Abstract
Two classes of artificially constructed defective RNAs (dRNAs) of Tobacco mosaic virus (TMV) were examined in planta with helper viruses that expressed one (183 kDa) or both (126 and 183 kDa) of the replicase-associated proteins. The first class of artificially constructed dRNAs had the helicase and polymerase (POL) domains deleted; the second had an intact 126-kDa protein open reading frame (ORF). Despite extremely high levels of replication in protoplasts, the first class of dRNAs did not accumulate in plants. The dRNAs with an intact 126-kDa protein ORF were replicated at moderate levels in protoplasts and in planta when supported by a TMV mutant that expressed the 183-kDa protein but not the 126-kDa protein (183F). These dRNAs were not supported by helper viruses expressing both replicase-associated proteins. De novo dRNAs were generated in plants infected by 183F but not in plants infected with virus with the wild-type replicase. These novel dRNAs each contained a new stop codon near the location of the wild-type stop codon for the 126-kDa protein and had most of the POL domain deleted. The fact that only dRNAs that contained a complete 126-kDa protein ORF moved systemically suggests that expression of a functional 126-kDa protein or the presence of certain sequences and/or structures within this ORF is required for movement of dRNAs. At least two factors may contribute to the lack of naturally occurring dRNAs in association with wild-type TMV infections: an inability of TMV to support dRNAs that can move in plants and the inability of dRNAs that can be replicated by TMV to move in plants.
Defective RNAs (dRNAs) are RNA molecules that arise through deletion of internal sequences from the genome of a virus. Since dRNAs generally are incapable of autonomous replication, they depend on trans-acting factors from a helper virus for replication (12, 15, 34, 40). Some dRNAs interfere with replication of the helper virus and thus are referred to as defective interfering (DI) viruses. dRNAs are an intrinsic and common feature among animal RNA viruses (reviewed in references 19, 23, and 27). However, the majority of plant virus genera have not been associated with dRNAs in nature. There is no information concerning the principles of propagation of dRNAs among different viral groups. It has been suggested that certain viral mechanisms that act to suppress the formation and/or accumulation of defective genomes have likely evolved (40). High-multiplicity transfers foster accumulation of dRNAs. Thus, low-multiplicity infections of plant cells resulting from cell-to-cell and long-distance movement of the virus throughout the plants might prevent the survival of dRNAs in populations (3, 40). However, there might also be differences in replication strategies of different dRNAs that affect their ability to compete and survive in populations.
Although the Tobamovirus genus is one of the most examined groups of viruses, dRNAs have never been reported in a natural tobamovirus isolate. However, a naturally occurring satellite virus is common in natural populations of tobacco mild green mosaic virus strains in Nicotiana glauca, thus demonstrating that tobamovirus replicases have evolved the ability to support replication of RNA molecules in trans (37). Tobacco mosaic virus (TMV), the type member of the genus, expresses its genetic information from a positive-sense monopartite RNA molecule 6,395 nucleotides (nt) in length (Fig. 1A). The 126-kDa protein is translated from the first open reading frame (ORF) within the genomic RNA, and the 183-kDa protein is expressed via occasional readthrough of an amber stop codon (33). The 126-kDa protein is produced at levels 10 to 20 times higher than those at which the 183-kDa protein is produced. However, mutation of the stop codon to only allow the translation of the 183-kDa protein did not increase accumulation of the 183-kDa protein (18, 21). Both proteins associate at a 1:1 ratio to form a viral replication complex that retains activity in vitro, suggesting that a heterodimer is the active form (39). Nonetheless, the 183-kDa protein alone is sufficient for replication in protoplasts (18, 21). The movement protein (MP) and coat protein (CP) are expressed via individual 3′-coterminal subgenomic RNAs from the two 3′-proximal ORFs. TMV MP and CP are dispensable for replication in protoplasts (18, 21) but are necessary for cell-to-cell and long-distance movement in tobacco (6, 41).
FIG. 1.
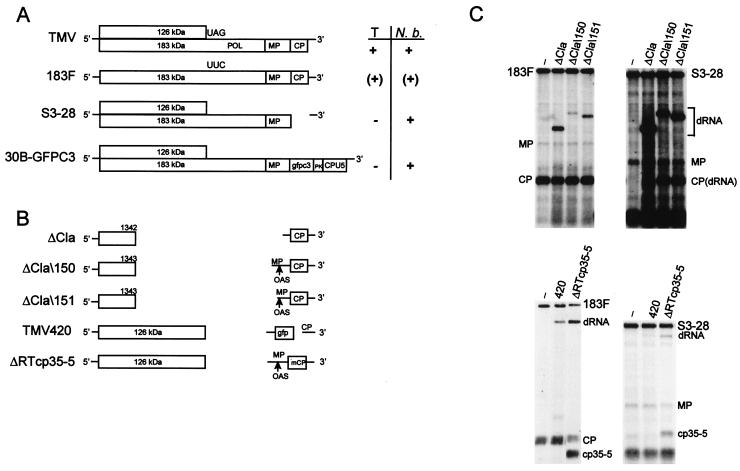
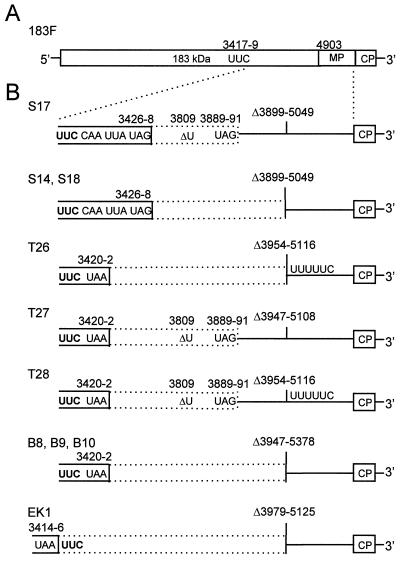
(A) Schematic diagram of the genomic organization of TMV-based helper viruses. Locations of POL domain in TMV and stop codon replacement in 183F are indicated. The movement phenotype of helper virus (right) is indicated as follows: +, wt; (+), delayed; −, no long-distance movement. T, tobacco; N. b., N. benthamiana; PK, TMV pseudoknots; CPU5, subgenomic promoter and CP from TMGMV strain U5. (B) Artificially constructed dRNAs of TMV. Boxes, ORFs. Translation products encoded by the respective ORFs are indicated. Lines, nontranslatable sequences; gaps, deleted sequences; mCP, mutated coat protein (cp35–5); gfp, GFP (cycle 3); arrows, retention of complete OAS. (C) Replication of artificially constructed dRNAs in protoplasts. Tobacco suspension cell protoplasts (2 × 106) were transfected with helper virus alone or in combination with a dRNA. Total RNA extracted at 22 h p.i. was analyzed by Northern blot hybridization with a riboprobe complementary to the 3′ untranslated region (20). Positions of helper virus (183F or S3–28) and dRNA genomic RNAs and MP and CP and mutated CP (cp35–5) subgenomic mRNAs are indicated. Lane −, helper virus without dRNA.
Tobamovirus-based dRNAs that can be replicated efficiently have been created artificially. However, sequences removed or retained greatly affected their ability to be replicated. Removal of the polymerase (POL) domain from the dRNA genome appears to be a minimum requirement to obtain viable dRNAs from TMV and tomato mosaic virus (20, 25, 26). When sequences in addition to the POL domain were removed, the levels of accumulation of artificial dRNAs varied considerably (20, 26). dRNAs containing the entire 126-kDa protein ORF sequences appeared to require the functional 126-kDa protein in cis for replication (21). Wild-type (wt) TMV and replication-competent TMV mutants can support the artificially constructed dRNAs without notable interference with replication of the helper virus in protoplasts (20).
We investigated the performance of two classes of artificially constructed TMV-based dRNAs in planta. dRNAs that accumulated to high levels in protoplasts in combination with wt TMV-like viruses which produce both replicase-associated proteins lacked the ability to move in plants. dRNAs that were able to move in plants were not efficiently replicated by the wt virus. A mutant helper virus deficient in production of the 126-kDa protein replicated and moved the class of dRNAs that encoded a functional 126-kDa protein. Most importantly, the full-length TMV mutant deficient in the 126-kDa protein also generated dRNAs de novo in plants. This is the first report of dRNAs arising de novo within the genus Tobamovirus. These data provide an explanation for the absence of dRNAs associated with tobamovirus infections, namely, that in wild populations dRNAs supported by the helper virus cannot move in plants and that dRNAs that can move are not efficiently replicated by the wt virus.
MATERIALS AND METHODS
Plasmid DNA of helper viruses and dRNAs.
All constructs were built using standard recombinant DNA techniques (2) and are derivatives of an infectious wt TMV cDNA clone (5). pTMV004, pT7S3–28, pTMV183F, and p30B-GFPC3 have been described (20, 32).
pTMV150 and pTMV151 are full-length TMV clones that contain a SalI site at positions 5017 and 5179, respectively (S. N. Chapman, unpublished data). To create pTMVΔCla\150 and pTMVΔCla\151, pTMV150 and pTMV151 were digested with SalI, end filled with T4 DNA polymerase, and subsequently digested with KpnI and the resulting 1.4- and 1.2-kb fragments, respectively, were ligated into pTMVΔCla (20) digested with ClaI, end filled with T4 DNA polymerase, and digested with KpnI.
The construction of pTMV420 is described elsewhere (D. J. Lewandowski et al., unpublished data). Briefly TMV420 contains the 126-kDa protein ORF followed by the jellyfish green fluorescent protein (GFP) ORF under the control of the TMV CP subgenomic RNA promoter. pTMVΔRTcp35–5 was constructed by ligating the NcoI/BsiWI fragment from pTMVcp35–5 (6) into similarly digested pTMVΔRT (20).
In vitro transcription and inoculation.
In vitro transcripts of KpnI- or PstI-linearized plasmid DNA were electroporated into Nicotiana tabacum cv. Xanthi suspension cells as previously described (20).
Plant inoculation and harvest of infected tissue.
Nicotiana benthamiana and tobacco plants with at least three fully expanded leaves were coinoculated with a mixture of transcripts containing one-half (0.5×) of a 25-μl in vitro transcription reaction mixture (20) with a single helper virus and a single dRNA clone. Experiments were repeated using the 0.5× reaction mixture of transcripts. In additional experiments, 1.5× TMV, S3–28, and 183F transcription reactions alone and in combination with 3× reaction of ΔCla, ΔCla\150, ΔCla\151, or ΔRTcp35–5 were used to inoculate N. benthamiana plants. Inoculation buffer (5) was mixed with in vitro transcripts at 30 to 50% of the total volume immediately before inoculation. For sequential inoculations, plants that had developed systemic symptoms following inoculation with either S3–28, 30B-GFPC3, or 183F in vitro transcripts were subsequently inoculated with a 0.5× transcription reaction of a dRNA.
Upper leaves were powdered in liquid nitrogen and ground in inoculation buffer (100 mM glycine, 60 mM K2HPO4, 1% Celite, pH 9.2) for passage of dRNAs between plants. Tissue from inoculated and noninoculated upper leaves was harvested at 7 days and at 1, 2, or 3 months postinoculation (p.i.).
Analysis of viral RNA.
Extraction and analysis of total RNA from protoplasts were done as described previously (20). Inoculated leaf halves (N. benthamiana) or quarters (tobacco) or upper not fully expanded leaf halves were powdered in liquid nitrogen and ground in 900 μl of lysis buffer (22). An aliquot of 750 μl of crude extract was extracted twice with 750 μl of phenol-chloroform-isoamyl alcohol (25:24:1) and ethanol precipitated at −20°C. Total RNA was resuspended in 500 μl of diethyl pyrocarbonate-treated H2O and analyzed by Northern blot hybridization using TMV- (20) or GFP-specific probes (32).
Virion RNA was obtained following the protocol of Gooding and Hebert (10) in a scaled-down version. Double-stranded (ds) RNA was isolated from upper leaves of Nicotiana sylvestris by phenol extraction and CF-11 column chromatography (8).
Cloning of de novo-generated dRNAs.
Reverse transcription (RT) and PCR amplification of dRNAs were carried out with the Titan one-tube RT-PCR system (Boehringer GmbH, Mannheim, Germany) essentially following instructions given by the manufacturer. Template RNA was denatured for 10 min at 70°C in the presence of the reverse primer and chilled on ice for 2 min prior to addition of the RT-PCR cocktail. T26, T27, and T28 were amplified from virion RNA isolated from N. tabacum cv. Xanthi; S14, S17, and S18 were amplified from ds RNA isolated from N. sylvestris using forward primer T2 (TMV nt 3157 to 3178), 5′-CAAGAGGGTATTCAGATGTTCA-OH, and reverse primer M28 (complementary to TMV nt 6381 to 6395), 5′-TTCGACCTCGGTACCTGGGCCCCTACCGGG-OH, which had a 5′ KpnI restriction site (boldface) followed by nine nonviral nucleotides (italics).
RT-PCR products B8, B9, B10, and EK1 were amplified from total RNA extracted from N. benthamiana using forward primer T2 and reverse primer T107 (complementary to TMV nt 5440 to 5469), 5′-AAGTTCCATGGGCCCTCCGTCTCTCACGTT-OH.
RT-PCR fragments were separated on 1% agarose gels, gel purified, and ligated into pCR2.1 (Invitrogen) or pGEM-T-Easy (Promega). Because restriction mapping identified a KpnI site near the readthrough stop codon (nt 3417 to 3419), cDNAs were digested with BamHI (nt 3332) and BsiWI (nt 6245) and the fragments were substituted into similarly digested pTMV183F, resulting in plasmids that could be linearized with PstI. The DNA sequences for the exchanged fragments (T26, T27, T28, S14, S17, and S18) and for the amplified RT-PCR products cloned into pGEM-T-Easy (B8, B9, B10, and EK1) were obtained.
RESULTS
dRNAs replicated and moved in plants only when helped by a 126-kDa protein-deficient virus.
To investigate the requirements for dRNA accumulation in planta, we tested the two classes of TMV-based dRNAs in combination with different helper viruses. The first class of dRNAs is represented by ΔCla, containing a deletion of the helicase (HEL) and POL domains of the 183-kDa protein ORF and most of the MP ORF (nt 1343 to 5664) (Fig. 1B). ΔCla is replicated efficiently by wt TMV, usually exceeding the level produced by the helper virus (20). dRNAs ΔCla\150 and ΔCla\151 (deletions of nt 1344 to 5017 and 1344 to 5179, respectively) (Fig. 1B) were created to contain the complete origin of assembly (OAS) to allow encapsidation (42).
The second class of dRNAs contained the complete ORF for expression of a functional 126-kDa protein. TMV420 lacks the entire POL domain and most of the MP ORF, including a substantial portion of the OAS. To enable TMV420 to express a foreign gene, the jellyfish GFP ORF was inserted behind the CP subgenomic promoter followed by the 3′-proximal 100 nt of the CP ORF (D. J. Lewandowski et al., unpublished data) (Fig. 1B). ΔRTcp35–5 was created by deletion of the POL domain from mutant TMVcp35–5, whose truncated coat protein provides a visual yellow phenotype in infected plants (6) (Fig. 1B).
The helper viruses examined were wt TMV and TMV derivatives that expressed both 126- and 183-kDa replicase-associated proteins or a full-length TMV mutant (183F) that expressed the 183-kDa but not the 126-kDa protein. TMV derivative S3–28 has a precise deletion of the CP ORF (6) (Fig. 1A). 30B-GFPC3 is a TMV and tobacco mild green mosaic virus strain U5 chimeric vector that expresses GFP in infected cells (32) (Fig. 1A). S3–28 and 30B-GFPC3 were restricted to the inoculated leaf in tobacco but moved systemically in N. benthamiana, a host killed by TMV (Fig. 1A). 183F was created through replacement of the amber stop codon (UAG) with a Phe (UUC) codon (20) (Fig. 1A). Systemic movement of 183F was delayed for ≈4 days or up to 2 weeks compared to that of wt TMV in N. benthamiana and tobacco, respectively.
In protoplasts, the ΔCla-like dRNAs were replicated to high levels by TMV, S3–28, and 30B-GFPC3 but were replicated only to moderate levels by 183F (Fig. 1C and data not shown). ΔRTcp35–5 and TMV420 were replicated to moderate levels by 183F but to low (ΔRTcp35–5) or barely detectable (TMV420) levels by TMV, S3–28, and 30B-GFPC3 (Fig. 1C and data not shown).
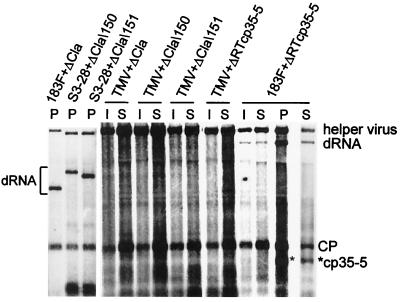
N. benthamiana and tobacco plants were coinoculated with combinations of dRNAs and TMV, S3–28, or 183F helper viruses. No detectable ΔCla, ΔCla\150, or ΔCla\151 dRNA accumulated in inoculated leaves in combination with any of these helper viruses, based on Northern blot hybridization (Fig. 2 and data not shown). Although TMV420 and ΔRTcp35–5 did not accumulate to detectable levels when coinoculated with either TMV or S3–28, 183F supported TMV420 and ΔRTcp35–5 in inoculated leaves (Fig. 2 and data not shown). 183F plus TMV420 expressed GFP in isolated infection foci as seen under long-wave UV illumination in inoculated leaves of both N. benthamiana and tobacco (data not shown). ΔRTcp35–5 and its CP subgenomic RNA (cp35–5) were identified in RNAs from inoculated leaves by Northern blot hybridization (Fig. 2). ΔCla, ΔCla\150, and ΔCla\151 RNAs did not accumulate in upper leaves of N. benthamiana or tobacco coinoculated with any of the helper viruses (Fig. 2 and data not shown). Although TMV420 accumulated and moved cell to cell when supported by 183F, it did not accumulate in noninoculated leaves. ΔRTcp35–5 spread efficiently into upper leaves of N. benthamiana when supported by 183F but not when in combination with wt TMV, S3–28, or 30B-GFPC3 (Fig. 2 and data not shown).
FIG. 2.
Replication and movement of artificially constructed dRNAs in plants. Shown is Northern blot hybridization of total RNA extracted from tobacco protoplasts (P) at 22 h p.i. and from inoculated (I) and systemically infected (S) leaves of N. benthamiana plants 7 days p.i. Protoplasts and plants were inoculated with in vitro transcripts of the helper virus-dRNA combinations indicated. Positions of CP and mutated CP (*cp35–5) subgenomic mRNAs are indicated. Total RNA was analyzed as described for Fig. 1C.
Attempts were made to coinfect a higher number of cells by mechanically inoculating ΔCla or ΔRTcp35–5 in vitro transcripts onto N. benthamiana plants already systemically infected with S3–28, 30B-GFPC3, or 183F. Analysis of total RNA extracted from inoculated leaves at 4 or 17 days p.i. showed that neither ΔCla nor ΔRTcp35–5 was amplified by any of the three helper viruses nor was TMV420 replicated by 183F under this inoculation regimen (data not shown).
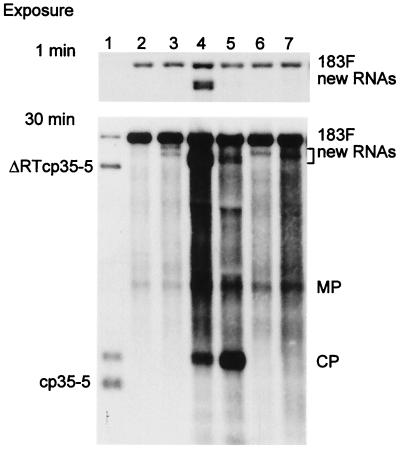
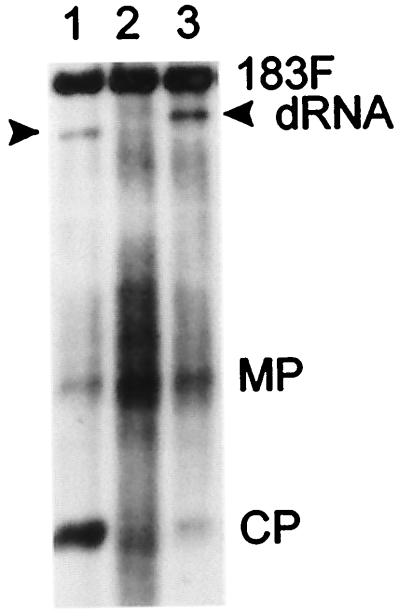
Interestingly, some 183F-infected N. benthamiana and tobacco plants that were subsequently inoculated with a dRNA contained one or more additional hybridization signals that were ≈1.0 to 1.5 kb smaller than that for 183F but larger than those for the input dRNAs (Fig. 3). These additional RNAs were found regardless of whether the dRNA inoculum was ΔCla, TMV420, or ΔRTcp35–5. They were larger than ΔCla, did not hybridize with a GFP-specific probe (data not shown), and did not produce the smaller-than-full-length CP subgenomic mRNA associated with ΔRTcp35–5. The abundances of these RNAs differed and sometimes exceeded the level of accumulation of helper virus 183F (Fig. 3, lane 4). Additional hybridization signals were not detected in Northern blots of RNA extracted from plants infected with S3–28 or 30B-GFPC3 plus a dRNA. Prolonged incubation periods of at least 2 months did not result in formation of additional smaller-than-full-length genomic RNAs in plants infected with helper viruses expressing both the 126- and 183-kDa proteins (data not shown).
FIG. 3.
Accumulation of additional new RNAs in upper leaves of 183F-infected tobacco. Lane 1, 183F plus ΔRTcp35–5 protoplast control; lanes 2 to 7, tobacco plants containing no (lane 2), one (lanes 6 and 7), two (lanes 3 and 5), or more than two (lane 4) additional new RNAs. Total RNA was analyzed by Northern blot hybridization as described for Fig. 1C. Films were exposed for 1 or 30 min. Positions of MP, CP, and mutated CP (cp35–5) subgenomic mRNAs are indicated.
Analysis of novel dRNAs from systemically infected leaves.
Upper leaves from two tobacco plants harvested 3 to 4 weeks p.i. were independently homogenized and used to inoculate several different Nicotiana species. Total RNA was extracted from inoculated and upper noninoculated leaves of the various Nicotiana species and analyzed by Northern blot hybridization. Some of these plants contained the additional new RNAs (data not shown). The additional RNAs were cloned from several of these newly infected plants and from one of the original tobacco plants used for the inoculum.
dsRNA containing a new RNA isolated from an N. sylvestris plant 10 weeks after virus transfer (Fig. 4) was used in RT-PCRs to amplify the 3′ portion of the new RNA (nt 3157 to 6395). The major products obtained were ≈1 kb smaller than the 3.2-kb fragments obtained from control RT-PCRs with 183F or wt TMV in vitro transcripts (data not shown). PCR products were cloned, and the approximate location and length of an internal deletion were determined through restriction pattern analysis. cDNA clones were digested with BamHI (nt 3332) and BsiWI (nt 6245), and the fragments were substituted into similarly digested pTMV183F to reconstruct dRNAs with wt TMV termini (designated S14, S17, and S18). The entire substituted region was sequenced for each clone.
FIG. 4.
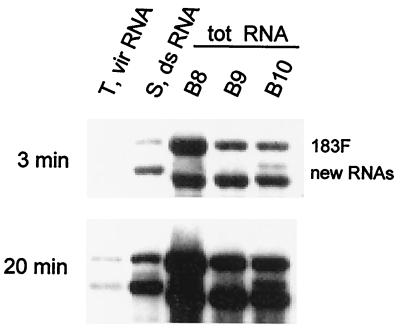
Northern blot of RNAs used to clone the new RNAs present in systemically infected plants. Shown are virion (vir) RNA obtained from tobacco (T), ds RNA from N. sylvestris (S), and total (tot) RNA extracted from three different N. benthamiana plants (B8, B9, and B10). Northern blot hybridization was carried out as described for Fig. 1C. Films were exposed for 3 or 20 min.
We also cloned the additional RNAs from one of the original tobacco plants that was used for virus transfer. This tobacco plant was different from the one used as the inoculum for the above-characterized N. sylvestris plant. We obtained virion RNA that contained two hybridization signals, showing that at least some new RNAs were encapsidated (Fig. 4). This virion RNA was used in RT-PCRs, and the same region as that described for N. sylvestris was cloned. cDNA clones were restriction mapped and substituted into pTMV183F (designated T26, T27, and T28), and the entire substituted region was sequenced.
Passage of the inoculum from the tobacco plant from which the T clones were derived also resulted in several N. benthamiana plants that contained new RNAs in the upper leaves (Fig. 4). Based on the sequences of the T and S cDNA clones and the similar sizes, we amplified a smaller region (≈0.9 kb) from N. benthamiana total RNA comprising nt 3157 to 5469. RT-PCR products from three independent plants were cloned and restriction mapped. One cDNA clone from each plant, designated B8, B9, or B10 to reflect the source of the RNA, was sequenced.
None of the sequences of the novel dRNAs appeared to be derived from the artificially constructed dRNAs. Each cDNA clone contained a single continuous deletion that extended from within the POL domain to within the MP ORF (Fig. 5B). S14, S17, and S18 had a deletion of 1,151 nt (Δ[3899 to 5049]). T27 had 1,162 nt deleted (Δ[3947 to 5108]). T26 and T28 had 1,163 nt (Δ[3954 to 5116]) deleted but contained an insertion of UUUUUC at the junction site (Fig. 5B). The helper virus genome contained UUUUUC sequences at nt 4141 to 4146 and 6265 to 6270. B8, B9, and B10 had a deletion of 1,432 nt (Δ[3947 to 5378]).
FIG. 5.
Schematic diagram of the genomic organization of 183F (A) and novel dRNAs (B). (A) The position of the Phe codon (UUC) and the start of the MP ORF are indicated. (B) Open solid boxes, 126-kDa ORF; dotted boxes, translational readthrough of the 5′ POL ORF; lines, nontranslated sequences. The positions of the new stop codons and the additional U deletion and nucleotides flanking the internal deletions are indicated. The UUUUUC insertions in T26 and T28 are shown. Numbering corresponds to TMV (9).
All these novel dRNAs contained the Phe codon (UUC) in place of the amber stop codon at nt 3417 to 3419 characteristic of 183F. However, each of these dRNAs also acquired a new stop codon (Fig. 5B). T26, T27, T28, B8, B9, and B10 had a C-to-U substitution within the first codon (CAA) downstream of the Phe codon, creating an ochre stop codon (UAA). S14, S17, and S18 had the third codon downstream of UUC changed from CAG into an amber stop codon (UAG). The new stop codons were in frame, thereby allowing the dRNAs to encode a 126-kDa protein with a slightly modified C terminus.
Each of the novel dRNAs had a potential partial-readthrough ORF containing partial POL domain sequences. The readthrough domain was terminated by either an out-of-frame junction with MP sequences (T26, B8, B9, B10, S14, and S18) or an amber stop codon at positions 3889 to 3891 created by a frameshift (T27, T28, and S17) (Fig. 5B).
Thus, the dRNAs found in upper leaves appeared to be derived de novo from the 183F helper virus instead of the dRNAs used to inoculate the lower leaves of the plants.
183F generates dRNAs de novo.
To examine whether 183F alone could produce dRNAs, N. benthamiana plants were inoculated with in vitro transcripts of 183F. Analysis of total RNA extracted at 8 days p.i. from noninoculated systemically infected leaves revealed that 1 of 12 plants contained a dRNA in addition to the full-length 183F. dRNA molecules were not detected in the corresponding inoculated leaves at 15 days p.i. dRNA-negative plants did not acquire dRNAs within 70 days p.i., whereas a dRNA was still present in the original dRNA-positive plant, but this dRNA was larger than the dRNA isolated at 8 days p.i. (Fig. 6). An RT-PCR product, which was cloned and sequenced, comprising the dRNA genome from nt 3157 to 5469 was obtained from total RNA extracted at 8 days p.i. The sequence was similar to those of the other novel dRNAs. This dRNA, referred to as EK1, retained the Phe codon of 183F but had a new ochre stop codon at nt 3414 to 3416. EK1 had 1,147 nt deleted (Δ[3979 to 5125]) and the 5′ and 3′ borders of the deletion joined out of frame (Fig. 5B). These data confirm that dRNAs can arise de novo from the 183F virus in planta.
FIG. 6.
De novo-generated dRNAs in N. benthamiana inoculated with in vitro transcripts of 183F. Total RNA was extracted from a noninoculated upper leaf at 8 days p.i. (lane 1), an inoculated leaf at 15 days p.i. (lane 2), and a different upper leaf at 70 days p.i. (lane 3). RNA was analyzed by Northern blot hybridization as described for Fig. 1C. Positions of MP and CP subgenomic mRNAs are indicated.
Comparison of de novo-formed dRNAs to the artificially constructed dRNAs in protoplasts.
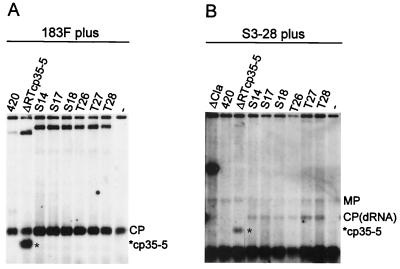
None of the reconstructed de novo dRNAs replicated when electroporated into protoplasts without a helper virus (data not shown). However, all were replicated in trans by 183F as the helper virus, thus confirming that they were replication-defective, helper-dependent RNAs. The levels of accumulation of the S dRNAs with the amber stop codon (UAG) were slightly higher than those of the T dRNAs with the ochre stop codon (UAA). S and T dRNAs accumulated to levels similar to those of ΔRTcp35–5 or to levels higher than those of TMV420 when replicated by 183F (Fig. 7A). With TMV or S3–28 as the helper virus, there was barely detectable replication of TMV420, whereas ΔRTcp35–5 and the de novo-generated dRNAs accumulated to extremely low levels. In comparison, ΔCla accumulated to high levels with TMV or S3–28 as the helper virus (Fig. 7B and data not shown).
FIG. 7.
Comparative replication levels of de novo-formed and artificially constructed dRNAs. Tobacco suspension cell protoplasts were transfected with 183F (A) or S3–28 (B) alone or in combination with in vitro transcripts of artificially constructed dRNA TMV420 or ΔRTcp35–5 or cDNA clones of de novo-formed dRNAs. Total RNA extracted at 22 h p.i. was analyzed as described for Fig. 1C. Positions of MP, CP, and mutated CP (*cp35–5) subgenomic mRNAs are indicated. Lanes −, helper virus without dRNAs.
DISCUSSION
Previously, we characterized two classes of artificially constructed dRNAs for their ability to be replicated in protoplasts. The smaller, ΔCla-like dRNAs with the HEL and POL domains deleted were replicated to very high levels with two types of helper viruses (20, 21). One type of helper virus produced both the 126- and 183-kDa replicase-associated proteins, whereas the other type of helper virus produced only the 183-kDa protein (21). The larger dRNAs with the intact 126-kDa protein ORF were replicated efficiently by only the 183-kDa protein-producing helper virus that was deficient in the 126-kDa protein (21). Despite their ability to be efficiently replicated in protoplasts, the ΔCla-like dRNAs did not accumulate detectably in plants with any of the helper viruses tested. Since these dRNAs were efficiently replicated in single cells, this suggests an impairment in the ability of the dRNA to move within the plant. Movement deficiencies caused by a nonfunctional MP can be complemented in trans as shown for replication-competent TMV molecules (17, 38) or for natural dRNAs of cucumo- and alfalmoviruses (11, 13). In contrast to the ΔCla-like dRNAs, artificially constructed and de novo-formed dRNAs containing the 126-kDa protein ORF moved from cell to cell. These data suggest that either the sequence difference between these two classes of dRNAs (HEL domain or adjacent sequences) or the cis-produced 126-kDa protein is involved in movement of dRNAs.
Evidence for a possible involvement of the replicase in movement comes from several lines of research, including complementation studies with hybrid viruses expressing a heterologous MP. Interestingly, MP transgenic plants could not complement movement deficiencies of some MP hybrid viruses, yet MP expressed in trans by a replicating virus could, suggesting that elements in addition to the MP are needed for movement (reviewed in reference 1). The masked strain of TMV (16) produces mild (attenuated) symptoms in tobacco that coincide with a reduction in phloem-dependent accumulation of the virus (24). The differences between the masked strain of TMV and TMV U1 were 8 amino acid changes in the region between the MT and HEL domains of the 126- and 183-kDa proteins (7). Deletion analysis of brome mosaic virus (BMV) 2a protein also pointed to a possible role for the replicase-associated proteins in movement. BMV RNA 2 mutants producing a truncated replicase-associated protein replicated efficiently in protoplasts yet did not accumulate in plants (36).
It is noteworthy that de novo-generated TMV dRNAs retained most of the MP sequences, although these were not expressed. Movement of artificially constructed dRNAs containing the 126-kDa protein ORF in N. benthamiana was enhanced by the inclusion of additional MP sequences (D. J. Lewandowski et al., unpublished data). Retention of most of the MP sequences was also a common feature of a heterogeneous population of replicons that arose through recombination between an artificially constructed TMV-based dRNA and its helper virus in tobacco plants (28).
Encapsidation is required for efficient long-distance movement of TMV in tobacco (6, 30, 31). Lack of encapsidation is also a factor preventing accumulation of broad bean mottle bromovirus dRNAs in certain hosts (29). De novo-formed TMV dRNAs expressed CP as did other movement-competent dRNAs (4, 11, 14, 28, 29). ΔRTcp35–5 and TMV420 moved from cell to cell, but, in contrast to ΔRTcp35–5, TMV420 did not move long distances. However, since these dRNAs differed considerably in design, it is unclear whether a reduced encapsidation efficiency of TMV420 due to an incomplete OAS caused the movement defect of TMV420. Other factors, such as expression of a foreign gene, might have contributed to the movement defect of TMV420.
De novo-produced dRNAs isolated from different Nicotiana species had strikingly similar deletions. They apparently arose from 183F through deletion of a continuous large segment from within the POL domain and the MP ORF. The Phe codon that replaced the amber stop codon in 183F was retained in each dRNA, but most remarkably all the characterized de novo-formed dRNAs acquired a new ochre or amber stop codon near the mutated readthrough position. The new stop codons terminated the ORF to allow the expression of a 126-kDa protein with a slightly modified C terminus. We showed previously that a functional 126-kDa ORF was required for replication of this class of dRNAs (21). In contrast to the artificially constructed dRNAs, de novo-produced dRNAs also retained a 5′ portion of the POL domain that encoded 156 to 186 amino acids. Because the new stop codons were not flanked by the 3′ consensus sequence of CAR YYA codons found to confer leakiness (35), no or only infrequent readthrough resulting in a truncated 183-kDa protein should occur. Earlier data suggested that POL sequences were inhibitory to in trans replication of dRNAs (20, 26). The ability of de novo-formed dRNAs with partial POL sequences to replicate is likely due to the expression of a functional 126-kDa protein, a characteristic that distinguishes de novo-formed dRNAs from dRNAs of earlier studies. The POL sequences are not required for replication, as evidenced by the fact that TMV420 and ΔRTcp35–5 were replicated (Fig. 1C and 7), but might make dRNAs more competitive under selection.
The Phe codon replacement of the amber stop codon in 183F was stable in protoplasts and appeared to be stable in inoculated leaves (21). Slow progression and mildness of symptoms suggested that the virus did not revert to wt TMV. However, the emergence of dRNAs with stop codons suggested that some 183F molecules accumulated an in-frame stop codon, which then provided the template for dRNA formation. The argument for acquiring a stop codon before dRNA formation is supported by several lines of evidence. dRNAs that contain the 126-kDa protein ORF appear to need a functional 126-kDa protein to be replicated (20, 21). Without the stop codon, dRNAs containing the type of POL/MP deletion that we observed would express a protein of intermediate length between 126 and 183 kDa. It is unknown whether this protein could function like the 126-kDa protein, but frameshift mutations at a similar position within the POL domain of the 183-kDa protein were inhibitory to dRNA replication (20). If generation of a stop codon is a prerequisite to formation of viable dRNAs encoding the 126-kDa protein, maintenance of these dRNAs would require a helper virus population containing some unreverted 183F, because wt-like viruses were found to be unsuitable helper viruses for this type of dRNA.
dRNAs have not been found in naturally occurring populations of tobamoviruses or even laboratory-maintained stocks. We showed that survival of dRNAs in plants depended on the ability of the helper virus to support the dRNA and on the ability of dRNAs to move in plants. TMV dRNAs replicated by wt TMV did not move in plants, while dRNAs that could move in plants were not replicated by wt TMV. Although dRNAs are produced during propagation of the 183F virus, after longer periods wt-like viruses containing a new stop codon and producing both the 126- and 183-kDa proteins overtake the population. These revertants would be expected no longer to replicate de novo-formed dRNAs, resulting in the depletion of these dRNAs from the population. For whatever reason, tobamoviruses apparently have evolved a system that excludes dRNAs that surely are produced by recombination. One type fails to be replicated, and the other type fails to be moved.
ACKNOWLEDGMENTS
We thank C. J. Robertson, C. R. Sine, and G. M. Danyluk for technical assistance; S. Rabindran and M. Mawassi for helpful discussions; and M. A. Ayllón, G. M. Danyluk, and J. Cook for critical reading of the manuscript.
This research was supported by the Florida Agricultural Experiment Station, a grant from USDA (98–35505-6752), and a “Schrödinger” postdoctoral fellowship (J1651-GEN, J1833-GEN) from the Austrian Science Fund to E. Knapp.
Footnotes
Approved for publication as Journal Series no. R-08043.
REFERENCES
- 1.Atabekov J G, Malyshenko S I, Morozov S Y, Taliansky M E, Solovyev A G, Agranovsky A A, Shapka N A. Identification and study of tobacco mosaic virus movement function by complementation tests. Phil Trans R Soc Lond Ser B. 1999;354:629–635. doi: 10.1098/rstb.1999.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. [Google Scholar]
- 3.Celix A, Rodriguez-Cerezo E, Garcia-Arenal F. New satellite RNAs but not DI RNAs are found in natural populations of tomato bushy stunt virus. Virology. 1997;239:277–284. doi: 10.1006/viro.1997.8864. [DOI] [PubMed] [Google Scholar]
- 4.Damayanti T A, Nagano H, Mise K, Furusawa I, Okuno T. Brome mosaic virus defective RNAs generated during infection of barley plants. J Gen Virol. 1999;80:2511–2518. doi: 10.1099/0022-1317-80-9-2511. [DOI] [PubMed] [Google Scholar]
- 5.Dawson W O, Beck D L, Knorr D A, Grantham G L. cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc Natl Acad Sci USA. 1986;83:5043–5047. doi: 10.1073/pnas.83.6.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson W O, Bubrik P, Grantham G L. Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement, and symptomatology. Phytopathology. 1988;78:783–789. [Google Scholar]
- 7.Derrick P M, Carter S A, Nelson R S. Mutation of the tobacco mosaic tobamovirus 126- and 183-kDa proteins: effects on phloem-dependent virus accumulation and synthesis of viral proteins. Mol Plant-Microbe Interact. 1997;10:589–596. [Google Scholar]
- 8.Dodds J A, Bar-Joseph M. Double-stranded RNA from plants infected with closteroviruses. Phytopathology. 1983;73:419–423. [Google Scholar]
- 9.Goelet P, Lomonossoff G P, Butler G J P, Akam M E, Gait M J, Karn J. Nucleotide sequence of tobacco mosaic virus RNA. Proc Natl Acad Sci USA. 1982;79:5818–5822. doi: 10.1073/pnas.79.19.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gooding G V, Hebert T T. A simple technique for purification of tobacco mosaic virus in large quantities. Phytopathology. 1967;57:1285. [PubMed] [Google Scholar]
- 11.Graves M V, Roossinck M J. Characterization of defective RNAs derived from RNA 3 of the Fny strain of cucumber mosaic cucumovirus. J Virol. 1995;69:4746–4751. doi: 10.1128/jvi.69.8.4746-4751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves M V, Pogany J, Romero J. Defective-interfering RNAs and defective viruses associated with multipartite RNA viruses of plants. Semin Virol. 1996;7:399–408. [Google Scholar]
- 13.Hajimorad M R, Kurath G, Randles J W, Francki R I. Change in phenotype and encapsidated RNA segments of an isolate of alfalfa mosaic virus: an influence of host passage. J Gen Virol. 1991;72:2885–2893. doi: 10.1099/0022-1317-72-12-2885. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez C, Carette J E, Brown J F D, Bol J F. Serial passage of tobacco rattle virus under different selection conditions results in deletion of structural and nonstructural genes in RNA 2. J Virol. 1996;70:4933–4940. doi: 10.1128/jvi.70.8.4933-4940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holland J. Defective viral genomes. In: Fields B N, Knipe D M, editors. Fields virology. 2nd ed. Vol. 1. New York, N.Y: Raven Press; 1990. pp. 151–165. [Google Scholar]
- 16.Holmes F O. A masked strain of tobacco mosaic virus. Phytopathology. 1934;24:845–873. [Google Scholar]
- 17.Holt C A, Beachy R N. In vivo complementation of infectious transcripts from mutant tobacco mosaic virus cDNAs in transgenic plants. Virology. 1991;181:109–117. doi: 10.1016/0042-6822(91)90475-q. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa M, Meshi T, Ohno T, Okada Y. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J Virol. 1991;65:861–868. doi: 10.1128/jvi.65.2.861-868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazzarini R A, Keene J D, Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981;26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 20.Lewandowski D J, Dawson W O. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology. 1998;251:427–437. doi: 10.1006/viro.1998.9420. [DOI] [PubMed] [Google Scholar]
- 21.Lewandowski D J, Dawson W O. Functions of the 126-kDa and 183-kDa proteins of tobacco mosaic virus. Virology. 2000;271:90–98. doi: 10.1006/viro.2000.0313. [DOI] [PubMed] [Google Scholar]
- 22.Navas-Castillo J, Albiach-Marti M R, Gowda S, Hilf M E, Garnsey S M, Dawson W O. Kinetics of accumulation of citrus tristeza virus RNAs. Virology. 1997;228:92–97. doi: 10.1006/viro.1996.8369. [DOI] [PubMed] [Google Scholar]
- 23.Nayak D P, Chambers T M, Akkina R K. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol. 1985;114:103–151. doi: 10.1007/978-3-642-70227-3_3. [DOI] [PubMed] [Google Scholar]
- 24.Nelson R S, Li G, Hodgson R A J, Beachy R N, Shintaku M H. Impeded phloem-dependent accumulation of the masked strain of tobacco mosaic virus. Mol Plant-Microbe Interact. 1993;6:45–54. [Google Scholar]
- 25.Ogawa T, Watanabe Y, Meshi T, Okada Y. Trans complementation of virus-encoded replicase components of tobacco mosaic virus. Virology. 1991;185:580–584. doi: 10.1016/0042-6822(91)90528-j. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa T, Watanabe Y, Okada Y. Cis-acting elements for in trans complementation of replication-defective mutants of tobacco mosaic virus. Virology. 1992;191:454–458. doi: 10.1016/0042-6822(92)90209-8. [DOI] [PubMed] [Google Scholar]
- 27.Perrault J. Origin and replication of defective-interfering particles. Curr Top Microbiol Immunol. 1981;93:151–207. doi: 10.1007/978-3-642-68123-3_7. [DOI] [PubMed] [Google Scholar]
- 28.Raffo A J, Dawson W O. Construction of tobacco mosaic virus subgenomic replicons that are replicated and spread systemically in tobacco plants. Virology. 1991;184:277–289. doi: 10.1016/0042-6822(91)90844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero J, Huang Q, Pogany J, Bujarski J J. Characterization of defective-interfering RNA components that increase symptom severity of broad bean mottle virus infections. Virology. 1993;194:576–584. doi: 10.1006/viro.1993.1297. [DOI] [PubMed] [Google Scholar]
- 30.Saito T, Meshi T, Takamatsu N, Okada Y. Coat protein gene sequence of tobacco mosaic virus encodes a host response determinant. Proc Natl Acad Sci USA. 1987;84:6074–6077. doi: 10.1073/pnas.84.17.6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito T, Yamanaka K, Okada Y. Long-distance movement and viral assembly of tobacco mosaic virus mutants. Virology. 1990;176:329–336. doi: 10.1016/0042-6822(90)90002-9. [DOI] [PubMed] [Google Scholar]
- 32.Shivprasad S, Pogue G P, Lewandowski D J, Hidalgo J, Donson J, Grill L K, Dawson W O. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology. 1999;255:312–323. doi: 10.1006/viro.1998.9579. [DOI] [PubMed] [Google Scholar]
- 33.Siegel A, Hari V, Kolacz K. The effect of tobacco mosaic virus infection on host and virus-specific protein synthesis in protoplasts. Virology. 1978;85:494–503. [Google Scholar]
- 34.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in virus-infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 35.Skuzeski J M, Nichols L M, Gesteland R F, Atkins J F. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- 36.Traynor P, Young B M, Ahlquist P. Deletion analysis of brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J Virol. 1991;65:2807–2815. doi: 10.1128/jvi.65.6.2807-2815.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valverde R A, Dodds J A. Some properties of isometric virus-particles which contain the satellite RNA of tobacco mosaic virus. J Gen Virol. 1987;68:965–972. [Google Scholar]
- 38.Waigmann E, Chen M-H, Bachmaier R, Ghoshroy S, Citovsky V. Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 2000;19:4875–4884. doi: 10.1093/emboj/19.18.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe T, Honda A, Iwata A, Ueda S, Hibi T, Ishihama A. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J Virol. 1999;73:2633–2640. doi: 10.1128/jvi.73.4.2633-2640.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White K A, Morris T J. Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. Curr Top Microbiol Immunol. 1999;239:1–17. doi: 10.1007/978-3-662-09796-0_1. [DOI] [PubMed] [Google Scholar]
- 41.Wolf S, Deom C M, Beachy R N, Lucas W J. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science. 1989;246:377–379. doi: 10.1126/science.246.4928.377. [DOI] [PubMed] [Google Scholar]
- 42.Zimmern D. Nucleotide sequence at the origin for assembly on tobacco mosaic virus RNA. Cell. 1977;11:463–481. doi: 10.1016/0092-8674(77)90065-4. [DOI] [PubMed] [Google Scholar]