivo are also mediated by a downregulation of PHOX2B expression, resulting in a decreased amount of misfolded proteins and thus a reduction in their pathogenic consequences. Relevant to this possibility is that, in cellular models, SAHA was shown to halve both mRNA and PHOX2B protein levels,34 a reduction that is likely sufficient to allow a more efficient protein quality control. In addition it has been shown that PHOX2B mRNA levels, PHOX2B protein expression, and PHOX2B target gene expression can be regulated and changed in a range of hours.50,51 Moreover, in a very recent publication,52 it has been revealed that the +7Ala expansion induces specific conformational properties in PHOX2B protein in solution and a strong propensity to aggregation. Thus, in our ex vivo model, the formation of PHOX2B aggregates very probably occurs, and we suspect that the underlying mechanisms of action of the tested drugs might, at least in part, include an effect on these aggregates. Interestingly we have previously shown that degradation rates of expanded PHOX2B protein by the ubiquitin proteasome system could be effective in a few hours.53 Thus, the fact that we can detect ex vivo an effect of drug application on the respiratory command activity after 8 h is fully compatible with an action of the tested compounds on PHOX2B mRNA and PHOX2B protein level regulation. Taken altogether, these data are in agreement with the possibility that the effects observed ex vivo are mediated by one or a combination of the two mechanisms described above.
Additional cellular models could further confirm our present findings via evaluation in a more physiologically relevant context. This could include the recently described brainstem organoids, generated from PHOX2B+7Ala human pluripotent stem cells, which resemble the cells of the RTN/pFRG-respiratory center.54 Transcriptomic analysis has illustrated their potential utility for high-throughput drug screening and the validation of selected drugs.54 Similarly, dental pulp stem cells recently generated from patients affected with neurological disorders, including CCHS associated with different polyAla mutations, offer promising subjects for drug validation assessment,55 as do several different induced pluripotent stem cell (iPSC) lines from CCHS patients carrying heterozygous +5Ala expansion mutations that have recently been established.56,57
SAHA is currently used to treat cutaneous T cell lymphoma in patients where systemic therapies have failed. Importantly, if the SAHA safety profile is known, it is also known to pass through the blood-brain barrier poorly. Indeed, “rapid clearance and <5% brain penetration, compared with plasma” were reported in mice.58 However, despite poor brain penetrance, SAHA has been shown to reach biological activity doses in the brain after either oral or intraperitoneal (IP) administration.59 Therefore, formulation and administration routes would need to be carefully chosen when moving to CCHS in vivo validation.
In conclusion, our data have identified SAHA as a promising drug for the treatment of CCHS. Although this agent, under the name Vorinostat, is currently ready for use, an in vivo investigation in mouse or other animal models, as well as a thorough clinical evaluation, is now required before clinical application for CCHS.
Materials and methods
Cell lines and cell culture
Human neuroblastoma SK-N-BE cells were maintained in RPMI medium, HeLa cells were grown in minimal essential medium (MEM, Euroclone, Italy), and COS-7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Merck, Germany). Each medium was supplemented with 10% fetal bovine serum (Gibco, New Zealand), 1% L-glutamine, 100 U/mL penicillin, and 100 ng/mL streptomycin. Cells were incubated at 37°C in 5% CO2.
Transient transfection and RNA extraction
Human neuroblastoma SK-N-BE cells were used to carry out PHOX2B transfection experiments and subsequent gene expression analysis. To this end, 250,000 cells were plated in 35-mm-diameter dishes in triplicate, and the plasmid pcDNA3.1Myc PHOX2B WT or pcDNA3.1Myc PHOX2B+13Ala was transfected, as already reported.16 After 24 h, the cells were treated with 300 nM 17AAG (Merck, Germany) or were left untreated, and 24 h later RNAs were extracted from cell cultures for transcriptome analysis.
For the RNA extraction, total RNA was extracted and purified with an RNeasy Plus mini kit (Qiagen, Italy), and RNA samples thus obtained were quantified by NanoDrop (Thermo Scientific, Rockford, USA).
RNA-seq and transcriptome analysis
After verifying the amount and quality of RNA samples, mRNAs were isolated and processed using an Illumina TruSeq stranded mRNA library preparation kit. Individual libraries were then loaded on a single-end flow cell of a HiSeq 2500 instrument (Illumina, USA) to generate 30 million 75 bp single-end reads per sample.
The quality of the RNA-seq experiments was evaluated using RSeQC60; transcript abundance was estimated with Kallisto using Ensembl transcripts GRCh38,61 including the count of PHOX2B reads; transcript abundances were summarized at the gene level; and DE genes were identified using the DeSeq2 R package.62 Batch effects were also checked and corrected. DE genes were determined considering false discovery rate (FDR)-corrected p < 0.05. Gene set enrichment analysis (GSEA)63 was applied to identify gene sets and pathways that were significantly perturbed across conditions, using FDR < 0.1 as the significant threshold. Details on these procedures have already been reported.64
Gene fold changes and gene network analysis
The Cytoscape plug-in ClueGO v.2.5.9 was used to perform gene set enrichment analysis among the different groups,65 using the lists of DE genes as input (FDR-corrected p < 0.05, Table S1).
The following selected ontology reference sets were used: GO_Biological processes, GO_Molecular Function, and GO_Immune System Process, containing, in EBI-UniProt-GOA-ACAP-ARAP_25.05.2022, 18,085, 18,417, and 3,044 genes, respectively. The following statistical filters were applied: p < 0.05, Bonferroni step-down correction, KScore threshold = 0.3, GO tree level interval 1–20, and medium level of network specificity. Statistically significant GO pathways and terms were then ranked for significance.
Connectivity Map analysis
CMap is an online tool that allows the analysis of large RNA-seq datasets, especially to accelerate the discovery of novel therapeutics. CMap includes a 1.5-million-gene expression profile library from roughly 5,000 small molecules that have been tested in nine cell lines, thus making CMap especially designed for drug repurposing. The cloud-based compute infrastructure termed CLUE (CMap and LINCS Unified Environment) enables users to access and manipulate CMap data and integrate them with their own datasets.21 The connectivity score (tau) ranges between +100 and −100 and determines the connection between a query signature and a given perturbagen by comparing the input signature with the library gene expression profiles. A molecule that would have a gene expression profile highly connected with one of the queries would have a positive median tau score. The gene expression profiles of the CMap library were generated by the L1000 gene expression assay that measures the expression of 978 genes, referred to as landmark genes, in cells treated with perturbagens (small molecule here). We used the online cMAP software: https://clue.io/, using the Query tool to find small molecules that generate gene expression profiles similar to the one of PHOX2B+13Ala-expressing cells treated with 17AAG.
Compounds and treatment
17AAG, (Z)-guggulsterone, TPCA-1, CGP-57380, SAHA, TSA, and parthenolide were from Merck (Germany). All the compounds were dissolved in DMSO (final concentration at 0.001%). The treatments were performed in cell culture, 6 h after transfection with appropriate constructs, and continued for 24 or 42 h until the execution of the functional tests.
Cell viability assay
Eight thousand HeLa cells were plated in white 96 well plates (Corning, USA) and incubated at 37°C in 5% CO2. After 24 h, the cells were treated in sextuplets with different doses of each drug, namely (1) the doses used for the subcellular localization assays and (2) eight times the highest concentration tested as the toxic dose. After 48 h, drug toxicity/cell viability was evaluated by the MTT assay (Merck, Germany). The cells were washed with a complete medium and then incubated for 2 h at 37°C with 0.5 mg/mL MTT solution (Sigma-Aldrich) diluted in complete DMEM. After the medium was removed, carefully avoiding discard of crystals, 100 μL of DMSO was added and incubated for 15 min. When formazan crystals were dissolved, absorbance was detected at 560 nm. Cell viability was evaluated as the percentage of live cells compared to the control group (untreated cells).
Subcellular localization assay
Forty thousand COS-7 cells were plated in 0.8 cm2/well Nunc Lab-Tek chamber slides, and the day after, the cells were transiently transfected with 300 ng of pcDNA3.1/CT-GFP-TOPO PHOX2B WT or pcDNA3.1/CT-GFP-TOPO PHOX2B+13Ala. The transfection was performed using Fugene HD transfection reagent (Promega, USA), and after 6 h the cells were treated with drugs at their proper concentration.
Forty-eight hours posttransfection, the cells were washed with PBS 1× (Dulbecco’s phosphate-buffered saline, Euroclone, Italy), fixed with methanol (MeOH)/acetone 1:1 for 5–10 min at room temperature, and covered with ProLong antifade reagent (Thermo Fisher Scientific, USA) and DAPI (Roche, Swiss). The PHOX2B subcellular localization was examined for more than 50 cells using the Zeiss Axiophot fluorescence microscope.
For the PHOX2B subcellular localization experiment in Figure 5A, 40,000 COS-7 cells were plated in 0.8 cm2/well Nunc Lab-Tek chamber slides. The transfection was performed after 24 h with pcDNA3.1 PHOX2B WT, pcDNA3.1 PHOX2B+7Ala, and pcDNA3.1 PHOX2B+13Ala using Lipofectamine 2000. After 6 h the cells were treated with 10 nM SAHA or DMSO as control. The day after, the cells were washed with PBS 1×; fixed 15 min in 4% paraformaldehyde; permeabilized 20 min with 0.1% Triton X-100 in PBS; blocked 45 min in 0.1% Tween 20, 2% BSA in PBS; and incubated 1 h at room temperature with PHOX2B C-3 primary antibody (Santa Cruz, sc-376993 X, 1:500). Then they were stained 30 min with the anti-mouse secondary antibody A488 (abcam, ab150113, 1:1,000). The slides were covered with ProLong antifade reagent (Thermo Fisher Scientific, USA) and DAPI (Roche, Swiss) for nuclear staining. Subcellular localization was examined using a Leica SP5 confocal microscope.
To thoroughly investigate the SAHA (10 nM) effects, a subcellular localization assay was performed on 600,000 cells plated in 60-mm-diameter dishes (Euroclone, Italy). The cells were transfected and treated as reported before. Forty-eight hours posttransfection, the COS-7 cells were washed with PBS, and nuclei were stained with Hoechst 33342 (Merck, Germany) and analyzed using a three-laser (405, 488, and 642 nm) ImageStream X Mark II (Amnis, Cytek, Italy) with a 20×, 40×, and 60× magnification system. Single-color controls were acquired to create the compensation matrix. Samples were analyzed using IDEAS software (v.6.2.18, Amnis, USA).
Transcriptional activity assay
One day before transfection, 7.5 × 103 HeLa cells were plated in 15.6-mm-diameter dishes in complete medium; co-transfection was performed using Fugene HD transfection reagent (Roche, Italy) with 750 ng of expression plasmids (pcDNA3.1 PHOX2B WT or pcDNA3.1 PHOX2B+13Ala) and 250 ng of the DBH promoter-reporter plasmid (pGL3basic-DBH promoter, a kind gift from Prof Diego Fornasari, Milan, Italy). In particular, the 4×TK-luc construct was obtained by cloning into the pGL3-basic vector a synthetic double-stranded oligonucleotide containing four copies (three in the right orientation and one in the opposite orientation) of domain II of the DBH promoter, upstream of the TK promoter. Seventy nanograms of plasmid RLuc-SV40, expressing the Renilla luciferase gene, was used as an internal control. Six hours later, transfected cells were treated with drugs and assayed, 48 h after transfection, for luciferase activity (Dual-Luciferase Reporter Assay System, Promega, Italy) using a TD-20/20 luminometer. For the transcriptional activity graph, an inverse transfection was applied on 12,000 HeLa cells plated in white 96 well plates (Corning, USA). The cells were co-transfected with 50 ng of pcDNA3.1 PHOX2B WT, pcDNA3.1 PHOX2B+7Ala, or pcDNA3.1 PHOX2B+13Ala; 50 ng of pGL3basic-DBH promoter; and 3 ng of Rluc-SV40 plasmid using Fugene (Promega). Six hours later, the cells were treated with 10 nM SAHA or DMSO, and 48 h after the transfection, luciferase assay was done with the Dual -Luciferase Reporter Assay System a using Glomax (Promega) instrument.
Animals
All animal procedures were performed in accordance with the University of Bordeaux and European animal care committee’s regulation (2010/63/UE). Mice were raised and housed in our animal facility with a 12/12 h dark/light cycle and food and water provided ad libitum. All efforts were made to minimize animal suffering and to reduce the number of animals used, in accordance with the European Communities Council Directive.
The Phox2b27Ala/+ mouse lineage expressing the most frequent CCHS-causing expansion of the 20-residue polyAla tract in PHOX2B was maintained and bred in a homozygous Phox2b27Ala/27Ala state.18 Mutant and WT pups were generated by mating conditional Phox2b27Ala/27Ala females with Pgk:Cre males.18 Since mutant newborns die rapidly after birth, all experiments were performed blind on E18.5 embryos of either sex, and the genotype of each embryo was determined a posteriori on tail DNA. To ensure that experiments were performed at a specific embryonic stage, male and female mice were mated in the same cage for a single night. Observation of a vaginal plug the following morning attested the coupling, and this day was considered to be E0.5.
Isolated brainstem preparations
Pregnant mice were killed by cervical dislocation. E18.5 embryos were rapidly excised from uterine horns and placed in oxygenated aCSF at 18°C–20°C until dissection. The aCSF solution composition was (all from Merck, France) 120 mM NaCl, 8 mM KCl, 0.58 mM NaH2PO4, 1.15 mM MgCl2, 1.26 mM CaCl2, 21 mM NaHCO3, and 30 mM glucose (pH 7.4) and equilibrated with 95% O2-5% CO2. To induce acidosis, the pH of the bath aCSF was lowered to 7.2 by decreasing NaHCO3 concentration to 10 mM and adjusting NaCl to 130 mM.
Embryos were decerebrated and decapitated, and ventral tissues were removed. The brainstem and spinal cord were then carefully disassociated from the surrounding tissues, paying attention to keep ventral cervical motor roots intact for electrophysiological recordings. Then, the brainstem was completely isolated by a rostral section made at the mesorhombencephalic limit, and a caudal section was made below the C4 roots. Isolated brainstem preparations were positioned ventral side up in the recording chamber and continuously bathed in circulating oxygenated aCSF at 30°C. A recovery period of 30 min was respected before starting any recordings.
Electrophysiological recordings of nerve roots
Phrenic nerve ventral root (C4) activity was recorded using a suction electrode fabricated from glass tubing (Harvard Apparatus GC150F, Germany) broken at the tip to match the diameter of the recorded C4 root. The recording pipettes were filled with aCSF solution and connected to a silver wire to a high gain (AM Systems, USA). Signals were filtered (bandwidth 3 Hz to 3 kHz), rectified and integrated (time constant 100 ms; Neurolog, Digitimer, England), recorded, and analyzed offline on a computer through a Digidata 1440 interface and PClamp 10 software (Molecular Devices, USA). Fictive respiratory activities were continuously monitored for 8 h, allowing drug effect analysis over a relatively long period that was in accordance with preparation survival in vitro. Measurements of fictive respiratory frequency in control conditions (pH 7.4) and in response to acidosis (pH 7.2) were performed at time 0 (T0) and 7 h 30 min (T7h30). Each recording session was divided into three periods: 15 min control (pH 7.4), 15 min of acidosis (pH 7.2), and 15 min return to pH 7.4. Frequency values are given as means ± standard error of the mean, and statistical significance was estimated using two-way ANOVA. Differences were considered to be statistically significant at p < 0.05.
Pharmacological treatment
SAHA (Merck, St. Louis, USA) was dissolved in DMSO (final concentration of 0.001%) and bath-applied continuously during 8 h at a final concentration of 1 μM in oxygenated aCSF solution.
Statistical analysis
For the in vitro experiments, we performed Welch’s t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001) to evaluate drug effects on PHOX2B transcriptional activity (Figure 3). A paired t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001) was used to investigate the nuclear relocalization of PHOX2B after SAHA treatment (Figures 3, 4, and 5). To evaluate the in vitro toxicity of compounds and doses to be used for treatments, an unpaired t test was applied (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
For ex vivo experiments, values were compared using two-way ANOVA, followed by a post hoc Sidak’s comparison test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (Figure 6).
Data and code availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. The accession number for RNA-seq data is Gene Expression Omnibus (GEO): GSE250244.
Acknowledgments
We would like to acknowledge Anne Fayoux, for taking care of the animals, and Dr. John Simmers, for editing the manuscript. C.A. was supported by a PhD fellowship from the Associazione Italiana per la Sindrome da Ipoventilazione Centrale Congenita (AISICC). G.P. was supported by a CIFRE PhD fellowship obtained by M.T.-B. with AtmosR. This work was also supported by the Italian Ministry of Health through both Ricerca Corrente no. RC2024 to the Gaslini Institute and Ricerca Corrente no. U733A to T.B. and by a 2016 CCHS Pilot Grant awarded by the CCHS Family Network to I.C.
Author contributions
T.B., P.U., N.B., M.T.-B., and I.C. contributed to the study conception and design. T.B. and F.G. generated the RNA samples to be sequenced, while P.U., T.B., and I.C. analyzed the raw data. N.L. and N.B. carried out the CMap analysis, and N.B. selected the molecules to be tested. C.A., G.D.Z., and G.R. performed the in vitro tests, while G.P., A.M., and M.T.-B. performed and analyzed the ex vivo experiments. The first draft of the manuscript was written by C.A., N.B., M.T.-B., and I.C. All authors contributed to specific parts of the manuscript’s writing and commented on previous versions of the manuscript and the discussion. All authors read and approved the final manuscript. Funding acquisition: N.B., M.T.-B., and I.C.
Declaration of interests
The authors declare that they have no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102319.
Contributor Information
Muriel Thoby-Brisson, Email: muriel.thoby-brisson@u-bordeaux.fr.
Isabella Ceccherini, Email: isabellaceccherini@gaslini.org.
Supplemental information
References
- 1.Coleman M., Boros S.J., Huseby T.L., Brennom W.S. Congenital central hypoventilation syndrome. A report of successful experience with bilateral diaphragmatic pacing. Arch. Dis. Child. 1980;55:901–903. doi: 10.1136/adc.55.11.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weese-Mayer D.E., Silvestri J.M., Marazita M.L., Hoo J.J. Congenital central hypoventilation syndrome: inheritance and relation to sudden infant death syndrome. Am. J. Med. Genet. 1993;47:360–367. doi: 10.1002/ajmg.1320470313. [DOI] [PubMed] [Google Scholar]
- 3.Trang H., Samuels M., Ceccherini I., Frerick M., Garcia-Teresa M.A., Peters J., Schoeber J., Migdal M., Markstrom A., Ottonello G., et al. Guidelines for diagnosis and management of congenital central hypoventilation syndrome. Orphanet J. Rare Dis. 2020;15:252. doi: 10.1186/s13023-020-01460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pattyn A., Hirsch M., Goridis C., Brunet J.F. Control of hindbrain motor neuron differentiation by the homeobox gene Phox2b. Development. 2000;127:1349–1358. doi: 10.1242/dev.127.7.1349. [DOI] [PubMed] [Google Scholar]
- 5.Amiel J., Laudier B., Attié-Bitach T., Trang H., de Pontual L., Gener B., Trochet D., Etchevers H., Ray P., Simonneau M., et al. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 2003;33:459–461. doi: 10.1038/ng1130. [DOI] [PubMed] [Google Scholar]
- 6.Bachetti T., Ceccherini I. Causative and common PHOX2B variants define a broad phenotypic spectrum. Clin. Genet. 2020;97:103–113. doi: 10.1111/cge.13633. [DOI] [PubMed] [Google Scholar]
- 7.Matera I., Bachetti T., Puppo F., Di Duca M., Morandi F., Casiraghi G.M., Cilio M.R., Hennekam R., Hofstra R., Schöber J.G., et al. PHOX2B mutations and polyalanine expansions correlate with the severity of the respiratory phenotype and associated symptoms in both congenital and late onset Central Hypoventilation syndrome. J. Med. Genet. 2004;41:373–380. doi: 10.1136/jmg.2003.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki A., Kanai M., Kijima K., Akaba K., Hashimoto M., Hasegawa H., Otaki S., Koizumi T., Kusuda S., Ogawa Y., et al. Molecular analysis of congenital central hypoventilation syndrome. Hum. Genet. 2003;114:22–26. doi: 10.1007/s00439-003-1036-z. [DOI] [PubMed] [Google Scholar]
- 9.Weese-Mayer D.E., Berry-Kravis E.M., Zhou L., Maher B.S., Silvestri J.M., Curran M.E., Marazita M.L. Idiopathic congenital central hypoventilation syndrome: analysis of genes pertinent to early autonomic nervous system embryologic development and identification of mutations in PHOX2b. Am. J. Med. Genet. 2003;123A:267–278. doi: 10.1002/ajmg.a.20527. [DOI] [PubMed] [Google Scholar]
- 10.Bachetti T., Borghini S., Ravazzolo R., Ceccherini I. An in vitro approach to test the possible role of candidate factors in the transcriptional regulation of the RET proto-oncogene. Gene Expr. 2005;12:137–149. doi: 10.3727/000000005783992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borghini S., Bachetti T., Fava M., Di Duca M., Cargnin F., Fornasari D., Ravazzolo R., Ceccherini I. The TLX2 homeobox gene is a transcriptional target of PHOX2B in neural-crest-derived cells. Biochem. J. 2006;395:355–361. doi: 10.1042/BJ20051386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardani S., Janes T.A., Saini J.K., Di Lascio S., Benfante R., Fornasari D., Pagliardini S. Etonogestrel Administration Reduces the Expression of PHOX2B and Its Target Genes in the Solitary Tract Nucleus. Int. J. Mol. Sci. 2022;23:4816. doi: 10.3390/ijms23094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Lascio S., Bachetti T., Saba E., Ceccherini I., Benfante R., Fornasari D. Transcriptional dysregulation and impairment of PHOX2B auto-regulatory mechanism induced by polyalanine expansion mutations associated with congenital central hypoventilation syndrome. Neurobiol. Dis. 2013;50:187–200. doi: 10.1016/j.nbd.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Di Zanni E., Bianchi G., Ravazzolo R., Raffaghello L., Ceccherini I., Bachetti T. Targeting of PHOX2B expression allows the identification of drugs effective in counteracting neuroblastoma cell growth. Oncotarget. 2017;8:72133–72146. doi: 10.18632/oncotarget.19922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachetti T., Bocca P., Borghini S., Matera I., Prigione I., Ravazzolo R., Ceccherini I. Geldanamycin promotes nuclear localisation and clearance of PHOX2B misfolded proteins containing polyalanine expansions. Int. J. Biochem. Cell Biol. 2007;39:327–339. doi: 10.1016/j.biocel.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Di Zanni E., Bachetti T., Parodi S., Bocca P., Prigione I., Di Lascio S., Fornasari D., Ravazzolo R., Ceccherini I. In vitro drug treatments reduce the deleterious effects of aggregates containing polyAla expanded PHOX2B proteins. Neurobiol. Dis. 2012;45:508–518. doi: 10.1016/j.nbd.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Pattyn A., Morin X., Cremer H., Goridis C., Brunet J.-F. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 18.Dubreuil V., Ramanantsoa N., Trochet D., Vaubourg V., Amiel J., Gallego J., Brunet J.F., Goridis C. A human mutation in Phox2b causes lack of CO2 chemosensitivity, fatal central apnea, and specific loss of parafacial neurons. Proc. Natl. Acad. Sci. USA. 2008;105:1067–1072. doi: 10.1073/pnas.0709115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanantsoa N., Hirsch M.R., Thoby-Brisson M., Dubreuil V., Bouvier J., Ruffault P.L., Matrot B., Fortin G., Brunet J.F., Gallego J., Goridis C. Breathing without CO(2) chemosensitivity in conditional Phox2b mutants. J. Neurosci. 2011;31:12880–12888. doi: 10.1523/JNEUROSCI.1721-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb J., Crawford E.D., Peck D., Modell J.W., Blat I.C., Wrobel M.J., Lerner J., Brunet J.P., Subramanian A., Ross K.N., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K., et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubreuil V., Hirsch M.R., Pattyn A., Brunet J.F., Goridis C. The Phox2b transcription factor coordinately regulates neuronal cell cycle exit and identity. Development. 2000;127:5191–5201. doi: 10.1242/dev.127.23.5191. [DOI] [PubMed] [Google Scholar]
- 23.Alam Q., Alam M.Z., Sait K.H.W., Anfinan N., Noorwali A.W., Kamal M.A., Khan M.S.A., Haque A. Translational Shift of HSP90 as a Novel Therapeutic Target from Cancer to Neurodegenerative Disorders: An Emerging Trend in the Cure of Alzheimer's and Parkinson's Diseases. Curr. Drug Metabol. 2017;18:868–876. doi: 10.2174/1389200218666170728115606. [DOI] [PubMed] [Google Scholar]
- 24.Stockert J.C., Horobin R.W., Colombo L.L., Blázquez-Castro A. Tetrazolium salts and formazan products in Cell Biology: Viability assessment, fluorescence imaging, and labeling perspectives. Acta Histochem. 2018;120:159–167. doi: 10.1016/j.acthis.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Pareek A., Suthar M., Rathore G.S., Bansal V. Feverfew (Tanacetum parthenium L.): A systematic review. Phcog. Rev. 2011;5:103–110. doi: 10.4103/0973-7847.79105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sztiller-Sikorska M., Czyz M. Parthenolide as Cooperating Agent for Anti-Cancer Treatment of Various Malignancies. Pharmaceuticals. 2020;13:194. doi: 10.3390/ph13080194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu S., Sun P., Bennett S., Charlesworth O., Tan R., Peng X., Gu Q., Kujan O., Xu J. The therapeutic effect and mechanism of parthenolide in skeletal disease, cancers, and cytokine storm. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1111218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada T., Sugimoto K. Guggulsterone and Its Role in Chronic Diseases. Adv. Exp. Med. Biol. 2016;929:329–361. doi: 10.1007/978-3-319-41342-6_15. [DOI] [PubMed] [Google Scholar]
- 29.Bouyahya A., El Omari N., Bakha M., Aanniz T., El Menyiy N., El Hachlafi N., El Baaboua A., El-Shazly M., Alshahrani M.M., Al Awadh A.A., et al. Pharmacological Properties of Trichostatin A, Focusing on the Anticancer Potential: A Comprehensive Review. Pharmaceuticals. 2022;15:1235. doi: 10.3390/ph15101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vigushin D.M., Ali S., Pace P.E., Mirsaidi N., Ito K., Adcock I., Coombes R.C. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin. Cancer Res. 2001;7:971–976. [PubMed] [Google Scholar]
- 31.Grabarska A., Łuszczki J.J., Nowosadzka E., Gumbarewicz E., Jeleniewicz W., Dmoszyńska-Graniczka M., Kowalczuk K., Kupisz K., Polberg K., Stepulak A. Histone Deacetylase Inhibitor SAHA as Potential Targeted Therapy Agent for Larynx Cancer Cells. J. Cancer. 2017;8:19–28. doi: 10.7150/jca.16655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q., Zhou D., St John W.M. Lesions of regions for in vitro ventilatory genesis eliminate gasping but not eupnea. Respir. Physiol. 1997;107:111–123. doi: 10.1016/s0034-5687(96)02513-3. [DOI] [PubMed] [Google Scholar]
- 33.Wawruszak A., Borkiewicz L., Okon E., Kukula-Koch W., Afshan S., Halasa M. Vorinostat (SAHA) and Breast Cancer: An Overview. Cancers. 2021;13:4700. doi: 10.3390/cancers13184700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Zanni E., Fornasari D., Ravazzolo R., Ceccherini I., Bachetti T. Identification of novel pathways and molecules able to down-regulate PHOX2B gene expression by in vitro drug screening approaches in neuroblastoma cells. Exp. Cell Res. 2015;336:43–57. doi: 10.1016/j.yexcr.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Athira K.V., Sadanandan P., Chakravarty S. Repurposing Vorinostat for the Treatment of Disorders Affecting Brain. NeuroMolecular Med. 2021;23:449–465. doi: 10.1007/s12017-021-08660-4. [DOI] [PubMed] [Google Scholar]
- 36.Verrillo L., Di Palma R., de Bellis A., Drongitis D., Miano M.G. Suberoylanilide Hydroxamic Acid (SAHA) Is a Driver Molecule of Neuroplasticity: Implication for Neurological Diseases. Biomolecules. 2023;13:1301. doi: 10.3390/biom13091301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weese-Mayer D.E., Berry-Kravis E.M., Ceccherini I., Keens T.G., Loghmanee D.A., Trang H., ATS Congenital Central Hypoventilation Syndrome Subcommittee An official ATS clinical policy statement: Congenital central hypoventilation syndrome: genetic basis, diagnosis, and management. Am. J. Respir. Crit. Care Med. 2010;181:626–644. doi: 10.1164/rccm.200807-1069ST. [DOI] [PubMed] [Google Scholar]
- 38.Bayliss D.A., Barhanin J., Gestreau C., Guyenet P.G. The role of pH-sensitive TASK channels in central respiratory chemoreception. Pflugers Arch. 2015;467:917–929. doi: 10.1007/s00424-014-1633-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bourgeois T., Ringot M., Ramanantsoa N., Matrot B., Dauger S., Delclaux C., Gallego J. Breathing under Anesthesia: A Key Role for the Retrotrapezoid Nucleus Revealed by Conditional Phox2b Mutant Mice. Anesthesiology. 2019;130:995–1006. doi: 10.1097/ALN.0000000000002675. [DOI] [PubMed] [Google Scholar]
- 40.Dubreuil V., Thoby-Brisson M., Rallu M., Persson K., Pattyn A., Birchmeier C., Brunet J.F., Fortin G., Goridis C. Defective respiratory rhythmogenesis and loss of central chemosensitivity in Phox2b mutants targeting retrotrapezoid nucleus neurons. J. Neurosci. 2009;29:14836–14846. doi: 10.1523/JNEUROSCI.2623-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goridis C., Dubreuil V., Thoby-Brisson M., Fortin G., Brunet J.F. Phox2b, congenital central hypoventilation syndrome and the control of respiration. Semin. Cell Dev. Biol. 2010;21:814–822. doi: 10.1016/j.semcdb.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Madani A., Pitollat G., Sizun E., Cardoit L., Ringot M., Bourgeois T., Ramanantsoa N., Delclaux C., d’Ortho M.P., d'Ortho M.P., et al. Obstructive Apneas in a Mouse Model of Congenital Central Hypoventilation Syndrome. Am. J. Respir. Crit. Care Med. 2021;204:1200–1210. doi: 10.1164/rccm.202104-0887OC. [DOI] [PubMed] [Google Scholar]
- 43.Bali P., Pranpat M., Bradner J., Balasis M., Fiskus W., Guo F., Rocha K., Kumaraswamy S., Boyapalle S., Atadja P., et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 44.Kovacs J.J., Murphy P.J.M., Gaillard S., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Rao R., Fiskus W., Yang Y., Lee P., Joshi R., Fernandez P., Mandawat A., Atadja P., Bradner J.E., Bhalla K. HDAC6 inhibition enhances 17-AAG--mediated abrogation of hsp90 chaperone function in human leukemia cells. Blood. 2008;112:1886–1893. doi: 10.1182/blood-2008-03-143644. [DOI] [PubMed] [Google Scholar]
- 46.Scroggins B.T., Neckers L. Post-translational modification of heat-shock protein 90: impact on chaperone function. Expet Opin. Drug Discov. 2007;2:1403–1414. doi: 10.1517/17460441.2.10.1403. [DOI] [PubMed] [Google Scholar]
- 47.Laederich M.B., Degnin C.R., Lunstrum G.P., Holden P., Horton W.A. Fibroblast growth factor receptor 3 (FGFR3) is a strong heat shock protein 90 (Hsp90) client: implications for therapeutic manipulation. J. Biol. Chem. 2011;286:19597–19604. doi: 10.1074/jbc.M110.206151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider C., Sepp-Lorenzino L., Nimmesgern E., Ouerfelli O., Danishefsky S., Rosen N., Hartl F.U. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl. Acad. Sci. USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitesell L., Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol. Endocrinol. 1996;10:705–712. doi: 10.1210/mend.10.6.8776730. [DOI] [PubMed] [Google Scholar]
- 50.Bachetti T., Di Zanni E., Ravazzolo R., Ceccherini I. miR-204 mediates post-transcriptional down-regulation of PHOX2B gene expression in neuroblastoma cells. Biochim. Biophys. Acta. 2015;1849:1057–1065. doi: 10.1016/j.bbagrm.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 51.Cardani S., Di Lascio S., Belperio D., Di Biase E., Ceccherini I., Benfante R., Fornasari D. Desogestrel down-regulates PHOX2B and its target genes in progesterone responsive neuroblastoma cells. Exp. Cell Res. 2018;370:671–679. doi: 10.1016/j.yexcr.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 52.Diana D., Pirone L., Russo L., D'Abrosca G., Madheswaran M., Benfante R., Di Lascio S., Caldinelli L., Fornasari D., Acconcia C., et al. Structural characterization of PHOX2B and its DNA interaction shed light on the molecular basis of the +7Ala variant pathogenicity in CCHS. Chem. Sci. 2024;15:8858–8872. doi: 10.1039/d3sc06427a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parodi S., Di Zanni E., Di Lascio S., Bocca P., Prigione I., Fornasari D., Pennuto M., Bachetti T., Ceccherini I. The E3 ubiquitin ligase TRIM11 mediates the degradation of congenital central hypoventilation syndrome-associated polyalanine-expanded PHOX2B. J. Mol. Med. 2012;90:1025–1035. doi: 10.1007/s00109-012-0868-1. [DOI] [PubMed] [Google Scholar]
- 54.Lui K.N.C., Li Z., Lai F.P.L., Lau S.T., Ngan E.S.W. Organoid models of breathing disorders reveal patterning defect of hindbrain neurons caused by PHOX2B-PARMs. Stem Cell Rep. 2023;18:1500–1515. doi: 10.1016/j.stemcr.2023.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Victor A.K., Hedgecock T., Donaldson M., Johnson D., Rand C.M., Weese-Mayer D.E., Reiter L.T. Analysis and comparisons of gene expression changes in patient- derived neurons from ROHHAD, CCHS, and PWS. Front. Pediatr. 2023;11 doi: 10.3389/fped.2023.1090084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cuadros Gamboa A.L., Benfante R., Nizzardo M., Bachetti T., Pelucchi P., Melzi V., Arzilli C., Peruzzi M., Reinbold R.A., Cardani S., et al. Generation of two hiPSC lines (UMILi027-A and UMILi028-A) from early and late-onset Congenital Central hypoventilation Syndrome (CCHS) patients carrying a polyalanine expansion mutation in the PHOX2B gene. Stem Cell Res. 2022;61 doi: 10.1016/j.scr.2022.102781. [DOI] [PubMed] [Google Scholar]
- 57.Falik D., Rabinski T., Zlotnik D., Eshel R., Zorsky M., Garin-Shkolnik T., Ofir R., Adato A., Ashkenazi A., Vatine G.D. Generation and characterization of iPSC lines (BGUi004-A, BGUi005-A) from two identical twins with polyalanine expansion in the paired-like homeobox 2B (PHOX2B) gene. Stem Cell Res. 2020;48 doi: 10.1016/j.scr.2020.101955. [DOI] [PubMed] [Google Scholar]
- 58.Munkacsi A.B., Hammond N., Schneider R.T., Senanayake D.S., Higaki K., Lagutin K., Bloor S.J., Ory D.S., Maue R.A., Chen F.W., et al. Normalization of Hepatic Homeostasis in the Npc1(nmf164) Mouse Model of Niemann-Pick Type C Disease Treated with the Histone Deacetylase Inhibitor Vorinostat. J. Biol. Chem. 2017;292:4395–4410. doi: 10.1074/jbc.M116.770578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hockly E., Richon V.M., Woodman B., Smith D.L., Zhou X., Rosa E., Sathasivam K., Ghazi-Noori S., Mahal A., Lowden P.A.S., et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc. Natl. Acad. Sci. USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L., Wang S., Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 61.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 62.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertoni A., Penco F., Mollica H., Bocca P., Prigione I., Corcione A., Cangelosi D., Schena F., Del Zotto G., Amaro A., et al. Spontaneous NLRP3 inflammasome-driven IL-1-beta secretion is induced in severe COVID-19 patients and responds to anakinra treatment. J. Allergy Clin. Immunol. 2022;150:796–805. doi: 10.1016/j.jaci.2022.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. The accession number for RNA-seq data is Gene Expression Omnibus (GEO): GSE250244.