SUMMARY
Calorie restriction (CR) extends lifespan and healthspan in diverse species. Comparing ad libitum (AL) and CR-fed mice is challenging due to their significantly different feeding patterns, with CR-fed mice consuming their daily meal in 2 hours and then subjecting themselves to a prolonged daily fast. Here, we examine how AL and CR-fed mice respond to tests performed at various times and fasting durations, and find that effects of CR – insulin sensitivity, circulating metabolite levels and mTORC1 activity – result from the specific temporal conditions chosen, with CR-induced improvements in insulin sensitivity observed only after a prolonged fast, and the observed differences in mTORC1 activity between AL and CR-fed mice dependent upon both fasting duration and the specific tissue examined. Our results demonstrate that much of our understanding of the effects of CR are related to when, relative to feeding, we choose to examine the mice.
Keywords: calorie restriction, fasting, dietary restriction, time-restricted feeding, mTOR, aging, lifespan
Graphical Abstract
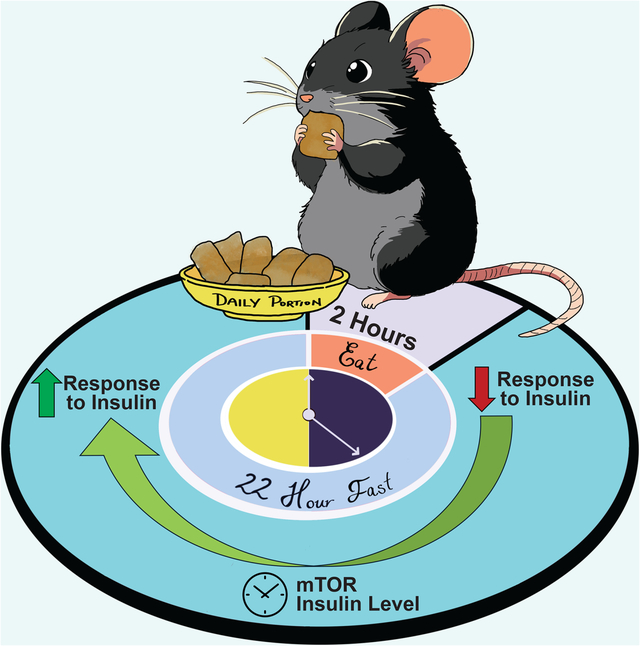
Introduction
Calorie restriction (CR) is the gold standard for geroprotective interventions1. Despite its efficacy, adherence challenges have prompted research into the underlying physiological and molecular mechanisms mediating the beneficial effects of CR. Many different molecular pathways have been proposed to mediate CR’s impact on healthspan and lifespan; however, a complete understanding of the mechanisms that lead to the benefits of CR remains elusive. A complicating factor in our quest to understand the mechanisms of CR is that CR- and AL-fed mice have dramatically different eating patterns, with CR-fed mice consuming their entire daily allotment of food within approximately ~2 hours and then fasting until the following day2,3.
We recently demonstrated that this prolonged fast between meals is necessary for the metabolic and geroprotective effects of a CR diet, and the imposition of a prolonged daily fast alone without decreased calorie consumption was sufficient to recapitulate many of the metabolic and molecular effects of a CR diet3. However, the distinctly different eating patterns of CR mice makes interpreting physiological and molecular differences between CR- and AL-fed mice difficult due to the challenge of aligning the assessment time of mice with such disparate feeding schedules. Further, CR-fed mice may have adapted to prolonged daily fasts, while AL-fed mice rarely experience fasting.3,4
For example, when examining the response to a glucose or insulin challenge, researchers routinely fast animals for a period between 6–16 hrs to minimize variability and bias in blood glucose measurements5–7. This approach not only assesses the animal’s response to the administration of glucose or insulin, but also their response to the preceding fast. Furthermore, CR-fed animals, which are subjected to a prolonged fast each day, may have adapted to a fast, whereas AL-fed mice are rarely subjected to fasting. Thus far, no study has comprehensively examined how the phenotypes of AL and CR-fed mice evolve as the time since their most recent meal increases, with most studies focusing on single-timepoint comparisons.
Here, we investigated the physiological and molecular responses of mice AL or CR diets, varying the time since their most recent meal from 4 to 24 hours. While CR-fed mice exhibited an overall improved response to glucose, an improved response to insulin relative to AL-fed controls was only evident at specific timepoints following their last meal. Specifically, CR-fed mice exhibited significant variations in response to insulin based on the length of the preceding fast, a pattern not observed in AL-fed mice. We also identify a crucial role for insulin in suppressing hepatic glucose production during an extended fast. Lastly, contrary to the prevailing theory that CR promotes health and longevity by reducing mTORC1 activity8, our findings indicate that hepatic mTORC1 activity is strongly regulated by feeding-fasting status, and is not constitutively suppressed by CR, while muscle mTORC1 activity is elevated in a time-dependent manner under a CR feeding regimen. Our study highlights the significance of considering temporal conditions in comprehending the physiological, metabolic, and molecular effects of CR, and underscores the necessity to consider these factors when evaluating the effects of dietary interventions.
Results
Fasting duration produces differential response to exogenous insulin in mice fed a CR diet
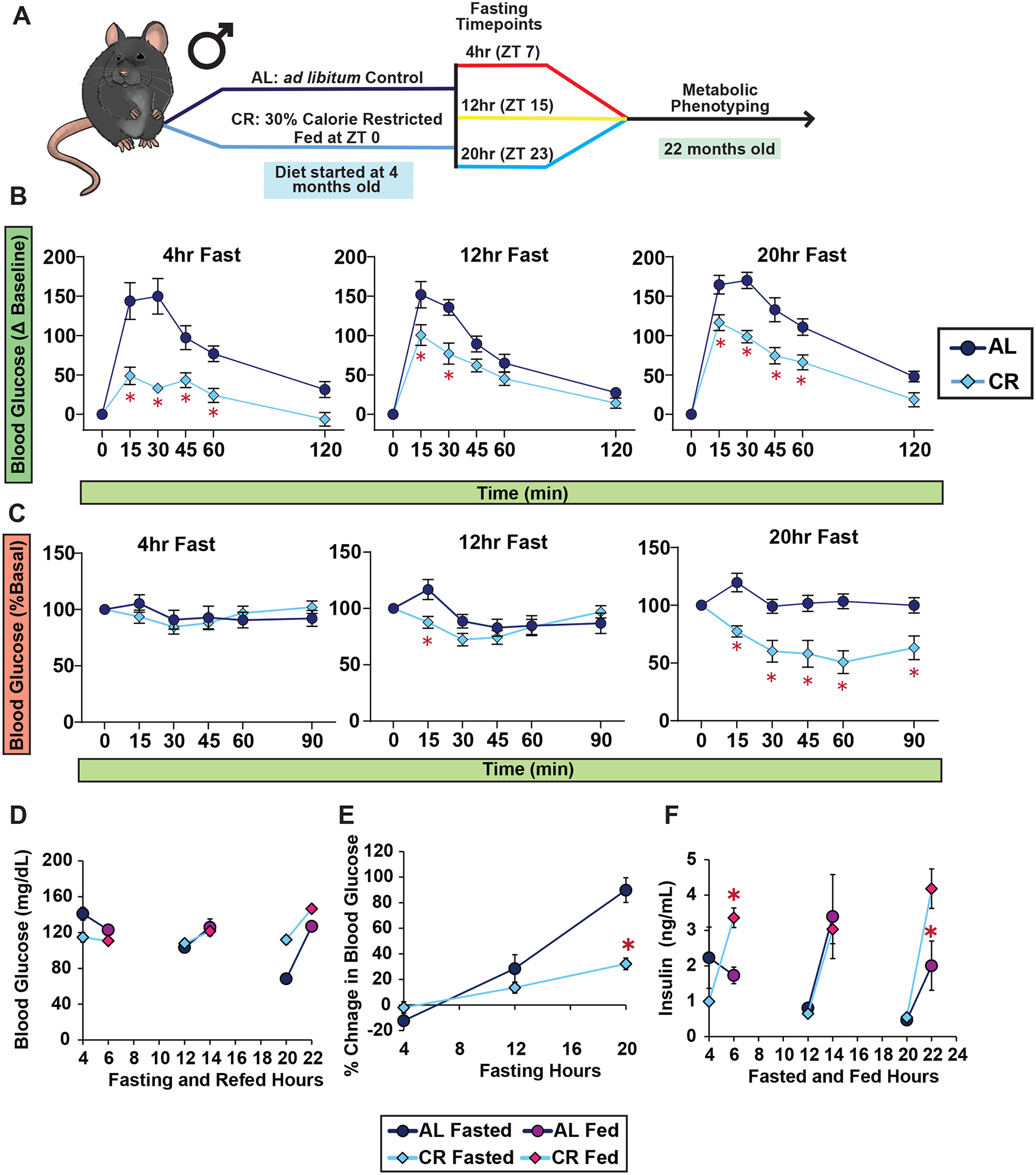
We initiated a study with 9-week-old male C57BL/6J mice, dividing them into two groups: AL (never subjected to fasting) or 30% CR (exposed to daily self-imposed fasting). CR mice were fed once per day at the beginning of the light cycle (Zeitgeber Time, ZT 0) a common practice in CR studies, including the National Institute on Aging (NIA) aged rodent colony9–12. We measured the bodyweight for the efficacy of the diet, and as expected, male mice on CR had decreased weight gain and decreased adiposity, with maintenance of lean mass (Figures 1A–D).
Figure 1: Response to insulin is dependent upon the length of the preceding fast in CR-fed mice.
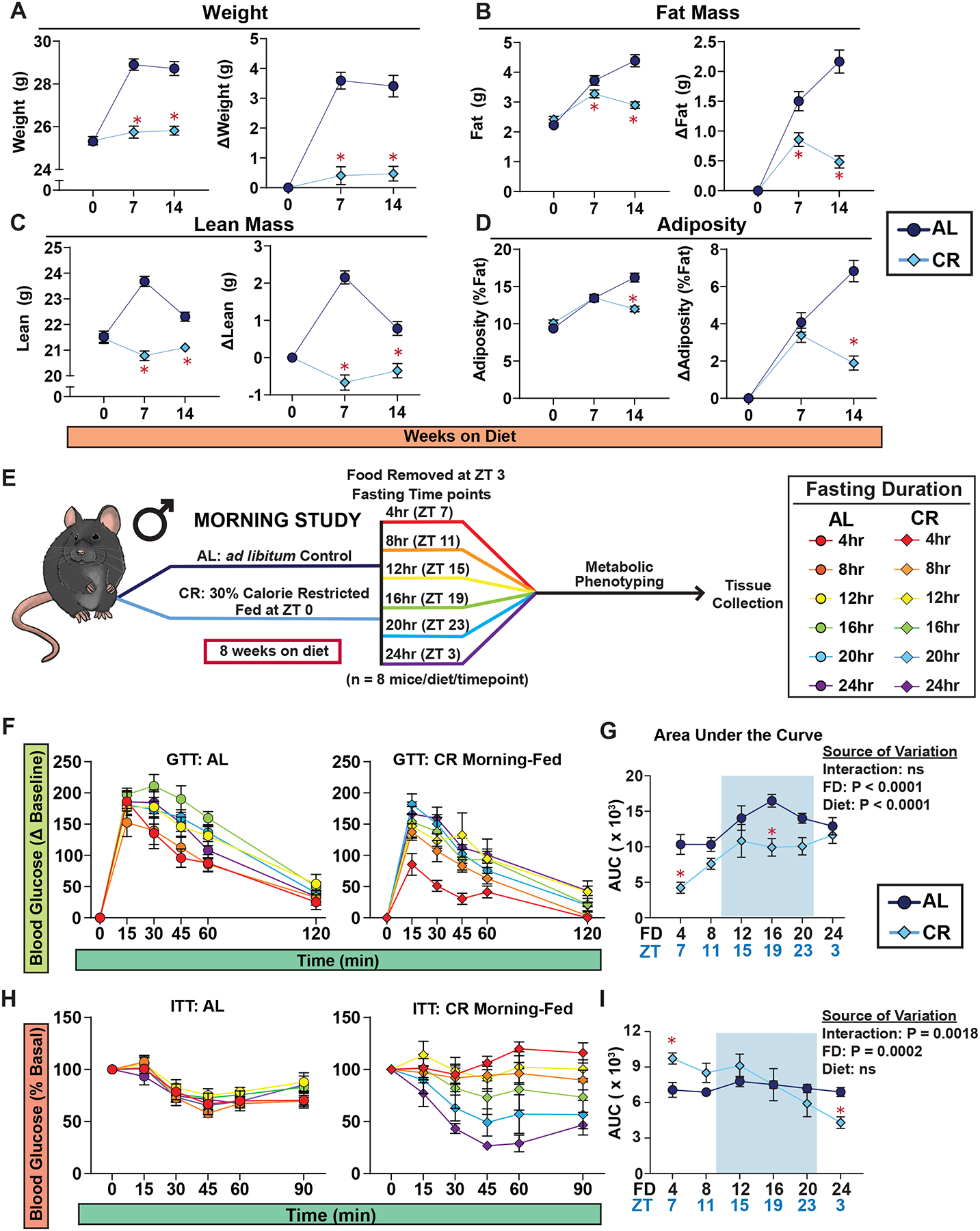
(A-D) Body composition of male mice under Morning-Fed conditions (n = 24 biologically independent mice per diet). (A) Total body weight with change in body weight from baseline. (B) Fat mass with change in fat mass from baseline. (C) Lean mass with change in fat mass from baseline. (D) Adiposity with change in adiposity from baseline. (E) Experimental design of Morning-Fed study. (F-G) GTTs of AL and CR-fed male mice (F) and AUC (G). (H-I) ITTs of AL and CR-fed male mice (H) and AUC (I). (F-I) n = 8 mice per diet per time point. (A-D, G and I) *p<0.05 AL-fed vs. CR-fed mice at each time point, Sidak’s test post two-way repeated measures ANOVA. (G, I) The overall effect of diet, fasting duration (FD), and the interaction represent the p-value from a two-way ANOVA. Data represented as mean ± SEM. AUC, area under the curve. See also Supplementary Figures 1–4.
We examined the metabolic response to fasting by conducting glucose and insulin tolerance tests (GTT and ITT) after fasts of different lengths. For each test, both AL and CR-fed mice were given access to food from ZT 0–3, and at the conclusion of this period, mice were placed in new cages without food. We then randomized the fasted AL and CR-fed mice into 6 groups, with approximately the same weight and body composition and performed the indicated assay, with the first group examined at ZT 7, and a different group examined every four hours (Figure 1E).
During the GTT, we noticed that the fasting blood glucose varied between diet groups and fasting timepoints, contributing to an observed difference between groups (Figure S1A–C), prompting us to calculate the delta from the baseline to calculate the AUC (Figure S1D). Both AL and CR-fed males had the lowest area under the curve (AUC) after 4 hrs of fasting as compared to other fasting timepoints (Figures 1F–G). While CR-fed mice consistently exhibited a lower AUC than AL-fed mice, this was only statistically significant after a fast of 4 or 16 hours (Figure 1G).
In contrast, while the response to insulin of AL-fed male mice remained consistent regardless of the length of the fast, the response of CR-fed mice to insulin was highly dependent on the length of the fasting period (Figures 1H–I and S2). While the literature broadly notes that CR improves insulin sensitivity in mammals1, CR-fed mice fasted for only 4–12 hrs had blood glucose levels that were essentially resistant to insulin administration. Only after 20 or 24 hrs of fasting did CR-fed mice exhibit improved insulin sensitivity relative to AL-fed mice (Figure 1H and S2).
Recognizing sex as a critical determinant in dietary interventions, including CR1,11, we extended our study to examine the fasting response of female C57BL/6J mice to glucose and insulin. Consistent with our observations in males, CR-fed females exhibited reduced weight gain, primarily due to diminished lean mass accrual (Figures S3A–D). CR-fed females had improved glucose tolerance across all fasting intervals relative to AL-fed counterparts (Figures S3E–K). Additionally, the response to insulin in CR-fed female mice paralleled that of males, with resistance to insulin during a 4–12 hour fast, both in absolute terms and relative to AL-fed mice, and improved insulin sensitivity only after 20 hours of fasting (Figure S4).
CR mice tightly maintain glucose homeostasis during a prolonged daily fast
To investigate the unique response to insulin in CR mice, we conducted a meal-stimulated insulin secretion (MSIS) test, measuring insulin levels post feeding rather than after glucose stimulation. This approach aimed to assess insulin levels under natural feeding conditions. Both AL and CR groups had access to food from ZT 0–3. Subsequently, we divided the mice into six groups, subjecting them to one of the fasting periods of either 4, 8, 12, 16, 20 or 24 hrs as in the experiments presented in Figure 1. Following the fast, we collected fasting blood and then provided the mice with food for two hours, after which we then collected fed blood (Figure 2A).
Figure 2. Plasma insulin level after feeding is dependent on time of day, not fasting duration.
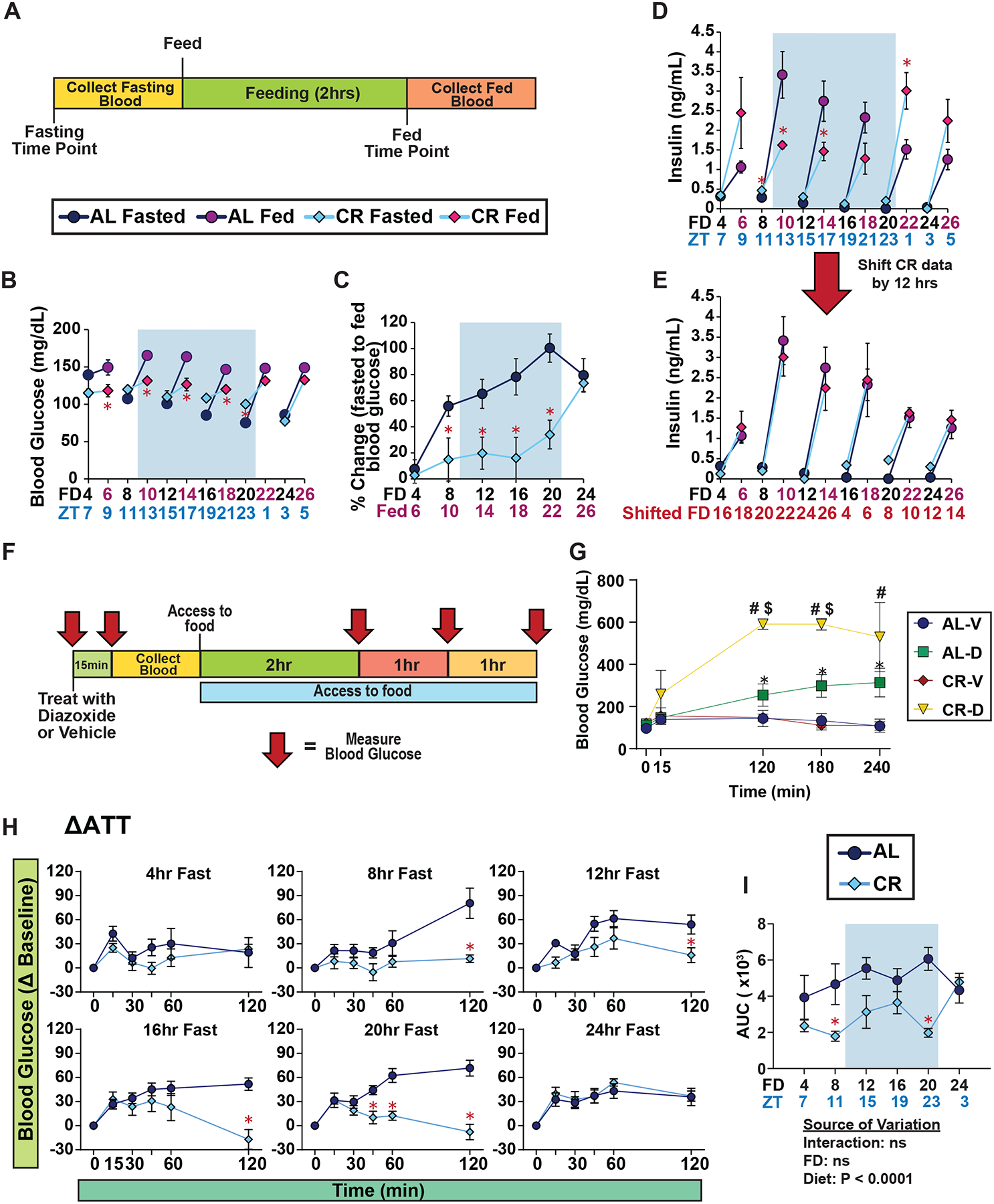
(A) Experimental design of MSIS test. (B-C) Blood glucose level of male mice fasted for varying lengths of time, prior to and subsequent to feeding; FD axis indicates length of time since fasting initiation; red axis label indicates mice refed for 2 hours (B), and percent change in blood glucose level from fasted to fed state (C). (D-E) Plasma insulin level of male mice fasted for varying lengths of time, prior to and subsequent to feeding (D), and data presented with CR data shifted by 12 hours (E). (F) Experimental design of acute diazoxide treatment experiment. (G) Blood glucose level of male mice treated with either vehicle or diazoxide after a 12 hr fast (H-I) ATTs of ad libitum and CR-fed male mice (H) and AUC (I) after fasts of the indicated length. (B-E, G-I) n = 6–8 mice per diet per time point. (B-D) *p<0.05 AL-fed vs. CR-fed mice at each time point, (G) *p<0.05 AL-D vs. AL-V, #p<0.05 CR-D vs. CR-V, and $p<0.05 CR-D versus AL-D; Sidak’s test post two-way repeated measures ANOVA. (I) The overall effect of diet, fasting duration (FD), and the interaction represent the p-value from a two-way ANOVA. Data represented as mean ± SEM. AUC, area under the curve. See also Supplementary Figures 5.
Intriguingly, CR-fed male mice exhibited resistance to fasting-induced changes in blood glucose. While AL-fed males displayed ~46% reduction in fasting blood glucose over 20 hours, CR-fed males showed only an ~15% decrease during such a fast (Figure 2B). Following a meal challenge, CR-fed males were able to maintain a stable blood glucose level, while AL-fed mice exhibited 50–100% increase in blood glucose level following feeding (Figures 2B–C). After 24 hours of fasting – which exceeds the normal length of time between meals for CR-fed mice – AL-fed and CR-fed males had almost identical fasting and refed blood glucose levels (Figure 2B–C).
Contrary to our expectations that insulin levels would parallel blood sugar levels, plasma insulin levels of both AL and CR mice were independent of fasting duration and diet (Figure 2D). Although we anticipated lower fasting and refed insulin levels in CR-fed mice compared to AL-fed mice, this was only evident at certain time points. CR-fed mice refed after 4, 20, or 24 hours of fasting had higher refed insulin levels than their AL-fed counterparts. Closer examination revealed that the refed insulin pattern of CR-fed mice appeared to be offset by 12 hours from that of AL-fed mice. When we shifted the CR data by 12 hrs, aligning the data based on the start time of the major eating periods, we observed identical refed insulin patterns in CR and AL-fed mice (Figures 2E and S5A–B).
To determine if the stable blood glucose level in CR-fed mice was driven in part by differences in insulin secretion, we tested the impact of inhibiting insulin secretion using diazoxide, a well-established drug that inhibits insulin secretion13–15. We performed this test after a 12 hour fast because at this timepoint both AL-fed and CR-fed mice had similar fasting blood glucose levels and insulin response while having different refed insulin levels. Mice were either treated with vehicle or diazoxide, and blood glucose level was measured pre- and post-treatment (Figure 2F). We expected to observe similar level of blood glucose level between AL- and CR-fed mice with the initial administration of diazoxide; however, even without the consumption of food or glucose administration, diazoxide-treated CR mice showed a sharp increase in blood glucose, exceeding 600 mg/dL, compared to a moderate rise in diazoxide-treated AL mice (Figure 2G). This data suggests that during a fast insulin plays a critical role in inhibiting hepatic glucose production in CR-fed animals.
To confirm that hepatic gluconeogenesis is suppressed in CR-fed mice, we performed an alanine tolerance test (ATT), which measures glucose production from this substrate16 (Figures 2H–I and S5C). Supporting our hypothesis, CR-fed mice exhibited reduced glucose production from alanine compared to AL-fed mice at all time points up to 20 hours (Figures 2H–I and S5D). For CR-fed females, we also observed tight blood glucose control and elevated insulin levels after refeeding following a 20 hr fast when conducting the MSIS test (Figures S5E–G). Furthermore, CR-fed females also exhibited a reduction in hepatic glucose production from alanine (Figures S5H–K). These results suggest that CR-fed mice of both sexes rewire their metabolism to maintain tight blood glucose levels, in part by suppressing endogenous glucose production, when subjected to extended periods of daily fasting.
The response of CR-fed mice to insulin is independent of time of feeding
Our initial observations suggested a 12-hr offset in insulin levels between AL- and CR-fed mice (Figure 2D–E). This led us to hypothesize that plasma insulin levels might be influenced by the time of feeding, a theory supported by previous studies showing the circadian regulation of insulin secretion and its dependency on feeding schedule17,18. To explore this further, we adjusted the feeding time for male mice to the onset of the dark cycle (ZT12) instead of the light cycle (ZT 0) and we examined the metabolic response to a fast (Figure 3A). AL-fed control mice remained in their natural feeding state. For simplicity, we refer to our previous studies with male mice as Morning-Fed (CR mice fed at ZT 0), and these new experiments as Night-Fed (CR mice fed diet at ZT 12).
Figure 3. The response to insulin in CR feeding regimen is highly dependent on fasting duration and not time of feeding.
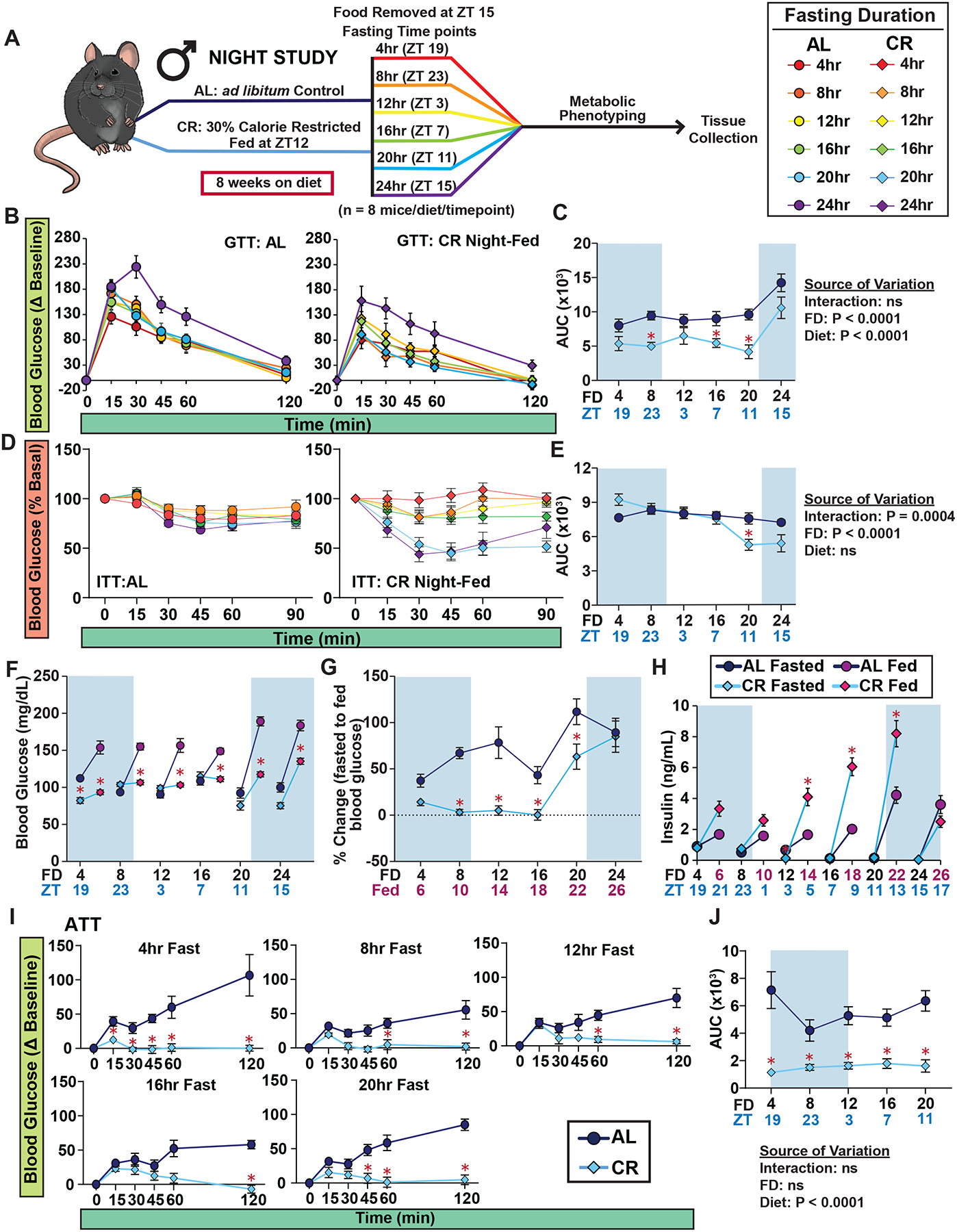
(A) Experimental design of Night-Fed study. (B-C) GTTs of male mice (B) and AUC (C); mice were placed in new cages without food from all groups at ZT 15, and each group was then fasted for varying lengths of time. (D-E) ITTs of AL and CR-fed male mice (D) and AUC (E). (F-H) Blood glucose level of mice fasted for 4, 8, 12, 16, 20 and 24hrs, prior to and subsequent to feeding (F), and percent change in blood glucose level from fasted to fed state (G). (H) Plasma insulin level of mice fasted for varying lengths of time, prior to and subsequent to feeding. (I-J) ATTs of C57BL/6J male mice (I) and AUCs. (B-J) n = 7–16 mice per diet at each time point. (C, E, F-J) *p<0.05 AL-fed vs. CR-fed mice at each time point, Sidak’s test post two-way repeated measures ANOVA. (C, E, J) The overall effect of diet, fasting duration (FD), and the interaction represent the p-value from a two-way ANOVA. Data represented as mean ± SEM. AUC, area under the curve. See also Supplementary Figures 6–9.
In line with our Morning-Fed CR studies, the Night-Fed CR group also exhibited reduced weight, fat mass, and lean mass gain, with an overall effect of decreased adiposity (Figures S6A–D). Mice show a diurnal pattern in their body weight, increasing during the active phase (i.e. dark cycle) and decreasing during inactive phase (i.e. light cycle)19, we evaluated body composition changes in AL- and CR-fed mice during a 24hr fast (Figures S6E–H). While there were no significant differences in body weight, fat mass and lean mass between the two groups during the fast, we observed a more rapid decrease in adiposity in CR-fed mice due to their lower absolute fat mass (Figures S6E–H).
Furthermore, like their Morning-Fed CR counterparts, Night-Fed CR mice had overall improved glucose tolerance compared to AL-fed mice, irrespective of the fasting duration, and this improvement reached statistical significance after 8, 16, or 20 hrs of fasting (Figures. 3B–C and S7A–D). Similar to what we observed with Morning-Fed CR males, Night-Fed CR males did not exhibit improved insulin sensitivity until fasting for at least 20 hours (Figures 3D–E and S8A–D), suggesting that the response to insulin in CR-fed mice is dependent on fasting duration rather than the time of day.
We next measured fasted and fed blood glucose and serum insulin levels in the Night-Fed study (Figures 3F–H). Initially, we found that Night-Fed CR mice maintained tight control over both their fasted and refed blood levels during the first 16 hours, similar to the Morning-Fed CR mice (Figures 3F–G). However, the Night-Fed CR mice showed a 2–3 fold increase in refed insulin levels after 12, 16, or 20 hours of fasting (Figure 3H and S9A–C). Lastly, examining hepatic glucose production post-alanine administration revealed that Night-Fed CR mice, like their Morning-Fed counterparts, suppressed glucose production from alanine (Figures 3I–J and S9D–E).
Collectively, these findings demonstrate that CR mice effectively resist changes in blood glucose induced by fasting and refeeding, irrespective of their feeding times. Importantly, while many of the metabolic effects of CR are linked to fasting duration, we find that insulin levels are primarily influenced by the time of feeding rather than the fasting duration.
AL and CR feeding have distinct and time-dependent effects on plasma metabolites and proteins
To gain additional insight into the effects of fasting and time-of-day on whole body metabolism, we used an untargeted metabolomics approach to examine plasma metabolite levels after either 8, 12, 16 or 24 hrs of fasting in AL- and CR-fed male mice from the Morning Study. We identified a total of 170 plasma metabolites; from this we utilized principal component analysis (PCA), plotting the first two principal components of each time point as well within each diet (Figures 4A and S9). Intriguingly, AL-fed and CR-fed mice displayed distinct metabolite profiles after 8 hours of fasting (blood collected at ZT 11) and 16 hours of fasting (blood collected at ZT 19). In contrast, the metabolite profiles of AL and CR mice were largely overlapping after either 12 hours or 24 hours of fasting (blood collected at ZT 15 and ZT3, respectively) (Figure 4A).
Figure 4. Plasma metabolite profile is dependent on time-of-day and diet.
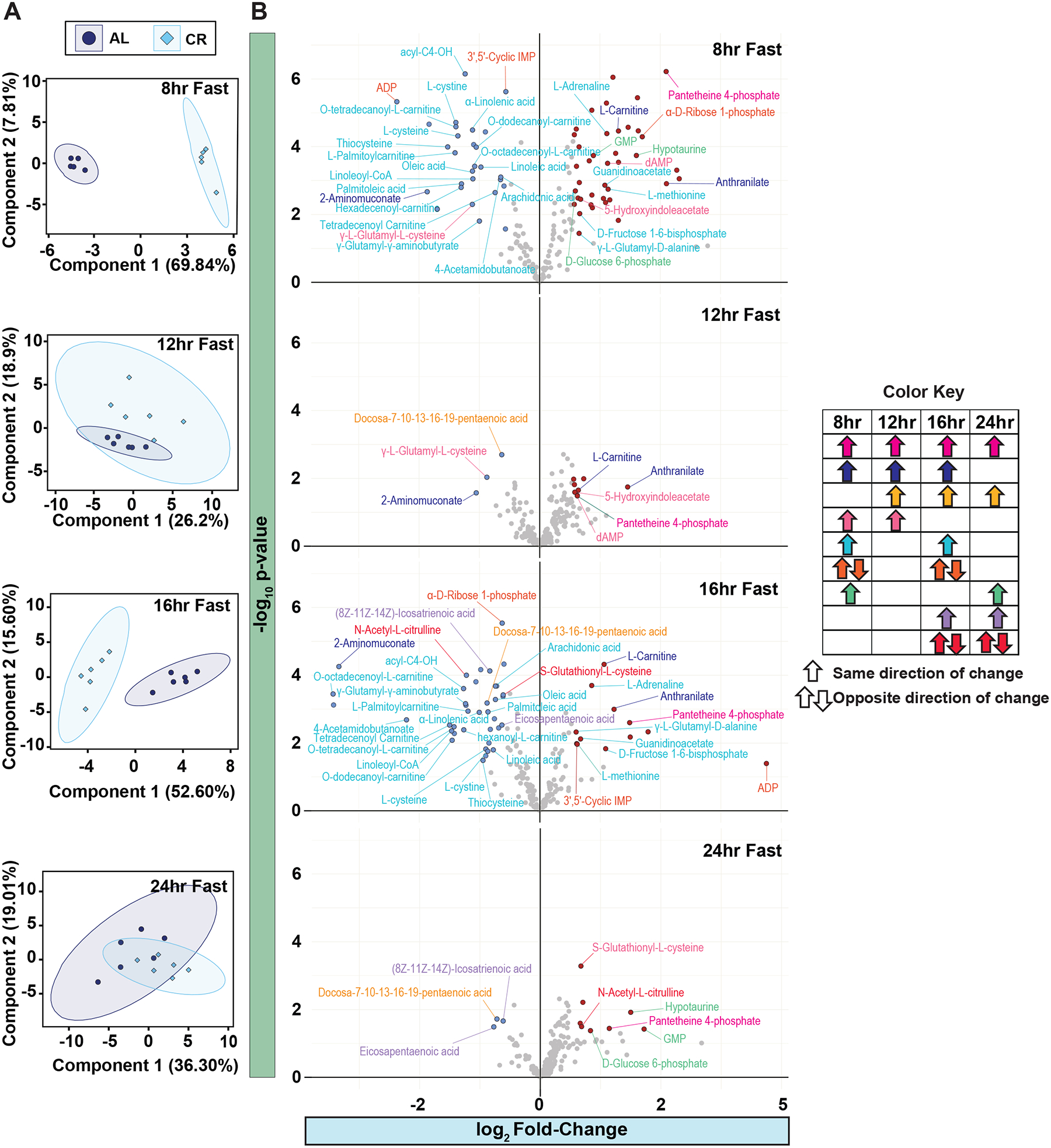
(A) Untargeted metabolomics of plasma from male mice fed AL and CR diet from the Morning Study. PCA of plasma metabolites with AL and CR mice collected after an 8hr, 12hr, 16hr and 24hr fast. (B) Volcano plots of significantly increased plasma metabolites. Significantly decreased metabolites are colored blue and significantly increased metabolites are colored red. Metabolite names are colored coded based on significant change between AL and CR at more than one time point (color key). n = 5–6 mice per diet per time point. See also Supplementary Figures 10.
We next focused on metabolites that showed significant changes (p-value ≤ 0.05) at more than one time point (Figures 4B). One standout finding was the consistent elevation of pantetheine-4-phosphate at all examined timepoints in CR-fed mice (Figure 4B). Four metabolites were significantly altered at three of the fasting durations, with elevation of Anthranilate and L-Carnitine and decreased levels of 2-Aminomuconate; the remainder of the metabolites were altered only at two of the fasting durations (Figure 4B).
In parallel with the metabolomics study, we examined the plasma proteome of AL- and CR-fed animals, identifying a total of 294 proteins. Particularly at the 8 and 16 hr fasting period, distinct protein profiles were apparent between AL and CR-fed mice (Figure S10A). However, to our surprise, less than 4% of proteins were significantly different between AL and CR-fed mice after 8 hours of fasting – with the differences becoming increasingly minimal as the fasting duration extended. Fasting duration or time of day were the main contributor to differences in plasma protein (Figure S10B–C). Overall, our results suggest that CR feeding produces a unique rhythmic pattern in plasma metabolite levels without external nutrient influx, while circulating proteins are minimally influenced by CR feeding.
Hepatic and skeletal muscle mTORC1 activity is paradoxically not repressed by CR
The protein kinase mTORC1 (mechanistic Target Of Rapamycin) is a critical regulator of insulin sensitivity via S6K1-mediated feedback inhibition of insulin receptor substrate20,21. We hypothesized that high post-prandial mTORC1 signaling might explain the insulin resistance of CR-fed animals due to the negative feedback regulation of mTORC1 on insulin action. To test our hypothesis, we sacrificed mice at multiple time points, matching the schedule from our in vivo metabolic phenotyping studies described above (Figures 5A–B). We measured the phosphorylation of mTORC1 substrates and downstream readouts, as well as phosphorylation of the mTORC2 substrate AKT S473 and the PDK1 substrate AKT T308 in the liver and skeletal muscle (Figures 5C–F and S11–12). We found minimal differences in hepatic mTORC1 activity between AL- and CR-fed mice in either our Morning-Fed or Night-Fed studies (Figures 5C–D). However, post-prandial S6 phosphorylation, a readout of mTORC1 activity, was higher in the liver of CR-fed mice than in AL-fed mice, reaching statistical significance in Morning-Fed CR mice at the 4 hour time point (Figures 5C–D). In the skeletal muscle, we observed similar trends with heightened post-prandial S6 S240/S244 and S6K1 T389 phosphorylation in CR-fed mice at multiple time points (Figures 5E–F). Additionally, the phosphorylation of other mTORC1 substrates, including 4E-BP1 T37/S46 and S757 ULK1, was elevated in the skeletal muscle of CR-fed mice at multiple time points (Figures 5E–F).
Figure 5. Liver and muscle mTORC1 activity is not constitutively repressed by CR.
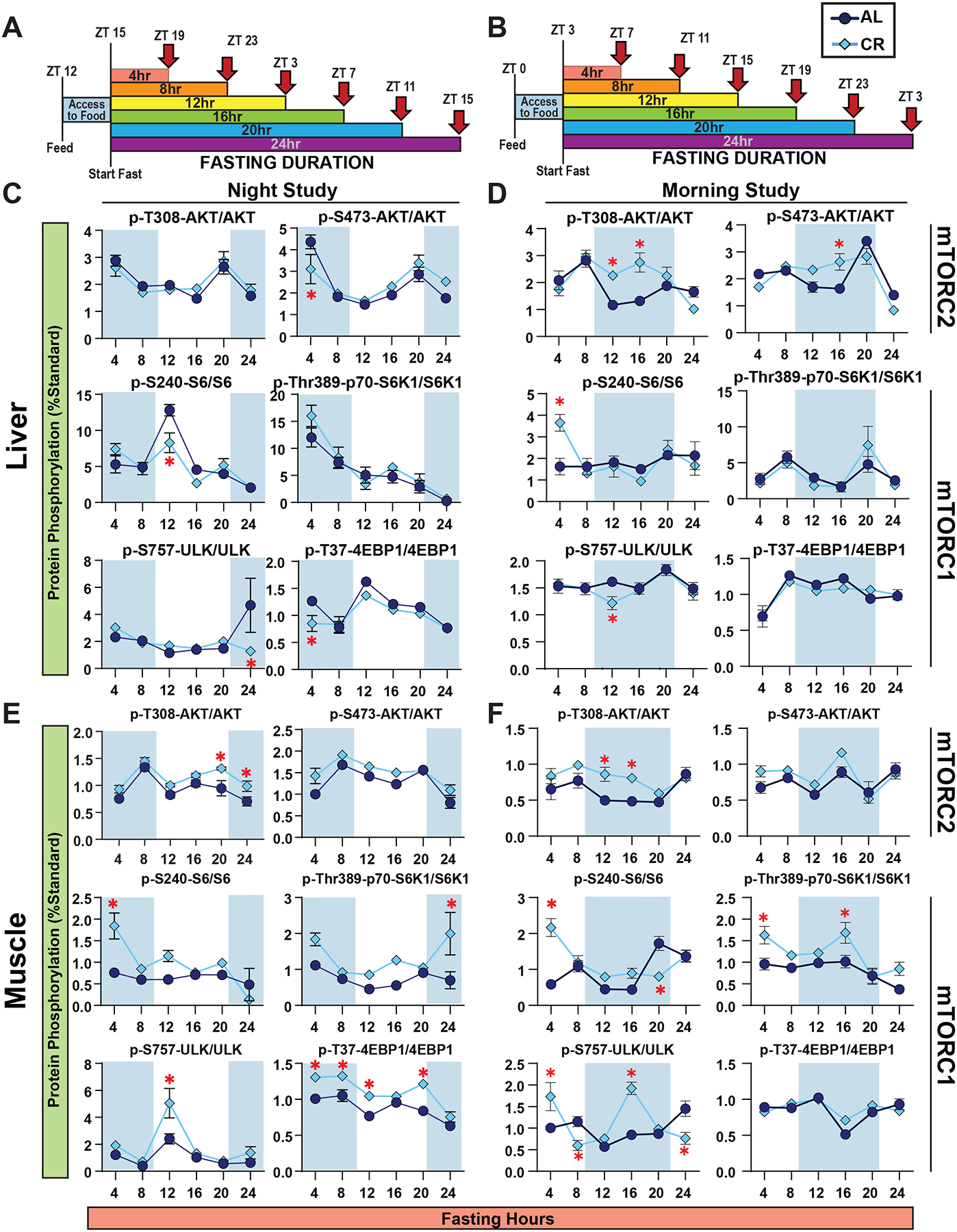
(A-B) Experimental design. Night-fed male mice were placed in new cages without food at ZT15 (A) and Morning-Fed mice at ZT3 (B). (C-D) Western analysis of Liver Night-Fed (C) and Morning-Fed (D)male mice. (E-F) Western analysis of Muscle Night-Fed (E) and Morning-Fed (F) male mice. (C-F) n = 6 mice per diet per time point; *p<0.05, Sidak’s test post 2-way ANOVA). Data represented as mean ± SEM. See also Supplementary Figures 11–13.
The significant differences in phosphorylation of S6 in the liver and muscle at the earliest fasting timepoint (4hrs) (Figure 5C–F) prompted us to consider that differences in mTOR signaling between AL- and CR-fed mice might be noticeable immediately after feeding. We synchronized the feeding times of all mice by fasting the mice at 6AM in the morning to encourage feeding at the time of the test and then refeeding the mice starting at 6PM; we staggered the feeding allowing us to time the collection to the minute when each mice took its first bite (Figure S13A). When feeding was synchronized, AL and CR mice exhibited similar levels of mTORC1 activity in the liver (Figure S13B–C). Together, our results suggest that hepatic mTORC1 activity is not constitutively repressed in CR-fed mice, but instead results from the prolonged fasting between meals, and post-prandial mTORC1 activity is elevated in some tissues in CR-fed mice.
Suppression of hepatic mTORC1 is required for CR-induced improvements to insulin administration in male mice
To investigate whether the suppression of hepatic mTORC1 activity is required to elicit the CR effect we utilized mice lacking hepatic TSC1 (TSC1-LKO; Tsc1 Liver Knock Out). TSC1-LKO mice are viable and have constitutively active hepatic mTORC1 signaling, even during fasting22, and we performed a series of metabolic measurements similar to those performed in Morning-Fed animals at two fasting durations (8hr and 21hr in males and females (Figures 6 and S14). There were no differences in glucose tolerance between the WT and TSC1-LKO mice within each respective diet for males; however, TSC1-LKO females on AL had impaired glucose tolerance relative to WT after a 21 hour fast (Figures 6A and 6B). In both sexes and genotypes, CR-fed mice had improved glucose tolerance relative to AL following a 21hr, but not an 8 hr fast (Figures 6A–B and S14A–B).
Figure 6. Constitutive suppression of hepatic mTORC1 activity is not required to produce the effect of CR.
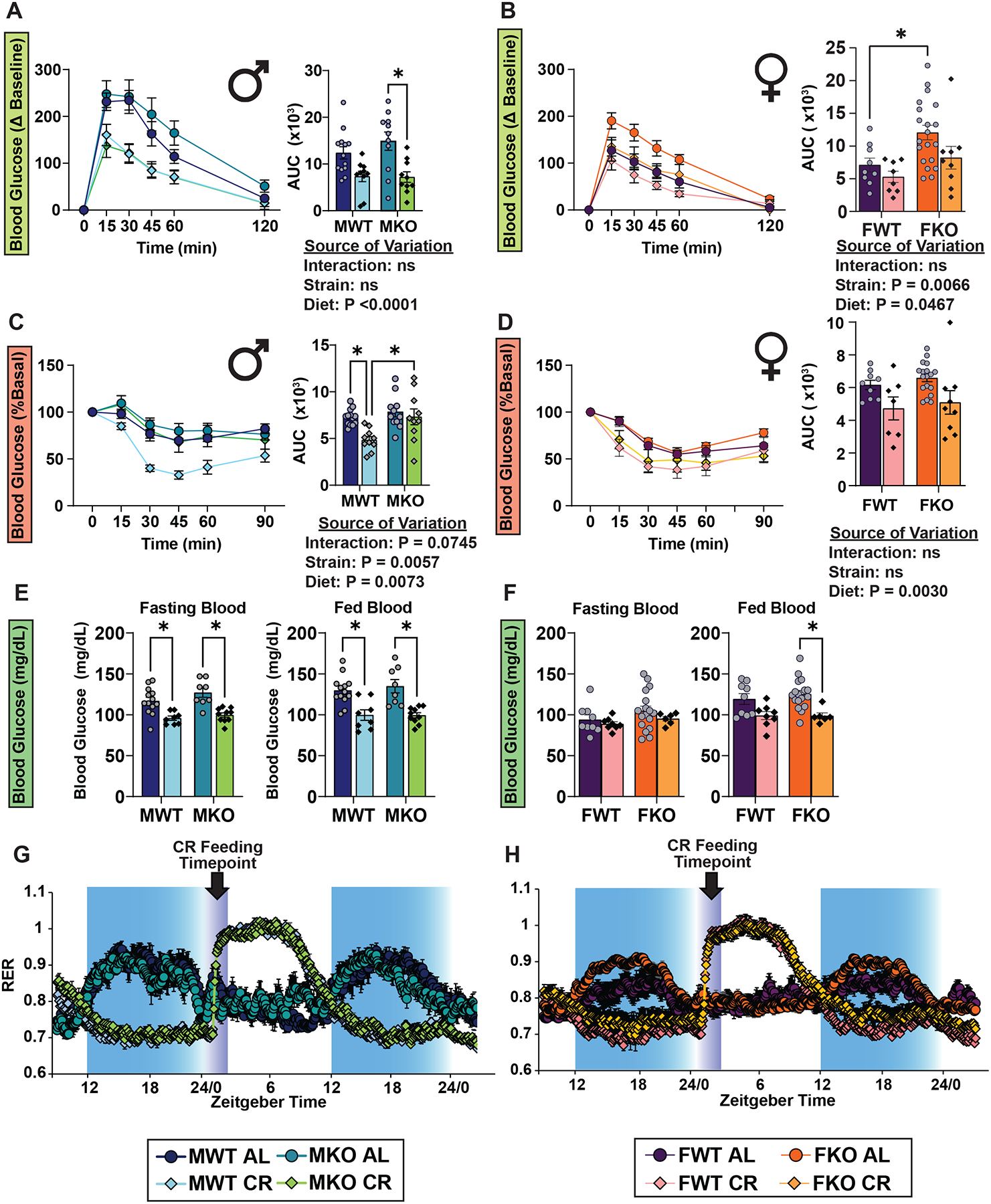
(A-B) GTTs in male (A) and female (B) mice lacking hepatic Tsc1 (KO) and their wild-type (WT) littermates. (C-D) ITTs in male (C) and female (D) mice lacking hepatic Tsc1 (KO) and their wild-type (WT) littermates. (E-F) Blood glucose level of male (E) and female (F) mice fasted for 8hrs starting at ZT3 then refed at ZT12, and blood glucose level at ZT 14 after 2 hr refed state. (A-F) n = 6–20 mice per diet per strain per sex, *p<0.05, from a Sidak’s post-test examining the effect of parameters identified significant in the two-way ANOVA. The overall effect of diet, strain, and the interaction represent the p value from a two-way ANOVA. (G-H) Respiratory exchange ratio from metabolic chamber analysis of male (G) and female (H) mice; n = 4 mice per diet per strain per sex. Data represented as mean ± SEM. See also Supplementary Figure 14.
As expected, we found no significant improvements in insulin sensitivity for either strain after an 8hr fast; in fact, response to insulin was significantly impaired by CR in TSC1-LKO mice of both sexes (Figures S14C–D). While CR diet significantly improved the response to insulin in WT males after a prolonged fast, unexpectedly we found that there was an interaction between diet and genotype (p=0.0745), with AL-fed and CR-fed TSC1-LKO male mice responding identically to insulin administration (Figure 6C). In contrast, CR-fed female mice showed an improved response to insulin regardless of genotype (Figure 6D). Furthermore, we found that both male and female mice fed a CR diet, regardless of genotype, maintained tight control of blood glucose levels even when challenged with refeeding (Figures 6E–F). Lastly, we observed no effect of genotype on the CR-mediated changes in fuel utilization in either sex (Figures 6G–H). Our data suggest that the metabolic effects of CR are largely independent of hepatic mTORC1 signaling. However, the effects of CR on whole body sensitivity to insulin administration are blocked by constitutive activation of hepatic mTORC1 in male mice.
Aged male mice placed on a long-term CR diet have similar temporal responses as young mice on CR
Our previous studies showed that aged mice maintained on a CR diet from a young age (long-term CR) retained a healthy phenotype3. Therefore, to examine if the variation in temporal conditions extended to long-term CR, we examined aged male mice (22 months old) which had been fed either an AL or CR diet starting at 4 months of age and were fed in the morning (Figure 7A). We found that as in young mice, long-term CR-fed mice had improved glucose tolerance after 4-, 12-, and 20-hour fasts, and only showed improved response to insulin after a 20 hr fast (Figure 7B–C). Furthermore, long-term CR-fed mice similarly maintained tight control over both fasting and meal-stimulated blood glucose levels (Figure 7D–E) and displayed a comparable trend in plasma levels of insulin as seen in younger mice (Figure 7F). From these observations, we conclude that long-term CR-fed mice to old age maintain a similar response to young mice, underscoring the critical role of fasting duration in the apparent response to a CR diet.
Figure 7. Morning-Fed aged males have similar response to glucose and insulin as young males.
(A) Schematic of GTTs and ITTs for 22-month-old male mice. (B-C) GTTs (B) and ITTs (C) conducted after fasts of varying lengths. (D-E) Blood glucose level of mice fasted for 4, 12, or 20 hrs, prior to and subsequent to feeding (D), and percent change in blood glucose level from fasted to fed state (E). (F) Plasma insulin level of mice fasted for varying lengths of time, prior to and subsequent to feeding. (B-F) n = 10 mice per diet per time point; *p<0.05 AL-fed vs. CR-fed mice at each time point, Sidak’s test post two-way repeated measures ANOVA. Data represented as mean ± SEM.
Discussion
There has been a long-standing interest in understanding the physiological and molecular basis by which CR promotes health and extends lifespan, with the ultimate goal of identifying CR mimetics23,24. Numerous signaling pathways have been proposed to mediate the benefits of CR, but as a whole, our understanding of the mechanisms by which CR promotes lifespan and metabolic health remains the subject of debate. A widely used approach to understand how CR functions is to identify physiological and molecular differences between AL and CR-fed animals. This approach has shown that CR has a range of physiological effects, including improved insulin sensitivity25 and protection from cancer26 in many mammalian species, and its modulation of numerous signaling pathways.
Why has this systematic approach to identifying mechanisms failed? CR feeding, at least in the context of once-per-day feeding, causes mice to rapidly consume their daily allotment of food within 2–3 hours, resulting in a self-imposed daily fast2,3,27. Critical consequences of this collaterally imposed fast is the misalignment of feeding status between AL and CR-fed animals, as well as potentially the induction of an adaptive response to fasting, confounding our understanding of the physiological and molecular effects of CR. Furthermore, the optimal fasting duration before a test, which typically ranges from 6 to 16hrs, remains a subject of debate.
Here, we gained insight into how experimental conditions – the length of the fast and the time the test is conducted – affect the observed metabolic response to CR. Notably, we discovered that one of the key physiological hallmarks of CR in mammals – improved insulin sensitivity – was only observed in mice that had been fasted for at least 20 hours. In fact, CR-fed animals appeared to be insulin resistant when tested after a 4 hr fast, a response seen in both males and females and in both young and old mice. Our results would suggest that under different paradigms, the literature would reflect that CR has minimal or even detrimental effects on insulin sensitivity.
While our studies do not say that the insulin-sensitizing effects of CR are unimportant, they highlight the discrepant findings between studies and call to attention the importance of timing and experimental conditions in assessing metabolic phenotypes and even changes at the molecular level. It will be interesting to learn if primates and humans on once-per-day CR regimens undergo similar daily periods of post-prandial insulin resistance and other metabolic shifts, and if the period of insulin sensitivity is limited to specific times of the day. A previous study with humans found a subset of participants on CR to have impaired glucose tolerance28, and this may be due to differences in feeding pattern between the CR participants. We have previously shown that calorie restriction alone is not sufficient to produce the same response to insulin, and this once-per-day feeding was necessary in mice3. And while other human studies have suggested the beneficial effects of fasting on health outcomes, controlled clinical trials remain to be explored29–31.
Deconvoluting the dynamic nature of blood glucose regulation during a fast is very complex and cannot be disentangled with the studies provided here. However, with the studies conducted here, we gained valuable insights as to how CR-fed mice may regulate their blood glucose during a prolonged fast. 1) CR mice are able to maintain tight blood glucose control during a prolonged fast of up to 20hrs, even after feeding, and insulin is required for the maintenance of blood glucose level in CR-fed mice. 2) Interaction between time and insulin levels were dependent on feeding schedule and this finding fit with previous research showing that insulin secretion is highly circadian regulated and this mechanism is gated within a limited timeframe32,33, with the response shifting dependent on feeding schedule17,18. Together, these findings suggest that a CR feeding regimen allows mice to adapt to a daily prolonged fast by rewiring their metabolism to preserve glucose stores.
A potential mechanism for the increased insulin sensitivity of CR-fed mice is the mechanistic Target of Rapamycin (mTOR) pathway, which regulates of insulin sensitivity through S6K1 mediated feedback inhibition of insulin receptor substrate20,21,34, and proposed as a key mediator of the effects of CR35–37. In yeast, deletion of TOR1 is epistatic with CR; that is, CR does not further extend the lifespan of yeast lacking TOR138. However, in higher species the relationship between CR and mTOR signaling is less clear; the lifespan of flies is extended by the mTORC1 inhibitor rapamycin at every level of calorie intake, pointing to the CR and mTOR inhibition working in part through parallel mechanisms39–44, and mice “omics” studies suggest that rapamycin and CR have distinct, largely non-overlapping effects45–49, suggesting that CR and rapamycin do not function through the same molecular mechanism. While extensive work has been done using methods of genetic and pharmacogenetic inhibition of mTOR, research on how CR feeding regimen affects mTOR activity in mammals is scarce.
Contrary to our initial hypothesis, mTORC1 activity in CR-fed mice was not constitutively reduced compared to AL-fed mice. Hepatic mTORC1 activity was primarily mediated by the fed and fasted state irrespective of diet, and this activity was synchronized when feeding was initiated at the same time. Furthermore, using TSC1-LKO mice, we found that hepatic mTORC1 suppression was not necessary for many of the phenotypes observed in CR animals. The nonlinear pattern of mTOR activity with fasting duration aligns with previous studies suggesting the circadian regulation of hepatic mTORC1 activity50–53. Interestingly, we observed increased muscle mTORC1 activity in CR-fed mice, potentially explaining why CR mice eventually regain lean mass. Other studies have also reported increased mTORC1 activity with CR in specific contexts54,55. These findings suggest that hepatic mTORC1 inhibition may not be a key regulator of the response to CR, and demonstrate that CR-fed mice do not exhibit a global decrease in mTORC1 activity. Our results add to accumulating evidence that mTORC1 inhibition is not the primary mechanism by which CR promotes healthy aging and longevity.
Circadian rhythms may be a key player in this phenomenon as insulin secretion is regulated by the hypothalamic suprachiasmatic nucleus (SCN), which houses the master clock56–58. Glucose homeostasis is also circadian regulated as shown by mouse models deficient in core circadian rhythms genes which exhibit hyperglycemia, impaired glucose tolerance, and hypoinsulinemia59–62. Furthermore, altered feeding times have been shown to impact the levels of hormone such as cortisol, which acts as a mediator for central and peripheral circadian rhythms63,64. While our study primarily focuses on glucose homeostasis, it is also important to note other aspects of energy metabolism, such as the metabolism of fat and ketone bodies, are highly circadian regulated65–67 and may play a role in some of the effects we observed in this study.
To gain a global view as to what may be happening during a fast, we examined metabolite and protein levels during the various fasting intervals. Vastly more metabolites were significantly altered after 8 and 16 hours of fasting than after 12 or 24 hours. Related to this, the majority of metabolites were significantly altered at only one or two time points. In addition to suggesting that there may be circadian or feeding-pattern dependent biological process affected by CR, our results highlight the possibility that a few key metabolites – such as pantetheine-4-phosphate – may serve as a timing independent serum marker of CR. Intriguingly, pantothenic acid (vitamin B5), a precursor of pantetheine-4-phosphate, has been found to be elevated in the blood of CR-fed mice in other studies68. Together our data suggest that plasma metabolome is greatly affected not only by CR, but also reflects the time of day and the duration of the fast preceding sample collection.
In conclusion, our work demonstrates that a number of the effects, such as response to insulin, mTOR activity, and metabolite levels, of a CR diet are the result of (or, at a minimum, affected by) the specific temporal conditions, including the time of day CR-fed mice have eaten and the length of the fast prior to testing. In particular, many of the phenotypes we observe are the result of when the mice are first fed, and this response may be an adaptive response to the feeding paradigm and schedule. We suggest that researchers should accurately record and report the time of feeding, the time at which the fast starts, and the duration of the fast to enable researchers to consistently reproduce data from studies on CR, as well as other dietary interventions.
Limitations of the study
We evaluated glucose homeostasis using GTTs and ITTs, and we acknowledge that there are alternative methods to assess glucose and insulin sensitivity. We considered using hyperinsulinemic-euglycemic clamps; however, this method cannot observe the dynamic state that occurs under normal postprandial conditions.69 Additionally, GTTs and ITTs are widely used to assess glucose homeostasis in CR studies70–73. Another limitation here was the reliance on food intake for the MSIS test, with CR mice consuming more than AL-fed mice. However, insulin levels did not correlate with food consumption, suggesting food load did not contribute to the overall effect we observed here. Additionally, we only have food consumption records for the Night-Study, but from our observations, Morning-Fed CR mice consumed a similar amount. Due to the large number of groups, we were not able to compare morning versus night fed in females or aged animals. Furthermore, we only utilized C57BL/6J mice, and as strain impacts the response to dietary interventions, additional studies will be needed to confirm our results’ applicability to all genetic backgrounds11,74,75. Lastly, while our study included a time-of-day component, this was not a circadian rhythms study, and additional studies will need to be done with proper controls to dissociate the contribution of circadian rhythms from the response we observed in this study.
STAR★Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dudley W. Lamming (dlamming@wisc.medicine.edu)
Materials availability
This study did not generate new unique reagents
Data and code availability
Metabolomics and proteomics datasets are publicly available at the following: Metabolomics data have been deposited to the EMBL-EBI MetaboLights database (DOI: 10.1093/nar/gkad1045, PMID:37971328) with the identifier MTBLS10795 and can be accessed in the following https://www.ebi.ac.uk/metabolights/MTBLS10795. Proteomics data have been deposited to MassIVE with the identifier MSV000095510.
The custom R code that was written for this publication is publicly available at the following dedicated GitHub repository: https://github.com/linsemanlab/Pak-et-al-2023.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Animals, Diets, and Feeding Regimens
All procedures were performed in conformance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee of the William S. Middleton Memorial Veterans Hospital (Assurance ID: D16–00403) (Madison, WI, USA). Male and female C57BL/6J mice (stock number 000664) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) at 8 weeks of age and acclimated to the animal research facility for at least one week before entering studies. Liver specific Tsc1 knockout mice (Tsc1-LKO) were generated by crossing mice expressing Albumin-Cre (stock number 003574) with mice expressing a conditional allele of Tsc1 (Tsc1LoxP/LoxP), stock number 005680 as previously described22,76,77. Aged male C57BL/6J mice were those used in our previously reported lifespan study3. All animals were housed in a specific pathogen free (SPF) mouse facility with a 12:12 hour light/dark cycle maintained at 20°−22°C. All animals were placed on 2018 Teklad Global 18% Protein Rodent Diet for one week before randomization. Mice were randomized to either AL, ad libitum diet or CR, animals in which calories were restricted by 30% and fed once per day. Animals fed an AL and CR were fed 2018 Teklad Global 18% Protein Rodent Diet, Envigo Teklad. A stepwise reduction in food intake by increments of 10% per week, starting at 20% was carried out for mice in the CR group. Bodyweight and food intake were monitored weekly. Morning-Fed CR mice were fed daily at 6:00 – 7:00 a.m. and Night-Fed CR mice were fed daily at 6:00 – 7:00 p.m. The caloric intake of the mice in the AL group was calculated weekly to determine the appropriate number of calories to feed the mice in the CR group during the following week.
Method details
Metabolic Phenotyping – Morning Fed
Glucose, insulin and alanine tolerance tests (GTT, ITT and ATT) were performed by feeding male mice at 6 a.m. and removing food at 9 a.m. We then tested the respective fasting durations: 4, 8, 12, 16, 20 or 24 hrs. At each time point, we injected a subset of mice with either glucose (1 g/kg), insulin (0.5U/kg) or alanine (2g/kg) intraperitoneally78; no mouse was injected twice during the course of the 24 hr cycle. Glucose measurements were taken using a Bayer Contour blood glucose meter and test strips7. Meal stimulated insulin tolerance (MSIS) test was performed by feeding mice at 6:00 a.m. and removing food at 9:00 a.m. We then measured and collected blood during their respective fasting durations: 4, 8, 12, 16, 20 or 24 hrs starting at 1 p.m. After the collection of fasted blood mice were allowed to feed for 2 hours which we then measured collected the fed blood. Insulin levels were assessed with Crystal Chem Insulin ELISA. Mouse body composition was determined using an EchoMRI Body Composition Analyzer. Adiposity was calculated by taking the fat mass and dividing by the sum of the fat mass and lean mass. For assay of multiple metabolic parameters (O2, CO2, food consumption and activity tracking), mice were acclimated to housing in a Columbus Instruments Oxymax/CLAMS metabolic chamber system overnight. AL-fed mice had access to food overnight and Morning-Fed CR mice were given their daily pellet the following day at 7 a.m. Then we removed food at 10 a.m. for both AL- and CR-fed mice and collected data from a continuous 24-hr period was then recorded and analyzed. Female mice were examined under similar conditions as male mice except ITTs were conducted on two separate weeks so that no mice were injected twice on a single day. Female mice were tested in the respective fasting durations – first week: 4, 12 or 20 hrs and second week: 8, 16, or 24hrs.
Acute loss of insulin function with diazoxide
Acute diazoxide treatment study was performed by feeding mice at 6:00 a.m. and removing food at 9:00 a.m. We then injected diazoxide (0.75mg/bw) at 5:45 p.m. and then measured, collected fasted blood glucose, and allowed access to food for the mice. We initially set up the experiment for an MSIS (described above); however, the mice did not consume food. Therefore, we only measured blood glucose levels after 2 hr, 3hr and 4hr post-feeding.
Metabolic Phenotyping – Night Fed
Glucose, insulin and alanine tolerance tests (GTT, ITT and ATT) were performed by feeding mice at 6:00 p.m. and removing food at 9:00 p.m. We then tested the respective fasting durations: 4, 8, 12, 16, 20 or 24 hrs and then injecting either glucose (1 g/kg), insulin (0.5U/kg) or alanine (2g/kg) intraperitoneally78. Glucose measurements were taken using a Bayer Contour blood glucose meter and test strips. Meal stimulated insulin tolerance (MSIS) tests were performed by feeding mice at 6:00 p.m. and removing food at 9:00 p.m. We then measured and collected blood during their respective fasting durations: 4, 8, 12, 16, 20 or 24 hrs starting at 1 a.m. After the collection of fasted blood mice were allowed to feed for 2 hours which we then measured and collected the fed blood. Insulin levels were assessed with Crystal Chem Insulin ELISA. Mouse body composition was determined using an EchoMRI Body Composition Analyzer. For assay of multiple metabolic parameters (O2, CO2, food consumption and activity tracking), mice were acclimated to housing in a Columbus Instruments CLAMS-HC metabolic chamber system for ~24 hours. During this time AL-fed mice had continual access to food while CR mice were given their daily pellet at 6 p.m. Then we removed food at 9 p.m. for both AL- and CR-fed mice and collected data from a continuous 24 hr period was then recorded and analyzed.
Metabolic Phenotyping – TSCKO
Glucose and insulin tolerance tests (GTT and ITT) were performed by feeding mice at 7 a.m. and removing food at 10 a.m. We then tested after either 8 or 21 hrs of fasting. At each time point, we injected a subset of mice with either glucose (1 g/kg or 2g/kg) or insulin (0.75U/kg for 8hr fast or 0.5U/kg for 21hr Fast) intraperitoneally78.Glucose measurements were taken using a Bayer Contour blood glucose meter and test strips7. Mouse body composition was determined using an EchoMRI Body Composition Analyzer. For assay of multiple metabolic parameters (O2, CO2, food consumption and activity tracking), mice were acclimated to housing in a Columbus Instruments Oxymax/CLAMS metabolic chamber system overnight. AL-fed mice had access to food overnight and Morning-Fed CR mice were given their daily pellet the following day at 7 a.m. and collected data from a continuous 24-hr period was then recorded and analyzed.
Sacrifice and Collection of Tissues
Mice were sacrificed after 15 weeks on diet. Mice in the Morning-Fed studies were fed at 6 a.m. then mice were placed in new cages without food starting at 9 a.m. and sacrificed in the respective fasting durations: 4, 8, 12, 16, 20 or 24 hrs. Mice in the Night-Fed studies were fed at 6 p.m. then mice were placed in new cages without foodstarting at 9 p.m. and sacrificed in the respective fasting durations: 4, 8, 12, 16, 20 or 24 hrs. Following blood collection via submandibular bleeding, mice were euthanized by cervical dislocation and tissues (liver, muscle, iWAT, eWAT, BAT, and cecum) were rapidly collected, weighed, and then snap frozen in liquid nitrogen.
Immunoblotting
20–50 mg tissue samples from liver and muscle were lysed in cold RIPA buffer supplemented with phosphatase inhibitor and protease inhibitor cocktail tablets using a FastPrep 24 (M.P. Biomedicals) with bead-beating tubes (Stellar Scientific #BS-D1031-T20) and zirconium ceramic oxide bulk beads (VWR #10032–374). Protein lysates were then centrifuged at 13,300 rpm for 10 min and the supernatant was collected. Protein concentration was determined by Bradford (Pierce Biotechnology). 20 μg protein was separated by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) on 8%, 10%, or 16% resolving gels and transferred to PVDF membrane. To account for the variability between different western runs, we made a stock standard liver lysate sample which was run in duplicate on every gel. Membranes were blocked in 5% non-fat dry milk dissolved in TBST for 10 minutes and were then incubated in primary antibody diluted in 5% BSA (for phospho antibodies) or 5% milk (for non-phospho antibodies) overnight. The following commercial primary antibodies were used for immunoblot analysis p-S473 AKT (1:1,000; Cell Signaling Technology #4060), p-T308 AKT (1:1,000; Cell Signaling Technology # 2965), AKT (1:1,000; Cell Signaling Technology #4691), p-p70 S6K1 (1:500; Cell Signaling Technology #9234L), S6K (1:1,000; Cell Signaling Technology #2708S), p-S757-ULK (1:500; Cell Signaling Technology #5869S), ULK (1:500; Cell Signaling Technology #8054S), HSP90 (1:1,000; Cell Signaling Technology #4877), p-S240/244 S6 ribosomal protein (1:1,000; Cell Signaling Technology #2215), S6 ribosomal protein (1:1,000; Cell Signaling Technology #2217), p-T37/S46 4E-BP1 (1:1,000; Cell Signaling Technology #2855), 4E-BP1 (1:1,000; Cell Signaling Technology #9452). HRP-linked anti-rabbit (1:2,000; Cell Signaling Technology #7074S was used as secondary antibody. Imaging was performed using a GE ImageQuant LAS 4000 imaging station (GE Healthcare). Quantification was performed by densitometry using NIH ImageJ software.
Metabolomics
Plasma samples were thawed on ice then a 20 uL aliquot was treated with 480 uL of ice cold 5:3:2 MeOH:MeCN:water (v/v/v) then vortexed for 30 min at 4 degrees C. Supernatants were clarified by centrifugation (10 min, 12,000 g, 4 C). The resulting metabolite extracts were analyzed (20 uL per injection) by ultra-high-pressure liquid chromatography coupled to mass spectrometry (UHPLC-MS — Vanquish and Q Exactive, Thermo). Metabolites were resolved on a Kinetex C18 column (2.1 × 150 mm, 1.7 um) using a 5-minute gradient method in positive and negative ion modes (separate runs) exactly as previously described79. Following data acquisition, .raw files were converted to .mzXML using RawConverter then metabolites assigned, and peaks integrated using Maven (Princeton University) in conjunction with the KEGG database and an in-house standard library. Quality control was assessed as using technical replicates run at beginning, end, and middle of each sequence as previously described80.
Proteomics
Plasma samples were digested in the S-Trap filter (Protifi, Huntington, NY). It was performed following the manufacturer’s procedure. Briefly, around 50 μg of plasma proteins were first mixed with 5% SDS. Samples were reduced with 10 mM DTT at 55 °C for 30 min, cooled to room temperature, and then alkylated with 25 mM iodoacetamide in the dark for 30 min. Afterward, to the samples was added a final concentration of 1.2% phosphoric acid and then six volumes of binding buffer (90% methanol; 100 mM triethylammonium bicarbonate, TEAB; pH 7.1). After gentle mixing, the protein solution was loaded to an S-Trap filter, spun at 2000 rpm for 1 min, and the flow-through collected and reloaded onto a filter. This step was repeated three times, and then the filter was washed with 200 μL of binding buffer 3 times. Finally, 1 μg of sequencing-grade trypsin and 150 μL of digestion buffer (50 mM TEAB) were added into the filter and digested at 47 °C for 1 h. The system was coupled to the timsTOF Pro mass spectrometer (Bruker Daltonics, Bremen, Germany) via the nano-electrospray ion source (Captive Spray, Bruker Daltonics) as previously describe81. Raw data files conversion to peak lists in the MGF format, downstream identification, validation, filtering and quantification were managed using FragPipe version 13.0. MSFragger version 3.0 was used for database searches against a mouse database with decoys and common contaminants added. The identification settings were as follows: Trypsin, Specific, with a maximum of 2 missed cleavages, up to 2 isotope errors in precursor selection allowed for, 10.0 ppm as MS1 and 20.0 ppm as MS2 tolerances; fixed modifications: Carbamidomethylation of C (+57.021464 Da), variable modifications: Oxidation of M (+15.994915 Da), Acetylation of protein N-term (+42.010565 Da), Pyrolidone from peptide N-term Q or C (−17.026549 Da).
Randomization
All studies were performed on animals or tissues collected from animals. Animals of each sex and strain were randomized into groups of equivalent weight prior to the beginning of the in vivo studies. Additionally, mice were randomized during each test so that no mice in the AL-group would encounter a weekly 16+ hour fast.
Quantification and statistical analysis
Metabolomics and Proteomics Data Cleaning and Normalization
Plasma samples were analyzed via UHPLC-MS to determine plasma levels of 170 different metabolites, and plasma samples were analyzed via LC-MS/MS as described to determine levels of 856 distinct proteins. Metabolite peak intensity values that were equal to zero were removed from the dataset and missing data were imputed using a multiple imputation methodology (classification and regression tree (CART))82. Due to the large number of zero values in the proteomics dataset, the data were filtered so that data columns with >75% zeroes were dropped. Then the data were filtered further based on standard deviation (variables with SD <3 were dropped). After imputation of missing values and/or filtering, a log (base 10) transformation was applied to both the metabolomics and proteomics datasets. Finally, the datasets were both scaled and centered using pareto scaling before further analyses were conducted. Final analysis of 170 metabolites and 294 proteins were conducted as described below. Fold-changes between each study group (CR-8hr/AL-8hr; CR-12hr/AL-12hr; CR-16hr/AL-16hr; CR-24hr/AL-24hr) were calculated using the raw data (non-normalized) for both the metabolomics and proteomics datasets. Then, Welch Two Sample t-tests were used to determine which metabolites/proteins were significantly different between each of the study groups. Volcano plots were generated to show significantly increased and decreased metabolites/proteins for each of the four study group pairs.
Principal Components Analysis
Principal Components Analysis (PCA) was conducted to reduce dimensionality in the dataset for downstream analyses and to understand whether diet (AL vs. CR) and/or fasting duration (4, 8, 12, or 16 hours) resulted in distinct data clusters corresponding to specific metabolites/proteins. To determine which factors to retain, we used the standard Kaiser criterion, which suggests that factors with eigenvalues >1 should be extracted83.
General statistical analysis
Data are presented as mean ± SEM unless otherwise specified. Analyses were performed using Excel (2010 and 2016, Microsoft) or Prism 9 (GraphPad Software). Statistical analyses were performed using one or two-way ANOVA followed by Sidak or Tukey-Kramer post hoc test specified in the figure legends. Other statistical details can also be found in the figure legends; in all figures, n represents the number of biologically independent animals. Sample sizes for metabolic studies were determined based on our previously published experimental results with the effects of dietary interventions84, with the goal of having > 90% power to detect a change in area under the curve during a GTT (p<0.05). Data distribution was assumed to be normal, but this was not formally tested. All statistical analyses and data visualization of the metabolomics and proteomics data were performed using RStudio for Mac (RStudio Team [2022]. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA URL). The alpha level for null hypothesis rejection was set at 0.05. Data are presented as mean ± standard error of the mean (SEM), unless otherwise noted.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-p-S6 ribosomal protein (Ser240/244) | Cell Signaling Technology | Cat# 2215; RRID: AB_331682 |
| Rabbit anti-S6 ribosomal protein | Cell Signaling Technology | Cat#2217; RRID: AB_331355 |
| Rabbit anti-4E-BP1 | Cell Signaling Technology | Cat# 9644; RRID: AB_2097841 |
| Rabbit Anti-p-4E-BP1 (Thr37/46) | Cell Signaling Technology | Cat# 2855; RRID: AB_560835 |
| Rabbit anti-HSP90 | Cell Signaling Technology | Cat# 4877; RRID: AB_2233307 |
| Rabbit anti-ATF-4 | Cell Signaling Technology | Cat# 11815; RRID: AB_2616025 |
| Rabbit anti-p- AKT (Ser473) | Cell Signaling Technology | Cat# 4060; RRID: AB_2315049 |
| Rabbit anti-p-AKT (Thr308) | Cell Signaling Technology | Cat#2965; RRID: AB_2255933 |
| Rabbit anti-AKT | Cell Signaling Technology | Cat# 4691; RRID: AB_915783 |
| Rabbit anti-p-ULK (Ser757) | Cell Signaling Technology | Cat# 5869; RRID: AB_10707365 |
| Rabbit anti-ULK | Cell Signaling Technology | Cat# 8054; RRID: AB_11178668 |
| HRP-linked anti-rabbit | Cell Signal Technology | Cat# 7074 RRID: AB_2099233 |
| Chemicals, peptides, and recombinant proteins | ||
| Human insulin | Eli Lilly | NDC 0002-8215-17 (Humulin R U-100) |
| Diazoxide | Sigma Aldrich | Cat# D9035–250MG |
| Critical commercial assays | ||
| Ultra-sensitive mouse insulin ELISA | Crystal Chem | Cat# 90080 |
| Deposited data | ||
| Metabolomics Data Set | This paper | MetaboLights: MTBLS10795 |
| Proteomics Data Set | This Paper | MassIVE: MSV000095510 |
| Experimental models: Organisms/strains | ||
| Mouse strain: C57BL/6J | The Jackson Laboratory | Cat# JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse strain:C57BL/6J; Albumin-Cre Tsc1loxP/loxP | Yu et al.76 | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | http://www.graphpad.com/scientific-software/prism |
| RStudio for Mac (Version 2024.04.1 + 748) | N/A | https://www.r-project.org/ |
| ImageJ | National Institutes of Health, (Schneider et al., 2012) | https://imagej.nih.gov/ij |
| Other | ||
| 2018 Teklad Global 18% Protein Rodent Diet | Envigo Teklad | 2018 |
ACKNOWLEDGEMENTS
We would like to thank all members of the Lamming lab. The Lamming laboratory is supported in part by the NIH/NIA (AG056771, AG062328, AG081482, and AG084156 to D.W.L.), NIH/NIDDK (DK125859 to D.W.L.) and startup funds from the University of Wisconsin-Madison School of Medicine and Public Health and Department of Medicine to D.W.L. H.H.P. was supported by F31AG066311. R.B. is supported by F31AG081115. CLG was supported in part by Dalio Philanthropies, the Glenn Foundation for Medical Research Postdoctoral Fellowship, and by grant HF-AGE AGE-009 from Hevolution Foundation. Support for this research was provided by the University of Wisconsin - Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. This work was supported in part by the U.S. Department of Veterans Affairs (I01-BX004031 and IS1-BX005524) and used facilities and resources from the William S. Middleton Memorial Veterans Hospital. We would also like to thank Abby Grier for her contributions to data acquisition, analysis, and initial statistical evaluation of the metabolomics and proteomics datasets. The Paredes laboratory was supported in part by the Movement Disorder Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work does not represent the views of the Department of Veterans Affairs or the United States Government.
COMPETING INTERESTS
D.W.L has received funding from, and is a scientific advisory board member of, Aeovian Pharmaceuticals, which seeks to develop novel, selective mTOR inhibitors for the treatment of various diseases. The remaining authors declare no competing interests.
REFERENCES
- 1.Green CL, Lamming DW, and Fontana L (2022). Molecular mechanisms of dietary restriction promoting health and longevity. Nat Rev Mol Cell Biol 23, 56–73. 10.1038/s41580-021-00411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acosta-Rodriguez VA, de Groot MHM, Rijo-Ferreira F, Green CB, and Takahashi JS (2017). Mice under Caloric Restriction Self-Impose a Temporal Restriction of Food Intake as Revealed by an Automated Feeder System. Cell Metab 26, 267–277 e262. 10.1016/j.cmet.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pak HH, Haws SA, Green CL, Koller M, Lavarias MT, Richardson NE, Yang SE, Dumas SN, Sonsalla M, Bray L, et al. (2021). Fasting drives the metabolic, molecular and geroprotective effects of a calorie-restricted diet in mice. Nat Metab 3, 1327–1341. 10.1038/s42255-021-00466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell SJ, Bernier M, Mattison JA, Aon MA, Kaiser TA, Anson RM, Ikeno Y, Anderson RM, Ingram DK, and de Cabo R (2019). Daily Fasting Improves Health and Survival in Male Mice Independent of Diet Composition and Calories. Cell Metab 29, 221–228 e223. 10.1016/j.cmet.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedrich HJ (2012). Preface. In The Laboratory Mouse (Second Edition), Hedrich HJ, ed. (Academic Press; ), pp. xxi. 10.1016/B978-0-12-382008-2.05001-7. [DOI] [Google Scholar]
- 6.Ebert B (2009). Automation of rodent fasting. In 5. (AMER ASSOC LABORATORY ANIMAL SCIENCE 9190 CRESTWYN HILLS DR, MEMPHIS, TN …), pp. 537–538. [Google Scholar]
- 7.Bellantuono I, de Cabo R, Ehninger D, Di Germanio C, Lawrie A, Miller J, Mitchell SJ, Navas-Enamorado I, Potter PK, Tchkonia T, et al. (2020). A toolbox for the longitudinal assessment of healthspan in aging mice. Nature protocols 15, 540–574. 10.1038/s41596-019-0256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamming DW (2014). Diminished mTOR signaling: a common mode of action for endocrine longevity factors. SpringerPlus 3, 735. 10.1186/2193-1801-3-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, and Hart RW (1999). Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci 54, B492–501. 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- 10.Solon-Biet SM, Mitchell SJ, Coogan SC, Cogger VC, Gokarn R, McMahon AC, Raubenheimer D, de Cabo R, Simpson SJ, and Le Couteur DG (2015). Dietary Protein to Carbohydrate Ratio and Caloric Restriction: Comparing Metabolic Outcomes in Mice. Cell reports 11, 1529–1534. 10.1016/j.celrep.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, Gonzalez-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, et al. (2016). Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab 23, 1093–1112. 10.1016/j.cmet.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell SE, Tang Z, Kerbois C, Delville C, Konstantopedos P, Bruel A, Derous D, Green C, Aspden RM, Goodyear SR, et al. (2015). The effects of graded levels of calorie restriction: I. impact of short term calorie and protein restriction on body composition in the C57BL/6 mouse. Oncotarget 6, 15902–15930. 10.18632/oncotarget.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S (1981). Effects of diazoxide on insulin secretion and metabolic efficiency in the db/db mouse. Life Sciences 28, 1829–1840. [DOI] [PubMed] [Google Scholar]
- 14.Bergsten P, Gylfe E, Wesslen N, and Hellman B (1988). Diazoxide unmasks glucose inhibition of insulin release by counteracting entry of Ca2+. American Journal of Physiology-Endocrinology and Metabolism 255, E422–E427. [DOI] [PubMed] [Google Scholar]
- 15.Bergsten P, and Hellman B (1987). Demonstration of glucose inhibition of insulin release in the presence of diazoxide. European Journal of Endocrinology 115, 170–174. [DOI] [PubMed] [Google Scholar]
- 16.Mutel E, Gautier-Stein A, Abdul-Wahed A, Amigo-Correig M, Zitoun C, Stefanutti A, Houberdon I, Tourette JA, Mithieux G, and Rajas F (2011). Control of blood glucose in the absence of hepatic glucose production during prolonged fasting in mice: induction of renal and intestinal gluconeogenesis by glucagon. Diabetes 60, 3121–3131. 10.2337/db11-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalsbeek A, and Strubbe JH (1998). Circadian Control of Insulin Secretion Is Independent of the Temporal Distribution of Feeding. Physiology & Behavior 63, 553–560. 10.1016/S0031-9384(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 18.Reznick J, Preston E, Wilks DL, Beale SM, Turner N, and Cooney GJ (2013). Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 1832, 228–238. [DOI] [PubMed] [Google Scholar]
- 19.Tucci V, Hardy A, and Nolan PM (2006). A comparison of physiological and behavioural parameters in C57BL/6J mice undergoing food or water restriction regimes. Behav Brain Res 173, 22–29. 10.1016/j.bbr.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 20.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. (2011). The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322. 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, and Blenis J (2011). Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326. 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 23.Speakman JR, Mitchell SE, and Mazidi M (2016). Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp Gerontol 86, 28–38. 10.1016/j.exger.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Fontana L, and Partridge L (2015). Promoting health and longevity through diet: from model organisms to humans. Cell 161, 106–118. 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Most J, Tosti V, Redman LM, and Fontana L (2017). Calorie restriction in humans: An update. Ageing Res Rev 39, 36–45. 10.1016/j.arr.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalaany NY, and Sabatini DM (2009). Tumours with PI3K activation are resistant to dietary restriction. Nature 458, 725–731. 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, and Hellerstein MK (2010). Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am J Physiol Endocrinol Metab 298, E108–116. 10.1152/ajpendo.00524.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontana L, Klein S, and Holloszy JO (2010). Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age (Dordr) 32, 97–108. 10.1007/s11357-009-9118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horne BD, Muhlestein JB, and Anderson JL (2015). Health effects of intermittent fasting: hormesis or harm? A systematic review. Am J Clin Nutr 102, 464–470. 10.3945/ajcn.115.109553. [DOI] [PubMed] [Google Scholar]
- 30.Rynders CA, Thomas EA, Zaman A, Pan Z, Catenacci VA, and Melanson EL (2019). Effectiveness of Intermittent Fasting and Time-Restricted Feeding Compared to Continuous Energy Restriction for Weight Loss. Nutrients 11. 10.3390/nu11102442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowosad K, and Sujka M (2021). Effect of Various Types of Intermittent Fasting (IF) on Weight Loss and Improvement of Diabetic Parameters in Human. Curr Nutr Rep 10, 146–154. 10.1007/s13668-021-00353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. (2015). Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350, aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saini C, Petrenko V, Pulimeno P, Giovannoni L, Berney T, Hebrok M, Howald C, Dermitzakis ET, and Dibner C (2016). A functional circadian clock is required for proper insulin secretion by human pancreatic islet cells. Diabetes, Obesity and Metabolism 18, 355–365. 10.1111/dom.12616. [DOI] [PubMed] [Google Scholar]
- 34.Simcox J, and Lamming DW (2022). The central moTOR of metabolism. Dev Cell 57, 691–706. 10.1016/j.devcel.2022.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadopoli D, Boulay K, Kazak L, Pollak M, Mallette F, Topisirovic I, and Hulea L (2019). mTOR as a central regulator of lifespan and aging. F1000Res 8. 10.12688/f1000research.17196.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichhart T (2018). mTOR as Regulator of Lifespan, Aging, and Cellular Senescence: A Mini-Review. Gerontology 64, 127–134. 10.1159/000484629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan H, and Finkel T (2017). Key proteins and pathways that regulate lifespan. J Biol Chem 292, 6452–6460. 10.1074/jbc.R116.771915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, and Kennedy BK (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196. 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 39.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, and Partridge L (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11, 35–46. 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greer EL, and Brunet A (2009). Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127. 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syntichaki P, Troulinaki K, and Tavernarakis N (2007). eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445, 922–926. 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 42.Henderson ST, Bonafe M, and Johnson TE (2006). daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci 61, 444–460. 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- 43.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, and Kenyon C (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110. 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 44.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fok WC, Bokov A, Gelfond J, Yu Z, Zhang Y, Doderer M, Chen Y, Javors M, Wood WH 3rd, Zhang Y, et al. (2014). Combined treatment of rapamycin and dietary restriction has a larger effect on the transcriptome and metabolome of liver. Aging Cell 13, 311–319. 10.1111/acel.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fok WC, Chen Y, Bokov A, Zhang Y, Salmon AB, Diaz V, Javors M, Wood WH 3rd, Zhang Y, Becker KG, et al. (2014). Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS One 9, e83988. 10.1371/journal.pone.0083988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fok WC, Livi C, Bokov A, Yu Z, Chen Y, Richardson A, and Perez VI (2014). Short-term rapamycin treatment in mice has few effects on the transcriptome of white adipose tissue compared to dietary restriction. Mech Ageing Dev 140, 23–29. 10.1016/j.mad.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fok WC, Zhang Y, Salmon AB, Bhattacharya A, Gunda R, Jones D, Ward W, Fisher K, Richardson A, and Perez VI (2013). Short-term treatment with rapamycin and dietary restriction have overlapping and distinctive effects in young mice. J Gerontol A Biol Sci Med Sci 68, 108–116. 10.1093/gerona/gls127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Z, Wang R, Fok WC, Coles A, Salmon AB, and Perez VI (2015). Rapamycin and dietary restriction induce metabolically distinctive changes in mouse liver. J Gerontol A Biol Sci Med Sci 70, 410–420. 10.1093/gerona/glu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, Brenner C, Masri S, Benitah SA, and Sassone-Corsi P (2017). Circadian Reprogramming in the Liver Identifies Metabolic Pathways of Aging. Cell 170, 664–677 e611. 10.1016/j.cell.2017.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, and Hall MN (2014). Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci U S A 111, 11592–11599. 10.1073/pnas.1412047111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, and Kondratov RV (2014). BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 6, 48–57. 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khapre RV, Patel SA, Kondratova AA, Chaudhary A, Velingkaar N, Antoch MP, and Kondratov RV (2014). Metabolic clock generates nutrient anticipation rhythms in mTOR signaling. Aging (Albany NY) 6, 675–689. 10.18632/aging.100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Igarashi M, and Guarente L (2016). mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell 166, 436–450. 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 55.Tulsian R, Velingkaar N, and Kondratov R (2018). Caloric restriction effects on liver mTOR signaling are time-of-day dependent. Aging (Albany NY) 10, 1640–1648. 10.18632/aging.101498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.La Fleur SE, Kalsbeek A, Wortel J, and Buijs RM (1999). A suprachiasmatic nucleus generated rhythm in basal glucose concentrations. Journal of neuroendocrinology 11, 643–652. 10.1046/j.1365-2826.1999.00373.x. [DOI] [PubMed] [Google Scholar]
- 57.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, and Buijs RM (2001). A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 50, 1237–1243. 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- 58.la Fleur SE, Kalsbeek A, Wortel J, van der Vliet J, and Buijs RM (2001). Role for the pineal and melatonin in glucose homeostasis: pinealectomy increases night-time glucose concentrations. Journal of neuroendocrinology 13, 1025–1032. 10.1046/j.1365-2826.2001.00717.x. [DOI] [PubMed] [Google Scholar]
- 59.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631. 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. (2005). Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308, 1043–1045. 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, and Fitzgerald GA (2004). BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2, e377. 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. (2010). Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16, 1152–1156. 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honma KI, Honma S, and Hiroshige T (1984). Feeding-associated corticosterone peak in rats under various feeding cycles. The American journal of physiology 246, R721–726. 10.1152/ajpregu.1984.246.5.R721. [DOI] [PubMed] [Google Scholar]
- 64.Chan S, and Debono M (2010). Replication of cortisol circadian rhythm: new advances in hydrocortisone replacement therapy. Ther Adv Endocrinol Metab 1, 129–138. 10.1177/2042018810380214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, Hall D, Davis S, Nelson CS, Brem RB, et al. (2016). Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell metabolism 23, 143–154. 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mezhnina V, Ebeigbe OP, Velingkaar N, Poe A, Sandlers Y, and Kondratov RV (2022). Circadian clock controls rhythms in ketogenesis by interfering with PPARalpha transcriptional network. Proc Natl Acad Sci U S A 119, e2205755119. 10.1073/pnas.2205755119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mezhnina V, Pearce R, Poe A, Velingkaar N, Astafev A, Ebeigbe OP, Makwana K, Sandlers Y, and Kondratov RV (2020). CR reprograms acetyl-CoA metabolism and induces long-chain acyl-CoA dehydrogenase and CrAT expression. Aging cell 19, e13266. 10.1111/acel.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Green CL, Soltow QA, Mitchell SE, Derous D, Wang Y, Chen L, Han JJ, Promislow DEL, Lusseau D, Douglas A, et al. (2019). The Effects of Graded Levels of Calorie Restriction: XIII. Global Metabolomics Screen Reveals Graded Changes in Circulating Amino Acids, Vitamins, and Bile Acids in the Plasma of C57BL/6 Mice. J Gerontol A Biol Sci Med Sci 74, 16–26. 10.1093/gerona/gly058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kowalski GM, and Bruce CR (2014). The regulation of glucose metabolism: implications and considerations for the assessment of glucose homeostasis in rodents. Am J Physiol Endocrinol Metab 307, E859–871. 10.1152/ajpendo.00165.2014. [DOI] [PubMed] [Google Scholar]
- 70.Matyi S, Jackson J, Garrett K, Deepa SS, and Unnikrishnan A (2018). The effect of different levels of dietary restriction on glucose homeostasis and metabolic memory. Geroscience 40, 139–149. 10.1007/s11357-018-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Velingkaar N, Mezhnina V, Poe A, Makwana K, Tulsian R, and Kondratov RV (2020). Reduced caloric intake and periodic fasting independently contribute to metabolic effects of caloric restriction. Aging cell 19, e13138. 10.1111/acel.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitchell SE, Delville C, Konstantopedos P, Hurst J, Derous D, Green C, Chen L, Han JJ, Wang Y, Promislow DE, et al. (2015). The effects of graded levels of calorie restriction: II. Impact of short term calorie and protein restriction on circulating hormone levels, glucose homeostasis and oxidative stress in male C57BL/6 mice. Oncotarget 6, 23213–23237. 10.18632/oncotarget.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Velingkaar N, Mezhnina V, Poe A, and Kondratov RV (2021). Two-meal caloric restriction induces 12-hour rhythms and improves glucose homeostasis. FASEB J 35, e21342. 10.1096/fj.202002470R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green CL, Pak HH, Richardson NE, Flores V, Yu D, Tomasiewicz JL, Dumas SN, Kredell K, Fan JW, Kirsh C, et al. (2022). Sex and genetic background define the metabolic, physiologic, and molecular response to protein restriction. Cell Metab 34, 209–226 e205. 10.1016/j.cmet.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barrington WT, Wulfridge P, Wells AE, Rojas CM, Howe SYF, Perry A, Hua K, Pellizzon MA, Hansen KD, Voy BH, et al. (2018). Improving Metabolic Health Through Precision Dietetics in Mice. Genetics 208, 399–417. 10.1534/genetics.117.300536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu D, Richardson NE, Green CL, Spicer AB, Murphy ME, Flores V, Jang C, Kasza I, Nikodemova M, Wakai MH, et al. (2021). The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab 33, 905–922 e906. 10.1016/j.cmet.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu D, Yang SE, Miller BR, Wisinski JA, Sherman DS, Brinkman JA, Tomasiewicz JL, Cummings NE, Kimple ME, Cryns VL, and Lamming DW (2018). Short-term methionine deprivation improves metabolic health via sexually dimorphic, mTORC1-independent mechanisms. FASEB J 32, 3471–3482. 10.1096/fj.201701211R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu D, Tomasiewicz JL, Yang SE, Miller BR, Wakai MH, Sherman DS, Cummings NE, Baar EL, Brinkman JA, Syed FA, and Lamming DW (2019). Calorie-Restriction-Induced Insulin Sensitivity Is Mediated by Adipose mTORC2 and Not Required for Lifespan Extension. Cell reports 29, 236–248 e233. 10.1016/j.celrep.2019.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nemkov T, Reisz JA, Gehrke S, Hansen KC, and D’Alessandro A (2019). High-Throughput Metabolomics: Isocratic and Gradient Mass Spectrometry-Based Methods. Methods Mol Biol 1978, 13–26. 10.1007/978-1-4939-9236-2_2. [DOI] [PubMed] [Google Scholar]
- 80.Nemkov T, Hansen KC, and D’Alessandro A (2017). A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun Mass Spectrom 31, 663–673. 10.1002/rcm.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas T, Stefanoni D, Dzieciatkowska M, Issaian A, Nemkov T, Hill RC, Francis RO, Hudson KE, Buehler PW, Zimring JC, et al. (2020). Evidence of Structural Protein Damage and Membrane Lipid Remodeling in Red Blood Cells from COVID-19 Patients. J Proteome Res 19, 4455–4469. 10.1021/acs.jproteome.0c00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van Buuren S, and Groothuis-Oudshoorn K (2011). mice: Multivariate Imputation by Chained Equations in R. Journal of Statistical Software 45, 1–67. 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 83.Braeken J, and van Assen MALM (2017). An empirical Kaiser criterion. Psychological Methods 22, 450–466. 10.1037/met0000074. [DOI] [PubMed] [Google Scholar]
- 84.Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. (2016). Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell reports 16, 520–530. 10.1016/j.celrep.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metabolomics and proteomics datasets are publicly available at the following: Metabolomics data have been deposited to the EMBL-EBI MetaboLights database (DOI: 10.1093/nar/gkad1045, PMID:37971328) with the identifier MTBLS10795 and can be accessed in the following https://www.ebi.ac.uk/metabolights/MTBLS10795. Proteomics data have been deposited to MassIVE with the identifier MSV000095510.
The custom R code that was written for this publication is publicly available at the following dedicated GitHub repository: https://github.com/linsemanlab/Pak-et-al-2023.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


