Abstract
To assess the antigenicity of envelope glycoproteins derived from primary human immunodeficiency virus type 1 populations, their interactions with the receptor CD4, and their coreceptor usage, we have cloned and expressed multiple gp120 proteins from a number of primary virus isolates. Characterization of these proteins showed a high degree of antigenic polymorphism both within the CD4 binding site and in defined neutralization epitopes, which may partially account for the general resistance of primary isolates to neutralizing agents. Furthermore, chimeric viruses expressing gp120 proteins with reduced CD4 binding abilities are viable, suggesting that primary viruses may require a less avid interaction with the receptor CD4 to initiate infection than do their laboratory-adapted counterparts. The coreceptor usage of chimeric viruses was related to the ability of the virus to bind CD4, with reduced CD4 binding correlating with preferential usage of CXCR4. Changes in coreceptor usage mapped to sequence changes in the C2 and V4 regions, with no changes seen in the V3 region.
Human immunodeficiency virus type 1 (HIV-1) predominantly infects T-helper cells and macrophages, inducing profound immunosuppression. This tropism correlates with expression of the viral receptor CD4 (9, 29) and one of several chemokine receptors which act as strain-dependent coreceptors (1, 5, 11, 14–16, 44; reviewed in reference 7). The binding determinants for both CD4 and coreceptor reside in the external gp120 subunit of the envelope glycoprotein (Env). Much evidence suggests that entry occurs through an initial gp120-CD4 binding event which induces conformational changes in gp120, facilitating the formation of a ternary gp120-CD4-coreceptor complex and culminating in activation of the fusogenic gp41 subunit of Env (39, 45, 47). The molecular interaction of monomeric HIV-1 glycoprotein with CD4 and its coreceptors has been defined (25, 43, 62) and has been modeled for oligomeric gp120 (26); however, this is complicated by the extensive degree of variation present within the env gene.
The coreceptor usage of a primary virus isolate correlates well with tropism for macrophages (CCR5 utilizing [R5]) and T-cell lines (CXCR4 utilizing [X4]). Viral isolates obtained from 40% of infected individuals during the course of disease progression change their in vitro properties from the R5 to the X4 phenotype (8, 57), suggesting that the appearance of X4-utilizing virus is associated with a more rapid CD4 cell decline and onset of symptoms (stage IV disease) (3, 8, 23, 49). These late-stage isolates are often dualtropic (R5X4), utilizing both CCR5 and CXCR4, and even at the molecular level, dualtropic isolates are composed predominantly of dualtropic R5X4 variants, as opposed to a mix of monotropic R5 and X4 viruses (54). The change from R5 to X4 has been associated with the occurrence of basic residues in the V3 region of gp120 (4, 19, 37) and changes elsewhere (38, 55).
Several studies have shown that intrasample sequence diversity increases during infection until the onset of stage IV disease and subsequently fails to increase or may even decline (10, 31, 32, 42, 50, 51, 64). All isolates obtained for this study were from patients showing signs of immunodeficiency and from whom X4 viruses were isolated either between the time points sampled or immediately thereafter. Hence, the level of genetic polymorphism in these samples is expected to be high. We characterized multiple gp120 glycoproteins derived from a number of paired isolates and demonstrated that each isolate consisted of a polymorphic population of phenotypically distinguishable variants. This antigenic variation may contribute to the resistance of primary viruses to neutralization by various ligands. Significantly, considerable variation was observed in gp120 affinity for CD4, and such polymorphism associated with the coreceptor usage of chimeric viruses expressing the heterologous gp120s.
MATERIALS AND METHODS
PCR amplification and cloning of gp120-encoding regions.
RNA was prepared from 200 μl of cell supernatant by the method of Chomezynski and Saachi (6). The RNA pellet was resuspended in 20 μl distilled water, and 10 μl was used to set up a cDNA synthesis. Each reaction consisted of 40 U of Superscript reverse transcriptase (Life Technologies, Paisley, Scotland) in a volume of 20 μl, 1× reverse transcription (RT) buffer, 1 mM each of the four deoxynucleoside triphosphates (dNTPs), and 20 ng of the antisense primer 631L (CCA GAC YGT GAG TTG CAA CAG ATG C, where Y is C or T). The reactions were incubated at 42°C for 90 min and then frozen at −20°C. A first-round PCR was performed in a volume of 50 μl, using 5 μl of either cDNA or isolate DNA containing provirus. The PCR contained 1.25 U of the proofreading polymerase Pfu (Stratagene, Cambridge, United Kingdom) and 0.125 U of Tbr polymerase (NBL, Cramlington, United Kingdom) in 1× Pfu buffer, with 200 μM each of the four dNTPs and 400 ng each of 627L (GAT GTT GAT GAT CTG TAG TGC, sense), 989L (TCA TCA AGT TTC TCT AYC AAA GC, sense), 632L (GCG CCC ATA GTG CTT CCT GCT GC, antisense), and 631L (CCA GAC TGT GAG TTG CAA CAG ATG C, antisense) per ml. The inclusion of multiple primers allowed the amplification of templates even when one of the primers failed to match the template. The PCR consisted of 30 cycles of 94°C for 45 s, 45°C for 45 s, and 72°C for 210 s and was performed in a Perkin-Elmer TC1 thermal cycler. Of the completed PCR, 2 μl was used to initiate a second reaction in a volume of 20 μl. This second-round PCR mixture contained 0.5 U of Pfu polymerase (Stratagene, Cambridge, United Kingdom) and 0.05 U of Tbr polymerase in 1× Pfu buffer, with 200 μM each of the four dNTPs and 400 ng each of primers 626L (GTG GGT CAC CGT CTA TTA TGG G, sense, BstEII restriction site underlined) and E524 (B-CAC CAC GCG TCT CTT TGC CTT GGT GGG, antisense, MluI restriction site underlined; B, biotin) per ml. The second-round PCR consisted of 25 cycles of 94°C for 35 s, 55°C for 35 s, and 72°C for 150 s. Of the completed PCR, 10 μl was visualized by agarose gel electrophoresis to assess the concentration of the PCR product. The PCR product was cloned into the expression vector pCDNA3-tpa-CA (Invitrogen), modified by insertion of the RER-targeting leader sequence of tissue plasminogen activator at the HindIII and BamHI restriction sites and by the addition of uracilated tails (Life Technologies) to the linearized vector to facilitate PCR cloning. A tertiary PCR was set up with 100 ng of the PCR product in a volume of 50 μl. This reaction mixture contained 1.25 U of Tbr polymerase in 1× Tbr buffer, with 200 μM each of the dNTPs and 400 ng of uracilated versions of 626L and E524 per ml. In order to minimize PCR-induced error, this reaction was performed using the same reaction temperatures and times as for the second-round amplification, but with a reduced cycle number (10 cycles). Together, these steps act to reduce the effective number of amplification cycles. Screening of transformants typically showed 60 to 100% of colonies to contain inserts.
Expression and characterization of soluble gp120.
For each isolate, 8 to 26 plasmids were transiently transfected using calcium phosphate into the human embryonic kidney cell line 293 previously infected with vTF7-3, a recombinant vaccinia virus expressing T7 polymerase (17). Extracellular supernatants were collected after 72 h, clarified by centrifugation, and assayed for gp120 levels in a quantitative capture enzyme immunoassay (EIA) (35). Saturating concentrations (500 ng/ml) of gp120 were tested for their ability to bind a polyclonal human serum (QC256), soluble CD4-immunoglobulin (sCD4-Ig) (5 μg/ml) (Genentech, San Francisco, Calif.), and a panel of neutralizing monoclonal antibodies (MAbs). The MAbs studied were 38.1a (34), recognizing a linear epitope in the C4 region (amino acids [a.a.] 427 to 436); 728, 589, and 654 (18, 27), human MAbs specific for the discontinuous CD4 binding site (b.s.); 39.3b (34) and 10/46c (unpublished), rat MAbs specific for the CD4 b.s.; 12b (35), recognizing a linear epitope in V2 (a.a. 181 to 191); 697 (18) and 55/39 (52), recognizing conformation-dependent V2 epitopes; 268 and 257 (60), human MAbs specific for linear epitopes in V3 (HIGPGR and KRIHI, respectively); and 2G12 (58), recognizing a discontinuous epitope dependent on N-linked glycosylation in C2, C3, V4, and C4. Proteins were also tested for their ability to bind a nonneutralizing MAb (58/30; unpublished), recognizing a conserved nonlinear gp120 epitope independent of the CD4 b.s. Bound ligands were visualized with either anti-human Ig-horseradish peroxidase (HRP) or anti-rat Ig-HRP (Harlan SeraLabs, Crawley, United Kingdom). Results were expressed as the ratio of the optical density (OD) of the expressed protein to BH10 protein (or MN gp120 for V3 MAbs) and termed the relative binding index (B.I.).
Construction of chimeric molecular clones.
Four isolates (669 and 1629 from individual A, 2401 from individual D, and 5015 from individual C) were chosen from the eight isolates described previously. Isolates 669 and 1629 were selected as the only confirmed R5-to-X4 switch pair. From each isolate, a number of gp120 open reading frames (ORFs) previously shown to display a range of CD4-binding abilities were chosen for transfer into chimeric viruses.
The vector used to provide all of the HIV-1 genes except the gp120 ORF was a molecular clone containing a truncated gp120 ORF with a 299-bp deletion in the V4 and V5 regions (HXB2Δenv) (33). The gp120 region of the env ORF was removed from this construct by digestion with the restriction enzymes BstEII and MluI. The chosen primary virus-derived gp120 ORFs were excised from the vector pCDNA3-tpa-CA, using BstEII and MluI, and ligated with the prepared pHXB2-Δenv. Colonies were screened for the presence of the gp120 insert by PCR, and positive colonies were confirmed by restriction digestion of plasmid DNA. Use of HXB2-Δenv rather than “full-length” HXB2 as the vector backbone allowed any bacterial transformants containing contaminating vector to be distinguished by size from those containing the desired insert. Chimeric viruses were designated by adding the gp120 clone number to the patient and isolate time; thus, chimera A2.172 is derived from gp120 clone 172, which was obtained from the second isolate (1629) of patient A.
Chemokine receptor usage of chimeric viruses.
Chimeric molecular clone DNAs were transfected into HEK-293 cells using Lipofectamine (Life Technologies, Paisley, Scotland), and production of p24 antigen in the extracellular supernatant was measured by capture EIA (35). The clarified supernatant was used to infect U87-CD4 cells expressing a range of chemokine receptors. Cells were fixed with methanol-acetone 3 days postinfection, and infection was identified by visualizing intracellular p24 antigen using two mouse anti-p24 MAbs (EF7.1 and 38.96K; Medical Research Council [MRC] AIDS Reagent Programme), followed by an anti-mouse Ig β-galactosidase-conjugated antibody. Inhibition of infection by an anti-CD4 MAb was assessed by treating cells with 1 μg of ADP 318 (MRC AIDS Reagent Programme) per ml for 1 h prior to incubation with virus-containing extracellular supernatant, as described above. Levels of MAb were maintained throughout the infection, and infected cells were visualized by staining with the anti-gp120 MAb 2G12, followed by an anti-human Ig β-galactosidase-2 conjugated antibody.
Sequencing.
Plasmid DNA was prepared from the pCDNA3-tpa clones using a Wizard miniprep kit (Promega, Southampton, United Kingdom) and sequenced using an Amersham Vistra 725 semiautomated sequencer and cycle sequencing protocol (RPN2444; Amersham UK). The V3 sense strand was sequenced using 2 pM primer 619L-TR (5′ TR-T GGC AGT CTA GCA GAA GAA G-3′; TR, Texas Red) and 1 μg of plasmid; the antisense strand was similarly obtained using primer 307D-TR (5′TR-CTG GGT CCC CTC CTG AGG-3′). For direct sequencing of proviral DNA, PCR products derived from infected cells were purified (Qiagen PCR cleanup column), and 200 ng was used in the sequencing reaction. The two sequenced strands were aligned using Sequencher 3.0 software (Gene Codes Corp., Ann Arbor, Mich.), and the consensus was translated to give the amino acid sequence shown. For the four full-length gp120 sequences, eight additional Texas red-labeled primers were used to generate complete sequences of both sense and antisense strands as described previously (28).
RESULTS
Source of primary isolates.
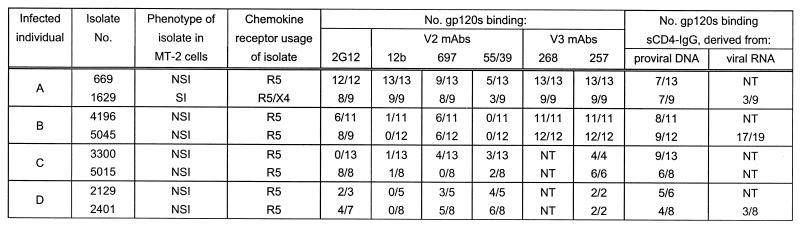
Eight virus isolates from four Swedish homosexual men (A to D) were used in the study. All individuals suffered from moderate immunodeficiency. Two subjects (A and B) were classified as B2 according to the Centers for Disease Control classification system (22); subjects C and D were classified as C2 and A2, respectively. The subjects and the virus isolates were selected from a larger cohort of well-characterized individuals (23). HIV-1 was isolated from peripheral blood mononuclear cells (PBMC), and the isolates were tested for their ability to infect U87-CD4 cells expressing various chemokine receptors and to replicate in the immortalized cell line MT-2 (Table 1) (23). Consistent with previous reports (8), the ability to use R4 correlated with syncytium induction in the MT-2 cell line. All isolates were resistant to neutralization by sCD4-Ig (20 μg/ml) and by MAbs (20 μg/ml) specific for the V3 (268 and 257) and CD4 b.s. (39.3b), compared with a >90% reduction in p24 antigen from MN-infected PBMC. However, MAb 2G12 (10 μg/ml) neutralized isolates 1629, 5015, and 2401 by 44, 33, and 50%, respectively (data not shown).
TABLE 1.
Characterization of primary virus cultures and recombinant soluble gp120 proteinsa
The non-syncytium-inducing (NSI)/syncytium-inducing (SI) and R5/X4 phenotypes of the original PBMC-derived isolate in MT-2 and U87-CD4 coreceptor cells, respectively, are shown. All expressed gp120 proteins shown were able to bind the conformation-dependent MAb 58/30 and were tested for their ability to bind linear (12b) and conformation-dependent (697 and 55/39) anti-V2 MAbs, linear anti-V3 MAbs (268 and 257), 2G12, and sCD4-IgG. Also shown is the ability of clones derived from viral RNA to bind sCD4-IgG. A gp120 is considered to bind the respective ligand if its binding index relative to BH10 gp120 is greater than 0.2. NT, not tested. Data are presented as number positive/number tested.
Antigenic variation within CD4 b.s.
To study the level of antigenic variation present within the CD4 b.s., the gp120-encoding region was amplified from proviral DNA obtained from all of the primary virus-infected PBMC cultures and cloned into the vector pCDNA3-tpa-CA. In addition, the gp120-encoding region was amplified from cDNA syntheses of viral RNA obtained from isolates 5045, 1629, and 2401. At least 20 clones containing a gp120 ORF were obtained from each sample, and plasmid DNA was prepared and transfected into vTF7.3-infected 293 cells. Multiple gp120 proteins, derived from the same PBMC culture, were tested by EIA for their ability to bind sCD4-Ig and a panel of neutralizing MAbs specific for epitopes within the CD4 b.s. In addition, the proteins were tested for their ability to bind the nonneutralizing conformation-dependent MAb 58/30, MAb 2G12, and a panel of neutralizing MAbs specific for epitopes within V2, V3, and C4. The results are expressed as the ratio of ligand binding to primary recombinant gp120 relative to the BH10 standard (measured as OD at 450 nm) and are termed the relative binding index. Levels of proteins tested were similar, as determined by comparable reactivity with HIV-positive human sera (QC256). The results are shown for proteins derived from sequential PBMC isolates obtained from two infected individuals, A and B (Tables 2 and 3, respectively); comparable results were also obtained from individuals C and D (summarized in Table 1).
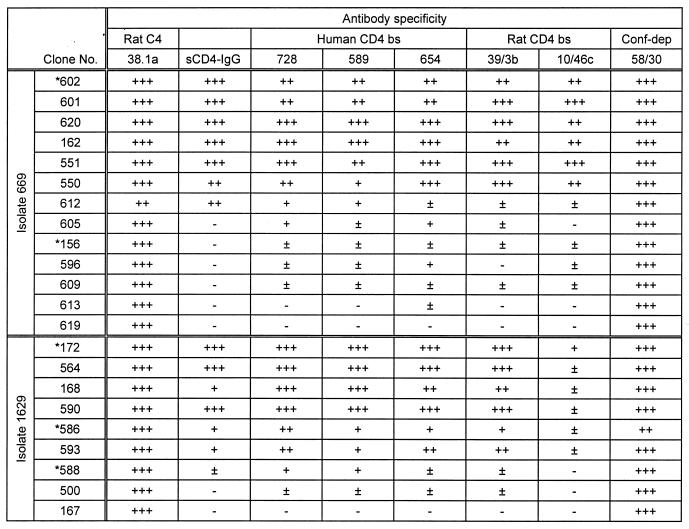
TABLE 2.
Antigenic variation within gp120 proteins cloned from proviral DNAa
Recognition of gp120 proteins by 38.1a, sCD4-IgG, MAbs specific for the CD4 b.s. and MAb 58/30, specific for a conformational CD4-independent (Conf-dep) epitope. Proteins analyzed were cloned from individual A, isolates 669 and 1629. Clones marked with an asterisk were also cloned and expressed as recombinant viruses. Results were calculated as a binding index relative to BH10 gp120: +++ ≡ >0.8; ++ ≡ 0.61 to 0.8; + ≡ 0.41 to 0.6; ± ≡ 0.21 to 0.4; — ≡ <0.21.
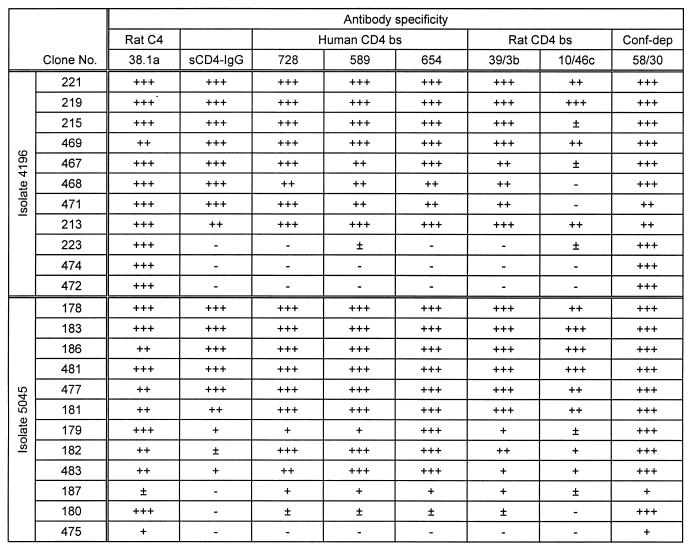
TABLE 3.
Antigenic variation in a second individuala
Data are for isolates from individual B. See Table 2, footnote a, for details.
Proteins derived from a single primary isolate showed a spectrum of binding indices for sCD4-Ig ranging from 0 to 1.5 relative to BH10 gp120 (Tables 2 and 3). The majority of proteins tested bound MAb 58/30, specific for a conformational epitope independent of the CD4 b.s., suggesting that the expressed proteins were correctly folded. Generally, a strong correlation between binding to sCD4-Ig and the CD4 b.s. MAbs (728, 589, 654, 39.3b, and 10/46c) existed; however, binding to the C4 epitope 38.1a was conserved for all proteins (Tables 2 and 3). The percentage of proteins obtained from a single isolate showing negligible binding to sCD4-Ig (B.I. < 0.2) ranged from 15 to 50% (Table 1). The frequency of such proteins changed over time when analyzing the second isolate from the same individual, suggesting that they were derived from a nonstatic population. Furthermore, similar numbers of proteins expressed from gp120 ORFs amplified from cDNA syntheses of viral RNA preparations (isolates 5045, 1629, and 2401) demonstrated a comparable variation in binding to sCD4-IgG (Table 1) and the CD4 b.s. MAb 39.3b (data not shown) while maintaining recognition of the 58/30 antibody, suggesting expression of the variant gp120s in infected cultures.
In addition to the variation within the CD4 b.s., proteins derived from a single isolate showed a range of binding indices for MAbs specific for a number of neutralizing epitopes within V2 (12b, 697, and 55/39) and the discontinuous 2G12 epitope, with no correlation to sCD4-Ig binding indices. However, binding to MAbs specific for linear epitopes within the V3 region (268 and 257) was conserved for the majority of proteins tested (summarized in Table 1).
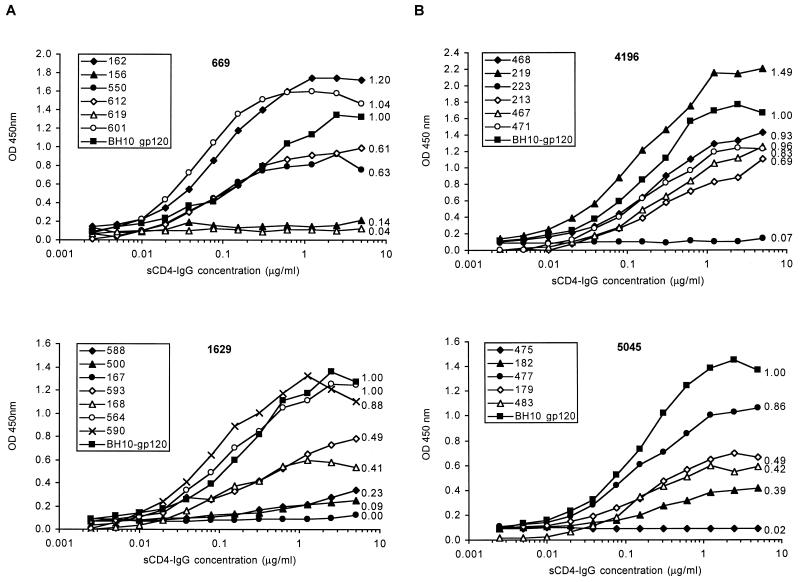
The data presented in Tables 2 and 3 represent a single (saturating) concentration of ligand binding to the various proteins. In order to study relative binding affinities, increasing concentrations of sCD4-Ig were tested for their ability to bind a number of proteins (Fig. 1). These data confirm that a range of binding affinities exist within a single isolate and, furthermore, that some proteins demonstrate a higher relative affinity for sCD4-Ig than the BH10-gp120 standard. The binding of gp120 to the dimeric sCD4-Ig molecule was shown to represent the interaction with cellular CD4, since the ability of a given protein to bind CD4 at the cell surface (measured by fluorescence-activated cell sorting analysis) correlated well with its binding index for sCD4-Ig obtained by EIA and was inhibited by the anti-CD4 MAb 337 (data not shown).
FIG. 1.
Affinity of gp120 proteins cloned from proviral DNA for sCD4-IgG. BH10 gp120 and a number of proteins cloned from individual A, isolates 669 and 1629 (A), and individual B, isolates 4196 and 5045 (B), were analyzed for their ability to bind increasing concentrations of sCD4-Ig. The previously determined sCD4-IgG binding index is shown next to each curve.
Assessment of PCR-induced error.
To address whether the antigenic polymorphism observed was partially due to PCR-induced error(s), we simultaneously performed a series of amplifications using a clonal template. First, we determined the viral copy number in the proviral DNA from isolates 1629 and 669 and in the cDNA derived from viral RNA of isolates 1629 and 2401 by the limiting dilution method (53) and found 2.6 × 106, 2.6 × 105, 4.3 × 105, and 1.1 × 105 viral genome copies per μl of template, respectively. Thus the original PCR assays, i.e., 5 μl of template, contained a minimum of 5.5 × 105 copies per reaction (for 2401 cDNA). We therefore diluted pHXB2 in uninfected cellular DNA to 2.6 × 105 and 4.3 × 105 copies per μl for use as the template in a nested gp120 amplification using the same conditions and enzyme as used for the other reactions. The gp120 PCR products obtained were cloned into pCDNA3-tpa-CA, and plasmids were prepared and transfected into vTF7.3-infected 293 cells. All of the 20 expressed gp120 proteins bound QC256, 39.3b, 58/30, and sCD4-Ig similarly, with binding indices of 0.92 to 1.23 (data not shown). These data suggest that the antigenic polymorphism of proteins derived from primary viruses was unlikely to be an artifact of PCR amplification.
Construction of chimeric molecular clones.
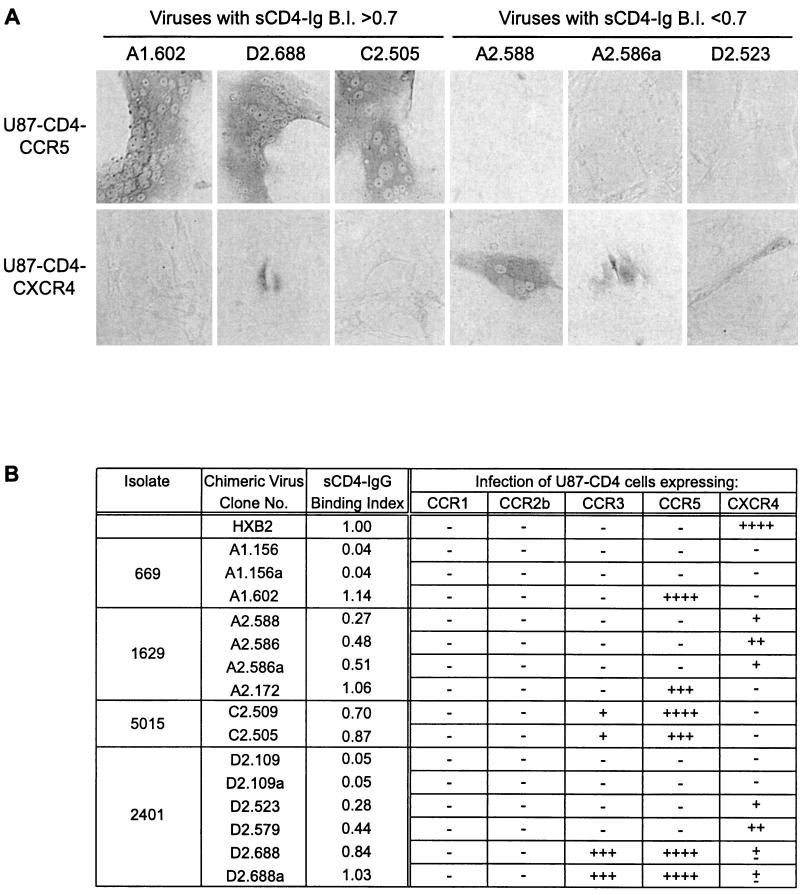
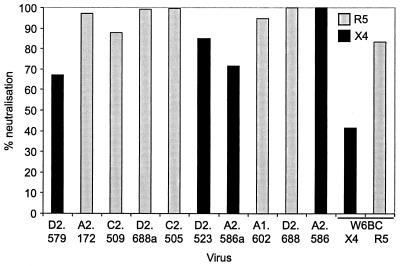
To study the effect of gp120 CD4-binding ability on virus viability and chemokine receptor usage, a number of gp120 ORFs encoding proteins with various abilities to bind sCD4-IgG were transferred into the full-length infectious molecular clone pHXB2. We have previously reported that HXB2 viruses expressing gp120 ORFs of several molecularly cloned laboratory-adapted viruses maintain the tropism of the gp120-derived clone and demonstrate the same sensitivity to neutralizing agents as the gp120-derived clone, validating the generation and characterization of such chimeric viruses (36). Chimeric constructs were transfected into 293 cells, and the recovered virus was used to infect U87-CD4 cells expressing CCR1, CCR2b, CCR3, CCR5, and CXCR4. All chimeric viruses with sCD4-Ig binding indices of 0.2 or greater were able to infect at least one of the U87-CD4 coreceptor cell lines, demonstrating their viability. This agrees with previous data from our laboratory (20, 28, 36) demonstrating that chimeric viruses expressing gp120 proteins with CD4 binding indices of <0.2 were nonviable. Significantly, chimeric viruses with sCD4-Ig binding indices between 0.2 and 0.7 infected only U87-CD4-CXCR4 cells, while viruses expressing gp120 proteins with sCD4-Ig binding indices of greater than 0.7 preferentially infected U87-CD4-CCR5 and -CCR3 cells (Fig. 2). No infection via CCR1 or CCR2b was observed, and the presence of Bonzo on the U87 cell line (12) also demonstrates that none of the chimeric viruses were able to use this coreceptor. The cytopathic effect (CPE) induced by viruses in the U87-CD4 coreceptor cell lines was far less pronounced for viruses with sCD4-Ig binding indices of less than 0.7 than for viruses with binding indices of 0.7 or greater, foci of infection being less frequent and containing fewer nuclei. This was not merely a reflection of a differential susceptibility to virus-mediated cytopathicity between U87-CD4-CCR5 and -CXCR4 cells, since infection of both cell lines with equivalent titers of a dualtropic virus, HIVW6BC, resulted in comparable CPE (data not shown). Infection of U87-CD4-CCR5 or -CXCR4 cells by all chimeric viruses tested could be blocked by the anti-CD4 MAb ADP 318, irrespective of sCD4-IgG binding index, indicating a CD4-dependent route of infection in all cases (Fig. 3). Furthermore, the characteristic neutralization resistance of primary viruses was retained in the chimeric viruses, as demonstrated by the resistance to neutralization by sCD4-Ig of four chimeras with high binding indices for this ligand (chimeras A1.602, C2.509, C2.505, and D2.688 were neutralized 49, 19, 20, and 25%, respectively, compared with 89% for HXB2). These data suggest that primary viruses retain viability despite a reduced ability to bind the receptor CD4, that the variant gp120s were likely derived from replication-competent virus, and that the chimeras generated maintain primary virus-like characteristics.
FIG. 2.
Chimeric virus infection of U87-CD4-coreceptor-expressing cells. Each cell-free transfection supernatant (200 μl, containing comparable levels of p24 antigen) was used to infect U87-CD4-coreceptor cells at ∼20% confluency in 1-cm2 wells. Virus was left for 3 days to infect. The cells were then stained for intracellular p24 antigen. (A) Representative fields of view shown for selected viruses. (B) Infection of all chimeras was measured by scoring the number of blue foci (both single blue cells and syncytia) per well: — ≡ 0 blue foci; ± ≡ <5 foci; + ≡ 5 to 10 foci; ++ ≡ 11 to 30 foci; +++ ≡ 31 to 100 foci; ++++ ≡ >100 foci.
FIG. 3.
CD4 dependence of chimeric viruses. U87-CD4-CXCR4 or -CCR5 cells at ∼20% confluency in 1-cm2 wells were treated with anti-CD4 MAb 318 (1 μg/ml) prior to infection with 200 μl of each cell-free transfection supernatant (containing comparable levels of p24 antigen). Virus was left for 3 days to infect. Cells were then stained for intracellular gp120 antigen, and the number of blue foci per well was counted and expressed as percent neutralization. The coreceptor preference of each chimeric virus is distinguished.
Sequence analysis of gp120 ORFs.
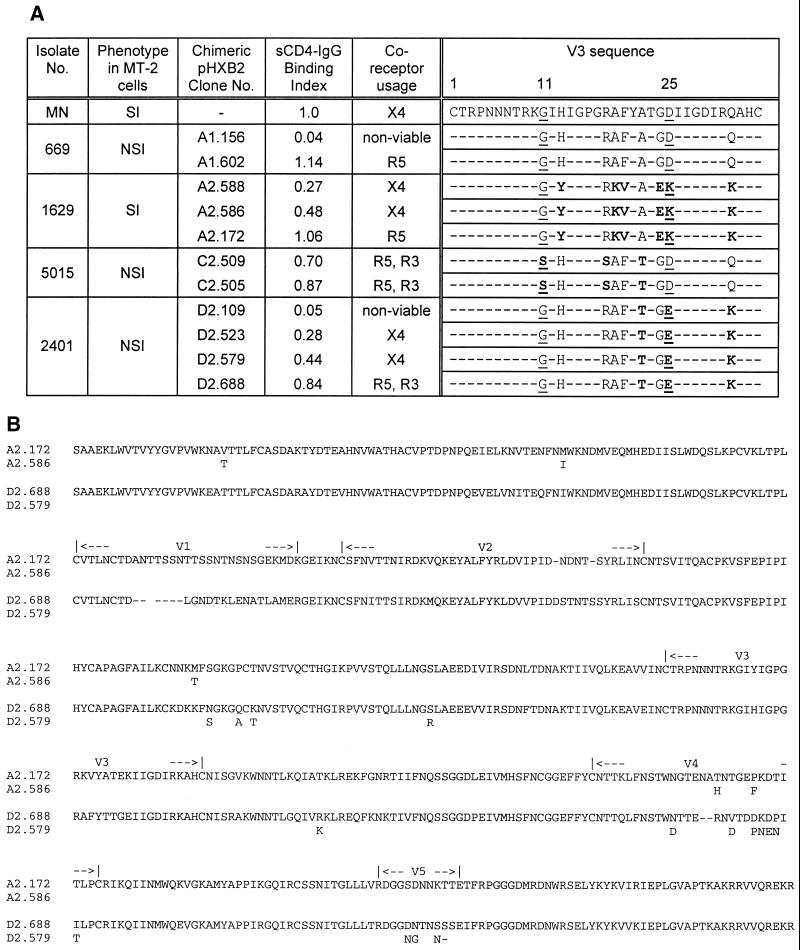
Since the sequence of the V3 region has been reported to determine both viral tropism and coreceptor usage, this region of each chimeric virus was sequenced. Sequencing of the V3 and surrounding region confirmed that the proviral DNA sequence was identical to the pcDNA3 sequence in all cases. Previous reports suggested that an uncharged residue (S or G) at position 11 and an uncharged (A or Q) or negatively charged (E or D) residue at position 25 conferred a macrophage R5-tropic phenotype, while a T-cell X4-tropic phenotype was associated with substitution of basic amino acids at either position (19, 56). However, in this study, no correlation was found between V3 sequence and coreceptor usage, with both R5- and X4-using clones derived from the same parental primary isolate displaying identical V3 sequences (Fig. 4A). These data suggest that regions of gp120 other than the V3 loop may influence viral coreceptor preference. Consistent with previous work, all of these chimeras expressing primary virus-derived gp120s carried the consensus N-linked glycosylation signal at position 8 of the V3, a glycosylation associated with neutralization resistance (30), and all carried the highly conserved arginine at position 3 reported to contribute to both X4 and R5 usage (61).
FIG. 4.
Chimeric virus V3 sequences and alignment of A2 and D2 gp120 proteins. (A) V3 regions of a number of chimeric virus gp120 ORFs were sequenced from the pHXB2 constructs, shown in the single-letter amino acid code. The consensus sequence was derived from the V3 sequences of the pCDNA3-gp120 ORFs (data not shown), and numbering is based on MN gp120. Residues 11 and 25, previously implicated in determining the non-syncytium-inducing (NSI)/R5 syncytium-inducing (SI)/X4 phenotype, are underlined, and any residues which vary from the consensus are highlighted in bold. Residues which do not vary from the consensus are shown as dashes. (B) Peptide translation (single-letter amino acid code) of two pairs of gp120 sequences with different coreceptor usage, two clones obtained from the A2 sample and two from D2. For the second sequence in each pair, only the different positions are shown. The upper sequence in each pair is R5 tropic, and the lower is X4 tropic.
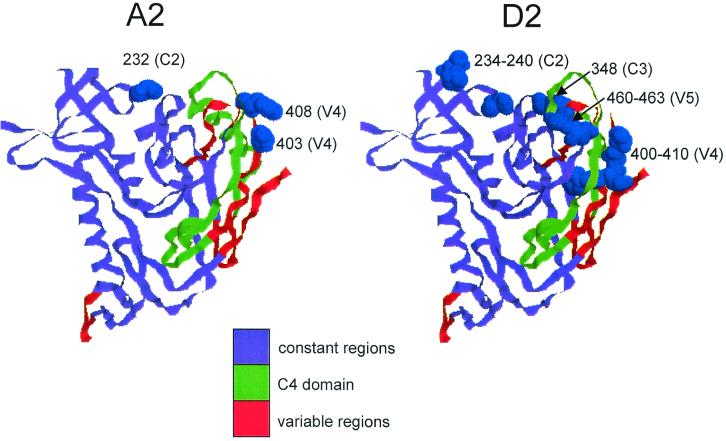
Complete sequences of the region transferred into the pHXB2 chimaeras were derived from both proviral DNA and pHXB2 sources for two pairs of clones for which coreceptor usage differed; each pair derived from the same parental isolate. The clones chosen were A2.172 (R5 utilizing, CD4 B.I. = 1.06) and A2.586 (X4, CD4 B.I. = 0.48), and D2.688 (R5/R3, CD4 B.I. = 0.84) and D2.579 (X4, CD4 B.I. = 0.44). Five amino acid differences were found between the two clones from isolate A2 and 15 between those from isolate D2 (Fig. 4B). Common to both pairs were changes in a region of C2 (a.a. 232 in A2 and 234 to 240 in D2) and in the V4 domain. In contrast, changes found in the C1 region between A2.172 and A2.586 and in both C3 and V5 regions between D2.579 and D2.688 were not shared by the other pair of clones studied. The positions of the changes identified are shown projected onto the crystal structure for HIV-1HXB2 gp120 (Fig. 5). Modeling of the M232T change in C2 using the Swiss-Model procedure showed no global disturbance in the overall structure as a result of the substitution (data not shown).
FIG. 5.
Positions of changes seen in A2 and D2 in the predicted gp120 structure. The crystal structure of gp120 (cartoon format) was drawn using the RasMol program (48) in the same orientation as in Wyatt et al. (62). Positions at which polymorphisms are seen are highlighted in blue by space-filling the amino acids on the ribbon structure.
DISCUSSION
The data presented in this study demonstrate that antigenic variation exists within the envelope glycoproteins of primary HIV-1 isolates with respect to recognition by sCD4-IgG and by MAbs specific for the discontinuous CD4 b.s. (Tables 2 and 3). No differences in these antigenicity patterns were observed between gp120 proteins cloned from R5 (virus 669) and X4 (virus 1629) viruses isolated from a single individual over time. However, the ability of a given gp120 to bind sCD4-Ig appeared to associate with the coreceptor usage of chimeric virus expressing the heterologous protein.
The majority of the glycoproteins tested were capable of binding MAb 58/30, which recognizes a discontinuous epitope independent of the CD4 b.s. (Tables 2 and 3), suggesting that the proteins were correctly folded and that the observed polymorphisms were not due merely to a loss of conformational structure but to specific variation in the epitopes tested. These data also support the conclusion that the observed polymorphisms were not PCR derived, since if this were the case, variation may be observed in the epitope(s) recognized by this nonneutralizing, conformation-dependent MAb. Confirming this, no variation was seen in gp120s obtained from a clonal template using the same PCR conditions as for the primary virus-derived gp120s.
The degree of variation observed within the CD4 b.s. of primary virus-derived gp120 proteins is surprising given the reportedly conserved nature of this region (40, 46), although the presence of islands of variability in this region is now known (25, 62). Several reports considering the resistance of primary viruses to neutralization by sCD4 have proposed that this is due to a reduced affinity of the envelope for CD4, apparent in the oligomeric form of the glycoprotein but not in the monomer (2, 59). Our results suggest that a reduced affinity for CD4 is also apparent in monomeric gp120 proteins derived from a primary virus isolate and that since replicate gp120 proteins derived from single primary virus isolates display a wide range of relative binding affinities for CD4, the overall population may demonstrate a reduced net affinity.
The ORFs encoding gp120 proteins demonstrating reduced or negligible binding to sCD4-Ig were cloned from both proviral DNA and viral RNA sources, suggesting that these variants were not derived from a latent proviral population. To determine whether these proteins may have been derived from replication-competent viruses and to address the possibility that they may mediate infection via CD4-independent routes, a number of gp120 ORFs were transferred into the full-length infectious molecular clone pHXB2. Infection of U87-CD4-CXCR4 cells demonstrated that chimeric viruses expressing gp120 proteins with reduced CD4-binding abilities are viable (Fig. 2); furthermore, their infectivity was shown to be CD4 dependent (Fig. 3). This suggests that the variant gp120 proteins were derived from infectious virus and that primary viruses are capable of CD4-dependent infection despite a lower ability to bind the receptor, albeit at a reduced efficiency. These data are in agreement with the work of Kabat and colleagues (21, 24, 41; reviewed in reference 13), who demonstrated that, while CXCR4-using primary viruses infect cells in direct proportion to the amount of cell surface-expressed CD4, infection by laboratory-adapted viruses is independent of CD4 levels, suggesting that adaptation to growth in T-cell lines involves the acquisition of a more efficient CD4-binding mechanism.
Chimeras expressing gp120s derived from an R5-using isolate were a mix of R5/R3- and X4-dependent viruses. Similarly, chimeras derived from an isolate defined as R5/X4-using (1629) also demonstrated this mix, although a higher proportion were X4-using. Numbers in each group are, however, too small to make statistically valid conclusions. However, it is clear that an isolate with a defined coreceptor preference contains a mix of envelope glycoproteins able to confer entry not only by the coreceptor preferred by the isolate, but also by others. This is not unexpected, since all infected individuals in this study initially displayed R5-using isolates and subsequently developed X4-using isolates. No correlation was observed between coreceptor usage of a given isolate and the proportion of envelope glycoproteins derived from it with high or low sCD4-IgG binding abilities.
The observation that viruses with reduced CD4 binding indices (B.I. between 0.2 and 0.7) preferentially used CXCR4 as a coreceptor, while those with a greater ability to bind the receptor (B.I. > 0.7) used CCR5 and sometimes CCR3 as a coreceptor (summarized in Fig. 3) requires further investigation. To date, however, all chimeric viruses expressing primary virus-derived gp120 proteins studied in our laboratory have conformed to this pattern, suggesting a possible mechanistic difference for infection via CXCR4. Previous work shows that for infection in vitro, levels of CD4 and CCR5 are interdependent, such that when CD4 is abundant, only trace amounts of CCR5 are required for maximal susceptibility to infection, whereas cells expressing low levels of CD4 require a much larger amount of CCR5 for infection (41). Mondor and colleagues reported that the ability of anti-CXCR4 MAbs to block gp120-CXCR4 interaction associated with cell surface CD4 expression levels. Furthermore, the gp120-CXCR4 interaction could be abolished by MAbs specific for a conformation-dependent CD4-induced binding site (38). This site is normally only exposed on gp120 which is bound to CD4; however, Wyatt and colleagues have shown that removal of the V1/V2 region leads to exposure of this epitope and proposed that the C2 region forms a part of this site (63). Thus, a complex interaction between CD4, coreceptor, and gp120 is a critical determinant of efficient entry; it is plausible that changes in affinity of the gp120-CD4 component of this interaction may impact on the coreceptor selected to achieve the most fusion-competent ternary complex. It will be interesting to assess whether other sample sets also display the association found here between CD4 affinity and chemokine receptor usage.
Complete gp120 sequences from pairs of chimeras with different coreceptor usage and CD4 affinity are valuable in mapping the genetic determinants of the phenotypes observed. In the pair of sequences derived from isolate 1629 (patient A, sample 2), only five differences in the deduced peptide sequence were found. In the C1 region two changes were seen (V49T and M95I; numbering by alignment to the HXB2 sequence); however, several authors have shown that this region is not required for CD4 or coreceptor binding. Indeed, this region of gp120 is absent from the recently published gp120-CD4 crystal structure (25, 38, 43, 62). In addition, the other pair of cloned sequences showed no differences in this region, suggesting that these mutations play little role in the phenotypic changes seen. In the V4 region, two changes were apparent between the A2 clones (T403H and P408F). The fifth change apparent between these clones is located in the C2 region (M232T). For the second pair of clones (patient D, sample 2), many more polymorphic sites were seen. Of the 15 nonsilent differences, 7 were found in the V4 region, 3 in the V5 region, 1 in the C3 domain (R348K), and 4 in C2 (N234S, Q238A, K240T, and S264R). Therefore, changes in the V4 and C2 regions were the only common features between the paired clones.
Given the proximity of the changes seen in the V4 to C4 region, which has been shown to be a major determinant of CD4 affinity, it is possible that such changes may affect the exposure of this part of the CD4 binding site and thereby modulate CD4 binding (33, 40, 43). The changes in the C2 region may also be associated with a change in coreceptor usage. This region is reported to form part of the CD4-induced epitope which may affect the gp120-CXCR4 interaction (38, 63). Furthermore, Rizzuto and colleagues reported that mutations in the C2 region abolished both CD4 and CXCR4 binding, with one mutation at position 257, T to D, being close to the polymorphic site in the D2 clones (S264R) (43).
The precise mechanism(s) of envelope glycoprotein-CD4-coreceptor interaction awaits further investigation. The gp120 proteins identified in this study, which are capable of mediating viral infection despite their reduced ability to bind CD4, will be useful in the further elucidation of these interactions.
ACKNOWLEDGMENTS
We thank A. Karlsson (Karolinska Hospital) for the provision of clinical material; H. Holmes, MRC AIDS Reagent Programme Repository (NIBSC, Potters Bar, Hertfordshire, United Kingdom) for MAbs and p24 reagents; and Susan Zolla-Pazner (New York University Hospital) for MAbs 728, 589, 654, 257, and 268.
This work was supported by the MRC, the Edward Jenner Institute for Vaccine Research, and the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR 5—a RANTES, MIP-1-alpha, MIP-1-beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Smith D H, Marsters S A, Riddle L, Gregory T J, Ho D D, Capon D J. Resistance of primary isolates of human immunodeficiency virus type-1 to soluble CD4 is independent of CD4-rgp120 binding affinity. Proc Natl Acad Sci USA. 1991;88:7056–7060. doi: 10.1073/pnas.88.16.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 4.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe H, Farzun M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1136–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Saachi N. Single step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Ann Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Clapham P R, Weiss R A. Spoilt for choice of co-receptors. Nature. 1997;388:230–231. doi: 10.1038/40758. [DOI] [PubMed] [Google Scholar]
- 8.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 10.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of human immunodeficiency virus type I quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Dimarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major coreceptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 13.Doms R W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 14.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 15.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–671. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor— functional cDNA cloning of a seven-transmembrane, G-protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesises bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorny M K, Moore J P, Conley A J, Karwowska S, Sodroski J, Williams C, Burda S, Boots L J, Zolla-Pazner S. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol. 1994;68:8312–8320. doi: 10.1128/jvi.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung C S, Vander Heyden N, Ratner L. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J Virol. 1999;73:8216–8226. doi: 10.1128/jvi.73.10.8216-8226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones A. Ph.D. thesis. Reading, England: University of Reading; 1997. [Google Scholar]
- 21.Kabat D, Kozak S L, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamps B S, Brodt H R, Staszewski S, Bergmann L, Helm E B. AIDS-free survival and overall survival in HIV-infection — the new CDC classification system (1993) for HIV disease and AIDS. Clin Investig. 1994;72:283–287. doi: 10.1007/BF00180041. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson A, Parsmyr K, Sandstrom E, Fenyo E M, Albert J. MT-2 cell tropism as a prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol. 1994;32:364–370. doi: 10.1128/jcm.32.2.364-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak S L, Platt E J, Madani N, Ferro F E, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwong P D, Wyatt R, Sattentau Q J, Sodroski J, Hendrickson W A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1961–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laal S, Burda S, Gorny M K, Karwowska S, Buchbinder A, Zolla-Pazner S. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J Virol. 1994;68:4001–4008. doi: 10.1128/jvi.68.6.4001-4008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis J, Balfe P, Arnold C, Kaye S, Tedder R S, McKeating J A. Development of a neutralizing antibody response during acute primary human immunodeficiency virus type 1 infection and the emergence of antigenic variants. J Virol. 1998;72:8943–8951. doi: 10.1128/jvi.72.11.8943-8951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddon P J, McDougal J S, Clapham P R, Dalgleish A G, Jamal S, Weiss R A, Axel R. HIV infection does not require endocytosis of its receptor, CD4. Cell. 1988;54:865–874. doi: 10.1016/s0092-8674(88)91241-x. [DOI] [PubMed] [Google Scholar]
- 30.Malenbaum S E, Yang D, Cavacini L, Posner M, Robinson J, Cheng-Mayer C. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J Virol. 2000;74:11008–11016. doi: 10.1128/jvi.74.23.11008-11016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markham R B, Wang W C, Weisstein A E, Wang Z, Munoz A, Templeton A, Margolick J, Vlahov D, Quinn T, Farzadegan H, Yu X F. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cells decline. Proc Natl Acad Sci USA. 1998;95:12568–12573. doi: 10.1073/pnas.95.21.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald R A, Mayers D L, Chung R C Y, Wagner K F, RattoKim S, Birx D L, Michael N L. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J Virol. 1997;71:1871–1879. doi: 10.1128/jvi.71.3.1871-1879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKeating J A, Bennett J, Zolla-Pazner S, Schutten M, Ashelford S, Brown A L, Balfe P. Resistance of a human serum-selected human immunodeficiency virus type 1 escape mutant to neutralization by CD4 binding site monoclonal antibodies is conferred by a single amino acid change in gp120. J Virol. 1993;67:5216–5225. doi: 10.1128/jvi.67.9.5216-5225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeating J A, Moore J P, Ferguson M, Marsden H S, Graham S, Almond J W, Evans D J, Weiss R A. Monoclonal antibodies to the C4 region of human immunodeficiency virus type 1 gp120 — use in topological analysis of a CD4 binding site. AIDS Res Hum Retroviruses. 1992;8:451–459. doi: 10.1089/aid.1992.8.451. [DOI] [PubMed] [Google Scholar]
- 35.McKeating J A, Shotton C, Cordell J, Graham S, Balfe P, Sullivan N, Charles M, Page M, Bolmstedt A, Olofsson S, et al. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J Virol. 1993;67:4932–4944. doi: 10.1128/jvi.67.8.4932-4944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKeating J A, Zhang Y J, Arnold C, Frederiksson R, Fenyo E M, Balfe P. Chimeric viruses expressing primary envelope glycoproteins of human immunodeficiency virus type I show increased sensitivity to neutralization by human sera. Virology. 1996;220:450–460. doi: 10.1006/viro.1996.0332. [DOI] [PubMed] [Google Scholar]
- 37.Milich L, Margolin B H, Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology. 1997;239:108–118. doi: 10.1006/viro.1997.8821. [DOI] [PubMed] [Google Scholar]
- 38.Mondor I, Moulard M, Ugolini S, Klasse P J, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau Q J. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology. 1998;248:394–405. doi: 10.1006/viro.1998.9282. [DOI] [PubMed] [Google Scholar]
- 39.Moore J P, Cao Y Z, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platt E J, Madani N, Kozak S L, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J Virol. 1997;71:883–890. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poss M, Rodrigo A G, Gosink J J, Learn G H, de Vange Panteleeff D, Martin H L, Jr, Bwayo J, Kreiss J K, Overbaugh J. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 44.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y J, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salzwedel K, Smith E D, Dey B, Berger E A. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J Virol. 2000;74:326–333. doi: 10.1128/jvi.74.1.326-333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sattentau Q J, Dalgleish A G, Weiss R A, Beverley P C. Epitopes of the CD4 antigen and HIV infection. Science. 1986;234:1120–1123. doi: 10.1126/science.2430333. [DOI] [PubMed] [Google Scholar]
- 47.Sattentau Q J, Moore J P. Human immunodeficiency virus type-1 neutralization is determined by epitope exposure on the gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayle R A, Milnerwhite E J. RASMOL—biomolecular graphics for all. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 49.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, Degoede R E Y, Vansteenwijk R P, Lange J M A, Schattenkerk J, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection; progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shankarappa R, Gupta P, Learn G H, Rodrigo A G, Rinaldo C R, Gorry M C, Mullins J L, Nara P L, Ehrlich G D. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–259. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- 51.Shankarappa R, Margolick J B, Gange S J, Rodrigo A G, Upchurch D, Farzadegan H, Gupta P, Rinaldo C R, Learn G H, He X, Huang X L, Mullins J I. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shotton C, Arnold C, Sattentau Q J, Sodroski J, McKeating J A. Identification and characterization of monoclonal antibodies specific for polymorphic antigenic determinants within the V2 region of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1995;69:222–230. doi: 10.1128/jvi.69.1.222-230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Leigh-Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral blood mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh A, Collman R G. Heterogeneous spectrum of coreceptor usage among variants within a dual-tropic human immunodeficiency virus type 1 primary isolate quasispecies. J Virol. 2000;74:10229–10235. doi: 10.1128/jvi.74.21.10229-10235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth R J, Yi Y J, Singh A, Collman R G. Determinants of entry cofactor utilization and tropism in a dual-tropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72:4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tersmette M, Gruters R A, De Wolf F, de Goede R E, Lange J M, Schellekens P T, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner S, Tizard R, Demarinis J, Pepinsky R B, Zullo J, Schooley R, Fisher R. Resistance of primary isolates of human immunodeficiency virus type to neutralization by soluble CD4 is not due to lower affinity with the viral envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1992;89:1335–1339. doi: 10.1073/pnas.89.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Cott T C, Bethke F R, Polonis V R, Gorny M K, Zolla-Pazner S, Redfield R R, Birx D L. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. J Immunol. 1994;153:449–459. [PubMed] [Google Scholar]
- 61.Wang W K, Lee C N, Dudek T, Chang S Y, Zhao Y J, Essex M, Lee T H. Interaction between HIV type 1 glycoprotein 120 and CXCR4 coreceptor involves a highly conserved arginine residue in hypervariable region 3. AIDS Res Hum Retroviruses. 2000;16:1821–1829. doi: 10.1089/08892220050195784. [DOI] [PubMed] [Google Scholar]
- 62.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 63.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang L Q, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]