Abstract
Cucumber necrosis virus (CNV) is naturally transmitted in the soil by zoospores of the fungal vector Olpidium bornovanus. Successful transmission requires that virus particles attach to the surface of zoospores prior to zoospore encystment on host roots. Mechanically passaged CNV was screened for mutants deficient in fungus transmission. We found six such mutants, exhibiting transmission efficiencies ranging from approximately 14 to 76% of that of wild-type (WT) CNV. Results of in vitro virus-zoospore binding assays show that each mutant binds to zoospores less efficiently than WT CNV (21 to 68%), suggesting that defects in transmission for these mutants are at least partially due to inefficient zoospore binding. Analysis of the structure of the CNV coat protein subunit and trimer indicates that affected amino acids in all of the mutants are located in the shell or protruding domain and that five of six of them are potentially exposed on the surface of the virus particle. In addition, several of the mutated sites, along with a previously identified site in a region of subunit-subunit interaction in the coat protein shell domain (M. A. Robbins, R. D. Reade, and D. M. Rochon, Virology 234:138–146, 1997), are located on the particle quasi-threefold axis, suggesting that this region of the capsid may be important in recognition of a putative zoospore receptor. The individual sites may directly affect attachment to a receptor or could indirectly affect attachment via changes in virion conformation.
Efficient transmission of the majority of plant viruses requires distinct invertebrate or fungal vectors. In most cases, transmission has been shown to be a highly specific process in which only certain vectors can transmit certain viruses (for reviews, see references 4, 6, 13, 14, 23, and 35). These observations suggest that virus particles as well as vectors contain specific sites that mediate their recognition. The coat protein (CP) of a plant virus has been shown to play an important role in transmission, and particular amino acids within the CP have been shown to be essential for this process (for reviews, see references 4, 6, 13, 14, 23, and 35). However, for the most part, the exact role of these amino acids in transmission including their potential role in vector attachment, is not known. Recent work with cucumber necrosis virus (CNV) has suggested that attachment of virions to vector zoospores is an important aspect of the transmission process (24).
CNV, a member of the genus Tombusvirus, is a 30-nm spherical virus with a monopartite positive-sense RNA genome (25). Transmission of CNV in nature occurs via zoospores of the Chytridiomycete fungus, Olpidium bornovanus (6, 9, 24). Zoospores and virus particles are released independently into the soil from the roots of infected plants. Virus is adsorbed onto the plasma membrane of zoospores and then enters into roots upon zoospore encystment. Studies of CNV transmission by O. bornovanus, and Olpidium transmission of several other small spherical plant viruses, have shown that the transmission process is highly specific (1, 6). For example, O. bornovanus transmits CNV but not Tobacco necrosis necrovirus (TNV), and conversely, O. brassicae transmits TNV but not CNV (10, 34). Moreover, different isolates of O. bornovanus transmit different viruses with varying efficiency (7), and different strains of TNV are transmitted with varying efficiency by the same O. brassicae isolate (17, 33, 34). Electron microscopy studies have shown that adsorption of virus to the zoospore plasmalemma is specific and reflects the virus-vector associations observed in nature (34). Together, these studies indicate the existence of a specific recognition mechanism between virus and vector zoospores.
Previous work has shown that the CNV CP contains determinants that specify its interaction with zoospores of O. bornovanus (20, 24). Reciprocal exchanges between the CP gene of CNV and that of the nontransmissible cherry strain of Tomato bushy stunt virus (TBSV) in infectious full-length cDNA clones showed that particles obtained from the TBSV genome containing the CNV CP were transmissible but particles from the CNV genome containing the TBSV CP were not. Also, a single amino acid mutation (Glu to Lys) in the CNV CP shell domain results in lowered transmission efficiency of CNV by O. bornovanus. In vitro binding studies demonstrated that this mutant bound zoospores less efficiently than CNV, indicating that specific regions of the CNV coat protein can mediate zoospore adsorption (24). In this study, we have isolated and characterized several distinct naturally occurring CNV transmission mutants. In each mutant, transmission deficiency was found to be due to a single amino acid substitution in the CNV CP. Moreover, each transmission mutant bound zoospores less efficiently than CNV, suggesting that the altered amino acids affect features of the CNV capsid involved in vector attachment.
MATERIALS AND METHODS
Isolation of CNV transmission mutants.
CNV transmission mutants were obtained essentially as described previously (24) except that cucumber cotyledons were used as the local lesion host for isolation of individual mutants.
Virus purification.
A miniprep procedure (24) was employed to partially purify CNV and CNV mutants for use in the initial screenings for transmission mutants. For all other experiments, virus was purified by differential centrifugation as follows. Infected leaves were ground in 2 volumes of 100 mM sodium acetate (pH 5.0) containing 5 mM β-mercaptoethanol and allowed to stand on ice for 30 to 60 min. The slurry was then filtered with Miracloth (Calbiochem) and centrifuged at 8,000 rpm in a GSA rotor. The supernatant was adjusted to 8% polyethylene glycol (molecular weight, 8,000; Sigma) and stirred at 4°C for 1 to 2 h. Virus was pelleted by centrifugation at 8,000 rpm in a GSA rotor, resuspended in 10 mM sodium acetate (pH 5.0), and subjected to high-speed centrifugation (40,000 rpm for 2.5 h in a Ti 50.2 rotor) at 4°C. Virus pellets were resuspended as before and centrifuged at 14,000 rpm in an Eppendorf microcentrifuge. The supernatant was collected and passed through a 0.2-μm-pore-size filter. Concentration of virus was determined spectrophotometrically. The concentration of virus purified by the miniprep procedure was determined by electrophoresis of several dilutions of virions through 1% agarose gels buffered in 45 mM Tris–45 mM borate, (pH 8.3) followed by ethidium bromide staining in buffer containing 1 mM EDTA.
Fungus transmission assay.
Purified virions were tested for transmission by O. bornovanus zoospores as previously described (5, 7, 20). Virus (1 μg) was incubated with 10 ml of zoospores (104/ml in 50 mM glycine, pH 7.6). After a 15-min acquisition period, the mixture was poured onto pots containing 12- to 16-day-old cucumber seedlings. Five days later, roots of cucumber seedlings were tested for the presence of virus by double-antibody sandwich (DAS) enzyme-linked immunosorbent assay (ELISA) using polyclonal antisera raised to CNV particles (20). Absorbance readings greater than fivefold over background (i.e., 0.1 at A405) were considered positive. Each transmission experiment included a wild-type (WT) CNV control, a test to determine any background level of CNV transmission in the absence of zoospores and a test for the presence of contaminating virus in zoospore preparations. Transmission in the absence of zoospores was not detectable in any of the experiments.
Cloning and sequence analysis of transmission mutants.
Double-stranded cDNA copies of the CP coding regions of transmission mutants were obtained by reverse transcription-PCR (RT-PCR) (30). The template was total RNA extracted from either infected leaves or purified virus particles. The plus-sense primer (CNV oligonucleotide 81, 5′AAGAGGTTGAATTCTGTCAGG3′) corresponded to CNV nucleotides 2148 to 2168 upstream up the CNV CP open reading frame (ORF) and included a unique EcoRI site (underlined). The minus-sense primer (CNV oligonucleotide 7, 5′TGTTCCCTAGCGTCGC3′) corresponded to the complement of CNV nucleotides 3854 to 3869 and lies downstream of the CP ORF. Following amplification, the RT-PCR product was digested with EcoRI and NcoI (both enzymes cut at regions flanking the CP ORF) and ligated into similarly digested pK2/M5, a full-length cDNA clone of WT CNV (26). The sequence of the transferred region of each transmission mutant was determined by cycle sequencing using dye-labeled terminators and AmpliTaq DNA polymerase FS (Perkin-Elmer Applied Biosystems). Samples were sequenced using an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer).
The double mutant LL5K8 was prepared by digestion of LLK8 with BglII and NcoI (which cleave at unique sites surrounding the LLK8 mutation) followed by insertion of the gel-purified fragment into BglII/NcoI-digested LL5 (24). The presence of both the LLK8 and LL5 mutations was verified by sequence analysis.
In vitro transcription and inoculation of plants.
Preparation of T7 polymerase runoff transcripts and inoculation of plants were as described previously (26).
In vitro binding assay.
The assay used was a modification of the one described by Robbins et al. (24). One hundred micrograms of purified virus was incubated with 5 × 105 O. bornovanus zoospores in 1 ml of 50 mM sodium phosphate buffer (pH 7.6) for 1 h. Following incubation, zoospores were pelleted by centrifugation at 5,000 rpm for 7 min in an Eppendorf microcentrifuge. The pellet was washed with 1.5 ml of binding buffer and then resuspended in sterile water. The zoospore pellet was assayed for the presence of virus by either Western blot or slot blot analysis using CNV monoclonal antibody 57-2 (24) and an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech). The quantity of virus in the pellet was determined by densitometric analysis of exposed film using the ImageQuant program (Molecular Dynamics). The amount of CNV that pelleted in the absence of fungus was subtracted from the amount of CNV that pelleted in the presence of fungus. Antibody 57-2 was confirmed to react equally to WT virus and mutants in slot blot analysis using denatured virus.
Homology modeling.
The three-dimensional coordinates of the CNV proteins were modeled after the published coordinates of TBSV, a virus with a known similar structure (PDB entry 2TBV) (22). The virus has icosahedral symmetry with three, independent quasi-equivalent structural positions, A, B, and C. Each protein was modeled after its cognate structural homolog with the program Modeler (29) on a Silicon Graphics computer (Silicon Graphics Inc., Mountain View, Calif.). Images of the modeled CNV subunit and trimer were manipulated using WebLab ViewerLite software (Molecular Simulations, Inc.). Surface representations were obtained using the “solvent surface” option. The CNV-TBSV alignment was from a multiple alignment using the CPs of several members of the Tombusviridae, including artichoke mottled crinkle virus (PIR2:S24926), carnation Italian ringspot virus (PIR2:S52718), cucumber leafspot virus (21), cymbidium ringspot virus (PIR1:VCVGCR), melon necrotic spot virus (PIR1:VCVEMN), pelargonium leaf curl virus (PIR1:A48355), pothos latent virus (SP_VI:Q84832), type TBSV PIR2:S07259, and the cherry strain of TBSV (PIR1:VCVGTB). The program Pileup (version 10.1; Genetics Computer Group) (8) was used to create the multiple alignment.
RESULTS
Isolation of transmission mutants from mechanically passaged CNV.
CNV was mechanically passaged 12 times through Nicotiana clevelandii, and individual local lesions were isolated following inoculation of cucumber cotyledons. The CP ORFs and flanking regions of six putative transmission mutants (as determined by reduced transmissibility [data not shown]) were amplified by RT-PCR and cloned in place of the WT CNV CP ORF in an infectious CNV cDNA clone. The cloned region was then sequenced to determined the presence of mutations. Transcripts of each of the clones were inoculated onto plants, and purified virus from infected plants was tested for transmissibility. Of 87 local lesions analyzed, 7 were ultimately found to contain virus with reduced transmission. Results of the transmission tests (Table 1) show that cloned mutants designated LLK8, LLK10, LLK63, LLK82, LLK84, and LLK85 were less transmissible than WT CNV (transmission efficiency, 96%). LLK8, LLK10, and LLK63 transmitted at lower efficiencies (i.e., 21, 27, and 14%, respectively), whereas LLK82, LLK84, and LLK85 transmitted at higher efficiencies (75, 50, and 76%). An uncloned mutant (LLK26) also transmitted with reduced efficiency (10%). Sequence analysis of LLK26 showed that it is identical to LLK8 (see below).
TABLE 1.
Transmission and in vitro binding efficiencies of CNV mutants
| Virus | Transmissiona | % Bindingb |
|---|---|---|
| WT CNV | 49/51 (96) | 100.0 |
| LLK00 | 10/10 (100) | 130.0 ± 8.5 |
| LLK8 | 3/14 (21) | 68.3 ± 17.5 |
| LLK10 | 4/15 (27) | 39.0 ± 22.3 |
| LLK26 | 1/10 (10) | ND |
| LLK63 | 3/21 (14) | 20.8 ± 17.2 |
| LLK82 | 15/20 (75) | 64.3 ± 24.11 |
| LLK84 | 10/20 (50) | 53.0 ± 7.5 |
| LLK85 | 19/25 (76) | 53.0 ± 12.3 |
| LL5K8 | 0/8 (0) | 21.7 ± 6.6 |
Number of pots showing transmission/number of pots tested. Values in parentheses indicate percentages of pots showing transmission. The data represent a compilation of at least three separate experiments for each virus.
Percentage of virus bound in an in vitro binding assay relative to the amount of WT CNV binding (average ± standard deviation of three separate experiments for each mutant). ND, not determined.
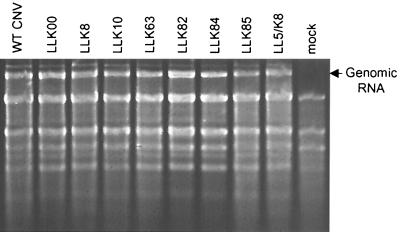
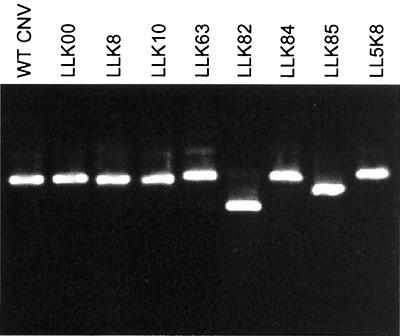
We wished to examine the infectivity and level of accumulation of each mutant in order to determine whether the reduced transmission efficiency was due to reduced ability of virus to accumulate in plants following transmission. LLK8, LLK10, LLK63, LLK82, LLK84, and LLK85 virions were inoculated onto N. clevelandii, and plants were monitored for symptoms and for RNA and virion accumulation. All mutants produced symptoms typical of WT CNV on N. clevelandii, resulting in large necrotic lesions on inoculated leaves and subsequent systemic necrosis and death of the plants within 10 to 14 days postinoculation (dpi) (data not shown). Agarose gel electrophoresis of total RNA extracts of infected plants at 3 dpi indicated that each mutant accumulated to approximately the same level as WT CNV (Fig. 1). In addition, in three separate experiments, DAS-ELISA of leaf extracts at 5 dpi indicated that, on average, virions of LLK8, LLK10, LLK63, LLK82, and LLK85 accumulated to approximately the same level as WT CNV (data not shown). LLK84 virions accumulated to approximately 50% of the WT CNV level. All mutants were also capable of infecting cucumber and produced equivalent-sized necrotic lesions on inoculated cotyledons (data not shown). In addition, virion accumulation in cucumber was monitored by agarose gel electrophoresis, and all mutants, including LLK84, accumulated to approximately the same level as WT CNV (data not shown). The integrity of virus particles used for transmission tests was also assessed. Virus particles of each of the transmission mutants were analyzed by agarose gel electrophoresis and found to migrate as discrete bands (Fig. 2). LLK8 and LLK10 particles comigrated with WT CNV, whereas particles of LLK63 and LLK84 migrated slightly slower than WT CNV and those of LLK82 and LLK85 migrated faster. The greater mobility of LLK82 and LLK85 particles is likely due to the higher net negative charge of the mutated CP (Gly to Glu and Asn to Asp, respectively [see below]). The basis for the slightly slower mobility of LLK63 and LLK84 is not known, but possibly these particles have a slightly expanded conformation, as previously suggested for the CNV LL5 transmission mutant (24). The ability of LLK8, LLK10, LLK63, LLK82, and LLK85 to accumulate to approximate WT levels in infected plants suggest that factors other than transmissibility do not likely contribute substantially to their reduced transmission frequencies. DAS-ELISA values for LLK84-infected leaves were approximately twofold less than that of WT CNV. As discussed below, it is possible that the lower accumulation of LLK84 may contribute to the lower transmission frequency of this mutant.
FIG. 1.
Agarose gel electrophoresis of total leaf RNA extracts from plants infected with CNV transmission mutants. N. clevelandii plants were inoculated with equal amounts of WT CNV or the indicated mutant virions, and total RNA was extracted from inoculated leaves 3 dpi. Equal amounts of total RNA were loaded onto a nondenaturing 1% agarose gel. The gel was stained with ethidium bromide.
FIG. 2.
Agarose gel electrophoresis of particles of CNV fungus transmission mutants. The indicated viruses (500 ng of each) were electrophoresed through a 1% agarose gel buffered in Tris-borate (pH 8.3). Virions were visualized by ethidium bromide staining in the presence of 1 mM EDTA.
Mutations in CNV transmission mutants map to either the CP shell or protruding domain.
Based on the structure of the related TBSV CP, the CNV CP contains three major structural domains: the R domain, which in the capsid faces the interior; the S domain, which forms the shell of the capsid; and the P domain, which projects outward from the capsid. The linear arrangement of these domains on the CNV CP as well as their predicted structures in the particle subunit and capsid are shown in Fig. 3D. The CP ORFs as well as flanking sequences used in the construction of cloned transmission mutants described above were sequenced to determine the location and nature of the mutations responsible for the reduced fungus transmission (Fig. 4). In addition to the unique mutations present in each clone, all transmission mutants also contained a T-to-G mutation at CNV nucleotide 2824, which results in a Phe-to-Cys change at amino acid position 66 in the CP arm domain, and a silent G-to-T mutation at nucleotide 3674 in the coding region of the CP protruding domain (LLK00 [Fig. 4]). These same mutations were noted in the previously described CNV transmission mutant LL5 (24), and studies ruled out any effect of the amino acid substitution in the arm domain mutation in the low transmission efficiency of LL5. In addition, these studies showed that the LL5 shell domain mutation was sufficient to induce the loss of transmissibility. To determine if the arm and protruding domain mutations together affect CNV transmission, particles produced from transcripts of a cDNA clone containing only these two mutations (LLK00) were tested for transmission. The results (Table 1) demonstrated that these mutations do not affect transmission efficiency. Subsequent sequence analysis of two other CNV clones from passaged virus showed that both mutations were present in both clones (data not shown). Therefore, it appears that these two mutations arose spontaneously following mechanical passage of the original full-length CNV cDNA clone and probably represent the predominant form of the WT transmissible virus from which subsequent transmission mutants arose. The following discussion of the transmission mutants is based on mutations unique to these viruses.
FIG. 3.
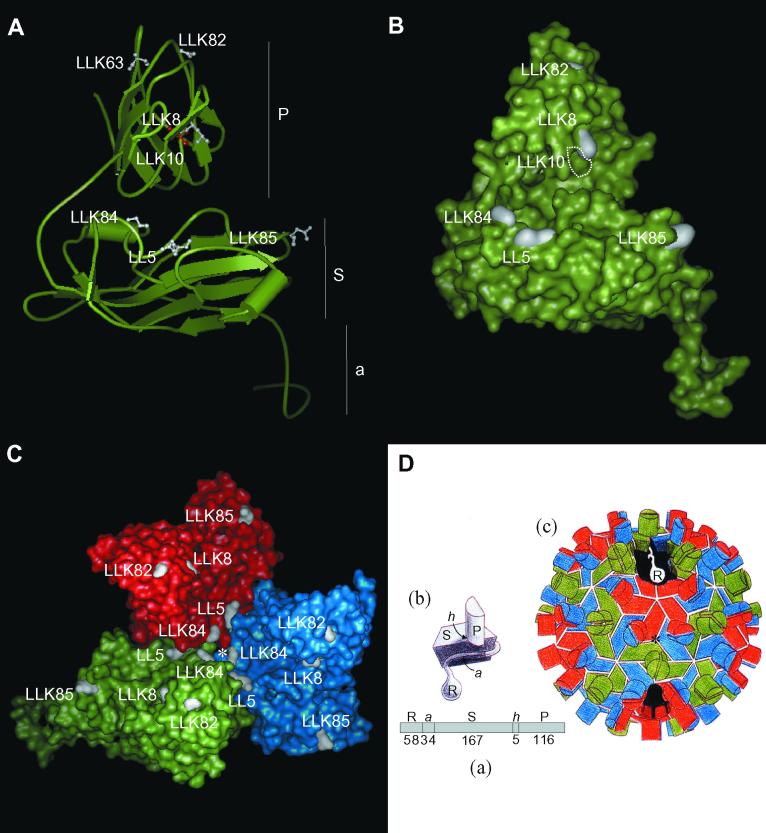
Locations of mutated amino acids on the CNV CP subunit and trimer in CNV transmission mutants. (A) Ribbon diagram of the homology modeled CNV CP subunit (subunit C) showing locations of mutated sites (in white in ball-and-stick form) in each of the transmission mutants. The mutated site in LLK10 is shown in red to distinguish it from the adjacent LLK8 mutation. Locations of the P, S, and a domains are indicated (see panel D for details). The disordered R domain is not shown. (B) Surface representation of the CNV CP subunit (subunit C) showing locations of mutated sites in white. The position of the buried LLK10 mutation is indicated by the white dotted lines. The LLK63 mutation is not visible in this orientation. (C) Surface representation of the CNV trimer (asymmetric unit) showing locations of mutated sites in each transmission mutant. The red, blue, and green areas correspond to the A, B, and C subunits. The asterisk shows the quasi-threefold axis of symmetry (D). (D) Diagrammatic representation of the structure of TBSV used for reference to the CNV structure. (a) Linear order of the different CP domains is shown along with the number of amino acids comprising each CNV domain (R, RNA binding domain; a, arm; S, shell domain; h, hinge; P, protruding domain). (b) Subunit structure with locations of indicated domains. (c) Particle structure with the A subunit in red, B in blue, and C in green. The cutaway section shows the region that the disordered R domain may occupy in the particle interior. (This diagram was adapted from reference 3).
FIG. 4.
Locations of mutations in CNV fungus transmission mutants. The portion of the mutant genome analyzed for mutation is shown. EcoRI and NcoI restriction enzyme sites used for cloning the mutant CP gene and flanking sequences are indicated for WT CNV. R, a, S, h, and P correspond to the different structural domains of the CNV CP (Fig. 3D). p92, p20, and p21 indicate flanking CNV ORFs. The two mutations present in the transmissible LLK00 and in all CNV transmission mutants are shown by asterisks and are described in detail for LLK00. Mutations in LL5 are also shown. LL5 was made by in vitro mutagenesis of our WT CNV infectious clone and does not contain the two substitutions present in the other mutants. Details of mutations including nucleotide position in the CNV genome, nucleotide change, amino acid position in the CNV CP, and amino acid change are given for each mutant.
Figure 4 shows that each transmission mutant (LLK8, LLK10, LLK63, LLK82, LLK84, and LLK85) contains a single amino acid substitution in the CP and that these occur in either the CNV CP shell or protruding domains; no amino acid changes were found in the R and arm domains, which are located in the particle interior. Two of the transmission mutants, LLK85 and LLK84, contain single amino acid substitutions in the shell domain, whereas the remaining transmission mutants contain single changes in the protruding domain (Fig. 4). As described above, mutants LLK26 and LLK8 contained identical protruding domain mutations.
Additional nucleotide substitutions that do not affect the CP amino acid sequence were found in LLK10, LLK82, and LLK85. In LLK10, two silent substitutions were found: one in the 3′-terminal region of CNV p92 ORF (the putative RNA-dependent RNA polymerase) (25) and the other in the arm region of the CNV CP ORF. LLK85 contained a silent substitution in the coding region of the CNV CP protruding domain. These mutations were not further investigated since they do not affect the protein sequence and are not present in areas of the genome which have known regulatory nucleotide sequences. LLK82 contains a T-to-C change in the core promoter for the subgenomic mRNA2 that encodes proteins involved in cell-to-cell movement (p21) and symptom induction (p20) (16, 26). However, as described above, several analyses of LLK82 accumulation levels failed to indicate that the T-to-C change affects virus accumulation (see above).
CNV transmission mutants show decreased binding to zoospores in vitro.
We have previously shown that CNV binds zoospores in vitro and that the CNV, transmission mutant LL5 shows reduced in vitro zoospore binding (24). These data suggested that the LL5 CP lacks an important determinant for attachment to zoospores. We wished to assess the possibility that reduced transmission of LLK8, LLK10, LLK63, LLK82, LLK84, and LLK85 is due to inefficient ability of mutant particles to bind zoospores. One hundred micrograms of each transmission mutant was incubated with 5 × 105 zoospores for 1 h, followed by low-speed centrifugation to pellet zoospores and washing to remove unbound or nonspecifically bound virus. The amount of bound virus in the pellet was determined by Western blot or slot blot analysis. Table 1 shows that each transmission mutant binds to zoospores less efficiently than WT CNV, with binding efficiencies ranging from approximately 21 to 68% of that of WT CNV. These results suggest that the reduced transmission of CNV mutants is at least partly due to their reduced abilities to attach to zoospores during the transmission process.
An artificial double mutant transmits and binds to zoospores at a lower efficiency than either of the individual mutants.
An artificial double mutant (LL5K8 [Fig. 4]) containing the mutations present in both LLK8 and the previously described LL5 mutant (24) was constructed and assessed for transmission. Table 1 shows that this mutant is less transmissible (0%) than either LLK8 (21% transmission) or LL5 (20% transmission) (24). Corresponding results were obtained in in vitro binding studies, i.e., LL5K8 binds zoospores less efficiently (22%) than either LLK8 (68%) (Table 1) or LL5 (50%) (24). When the double mutant was tested for its ability to infect and accumulate in N. clevelandii and cucumber, no substantial decrease in the level of RNA accumulation (Fig. 1) or particle accumulation as determined by ELISA (data not shown) was observed. In addition, particles appeared intact, as determined by agarose gel electrophoresis (Fig. 2). These results reinforce the role of both the LLK8 and LL5 mutations in the attachment and transmission processes.
DISCUSSION
We have isolated and characterized several naturally occurring CNV mutants deficient in transmission by O. bornovanus. Each mutant contains amino acid substitutions in the CP, reinforcing previous studies on the role of this protein in fungus transmission (20, 24). All of the CP mutations occurred in either the shell or protruding domain. These portions of the CP, unlike the R and arm domains, form the surface of the particle, which raises the possibility that the affected amino acids may serve as attachment sites for interaction of CNV with a putative zoospore receptor (see below).
In vitro binding studies showed that each transmission mutant bound to zoospores less efficiently than WT CNV (Table 1). These data suggest that zoospore binding plays an important role in transmission of these mutants, although other unidentified viral or host factors likely contribute to the transmission process.
All transmission mutants accumulated in cucumber to approximately the same level as WT CNV, indicating that virus particles are stable and that defects in transmission cannot be attributed to an inability of particles to accumulate in cucumber following transmission. With the exception of LLK84, which accumulated to approximately 50% of the WT CNV level, all transmission mutants also accumulated to WT CNV levels in N. benthamiana (Fig. 1 and 2). The basis for the slightly reduced accumulation of LLK84 in this host is not known, but considering the location of the LLK84 mutation in the trimer interface, it is possible that the particles are partly defective in assembly or disassembly. We note that accumulation data were taken from both inoculated and systemic tissue of infected N. benthamiana but only from inoculated leaves of cucumber. It is possible that the lower accumulation levels observed in N. benthamiana are due to decreased ability of LLK84 to move systemically.
LLK8 and LLK10 contain mutations corresponding to amino acids that are immediately next to each other in the linear structure of the CP P domain (amino acids 294 and 295, respectively [Fig. 4]). Amino acids from other mutants did not cluster on the primary CP structure. However, it was of interest to assess whether the other mutations clustered in the secondary or tertiary structure of the subunit or capsid and whether these sites are potentially exposed on the surface. To do this, homology modeling of the CNV CP subunit was conducted using the known high-resolution X-ray crystal structure of the related TBSV CP subunit (15). Figures 3A and B show ribbon and surface representations, respectively, of the modeled CNV subunit, and Fig. 3C shows a surface representation of the modeled CNV CP trimer (the asymmetric unit). The surface representation models predict that with the exception of LLK10, all of the mutated sites (including the previously identified site in LL5) are exposed on the surface of the subunit or trimer. In addition, six of seven of the mutated sites (i.e., LLK82, LLK8, LLK84, LL5, and LLK85) are preferentially located on one side of the CP subunit (Fig. 3B). Mutated amino acids in LLK8, LLK10, and LLK82 are all located on the outer wall of the protruding domain dimer, and those in LLK84 and LL5 are near each other in a region of subunit-subunit interaction in the trimer (Fig. 3A and B). The fact that the mutations map to distinct regions on the capsid is compatible with multiple mechanisms for transmission and binding defects. Nevertheless, the modeled CNV CP trimer predicts that most of the mutated sites (LL5, LLK8, LLK10, LLK82, and LLK84) are in or near a cavity formed by the trimer on the particle quasi-threefold axis. It is therefore possible that the trimer cavity represents an important site for recognition of a putative zoospore receptor. If these mutations disrupt binding to a receptor, it would suggest that the receptor has complementary symmetry. Alternatively, the affected amino acids in these mutants may affect subunit-subunit interactions and virion conformation, thereby indirectly affecting virion attachment and subsequent transmission. The slower electrophoretic mobility of mutants LL5 (24), LLK63, and LLK84 (and LL5K8) (Fig. 2) is consistent with the notion that reduced binding and transmission efficiencies may be due to conformational changes in particle structure as a result of the amino acid substitution. As stated above and shown in Fig. 3C, LLK84 and LL5 mutations lie in a region of subunit contact and could therefore affect subunit interactions. Similarly, the mutation in LLK63 lies in a region of protruding domain dimer interactions and could affect particle conformation by interfering with protruding domain contacts.
The mutation in LLK10 reduces transmission to about 27% of the WT CNV level and decreases binding to 39% as a result of a Val-to-Ala change at amino acid 295 in the CP protruding domain. This substitution lies immediately next to the mutated site in LLK8. The modeled CNV subunit does not predict that the affected LLK10 amino acid is exposed on the particle surface. It is possible that replacement of Val by Ala indirectly affects transmission and binding by changing the accessibility of other exposed amino acids in this region.
The structure of the shell domain of the tombusvirus CP subunit is similar to that of the picornavirus particle (27), which raises the question as to whether the putative zoospore attachment sites on CNV correspond to any of the known cellular receptor attachment sites on picornaviruses. In foot-and-mouth disease virus, an RGD motif in the G-H loop of VP1 has been implicated in receptor attachment (11, 19). Interestingly, the Gly-to-Val mutation in the CNV mutant LLK84 lies within the structurally analogous G-H loop and is located within an SGD triplet. Mutagenesis studies may help in the final identification of this region of the CNV capsid in zoospore attachment.
Virus attachment sites on animal viruses are for recognition of receptors that lie on host cells infected by the virus. Plant viruses do not recognize receptors for host cell attachment, but certain plant viruses are likely to possess attachment sites for recognition of the vector that transmits the virus to its host. In other cases, a virus-encoded helper factor is believed to mediate interaction between the vector and the virus particle (23). Specific virus attachment sites for cellular receptors have been identified for several animal viruses, including poliovirus, foot-and-mouth disease virus, and influenza virus (12, 28, 31). However, such sites have not yet been identified in plant viruses, despite their importance in the establishment and dissemination of plant virus disease. Specific regions of the capsid involved in transmission have been identified in several plant viruses (4, 6, 13, 14, 23, 32, 35), but to our knowledge no experiments have been conducted to determine if these sites are involved in the vector attachment stage of transmission. In tomato spotted wilt virus, an RGD motif has been identified in one of the viral structural proteins (18) and has been implicated but not proven to be involved in vector attachment. Our studies represent the initial stages of work that aims to identify features of virion architecture required for attachment to a vector. It is hoped that further work will provide information on evolutionarily conserved features of virus particles that are involved in receptor attachment. In addition, virus attachment mutants should aid in the identification of virus vector receptors about which very little is known.
ACKNOWLEDGMENTS
This work was partially supported by NSERC Operating Grant OGP0043840.
We thank Jack Johnson and Dave Theilmann for helpful comments on the manuscript. We also thank Ron Reade and Marjorie Robbins for many helpful discussions.
Footnotes
AAFC contribution no. 2101.
REFERENCES
- 1.Adams M J. Transmission of plant viruses by fungi. Ann Appl Biol. 1991;118:479–492. [Google Scholar]
- 2.Blanc S, Ammar E D, Garcia-Lampasona S, Dolja V V, Llave C, Baker J, Pirone T P. Mutations in the potyvirus helper component protein: effects on interactions with virions and aphid stylets. J Gen Virol. 1998;79:3119–3122. doi: 10.1099/0022-1317-79-12-3119. [DOI] [PubMed] [Google Scholar]
- 3.Branden C, Tooze J. Introduction to protein structure. New York, N.Y: Garland Publishing, Inc.; 1991. [Google Scholar]
- 4.Brown D J F, Robertson W M, Trudgill D L. Transmission of viruses by plant nematodes. Annu Rev Phytopathol. 1995;33:223–249. doi: 10.1146/annurev.py.33.090195.001255. [DOI] [PubMed] [Google Scholar]
- 5.Campbell R N, Lecoq H, Wipf-Scheibel C, Sim S T. Transmission of cucumber leaf spot virus by Olpidium radicale. J Gen Virol. 1991;72:3115–3119. doi: 10.1099/0022-1317-72-12-3115. [DOI] [PubMed] [Google Scholar]
- 6.Campbell R N. Fungal transmission of plant viruses. Annu Rev Phytopathol. 1996;34:87–108. doi: 10.1146/annurev.phyto.34.1.87. [DOI] [PubMed] [Google Scholar]
- 7.Campbell R N, Sim S T, Lecoq H. Virus transmission by host-specific strains of Olpidium bornovanus and Olpdium brassicae. Eur J Plant Pathol. 1995;101:273–282. [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias H F. Transmission of cucumber necrosis virus by Olpidium cucurbitacaerum Barr & Dias. Virology. 1970;40:828–839. doi: 10.1016/0042-6822(70)90128-5. [DOI] [PubMed] [Google Scholar]
- 10.Dias H F. The relationship between cucumber necrosis virus and its vector, Olpidium cucurbitacaerum. Virology. 1970;42:204–211. doi: 10.1016/0042-6822(70)90253-9. [DOI] [PubMed] [Google Scholar]
- 11.Fox G, Parry N R, Barnett P V, McGinn B, Rowlands D J, Brown F. The cell attachment site on foot-and-mouth-disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid) J Gen Virol. 1989;70:625–637. doi: 10.1099/0022-1317-70-3-625. [DOI] [PubMed] [Google Scholar]
- 12.Fry E E, Lea S M, Jackson T, Newman J W I, Ellard R M, Blakemore W E, Abu-Ghazaleh R, Samuel A, King A M Q, Stuart D I. The structure of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 1999;18:543–554. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray S M. Plant virus protein involved in natural vector transmission. Trends Microbiol. 1996;4:259–264. doi: 10.1016/0966-842X(96)10040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray S M, Rochon D M. Vectors of plant viruses. In: Granoff A, Webster R, editors. Encyclopedia of virology. Vol. 1. London, England: Academic Press; 1999. pp. 1899–1910. [Google Scholar]
- 15.Harrison S C, Olson A J, Schutt C E, Winkler F K, Brigogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature (London) 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 16.Johnston J C, Rochon D M. Deletion analysis of the promoter for the cucumber necrosis virus 0.9 kb subgenomic RNA. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 17.Kassanis B, MacFarlane I. Interaction of virus strain, fungus isolate, and host species in the transmission of tobacco necrosis virus. Virology. 1965;26:603–612. doi: 10.1016/0042-6822(65)90323-5. [DOI] [PubMed] [Google Scholar]
- 18.Kormelink R, de Haan P, Meurs C, Peters D, Goldbach R. The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J Gen Virol. 1992;73:2795–2804. doi: 10.1099/0022-1317-73-11-2795. [DOI] [PubMed] [Google Scholar]
- 19.Mason P W, Reider E, Baxt B. RGD sequence of foot-and-mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed by an antibody-dependent enhancement pathway. Proc Natl Acad Sci USA. 1994;91:1932–1936. doi: 10.1073/pnas.91.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLean M A, Campbell R N, Hamilton R I, Rochon D M. Involvement of the cucumber necrosis virus coat protein in the specificity of fungus transmission by Olpidium bornovanus. Virology. 1994;204:840–842. doi: 10.1006/viro.1994.1604. [DOI] [PubMed] [Google Scholar]
- 21.Miller J S, Damude H, Robbins M A, Reade R D, Rochon D M. Genome structure of cucumber leaf spot virus: sequence analysis suggests it belongs to a distinct species within the Tombusviridae. Virus Res. 1997;52:51–60. doi: 10.1016/s0168-1702(97)00105-6. [DOI] [PubMed] [Google Scholar]
- 22.Olson A J, Bricogne G, Harrison S C. Structure of tomato bushy stunt virus. IV. The virus particle at 2.9 Å resolution. J Mol Biol. 1983;171:61–93. doi: 10.1016/s0022-2836(83)80314-3. [DOI] [PubMed] [Google Scholar]
- 23.Pirone T P, Blanc S. Helper dependent vector transmission of plant viruses. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- 24.Robbins M A, Reade R D, Rochon D M. A cucumber necrosis virus variant deficient in fungal transmissibility contains an altered coat protein shell domain. Virology. 1997;234:138–146. doi: 10.1006/viro.1997.8635. [DOI] [PubMed] [Google Scholar]
- 25.Rochon D M, Tremaine J H. Complete nucleotide sequence of the cucumber necrosis virus genome. Virology. 1989;71:251–259. doi: 10.1016/0042-6822(89)90150-5. [DOI] [PubMed] [Google Scholar]
- 26.Rochon D M, Johnston J C. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology. 1991;181:656–665. doi: 10.1016/0042-6822(91)90899-m. [DOI] [PubMed] [Google Scholar]
- 27.Rossmann M G. The evolution of RNA viruses. Bioessays. 1987;7:99–103. doi: 10.1002/bies.950070302. [DOI] [PubMed] [Google Scholar]
- 28.Rossman M G. Viral cell recognition and entry. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A, Blundell T L. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Skehel J J, Wiley D C. Receptor binding and membrane fusion in virus entry: the influenza virus hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 32.Smith T J, Chase E, Schmidt T, Perry K L. The structure of cucumber mosaic virus and comparison to cowpea chlorotic mottle virus. J Virol. 2000;74:7578–7586. doi: 10.1128/jvi.74.16.7578-7586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teakle D S, Hiruki C. Vector specificity in Olpidium. Virology. 1964;24:539–544. [Google Scholar]
- 34.Temmink J H M, Campbell R N, Smith P R. Specificity and site of in vitro acquisition of tobacco necrosiss virus by zoospores of Olpidium brassicae. J Gen Virol. 1970;9:201–213. [Google Scholar]
- 35.Van den Heuvel J F J M, Hogenhout S A, van der Wilk F. Recognition and receptors in virus transmission by arthropods. Trends Microbiol. 1999;7:71–76. doi: 10.1016/s0966-842x(98)01434-6. [DOI] [PubMed] [Google Scholar]