Abstract
Vif is a human immunodeficiency virus type 1 (HIV-1) protein that is essential for the production of infectious virus. Most of Vif synthesized during HIV infection localizes within cells, and the extent of Vif packaging into virions and its function there remain controversial. Here we show that a small but detectable amount of Vif remains associated with purified virions even after their treatment with the protease subtilisin. However, treatment of these virions with 1% Triton X-100 revealed that most of the virion-associated Vif segregated with detergent-resistant virus particles consisting of unprocessed Gag, indicating that detergent-soluble, mature virions contain very little Vif. To investigate the control of Vif packaging in immature virus particles, we tested its association with Gag-containing virus-like particles (VLPs) in a Vif and Gag coexpression system in human cells. Only a small proportion of Vif molecules synthesized in this system became packaged into VLPs, and the VLP-associated Vif was protected from exogenous protease and detergent treatment, indicating that it is stably incorporated into immature virion-like cores. About 10-fold more Vpr than Vif was packaged into VLPs but most of the VLP-associated Vpr was removed by treatment with detergent. Mutagenesis of the C-terminal sequences in Gag previously shown to be responsible for interaction with Vif did not reduce the extent of Vif packaging into Gag VLPs. Surprisingly, short deletions in the capsid domain (CA) of Gag (amino acid residues 284 to 304 and 350 to 362) increased Vif packaging over 10-fold. The 350 to 363 deletion introduced into CA in HIV provirus also increased Vif incorporation into purified virions. Our results show that Vif can be packaged at low levels into aberrant virus particles or immature virions and that Vif is not present significantly in mature virions. Overall, these results indicate that the Vif content in virions is tightly regulated and also argue against a function of virion-associated Vif.
Human immunodeficiency virus type 1 (HIV-1) is a complex retrovirus that contains a number of genes not present in oncogenic retroviruses. The protein product of one of these genes, Vif (virion infectivity factor), is essential for productive HIV-1 infection in primary T cells and macrophages in vitro and for pathogenesis in models of AIDS (10, 18, 22, 23, 29, 55, 63, 66, 68). In cell culture, vif-defective (vif−) HIV-1 is able to replicate in some T-lymphoblastoid cell lines termed permissive (CEM-SS, SupT1, C8166, and Jurkat) whereas Vif is required in other cell lines, such as H9 or MT-2, termed nonpermissive (18, 22, 37, 64, 66, 69). It is generally accepted that Vif acts in late steps of the viral life cycle to ensure production of infectious virions (3, 12, 53, 56, 61, 64, 69). vif− HIV-1 is impaired in endogenous reverse transcription and in its ability to form proviral DNA in newly infected cells (4, 25, 59, 64, 69). The underlying reason for these deficiencies is unknown. It has been proposed that Vif participates in the processing of viral structural proteins and in the assembly of virus particles. An influence of Vif on the processing of viral structural proteins (4, 28, 53, 56) as well as on the maturation and stability of virion cores has been observed (4, 30, 43) although not confirmed by all investigators (6, 19). Another model suggested that Vif is needed in nonpermissive cells to overcome an unknown cellular function preventing replication of vif− HIV-1 (38, 39, 58).
Vif is a phosphorylated 23-kDa protein, which is abundantly expressed in infected cells. Vif has been shown to interact or copurify with membranes (24, 26), intermediate filaments (32), HIV-1 Gag (5, 31, 57), HIV-1 protease (PR) (2), and most recently with viral RNA (14, 72). Vif can inhibit PR activity in several models of proteolysis, and Vif-derived peptides inhibit Gag cleavage in HIV-1-infected cells (2, 34, 49). Vif function has been ascribed to most of these interactions, but each of these activities remains controversial.
While Vif is predominantly an intracellular protein (54), several reports have suggested that a fraction of the protein becomes incorporated into virions (32, 36). Liu et al. (36) reported that virions contained approximately 60 to 100 molecules of Vif that were incorporated into viral cores. Others reported a range of 20 to 100 Vif molecules per virion (8, 60). Observations that the amount of virion-associated Vif depends on the level of its expression within virus-producing cells (32, 60) and that Vif can be packaged into murine leukemia virus particles (8) led to the conclusion that Vif is not specifically incorporated into HIV virions. Recently, it was reported that highly purified, infectious HIV-1 virions do not contain Vif (15).
Resolving the question of Vif incorporation into virions is important for understanding the site of Vif activity. Here we demonstrate that Vif associates stably, albeit at low levels, with highly purified virions produced from several cell lines with different permissivity to the replication of vif− HIV-1. Interestingly, the virion-associated Vif was found primarily in aberrant, Pr55Gag-containing virions resistant to detergent treatment, indicating that mature virions that are soluble in detergent contain little Vif. To analyze the specificity of the association of Vif with immature virions, we created a Gag and Vif coexpression model system to produce virus-like particles (VLPs) containing unprocessed Gag and Vif. We report here that compared to Vpr, Vif is packaged inefficiently in Gag VLPs, but certain deletions in the capsid domain (CA) of Gag increased the amount of encapsidated Vif about 10-fold in both Gag VLPs and virions. Our data suggest that the association of Vif with virions is tightly regulated and indicate that Vif may be actively excluded from virus during particle release and maturation.
MATERIALS AND METHODS
Plasmids.
Vectors used for expression of Gag and Vif were based on pCITE-4a(+) (Novagen, Madison, Wis.). All gag and vif constructs were based on the HIV-1 NL4-3 molecular clone (1). The Gag Pr55 intermediate construct [pCITE4a(+)/Gag] was created by cloning the Gag coding region into EcoRI and XhoI sites in pCITE-4a(+) after amplification of the gag open reading frame in a chain reaction using Pfu polymerase (Stratagene, La Jolla, Calif.) and primers jGMA5 (49) and p6R (5′ GCCCCCCTCGAGTTATTGTGACG 3′). The resulting construct, expressing Gag with an N-terminally attached S-tag (33), was further modified to express wild-type Gag by removing the pCITE4a(+) vector sequence 519 to 629 by full-vector reamplification using the Advantage PCR enzyme system (Clontech, Palo Alto, Calif.) and primers CITER (5′ GGATCCGCCATGGGTGCGAGAGCGTCAG 3′) and CITEL (5′ GGTGGTGGCCATGGTATCATCGTG 3′), followed by treatment with DpnI, NcoI, and T4 DNA ligase to obtain a construct designated pCITEΔ/Gag. Essentially, pCITEΔ/Gag is a vector containing a bacteriophage T7 RNA polymerase promoter and CITE sequence (16, 17), followed by a gag open reading frame in native configuration. All subsequent gag deletion mutants (see Fig. 7B) were constructed by PCR amplification of the pCITEΔ/Gag template using the Advantage PCR enzyme system and a set of primers encompassing a deletion and carrying an NheI recognition site, allowing subsequent vector recircularization by treatment of a PCR product with NheI and DpnI and ligation with T4 DNA ligase. Mutagenic primers were as follows: (i) for Gag ΔMHR, in which Ala-Ser replaced Gag amino acid residues 284 to 304 composing the Gag major homology region (MHR), reverse primer CADLF2 (5′ CTTGTCTCGATCGCAGAAGCTTTC 3′) and forward primer MHDR (5′ CTATAAAGCTAGCAGAGCCGAGC 3′); (ii) for Gag Δ350-362, in which 13 CA protein C-terminal amino acid residues were replaced by Ser, reverse CSLF (5′ CCACTCCCTGGCTAGCTGTCATC 3′) and forward CADRG (5′ AAAGCAGCTAGCTTGGCTGAAGCAATG 3′); (iii) for Gag Δ350-377, in which CA protein C-terminal amino acid residues and the spacer peptide p2 were replaced by Ser, reverse CSLF and forward P2DRG (5′ GCTACCGCTAGCATACAGAAAGGC 3′); (iv) for Gag ΔNC, in which the entire nucleocapsid protein (NC) was replaced by Ala-Ser, reverse NCDLF (5′ TTGCCTTTGCTAGCCATTATGGTAGC 3′) and forward NCDRG (5′ GACAGGCTAGCTTTTTAGGGAAG 3′); (v) for Gag ΔNC1-14 and ΔNC1-14∗, in which the first 14 amino acid residues in NC were replaced by Ala-Ser, reverse NCDLF and forward ZN1DRG (5′ GACTGTTGCTAGCTTCAATTGTGGC 3′); (vi) for Gag Δp2, in which the Gag sequence 363 to 377 including a p2 spacer peptide was replaced by Ala-Ser, reverse P2DLF (5′ TGCTTCGCTAGCAACTCTTGCTTTATGG 3′) and forward P2DRG. Gag mutant ΔNC1-14∗ also carried, besides the deletion, a termination codon in position 27 within the first Cys-His box of NC, effectively removing a part of NC and the entire p1 and p6 Gag domains. The Gag mutants Δ350-362, Δ350-377, and ΔNC were inserted in a functional HIV-1 NL4-3 proviral clone by using Sph and BglII restriction sites.
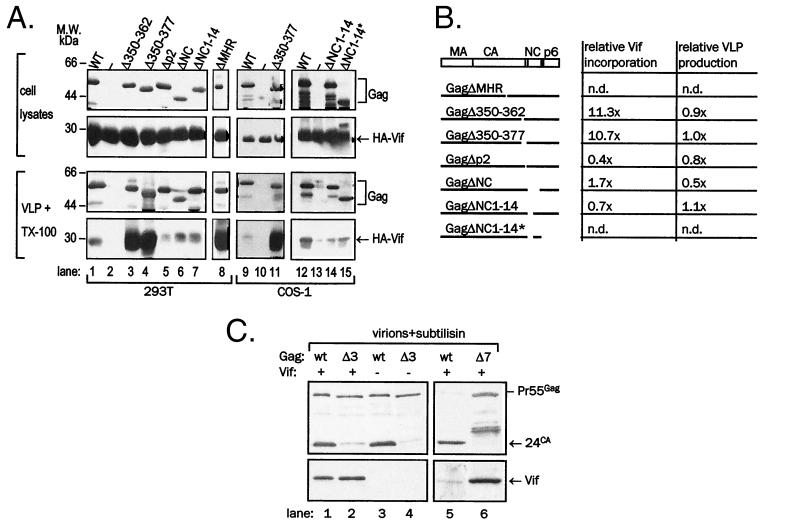
FIG. 7.
Association of Vif with mutant Gag VLPs and virions. (A) Gag was coexpressed with HA-Vif in 293T (lanes 1 to 8) or COS-1 (lanes 9 to 15) cells. VLPs sedimented from supernatants by ultracentrifugation were treated with 1% Triton X-100 to remove background Vif signal, resedimented, and resolved in SDS-PAGE together with a fraction of cell lysates. Vif was detected in immunoblotting with anti-HA monoclonal antibody (second and fourth panels from the top) and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top panel and third panel from the top). Gag ΔMHR in lane 8 was detected with AIDS patient sera. Differences in signal intensity among individual panels do not reflect differences in expression levels but rather the variability in film exposure. (B) Schematic representation of Gag deletion mutants used in the study and relative content of Vif in VLPs, normalized to Gag content. (C) Wild-type NL4-3 (wt) and GagΔ350-377 NL4-3 (Δ7) were produced in 293T cells upon transfection with proviral plasmid DNA and treated with subtilisin. Vif was detected in immunoblotting with anti-Vif rabbit antiserum (bottom panel), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top).
The vif gene was cloned in pCITE4(+) plasmid modification pCITEHA. pCITEHA/Vif expressed N-terminally tagged Vif with a sequence containing the influenza virus hemagglutinin epitope (HA; AYPYDVPDYA [27]). The construction of pCITEHA was carried out by amplifying pCITE4a(+) using Pfu polymerase from primers HACITER (5′ GGTTCCATGGCTTATCCTTATGACGTTCTGACTATGCTGGATCCGAATTCGAGCTCCGTCG 3′) and CITEL; the product of the amplification was digested by DpnI and NcoI and ligated. pCITEHA/Vif was constructed by inserting the vif gene from pGEX-2TVif (34) in BamHI and EcoRI cloning sites. pCITE/Vif expressing wild-type Vif without a tag was constructed by cloning the vif gene amplified by PCR using Pfu polymerase and primers VIFNCO1F (5′ CAGGGACCATGGAAAACAGATGGC 3′) and TRAP13 (5′ CGAGGAGATTCAGCTGATCACAGG 3′) in NcoI and SalI sites of pCITEHA, removing the HA tag. Vif was also expressed in S-tagged form from pCITE4a(+)/Vif. pCITEHA/Vpr was constructed by inserting the vpr gene amplified by PCR from forward primer VPRU (5′ CTGACAGAGGATCCATGGAACAAGCCCCAG 3′) and reverse primer VPRD (5′ CTTCCAGGGAATTCGTCTAGGATCTACTGG 3′) using pNL4-3 HIV-1 (1) as a template. Because vpr contains an internal EcoRI recognition site, the construction into pCITEHA/Vpr was carried out in two steps, using BamHI and EcoRI restriction enzymes.
The sequences of gag deletion mutants were confirmed by sequencing from primers CITE (Novagen), PS20 (5′ GGTCCAAAATGCGAACCCAG 3′), and PS21 (5′ TGGTTGGGGCTGTTTGGCTC 3′). The pCITEHA/Vif sequence was confirmed from primer CITE.
Cells and viruses.
The COS-1 cell line was obtained from American Type Culture Collection (ATCC; Rockville, Md.), and 293T cells were obtained from D. Littman. Cell cultures were maintained in Dulbecco modified Eagle medium (GIBCO/BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum and antibiotics. H9 and MT-2 cells were obtained from ATCC and maintained in RPMI 1640 (GIBCO/BRL) supplemented with 10% fetal calf serum and antibiotics. The HIV-1 NL4-3 molecular clone (1) was obtained from M. Martin through the National Institutes of Health (NIH) AIDS Research Reference and Reagent Program (Rockville, Md.). Construction of the HIV-1 molecular clone KS283 was described previously (54). The vaccinia virus recombinant clone vTF7-3 expressing the bacteriophage T7 RNA polymerase (20) was obtained from T. Fuerst and B. Moss through the NIH AIDS Research Reference and Reagent Program.
Transfection and expression using the vaccinia virus/T7 RNA polymerase expression system.
COS-1 (3 × 106 to 4 × 106) or 293T cells (5 × 106 to 8 × 106) plated in 100-mm petri dishes were transfected with plasmid DNA using a mixture of cationic lipid dimethyldioctadecylammonium bromide (DDAB; Sigma, St. Louis, Mo.) and phosphatidylethanolamine (Sigma) as described previously (52). Prior to transfection, cells were infected with vaccinia virus vTF7-3 (10 PFU/cell). Cells and supernatants were harvested approximately 24 h after transfection/infection.
Production of viruses.
293T cells were transfected with proviral plasmid DNA using Lipofectamine (GIBCO/BRL) and cocultivated with target T-lymphoblastoid cells; T cells were cultured until development of cytopathic effects, and then cell-free supernatants were harvested (wild-type HIV-1). Mutant gag HIV-1 recombinants were produced directly by transfection of 293T.
Preparation of virus and virus-like particles for analysis.
Harvested cells were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Culture supernatants of transfected or infected cells were clarified by low-speed centrifugation and filtration through a 0.45 μm-pore-size filter, and virion or VIPs were subsequently sedimented by centrifugation at 100,000 × g for 1 h through a 20% sucrose cushion. In some experiments, virions or VLPs were treated with subtilisin to digest contaminating cell debris and membrane vesicles (44). Briefly, concentrated virus suspension was supplemented with an equal volume of 2 mg of subtilisin (Sigma)/ml in 40 mM Tris-HCl (pH 8.0)–2 mM CaCl2 and incubated overnight at 37°C. Digestion was stopped by addition of phenylmethylsulfonyl fluoride to 5 μg/ml, and the virus or VLPs were recovered by centrifugation at 100,000 × g through a 20% sucrose cushion, resuspended in TNE (20 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA), and recentrifuged. In some experiments, VLPs or virions were treated with 1% Triton X-100 in TNE for 10 min at a specified temperature, followed by recentrifugation at 100,000 × g through a 30% sucrose cushion supplemented with 1% Triton X-100. The pelleted material was lysed by adding SDS-PAGE sample buffer.
Equilibrium sucrose gradient centrifugation.
VLPs or virions collected from supernatants with or without further treatment were placed on top of 10 ml of a 20 to 60% linear gradient of sucrose in phosphate-buffered saline or TNE and centrifuged for 18 h at 100,000 × g at 4°C. Fractions of 1 ml were collected from the bottom, and particles in each fraction were recovered by centrifugation at 100,000 × g, washed, and lysed in SDS-PAGE sample buffer.
Velocity Optiprep gradient centrifugation.
Optiprep (Iodixanol; GIBCO/BRL) gradient centrifugation was performed essentially as described by Dettenhofer and Yu (15). VLPs or virions were collected from supernatants through a sucrose cushion and washed free of sucrose. The pelleted particles were resuspended by repeated pipetting, and large aggregates were removed by centrifugation at 1,000 × g for 2 min. Particle suspension (100 to 250 μl) was placed on top of 11 ml of 6 to 18% linear Optiprep gradient in TNE and centrifuged for 1.5 h at 200,000 × g. Sixteen fractions of gradient were collected, and particles in each fraction were recovered by centrifugation at 100,000 × g and lysed in SDS-PAGE sample buffer.
Electrophoresis and immunoblotting.
Lysed cells or pelleted material were resolved by SDS-PAGE and transferred onto a 0.2-μm Trans-Blot nitrocellulose membrane (Bio-Rad, Hercules, Calif.) followed by immunoblotting as described elsewhere (37). Bound antibodies were visualized by using an enhanced chemiluminescence (ECL) kit (Amersham, Arlington Heights, Ill.). When needed, antibodies were stripped from nitrocellulose membranes by incubation in 0.2 M NaOH for 5 min. Antibodies used were as follows: anti-HA monoclonal antibody HA.11, clone 16B12 (BabCo, Berkeley, Calif.); anti-CA monoclonal antibody produced from Hybridoma 183, clone H12-5C (9), kindly provided by B. Chesebro and H. Chen through the NIH AIDS Research Reference and Reagent Program; and anti-Vif antibody (24), kindly provided by D. Gabuzda through the NIH AIDS Research Reference and Reagent Program. Densitometry was performed using the Kodak Digital Science system (Kodak, Rochester, N.Y.).
RESULTS
Vif is specifically associated with HIV-1 virions made in permissive and nonpermissive cells.
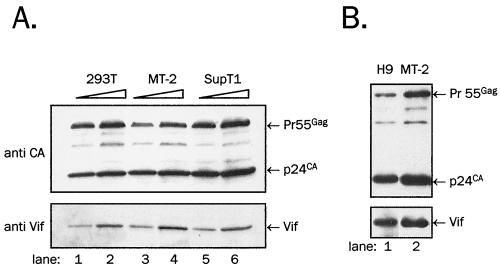
We first examined whether the level of Vif incorporation in HIV-1 virions depends on the ability of host cells to support replication of vif− HIV-1. We used permissive SupT1 cells (22), nonpermissive H9 and MT-2 cells (22, 37, 64), and 293T human embryonic kidney cells, which in our hands produce vif− HIV-1 virions with approximately twofold lower infectivity than that of wild-type HIV-1 (data not shown) and thus can be considered semipermissive. Virus from supernatants of infected or transfected cells (see Materials and Methods) was sedimented by centrifugation at 100,000 × g through a 20% sucrose cushion. Resuspended virions were treated with subtilisin to remove membrane vesicles and proteinaceous debris (44) and resedimented through a 20% sucrose cushion (Fig. 1). As shown in Fig. 1A, virions produced in either permissive or nonpermissive cells contained clearly detectable Vif after subtilisin treatment, and MT-2 cells carried the highest amount of Vif among the cell lines tested (compare Vif signals in lanes 1, 4, and 5, which contain an approximately equivalent amount of Gag). Virions produced in nonpermissive H9 cells contained Vif at a level similar to that found in MT-2-produced virions (Fig. 1B). Thus Vif associates with protease-treated virions made in permissive or nonpermissive cells.
FIG. 1.
Association of Vif with protease-treated virions. (A) NL4-3 virus was produced in cells as indicated and treated with subtilisin. Two aliquots (the second aliquot is a twofold amount of the first) of virus lysates from each producing line were resolved on SDS-PAGE. (B) KS283 virus produced from H9 and MT-2 cells was resolved on SDS-PAGE at a similar p24CA content. Vif was detected in immunoblotting with anti-Vif rabbit antiserum (bottom panels), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top panels).
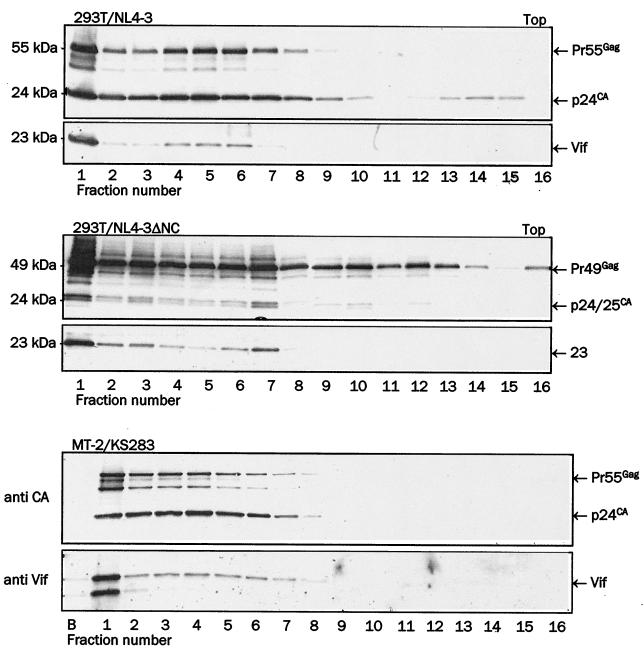
We further tested the association of Vif with virions purified through Optiprep gradients, the method that was shown to separate nonvirion particles containing Vif from bona fide virions (15). HIV-1 KS283 was produced by infection of MT-2 cells, and NL4-3 mutant in Gag lacking the NC domain were made by transfection of 293T cells with proviral plasmid DNA. The NC deletion mutant was used because NC was reported to mediate the interaction of Vif to Gag and the inclusion of Vif in VLPs (5, 31). Viruses collected from supernatants by ultracentrifugation were resolved in Optiprep velocity gradient centrifugation essentially as described previously (15). Individual gradient fractions were centrifuged to pellet the virions, and the Gag and Vif content of the pelleted material was analyzed by electrophoresis and immunoblotting (Fig. 2). We found the bulk of Gag in the bottom fraction (number 1) and the rest distributed through the lower half of gradients with a slight peak in higher fractions 4 to 7. Our interpretation of this result is that the bottom fraction contained virion aggregates while fractions 4 to 7 contained virion particles in suspension. Most importantly, Vif-specific signals aligned precisely with Gag signals, confirming that Vif copurifies with virions in Optiprep gradients. Deletion of the NC domain did not affect Vif incorporation. Similar results were obtained using HIV-1 KS283 produced from H9 cells (data not shown). We conclude that Vif is specifically associated with highly purified HIV-1 virions.
FIG. 2.
Association of Vif with virions purified by Optiprep gradient velocity centrifugation. Wild-type HIV-1 NL4-3 (top set of panels) and GagΔNC NL4-3 (middle set) produced in 293T cells or HIV-1 KS283 produced in MT-2 (bottom set) were resolved by velocity Optiprep gradient centrifugation. The gradient fractions were collected from the bottom (left, starting with number 1), and pelleted virions from each fraction were resolved on SDS-PAGE. In the HIV KS283 gradient, virions were also collected from the bottom of the centrifugation tube (B). Vif was detected by immunoblotting with anti-Vif rabbit antiserum (lower panel in each set), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top panel in each set).
Vif resides predominantly in virions composed of Pr55Gag that are insoluble in detergent.
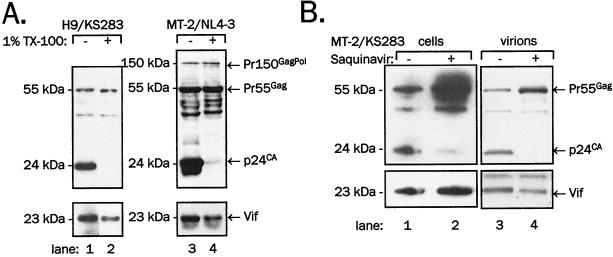
The virion samples from different cell lines used here contained relatively high amounts of unprocessed Pr55Gag in addition to p24CA (Fig. 1 and 2). The presence of both processed and unprocessed Gag observed in these experiments can be explained in two ways. Either individual virions contain both processed and unprocessed Gag, or virions consist of mixed populations of distinct particle types, one containing mature p24CA and no unprocessed Gag and the other containing Pr55Gag and no p24CA. To distinguish between these two alternatives, we treated purified virions with Triton X-100 and analyzed their composition compared to that of untreated virions (Fig. 3). It has been shown that mature but not immature retroviral cores dissolve in nonionic detergents such as Triton X-100 or Nonidet P-40 (7, 46, 47, 50, 65, 67, 70, 71). We reasoned that particles composed exclusively of Pr55Gag should remain intact and thus able to sediment after detergent treatment, while mature particles composed solely of processed Gag and presumably also particles containing a mixture of mature and unprocessed Gag should dissolve upon detergent treatment and fail to sediment. We treated KS283 and NL4-3 virions produced in H9 and MT2 cells with 1% Triton X-100 and centrifuged treated and untreated virions through 20% sucrose cushions (Fig. 3). As expected, the p24CA signal was almost entirely eliminated by Triton X-100 treatment (Fig. 3A; compare lanes 1 to 2 and 3 to 4 in the top panels), consistent with the solubilization of mature virions containing p24CA. In contrast, the signal of Pr55Gag was essentially unchanged, suggesting that detergent-stable particles containing unprocessed Gag contain no mature p24CA. This result suggests that virions contain either mature p24CA or unprocessed Pr55Gag but not both. Upon Triton X-100 treatment, most of Vif present in virion preparations cosedimented with Pr55Gag (Fig. 3A; compare lanes 1 to 2 and 3 to 4 in the bottom panels), indicating that Vif associates primarily with particles containing unprocessed Gag and not with mature virions. The slight decrease in the Vif signal following detergent treatment is consistent with removal of membrane vesicles or cell debris containing Vif as suggested by Dettenhofer and Yu (15) and our experiments with expression of Vif in the absence of Gag (see below).
FIG. 3.
Association of Vif with virions treated with detergent or viral protease inhibitor. (A) KS283 virus was harvested from H9 or MT-2 cells and treated with 1% Triton X-100 for 10 min at room temperature. Virions were resedimented through a 20% sucrose cushion and resolved on SDS-PAGE. (B) KS-283-infected MT-2 cells were treated with the viral protease inhibitor saquinavir 2 days after infection. Virus was collected 24 h later and centrifuged through a 20% sucrose cushion. Cell (left) or virus lysates were resolved on SDS-PAGE. Vif was detected in immunoblotting with anti-Vif rabbit antiserum (bottom), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top).
Effect of HIV-1 PR inhibitor on Vif incorporation in virions.
The result shown in Fig. 3A indicated that most of the virion-associated Vif resides in virus particles containing Pr55Gag, some of Vif is present in membrane vesicles or cell debris, but very little or no Vif becomes incorporated into mature virions. The apparent selective incorporation of Vif into Pr55Gag-containing particles and, conversely, its exclusion from mature virions was surprising. One possible explanation for Vif exclusion is that it may be degraded by viral protease in mature virions. However, this is unlikely because Vif was shown to be resistant to PR-mediated cleavage in bacteria, in mammalian cells, and in cell-free proteolysis assays (34, 49). Alternatively, Vif may be excluded from virions by some mechanism during budding and maturation while a fraction of the protein remains associated with defective Gag assembly complexes that are not destined for maturation into infectious virions but are released as Pr55Gag-containing particles. To investigate this possibility, we treated HIV-1-infected MT-2 with the HIV-1 protease inhibitor saquinavir (51) under conditions in which processing and infectivity are blocked (Fig. 3B). If Gag and Gag-Pol processing by viral protease is responsible for exclusion of Vif from virions or Vif is degraded by protease, its inhibition should increase the content of Vif in virions. As shown in Fig. 3B, inhibition of protease-mediated proteolysis eliminated the p24CA signal in both virions and cell extracts and slightly reduced—rather than increased—the amount of Vif associated with virions (Fig. 3B; compare lanes 3 and 4, bottom panels). This indicates that Gag processing does not lead to exclusion of Vif from mature virions and that Vif is not degraded by viral protease. Rather, this result suggests that Vif may associate with putative defective particles lacking other viral proteins or assuming a defective structure and subsequently be unable to process Gag and pass through maturation of the virion core.
Analysis of Vif encapsidation in Gag and Vif coexpression system.
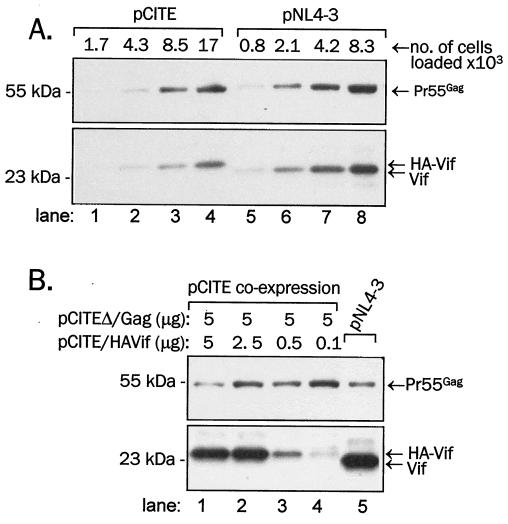
The association of Vif with Pr55Gag containing viral cores suggested a specific interaction between the two proteins. To determine the requirements for the association of Vif with immature virions, we adopted a versatile system for Gag and Vif mutagenesis and coexpression to produce Gag-containing VLPs that closely resemble immature virions (21). Gag, Vif, and Vpr were expressed from the pCITE vector, which permits efficient transcription of inserted genes from the T7 bacteriophage promoter, further facilitated by the presence of a cap-independent translational enhancer derived from encephalomyocarditis virus in cis (CITE [16, 17]). The T7 polymerase is supplied in trans by infection of cells with recombinant vaccinia virus vTF7-3 (20). Vif and Vpr were N-terminally tagged with an HA tag (27) to facilitate detection and to directly compare the efficiency of incorporation of Vif and Vpr in VLPs, and the S tag (33) was used as a control to the HA tag. Similar systems have been used previously to analyze the association of HIV-1 Vpr or HIV-2 Vpx with Gag and VLPs (40, 41, 45, 62). In the first set of experiments, we compared the relative efficiency of Gag and Vif expression from pCITE with the expression of these proteins during HIV-1 infection (Fig. 4). As shown in Fig. 4A, the overall expression of Gag and Vif from provirus in 293T cells was slightly more efficient than the expression of these proteins from pCITE (Fig. 4A; compare Gag and Vif signals in lanes 3 and 6). To interpret obtained data, it was crucial to maintain the ratio of Gag and Vif expressed from pCITE similar to that of the proviral expression. This was achieved when the pCITEΔ/Gag and pCITEHA/Vif DNAs were transfected at a 1:1 or 1:2 weight ratio (Fig. 4B; compare Gag and Vif signals in lanes 1, 2, and 5), and we therefore typically cotransfected 5 μg of pCITEΔ/Gag and 2.5 to 5 μg of pCITEHA/Vif per petri dish.
FIG. 4.
Comparison of Gag and Vif (HA-Vif) expression in 293T cells transfected with pCITE or with an NL4-3 proviral clone. (A) 293T cells cotransfected with pCITEΔ/Gag and pCITEHA/Vif or 293T cells transfected with pNL4-3 were loaded in the amounts indicated. (B) 293T cells were cotransfected with pCITEΔ/Gag and pCITEHA/Vif as indicated (in μg DNA per petri dish) or with pNL4-3. Gag and Vif in 17,000 pCITE-transfected or 8,300 pNL4-3-transfected cells were detected in immunoblotting with anti-Vif rabbit antiserum (bottom), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top). Note that exposure of Vif in panel B was longer than in panel A to detect weak signals in lanes 3 and 4.
Vif is packaged into VLPs less efficiently than Vpr.
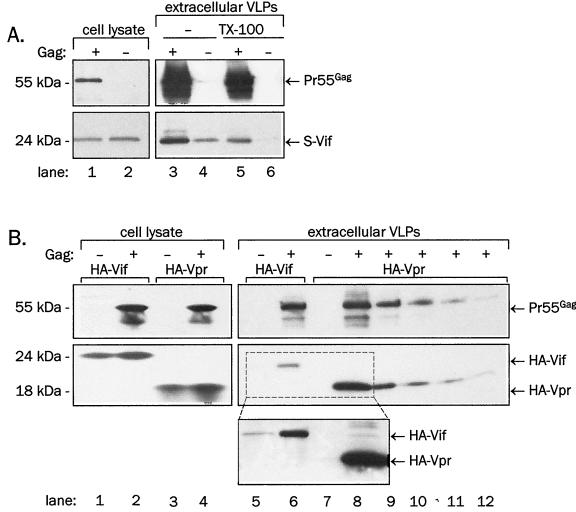
We next examined whether coexpression of Gag and Vif would lead to the production of VLPs containing Vif. As shown in Fig. 5, the expression of Gag in COS-1 or 293T cells directed the assembly and release of VLPs, which could be sedimented from culture supernatants by ultracentrifugation. This is shown by detection of a CA-specific 55-kDa band corresponding to the Gag precursor polyprotein in culture supernatant sediments (Fig. 5A, lanes 3 and 5; Fig. 5B, lanes 6 and 8 to 12; top panels). Vif coexpressed with Gag cosedimented with VLPs as attested by the presence of a Vif-specific protein of 24 kDa in Gag-containing lanes (Fig. 5A, lanes 3 and 5; Fig. 5B, lane 6; bottom panels). Wild-type Vif cosedimented with Gag equally as well as the N-terminally HA- or S-tagged Vif (Fig. 6B), indicating that Vif association with VLPs was not due to the presence of oligopeptide tags. To exclude the possibility that Vif itself forms extracellular sedimentable aggregates, we collected and analyzed particulate material present in the supernatants of cells expressing Vif in the absence of Gag. Although a small amount of Vif was pelletable, it dissolved upon treatment with detergent, indicating that Vif released from cells in the absence of Gag mostly associates with membrane vesicles or membranous debris (Fig. 5A, lanes 4 and 6). The amount of Vif in particles released from Gag and Vif coexpressing cells also slightly diminished after detergent treatment (Fig. 5A; compare lanes 3 and 5), confirming that a small portion of Vif sedimentable from supernatants of these cells associates with membrane vesicles while most of it represents Vif incorporated in VLPs. We conclude that Vif is packaged into Gag VLPs.
FIG. 5.
Association of Vif or Vpr with Gag VLPs. Lysates of cells coexpressing Gag with HA-Vif or HA-Vpr or lysates of supernatant sediments were resolved on SDS-PAGE. (A) S-tagged Vif was coexpressed with the wild-type Gag in COS-1 cells. An aliquot of sedimented VLPs was treated with 1% Triton X-100 for 15 min at room temperature and resedimented. S-Vif was detected in immunoblotting with anti-Vif rabbit antiserum (bottom), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top). (B) HA-Vif or HA-Vpr were coexpressed with wild-type Gag from the pCITE vector in 293T cells. Lanes 8 to 12: decreasing fractions of supernatant pellet lysate were loaded in the order 1/2, 1/5, 1/10, 1/20, and 1/50. HA-Vif and HA-Vpr were detected in immunoblotting with anti-HA monoclonal antibody (bottom and middle panels), and Gag was detected by reprobing of stripped membranes with anti-CA monoclonal antibody (top). Inset: longer exposure of lanes 5 to 8 indicates background levels of sedimentable Vif and Vpr.
FIG. 6.
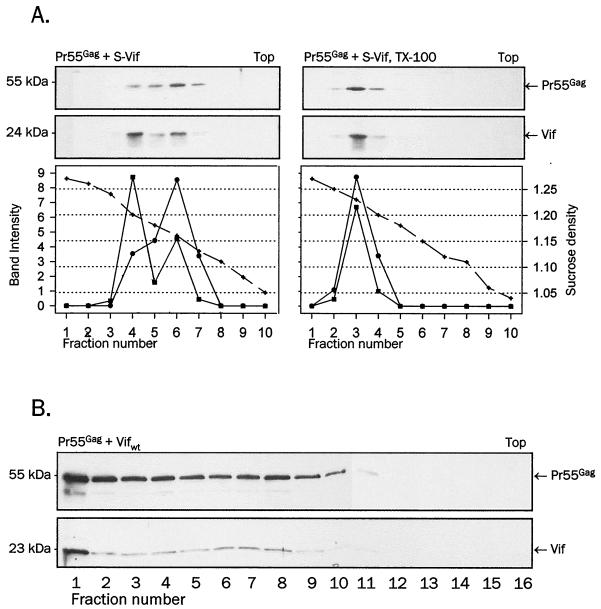
Association of Vif with VLPs purified by gradient centrifugation. (A) Gag was coexpressed with S-Vif from pCITE in COS-1 cells. Sedimented VLPs were treated with Triton X-100 for 15 min on ice or left untreated and centrifuged through a 20 to 60% sucrose gradient to equilibrium. Gradient fractions collected from the bottom (fraction number 1) were pelleted and probed for Vif with anti-Vif rabbit antiserum (bottom panels) and for Gag with anti-CA monoclonal antibody after stripping (top panels). The sucrose density of individual fractions or the intensity of bands obtained by densitometry are plotted below each panel: the solid line with square symbols represents Vif signal intensity; the solid line with circle symbols represents Gag signal intensity; the dashed line with crossed symbols represents sucrose density. (B) Wild-type Gag VLPs containing wild-type Vif were produced in 293T cells upon cotransfection with pCITEΔ/Gag and pCITE/Vif and resolved by velocity Optiprep gradient centrifugation. The gradient fractions were collected from the bottom (left, starting with number 1), and pelleted VLPs from each fraction were resolved on SDS-PAGE and immunoblotted with anti-Vif (bottom) and anti-CA (top) antibodies as in panel A.
This coexpression system allowed us to directly compare the efficiency of the incorporation of Vif into Gag VLPs with that of Vpr when both proteins were N-terminally tagged with HA epitope (Fig. 5B). Vpr is known to be efficiently encapsidated in HIV-1 virions or Gag VLPs (11, 41, 42, 62). Although the expression levels of HA-Vif or HA-Vpr were similar (Fig. 5B, lanes 2 and 4), extracellular Gag VLPs contained much higher levels of HA-Vpr than HA-Vif (compare lanes 6 and 8 in Fig. 5B, bottom panel). By loading decreasing amounts of VLPs collected from supernatants of cells coexpressing Gag and HA-Vpr (Fig. 5B, lanes 9 to 12), we estimated that HA-Vpr is approximately 10- to 20-fold more abundant in Gag VLPs than is HA-Vif. Thus, Vif is packaged into Gag VLPs much less efficiently than Vpr.
Vif is specifically incorporated in Gag VLPs resolved by gradient centrifugation.
We sought to obtain more conclusive evidence for the specificity of Vif packaging in VLPs by purifying particles through sucrose density gradients. Extracellular VLPs were isolated from supernatants of COS-1 cells expressing wild-type Gag and N-terminally tagged Vif and resolved by density equilibrium centrifugation through a 20 to 60% linear sucrose gradient. As shown in Fig. 6A, left panels, Gag VLPs were most abundant in fractions 4 to 6, corresponding to the density of 1.16 to 1.20 g/cm3 typical for retroviral particles. Detergent treatment of VLPs shifted the Gag signal to fraction 3 with a density of 1.26 g/cm3 (Fig. 6A, right panels), consistent with the removal of lipid envelopes. The peak signals of Vif aligned precisely with the peak Gag signals, indicating that Vif specifically associates as an integral component with Gag VLPs. VLPs composed of Gag alone or Gag and Vif migrated to fractions of similar density, indicating that the presence of Vif does not affect the density of VLPs (data not shown).
Using Optiprep velocity gradients, we demonstrated that highly purified HIV-1 virions contain Vif (Fig. 2). To confirm that VLPs reproduce this association, we examined the content of Vif in VLPs purified through Optiprep gradients (Fig. 6B). Gag and wild-type Vif were coexpressed in 293T cells, and particles from culture supernatant sedimented through a 20% sucrose cushion and washed free of sucrose were resolved in a 6 to 18% Optiprep gradient. The pelletable material in individual fractions was screened for Gag and Vif by immunoblotting (Fig. 6B). Strong signals of both Gag and Vif were detected in the bottom fraction (number 1), and Gag and Vif were further dispersed in identical fractions 2 through 10 with a slight increase in fractions 7 and 8. We conclude that coexpression of wild-type Gag with wild-type or N-terminally tagged Vif results in the assembly and release of VLPs that specifically incorporate Vif. A detectable amount of particulate Vif is also secreted, in addition to that present in Gag VLPs, but it can be distinguished from Vif within VLPs on the basis of its detergent sensitivity (Fig. 5A) but not by its density or shape as revealed by migration in density gradients (Fig. 6).
Pattern of Vif incorporation in VLPs and virions consisting of Gag with deletions.
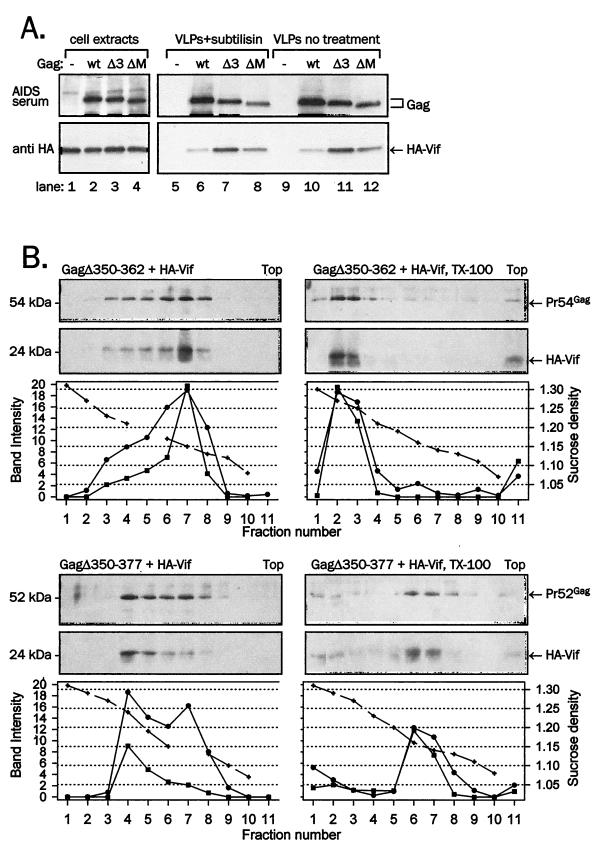
Sequences in the NC domain of Gag have been shown to mediate interaction with Vif as well as packaging of Vif in Gag VLPs isolated from insect cells (31). We sought to examine whether this C-terminal region of the Gag precursor polyprotein is required for Vif particle packaging in animal cells, anticipating that coexpression of Vif with Gag precursor polyprotein carrying C-terminal deletions will lead to secretion of VLPs lacking Vif. We constructed a set of deletion or truncation mutants spanning the C-terminal part of Gag including the CA and NC domains as well as the spacer peptide p2 (see Materials and Methods and Fig. 7B). Figure 7A shows the Gag and Vif content in VLPs obtained in three experiments in which Gag mutants were coexpressed with HA-Vif in 293T or COS-1 cells. Samples where Vif was expressed together with wild-type Gag (lanes 1, 9, and 12) or alone (lanes 2, 10, and 13) served as controls. As shown in the VLP Gag panels in Fig. 7A (third row from top) and summarized in the table in Fig. 7B, most constructs produced VLPs at levels similar to that of the wild-type Gag. Vif was present in VLPs formed of all Gag mutants but its amount strongly depended on the Gag structure. Significantly, Vif was present in VLPs formed of GagΔNC, GagΔNC1-14, and GagΔNC1-14∗, indicating that the nucleocapsid region and downstream C-terminal areas in Gag are not required for Vif packaging in VLPs (Fig. 7A, lanes 6, 7, 14, and 15, and the table in Fig. 7B). Surprisingly, VLPs consisting of Gag containing short deletions in the C-terminal region of CA protein (ΔMHR, Δ350-362, and Δ350-377) incorporated a significantly larger amount of Vif than the wild-type Gag VLPs (Fig. 7A, lanes 3, 4, 8, and 11), an approximately 10-fold excess (table in Fig. 7B for GagΔ350-362 and GagΔ350-377). In contrast to other deletion mutants, VLPs consisting of Δp2 Gag contained approximately 30 to 60% of the Vif present in wild-type Gag VLPs (Fig. 7A, lane 5, and the table in Fig. 7B).
To determine whether the effect of Gag structure upon Vif incorporation into VLPs applied also to its encapsidation in virions, we constructed an NL4-3 recombinant clone containing a deletion of the extreme CA C terminus and p2, (Δ350-377) and expressed it by transfection in 293T cells. Wild-type NL4-3 and Gag Δ350-377 virions were isolated, treated with subtilisin, and tested for Vif content (Fig. 7C). Gag Δ350-377 virus showed a higher ratio of Vif to Gag than the wild-type virus. Similarly, HIV-1 NL4-3 Gag mutant Δ350-362 carried an increased amount of Vif (data not shown), verifying that CA deletions cause increased packaging of Vif in bona fide virions.
To confirm that the increased content of Vif in VLPs made of Gag containing CA deletions was indeed specific, VLP fractions were subjected to overnight treatment with subtilisin to degrade vesicles and cellular debris (44). The treatment with subtilisin removed approximately 50% of Gag and Vif from sedimentable material (data not shown) but the relative amount of HA-Vif in VLPs treated with subtilisin was equivalent to that in untreated VLPs and the marked increase of Vif content in VLPs consisting of Gag with a deletion in the CA domain was preserved (Fig. 8A; compare lanes 6 to 8 to lanes 10 to 12).
FIG. 8.
Association of Vif with mutant Gag VLP purified by protease treatment or through a sucrose gradient. (A) HA-Vif was coexpressed with wild-type Gag (wt), GagΔ350-362 (Δ3), or GagΔMHR (ΔM) in 293T cells. VLPs collected from supernatants were treated with subtilisin and resolved together with cell lysates on SDS-PAGE. (B) Gag was coexpressed with HA-Vif in 293T cells, and VLPs pelleted from supernatant were treated with Triton X-100 (right panels) or left untreated and resedimented through a 20 to 60% density sucrose gradient overnight. Ten fractions of gradient, starting from the bottom (fraction number 1), as well as the sediment from the bottom of centrifugation tubes representing material denser than 60% sucrose (fractions 11), were collected, resedimented, and analyzed for Vif content in immunoblotting with anti-HA monoclonal antibody (bottom panels) and for Gag by reprobing of stripped membranes with anti-CA monoclonal antibody (top panels). The sucrose density of individual fractions obtained by refractometry or the intensity of Vif and Gag bands obtained by densitometry are plotted below each panel. The solid line with square symbols represents Vif signal intensity; the solid line with circle symbols represents Gag signal intensity; and the dashed line with crossed symbols represents sucrose density.
To further verify the association of increased amounts of Vif within mutant Gag VLPs, GagΔ350-362 and GagΔ350-377 VLPs containing HA-Vif were purified by centrifugation through a sucrose density gradient (Fig. 8B); VLPs were either treated with Triton X-100 (right panels) or left untreated and evaluated for the distribution of Vif and Gag. Similar to data shown in Fig. 6, the Vif signal colocalized with Gag, demonstrating that Vif was specifically incorporated in VLPs. For GagΔ350-362, the density of untreated VLPs ranged between 1.15 and 1.17 g/cm3 (Fig. 8B, top left set of panels, fractions 6 and 7), while the detergent-treated VLPs migrated to fractions with a density of 1.25 to 1.27 g/cm3 (Fig. 8B, top right set of panels, fractions 2 and 3), identical to the wild-type Gag VLPs (compare with Fig. 6). VLPs composed of GagΔ350-377 displayed much broader density distribution, with a peak between 1.13 and 1.24 g/cm3, indicating that the larger deletion of the C terminus of CA protein plus spacer peptide p2 disturbed the structure of VLPs (Fig. 8B, bottom left set of panels, fractions 4 to 8). The detergent treatment of GagΔ350-377 VLPs, unexpectedly, concentrated the peak Gag and Vif signals in lighter 1.14 to 1.16 g/cm3 density fractions (Fig. 8B, bottom right set of panels, fractions 6 and 7), although some Gag signal was also detected in bottom fractions with a density of 1.27 to 1.3 g/cm3. It is not clear why the density of detergent-treated GagΔ350-377 VLPs decreased upon detergent treatment that invariably increases the density of wild-type Gag VLPs (Fig. 6). We consider that the deletion in Gag (position 350 to 377) grossly affects the structure of the resulting VLPs and their conformation after detergent treatment. The presence of Vif did not affect the density of VLPs formed by Gag mutants Δ350-362 and Δ350-377 as these mutant Gag VLPs displayed similar densities and response to Triton X-100 treatment in the absence of Vif (data not shown). Specific comigration of HA-Vif with GagΔ350-362 was also seen in the Optiprep velocity gradient (data not shown). We conclude that two short deletions in the CA domain of the Gag precursor polyprotein that otherwise are not disruptive to assembly and release of Gag VLPs lead to a 10-fold increase in the packaging of Vif in VLPs.
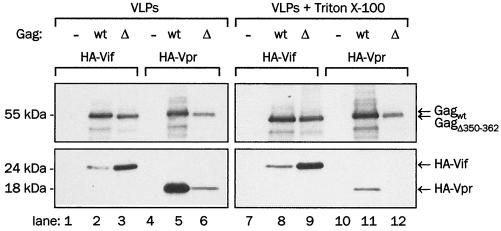
Comparison of Vif and Vpr incorporation in CA mutant and wild-type Gag VLPs.
In the next experiment, we sought to determine whether increased incorporation of Vif in VLPs formed by CA deletion mutants is a result of the general tendency of these mutant Gag VLPs to incorporate increased amounts of viral proteins. We coexpressed HA-Vif or HA-Vpr in the presence of wild-type Gag or GagΔ350-362 and determined the efficiency of incorporation of these proteins in VLPs and the stability of incorporation after detergent treatment (Fig. 9). Intracellularly, HA-Vif and HA-Vpr were expressed at similar levels, but the content of HA-Vpr was approximately 25-fold greater than the content of HA-Vif in untreated wild-type Gag VLPs, as determined by immunoblotting with anti-HA antibody and densitometry (Fig. 9; compare lanes 2 and 5). While the signal of HA-Vif markedly increased in Δ350-362 Gag VLPs compared to that in wild-type Gag VLPs (Fig. 9, lanes 2 and 3), the amount of HA-Vpr was lower in GagΔ350-362 VLPs than in the wild-type Gag VLPs (Fig. 9, lanes 5 and 6). Triton X-100 treatment removed most of the HA-Vpr but little or no HA-Vif (Fig. 9; compare lanes 2 and 3 to 8 and 9 and lanes 5 and 6 to 11 and 12, respectively). Thus the 350 to 362 deletion in the CA domain of Gag yields VLPs with an increased capacity to package Vif but not Vpr. This suggests that the molecular mechanisms of Vif and Vpr packaging in Gag VLPs are different and that Vpr is located at the periphery of VLPs, becoming soluble upon removal of the particle-enveloping membrane, while Vif is firmly integrated in the VLP structure.
FIG. 9.
Association of HA-Vif and HA-Vpr with wild-type (wt) or Δ350-362 Gag (Δ) VLPs. Gag was coexpressed with HA-Vif or HA-Vpr in 293T cells, and VLPs released to culture supernatants were collected by ultracentrifugation, with or without detergent treatment. Lysates of sedimented VLPs were analyzed with anti-HA monoclonal antibody for HA-Vif and HA-Vpr (bottom panels) and Gag was analyzed by reprobing of stripped membranes with anti-CA monoclonal antibody (top panels).
DISCUSSION
Besides viral structural proteins and enzymes, HIV-1 virions have been shown to contain Nef, Vif, and Vpr (13). The issue of Vif incorporation in virions remains unsettled; while the initial report suggested that virions contain 60 to 100 copies of Vif per virion (36), subsequent reports lowered estimated Vif content from 20 molecules per virion to none (8, 15, 60). The controversy about Vif incorporation into virions, its proposed involvement in late steps of virus assembly and maturation (22, 30, 53, 56, 69), and observations that Vif colocalizes or interacts with Gag (5, 31, 57) led us to investigate the forms of HIV-1 virions which incorporate Vif. Our results from both physical analyses and mutagenesis indicate that Vif incorporation is restricted to aberrant virions, with little or no Vif packaging into typical mature viral particles.
We believe that our results may explain the discrepancy between findings from different laboratories concerning the issue of Vif packaging into virions. Like some other investigators (32, 36), we have found that Vif associates tightly with viral particles in the presence of detergent (Fig. 1 to 3). Treatment of virus samples with protease subtilisin, known to remove membrane vesicles (44), only slightly diminished Vif content in virions (Fig. 1), in agreement with a report by Liu et al. (36), who observed the presence of Vif in virions treated with proteinase K. Also similar to the findings of Liu et al. (36), Vif association with virions was independent of virus-producing cells with respect to their permissivity to vif− HIV-1 infection. On the other hand, our findings contradict data published by other investigators showing that virions contain little or no Vif (8, 15, 60). An unexpected insight into the Vif packaging controversy came from our experiments in which we employed detergent treatment to distinguish between mature virions and those containing Pr55Gag, putatively of immature phenotype. We found that Vif associates predominantly with Pr55Gag-containing particles resistant to detergent treatment but associates insignificantly with mature virions sensitive to detergent treatment (Fig. 3A). The solubility of mature retroviral particles in detergent is well documented (7, 46, 47, 65, 67, 71), whereas cores of “stripped” immature or immature-like defective virions retain physical integrity and the ability to sediment upon high-speed centrifugation (50, 70). We suggest that the Pr55Gag in virus lysates such as those seen in Fig. 1 to 3 is derived from aberrant or immature particles, and Vif copurification with detergent-stable Pr55Gag-containing particles (Fig. 3) indicates that Vif is incorporated predominantly into these aberrant immature-like virions while it is excluded from mature virions. Thus, the inconsistency among studies on Vif encapsidation may be related to the differences in the ratio of mature and immature-like virions analyzed. Dettenhofer and Yu observed that extracellular particles containing Vif migrate more slowly during Optiprep gradient velocity centrifugation than do virions composed of Gag (15). We performed similar Optiprep gradient velocity centrifugation with virions produced from 293T and MT-2 cells (Fig. 2) or H9 cells (data not shown) and detected Vif and Gag in identical fractions of gradients, leading us to conclude that the bulk of Vif is specifically associated with virions. Dettenhofer and Yu (15) used cloned H9 cells chronically infected with HXB2NEO HIV-1 that seem to produce virions containing less Pr55Gag than the acutely infected cells used in our study, possibly contributing to disagreement with our data. The mechanism of Vif exclusion from mature virions is not clear. Although we did not exclude the possibility that Vif is degraded by viral protease upon virion maturation, processing of Gag or proteolytic cleavage of Vif is unlikely to be the main mechanism of its exclusion because treatment with HIV-1 protease inhibitor did not increase the content of Vif in virions (Fig. 3B). Rather, Vif may associate with defective particles consisting mostly of Pr55Gag that contain a suboptimal dose of Pol products, including viral protease, or with structurally aberrant particles. The properly assembled virions may exclude Vif entirely.
Since immature virions consist mostly of unprocessed Gag, we evaluated conditions influencing Vif packaging in VLPs, a model closely resembling immature virions. We show that a relatively minor fraction of Vif is incorporated into VLPs and that the association of Vif with VLPs remains stable upon detergent treatment. Under the same conditions, about 20-fold more Vpr than Vif is incorporated in VLPs but it can be extracted by detergent treatment. Limited mutagenesis of Gag failed to reveal any sequences required for Vif incorporation in VLPs; however, short deletions in the Gag CA domain were found to increase Vif packaging about 10-fold. Our data are similar to those reporting that Vif is incorporated in VLPs produced from insect cells coexpressing Gag and Vif (31); however, in contrast to that study, we have found no requirement for the presence of NC and downstream p1 and p6 domains for Vif encapsidation either in VLPs (Fig. 7) or in virions (Fig. 2). The use of insect versus animal expression systems may account for this difference.
The increased Vif incorporation in virus or virus-like particles consisting of Gag CA deletion mutants, compared to that in wild-type Gag particles, suggests that the CA or certain CA sequences (MHR and extreme C terminus) regulate the encapsidation of Vif (Fig. 7). We considered two possible explanations of this observation. It is possible that a mutant Gag may form VLPs or virions of aberrant size or shape that nonspecifically incorporate increased amounts of non-Gag proteins, including Vif. Alternatively, certain Gag mutants may be unable to assume the specific conformation of wild-type Gag in particles that is unfavorable for Vif encapsidation. While Gag Δ350-362 VLPs encapsidated excess Vif, Vpr incorporation was reduced (Fig. 7), suggesting that these VLPs have no increased tendency to package viral proteins in general. Further, we observed no consistent correlation between aberrant physical properties of particles formed by two Gag mutants and increased Vif content. Gag Δ350-377 has an abnormal sucrose gradient profile while Δ350-362 does not (Fig. 8B). Some of the Gag mutants that have been reported to produce virions with aberrant morphology, such as the Δp2 mutant (35), showed slightly decreased Vif content in VLPs (Fig. 7), and therefore the shape of VLPs itself is not likely to cause a greatly increased content of Vif. Rather, we suggest that Vif is excluded from wild-type Gag VLPs as well as from virions as a consequence of structural rearrangements during particle assembly.
The previous observation that Vif can be packaged into murine leukemia virus particles upon coexpression (8) indicates that Vif may have a natural propensity for inclusion into retroviral Gag particles. We have observed that feline immunodeficiency virus Vif (48) and C-terminal truncation mutants of HIV-1 Vif were more efficiently packaged in HIV-1 Gag VLPs than wild-type HIV-1 Vif (data not shown). It is possible that Vif has a natural tendency to incorporate in virus-like particles or in virions but is partially excluded during their assembly and release dependent on intact Vif and Gag structure. Further exclusion may occur secondarily during assembly and maturation of functional HIV-1 virions. It has previously been suggested that Vif is poorly incorporated in virions based on studies showing that Vif acts as an inhibitor of viral protease (34, 49) and must be excluded from virions to permit capsid maturation.
Involvement of Vif in late stages of the virus life cycle (3, 12, 53, 56, 61, 64, 69), either by its participation in regulation of the processing of viral structural proteins (4, 28, 34, 53, 56), in maturation of the virion core (4, 30, 43), or in folding and encapsidation of viral genomic RNA (14, 72) imply that Vif may be located at the site of virion assembly. Upon providing its function, further incorporation in virions may be unnecessary or detrimental, leading to evolution of a mechanism of its exclusion from mature virions.
ACKNOWLEDGMENTS
We thank Mary Jane Potash, Bill Grossman, Beda Brichacek, and Malgorzata Simm for assistance with part of this work. We acknowledge the help of the NIH AIDS Research and Reference Reagent Program in providing some reagents used in this study, specifically from B. Chesebro, H. Chen, T. Fuerst, B. Moss, and D. Gabuzda. We thank D. Littman for providing a 293T cell line.
This work was supported by Public Health Service grants to P.S. and D.J.V.
REFERENCES
- 1.Adachi H, Gendelman H, Koening S, Folks T, Willey R, Rabson A, Martin M. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baraz L, Friedler A, Blumenzweig I, Nussinuv O, Chen N, Steinitz M, Gilon C, Kotler M. Human immunodeficiency virus type 1 Vif-derived peptides inhibit the viral protease and arrest virus production. FEBS Lett. 1998;441:419–426. doi: 10.1016/s0014-5793(98)01602-0. [DOI] [PubMed] [Google Scholar]
- 3.Blanc D, Patience C, Schulz T, Weiss R, Spire B. Transcomplementation of VIF− HIV-1 mutants in CEM cells suggests that VIF affects late steps of the viral life cycle. Virology. 1993;193:186–192. doi: 10.1006/viro.1993.1114. [DOI] [PubMed] [Google Scholar]
- 4.Borman A M, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 Vif− mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouyac M, Courcoul M, Bertoia G, Baudat Y, Gabuzda D, Blanc D, Chazal N, Boulanger P, Sire J, Vigne R, Spire B. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouyac M, Rey F, Nascimbeni M, Courcoul M, Sire J, Blanc D, Clavel F, Vigne R, Spire B. Phenotypically Vif− human immunodeficiency virus type 1 is produced by chronically infected restrictive cells. J Virol. 1997;71:2473–2477. doi: 10.1128/jvi.71.3.2473-2477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinskaya A G, Sharova N K. Unusual features of protein interaction in human immunodeficiency virus (HIV) virions. Arch Virol. 1990;110:287–293. doi: 10.1007/BF01311297. [DOI] [PubMed] [Google Scholar]
- 8.Camaur D, Trono D. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury I H, Chao W, Potash M J, Sova P, Gendelman H E, Volsky D J. Vif-negative human immunodeficiency virus type 1 persistently replicates in primary macrophages, producing attenuated progeny virus. J Virol. 1996;70:5336–5345. doi: 10.1128/jvi.70.8.5336-5345.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courcol M, Patience C, Rey F, Blanc D, Harmache A, Sire J, Vigne A, Spire B. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles that are restricted for the preretrotranscription step. J Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 14.Dettenhofer M, Cen S, Carlson B A, Kleiman L, Yu X F. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J Virol. 2000;74:8938–8945. doi: 10.1128/jvi.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dettenhofer M, Yu X F. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73:1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duke G M, Hoffman M A, Palmenberg A C. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol. 1992;66:1602–1609. doi: 10.1128/jvi.66.3.1602-1609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elroy-Stein O, Fuerst T R, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci USA. 1989;86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher A G, Ensoli B, Ivanoff L, Chamberlain M, Petteway S, Ratner L, Gallo R C, Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. [DOI] [PubMed] [Google Scholar]
- 19.Fouchier R A, Simon J H, Jaffe A B, Malim M H. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller S D, Wilk T, Gowen B E, Krausslich H G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 22.Gabuzda D H, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine W A, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retrovir. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 24.Goncalves J, Jallepalli P, Gabuzda D H. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J Virol. 1994;68:704–712. doi: 10.1128/jvi.68.2.704-712.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goncalves J, Korin Y, Zack J, Gabuzda D. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:8701–8709. doi: 10.1128/jvi.70.12.8701-8709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goncalves J, Shi B, Yang X, Gabuzda D. Biological activity of human immunodeficiency virus type 1 Vif requires membrane targeting by C-terminal basic domains. J Virol. 1995;69:7196–7204. doi: 10.1128/jvi.69.11.7196-7204.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green N, Alexander H, Olson A, Alexander S, Shinnick T M, Sutcliffe J G, Lerner R A. Immunogenic structure of the influenza virus hemagglutinin. Cell. 1982;28:477–487. doi: 10.1016/0092-8674(82)90202-1. [DOI] [PubMed] [Google Scholar]
- 28.Guy B, Geist M, Dott K, Spehner D, Kieny M P, Lecocq J P. A specific inhibitor of cysteine proteases impairs a Vif-dependent modification of human immunodeficiency virus type 1 Env protein. J Virol. 1991;65:1325–1331. doi: 10.1128/jvi.65.3.1325-1331.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harmache A, Bouyac M, Audoly G, Hieblot C, Peveri R, Vigne R, Suzan M. The vif gene is essential for efficient replication of caprine arthritis and encephalitis virus in goat synovial membrane cells and affects the late steps of the virus replication cycle. J Virol. 1995;69:3247–3257. doi: 10.1128/jvi.69.6.3247-3257.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoglund S, Ohagen A, Lawrence K, Gabuzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- 31.Huvent I, Hong S S, Fournier C, Gay B, Tournier J, Carriere C, Courcoul M, Vigne R, Spire B, Boulanger P. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J Gen Virol. 1998;79:1069–1081. doi: 10.1099/0022-1317-79-5-1069. [DOI] [PubMed] [Google Scholar]
- 32.Karczewski M K, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J S, Raines R T. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 1993;2:348–356. doi: 10.1002/pro.5560020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotler M, Simm M, Zhao Y S, Sova P, Chao W, Ohnona S F, Roller R, Krachmarov C, Potash M J, Volsky D J. Human immunodeficiency virus type 1 (HIV-1) protein Vif inhibits the activity of HIV-1 protease in bacteria and in vitro. J Virol. 1997;71:5774–5781. doi: 10.1128/jvi.71.8.5774-5781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krausslich H G, Facke M, Heuser A M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Wu X, Newman M, Shaw G M, Hahn B H, Kappes J C. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J Virol. 1995;69:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma X Y, Sova P, Chao W, Volsky D J. Cysteine residues in the Vif protein of human immunodeficiency virus type 1 are essential for viral infectivity. J Virol. 1994;68:1714–1720. doi: 10.1128/jvi.68.3.1714-1720.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madani N, Kabat D. Cellular and viral specificities of human immunodeficiency virus type 1 vif protein. J Virol. 2000;74:5982–5987. doi: 10.1128/jvi.74.13.5982-5987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madani N, Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J Virol. 1998;72:10251–10255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahalingam S, Patel M, Collman R G, Srinivasan A. The carboxy-terminal domain is essential for stability and not for virion incorporation of HIV-1 Vpr into virus particles. Virology. 1995;214:647–652. doi: 10.1006/viro.1995.0079. [DOI] [PubMed] [Google Scholar]
- 42.Muller B, Tessmer U, Schubert U, Krausslich H G. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J Virol. 2000;74:9727–9731. doi: 10.1128/jvi.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohagen A, Gabuzda D. Role of Vif in stability of the human immunodeficiency virus type 1 core. J Virol. 2000;74:11055–11066. doi: 10.1128/jvi.74.23.11055-11066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott D E, Coren L V, Johnson D G, Sowder II R C, Arthur L O, Henderson L E. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res Hum Retrovir. 1995;11:1003–1006. doi: 10.1089/aid.1995.11.1003. [DOI] [PubMed] [Google Scholar]
- 45.Pancio H A, Ratner L. Human immunodeficiency virus type 2 Vpx-Gag interaction. J Virol. 1998;72:5271–5275. doi: 10.1128/jvi.72.6.5271-5275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J, Morrow C D. Mutations in the protease gene of human immunodeficiency virus type 1 affect release and stability of virus particles. Virology. 1993;194:843–850. doi: 10.1006/viro.1993.1328. [DOI] [PubMed] [Google Scholar]
- 47.Pepinsky R B. Localization of lipid-protein and protein-protein interactions within the murine retrovirus gag precursor by a novel peptide-mapping technique. J Biol Chem. 1983;258:11229–11235. [PubMed] [Google Scholar]
- 48.Phillips T R, Talbott R L, Lamont C, Muir S, Lovelace K, Elder J H. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64:4605–4613. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potash M J, Bentsman G, Muir T, Krachmarov C, Sova P, Volsky D J. Peptide inhibitors of HIV-1 protease and viral infection of peripheral blood lymphocytes based on HIV-1 Vif. Proc Natl Acad Sci USA. 1998;95:13865–13868. doi: 10.1073/pnas.95.23.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reicin A S, Ohagen A, Yin L, Hoglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts N A, Martin J A, Kinchington D, Broadhurst A V, Craig J C, Duncan I B, Galpin S A, Handa B K, Kay J, Krohn A, et al. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–360. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 52.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. Biotechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 53.Sakai H, Shibata R, Sakuragi J, Sakuragi S, Kawamura M, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J Virol. 1993;67:1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakai K, Ma X Y, Gordienko I, Volsky D J. Recombinational analysis of a natural noncytopathic human immunodeficiency virus type 1 (HIV-1) isolate: role of the vif gene in HIV-1 infection kinetics and cytopathicity. J Virol. 1991;65:5765–5773. doi: 10.1128/jvi.65.11.5765-5773.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shacklett B L, Luciw P A. Analysis of the vif gene of feline immunodeficiency virus. Virology. 1994;204:860–867. doi: 10.1006/viro.1994.1609. [DOI] [PubMed] [Google Scholar]
- 56.Simm M, Shahabuddin M, Chao W, Allan J S, Volsky D J. Aberrant Gag protein composition of a human immunodeficiency virus type 1 vif mutant produced in primary lymphocytes. J Virol. 1995;69:4582–4586. doi: 10.1128/jvi.69.7.4582-4586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon J H, Fouchier R A, Southerling T E, Guerra C B, Grant C K, Malim M H. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J Virol. 1997;71:5259–5267. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–1400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 59.Simon J H, Malim M H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon J H, Miller D L, Fouchier R A, Malim M H. Virion incorporation of human immunodeficiency virus type-1 Vif is determined by intracellular expression level and may not be necessary for function. Virology. 1998;248:182–187. doi: 10.1006/viro.1998.9296. [DOI] [PubMed] [Google Scholar]
- 61.Simon J H, Southerling T E, Peterson J C, Meyer B E, Malim M H. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J Virol. 1995;69:4166–4172. doi: 10.1128/jvi.69.7.4166-4172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh S P, Lai D, Cartas M, Serio D, Murali R, Kalyanaraman V S, Srinivasan A. Epitope-tagging approach to determine the stoichiometry of the structural and nonstructural proteins in the virus particles: amount of Vpr in relation to Gag in HIV-1. Virology. 2000;268:364–371. doi: 10.1006/viro.2000.0191. [DOI] [PubMed] [Google Scholar]
- 63.Sodroski J, Goh W C, Rosen C, Tartar A, Portetelle D, Burny A, Haseltine W. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science. 1986;231:1549–1553. doi: 10.1126/science.3006244. [DOI] [PubMed] [Google Scholar]
- 64.Sova P, Volsky D J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin M A. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 67.Stromberg K, Hurley N E, Davis N L, Rueckert R R, Fleissner E. Structural studies of avian myeloblastosis virus: comparison of polypeptides in virion and core component by dodecyl sulfate-polyacrylamide gel electrophoresis. J Virol. 1974;13:513–528. doi: 10.1128/jvi.13.2.513-528.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomonaga K, Norimine J, Shin Y S, Fukasawa M, Miyazawa T, Adachi A, Toyosaki T, Kawaguchi Y, Kai C, Mikami T. Identification of a feline immunodeficiency virus gene which is essential for cell-free virus infectivity. J Virol. 1992;66:6181–6185. doi: 10.1128/jvi.66.10.6181-6185.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Schwedler U, Song J, Alken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyma D J, Kotov A, Aiken C. Evidence for a stable interaction of gp41 with Pr55(Gag) in immature human immunodeficiency virus type 1 particles. J Virol. 2000;74:9381–9387. doi: 10.1128/jvi.74.20.9381-9387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshinaka Y, Luftig R B. Murine leukemia virus morphogenesis: cleavage of P70 in vitro can be accompanied by a shift from a concentrically coiled internal strand (“immature”) to a collapsed (“mature”) form of the virus core. Proc Natl Acad Sci USA. 1977;74:3446–3450. doi: 10.1073/pnas.74.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang H, Pomerantz R J, Dornadula G, Sun Y. Human immunodeficiency virus type 1 vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J Virol. 2000;74:8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]