Abstract
High-risk human papillomaviruses (HPVs) immortalize keratinocytes by disrupting the retinoblastoma protein (Rb)/p16 pathway and activating telomerase. The E7 oncoprotein targets Rb, while the E6 oncoprotein induces telomerase activity in human keratinocytes. This study has examined the mechanism by which E6 activates telomerase. Expression of human telomerase reverse transcriptase (hTERT), the catalytic subunit of telomerase, was found to be increased in keratinocytes stably expressing HPV type 16 E6, suggesting that E6 acts to increase hTERT transcription. hTERT expression and telomerase activity were activated to significantly higher levels in cells expressing both E6 and E7 than in cells expressing E6 alone. This indicates that E7 may augment E6-mediated activation of hTERT transcription. In transient-transfection assays using hTERT reporters, the induction of hTERT expression by E6 was found to be mediated by a 258-bp fragment of the hTERT promoter, proximal to the ATG initiation codon. Previous studies have demonstrated that overexpression of Myc can activate hTERT expression, suggesting that Myc may be a mediator of E6-mediated hTERT induction. However, in cells stably expressing E6, no strict correlation between the level of Myc and the activation of hTERT was found. Consistent with this observation, mutation of the two Myc binding sites in the hTERT promoter only modestly reduced responsiveness to E6 in transient reporter assays. This indicates that activation of Myc-dependent transcription is not essential for E6-mediated upregulation of hTERT expression. The hTERT promoter also contains five GC-rich elements that can bind Sp1. Mutation of these sites within the 258-bp fragment partially reduced hTERT induction by E6. However, when mutations in the Sp1 sites were combined with the mutated Myc binding sites, all activation by E6 was lost. This indicates that it is the combinatorial binding of factors to Myc and Sp1 cis elements that is responsible for hTERT induction by E6.
Human papillomaviruses (HPVs) are small double-stranded DNA viruses that induce hyperproliferative lesions in epithelial tissues (13). More than 85 distinct types of HPV have been identified and fully sequenced (5, 39). These types include some that infect cutaneous tissues and induce warts on the hands or feet, as well as others that infect the oral mucosa. Among the most well-characterized HPV types are those that infect genital epithelia, and these can be grouped on the basis of their association with cervical and other anogenital cancers. “High-risk” HPV types, such as HPV type 16 (HPV-16), HPV-18, HPV-31, and HPV-54, induce lesions which can lead to cancer, while “low-risk” types, such as HPV-6 and HPV-11, induce benign lesions that rarely progress to malignancy (2, 13, 21, 39).
The major transforming proteins of high-risk HPV types are the E6 and E7 proteins, and numerous modulatory functions have been attributed to them. Two key targets of E6 and E7 are the tumor suppressors p53 and the retinoblastoma protein (Rb) (13, 25, 39). Inactivation of the p53 and Rb pathways is thought to be a critical step in the progression to malignancy. For example, E7 can bind to Rb and alleviate repression of E2F-dependent target genes, thereby allowing rapid progression into S phase (8, 16, 29). Similarly, E6 facilitates the degradation of p53 through the actions of E6-associated protein (E6-AP), which results in the abrogation of the G1/S and G2/M checkpoints (15, 32, 33, 37). Each of these functions has been shown to be specific to the high-risk HPV types.
In addition to targeting p53 for degradation, E6 from high-risk HPV types activates telomerase (19). Telomerase is a multisubunit complex that is responsible for synthesizing the hexamer repeats which comprise the telomeres at the ends of chromosomes (3, 30). Telomerase is generally not active in adult cells, and this results in a gradual loss of telomeres through successive cell divisions. Such a process has been suggested to be a natural mechanism of aging (23). Tumors often exhibit uncontrolled proliferative capacity, so it is not surprising that telomerase activity is detected in virtually all tumors (23). Recent studies have shown that expression of human telomerase reverse transcriptase (hTERT), the catalytic subunit of telomerase, is sufficient to induce immortalization in a number of primary cell lines (1, 23). However, immortalization of human foreskin keratinocytes (HFKs) requires the inactivation of Rb via E7 or loss of expression of the cyclin-dependent kinase inhibitor p16, in addition to hTERT expression (18).
While telomerase activation is required for immortalization, recent studies suggest that immortalization of HFKs does not require p53 inactivation (7, 18). Earlier work in several laboratories demonstrated that expression of E6 and E7 is sufficient to induce cellular immortalization (12, 14, 28). Several E6 mutants that cannot degrade p53 are also unable to induce immortalization, suggesting that p53 is the key target of E6 (18). However, the finding that hTERT can substitute for E6 in immortalization of HFKs indicates that the critical function of E6 may be the activation of telomerase (18). It is more likely that both functions are relevant, with telomerase activation being important for the extension of life span and p53 degradation being critical for the progression to malignancy through the development of secondary mutations.
The mechanism by which E6 activates telomerase has not yet been elucidated. Through mutational analysis of E6, it has been determined that p53 degradation is not required for activation of telomerase (18). In addition, it has been demonstrated that overexpression of Myc can induce telomerase activity by directly activating the hTERT promoter (9, 20, 36, 38). Thus, it is possible that E6 activates telomerase via induction of Myc, although the exact mechanism by which this might occur is unclear. It has been reported that E6 activates the Myc promoter in transient assays and that overexpression of E6 in human mammary epithelial cells leads to increased Myc protein levels (17, 36). However, Myc mRNA levels are not upregulated in E6-expressing human mammary epithelial cells, suggesting that E6 induces Myc through a posttranscriptional mechanism. In an apparently contradictory study, E6 has been shown to induce the degradation of Myc (10). Thus, the true relationship between E6 and Myc remains to be resolved.
In the present study, we observed in long-term assays that E6 upregulates the expression of hTERT but that this does not strictly correlate with increased levels of Myc. Using transient- transfection assays with hTERT reporters, we found that activation by E6 was mediated by a combination of Myc and GC-rich Sp1 cis elements. Thus, E6 may activate or recruit factors that bind to these sites to induce hTERT transcription.
MATERIALS AND METHODS
Cell culture.
HFKs were derived from neonatal human foreskin epithelium and maintained in serum-free keratinocyte growth medium supplemented with bovine pituitary extract (Clonetics, San Diego, Calif.). C33A cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (Gibco BRL, Grand Island, N.Y.). Retrovirus-packaging cell lines expressing Myc were created by transfection of pLXSN-Myc into the ecotropic Bosc23 packaging cell line and subsequent infection of the amphotropic PA317 packaging cell line. PA317 cell lines expressing HPV-16 E6, E7, and E6/E7 were obtained from D. Galloway (University of Washington, Seattle). 3T3 J2 fibroblast feeders were maintained in Dulbecco modified Eagle medium supplemented with 10% calf serum. Retroviral infection of HFKs has been previously described (11, 35). Subsequent to infection, HFKs were maintained in E medium with mitomycin-treated J2 3T3 fibroblast feeders (26). Cells were selected for neomycin resistance by incubation with G418 for 4 days at 200 μg/ml and then for 4 days at 100 μg/ml. Prior to harvest, feeders were detached by treatment with EDTA (0.5 mM).
Plasmids.
TL 800 and TL DM hTERT promoter-reporter constructs and the PXP2 luciferase vector were obtained from R. Dalla-Favera (Columbia University, New York, N.Y.). Deletion mutants were generated by recombinant PCR. WT 181 contains 260 bp of the hTERT promoter upstream of the ATG initiation codon. Sp1 MT1, Sp1 MT2, Sp1 MT3, Sp1 MT4, Sp1 MT5, and Sp1 MT1–5 have been described elsewhere (20). Sp1 MT1–5/Myc MT was generated by PCR-based mutagenesis. Fos-luc was generated by insertion of 100 bp of the Fos promoter into the HindIII and KpnI sites of PXP2. pLXSN-Myc was created by insertion of the Myc coding region into the EcoRI and XhoI sites of pLXSN. pSG5-Myc was generated by insertion of the EcoRI-BamHI fragment from pLXSN-Myc into pSG5 (Stratagene, La Jolla, Calif.). pSG5-16E6 and pSG5-11E6 have been described previously (22). pSG5-16E6Δ9–13 was generated by insertion of the EcoRI-BamHI fragment from LXSN-16E6Δ9–13 into the EcoRI and BamHI sites of pSG5. pSG5-16E7 was obtained from D. McCance (University of Rochester, Rochester, N.Y.).
Telomeric repeat amplification protocol (TRAP) assays.
Telomerase activity was analyzed via the TRAPeze telomerase detection kit (Intergen, Purchase, N.Y.). Cell extracts were incubated with a [γ-32P]ATP-end-labeled telomere substrate (TS primer) for 45 min at 30°C and then subjected to a two-step PCR amplification. The products were electrophoresed on a nondenaturing polyacrylamide gel, and the gel was subsequently dried and subjected to autoradiography.
Reverse transcription (RT)-PCR.
Total RNA was harvested using Trizol reagent (Gibco BRL). Total RNA (200 ng) was reverse transcribed for 45 min at 60°C, after which a two-step PCR amplification was performed. hTERT primer pairs LT5 (5′-CGG AAG AGT GTC TGG AGC AA-3′, sense) and LT6 (5′-GGA TGA AGC GGA GTC TGG A-3′, antisense) amplified a 142-bp product. Glyceraldehyde-6-phosphate dehydrogenase (GAPDH) primer pairs K136 (5′-CTC AGA CAC CAT GGG GAA GGT GA-3′, sense) and K137 (5′-ATG ATC TTG AGG CTG TTG TCA TA-3′, antisense) amplified a 440-bp product.
Western analysis.
Whole-cell extracts were prepared using 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer (10 mM Tris-HCl [pH 7.5], 1 mM MgCl2, 1 mM EGTA, 0.1 mM benzamidine, 5 mM β-mercaptoethanol, 0.5% CHAPS, 10% glycerol) and quantitated with the Bradford assay (Bio-Rad, Hercules, Calif.). Equal amounts of protein were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel and subsequently transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.). The membrane was blocked in wash solution (0.1% Tween 20 in phosphate-buffered saline) containing 5% nonfat dry milk. Rabbit polyclonal anti-Myc (sc-764; Santa Cruz Biotechnology) was used as the primary antibody. Proteins were visualized via enhanced chemiluminescence (ECL; Amersham, Arlington Heights, Ill.)
Luciferase assays.
Cells were transfected with Fugene 6 transfection reagent (Roche, Indianapolis, Ind.) and harvested 48 h posttransfection. Total DNA was equalized in all transfections by adding pSG5 DNA. Luciferase activity was quantitated by using a luciferase assay kit (Tropix, Bedford, Mass.) and normalized to the protein concentration. Values, expressed as relative level of activation, are averages of data from at least three experiments.
RESULTS
Analysis of telomerase activity, hTERT expression, and Myc levels in cells expressing E6 and/or E7.
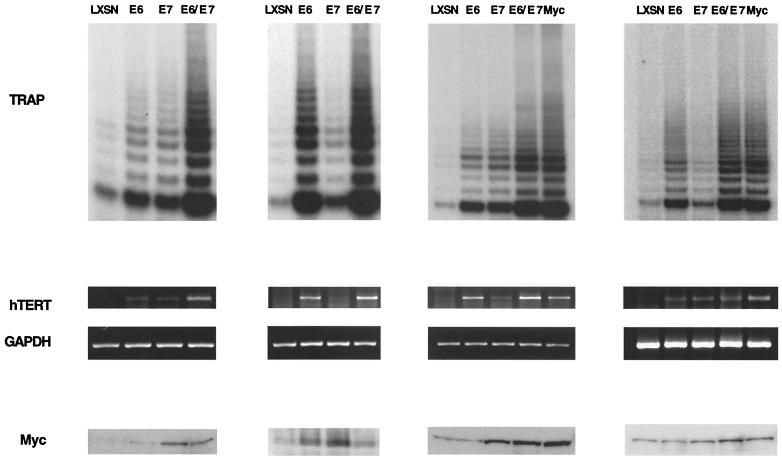
To investigate the mechanism of telomerase induction by E6, we first examined telomerase enzymatic activity in primary HFKs stably expressing HPV-16 E6 and/or E7. These cells were isolated after infection with LXSN-based retroviruses and selection for neomycin resistance. In addition, keratinocytes expressing high levels of Myc were generated after infection with recombinant LXSN-based retroviruses expressing Myc. HFKs infected with the empty pLXSN vector were used as a control cell line. Following retroviral infection and selection, cells were harvested at identical passage numbers and telomerase activity was determined using the TRAP assay. Consistent with previous reports, cells expressing E6 exhibited high but variable levels of telomerase activity, while normal cells had virtually no detectable activity (Fig. 1). We observed increased levels of telomerase activity in four independent experiments involving infections with retroviruses expressing E6 alone (Fig. 1). While there was variation in the average level of activation, we always observed an increase. In cells expressing E7 alone, we consistently detected a low level of telomerase activity that was approximately 10% of that seen in Myc-expressing cells. In cells expressing both E6 and E7, the telomerase activity was consistently found to be approximately three- to fivefold higher than that in keratinocytes expressing either oncoprotein alone. This suggests that although E6 is a primary activator of telomerase, E7 may augment this activity.
FIG. 1.
Analysis of telomerase activity, hTERT mRNA levels, and Myc protein levels in HFKs stably expressing HPV-16 E6, E7, E6/E7, or Myc. Panels represent four independently infected sets of HFKs. Telomerase activity in whole-cell extracts was measured with the TRAP assay. RT-PCR was performed on total RNA to determine hTERT and GAPDH mRNA levels. Myc protein levels in whole-cell extracts were determined via Western analysis.
We next determined whether the increase in telomerase activity induced by E6 correlated with an upregulation of hTERT transcription. For these studies, RT-PCR was performed on total RNA isolated from cells expressing E6 and/or E7. RT-PCR for GAPDH was used as an internal control. As expected, hTERT mRNA was detected in cells expressing E6 or E6/E7 but not in normal cells (Fig. 1). In addition, hTERT expression correlated well with overall telomerase enzymatic activity. This suggests that E6 activates telomerase by upregulating hTERT transcription.
Because Myc has been implicated as an activator of hTERT expression, we next examined whether Myc levels correlated with increased transcription of hTERT. For these studies, Western analysis of Myc protein levels in cells stably expressing E6 and/or E7 was performed. Analysis of multiple sets of infected cells indicated that the levels of Myc were variable and were not reproducibly enhanced by E6 expression (Fig. 1). Indeed, Myc levels were more consistently increased in cells expressing E7 alone or both E6 and E7. Interestingly, despite the presence of high levels of Myc in most E7-expressing cells, telomerase activity was only moderately induced. Mad family proteins are known to bind to the same DNA binding sites as Myc but repress transcription, so it is possible that E7 expression leads to higher levels of a Mad protein, thus offsetting the increased Myc levels. Overall, the levels of Myc did not correlate with telomerase activity, suggesting that although it may play a role, Myc may not be the sole mediator by which E6 upregulates hTERT expression.
E6-mediated activation of the hTERT promoter is partially dependent on Myc binding sites.
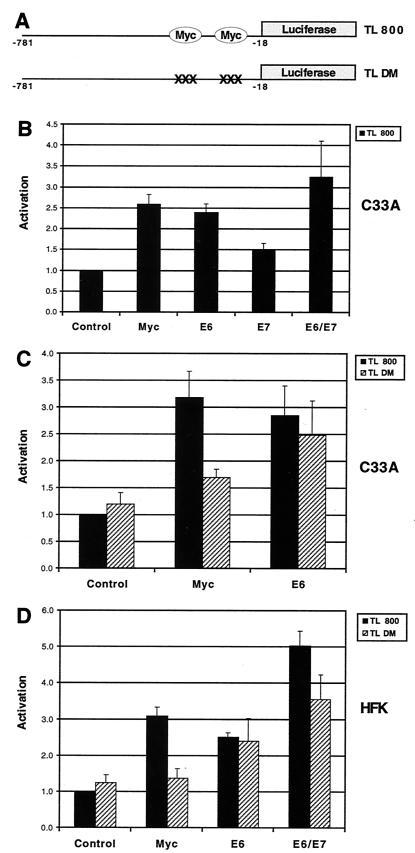
To investigate how E6 induces hTERT expression, we utilized two reporter plasmids, TL 800 and TL DM, which contain approximately 800 bp of the hTERT promoter fused to the firefly luciferase gene (Fig. 2A). TL DM contains point mutations (CACGTG→CACCTG) in each of the two Myc binding sites, located at −242 and −34 relative to the ATG initiation codon. These mutations have previously been shown to abrogate Myc binding and activation of the hTERT promoter (38). Cells from the HPV-negative cervical carcinoma cell line C33A were transfected with TL 800 and expression vectors for E6, E7, or E6/E7. In addition, we cotransfected an expression vector for Myc as a positive control for TL 800 activation. Transfection of expression vectors for either E6 or Myc resulted in approximately a threefold activation of the hTERT promoter (Fig. 2B). In contrast, transfection of an expression vector for E7 activated the reporter approximately 1.5-fold, while cotransfection of both E6 and E7 expression vectors resulted in a 3- to 4-fold activation. This enhancement of reporter activity by coexpression of E6 and E7 correlated with our TRAP analysis data and further supports the idea that E7 can enhance E6-mediated telomerase activation. We also conclude that E6 can activate hTERT transcription in transient reporter assays.
FIG. 2.
Activation of hTERT promoter-reporter plasmids by E6. (A) Structure of TL 800 and TL DM. Each reporter contains approximately 800 bp of the hTERT promoter fused to the firefly luciferase gene. TL DM contains a point mutation (CACGTG→CACCTG) in each of the two Myc binding sites, located at −242 and −34 relative to the ATG initiation codon. (B) C33A cells were transfected with TL 800 and either pSG5-Myc (Myc), pSG5-16E6 (E6), pSG5-16E7 (E7), or both pSG5-16E6 and pSG5-16E7 (E6/E7) (P < 0.05 for Myc, E6, E7, and E6/E7 when compared to control). (C) C33A cells were transfected with TL 800 or TL DM and either pSG5-Myc or pSG5-16E6 (P < 0.05 for Myc and E6 with TL 800 when compared to control; P < 0.05 for E6 with TL DM when compared to control). (D) HFKs were transfected with TL 800 or TL DM and either pSG5-Myc, pSG5-16E6, or both pSG5-16E6 and pSG5-16E7 (P < 0.05 for Myc, E6, and E6/E7 with TL 800 when compared to control; P < 0.05 for E6-E7 with TL DM when compared to control). Total DNA was equalized with pSG5 in each transfection. Cells were harvested 48 h posttransfection, and luciferase activity was normalized to total cellular protein concentration. Results are the means ± standard deviations of data from at least three experiments.
Next, we investigated the dependence of E6-mediated activation on the two Myc binding sites in the hTERT promoter. For these experiments, TL 800 or TL DM reporter was transfected into C33A cells and the activation mediated by Myc or E6 was examined. As expected, transfection of a Myc expression vector did not significantly activate the mutant reporter (Fig. 2C). In contrast, E6-mediated activation of TL DM was only slightly reduced from that of the wild-type reporter construct, indicating that E6 could still activate the hTERT promoter in the absence of functional Myc binding sites.
We next sought to confirm these observations in normal human keratinocytes. As shown in Fig. 2D, Myc-mediated activation of TL 800 in HFKs was entirely dependent on the presence of two functional Myc binding sites, while E6-mediated activation was at best slightly reduced by mutation of the two Myc binding sites. In addition, in the presence of both E6 and E7, a high degree of hTERT activation was retained even with TL DM. These findings were similar to the results for transfections in C33A cells and indicate that activation of the hTERT promoter via E6 can occur by a Myc-independent mechanism.
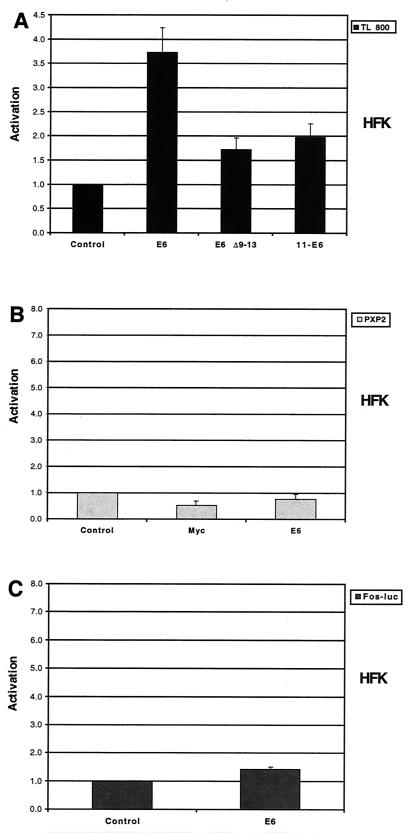
To ensure that E6-mediated activation of hTERT transcription was not due to a nonspecific effect, we examined the ability of a telomerase-defective E6 mutant (HPV-16 E6Δ9–13) and low-risk HPV-11 E6 to activate the hTERT promoter. Both HPV-16 E6Δ9–13 and low-risk HPV-6 E6 have been shown to be unable to activate telomerase enzymatic function in stable long-term assays (19). While wild-type HPV-16 E6 significantly activated the hTERT promoter, the telomerase-defective E6 mutant and HPV-11 E6 were significantly impaired in this ability (Fig. 3A). Thus, the activation of the hTERT promoter correlates with telomerase induction by E6 and is specific to high-risk E6. To confirm that E6-mediated activation was indeed specific to the hTERT promoter, we examined the ability of E6 to activate the PXP2 luciferase vector nonspecifically. Neither Myc nor E6 was capable of activating this reporter (Fig. 3B). In addition, we investigated the effects of E6 on a reporter containing the minimal Fos promoter. Similar fragments of the Fos promoter have previously been shown to be unresponsive to E6 (6, 27). Transfection of an expression vector for E6 did not significantly activate this reporter (Fig. 3C). These findings suggest that E6-mediated activation of the hTERT promoter in transient assays is specific and correlates with effects seen in stable cell lines.
FIG. 3.
Specificity of hTERT promoter activation by E6. (A) HFKs were transfected with TL 800 and either pSG5 (control), pSG5-16E6 (E6), pSG5-16E6Δ9–13 (E6 Δ9–13), or pSG5-11E6 (11-E6) (P < 0.05 for E6, E6 Δ9–13, and 11 E6 when compared to control; P < 0.05 for E6 Δ9–13 and 11 E6 when compared to E6). (B) HFKs were transfected with PXP2 and either pSG5 (control), pSG5-Myc (Myc), or pSG5-16E6. (C) HFKs were transfected with Fos-luc and either pSG5 or pSG5-16E6. Total DNA was equalized with pSG5 in each transfection. Cells were harvested 48 h posttransfection, and luciferase activity was normalized to total cellular protein concentration. Results are the means ± standard deviations of data from at least three experiments.
E6-responsive sequences in the hTERT promoter can be localized to a region containing 258 bp upstream of the ATG initiation codon.
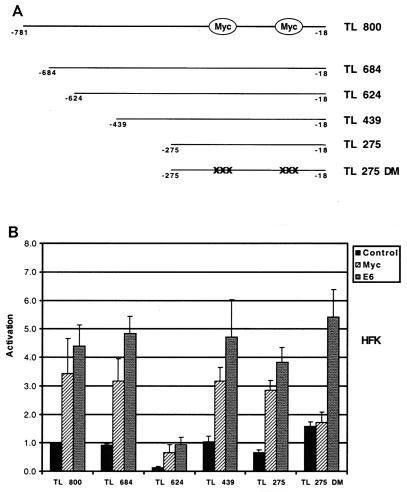
To localize the cis elements responsible for hTERT induction by E6, we generated a series of hTERT promoter deletions (Fig. 4A). These four derivative plasmids were generated by progressive deletions from the 5′ end of the hTERT promoter. Each of these constructs was transfected into HFKs, and their responsiveness to both Myc and E6 was determined. The basal activities of the reporters differed, suggesting that cis elements that positively or negatively regulate basal transcriptional activity may have been deleted. However, transfection of expression vectors for either Myc or E6 activated each reporter significantly relative to the negative control (Fig. 4B). One plasmid, TL 275 DM, which consists of 258 bp upstream of the ATG initiation codon, also contains point mutations in the two Myc binding sites. In transient assays, this reporter was no longer responsive to Myc but was still activated by E6. This suggests that hTERT induction by E6 can be localized to a small region containing 258 bp of the hTERT promoter and that this activation can occur by a Myc-independent pathway.
FIG. 4.
Deletion analysis of hTERT promoter activation by E6. (A) Schematic of hTERT promoter deletion mutants. Deletions were created by recombinant PCR. (B) HFKs were transfected with hTERT promoter-reporter plasmids and either pSG5 (control), pSG5-Myc (Myc), or pSG5-16E6 (E6). Cells were harvested 48 h posttransfection, and luciferase activity was normalized to total cellular protein concentration. Results are the means ± standard deviations of data from at least three experiments. P < 0.05 for Myc with TL 800, TL 684, TL 624, TL 439, and TL 275 when compared to control; P < 0.05 for E6 with each reporter when compared to control.
Loss of Sp1 binding sites partially impairs activation of the hTERT promoter by E6.
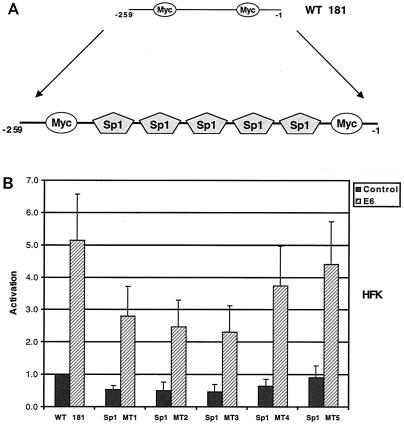
Five GC-rich Sp1 sites have previously been shown to be important for hTERT promoter activity (Fig. 5A). Each of these sites has previously been shown by gel shift analysis to bind Sp1(20). We next investigated whether mutation of these GC-rich sites would impair the responsiveness of this reporter to E6. For these studies, we utilized five different reporters in which each of the individual Sp1 sites has been mutated individually. These mutant reporters were previously constructed using the plasmid WT 181, which contains 260 bp of the hTERT promoter upstream of the luciferase gene (Fig. 5A). WT 181 and TL 275 were activated by E6 to similar levels (Fig. 5B). While mutation of any of the individual Sp1 sites resulted in a slight variation in luciferase activity, responsiveness to E6 was only modestly altered. This suggests that the five sites may act in a redundant fashion to activate hTERT expression in the absence or presence of E6.
FIG. 5.
Mutational analysis of individual Sp1 binding sites in the hTERT promoter. (A) Schematic of hTERT promoter region containing Sp1 binding sites. (B) HFKs were transfected with hTERT promoter-reporters and either pSG5 (control) or pSG5-16E6 (E6). Cells were harvested 48 h posttransfection, and luciferase activity was normalized to total cellular protein concentration. Results are means ± standard deviations of data from five experiments. P < 0.05 for E6 with each reporter when compared to control.
Mutation of Myc binding sites and Sp1 binding sites results in a loss of hTERT promoter activation by E6.
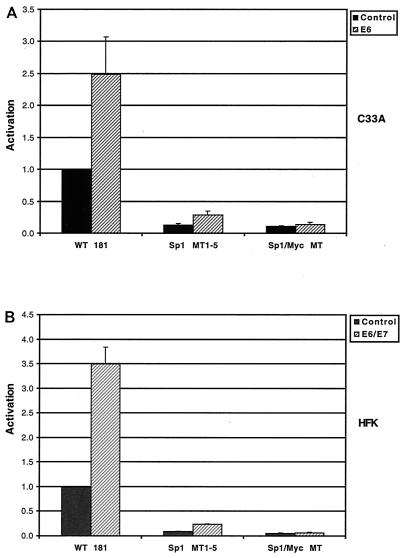
Given that the loss of any individual Sp1 site did not significantly alter the response of the hTERT promoter to E6, we next examined the effect of mutation of all five of the Sp1 sites. As shown in Fig. 6A, mutation of all of the Sp1 sites (Sp1 MT1–5) significantly reduced the basal activity of the reporter. However, while the fold induction was reduced from that seen with the wild-type construct, this reporter could still be activated approximately twofold by E6. We reasoned that a possible explanation for this residual activation might be the presence of the Myc binding sites in the hTERT promoter. We therefore constructed an additional reporter, Sp1/Myc MT, in which all five of the Sp1 sites as well as the Myc sites were mutated. When this reporter was tested in transient assays in C33a cells, it was found to be unresponsive to E6 (Fig. 6A) or E6/E7 (data not shown). We confirmed these findings in HFKs with E6/E7, and similar effects were seen (Fig. 6B). Thus, we conclude that activation of the hTERT promoter by E6 is mediated by both Myc and Sp1 cis elements. Loss of either of these elements only partially impairs activation, but when both sets of mutations are combined, all activation is lost.
FIG. 6.
Effects of mutations in Sp1 binding sites and Myc binding sites. (A) C33A cells were transfected with hTERT promoter-reporter plasmids and either pSG5 (control) or pSG5-16E6 (E6) (P < 0.05 for E6 with WT 181 and Sp1 MT1–5 when compared to control). (B) HFKs were transfected with hTERT promoter-reporter plasmids and either pSG5 or pSG5-16E6 and pSG5-16E7 (E6/E7) (P < 0.05 for E6-E7 with WT 181 and Sp1 MT1–5 when compared to control). Cells were harvested 48 h posttransfection, and luciferase activity was normalized to total cellular protein concentration. Results are means ± standard deviations of data from three experiments.
DISCUSSION
We have investigated the mechanism by which the high-risk HPV-16 E6 protein activates telomerase. Previous studies have shown that expression of hTERT, the catalytic subunit of telomerase, is the rate-limiting determinant of telomerase activity (23). In our studies, activation of telomerase enzymatic function by E6 correlated strongly with the induction of hTERT expression. In addition, we observed that E6 could activate the hTERT promoter in transient assays. This induction was significantly impaired with a telomerase-defective E6 mutant or the low-risk HPV-11 E6 protein. We conclude that HPV-16 E6 activates telomerase, at least in part, by increasing transcription from the hTERT promoter.
The levels of telomerase activity and hTERT expression in our studies varied in keratinocytes stably expressing E6 alone. In contrast, a consistently high level of telomerase activity was observed in all cell lines examined expressing both E6 and E7. These observations are similar to previous reports on E6-mediated telomerase activation (18, 19). Interestingly, we also observed a consistently low level of telomerase activity in cells expressing E7 alone. This is in contrast to previous reports and may be due to differences in culture conditions. Our observations in stable cell lines are in agreement with our transient assays in which we have observed moderate hTERT promoter activation by E7. In addition, E7 has been shown to immortalize primary keratinocytes at a low frequency, suggesting that E7 alone may also be capable of low-level activation of telomerase (11, 14). The consistently high levels of telomerase activity seen in cells expressing both E6 and E7 may be due to the synergistic action of these two proteins. This is in accord with the observation that E6 and E7 function cooperatively to immortalize human keratinocytes (12, 14, 28).
In our studies, both the telomerase-defective HPV-16 E6Δ9–13 mutant and the low-risk HPV-11 E6 protein were significantly impaired in their ability to activate the hTERT promoter. This indicates that the induction by E6 in our transient assays reflects effects seen in stable cell lines. We also found that the degree of induction by E6 in HFKs varied with the amount of DNA transfected as well as the donor from which the keratinocytes were derived. E6 has been reported to activate several heterologous promoters in a nonspecific manner in NIH 3T3 cells (4). In our studies, we did not observe activation of a control reporter lacking defined promoter sequences or a reporter containing a minimal Fos promoter. Therefore, we believe that the effects we observed with the hTERT promoter are indeed a specific activity of the high-risk HPV-16 E6 protein.
Although E6 has been shown to exhibit nonspecific DNA binding activities, no specific DNA binding sequences have been identified (24). Therefore, it is likely that the induction of hTERT transcription by E6 occurs via an intermediary protein. An obvious candidate is Myc, which has previously been shown to bind and activate the hTERT promoter (9, 20, 36, 38). Previous studies have shown that mutation of the two Myc binding sites in the hTERT promoter abrogates activation by Myc (9, 20, 38). In our studies, activation of the hTERT promoter by E6 was only partially reduced by mutations in the two Myc binding sites. Similar effects were seen in both C33A cells and HFKs, as well as with both the full-length (TL DM) and truncated (TL 275 DM) hTERT promoters. These findings suggest that activation of Myc is not sufficient for E6-mediated induction of hTERT transcription and are consistent with our observations that Myc levels in keratinocytes stably expressing E6 did not directly correlate with hTERT mRNA levels.
We also investigated the significance of five GC-rich Sp1 binding sites in the hTERT promoter. Mutation of these five sites reduced E6-mediated activation from approximately threefold to twofold. However, when both the Myc binding sites and the Sp1 binding sites were mutated, all activation was lost. Thus, we conclude that it is the cooperative action of the Myc and Sp1 cis elements that is responsible for E6-mediated hTERT induction. It is possible that E6 acts through Sp1 to activate hTERT transcription, but since numerous factors other than Sp1 are known to bind to these GC-rich sequences, it is possible that other cellular transcription factors may be involved (31, 34). In preliminary studies, we did not observe any alteration in Sp1 levels in E6-expressing cell lines (S. Oh, unpublished data), but we cannot exclude the possibility that Sp1 was posttranslationally activated by E6. To better understand the role of E6 in the activation of telomerase during the progression to malignancy, a more detailed analysis of the factors activated by E6 to induce hTERT transcription will be required.
ACKNOWLEDGMENTS
We thank S. Terhune and M. Vana for critical comments on the manuscript and members of the Laimins laboratory for technical advice.
S.T.O was supported by an NIH/NRSA Carcinogenesis Training Grant (5 T32 CA09560-15) to Northwestern University. This work was supported by a grant from the NAIAD (U19 AI31494) to L.A.L.
REFERENCES
- 1.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 2.Cannistra S A, Niloff J M. Cancer of the uterine cervix. N Engl J Med. 1996;334:1030–1038. doi: 10.1056/NEJM199604183341606. [DOI] [PubMed] [Google Scholar]
- 3.Colgin L M, Reddel R R. Telomere maintenance mechanisms and cellular immortalization. Curr Opin Genet Dev. 1999;9:97–103. doi: 10.1016/s0959-437x(99)80014-8. . (Erratum, 9:247.) [DOI] [PubMed] [Google Scholar]
- 4.Desaintes C, Hallez S, Van Alphen P, Burny A. Transcriptional activation of several heterologous promoters by the E6 protein of human papillomavirus type 16. J Virol. 1992;66:325–333. doi: 10.1128/jvi.66.1.325-333.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Villiers E M. Human pathogenic papillomavirus types: an update. Curr Top Microbiol Immunol. 1994;186:1–12. doi: 10.1007/978-3-642-78487-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Dey A, Atcha I A, Bagchi S. HPV16 E6 oncoprotein stimulates the transforming growth factor-beta 1 promoter in fibroblasts through a specific GC-rich sequence. Virology. 1997;228:190–199. doi: 10.1006/viro.1996.8363. [DOI] [PubMed] [Google Scholar]
- 7.Dickson A M, Hahn W C, Ino Y, Ronfard V, Wu J Y, Weinberg R A, Louis D N, Li F P, Rheinwald J G. Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson N, Howley P M, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg R A, O'Hagan R C, Deng H, Xiao Q, Hann S R, Adams R R, Lichtsteiner S, Chin L, Morin G B, DePinho R A. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 10.Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias K E, Schwartz A L, Kahana C, Ciechanover A. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc Natl Acad Sci USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbert C L, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 947–978. [Google Scholar]
- 14.Hudson J B, Bedell M A, McCance D J, Laimins L A. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus type 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones D L, Münger K. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin Cancer Biol. 1996;7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita T, Shirasawa H, Shino Y, Moriya H, Desbarats L, Eilers M, Simizu B. Transactivation of prothymosin alpha and c-myc promoters by human papillomavirus type 16 E6 protein. Virology. 1997;232:53–61. doi: 10.1006/viro.1997.8536. [DOI] [PubMed] [Google Scholar]
- 18.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 19.Klingelhutz A J, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 20.Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, Ariga H, Inoue M. Sp1 cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 22.Lechner S M, Mack D H, Finicle A B, Crook T, Vousden K H, Laimins L A. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–3052. doi: 10.1002/j.1460-2075.1992.tb05375.x. . (Erratum, 11:4248.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J P. Studies of the molecular mechanisms in the regulation of telomerase activity. FASEB J. 1999;13:2091–2104. doi: 10.1096/fasebj.13.15.2091. [DOI] [PubMed] [Google Scholar]
- 24.Mallon R G, Wojciechowicz D, Defendi V. DNA-binding activity of papillomavirus proteins. J Virol. 1987;61:1655–1660. doi: 10.1128/jvi.61.5.1655-1660.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantovani F, Banks L. The interaction between p53 and papillomaviruses. Semin Cancer Biol. 1999;9:387–395. doi: 10.1006/scbi.1999.0142. [DOI] [PubMed] [Google Scholar]
- 26.Meyers C, Laimins L A. In vitro systems for the study and propagation of human papillomaviruses. Curr Top Microbiol Immunol. 1994;186:199–215. doi: 10.1007/978-3-642-78487-3_11. [DOI] [PubMed] [Google Scholar]
- 27.Morosov A, Phelps W C, Raychaudhuri P. Activation of the c-fos gene by the HPV16 oncoproteins depends upon the cAMP-response element at −60. J Biol Chem. 1994;269:18434–18440. [PubMed] [Google Scholar]
- 28.Münger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Münger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 31.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 33.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 34.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 35.Thomas J T, Laimins L A. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J Virol. 1998;72:1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Xie L Y, Allan S, Beach D, Hannon G J. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 38.Wu K J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 39.zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]