Abstract
The constitutive transport element (CTE) of type D retroviruses serves as a signal of nuclear export of unspliced viral RNAs. The human TAP(NXF1) protein, a cellular mRNA export factor, directly binds to CTE and mediates nuclear export of CTE-containing RNAs. Here, we use genomic SELEX (systematic evolution of ligands by exponential enrichment) to show that the human genome encodes a family of high-affinity TAP ligands. These TAP-binding elements (TBE) are 15-bp minisatellite repeats that are homologous to the core TAP-binding sites in CTE. The repeats are positioned similarly in the RNA secondary structures of CTE and TBE. Like CTE, TBE is an active nuclear export signal. CTE elements of different species share sequence similarities to TBE in the regions that are neutral for CTE function. This conservation points to a possible common ancestry of the two elements, and in fact, TBE has properties expected from a primordial CTE. Additionally, a molecular fossil of a TBE-like minisatellite is found in the genome of a modern retroelement. These findings constitute direct evidence of an evolutionary link between TBE-related minisatellites and CTE.
Replication of many retroviruses requires that their mRNAs be posttranscriptionally regulated. This step is necessary for the nuclear export of the unspliced, full-length viral RNA and ensures the availability of genomic RNA for packaging into the progeny virions and for the production of Gag-Pol polyproteins. Among the best-studied nuclear export systems are those used by the simian type D retroviruses and the lentiviruses, such as human immunodeficiency virus type 1 (HIV-1) (for reviews, see references 8, 12, 15, 17, 25, 36, and 41). Type D retroviruses such as simian retrovirus type 1 (SRV-1), SRV-2, and Mason-Pfizer monkey virus (MPMV) contain a conserved cis-acting constitutive transport element (CTE) (5, 38, 46) that is necessary for virus replication. CTE is the binding site for the cellular mRNA export factor TAP(NXF1) (1, 14) and is essential for the export of the unspliced viral RNA (11, 38). Another factor that can specifically interact with the type D CTE is RNA helicase A, which is thought to participate in CTE function (27, 40). The CTE-containing RNAs and cellular mRNAs share a conserved nuclear export pathway that utilizes TAP(NXF1) (3, 14, 21, 26, 29). TAP's orthologues from Saccharomyces cerevisae (16, 21, 30, 31) and Caenorhabditis elegans (39) were also shown to be essential for the export of mRNAs from the nucleus. Since TAP can bind RNA in a non-structure-specific manner in vitro (4, 14, 21) and can be cross-linked to poly(A)+ RNA in vivo (21), it is thought to bind directly to cellular mRNAs during export. Additionally, several TAP-binding factors have been identified that may bridge or facilitate its association with cellular mRNAs. These factors include the hnRNP-like proteins E1B-AP5 (1) and REF (also known as Aly) (34, 35, 45). REF is one of the proteins that is thought to associate with cellular mRNAs as a result of splicing, and it has been recently proposed to link the mRNA splicing to the nuclear export (45). It is possible that the export of cellular mRNAs occurs only after the completion of splicing because the TAP-binding export signals such as Aly are deposited onto mRNAs as a result of splicing. Since the CTE-containing mRNAs are exported before splicing, it is plausible that CTE provides a constitutive TAP-binding export signal that bypasses the requirement for splicing-dependent protein signals, leading to the constitutive export.
The type D retroviruses are evolutionarily related to the rodent intracisternal A-type particle (IAP) retroelements (44). We have previously shown that some IAPs contain CTE elements that are closely related to these of type D retroviruses (37). The type A and the type D CTEs share a conserved secondary structure that is necessary for their function and contain four conserved motifs that are the core binding sites of TAP(NXF1). The motifs are arranged in two mirror-symmetrical pairs that form the internal loops of an extended hairpin loop structure (10, 11, 14, 20, 37, 38). Apart from these motifs, the primary structure of the type A and the type D CTEs is not conserved. Since the double-stranded regions of the CTEs are formed by the nonconserved sequences, they do not likely participate directly in TAP binding and may serve to maintain the overall secondary structure of CTE (37, 38). Since TAP(NXF1) is a cellular protein that has high affinity to viral RNA elements, there may be cellular counterparts of CTE. We have previously performed database searches for cellular sequences that have a CTE-like arrangement of TAP-binding repeated motifs. Using probes that were based on the conserved features of CTE structure, we have identified a variety of rodent CTEs that belong to the known or putative endogenous IAP retroelements, whereas no strong matches were found in the primate sequences (37). Since such database comparisons are limited to the close homologs of known CTEs, it is possible that cellular sequences exist that have lower degree of structural homology to modern CTEs.
Here, we used in vitro selection to identify the cellular targets of TAP(NXF1). We performed a genome-wide search for the natural RNA ligands that selectively bind to the recombinant TAP protein, using the genomic SELEX (systematic evolution of ligands by exponential enrichment) method (13, 32). This selection identified a family of human TAP-binding elements (TBE) that contain repeated motifs that are homologous to the TAP-binding sites in CTE. Like CTE, TBE can act as RNA nuclear export signal. Besides the structural and functional similarities, TBE and CTE share additional features that point to their common ancestry. We propose a model in which CTEs evolved from ancient TBE-like repeats that were present in the precursors of modern retroelements.
MATERIALS AND METHODS
Construction of RNA selection libraries.
Time course digestions of genomic DNA from human placenta tissue were performed using small amounts of DNaseI, and the conditions leading to high yield of 600- to 800-bp fragments were established. Fragments in this size range were excised from native agarose gels and further used for library construction. After being filled in by T4 DNA polymerase, the fragments were ligated to synthetic adapters AD1 and AD2 as described previously (9) and the adapters were filled in with Taq polymerase. An aliquot was filled in the presence of [α-P32]dCTP. As calculated from the specific radioactivity, the library contained ∼1011 amplifiable DNA molecules and therefore redundantly represented the human genome. These fragments were subjected to 10 PCR amplification cycles using Taq DNA polymerase with primers P1 and P2 (9), followed by 10 cycles with primers T7.PN1 and PN2 under the same conditions. Primer T7.PN1 (5′-GCGAAATTAATACGACTCACTATAGGGAGATCGAGCGGCCGCCCGGGCAGGT-3′) contained T7 RNA polymerase promoter followed by primer PN1 (9). The DNA was transcribed in vitro using T7 RNA polymerase, and the RNA was reverse transcribed with avian myeloblastosis virus reverse transcriptase, using primer PN2. The cDNA was amplified by PCR for 5 cycles (94°C for 30 min and 72°C for 3 min) and purified on 1% agarose gel.
In vitro RNA selection.
The genomic SELEX RNAs transcribed in vitro were incubated in binding buffer (15 mM HEPES [pH 7.7], 50 mM KCl, 200 mM NaCl, 0.2 mM EDTA, 5% glycerol, 0.5 mM dithiothreitol) for 15 min at room temperature, with recombinant glutathione-S transferase (GST)–TAP protein (14) that was immobilized on glutathione-Sepharose beads. Prior to selection, the endogenous Escherichia coli RNA was removed from GST-TAP by micrococal nuclease digestion. At steps 3 and 6, binding to naked beads was performed prior to TAP binding. Yeast tRNA (5 mg/ml) and/or heparin (0.05 to 20 μg/ml) was used as the competitor. The bound RNA was eluted with 1 M NaCl in binding buffer followed by isopropanol precipitation. After reverse transcription, the cDNAs were PCR amplified using primers T7.PN1 and PN2 (94°C for 30 min and 72°C for 3 min) for 5 to 10 cycles. The resulting DNAs were transcribed with T7 polymerase, and the RNA was used in the next selection step. At all steps, the binding conditions were adjusted to obtain ∼2 to 5% RNA retention.
DNA templates for in vitro RNA synthesis, Xenopus laevis oocyte microinjections, and RNA analysis.
The TBE was PCR amplified using the PN1 and PN2 primers containing SacII restriction sites and inserted into the SacII site of plasmid pBSAd1 (23). Radiolabeled RNA was transcribed from BamHI-digested DNA using T7 polymerase. U1ΔSm and U6Δss RNAs have been previously described (29). Xenopus oocyte injections and analysis of microinjected RNA by denaturing gel electrophoresis were performed as previously described (18, 29).
RNA binding assays.
For RNA competition binding, the reaction mixtures contained 2.5 ng of 32P-labeled SRV-1 CTE and unlabeled competitor CTE, TBE, or unselected genomic SELEX library RNA. Synthesis and purification of CTE RNA was as previously described (29). TBE and library RNAs were synthesized from DNA templates as described above. RNAs were incubated in 36 μl of binding buffer for 15 min at room temperature with recombinant GST-TAP protein (amino acids 61 to 610) that was immobilized on glutathione-Sepharose beads. The beads were washed three times with 500 μl of binding buffer, and the retained radioactivity was quantitated by Cerenkov counting. The competitor concentrations that reduced radioactive probe binding by 50% (50% inhibitory concentrations [IC50]) were determined.
RNA structure probing.
Transcription in vitro was performed with T7-MEGAshortscript kit (Ambion), in the presence of 3 mM guanosine. The transcripts were resolved on a denaturing 6% polyacrylamide gel, stained with Stains All dye (Fluka), and eluted with a solution containing 0.3 M sodium acetate (pH 5.2), 0.5 mM EDTA, and 0.1% (wt/wt) sodium dodecyl sulfate. After ethanol precipitation, the RNA was phosphorylated with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (NEB) under standard conditions. The labeled RNAs were purified on a denaturing 6% polyacrylamide gel and identified in the gel by autoradiography and recovered as described above. The unlabeled carrier RNA was added to the RNA solution to a final concentration of 1.25 μg/μl. The RNA was denatured in a solution containing 10mM Tris-HCl [pH 7.5], 10 mM MgCl2, and 40 mM NaCl, at 65°C, and allowed to renature at 25°C for 15 min. The reactions were carried out at 25°C for 10 min with the following treatments: S1 nuclease (312.5 U/ml; Pharmacia) in the presence of 1 mM ZnCl2, mung bean nuclease I (62.5 U/ml; Boehringer) in the presence of 1 mM ZnCl2, T2 ribonuclease (25 U/ml; Gibco BRL), T1 ribonuclease (62.5 U/ml; Pharmacia), lead acetate 0.35 mM; (Sigma). The reactions were stopped by mixing with equal volume of loading buffer (95% formamide–10 mM EDTA–dye) and frozen. The products of enzymatic and metal ion digestions were analyzed by polyacrylamide gel electrophoresis (6, 8, and 10% denaturing polyacrylamide) and autoradiography. The RNA products were run along with the products of formamide RNA hydrolysis and limited ribonuclease T1 digestion. The single-nucleotide ladders were generated by incubation of RNA with five-times-larger volumes of formamide–0.5 mM MgCl2 at 100°C for 9 min. The T1 nuclease ladder was obtained by digestion of denatured RNA in presence of a solution containing 50 U of enzyme per ml, 10 mM sodium citrate [pH 4.5], 0.5 mM EDTA, and 3.5 M urea at 55°C for 7.5 min.
Biocomputing.
Multiple sequence alignments, phylogeny, database similarity searches, and RNA folding were performed with the standard programs of the Genetics Computer Group (GCG) package. Construction of MEME (multiple expectation maximization for motif elicitation) profiles and the database searches were performed with MEME tools (2) implemented in GCG, using the default paramaters.
Nucleotide sequence accession number.
The sequence of clone no. 30 has been submitted to GenBank under accession no. AF260329.
RESULTS
Human genome encodes RNA elements that are selectively recognized by TAP(NXF1) protein.
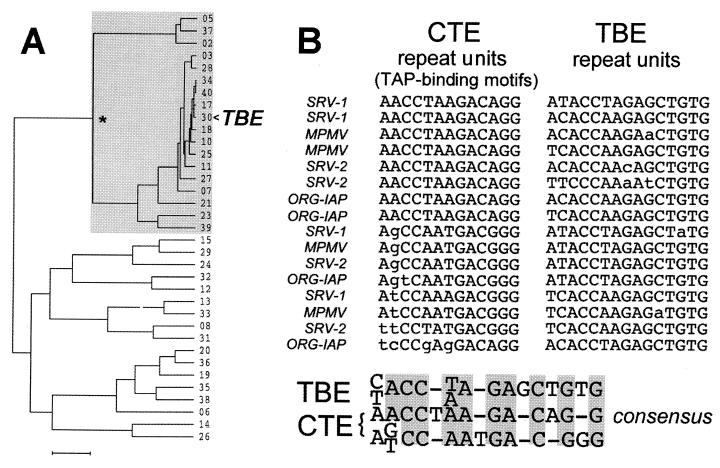
For the genomic SELEX search (13, 32), we constructed selection libraries from human genomic DNA. RNAs were subjected to successive rounds of protein binding, reverse transcription, and PCR amplification, following the procedures established for regular SELEX. As the protein bait, we used a recombinant protein spanning amino acids 61 to 610 of human TAP(NXF1) that was fused to a GST moiety. This protein was shown to bind to CTE with high affinity, like authentic TAP protein (14). After six rounds of binding and amplification, the library RNAs were sequenced and compared using multiple-sequence alignment. This analysis is illustrated in the dendrogram shown in Fig. 1A. We observed that about 50% of these RNAs had similar sequences and formed a distinct cluster in the dendrogram (Fig. 1A), whereas the others have divergent sequences. Thus, a single winning family was selected that we termed TBE. Further inspection revealed that these elements are encoded by a minisatellite containing from 2 to 19 copies of a 15-nucleotide (nt) imperfect direct repeat (Fig. 1B). Analysis of the remaining divergent sequences using a motif discovery algorithm (MEME) did not reveal detectable common features; they thus most likely represent low-affinity background binders, and they were not further addressed in this study. Figure 1B shows a sampling of 16 direct repeats from three representative TBE family members (clone no. 17, 30, and 34 in Fig. 1A). Since the TBE repeats are imperfect, several different variants are shown in Fig. 1B to illustrate this diversity. We chose clone no. 30 for further characterization because it most closely matches the TBE consensus (note its central position in the TBE cluster shown in Fig. 1A). This clone is 239 nt long and contains 16 direct repeats. Throughout this study, this sequence is referred to as TBE. After this work was completed, a related sequence (GenBank accession no. AC023524) which shares 92% homology with TBE, was identified on human chromosome 4.
FIG. 1.
Sequence analysis of TAP-binding elements. (A) Dendrogram illustrating the TAP-selected RNAs. After multiple-sequence alignment of 35 independent sequences (shown as clone numbers), a pseudophylogram was drawn using UPGMA (unweighted pair group method using arithmetic averages) and the default parameters. Bar, 100 substitutions per 100 residues. The node marked with an asterisk defines the TBE family (also indicated by grey shading), and the representative TBE (clone no. 30, GenBank accession no. AF260329) is shown by an arrowhead. (B) Multiple-sequence alignment of TAP-binding motifs in repeat units of CTE and repeat units of TBE. CTE motifs (11, 38) are from the published sequences of SRV-1 and SRV-2 (42), MPMV (33), and IAP-related retroelement ORG-IAP (37), as indicated to the left. TBE repeats were chosen from three representative variants of the family. The consensus sequences derived from the alignments are shown at the bottom. Two distinct consensus sequences were drawn for the CTE. The grey boxes show the conserved positions. Dashes indicate alignment gaps.
TBE and CTE share homologous sequence motifs that are arranged similarly in the secondary structure.
We asked whether TBE shares sequence homology with the prototype TBE, the CTE. A consensus was drawn from the alignment of TBE repeat units and compared to the consensus TAP-binding motifs of CTEs which had been identified previously using a combination of phylogenetic, biochemical, and structure-function studies (10, 11, 14, 20, 37, 38). This comparison revealed a compelling homology (Fig. 1B).
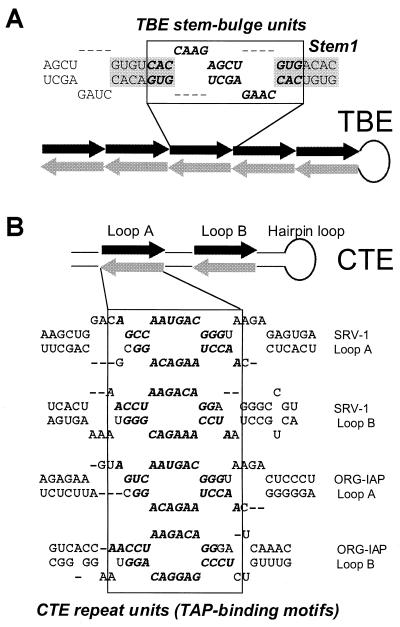
Since the secondary structure of the CTE is essential for its function, we also compared the secondary structures of the two elements. The CTEs of SRV-1 and a closely related MPMV have been studied in detail (10, 11, 19, 37, 38). They form an extended stem-loop structure with two internal loops (designated loops A and B [see Fig. 3B]). Each loop region contains two repeated TAP-binding motifs (see Fig. 3) that form mirror-symmetrical pairs and are partly single stranded (14, 37). These motifs and their arrangement are conserved between CTEs of different species (Fig. 1) and were shown to be essential for CTE function (37, 38). The secondary structures of the stem regions and the hairpin loop of the CTE (see Fig. 3) are also important for function, whereas the primary structures of these regions are not essential and are not conserved (37, 38).
FIG. 3.
The homologous repeat units of TBE (A) and CTE (B) form similar mirror-symmetrical pairs in the secondary structure. The juxtaposed repeats are shown by black and grey arrows in the schematic drawings, and by bold italics in the RNA sequences. Boxes that are aligned to the matching regions of the schematic drawing indicate the mirror-symmetrical pairs of repeats. The internal loops A and B and the hairpin loop of CTE are indicated. The secondary structures are shown for the portions of the CTE from SRV-1 (38) and of the IAP-related retroelement ORG-IAP (37), as indicated to the right. For TBE, a portion of the secondary structure model is shown, and its regions are marked as indicated for Fig. 2.
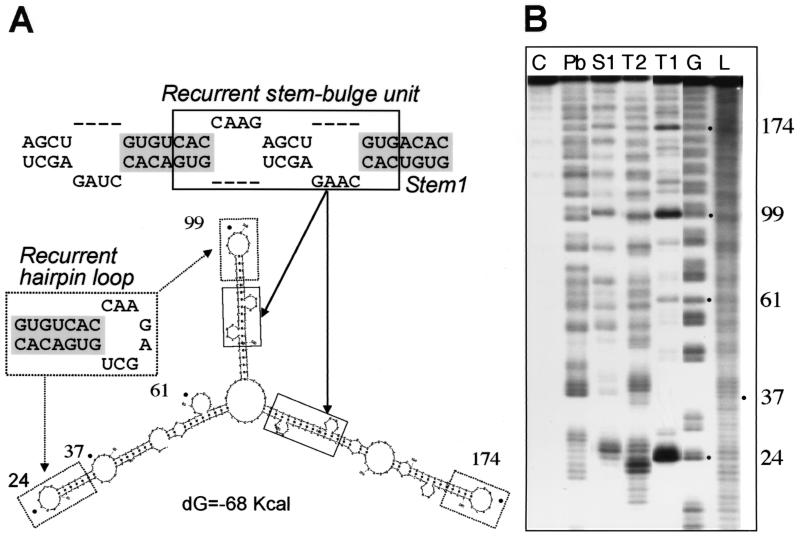
To study the secondary structure of TBE, we first compared the predicted foldings of six different members of this family. Due to the repeating nature of TBEs, all foldings included recurrent elements such as stem-bulge units (Fig. 2 and 3) that consisted of two mirror-symmetrical repeats (Fig. 3). These units formed extended structures that included a basic stem element, Stem1 (Fig. 2 and 3). Other recurrent elements included a hairpin loop (Fig. 2) and the bottom region of the stem (see Fig. 6A).
FIG. 2.
Enzymatic and chemical probing of TBE RNA structure. (A) A secondary structure model of TBE GenBank accession no. AF260329) showing the recurrent hairpin loop (dashed boxes), the recurrent stem-bulge unit (solid boxes), and the basic stem element, Stem1 (grey shading). (B) TBE RNA was radiolabeled on the 5′ end and treated with ribonuclease T1 or T2, S1 nuclease, or lead acetate (Pb), and the cleavage products were analyzed as described in Materials and Methods. The respective cleaving agents are indicated above each lane. The three remaining lanes contain results for the G sequencing reaction (G), untreated control RNA (C), and RNA nucleotide ladder (L), as indicated. The relevant nucleotide positions of the TBE are shown by dots and numbers to the right of the gel. The assignment was confirmed by numerous independent experiments.
FIG. 6.
Evolutionary relationship between CTE and TBE. (A) Conservation of neutral sequences between TBE and individual CTEs. Different portions of TBE secondary structure model are shown. The portions of the secondary structure models of CTEs from SRV-1, ERA-IAP, intracisternal A-particle-related retroelement ORG-IAP, and IAPE-related CTE-like sequence (CTE IAPE-10, accession no. U53819) (37) are shown, as indicated. Red letters denote the conserved TAP-binding motifs in CTEs, as shown in Fig. 1 and 2. The recurrent Stem1 structure of TBE and similar stem regions of CTEs are indicated by green shading. Shadings in the hairpin loop regions show the residues that are conserved between TBE and CTEs of SRV-1 (yellow) and ORG-IAP (blue). Italics show the conserved primary structure. Dashes indicate virtual gaps in RNA folding; the downward-pointing arrow indicates the start of stem 1. (B) TBE-related tandem repeats in ERA-IAP retroelement. At the right, the alignment of TBE and a region of the ERA-IAP LTR is shown. Arrows show the 15-nt tandem repeat units in ERA-IAP (R1 through R4). At the left, the alignment of the ERA-IAP repeat units and the consensus TBE repeat unit is shown. The conserved residues are shaded.
The predicted models were assessed by probing of TBE RNA (239 nt) with nucleases T1, T2, and S1 and lead acetate (Pb) (6, 22) (Fig. 2B), as well as mung bean nuclease and nuclease P1 (data not shown). We chose 5′ labeling as the detection method, because the use of internal primers and reverse transcription was precluded due to the repeated nature of TBE. Figure 2 shows that the RNA showed extensive protection from single-stranded cleavage, suggesting that it is predominantly double-stranded. We identified three major regions of single-strandedness that matched the predicted recurrent hairpin loops (Fig. 2) Other susceptible positions were found in the regions that were predicted to contain internal loops and bulges, as indicated in Fig. 2. In summary, the experimental data and the predicted structure were in good agreement. We note that five closely related structures were predicted that had different topologies but similar free energies of formation, whereas only one of them (Fig. 2) matched the experimental data.
We summarized these results in a generic secondary structure model made of consensus TBE sequence motifs (Fig. 3A). In this model, the motifs form mirror-symmetrical pairs in partially single-stranded regions. This arrangement is analogous to that of the repeated TAP-binding motifs in CTE (10, 11, 14, 20, 37, 38) (Fig. 3B).
In summary, we showed that TBE contains repeated sequence motifs that are homologous to the repeated TAP-binding motifs in CTE. In both elements, these motifs are also positioned similarly in the secondary structure. Since these features of CTE are crucial for its function, we concluded that their conservation in TBE may reflect functional similarities between the two elements.
TBE and CTE use the same binding sites on TAP(NXF1).
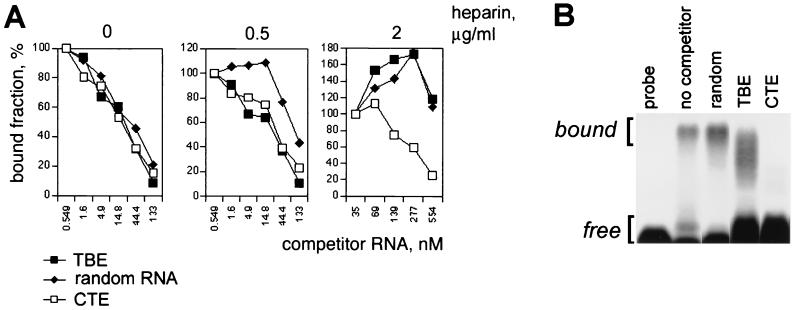
We examined whether CTE and TBE use the same binding determinants on TAP. The binding of TBE to TAP was studied in competition assays using radiolabeled SRV-1 CTE RNA in vitro (Fig. 4A). The nonselected genomic SELEX library RNA (random RNA) was used as a non-structure-specific control. The binding assays were performed using recombinant GST-TAP61-610 immobilized on glutathione-Sepharose. For each competitor RNA, the IC50 was determined. Under mild binding conditions (200 mM NaCl), CTE, TBE, and the nonselected library RNA competed for TAP with the similar IC50 of ∼10−8 M (Fig. 4A, left panel), suggesting that the respective binding determinants are overlapping. This result indicated that under these conditions TAP binds RNA in a non-structure-specific manner. However, at higher binding stringency (0.5 μg of heparin per ml), TBE and CTE competed equally well, whereas the nonselected library RNA competed poorly (IC50, ∼10−7 M) (Fig. 4A, middle panel). Taken together with the efficient selection of the TBE family by the genomic SELEX method, these data confirm that TBE binds to TAP in a structure-specific manner. At 2 μg of heparin per ml (Fig. 4A), the CTE RNA competed efficiently, whereas the TBE and the nonselected library RNA competed poorly, showing that TBE binds less efficiently than CTE. Under these conditions, the presence of poor competitors reproducibly led to an initial increase of CTE RNA binding (Fig. 4A, right panel). This effect could be due to complex stoichiometry of the reaction and was not further studied. In summary, the following order of affinity was observed: CTE > TBE > random RNA. Similar results were obtained using gel mobility shift assays (Fig. 4B). These results showed that TBE and CTE compete for binding to a shared determinant(s) on TAP in a sequence-specific manner. Taken together with the structural similarities between the two RNA elements, this finding led us to suggest that TBE and CTE are recognized by TAP in a similar fashion, although TBE has a lower affinity to TAP, as expected.
FIG. 4.
Competition for TAP binding between CTE and TBE. (A) Pulldown assays with GST-TAP61-610 were performed using 32P-labeled CTE of SRV-1 in the absence of heparin (left panel) or in the presence of 0.5 μg of heparin per ml (middle panel) or 2 μg of heparin per ml (right panel). CTESRV−1 RNA (open rectangles), TBE RNA (filled rectangles), and random RNA from the unselected genomic library (closed diamonds) were used as unlabeled competitors. The fraction of 32P-labeled CTE RNA bound to TAP was plotted against the final concentration of unlabeled competitor RNAs. (B) Gel mobility shift assays with GST-TAP61-610 were performed using binding conditions that were the same as those used to obtain the results shown as in panel A, in the presence of 0.5 μg of heparin per ml. The unlabeled competitor RNAs were present at 333 nM, as indicated. The complexes (bound) and the probe (free) were separated on 1% agarose gels and detected by autoradiography. Similar results were obtained in three independent experiments.
TBE is a functional RNA export element.
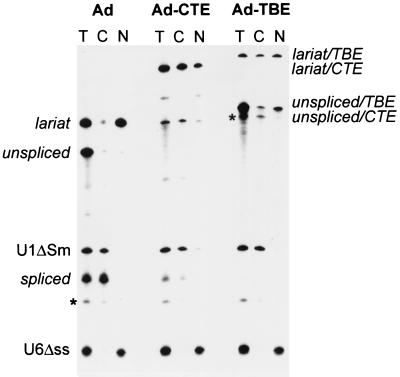
We tested the export ability of TBE from the X. laevis oocyte nuclei. When adenovirus pre-mRNA derivative is microinjected in the nuclei, it is spliced and the excised intron lariat is retained in the nucleus. However, if a nuclear export element such as CTE is present in the intron, the intron lariat is efficiently exported to the cytoplasm (26, 29). We therefore inserted TBE into Ad pre-mRNA intron and found that the presence of TBE led to efficient export of the intron lariat (Fig. 5). In parallel experiments, the empty Ad lariat was retained in the nucleus, whereas the CTE-containing lariat was exported efficiently (Fig. 5). We used U1ΔSm RNA as a positive internal control for nuclear export and U6Δss RNA as a negative control (29). As shown in Fig. 5, U1ΔSm is exported efficiently, whereas U6Δss is confined to the nucleus, confirming the quality of cells and nuclear injections. These data showed that TBE can act as a functional RNA export element, similarly to CTE (Fig. 5). We further compared the two elements using a cell culture assay based on the ability of strong elements like CTE to replace the Rev/RRE regulatory system of HIV (5, 37, 46). As a reporter, we used a Rev-dependent subgenomic mRNA of HIV-1 that produced Gag protein (p37gag reporter mRNA, [38]). When TBE was inserted into this mRNA, it did not activate gag expression, whereas in parallel experiment, the presence of SRV-1 CTE resulted in efficient activation (data not shown). In this assay, TBE scores like CTE mutants that have reduced affinity to TAP. Similarly to TBE, such CTE mutants can promote efficient export of Ad intron lariat in oocytes but are inactive in the p37gag assay in mammalian cells (14). We note that the p37gag assay is more stringent, because it measures the activation of the complete mRNA utilization pathway leading to protein production, whereas the intron lariat export assay only measures the activation of one metabolic step, the nuclear export. Collectively, our data show that TBE can function as an RNA export element in Xenopus oocytes but does not exhibit the full CTE function.
FIG. 5.
Nuclear export of excised intron lariats harboring TBE in Xenopus laevis oocytes. Xenopus oocyte nuclei were injected with a mixture of 32P-labeled U6Δss and U1ΔSm RNAs transcribed in vitro and the precursor RNAs as indicated on the top. RNA samples from total oocytes (T) or cytoplasmic (C) or nuclear (N) fractions were collected 3 h after injection. Products of the splicing reaction were resolved on 10% acrylamide–7 M urea denaturing gels. The positions of the mature products and intermediates of the splicing reaction are indicated. The asterisks indicate the positions of RNA molecules that are likely to be originated by degradation of the intron lariat and of the precursor RNA. Similar results were obtained in three independent experiments.
CTE and TBE are evolutionarily related.
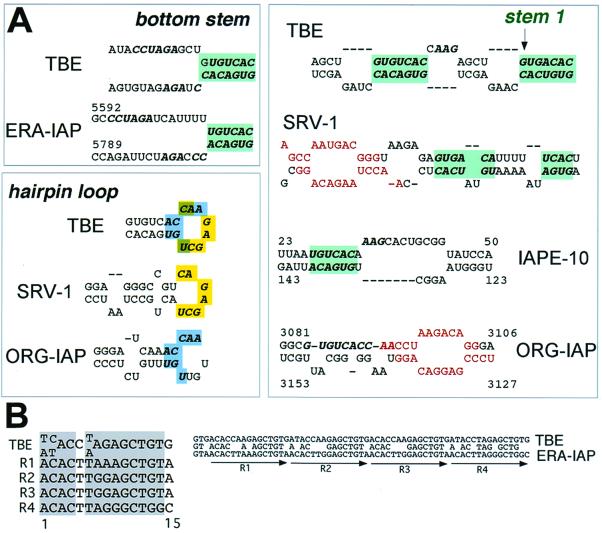
We demonstrated that CTE shares structural similarities with TBE in the conserved regions that are essential for its function (Fig. 2 and 3). In addition, we found that individual CTEs of different species contain sequence homologies to TBE in the regions that are not phylogenetically conserved (Fig. 6A). Some CTEs also contain secondary structure features similar to the Stem1 of TBE (Fig. 6A). The sequences of hairpin loops in some CTEs are conserved in the recurrent hairpin loop of TBE. The predicted bottom stem region of Eker rat-associated IAP (ERA-IAP) CTE (Fig. 6A) and a stem region of a murine CTE-like sequence (IAPE-10, GenBank accession no. U53819) show remarkable similarity to TBE, both in the primary and the predicted secondary structures (Fig. 6A). These similarity regions in CTEs are located outside the TAP-binding motifs (10, 11, 14, 20, 37, 38) (Fig. 6A). For the SRV-1 CTE, the primary structures of such regions were shown to be nonessential or neutral for function (37). The respective regions in other CTEs are also predicted to be neutral, both by analogy to the known secondary structure of SRV-1 CTE and by the lack of their sequence conservation among CTEs. We therefore concluded that the sequences of the CTE and TBE similarity regions shown in Fig. 6A are neutral for CTE function.
In order to assign a probability that these similarities are significant, we compared the nucleotide databases with sequence motif profiles of TBE, using the MEME algorithm (2). Remarkably, the TBE profile revealed two strong independent matches in IAPE CTEs (P value of ∼10−5 for each) corresponding to the two predicted complementary regions in IAPE-10 that form a Stem1-like structure (Fig. 6A). This analysis demonstrated that the similarities between IAPE-10 CTE and TBE shown in Fig. 6A are not likely to occur by chance. Collectively, these data indicated that several neutral structural features are conserved between TBE and CTEs.
By database comparisons with TBE-derived MEME profiles, the closest homologue of TBE (P, ∼10−19) among the known nucleotide sequences is the CTE-containing ERA IAP retroelement (43). The similarity is located in the ERA IAP long terminal repeat (LTR) region, 274 nt downstream of its CTE. This match (68% identity within the 79-nt overlap) includes four tandem 15-nt repeats that show high homology (13 of 15 residues conserved) to those found in TBE (Fig. 6B). Thus, besides the similarities in the bottom stem region of its CTE (Fig. 6A), the ERA IAP contains an apparent molecular fossil of TBE-related minisatellite in the vicinity of its CTE. We further detected a strong TBE homology (55% identity in 211-nt overlap; Z score = 155) (Fig. 7) in a solo IAP LTR (mouse IE36 insertion element, GenBank accession no. X05354 [24]). Taken together, these data suggest that the TBE similarity regions in type A LTRs were derived from TBE-related minisatellites.
FIG. 7.
Multiple-sequence alignment of TBE and the homologous sequences in ERA-IAP and IE36 retroelements. The positions that are identical in at least two sequences are shaded. The IE36 homology to TBE was identified using the SSearch program (Z score = 155).
In summary, the TBE shares several independent structural features with CTE-containing retroelements. Some functionally important aspects of CTE structure are conserved in TBE and likely account for the TBE recognition by TAP. In support of this idea, TBE and CTE were found to share functional activity and they bind to TAP in a similar fashion. Other conserved features are found in the regions where the primary structure is known or predicted to be neutral for CTE function. Therefore, their conservation points to a common ancestry rather than to shared functional or structural constraints. The finding of additional TBE-related sequences in ERA IAP and IE36 provides independent, direct evidence of a common ancestry for TBE-like repeats and the type A retroelements.
DISCUSSION
Simian type D retroviruses and rodent IAP retroelements are evolutionarily related (44). Although all known IAPs are replication incompetent, there is a subset of relatively intact IAP-related proviruses termed IAPE (28) that contain well-preserved CTE regulatory elements (37). We have previously described a functionally active CTE from the IAPE-related osteocalcin-related gene IAP (ORG-IAP) provirus and identified CTE elements in other known and putative retroelements (37). These CTEs share a highly conserved architecture, indicating that they are recognized by a common conserved factor. The type D CTE is a direct recognition site for cellular TAP(NXF1) protein that is conserved in eukaryotes, and the rodent TAP proteins have been identified. TAP(NXF1) is a general mRNA export factor that was proposed to associate with cellular mRNA either directly or through bridging proteins such as E1B-AP5 (1) and REF (34, 35). Therefore, the CTEs apparently evolved as direct, high-affinity TAP ligands. Since the evolution of many retroelements has likely included the acquisition of host genetic modules (7), we proposed that CTEs were derived from preexisting cellular TAP-binding elements. In support of this idea, TAP's ability to directly bind RNA is conserved across species and is therefore essential (14, 31, 39), pointing to the existence of cellular high-affinity RNA ligands. In order to identify such ligands, we performed a genome-wide search for RNAs that selectively bind to TAP in vitro. We chose the human genome because unlike these of rodents, it is not known to contain the endogenous CTE-containing retroelements. Here, we describe a human sequence termed TBE that has properties expected from a primitive CTE. Like CTE, TBE selectively binds to TAP and acts as an active signal of RNA nuclear export, indicating a remarkable degree of functional similarity. We demonstrate that this functional conservation is likely due to the common features of both the primary and the secondary structures of CTE and TBE elements. These features include the conserved sequence motifs and their analogous arrangement in the secondary structure. Both RNAs bind to TAP competitively and in the same fashion, further pointing to their shared structural and functional constraints. Additionally, we show that several modern CTEs contain TBE homologies in their functionally neutral regions, and that TBE-like repeats are present in the genome of a modern IAPE retroelement. Although the CTEs are far divergent from TBE, these similarities are sufficient to assign statistical probabilities to their significance. Here, we report four unbiased probabilities, each independently attesting to the nonrandom similarities between the TBE and the genomes of IAP retroelements. Taken together with the structural and functional conservation, these results lead us to suggest that CTEs evolved from TBE-like minisatellite repeats. This model implies that some ancestral TBE-like repeat units evolved into the internal loops of CTE, while the others comprised CTE's stem- and hairpin-loop structures (as shown in Fig. 6) or remained relatively intact outside the CTE (like the four repeats found in ERA IAP retroelement). In this scenario, the TBE-like repeats that had been acquired by an ancient retroelement provided a weak TAP-binding scaffold that, due to the selective pressure of viral replication, evolved into CTE, that is, a strong TAP-binding element able to mediate efficient export and expression of unspliced viral mRNA.
ACKNOWLEDGMENTS
We thank Elisa Izaurralde for performing the preliminary oocyte microinjection experiments as well as for the reagents and discussions, Susan Lindtner and Alexander Gragerov for discussions and critical suggestions, and Theresa Jones for editorial assistance.
REFERENCES
- 1.Bachi A, Braun I C, Rodrigues J P, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey T L, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 3.Bear J, Tan W, Zolotukhin A S, Tabernero C, Hudson E A, Felber B K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol Cell Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciesiolka J, Michalowski D, Wrzesinski J, Krajewski J, Krzyzosiak W J. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs J. Mol Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 7.Coffin J M, Hughes S H, Varmus H. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 8.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 9.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felber B K. Posttranscriptional control: a general and important regulatory feature of HIV-1 and other retroviruses. In: Meyers G, editor. Viral regulatory structures and their degeneracies. Proc Vol XXVIII. Reading, Mass: Addison-Wesley; 1998. pp. 101–122. [Google Scholar]
- 13.Gold L, Brown D, He Y, Shtatland T, Singer B S, Wu Y. From oligonucleotide shapes to genomic SELEX: novel biological regulatory loops. Proc Natl Acad Sci USA. 1997;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 15.Hope T J. The ins and outs of HIV. Rev. Arch Biochem Biophys. 1999;365:186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- 16.Hurt E, Straser K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem. 2000;275:8361–8368. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- 17.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–634. [PMC free article] [PubMed] [Google Scholar]
- 18.Jarmolowski A, Boelens W C, Izaurralde E, Mattai I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang Y, Bogerd H P, Yang J, Cullen B R. Analysis of the RNA binding specificity of the human tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- 20.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung J U, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 23.Konarska M M, Padgett R A, Sharp P A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 24.Man Y M, Delius H, Leader D P. Molecular analysis of elements inserted into mouse gamma-actin processed pseudogenes. Nucleic Acids Res. 1987;15:3291–3304. doi: 10.1093/nar/15.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 26.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy T R, Tang H, Xu W, Wong-Staal F. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene. 2000;19:3570–3575. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- 28.Reuss F U, Schaller H C. cDNA sequence and genomic characterization of intracisternal A-particle-related retroviral elements containing an envelope gene. J Virol. 1991;65:5702–5709. doi: 10.1128/jvi.65.11.5702-5709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 30.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singer B S, Shtatland T, Brown D, Gold L. Libraries for genomic SELEX. Nucleic Acids Res. 1997;25:781–786. doi: 10.1093/nar/25.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 34.Strasser K, Hurt E. Yralp, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stutz F, Bachi A, Doerks T, Braun I C, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 37.Tabernero C, Zolotukhin A S, Bear J, Schneider R, Karsenty G, Felber B K. Identification of an RNA sequence within an intracisternal-A particle element able to replace Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Virol. 1997;71:95–101. doi: 10.1128/jvi.71.1.95-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan W, Zolotukhin A S, Bear J, Patenaude D J, Felber B K. The mRNA export in C. elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF1 and S cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 41.Tang H, Kuhen K L, Wong-Staal F. Lentivirus replication and regulation. Annu Rev Genet. 1999;33:133–170. doi: 10.1146/annurev.genet.33.1.133. [DOI] [PubMed] [Google Scholar]
- 42.Thayer R M, Power M D, Bryant M L, Gardner M B, Barr P J, Luciw P A. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology. 1987;157:317–329. doi: 10.1016/0042-6822(87)90274-1. [DOI] [PubMed] [Google Scholar]
- 43.Xiao G H, Jin F, Yeung R S. Germ-line Tsc2 mutation in a dominantly inherited cancer model defines a novel family of rat intracisternal-A particle elements. Oncogene. 1995;11:81–87. [PubMed] [Google Scholar]
- 44.Xiong Y, Eickbush T H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Z, Luo M-J, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metaxoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 46.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]