Abstract
CD4 and CCR5 mediate fusion and entry of R5 human immunodeficiency virus type 1 (HIV-1) strains. Sulfotyrosine and other negatively charged residues in the CCR5 amino-terminal domain (Nt) are crucial for gp120 binding and viral entry. We previously showed that a soluble gp120-CD4 complex specifically binds to a peptide corresponding to CCR5 Nt residues 2 to 18, with sulfotyrosines in positions 10 and 14. This sulfopeptide also inhibits soluble gp120-CD4 binding to cell surface CCR5 as well as infection by an R5 virus. Here we show that residues 10 to 18 constitute the minimal domain of the CCR5 Nt that is able to specifically interact with soluble gp120-CD4 complexes. In addition to sulfotyrosines in positions 10 and 14, negatively charged residues in positions 11 and 18 participate in this interaction. Furthermore, the CCR5 Nt binds to a CD4-induced surface on gp120 that is composed of conserved residues in the V3 loop stem and the C4 domain. Binding of gp120 to cell surface CCR5 is further influenced by residues in the crown of the V3 loop, C1, C2, and C3. Our data suggest that gp120 docking to CCR5 is a multistep process involving several independent regions of the envelope glycoprotein and the coreceptor.
Entry of human immunodeficiency virus type 1 (HIV-1) R5 isolates into target cells is mediated by the successive interaction of the envelope glycoprotein gp120 with CD4 and the CCR5 coreceptor (3). gp120-CD4 complex formation generates a large bonding energy that drives reordering of the gp120 core structure (21, 31, 48). Changes in the orientation of the V1/V2 and V3 loops, as well as the bridging sheet (composed of the V1/V2 stem and C4), cooperatively create and/or expose a coreceptor binding site on gp120 (21, 38, 48). The predicted coreceptor binding surface on gp120 has a hydrophobic core surrounded by a positively charged periphery and is composed of both conserved and variable residues located in the C4 domain and V3 loop, with lesser contributions from the V1/V2 stem (21, 37, 38).
We and others have demonstrated that specific amino acids within the CCR5 amino-terminal domain (Nt, amino acids 2 to 31), including negatively charged and tyrosine residues, are essential for CCR5-mediated fusion and entry of R5 and R5X4 HIV-1 strains (5, 12, 13, 15, 36). Farzan et al. demonstrated that the CCR5 Nt undergoes both O-linked glycosylation and tyrosine sulfation (16). It is presently not known whether O-linked glycosylation plays a role in coreceptor function, but this possibility is suggested by observations that serines in the Nt are important for viral entry (15, 36). Inhibition of cellular sulfation pathways, including tyrosine sulfation, greatly decreases gp120 binding to CCR5 as well as the entry of R5 and R5X4 HIV-1 strains into target cells (16; E. G. Cormier, unpublished data). Posttranslational sulfation of tyrosine residues in the CCR5 Nt, therefore, may critically modulate the susceptibility of target cells to HIV-1 infection in vivo.
We demonstrated that a CCR5 Nt-based peptide spanning residues 2 to 18 and containing sulfotyrosines in positions 10 and 14 specifically associates with soluble gp120-CD4 complexes containing envelope glycoproteins from R5 (HIV-1JR-FL) and R5X4 (HIV-1DH123) but not X4 (HIV-1LAI) strains (11). The tyrosine-sulfated CCR5 Nt therefore specifically interacts only with gp120 proteins from isolates that use this coreceptor to gain entry into target cells. Peptides containing unmodified tyrosines or phosphotyrosines do not bind soluble gp120-CD4 complexes, regardless of gp120 origin (11). Furthermore, only the CCR5 Nt-based sulfopeptide inhibits binding of soluble gp120JR-FL-CD4 to intact, cell surface-expressed CCR5 and blocks the entry of HIV-1JR-FL into target cells.
Here we report a novel enzyme-linked immunosorbent assay (ELISA) to detect binding of sulfopeptides to soluble gp120-CD4 complexes, as well as anti-CCR5 monoclonal antibodies (MAbs) and chemokines. ELISA and surface plasmon resonance (SPR) were used to further delineate the determinants of the gp120-CCR5 Nt interaction. In order to define the minimal domain of the CCR5 Nt capable of specifically binding to soluble gp120-CD4 complexes, we analyzed sulfopeptides corresponding to different regions of the Nt. To identify the gp120 domains involved in CCR5 binding, we studied inhibition of gp120-CD4 complex binding to CCR5 Nt sulfopeptides and cell surface CCR5 by anti-gp120 MAbs. In addition, residues in or near the epitopes of inhibitory MAbs were mutated to alanine, and the gp120 point mutants were compared for their ability to bind to CCR5 Nt sulfopeptides and cell surface CCR5. Our data suggest that a mostly conserved surface of gp120 binds to a 9-residue stretch of the CCR5 Nt, whereas more variable residues in the crown of the V3 loop—C1, C2, and C3—participate in binding to cell surface CCR5.
MATERIALS AND METHODS
Reagents.
CD4-immunoglobulin G2 (IgG2), soluble CD4 (sCD4), recombinant soluble gp120s from HIV-1LAI (X4), HIV-1DH123 (R5X4), and HIV-1JR-FL (R5) isolates, anti-gp120 MAb PA1 (directed against the V3 loop of HIV-1JR-FL), and anti-CCR5 MAbs PA8, PA10, PA11, PA12, and PA14 were produced by Progenics Pharmaceuticals, Inc. (Tarrytown, N.Y.), as described previously (1, 32). MAbs 133-290, 133-192, 135-9, A32, 17b, 19b, 48d, 9284, G3-42, C11, G45-60, and 2G12 were described previously (26). The small-molecule CCR5 antagonist TAK-779 was obtained as described previously (14).
Peptides corresponding to different segments of the CCR5 Nt were synthesized as described previously (11). Sulfo- or phosphotyrosines were incorporated in positions 10 and 14, and all peptides carried a carboxy-terminal GAG spacer preceding a biotinylated lysine. Residues were numbered according to their positions in the full-length CCR5 protein (Table 1).
TABLE 1.
Amino acid sequences of CCR5 Nt-based peptidesa
| Amino acid sequence | Name of peptide |
|---|---|
| Sulfated peptides | |
| D Y Q V S S P I Y DI N Y Y T S E | 2-18 |
| Y D I N Y Y T S E | 10-18 |
| Y A I N Y Y T S A | 10-18(11A/18A) |
| P I Y D I N Y Y | 8-15 |
| S P I Y D I N Y Y T | 6-16 |
| Y D I N Y Y | 10-15 |
| Phosphorylated peptide | |
| O Y Q V S S P I Y D I N Y Y T S E | 2-18[P] |
The peptides are named according to the positions of their first and last residues in the full-length sequence of CCR5. They contain either sulfotyrosines (Y, boldface) or phosphotyrosines (Y, italics) in positions 10 and 14. Residues D11 and E18 in peptide 10-18(11A/18A) are replaced with alanines (A, boldface). All peptides carry a carboxy-terminal GAG spacer followed by a biotinylated lysine.
SPR.
Binding of soluble gp120-CD4 complexes to CCR5 Nt-based peptides was measured as described previously (11). Briefly, streptavidin-coated sensor chips were divided into two surfaces, each with a separate flow chamber. The sensor chip was conditioned and equilibrated as recommended by the manufacturer. Biotinylated peptide (400 resonance units [RU]) was bound to the surface of the second chamber, whereas the first chamber of the chip was used as a negative control. gp120-CD4–IgG2 complex (200 nM/50 nM) was passed over the chip surface in the presence or absence of MAbs (150 nM). SPR was monitored and displayed in RU as a function of time using a Biacore X. After each measurement the chip was regenerated and equilibrated as recommended by the manufacturer.
Generation and characterization of gp120 alanine mutants.
Mutant gp120 proteins were generated using the QuickChange Kit from Stratagene (San Diego, Calif.) according to the manufacturer's instructions. gp120JR-FL, cloned into the PPI4 expression vector (4), served as the template for site-directed mutagenesis. Nucleotide sequencing was performed to ascertain the presence of the appropriate mutation in the gp120 coding sequence. Supernatants containing mutant gp120 proteins were harvested and cleared of debris by centrifugation 24 h after calcium phosphate transfection of 293T cells (4). Soluble gp120 in the supernatants was quantified by ELISA as described previously (41).
Briefly, 293T supernatants or purified gp120 in tissue culture medium were heated to 95°C for 5 min, and different dilutions were deposited on ELISA plates coated with the anti-gp120 MAb D7324 (International Enzymes Inc., Fallbrook, Calif.). Captured gp120 was detected by a mixture of anti-gp120 MAbs B12 and B13 (41), followed by incubation with a horseradish peroxidase-conjugated (HRP) anti-mouse IgG antibody (Amersham Pharmacia, Piscataway, N.J.). Optical density at 450 nm (OD450) was measured using the ImmunoPure TMB substrate kit (Pierce, Rockford, Ill.). All supernatants were adjusted to a final gp120 concentration of 20 nM.
CD4-IgG2 binding to native, mutant gp120 also was measured. ELISA plates coated with D7324 were contacted with different concentrations of gp120 (50 nM to 20 pM). CD4-IgG2 (50 nM) was added to the plates, and its binding to gp120 was detected using an HRP-conjugated goat anti-human IgG (ICN, Costa Mesa, Calif.) and TMB substrate as described above. The percentage of CD4-IgG2 binding to gp120 was calculated using the following formula: (OD450 with mutant gp120)/(OD450 with wild-type gp120) × 100%. OD450 values corresponding to the 50% maximal effective binding concentration (EC50) of CD4-IgG2 binding to wild-type gp120 were used for the calculations.
CCR5 Nt peptide ELISAs.
Streptavidin-coated ELISA plates (Pierce) were blocked with 5% bovine serum albumin in Dulbecco's phosphate-buffered saline for 2 h at room temperature and then were washed three times with assay buffer (Dulbecco's phosphate-buffered saline, 0.5% Tween 20, 1% fetal bovine serum, 2% bovine serum albumin). Plates were then contacted with sulfo- or phosphopeptides (∼500 nM) for 1 h at room temperature and were washed three times with assay buffer. Different dilutions of gp120-CD4–IgG2 (starting with 20 nM/5 nM) were added to the plates for 1 h at room temperature. Plates were washed three times, and HRP-conjugated goat anti-human IgG was used to detect the presence of bound CD4-IgG2. The plates were developed using the TMB substrate as described above. The percentage of gp120-CD4–IgG2 binding to the sulfopeptide was calculated using the following formula: (OD450 with mutant gp120)/(OD450 with wild-type gp120) × 100%. OD450 values corresponding to the EC50 of wild-type gp120 binding to the sulfopeptide were used for the calculations. To normalize binding of the complex to the sulfopeptide for differences in gp120 binding to CD4-IgG2, the following formula was used: (percentage of gp120-CD4–IgG2 binding to sulfopeptide)/(percentage of gp120 binding to CD4-IgG2).
In a competition ELISA, peptides were captured onto the plates as described above. Inhibitor or assay buffer was added for 1 h prior to addition of gp120JR-FL/CD4-IgG2 complex for an additional hour at room temperature. The assay was then completed as described above. Direct binding of anti-CCR5 murine MAbs to the peptides was examined as described above, except that a MAb (150 nM to 15 pM) was substituted for gp120-CD4–IgG2 complex and a goat anti-mouse HRP-coupled antibody was used for detection.
Binding of gp120-CD4–IgG2 complexes to cell surface CCR5.
L1.2-CCR5+ cells (106) were incubated with gp120-containing supernatant (20 nM) and biotinylated CD4-IgG2 (5 nM) for 1 h at 37°C in assay buffer, as previously described (32). gp120-CD4–IgG2 bound to the cells was revealed by fluorescence-activated cell sorter analysis of the mean fluorescence of intensity (MFI) after addition of streptavidin-phycoerythrin (Pharmingen, San Diego, Calif.). Binding was calculated using the formula (MFI of gp120 mutants)/(MFI of gp120 wild type) × 100% and normalized for CD4-IgG2 binding to the mutant gp120 proteins as described above.
RESULTS
ELISA to detect binding of soluble gp120-CD4 complexes to CCR5 Nt-based peptides.
We previously used SPR to show that gp120-sCD4 complexes specifically interact with a peptide spanning CCR5 residues 2 to 18 and containing sulfotyrosines in positions 10 and 14 (peptide 2-18) (Table 1) (11). The on and off rates of complex-peptide binding were extremely rapid and could not be measured precisely by SPR. The Kd was estimated to be in the range of 10−7 to 10−8 M. Replacing monovalent sCD4 with tetravalent CD4-IgG2, however, led to a dramatic shift in both on and off rates, lowering the Kd into the range of 10−8 to 10−9 M (Fig. 1a). gp120-CD4–IgG2 binding to the sulfopeptides continued to be dependent on gp120 from an R5 strain, the V3 loop, and the presence of sulfotyrosines in the peptide (data not shown). These observations prompted us to develop an ELISA to detect complex-sulfopeptide binding. An ELISA could not be developed using gp120-sCD4 because the low affinity of this complex for the sulfopeptide meant that all the complex was eliminated by the multiple washing steps required for this assay.
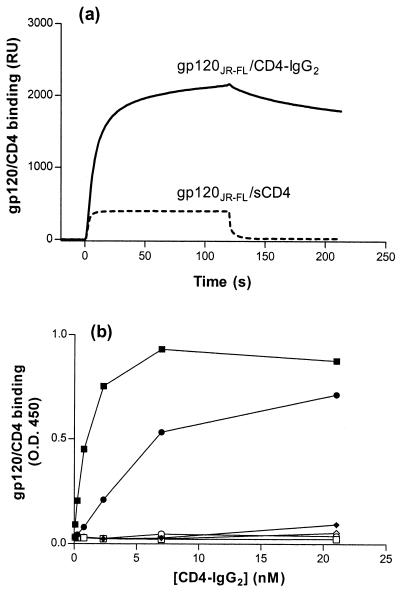
FIG. 1.
gp120-CD4 complex binding to CCR5 Nt sulfopeptides. (a) Peptide 2-18 was bound to streptavidin-coated biosensor chips, and gp120JR-FL-sCD4 or gp120JR-FL-CD4–IgG2 was directed over the sensor chip surface. RU were measured as a function of time using a Biacore X and reflect complex-peptide binding. (b) Sulfopeptide 2-18 (solid symbols) or phosphopeptide 2-18(P) (open symbols) was immobilized on streptavidin-coated ELISA plates and incubated with gp120-CD4–IgG2 complexes. gp120 proteins were derived from the R5 isolate HIV-1JR-FL (squares), the R5X4 isolate HIV-1DH123 (circles), and the X4 isolate HIV-1LAI (diamonds). Complex-peptide binding was detected by an HRP-conjugated goat anti-human IgG antibody. OD450 was measured after the addition of HRP substrate and is expressed as a function of CD4-IgG2 concentration. Results shown are from representative experiments.
Streptavidin-coated ELISA plates were used to capture biotinylated, CCR5 Nt-based peptides which were then further incubated with soluble gp120-CD4–IgG2 complexes. Complex binding was detected using an HRP-conjugated goat anti-human IgG antibody. Sulfopeptide 2-18 bound gp120JR-FL-CD4–IgG2 with an EC50 of ∼1 nM and bound gp120DH123-CD4–IgG2 with an EC50 of ∼5 nM (Fig. 1b). Sulfopeptide 2-18 did not measurably bind CD4-IgG2 alone or in complex with either gp120LAI or gp120JR-FL with a deletion of the V3 loop (Fig. 1b; data not shown). No binding was observed to an analogous CCR5 Nt phosphopeptide (peptide 2-18[P]) (Table 1) by any of the gp120-CD4–IgG2 complexes (Fig. 1b). Similar patterns of reactivity were observed for gp120s in complex with CD4-γ2, a divalent CD4-Ig fusion protein (1; data not shown). However, no binding was observed for gp120 in complex with anti-gp120 MAbs 2G12 and IgG1b12, even though the epitope of IgG1b12 overlaps the CD4 binding site on gp120 (data not shown). Thus, the ELISA reproduces previous observations on peptide-complex interactions measured by SPR (11).
Binding of CCR5 Nt peptides to anti-CCR5 MAbs, chemokines, and TAK-779.
ELISA was used to test the binding of a panel of anti-CCR5 MAbs to peptides 2-18 and 2-18[P]. It was previously demonstrated that MAbs PA8, PA11, and PA12 bind epitopes in the Nt, whereas PA10 and PA14 bind epitopes that span the Nt and the second extracellular loop (ECL2) of CCR5 (32). Here we show that PA8 avidly binds peptides 2-18 and 2-18[P] (Fig. 2a and b). PA10 binds avidly to 2-18 and moderately to 2-18[P]. PA11 binds moderately to 2-18 and weakly to 2-18[P]. Binding of PA12 is weak for 2-18 and undetectable for 2-18[P]. PA14 does not recognize either the sulfopeptide or the phosphopeptide (Fig. 2a and b). Furthermore, PA8 binds with similar avidity to all of the CCR5 Nt-based sulfopeptides listed in Table 1 (data not shown).
FIG. 2.
Binding of anti-CCR5 MAbs to CCR5 Nt peptides. Sulfopeptides (a) or phosphopeptides (b) were immobilized on streptavidin-coated ELISA plates and incubated with anti-CCR5 MAb PA8 (solid squares), PA10 (open circles), PA11 (solid circles), PA12 (solid diamonds), or PA14 (solid triangles). Binding of the MAbs to the peptides was detected by an HRP-conjugated goat anti-mouse IgG antibody. OD450 was measured after addition of HRP substrate and expressed as a function of MAb concentration. (c) Biotinylated sulfopeptide 2-18 was immobilized on streptavidin-coated plates and incubated with gp120-CD4–IgG2 complex in the presence of increasing concentrations of PA8 (solid squares), TAK-779 (triangles), RANTES (inverted triangles), MIP-1α (diamonds), MIP-1β (circles), or SDF-1α (open squares). Binding of the complexes to the peptide was detected by incubation with HRP-conjugated goat anti-human IgG antibody. OD450 was measured after addition of HRP substrate, and the percentage of binding was expressed as a function of inhibitor concentration. Results shown are from representative experiments. Some symbols are not visible due to overlay.
Using a competition ELISA, we were able to inhibit gp120JR-FL-CD4–IgG2 binding to the sulfopeptide 2-18 with the anti-CCR5 MAb PA8 (Fig. 2c). However, binding of soluble gp120-CD4–IgG2 complexes to the sulfopeptide was not inhibited by TAK-779 or by the CC chemokines MIP-1α, MIP-1β, and RANTES, even when used at supraphysiologic concentrations (Fig. 2c).
Binding of CCR5 Nt peptides to soluble gp120-CD4–IgG2 complexes.
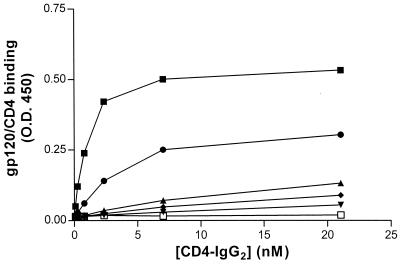
In order to delineate more precisely the minimal CCR5 Nt domain that specifically binds to soluble gp120-CD4 complexes, we synthesized a panel of sulfopeptides spanning different stretches of the CCR5 Nt (Table 1). All of the peptides carried sulfotyrosines in positions 10 and 14, since it was previously shown that these are required for complex-peptide binding (11). Binding of gp120JR-FL-CD4–IgG2 to the different sulfopeptides was tested by ELISA. Although the strongest binding was observed using the longest sulfopeptide, 2-18, we also observed significant binding to peptide 10-18, which demonstrated an avidity only approximately threefold lower than did peptide 2-18 for the soluble complex (Fig. 3). Peptides 8-15, 6-16, and 10-15 bound the soluble complex at least 10-fold less avidly than did 2-18. (It was previously shown that a sulfopeptide consisting of residues 10 to 14 did not bind soluble gp120-CD4 complexes [11].) Furthermore, the gp120JR-FL-CD4–IgG2 complex only weakly bound to peptide 10-18(11A/18A), which carries alanine mutations in positions 11 and 18. Previous mutagenesis studies of CCR5 have shown that residues D11 and E18 are important for fusion, entry, and gp120-CCR5 binding (12, 13, 15). Finally, it should be noted that similar patterns of binding to the different sulfopeptides were observed with soluble complexes containing gp120DH123 (data not shown).
FIG. 3.
Binding of gp120JR-FL-CD4–IgG2 to different CCR5 Nt-based peptides. Streptavidin plates were coated with sulfopeptide 2-18 (solid squares), 10-18 (solid circles), 8-15 (solid diamonds), 6-16 (solid stars), 10-15 (open squares), or 10-18(11A/18A) (solid triangles). Plate contents were then incubated with gp120JR-FL-CD4–IgG2 complex. Binding of the complex to the peptide was detected by an HRP-conjugated goat anti-human IgG antibody. OD450 was measured after addition of HRP substrate and expressed as a function of CD4-IgG2 concentration. Results shown are from a representative experiment.
Inhibition of gp120-CD4–IgG2 binding to CCR5 Nt sulfopeptides by anti-gp120 MAbs.
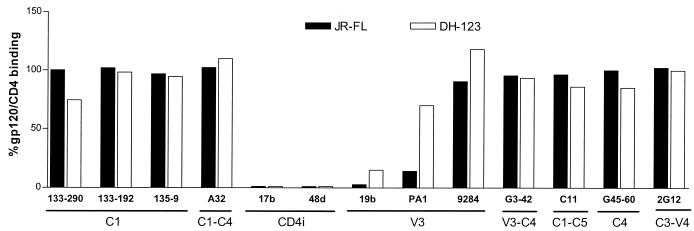
In order to determine which domains of gp120 are involved in binding to CCR5 Nt-based sulfopeptides, we used SPR to test the ability of a panel of well-characterized anti-gp120 MAbs (25) to inhibit binding of either the gp120JR-FL-CD4–IgG2 or the gp120DH123-CD4–IgG2 complex to the 2-18 sulfopeptide. Only MAbs directed against CD4-induced (CD4i) epitopes and the V3 loop were capable of inhibiting binding of the soluble complexes to the CCR5 sulfopeptide (Fig. 4). Inhibition of gp120JR-FL-CD4–IgG2 and gp120DH123-CD4–IgG2 binding by MAbs 17b and 48d was >90%. The anti-V3 loop MAb 19b, which recognizes an epitope in the crown of the V3 loop (sequence -I----G--FY-T) and reacts exclusively with R5 strains (28), inhibited gp120JR-FL binding by >90% and gp120DH123 binding by ∼80%. Anti-V3 loop MAb PA1, which was raised against gp120JR-FL (W. C. Olson, unpublished results), efficiently inhibited binding of gp120JR-FL-containing but not of gp120DH123-containing complexes. Finally, anti-V3 loop MAb 9284 (20), which recognizes an epitope spanning residues 307 to 330 in the V3 loop of X4 strains, was unable to inhibit binding of either gp120 protein to the sulfopeptide. MAbs directed against epitopes in other constant and variable regions of gp120 also had no effect on binding of the soluble complex to the peptides. Similar results were obtained when the anti-gp120 MAbs were used to inhibit soluble complex binding to cell surface CCR5 (data not shown).
FIG. 4.
Inhibition of gp120-CD4–IgG2 complex binding to sulfopeptides by anti-gp120 MAbs. Biotinylated sulfopeptide 2-18 was bound to streptavidin-coated biosensor chips, and solutions of either gp120JR-FL-CD4–IgG2 complex (solid bars) or gp120DH123-CD4–IgG2 complex (open bars) were directed over the surface of the chip in the presence of different anti-gp120 MAbs. The names of the MAbs and the locations of their epitopes are indicated along the x axis. RU were measured as a function of time using a Biacore X and reflect complex-peptide binding in the presence of the MAbs. gp120-CD4–IgG2 binding was calculated using the following formula: (RU in the presence of MAbs)/(RU in the absence of MAbs) × 100%. The values shown are averages of three independent experiments. Standard deviation was no more than ±20% for each data set; standard deviations are not represented for the sake of clarity.
Binding of mutant soluble gp120-CD4 complexes to CCR5 Nt sulfopeptides and cell surface CCR5.
Numerous studies have shown that residues in the V3 loop determine coreceptor usage and binding (6–10, 18–20, 22, 24, 27, 29, 33, 42–45, 47). The crystal structure of a gp120 lacking the V1/V2 and V3 loops in complex with sCD4 and the 17b MAb implicated a conserved, CD4i surface on gp120, adjacent to the V3 loop, in coreceptor binding (37, 38). We generated single alanine mutants of all of the residues near or within regions previously shown to be important for coreceptor usage. These gp120 mutants were tested for their ability to interact with the CCR5-based sulfopeptide 2-18 as well as with cell surface CCR5. Complex-peptide associations were normalized for gp120 mutant binding to CD4-IgG2. Binding of most of the gp120 mutants to CD4-IgG2 was similar to that of the wild-type envelope glycoprotein (data not shown). Only mutants W427A, R440A, and R469A bound CD4-IgG2 with 5- to 10-fold-lower but nonetheless measurable avidity (data not shown).
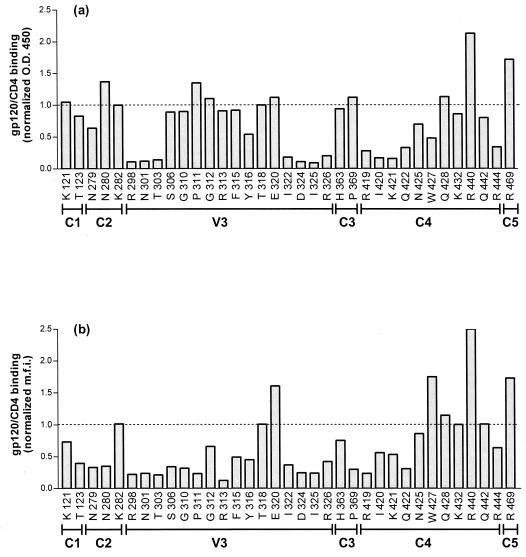
Residues in both strands of the V3 loop stem were found to be involved in gp120 binding to the 2-18 sulfopeptide: alanine mutants of residues R298, N301, T303, I322, D324, I325, and R326 were found to decrease complex binding to the peptide >10-fold (Fig. 5a). Residues in the crown of the V3 loop, including the GPGR motif, however, had no effect on soluble complex binding to the sulfopeptide. C4 residues in or adjacent to the two C-terminal β strands of the bridging sheet were also shown to participate in binding to the sulfopeptide: alanine substitutions of R419, I420, K421, Q422, and R444 decreased complex binding to the sulfopeptide 5- to 10-fold. None of the alanine substitutions introduced in the other regions of gp120 significantly affected complex-peptide interactions.
FIG. 5.
Binding of gp120 mutants to sulfopeptide and wild-type CCR5. (a) Sulfopeptide 2-18 was immobilized on streptavidin-coated plates and incubated with a mixture of gp120-containing supernatants and CD4-IgG2. Peptide-complex binding was detected by an HRP-conjugated goat anti-human IgG antibody. OD450 was measured after addition of HRP substrate and normalized for binding of the gp120 mutants to CD4-IgG2. The dotted line represents the normalized value for the binding of wild-type gp120 to the peptide. The mutated amino acids and their locations in gp120 are indicated along the x axis. (b) L12-CCR5+ cells were incubated with a mixture of gp120-containing supernatants and CD4-IgG2. Binding of the complex was detected by fluorescence-activated cell sorter analysis after addition of streptavidin-phycoerythrin. The percentage of gp120-CD4–IgG2 binding to CCR5 was normalized for gp120 binding to CD4-IgG2. The dotted line represents the normalized value for the binding of wild-type gp120 to the L12-CCR5+ cells. The mutated amino acids and their locations in gp120 are indicated along the x axis. The values shown are averages of three independent experiments. Standard deviation was no more than ±20% for each data set; standard deviations are not represented for the sake of clarity.
Additional gp120 residues appear to be involved in soluble complex binding to cell surface CCR5. Alanine substitutions of residues S306, G310, P311, R313, F315, and Y316 in the crown of the V3 loop decreased complex binding to CCR5 5- to 10-fold (Fig. 5b). Furthermore, alanine substitutions of several residues in C1, C2, and C3 also had a moderate effect on complex binding to cell surface CCR5. Finally, we note that alanine substitutions of R440 and R469 increased complex binding to both 2-18 and CCR5, whereas alanine substitutions of E320 and W427 increased complex binding to CCR5 only.
DISCUSSION
Tyrosine-sulfated CCR5 Nt peptides were studied for binding to soluble gp120-CD4 complexes as well as anti-CCR5 MAbs, CC chemokines, and TAK-779 using a novel solid-phase ELISA. Inhibition of peptide-complex interactions by anti-gp120 MAbs was explored by SPR. These MAbs were also tested for their ability to inhibit complex binding to cell surface CCR5. In addition, we generated a panel of gp120 point mutants and compared their reactivity with CCR5 Nt peptides and cell surface CCR5. Our principal conclusions are that (i) residues 10 to 18 of the CCR5 Nt may define the minimum recognition site for gp120, (ii) gp120 binding to the CCR5 Nt depends on highly conserved residues located in the C4 domain and the stem of the V3 loop, and (iii) gp120 binding to cell surface CCR5 depends on a broader region that includes residues in the crown of the V3 loop: C1, C2, and C3. Our findings suggest that distinct domains of gp120 and CCR5 bind in a multistep fashion and raise questions about the determinants of specificity of the coreceptor-gp120 interaction.
We developed an ELISA to detect complex-peptide binding based on the observation that the gp120-CD4–IgG2 complex binds to CCR5 Nt sulfopeptides 10- to 100-fold more avidly than does the gp120-sCD4 complex. This increase in avidity is probably due to an increase in the valence of the interaction. Complex-sulfopeptide binding was only observed with gp120 proteins derived from R5 and R5X4 but not from X4 HIV-1 strains. gp120JR-FL with a deletion of the V3 loop failed to bind to the sulfopeptides. Phosphopeptides did not bind to any of the soluble complexes. Thus, the ELISA reproduces the critical biological features of cell surface CCR5-gp120 interactions, including a dependence upon CD4, CCR5 tyrosine sulfation, the V3 loop, and the coreceptor usage patterns of the parental viruses.
CCR5 Nt phosphopeptides and sulfopeptides were differentially recognized by anti-CCR5 MAbs in ELISA. PA8 possessed equal avidity for sulfated and phosphorylated peptides, implying that its epitope does not include tyrosine side chains. PA10 and PA11 preferentially recognized the sulfopeptide, albeit with various efficiencies, suggesting that sulfotyrosines participate either directly in peptide-MAb interactions or indirectly by influencing epitope conformations. PA12 interacted only with the sulfopeptide, and PA14 did not bind either Nt peptide. We had previously shown that both of these MAbs recognize discontinuous epitopes comprising residues in the Nt and ECL2 of CCR5. Our observations now imply that ECL2 residues are marginal for PA12 binding and essential for PA14 binding to CCR5. Finally, binding of soluble gp120-CD4 complexes to CCR5 Nt peptides could be competed with an anti-CCR5 MAb but not with CC chemokines and TAK-779, whose binding sites have been mapped to other regions of CCR5 (14, 40). Both CC chemokines and TAK-779, however, are able to compete with gp120-CD4 binding to cell surface CCR5, perhaps through steric or conformational mechanisms. (14, 43, 47). We note that Farzan et al. reported that a CCR5 Nt sulfopeptide spanning residues 1 to 22 partially blocks MIP-1α binding to cell surface CCR5, and we attribute the discrepancy to differences in peptides and assays (17).
In order to more precisely delineate the gp120 binding site in the CCR5 Nt, we used ELISA to test binding of soluble complexes to several CCR5 Nt sulfopeptides. Sulfopeptide 10-18 was the smallest that avidly bound soluble gp120-CD4 complexes. In addition to the two sulfotyrosines in positions 10 and 14, negatively charged amino acids D11 and E18 were found to be critical for complex-peptide binding. We conclude that a cluster of negative charges within the CCR5 Nt residues 10 to 18 represents the minimal recognition motif for gp120, although residues 2 to 9 further contribute to binding. Similar patterns of peptide reactivity were observed for recombinant gp120s derived from HIV-1JR-FL (R5) and HIV-1DH123 (R5X4), suggesting that the CCR5 Nt sulfopeptides recognize conserved structures in the envelope glycoprotein. gp120DH123, however, bound about fivefold less than did gp120JR-FL to the sulfopeptides, which probably accounts for its less efficient usage of CCR5 (13).
Anti-gp120 MAbs were tested for their ability to inhibit gp120-CD4 binding to sulfopeptides or to cell surface CCR5. A number of anti-gp120 MAbs directed against conserved and variable regions of the envelope glycoprotein were not inhibitory. Only MAbs 48d and 17b, directed against CD4i epitopes, and 19b and PA1, directed against the V3 loop, efficiently inhibited gp120 binding to the 2-18 sulfopeptide and to cell surface CCR5. The CD4i epitope was previously shown to participate in coreceptor binding, and residues in the V3 loop primarily determine coreceptor specificity (37, 38). Our results now suggest that these regions of gp120 determine its association with the CCR5 Nt. We note that inhibition of peptide-complex binding by 19b, which recognizes an epitope in the V3 crown, is inconsistent with the finding by gp120 mutagenesis experiments that residues in the V3 loop crown do not participate in complex-peptide binding. This leads us to propose that the inhibitory effect of 19b may be due to steric hindrance. Also, MAbs G3-42 and G45-60 do not inhibit gp120-CD4 binding to the sulfopeptide even though their epitopes are in V3-C4 and C4, respectively (25). However, these MAbs do not cross-compete with 17b, 48d, 19b, and 9284 for binding to gp120, and therefore their epitopes probably also do not overlap the CCR5 Nt binding site (25).
In order to determine more precisely the regions of gp120 that modulate the gp120-CCR5 interaction, we tested the binding of a panel of gp120 point mutants to the CCR5 Nt sulfopeptide and to cell surface CCR5. The mutants were created by the introduction of single alanine substitutions near or within regions previously shown to be important for the integrity of the CD4i epitope and/or CCR5 binding (37, 38). Highly conserved residues in C4 and the V3 loop stem, including four arginines and a lysine, were found to affect binding of gp120 to the CCR5 Nt sulfopeptide (Fig. 6). These residues are located in two random coil segments of C4 that straddle the V3 loop stem and may constitute a positively charged CCR5 Nt binding domain (21). Furthermore, N-linked glycosylation at position 301 in the base of the V3 loop has been shown previously to influence coreceptor usage, and we find that the loss of this glycosylation site abolishes the gp120-CCR5 Nt sulfopeptide interaction (23, 35). Additional, conserved residues in the crown of the V3 loop, C1, C2, and C3, contribute to gp120 binding to cell surface CCR5 (Fig. 6). It remains to be determined whether these residues interact with other extracellular domains of CCR5 or whether they influence the conformation of C4 and the V3 loop stem in a way that is relevant only in the context of gp120-CD4 binding to cell surface CCR5. It is unlikely that these residues also interact with the Nt in the context of cell surface CCR5 because they are relatively distal from the C4 and V3 residues that were implicated in sulfopeptide binding (21).
FIG. 6.
Amino acid conservation among R5 isolates. The first column indicates the gp120 domains, the second column indicates the gp120 residues, and the third column indicates the percentage of conservation of the residues. Envelope sequences from 25 R5 strains described in the HIV Database and retrieved from the National Center for Biotechnology Information GenBank were aligned, and the percentage of conservation for the indicated residues was calculated and combined with results by Hwang et al. (19). Residues exhibiting a >50% decrease in binding to sulfopeptide 2-18 when changed to alanines are highlighted in gray. Residues exhibiting a >50% decrease in binding to cell surface CCR5 when changed to alanines are in boldface.
To date, several lines of evidence suggest that gp120 binds to more than one region of the CCR5 coreceptor: (i) the affinity of soluble gp120-CD4 complex for the CCR5 Nt sulfopeptide is approximately 10- to 100-fold lower than for the native, membrane-associated coreceptor (11, 43, 47); (ii) coreceptor chimera studies implicate the extracellular loops in viral fusion and entry (2, 12, 34, 39); and (iii) inhibitors of CCR5 coreceptor function, such as MAbs 2D7 and PA14, as well as TAK-779, do not bind to the CCR5 Nt yet block gp120-CD4 binding to CCR5 (14, 32). Our present findings could be interpreted to support a distributed model for gp120-CCR5 interactions that mirrors the two-site paradigm proposed for the interaction of certain chemokines with their receptors (30, 46). In this model, binding is initially driven by electrostatic interactions between negatively charged residues in the receptor Nt and basic surfaces on the chemokine ligand. This binding serves to orient the ligand and promote its interactions with other portions of the chemokine receptor. The V3 loop crown may form initial electrostatic interactions with the extracellular loops of CCR5, which would allow the CCR5 Nt to bind to a conserved region of gp120 comprising residues in C4 and the V3 loop stem. Alternatively, the CCR5 Nt could first bind the C4/V3 stem domain, which would then promote an interaction of the V3 loop with some other region of CCR5. All of these interactions may involve additional gp120 residues that we have yet to identify. The role of the putative second interaction is unclear, but it may further stabilize the gp120-CCR5 interaction, optimally orienting the fusion apparatus or triggering gp41 conformational changes that are required for fusion.
Our findings present us with a seeming paradox wherein 9 residues of the CCR5 Nt confer specificity on the CCR5-gp120 interaction by binding to gp120 residues that are highly conserved among clade B isolates, regardless of their coreceptor usage. However, although the C4 and V3 stem residues themselves are conserved, their precise placement may differ in R5 and X4 gp120s. Clearly, relatively minor differences in the orientation, exposure, or relative positioning of these residues could abrogate binding to a short peptide but not to a MAb (e.g., 17b) possessing a larger, more distributed binding site (36). In addition, more variable amino acids within or outside the C4/V3 loop stem may contribute to the specificity of the gp120-Nt interaction. Future studies employing additional gp120 mutants together with CCR5 mutants and CXCR4-based sulfopeptides will shed light on the specificity determinants of the gp120-coreceptor interaction.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI 43847 (T.D.) and by Progenics Pharmaceuticals, Inc.
REFERENCES
- 1.Allaway G P, Davis-Bruno K L, Beaudry G A, Garcia E B, Wong E L, Ryder A M, Hasel K W, Gauduin M C, Koup R A, McDougal J S, et al. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retrovir. 1995;11:533–539. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- 2.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 3.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 4.Binley J M, Sanders R W, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma D J, Maddon P J, Olson W C, Moore J P. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanpain C, Doranz B J, Vakili J, Rucker J, Govaerts C, Baik S S W, Lorthioir O, Migeotte I, Libert F, Baleux F, Vassart G, Doms R W, Parmentier M. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 env protein. J Biol Chem. 1999;274:34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- 6.Chan S Y, Speck R F, Power C, Gaffen S L, Chesebro B, Goldsmith M A. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavda S C, Griffin P, Han-Liu Z, Keys B, Vekony M A, Cann A J. Molecular determinants of the V3 loop of human immunodeficiency virus type 1 glycoprotein gp120 responsible for controlling cell tropism. J Gen Virol. 1994;75:3249–3253. doi: 10.1099/0022-1317-75-11-3249. [DOI] [PubMed] [Google Scholar]
- 8.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 11.Cormier E G, Persuh M, Thompson A D, Lin S W, Sakmar T P, Olson W C, Dragic T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfo-tyrosines with HIV-1 envelope glycoprotein gp120. Proc Natl Acad Sci USA. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon P J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragic T, Trkola A, Thompson D A, Cormier E G, Kajumo F A, Maxwell E, Lin S W, Ying W, Smith S O, Sakmar T P, Moore J P. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc Natl Acad Sci USA. 2000;97:5639–5644. doi: 10.1073/pnas.090576697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 17.Farzan M, Vasilieva N, Schnitzler C E, Chung S, Robinson J, Gerard N P, Gerard C, Choe H, Sodroski J. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275:33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- 18.Harrowe G, Cheng-Mayer C. Amino acid substitutions in the V3 loop are responsible for adaptation to growth in transformed T-cell lines of a primary human immunodeficiency virus type 1. Virology. 1995;210:490–494. doi: 10.1006/viro.1995.1367. [DOI] [PubMed] [Google Scholar]
- 19.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 20.Ivanoff L, Looney D, McDanal C, Morris J, Wong-Staal F, Langlois A, Petteway S, Matthews T. Alteration of HIV-1 infectivity and neutralization by a single amino acid replacement in the V3 loop domain. AIDS Res Hum Retrovir. 1991;7:595–603. doi: 10.1089/aid.1991.7.595. [DOI] [PubMed] [Google Scholar]
- 21.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong P D, Wyatt R, Sattentau Q J, Sodroski J, Hendrickson W A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malenbaum S E, Yang D, Cavacini L, Posner M, Robinson J, Cheng-Mayer C. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J Virol. 2000;74:11008–11016. doi: 10.1128/jvi.74.23.11008-11016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menzo S, Sampaolesi R, Vicenzi E, Santagostino E, Liuzzi G, Chirianni A, Piazza M, Cohen O J, Bagnarelli P, Clementi M. Rare mutations in a domain crucial for V3-loop structure prevail in replicating HIV from long-term non-progressors. AIDS. 1998;12:985–997. [PubMed] [Google Scholar]
- 25.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore J, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore J P, Nara P L. The role of the V3 domain of gp120 in HIV infection. AIDS. 1991;5(Suppl. 2):S21–S33. doi: 10.1097/00002030-199101001-00004. [DOI] [PubMed] [Google Scholar]
- 28.Moore J P, Trkola A, Korber B, Bouts L J, Kessler J A, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. A human monoclonal antibody to a complex epitope in the V3 region of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris J F, Sternberg E J, Gutshall L, Petteway S R, Jr, Ivanoff L A. Effect of a single amino acid substitution in the V3 domain of the human immunodeficiency virus type 1: generation of revertant viruses to overcome defects in infectivity in specific cell types. J Virol. 1994;68:8380–8385. doi: 10.1128/jvi.68.12.8380-8385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser B, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. Interleukin-8 antagonists generated by N-terminal modification. J Biol Chem. 1993;268:7125–7128. [PubMed] [Google Scholar]
- 31.Myszka D G, Sweet R W, Hensley P, Brigham-Burke M, Kwong P D, Hendrickson W A, Wyatt R, Sodroski J, Doyle M L. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson W C, Rabut G E E, Nagashima K A, Tran D N H, Anselma D J, Monard S P, Segal J P, Thompson D A D, Kajumo F, Guo Y, Moore J P, Maddon P J, Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page K, Stearns S, Littman D. Analysis of mutations in the V3 domain of gp160 that affect fusion and infectivity. J Virol. 1992;66:524–533. doi: 10.1128/jvi.66.1.524-533.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollakis G, Kang S, Kliphuis A, Chalaby M I M, Goudsmit J, Paxton W A. N-linked glycosylation of the HIV-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 co-receptor utilization. J Biol Chem. 2001;276:13433–13441. doi: 10.1074/jbc.M009779200. [DOI] [PubMed] [Google Scholar]
- 36.Rabut G E, Konner J A, Kajumo F, Moore J P, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retrovir. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 38.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 39.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 40.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 41.Sanders R W, Schiffner L, Master A, Kajumo F, Guo Y, Dragic T, Moore J P, Binley J M. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J Virol. 2000;74:5091–5100. doi: 10.1128/jvi.74.11.5091-5100.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trkola A, Dragic T, Arthos J, Binley J, Olson W, Allaway G, Cheng-Mayer C, Robinson J, Maddon P, Moore J. CD4-dependent, antibody sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;384:184–186. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 44.Verrier F, Borman A M, Brand D, Girard M. Role of the HIV type 1 glycoprotein 120 V3 loop in determining coreceptor usage. AIDS Res Hum Retrovir. 1999;15:731–743. doi: 10.1089/088922299310827. [DOI] [PubMed] [Google Scholar]
- 45.Wang W K, Dudek T, Essex M, Lee T H. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci USA. 1999;96:4558–4562. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells T N, Power C A, Lusti-Narasimhan M, Hoogewerf A J, Cooke R M, Chung C W, Peitsch M C, Proudfoot A E. Selectivity and antagonism of chemokine receptors. J Leukoc Biol. 1996;59:53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 47.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 48.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]