Abstract
PiT-1 and PiT-2 are related multiple transmembrane proteins which function as sodium-dependent phosphate transporters and as the cell receptors of several oncoretroviruses. Two copies of a homology domain that is found in distantly related species assign these proteins to a large family of phosphate transporters. A current membrane topology model of PiT-1 and PiT-2 predicts 10 transmembrane domains. However, the validity of this model has not been addressed experimentally. We addressed this issue by a comprehensive study of human PiT-2. Evidence was obtained for glycosylation of asparagine 81. Epitope tagging showed that the N- and C-terminal extremities are extracellular. The orientation of C-terminal-truncation mutants expressed in cell-free translation assays and incorporated into microsomal membranes was examined by immunoprecipitation. Data were interpreted with respect to previous knowledge about retrovirus binding sites, to the existence of repeated homology domains, and to predictions made in family members. A model in which PiT-2 has 12 transmembrane domains and extracellular N- and C-terminal extremities is proposed. This model, which differs significantly from previous predictions about PiT-2 topology, may be useful for further investigations of PiT-2 interactions with other proteins and for the understanding of PiT-2 transporter and virus receptor functions.
Cell infection with amphotropic murine leukemia virus (A-MLV) is mediated by the binding of viral envelope glycoproteins to a cell surface receptor called PiT-2 (27, 52). PiT-2 is expressed in all tissues of all mammalian species, conferring susceptibility to A-MLV infection on all known mammalian cells, with the exception of certain hamster cells, like the CHO cell line, which produces soluble-factor-impairing envelope-receptor interactions (29, 30, 54). Gibbon ape leukemia virus (GaLV) and feline leukemia virus subgroup B (FeLV-B) use a receptor called PiT-1 (19, 32, 49), which is highly homologous to PiT-2 (27). Although PiT-1 is also widely expressed in mammalian tissues (51), receptors are functional for GaLV and FeLV infection in certain species only. The 10A1 murine leukemia virus is a natural variant of A-MLV that recognizes both PiT-2 and PiT-1 in human, feline, and simian cells (28, 57). The ability of PiT-1 and PiT-2 proteins to function as retrovirus receptors has been attributed to amino acid residues that differ between the receptors of the various species (6, 8, 15, 17, 20, 24, 34, 35, 46–48).
Independent of their capacity to mediate retrovirus binding and entry, PiT-1 and PiT-2 proteins serve as sodium-dependent phosphate (NaPi) transporters (21, 33, 55). This finding led to the description of mammalian type III NaPi transporters. In contrast with type I and type II transporters, which are expressed in specialized structures, like the brush border membranes of kidney and intestinal cells, and which contribute to phosphate homeostasis (4), type III transporters likely represent a general phosphate exchange system between the cells and the extracellular medium. PiT-1 and PiT-2 do not share homology with the members of the other NaPi transporter families.
PiT-1 and PiT-2 activities are modulated in response to variations in extracellular inorganic phosphate concentrations ([Pi]). Phosphate starvation increases PiT-1 and PiT-2 gene expression (10, 11) and induces posttranslational modifications of PiT-2 that activate both phosphate transport and retrovirus entry (36). It was previously shown that only a fraction of PiT-2 molecules that are present at the cell surface are capable of processing virus entry, suggesting that the receptor may exist in an activated or inactivated form (2). It was found that PiT-2 is associated with the actin cytoskeleton network and forms high-molecular-weight complexes (36). As both interaction with actin and high-molecular-weight complex formation appeared inversely related to [Pi] (C. Salaün, unpublished data), and thus to receptor activity, an attractive hypothesis is that they participate in posttranslational changes important for function. Investigations that are necessary to better understand these molecular interactions require an unambiguous model of the membrane organization of PiT-2.
A topological organization of PiT-1 and PiT-2 has been proposed based on hydropathy plots. The model supposed 10 transmembranes (TMs), internal N- and C-terminal segments, and a large central intracytoplasmic domain (27, 34, 52). With the exception of the two extracellular domains that are involved in virus-envelope binding (reviewed in reference 44) and of the large intracellular loop that is recognized by specific antibodies (10), the predicted topology of PiT-1 and PiT-2 has not been confirmed experimentally. We examined this issue by a broad analysis of PiT-2-related sequences available in databanks and by a comprehensive experimental approach combining a study of protein glycosylation, the fusion of tagging epitopes, and the in vitro translation of truncation mutants. We propose a model of PiT-2 topology in the plasma membrane which accounts for the experimental findings and for the organization of the protein in several homology domains. This model differs from the predictions based on hydropathy plots and points out regions potentially important for PiT-2 functions.
MATERIALS AND METHODS
Computational analysis.
Multiple-sequence alignment was obtained based on the domain database collected by Corpet et al. (12), which is accessible at the University of Toulouse (http://protein.toulouse.inra.fr/cgi-bin/ReqProdomII.pl). Aligned sequences included the following (numbers in parentheses correspond to amino acid numbers in the aligned sequences): Y630 METJA, Methanococcus jannaschii unknown protein (17 to 131, 168 to 285); O29467 ARCFU, Archaeoglobus fulgidus permease (13 to 134, 174 to 296); O58858 PYRHO, Pyrococcus horikoshii transporter (13 to 124, 176 to 262); O54523 AAAAA, Halobacterium halobium permease (18 to 145); Q50684 MYCTU, Mycobacterium tuberculosis transporter (79 to 135, 405 to 533); YG04 HAEIN, Haemophilus influenzae permease HI1604 (21 to 155, 268 to 400); Q17454 CAEEL, Caenorhabditis elegans phosphate permease (44 to 118, 352 to 476); Q17455 CAEEL, C. elegans P4 phosphate permease (44 to 118, 342 to 477); P91840 CAEEL, C. elegans W05H5 protein (55 to 184); Q17404 CAEEL, C. elegans F09G2.3 protein (40 to 172, 348 to 483); O18697 CAEEL, C. elegans C48A7.2 protein (19 to 164, 348 to 483); O45220 CAEEL, C. elegans BO331.2 protein (29 to 92, 348 to 483); Q60421 CRIGR, Cricetulus griseus (Chinese hamster) PiT-2 (20 to 153, 503 to 632); Q63488 RAT, Rattus norvegicus PiT-1 (20 to 153, 501 to 633); Q08357 HUMAN, Homo sapiens PiT-2 (20 to 153, 500 to 632); Q61609 MOUSE, Mus musculus PiT-1 (39 to 172, 531 to 662); Q60422 CRIGR, C. griseus PiT-1 (35 to 168, 528 to 660); Q08344 HUMAN, H. sapiens PiT-1 (35 to 168, 528 to 660); O97596 FELCA, Felis silvestris catus PiT-1 (39 to 172, 532 to 664); YB8I YEAST, Saccharomyces cerevisiae permease YBR29C/PHO89 (21 to 155, 414 to 521); PHO4 NEUCR, Neurospora crassa PHO-4 (21 to 157, 436 to 568); O58374 PYRHO, P. horikoshii permease PHO 640 (18 to 114, 255 to 390); O28476 ARCFU, A. fulgidus permease (14 to 150, 190 to 317); PITB ECOLI, Escherichia coli phosphate transporter (26 to 124); PITA ECOLI, E. coli phosphate transporter (26 to 124); Q50173 MYCLE, Mycobacterium leprae transporter (23 to 114); O06411 MYCTU, Mycobacterium tuberculosis transporter (23 to 114); O34436 BACSU, Bacillus subtilis transporter (23 to 109, 231 to 284); O30499 BBBBB, Rhizobium meliloti permease (23 to 112, 228 to 315); Q38954 ARATH, Arabidopsis thaliana permease (167 to 301, 430 to 567); P93264 MESCR, Mesembryanthemum crystallinum permease (161 to 295, 428 to 529); Q9Z7M4 BBBBB, Chlamydia pneumoniae permease (14 to 148, 276 to 406); O84698 CHLTR, Chlamydia trachomatis permease (14 to 148, 276 to 406); O34734 BACSU, B. subtilis YLNA protein (11 to 102, 175 to 300); O37913 METTH, Methanobacterium thermautotrophicum sodium-dependent phosphate transporter (14 to 140, 186 to 307); Q9ZJC8 BBBBB, Helicobacter pylori permease (59 to 192, 301 to 405); and O26024 HELPY, H. pylori HP 1491 permease (59 to 192, 301 to 405).
TM predictions were performed by PredictProtein (38), which is accessible at the European Molecular Biology Laboratory (Heidelberg, Germany) (http: //www.embl-heidelberg.de/predictprotein/predictprotein.html). Input sequences used by the network for prediction were Y630 METJA, YG04 HAEIN, PHO4 NEUCR, and YB8I YEAST for the entire human PiT-2 sequence and the same plus PITB ECOLI for the C-PD1131 sequence. The expected accuracy was 72% for both predictions. TM predictions were also performed using the DAS server (http://www.biomedi.su.se/-server/DAS) (37) and the TMHMM server (http://genome.cbs.dtu.dk/htbin) (45).
Cell lines, materials, reagents, and transfections.
Restriction enzymes and antiprotease Complete tablets were purchased from Boehringer Mannheim. The TNT quick-coupled transcription-translation system and canine pancreatic microsomes were from Promega. N-glycosidase F was from New England Biolabs. Pro-Mix 35S labeling mix, horseradish peroxidase-coupled and cy3-coupled secondary antibodies, and the ECL+ kit were from Amersham Pharmacia Biotech. The 9E10-cy3 monoclonal antibody (MAb) was from Sigma. The 3F10 and 12CA5 MAbs were from Boehringer Mannheim. pCDNA3 and pCDNA3.1 Myc-His plasmids were from Invitrogen. The anti-PiT-2 rabbit serum was a generous gift from S. Kuhmann and D. Kabat (Oregon Health Sciences University, Portland).
CHO-K1 cells were obtained from the American Type Culture Collection. Cells were grown in minimal essential medium, alpha medium (Gibco BRL) with 10% fetal calf serum (HyClone). Transient transfections were performed with the Lipofectamine PLUS reagent (Gibco BRL). Subconfluent CHO cells were incubated with 1 μg of plasmid DNA for 2 h, washed, and analyzed 24 h later.
Immunofluorescence on living cells.
Cells were seeded on slides and cultured overnight. Slides were washed with cold phosphate-buffered saline (PBS) and placed on a 25-μl drop containing 12CA5 (1:500 dilution in culture medium containing 20 mM HEPES) or 9E10-cy3 (1:250 dilution) for 1 h at 4°C. After three washes with cold PBS, cells were fixed in 3% formaldehyde for 20 min at 4°C and quenched with 50 mM NH4Cl in PBS for 10 min. Slides were placed on a 25-μl drop containing the cy3-coupled secondary antibody for 1 h at room temperature. Images were acquired using a Zeiss confocal fluorescence microscope and contrast enhanced using Adobe Photoshop 4.0 software.
Immunoblotting.
After electrotransfer of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels to nitrocellulose membranes using a Bio-Rad apparatus; membranes were blocked for 1 h in a solution containing 5% lowfat milk, 0.1% Tween 20, and PBS (PBS-milk); and the first antibody (anti-PiT-2, 1:2,000 dilution) was added for 1 h in PBS-milk at room temperature before the membranes were washed in 0.1% Tween–PBS. Horseradish peroxidase-coupled secondary antibodies were added for 1 h at room temperature. After washes in PBS-Tween 20, peroxidase activity was revealed using the ECL+ reagent and each membrane was exposed.
Plasmid construction.
The PiT-2V plasmid contains a tagged version of human PiT-2 in which a peptide from the vesicular stomatitis virus G protein has been fused to the C terminus of the receptor (36). PiT-2-M was constructed by inserting human PiT-2 cDNA in the pCDNA3.1 Myc-His B plasmid vector, leading to in-frame ligation of the 3′ extremity with sequences encoding a Myc-polyhistidine tag. This construct was then inserted downstream of a sequence encoding an influenza virus hemagglutinin (HA) epitope (MYPYDVPDYA) in the pCEP4-HA plasmid (a gift from S. Michelson), giving rise to the HA-PiT-2M plasmid.
C-terminally truncated mutants of human PiT-2 were constructed by PCR. Amplification products were digested by HindIII and EcoRI and inserted into pCDNA3.1 Myc-His. The PiT-2 open reading frame encompasses codons 1 to 84 (tL-2), 114 (tL-3), 138 (tL-4), 180 (tL-5), 213 (tL-6), 258 (tL-7), 529 (tL-8), 567 (tL-9), and 616 (tL-10). The PiT-2 sequences were immediately followed by in-frame codons encoding a Myc-His tag and a translation termination signal.
The glycosylation mutant N81V was constructed by site-directed mutagenesis (QuikChange; Stratagene) with the following primers: GGTATCATTGACGTGAACCTGTACGTAGAGACGGTGGAGACTCTCATGG and a complementary strand.
Cell-free transcription-translation, immunoprecipitation, and posttranslational analysis.
Cell-free transcription-translations were performed using the TNT quick-coupled system (Promega). Reactions were carried out for 90 min at 30°C in a final volume of 25 μl containing 20 μl of rabbit reticulocyte lysate, 1 μg of plasmid DNA, and 10 μCi of Pro-Mix 35S label, with or without 2 μl of canine pancreatic microsomes. For glycosylation studies, vesicles were ultracentrifuged in a TL-100 rotor for 15 min, washed once, and resuspended in 25 μl of PBS before analysis by SDS-polyacrylamide gel electrophoresis. For immunoprecipitations, translation products were diluted in 150 μl PBS or PBS–0.5% Triton X-100 and incubated for 2 h with 0.5 μg of the 9E10 MAb. Magnetic beads coated with anti-mouse immunoglobulins (Igs) were then added and carefully washed with PBS using a magnetic tube holder. Recovered material was solubilized in loading buffer supplemented with 2% β-mercaptoethanol or in a solution containing 1% Triton X-100, 0.5% SDS, 1% β-mercaptoethanol, 1% NP-40, and 50 mM sodium phosphate (pH 7.5), and the material was incubated for 1 h at 37°C with 1,000 U of protein N-glycosidase F (PNGase F). N-glycosidase F treatment was similarly performed on cell lysates and on in vitro translation products.
RESULTS AND DISCUSSION
Homology domains and predicted TM segments in human PiT-2.
The sequences of human (32, 52), rat (27, 50), mouse (19, 42, 56), and hamster (9, 57) PiT-1 and PiT-2, as well as that of feline PiT-1 (39), have been determined. A search of the ProDom database (12) identified several domains within these proteins (Fig. 1A). They include domains specific for the mammalian PiT-1 and PiT-2 proteins, such as the N- and C-terminal extremities and a large central region, and domains for which homologies exist with nonmammalian proteins. These homology domains are referred to as PD1131 and PD7717 in the ProDom database.
FIG. 1.
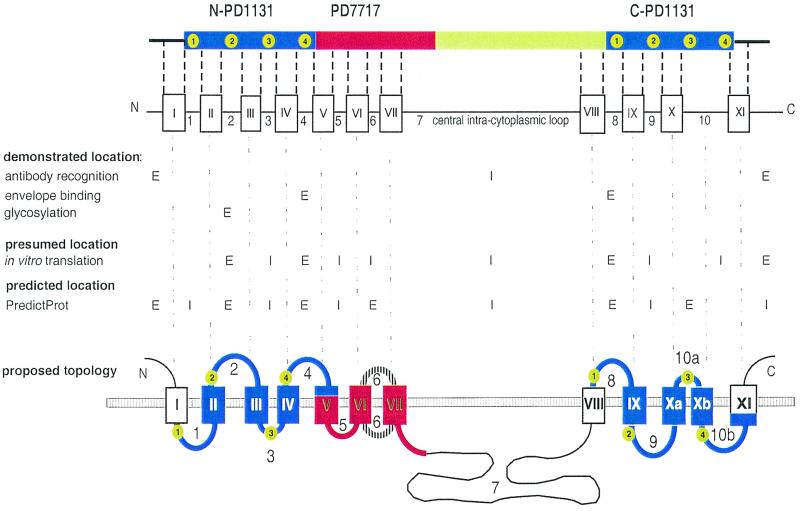
Human PiT-2 homology domains and predicted membrane organization. (A) Various domains of human PiT-2. The amino- and carboxy-terminal copies of homology domain PD1131, which can be recognized in the 37 members of the PiT family, are shown as blue boxes. The homology domain 7717, which is not found in bacteria or plants, is shown in red. Green boxes indicate domains that are found in mammalian PiT-1 and PiT-2 sequences only. They include the amino- and carboxy-terminal extremities and the large hydrophilic central domain. (B) Alignment of human PiT-2 N- and C-terminal PD1131 homology domains. Identical residues found at the same position are indicated by a bar. Residues shown in green boxes correspond to amino acids conserved at these positions in more than 85% of aligned PD1131 sequences (n = 51 to 68, depending on the location). Conserved residues define four conserved blocks, indicated in green circles. Regions predicted as potential TM domains are shown in bold blue letters. Numbering is that of the human PiT-2 sequence. (C) Alignment of sequences of 11 PD7717 homology domains. Sequence identifiers (SEQ ID), organisms of origin (org.), the start (st.) and the end of the aligned sequences, and the weight (w.) of the sequence in the alignment are indicated. Sequences are as specified in Materials and Methods. Hydrophobic amino acids are boxed in yellow. Blue lines indicate TM regions that are predicted in all shown sequences. They correspond to predicted TM-5, TM-6, and TM-7 in human PiT-2. (D) Location of human PiT-2 regions (shown as open boxes with roman numerals I to XI) which are predicted to form TM domains when the PredictProtein algorithm is run on the entire human PiT-2 sequence. Arabic numerals between boxes refer to loops (L-1 to L-10). Numbers in green boxes within PD1131 homology domains refer to highly conserved amino acid blocks.
Domain PD1131 is found in prokaryotes, fungi, yeasts, and plants. PD1131 homology defines the PiT family of proteins according to a classification proposed by the Transport Commission (41) (http://www.biology.ucsd.edu/∼msaier); the PiT family is also called the Pho-4 family (SwissProt). This family currently includes 37 members. In most members, including mammalian PiT-1 and PiT-2, PD1131 is duplicated so that proteins contain amino- and carboxy-terminal copies of PD1131 (hereafter referred to as N-PD1131 and C-PD1131, respectively). A total of 68 PiT-related sequences is available in databases. In addition to PiT-1 and PiT-2, low-affinity phosphate or sulfate transporter activity has been demonstrated for the related proteins E. coli PITA and PITB (18), B. subtilis YLNA-CysP (25), R. meliloti OrfA-Pit (1), A. thaliana Pht2;1 (13), N. crassa PHO-4 (53), and S. cerevisiae YBR29C/PHO89 (26).
Alignments of the 68 PD1131 homology domains revealed four blocks of highly conserved amino acids. Figure 1B shows the alignments of N-PD1131 and C-PD1131 of human PiT-2 (amino acids 130 and 131, respectively) and the locations of these blocks. The block 1 sequence is GΦNDΦ (amino acids 25 to 29 and 503 to 507 of human PiT-2), where Φ is a hydrophobic amino acid. Among the 68 aligned PD1131 sequences, GΦ was found in 67, ND was found in 61, and the second Φ was found in 58. In block 2, the conserved sequence is GxxxxGxxVxxT, where x is any amino acid (amino acids 58 to 69 and 547 to 558 in human PiT-2). G, G, V, and T were found at these positions in 67, 64, 60, and 61 of the 68 aligned PD1131 sequences, respectively. The block 3 sequence is PΦS (amino acids 111 to 113 and 591 to 593 of human PiT-2). It was found at this location in 67 of the 68 members. Amino acids forming block 4 (IxxxWΦ; 142 to 147 and 622 to 627 of human PiT-2) appeared at these locations in 44 (I) and 51 (WΦ) of the 51 PD1131 available sequences covering this region. In addition, other amino acids conserved between N-PD1131 and C-PD1131 were regularly found at a conserved location in more than 50% of the 68 aligned sequences. These motifs were not found in the sequences of type II NaPi transporters. Figure 1B also shows regions of human PiT-2 that correspond to domains for which a TM segment is predicted by the PredictProtein algorithm (38) (see Materials and Methods) in most members exhibiting the conserved amino acid blocks. Interestingly, blocks of highly conserved amino acids appear adjacent to predicted TMs. A high degree of conservation among distant species suggests that these regions may be important for transporter function. Similar conclusions were previously drawn from a study of the PiT family member Pht2;1, a low-affinity phosphate transporter of Arabidopsis (13).
In PiT-1 and PiT-2, domain PD7717 is located between N-PD1131 and the central intracellular loop. This 127-amino-acid domain is conserved in animals, worms, yeasts, and fungi, but it is absent in plants and bacteria. Multiple-sequence alignments showed a high frequency of conserved hydrophobic residues, which represent 80% of the 80 N-terminal amino acids of PD7717 in human PiT-2 (Fig. 1C). A high frequency of leucine, with several LxxL repeats, can be recognized. However, this domain is not predicted to form coiled coils. The PredictProtein algorithm indicates three potential TM segments, the locations of which are fully conserved among 11 aligned PD7717 sequences.
A prediction of potential TM regions was performed on the entire human PiT-2 sequence using the PredictProtein algorithm. Eleven TM segments were predicted at the following amino acid locations: TM-I, 11 to 25; TM-II, 45 to 62; TM-III, 90 to 106; TM-IV, 115 to 130; TM-V, 143 to 165; TM-VI, 185 to 199; TM-VII, 217 to 236; TM-VIII, 483 to 501; TM-IX, 534 to 552; TM-X, 572 to 590; and TM-XI, 625 to 642. TM segments assign 10 intramolecular loops, which we referred to as L-1 to L-10, plus the N- and C-terminal extremities. The predicted organization of human PiT-2 can be described as shown in Fig. 1D. The N-terminal domain consists of the first 22 amino acids, including 15 residues of TM-I. N-PD1131 encompasses the rest of TM-I, L-1, TM-II, L-2, TM-III, L-3, TM-IV, L-4, and 4 amino acids of TM-V. PD7717 encompasses the rest of TM-V, L-5, TM-VI, L-6, TM-VII, and 47 amino acids of L-7. The so-called large central domain contains the rest of L-7 and TM-VIII. C-PD1131 encompasses L-8, TM-IX, L-9, TM-X, L-10, and 10 amino acids of TM-XI. The C-terminal extremity consists of the rest of TM-XI up to amino acid 652.
Interestingly, predictions made using the PredictProtein algorithm about potential TMs in C-PD1131 differed depending on whether the analysis accounted for the entire human PiT-2 sequence or for the isolated human C-PD1131 sequence. The latter analysis predicted an additional TM at positions 594 to 609 (THCKVGSVVAVGWIRS), as shown in Fig. 1B. Although the reliability index was lower for this TM than for the other predicted TMs, a similar prediction was made for almost every N-PD1131 and C-PD1131 domain when analyzed apart from the protein in which they are contained. Other TM prediction algorithms, such as TMHMM (45) and DAS (37), performed similarly. Thus, predictions led to two alternative models of human PiT-2 organization, assuming either 11 TM segments when the entire sequence was considered (as shown in Fig. 1D) or 12 TM segments when PD1131 domains were considered separately.
Experimental investigations of human PiT-2 TM organization. (i) Glycosylation of asparagine 81.
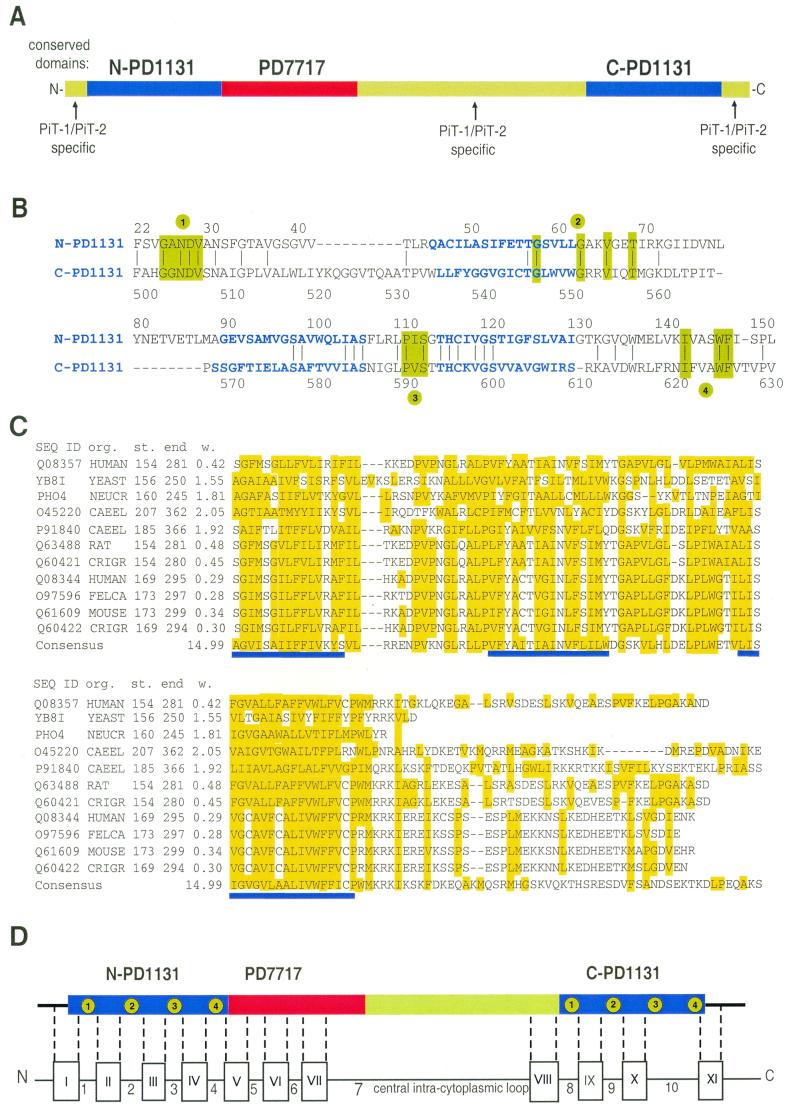
The currently used 10-TM model of PiT-2 topology predicts that all potential N-linked glycosylation sites (residues 81, 328, and 383) would be located in intracellular domains (27, 34, 52). Thus, according to this prediction, PiT-2 should not bear N-linked oligosaccharide chains. We examined this issue in CHO cells by expressing human PiT-2. Cell extracts were analyzed by Western blotting using a rabbit antiserum directed against the intracellular loop L-7 (Fig. 2A). A signal at 70-kDa was visible in naive CHO cells as well as in CHO–PiT-2 cells. Although the size of this signal is consistent with hamster PiT-2, we presume that it is nonspecific background, as, in contrast with PiT-2 signals, migration was not affected by heat (data not shown) (10). A 73-kDa human PiT-2–specific signal was detected in CHO–PiT-2 cells (Fig. 2A, PiT-2.gly+). The digestion of cell extracts with PNGase F, which removes N-linked oligosaccharide chains, induced a shift of this signal to 68-kDa species (Fig. 2A, PiT-2.gly−). This shift indicated that PiT-2 carries N-linked oligosaccharide chains. Similar observations were made with the tagged versions of human PiT-2 that are described below (data not shown). Since asparagines 328 and 383 are located in the intracellular L-7 segment, asparagine 81 appeared to be the only potential candidate for N-linked glycosylation. Site-directed mutagenesis was performed on this residue, such that asparagine was changed to valine (PiT-2.N81V). Expression of PiT-2.N81V in CHO cells conferred susceptibility to A-MLV infection (data not shown). Western blotting performed on CHO cells expressing PiT-2.N81V showed a signal at 68 kDa, the migration of which was not modified by treating cell extracts with PNGase F (Fig. 2A). Wild-type human PiT-2 and PiT-2.N81V proteins were also synthesized in vitro using reticulocyte lysates (Fig. 2B). The synthesis of wild-type human PiT-2 in the absence of microsomal vesicles produced a nonglycosylated protein migrating at 68 kDa (Fig. 2B, PiT-2.gly−). The addition of microsomes allowed for glycosylation and produced a 73-kDa species (Fig. 2B, PiT-2.gly+) that was susceptible to PNGase F digestion. Glycosylation was not observed for the PiT-2.N81V mutant. We concluded from these experiments that PiT-2 is a glycoprotein that carries an N-linked oligosaccharide on asparagine 81. Since this amino acid is located in L-2, we concluded that L-2 is located at the cell surface.
FIG. 2.
Glycosylation of human PiT-2 on asparagine 81. (A) Western blot analysis of cell extracts using rabbit serum directed against the central intracytoplasmic domain (L-7) of human PiT-2 (hPiT-2). Extracts were prepared from naive CHO cells (−), CHO–PiT-2 cells (wild type), and CHO cells expressing a mutant human PiT-2 in which asparagine 81 was changed to valine (N81V) and treated (+) or not treated (−) with an enzyme removing N-linked oligosaccharide (PNGase F). (B) Analysis of radiolabeled in vitro translation products. In vitro translation products of human PiT-2 (wild type) and the glycosylation mutant (N81V) were synthesized in either the presence (+) or absence (−) of microsomal vesicles and treated (+) or not treated (−) with PNGase F. Signals corresponding to glycosylated (PiT-2.gly+) and deglycosylated (PiT-2.gly−) species are indicated. Molecular mass markers are in kilodaltons.
(ii) Tagging of the N- and C-terminal extremities of human PiT-2.
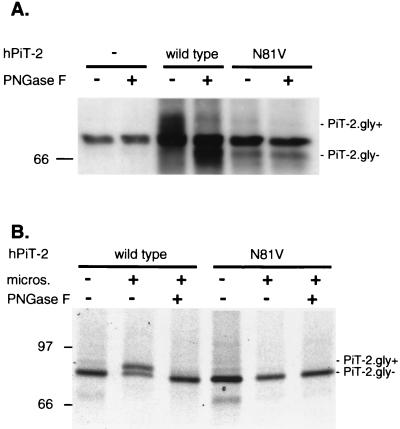
It was previously reported that an antigenic epitope fused at the C-terminal extremity of PiT-2 could be detected at the cell surface (36). This observation was made using a tag derived from the vesicular stomatitis virus G envelope glycoprotein. In order to confirm this result with a different tag and to extend the observation to the N-terminal extremity, human PiT-2 was fused to an HA tag at the N-terminal extremity and to a c-Myc tag at the C-terminal extremity (HA-PiT-2M). The construction was expressed in CHO cells, conferring A-MLV envelope binding and susceptibility to A-MLV infection (data not shown). Living cells were incubated at 4°C either with the anti-HA MAb 12CA5 or with the anti-Myc MAb 9E10-cy3, then fixed, and stained with fluorescent anti-IgG. Incubation at 4°C prevented receptor internalization. Both the HA and the c-Myc signals were detected at the cell margin (Fig. 3). As a control, cells transfected with an expression vector for an HA-tagged version of the intracellular Rho protein were not stained unless the plasma membrane was permeabilized (data not shown). These results indicate that the N- and C-terminal extremities of human PiT-2 are accessible to antibodies at the cell surface. Thus, both the N- and C-terminal extremities of epitope-tagged human PiT-2 fusion protein are extracellular.
FIG. 3.
Detection of N- and C-terminal tags at the surface of living cells. Naive CHO cells or CHO cells expressing human PiT-2 bearing an N-terminal fused influenza virus HA tag and a C-terminal fused c-Myc tag (CHO-HA-PiT-2M) were grown on glass slides and incubated at 4°C for 1 h with either a MAb against the HA tag (anti-HA 12CA5) or a MAb against the Myc tag (anti-Myc 9E10-cy3). Cells were then washed at 4°C and fixed before incubation with a fluorescent anti-mouse IgG secondary antibody.
(iii) In vitro translation of human PiT-2 C-terminal-truncation mutants.
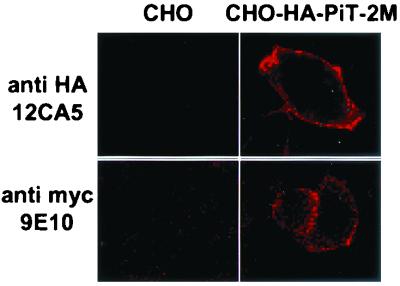
C-terminal-truncation mutants of PiT-2 were constructed with a c-Myc tag fused at the C-terminal extremity. Proteins were synthesized in vitro using rabbit reticulocyte lysates. The addition of microsomal vesicles allowed for processing, glycosylation, and incorporation into membranes. The presence of the C-terminal tag of truncation mutants at the surface of microsomal vesicles was examined with an aim to determine their orientation in the membrane. Tags were detected by incubating in vitro translation products with the anti-Myc 9E10 MAb. Vesicles were disrupted either before the addition of the antibody, giving access to both intra- and extravesicular epitopes, or after the addition of the antibody, which could bind to extravesicular tags only. Immune complexes were precipitated and analyzed for the presence of glycosylated products. As only a fraction of the translation products was incorporated into vesicles, analysis of glycosylated material allowed a focus on processed products that had been actually incorporated into vesicles. Thus, immunoprecipitation of glycosylated species indicated that the mutant protein was oriented with its C-terminal extremity at the surface of the vesicles, whereas failure to immunoprecipitate glycoslylated species indicated that the mutant protein was oriented with its C-terminal extremity inside the vesicles.
Truncations were performed according to the 11-TM prediction model, by inserting a termination codon at the junctions of L-2 and TM-III (tL-2), L-3 and TM-IV (tL-3), L-4 and TM-V (tL-4), L-5 and TM-VI (tL-5), L-6 and TM-VII (tL-6), L-7 and TM-VIII (tL-7), L-8 and TM-IX (tL-8), L-9 and TM-X (tL-9), and L-10 and TM-XI (tL-10). Wild-type receptors bearing a c-Myc tag fused at the C-terminal extremity provided a control. A prediction of the potential TM region was performed on the sequences of the full-length tagged and truncated tagged versions of PiT-2. The number and the location of predicted TMs were consistent with predictions performed on the sequence of the full-length nontagged protein.
The results of this experiment are shown in Fig. 4. Glycosylated products of tL-2 translation were accessible to 9E10 only after the disruption of the vesicles. Thus, the C-terminal tag of glycosylated tL-2 species was inside the vesicles. In contrast, the glycosylated products of tL-3 translation present in intact vesicles were accessible to 9E10. Thus, the C-terminal tag of glycosylated tL-3 species was at the surface of the vesicles. Analysis of truncation mutants revealed that the C termini of proteins truncated in L-3, L-5, L-6, L-7, L-9, and L-10 were accessible at the vesicle surface, whereas the C-terminal extremities of proteins truncated in L-2, L-4, and L-8 and of the full-length receptor were located inside the vesicles. Accounting for the fact that the surface of the vesicles is equivalent to the inner face of the plasma membrane, these results confirmed the observations made by other methods. Noticeably, they were consistent with the extracellular localization of L-2, L-4, L-8, and the C-terminal extremity, as well as with the intracellular localization of L-7. Additionally, they suggest that L-3, L-5, L-6, L-9, and L-10 may be intracellular.
FIG. 4.
Immunoprecipitation of in vitro translation products of human PiT-2 C-terminal-truncation mutants. Plasmids encoding C-terminally truncated forms of human PiT-2 and bearing a C-terminal Myc-His tag were transcribed in vitro, and transcription products were translated in a rabbit reticulocyte lysate assay in the presence of 35S-labeled methionine and cysteine and of microsomal vesicles. Vesicles were then either disrupted by a detergent (Triton X-100) (+) or kept intact (−). A MAb directed against the Myc tag (9E10) was added for 4 h at 4°C, and immune complexes were precipitated by binding to antimouse magnetic beads. After elution from beads, translation products were treated (+) or not treated (−) with PNGase F, an enzyme removing N-linked oligosaccharides. C-terminal truncations were performed at the C-terminal extremities of the predicted L-2, L-3, L-4, L-5, L-6, L-7, L-8, L-9, and L-10 loops (see Materials and Methods and Fig. 1D). A control reaction was performed with wild-type human PiT-2 (wild type). Signals corresponding to glycosylated (gly+) and deglycosylated (gly-) species are indicated. The deduced orientation of C-terminal-truncation mutants in vesicles is indicated. Molecular mass markers are in kilodaltons.
A widely accepted model states that polytopic protein topogenesis occurs by coupling membrane translocation to translation through the sequential action of alternating anchor and stop transfer sequences (5). According to this model, TMs sequentially translocate across the membrane as they emerge from the ribosome, and each topogenic determinant acts independently. Thus, shortened constructions of proteins would not behave differently from full-length molecules. However, this model is not universally applicable. Evidence has been provided that TM segments wait in close contact with the Sec61α and TRAM proteins of the translocon until the remainder of the protein is synthesized and released from the ribosome (7, 14). Interactions between neighbor TMs can occur during this transient stage preceding membrane insertion and affect topogenesis (23, 43). In those cases, C-terminally truncated mutants may behave differently from full-length molecules. Thus, the intracellular localization of L-3, L-5, L-6, L-9, and L-10 could be presumed from the translation of truncation mutants, but it was not unambiguously demonstrated.
Proposed topology of human PiT-2.
Two regions have been recognized as important for the interaction of PiT-1 or PiT-2 with their cognate retroviral envelopes. Amino acid 522 (8, 17, 20) and a surrounding region of 13 amino acids in L-8 (15, 35), often referred to as region A, together determine recognition by GaLV, A-MLV, or FeLV-B envelopes. This region is a presumed binding site for the VRA receptor-binding domain of the envelopes (46, 47). The other important region for envelope recognition is located in L-4, between amino acids 132 and 142 (34). This region is presumably a binding site for the VRB receptor-binding domain of the envelopes (46, 47). These published data unambiguously indicate that L-4 and L-8 are extracellular.
Figure 5 summarizes the available data about the TM organization of human PiT-2. The combination of data from envelope binding, antibody recognition, and glycosylation studies allows us to firmly designate the N terminus, L-2, L-4, L-8, and the C terminus as extracellular domains and L-7 as an intracellular loop. Thus, TM-VIII crosses the membrane. Assignment of other loops and TM segments is more speculative, as it involves the consideration of data from in vitro translation of truncated mutants, from secondary-structure prediction algorithms, and from the organization of PiT-2 as various homology domains.
FIG. 5.
Proposed model of human PiT-2 topology. The various domains of human PiT-2 and the predicted TMs and loops are shown in the top panel. The expected extracellular (E) or intracellular (I) localization of the N- and C-terminal extremities and of each predicted loop is indicated in the middle panel, according to the various investigation methods. The bottom panel shows the proposed topology of human PiT-2. Blue segments are the N-PD1131 and C-PD1131 homology domains, in which numbers in green circles indicate the conserved amino acid blocks. The red segment is PD7717. As the orientation of loop 6 is still unrevealed, this segment appears as a striped line. Black lines and open boxes correspond to segments for which homologous sequences were found in mammals only.
The TM organization of the N-PD1131 domain can be established with relative certitude. Since the N terminus and L-2 are extracellular and both TM-I and TM-II are highly liable to cross the membrane, L-1 is likely intracellular. L-3, which is located between extracellular L-2 and L-4, has only eight amino acids (FLRLPISG). Truncation in L-3 suggests that this segment is intracytoplasmic. However, this result does not exclude the possibility that L-3 is entirely located within the membrane.
We consider it reasonable to assume that the N- and C-terminal copies of the PD1131 homology domain fold similarly. Consequently, both the N-PD1131 and C-PD1131 domains should have three TM segments, as predicted for several members of the PiT family and shown when PD1131 sequences are analyzed apart from the protein in which they are contained (Fig. 1B). According to this hypothesis, TM-X should be split into two TM segments, TM-Xa (amino acids 572 to 586) and TM-Xb (amino acids 594 to 609), separated by a short loop (L-10a; amino acids 587 to 593). Subsequently, the former loop L-10 becomes loop L-10b (amino acids 610 to 621) (Fig. 5). This hypothesis is consistent with the presence of an asparagine (N587) and a proline (P591) in L-10a, which could promote the formation of a short loop between TM-Xa and TM-Xb. Indeed, these amino acids are strong turn promoting residues with a high propensity to induce helical hairpin formation in long hydrophobic stretches (31).
However, this model implies that N-PD1131 and C-PD1131 have inverted topologies. Indeed, loops L-2, L-4, and L-8, which are certainly extracellular, are, respectively, homologous to loops L-9, L-10b, and L-1, which are most likely intracellular; loop L-3, which is likely to be intracellular, is homologous to loop L-10a, in which the presence of N587 and P591 suggests the formation of an extracellular kink (31, 40). An inverted topology has been previously proposed for the N- and C-terminal homologous domains Pht2;1 of Arabidopsis (13), which are homologous to the PD1131 domains. The description of a dual orientation of ductin (16) suggests that similarly folded proteins may adopt either one or the other orientation in the lipid bilayer. It is important to consider that each loop in PD1131 bears highly conserved amino acid blocks, which are likely adjacent to the membrane or even partly embedded in the lipid bilayer. This may be especially relevant to L-3 and L-10a, which contain the PΦS motif that was found at this location in 67 out of 68 PD1131 sequences. It is very likely that the presence of conserved sequences at these locations is important for transporter functions.
PD7717 is a highly hydrophobic domain. As L-4 is extracellular and L-5 is likely intracellular, TM-V probably crosses the membrane. L-6 could not be assigned with certitude. All prediction algorithms assign L-6 outside the cell. Our in vitro translation data suggest that the L-6–TM-VII junction is intracellular. However, this result may be a consequence of the truncation of the molecule in loop L-6, leading to abnormal topogenesis. Attempts to solve this issue by the use of protease digestion, insertion of a glycosylation site, or insertion of a tagging epitope could not relieve the ambiguity. Hydrophobicity conferred by PD7717 may be important for its interactions with the intracellular actin network (36). Hydrophobicity of envelope-receptor complexes is also believed to be a crucial feature for processing the membrane fusion step of virus entry, as documented for the influenza virus and human immunodeficiency virus models (3, 22).
Altogether, these data suggest that PiT-2 would possess 12 TM domains. They point out regions that could be important for the function and the organization of the molecule. Conserved amino acid blocks in PD1131 are candidate targets for mutagenesis aimed at determining residues important for the phosphate transport activity. The large intracytoplasmic loop L-7, which bears potential phosphorylation sites, may be important for the regulation of this activity. The highly hydrophobic PD7717 domain could be important for determining the quaternary structure of the receptor. This model should be useful for designing further investigations of the involvement of these various domains in the fusogenic reaction that mediates retrovirus entry.
ACKNOWLEDGMENTS
We are grateful to S. Kuhmann for the generous gift of rabbit anti-PiT-2 serum and to D. Kabat for exciting discussions and help.
This work was supported by grants from the Agence National de Recherche contre le SIDA (ANRS) and Sidaction. P.R. is currently a fellow of Fundação para a Ciência e Tecnologia (Portugal), and C.S. is currently a fellow of the Ministère de l'Enseignement Supérieur et de la Recherche.
C.S. and P.R. contributed equally to this work.
REFERENCES
- 1.Bardin S D, Voegele R T, Finan T M. Phosphate assimilation in Rhizobium (Sinorhizobium) meliloti: identification of a pit-like gene. J Bacteriol. 1998;180:4219–4226. doi: 10.1128/jb.180.16.4219-4226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini J-L, Rodrigues P, Müller R, Danos O, Heard J M. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J Virol. 1996;70:4387–4393. doi: 10.1128/jvi.70.7.4387-4393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentz J. Membrane fusion mediated by coiled coils: a hypothesis. Biophys J. 2000;78:886–900. doi: 10.1016/S0006-3495(00)76646-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biber J, Custer M, Magagnin S, Hayes G, Werner A, Lotscher M, Kaissling B, Murer H. Renal Na/Pi-cotransporters. Kidney Int. 1996;49:981–985. doi: 10.1038/ki.1996.139. [DOI] [PubMed] [Google Scholar]
- 5.Blobel G. Intracellular protein topogenesis. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borel A, Simon S. Biogenesis of polytopic membrane proteins: membrane segments assemble within translocation channels prior to membrane integration. Cell. 1996;85:379–389. doi: 10.1016/s0092-8674(00)81116-2. [DOI] [PubMed] [Google Scholar]
- 8.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudry G J, Farrell K B, Ting Y-T, Schmitz C, Lie Y S, Petropoulos C J, Eiden M V. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J Virol. 1999;73:2916–2920. doi: 10.1128/jvi.73.4.2916-2920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien M-L, Foster J L, Douglas J L, Garcia J V. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J Virol. 1997;71:4564–4570. doi: 10.1128/jvi.71.6.4564-4570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien M L, O'Neill E, Garcia J V. Phosphate depletion enhances the stability of the amphotropic murine leukemia virus receptor mRNA. Virology. 1998;240:109–117. doi: 10.1006/viro.1997.8933. [DOI] [PubMed] [Google Scholar]
- 12.Corpet F, Gouzy J, Kahn D. The ProDom database of protein domain families. Nucleic Acids Res. 1998;26:323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M. Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell. 1999;11:2153–2166. doi: 10.1105/tpc.11.11.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do H, Falcone D, Lin J, Andrews D W, Johnson A E. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- 15.Dreyer K, Pedersen F S, Pedersen L. A 13-amino-acid Pit1-specific loop 4 sequence confers feline leukemia virus subgroup B receptor function upon Pit2. J Virol. 2000;74:2926–2929. doi: 10.1128/jvi.74.6.2926-2929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunlop J, Jones P C, Finbow M E. Membrane insertion and assembly of ductin: a polytopic channel with dual orientations. EMBO J. 1995;14:3609–3616. doi: 10.1002/j.1460-2075.1995.tb00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elvin C M, Dixon N E, Rosenberg H. Molecular cloning of the phosphate (inorganic) transport (pit) gene of Escherichia coli K12. Identification of the pit+ gene product and physical mapping of the pit-gor region of the chromosome. Mol Gen Genet. 1986;204:477–484. doi: 10.1007/BF00331028. [DOI] [PubMed] [Google Scholar]
- 19.Johann S V, Gibbons J J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johann S V, van Zeijl M, Cekleniak J, O'Hara B. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J Virol. 1993;67:6733–6736. doi: 10.1128/jvi.67.11.6733-6736.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leikina E, Chernomordik L V. Reversible merger of membranes at the early stage of influenza hemagglutinin-mediated fusion. Mol Biol Cell. 2000;11:2359–2371. doi: 10.1091/mbc.11.7.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Xiong X, Helm A, Kimani K, Bragin A, Skach W R. Co- and posttranslational translocation mechanisms direct cystic fibrosis transmembrane conductance regulator N terminus transmembrane assembly. J Biol Chem. 1998;273:568–576. doi: 10.1074/jbc.273.1.568. [DOI] [PubMed] [Google Scholar]
- 24.Lundorf M D, Pedersen F S, O'Hara B, Pedersen L. Amphotropic murine leukemia virus entry is determined by specific combinations of residues from receptor loops 2 and 4. J Virol. 1999;73:3169–3175. doi: 10.1128/jvi.73.4.3169-3175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansilla M C, de Mandoza D. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology. 2000;146:815–821. doi: 10.1099/00221287-146-4-815. [DOI] [PubMed] [Google Scholar]
- 26.Martinez P, Persson B L. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:628–638. doi: 10.1007/s004380050776. [DOI] [PubMed] [Google Scholar]
- 27.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller D G, Miller A D. Inhibitors of retrovirus infection are secreted by several hamster cell lines and are also present in hamster sera. J Virol. 1993;67:5346–5352. doi: 10.1128/jvi.67.9.5346-5352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller D G, Miller A D. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monné M, Hermansson M, von Heijne G. A turn propensity scale for transmembrane helices. J Mol Biol. 1999;288:141–145. doi: 10.1006/jmbi.1999.2657. [DOI] [PubMed] [Google Scholar]
- 32.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunne K J, Sass P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;3:119–127. [PubMed] [Google Scholar]
- 33.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 34.Pedersen L, Johann S V, van Zeijl M, Pedersen F S, O'Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen L, van Zeijl M, Johann S V, O'Hara B. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J Virol. 1997;71:7619–7622. doi: 10.1128/jvi.71.10.7619-7622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodrigues P, Heard J M. Modulation of phosphate uptake and amphotropic murine leukemia virus entry by posttranslational modifications of PIT-2. J Virol. 1999;73:3789–3799. doi: 10.1128/jvi.73.5.3789-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rost B, Sander C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 39.Rudra-Ganguly N, Ghosh A K, Roy-Burman P. Retrovirus receptor PiT-1 of the Felis catus. Biochim Biophys Acta. 1998;1443:407–413. doi: 10.1016/s0167-4781(98)00241-3. [DOI] [PubMed] [Google Scholar]
- 40.Sääf A, Hermansson M, von Heijne G. Formation of cytoplasmic turns between two closely spaced transmembrane helices during membrane protein integration into the ER membrane. J Mol Biol. 2000;301:191–197. doi: 10.1006/jmbi.2000.3933. [DOI] [PubMed] [Google Scholar]
- 41.Saier M H, Eng B H, Fard S, Garg J, Haggerty D A, Hutchinson W J, Jack D L, Lai E C, Liu H J, Nusinew D P, Omar A M, Pao S S, Paulsen I T, Quan J A, Sliwinski M, Tseng T, Wachi S, Young G B. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 42.Schneiderman R D, Farrell K B, Wilson C A, Eiden M V. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J Virol. 1996;70:6982–6986. doi: 10.1128/jvi.70.10.6982-6986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skach W R, Lingappa V R. Amino-terminal assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J Biol Chem. 1993;268:23552–23561. [PubMed] [Google Scholar]
- 44.Sommerfelt M A. Retrovirus receptors. J Gen Virol. 1999;80:3049–3064. doi: 10.1099/0022-1317-80-12-3049. [DOI] [PubMed] [Google Scholar]
- 45.Sonnhammer E L, vonHeijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 46.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tailor C S, Nouri A, Kabat D. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J Virol. 2000;74:237–244. doi: 10.1128/jvi.74.1.237-244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tailor C S, Takeuchi Y, O'Hara B, Johann S V, Weiss R A, Collins M K L. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi Y, Vile R G, Simpson G, O'Hara B, Collins M K L, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatsumi S, Segawa H, Morita K, Haga H, Kouda T, Yamamoto H, Inoue Y, Nii T, Katai K, Taketani Y, Myiamoto K, Takeda E. Molecular cloning and hormonal regulation of PiT-1, a sodium-dependent phosphate cotransporter from rat parathyroid glands. Endocrinology. 1998;139:1692–1699. doi: 10.1210/endo.139.4.5925. [DOI] [PubMed] [Google Scholar]
- 51.Uckert W, Willimsky G, Pedersen F S, Blankenstein T, Pedersen L. RNA levels of human retrovirus receptors PiT1 and PiT2 do not correlate with infectibility by three retroviral vector pseudotypes. Hum Gene Ther. 1998;9:2619–2627. doi: 10.1089/hum.1998.9.17-2619. [DOI] [PubMed] [Google Scholar]
- 52.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O'Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Versaw W K, Metzenberg R L. Repressible cation-phosphate symporters in Neurospora crassa. Proc Natl Acad Sci USA. 1995;92:3884–3887. doi: 10.1073/pnas.92.9.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson C A, Eiden M V. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J Virol. 1991;65:5975–5982. doi: 10.1128/jvi.65.11.5975-5982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson C A, Eiden M V, Anderson W B, Lehel C, Olah Z. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J Virol. 1995;69:534–537. doi: 10.1128/jvi.69.1.534-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson C A, Farrell K B, Eiden M V. Comparison of cDNAs encoding the gibbon ape leukaemia virus receptor from susceptible and non-susceptible murine cells. J Gen Virol. 1994;75:1901–1908. doi: 10.1099/0022-1317-75-8-1901. [DOI] [PubMed] [Google Scholar]
- 57.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]