Abstract
A human immunodeficiency virus type 1 (HIV-1) subtype E (CRF01_AE) variant (99JP-NH3-II) possessing an in-frame 33-nucleotide insertion mutation in the β3-β4 loop coding region of the reverse transcriptase (RT) gene was isolated from a patient who had not responded to nucleoside analogue RT inhibitors. This virus exhibited an extremely high level of multiple nucleoside analog resistance (MNR). Neighbor-joining tree analysis of the pol sequences indicated that the 99JP-NH3-II variant had originated from the swarm of drug-sensitive predecessors in the patient. Population-based sequence analyses of 82 independently cloned RT segments from the patient suggested that the variants with the insertion, three or four 3′-azido-3′-deoxythymidine resistance mutations, and a T69I mutation in combination had strong selective advantages during chemotherapy. Consistently, in vitro mutagenesis of a drug-sensitive predecessor virus clone demonstrated that this mutation set functions cooperatively to confer a high level of MNR without deleterious effects on viral replication capability. Homology modeling of the parental RT and its MNR mutant showed that extension of the β3-β4 loop by an insertion caused reductions in the distances between the loop and the other subdomains, narrowing the template-primer binding cleft and deoxynucleoside triphosphate-binding pocket in a highly flexible manner. The origin of the insert is elusive, as every effort to find a homologue has been unsuccessful. Taken together, these data suggest that (i) HIV-1 tolerates in vivo insertions as long as 33 nucleotides into the highly conserved enzyme gene to survive multiple anti-HIV-1 inhibitors and (ii) the insertion mutation augments multiple-drug resistance, possibly by reducing the biochemical inaccuracy of substrate-enzyme interactions in the active center.
Human immunodeficiency virus type 1 (HIV-1) is estimated to produce on the order of 1010 virions per day in a single infected individual (29). This high replication capacity of HIV-1 under highly error-prone (30, 32) and recombination-prone (18) replication conditions is likely to contribute to the rapid occurrence of variants that adapt to a given environmental change. For example, the variants that grow in the presence of a particular HIV-1 inhibitor readily emerge in a patient soon after its administration.
Thus far, three types of mutations have been reported to confer drug resistance on HIV-1: substitutions, insertions, and deletions (12). Amino acid substitution is the most common mechanism by which to generate resistance to a single drug. For example, high-level resistance to 3′-azido-3′-deoxythymidine (AZT), a nucleoside analogue reverse transcriptase (RT) inhibitor (NRTI), results from combinations of six amino acid substitutions (M41L, D67N, K70R, L210W, T215F, and K219Q) in the viral RT (11, 15, 20, 25). The insertion and deletion in RT were more recently identified in individuals who had not responded to combination drug therapy (5, 6, 8, 9, 19, 24, 31, 33, 34, 38–40, 43). The mutations uniformly occur in the β3-β4 loop of the RT finger subdomain, a region critical for NRTI resistance (41). Consistently, insertion contributes to multiple nucleoside analog resistance (MNR) (24, 43) while deletion increases the level of AZT resistance (17).
These findings suggest that the β3-β4 loop of HIV-1 RT is a hot spot of insertion and deletion mutations for virus adaptation to multiple NRTIs in vivo. In fact, an in vitro mutagenesis study has suggested that RT can accommodate the insertion of 15 amino acid residues into the β3-β4 loop with no deleterious effect on polymerase activity (21). However, the insertions identified in patients thus far are short, mostly one or two amino acids (5, 6, 8, 9, 19, 24, 31, 33, 34, 38–40, 43). Moreover, it is unclear whether insertion mutation itself can confer the MNR phenotype on the virus or whether it functions with other substitutions in RT (24, 43).
While the emergence of a new HIV-1 variant following an environmental shift is an important issue from both clinical and scientific viewpoints, current studies are largely confined to nucleotide sequence changes (genotypes) and thus do not fully explain to what extent these genetic changes are relevant to phenotypic changes for adaptation. This is particularly the case for drug resistance evolution of non-subtype B (D. L. Robertson, J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber, Letter, Science 288:55–56, 2000) HIV-1 strains, as their genotype-phenotype relationships have been largely deduced from those of HIV-1 subtype B from North America and Europe.
For this study, we chose HIV-1 subtype E (CRF01_AE) (Robertson et al., letter) MNR as a model with which to study the genetic, structural, and functional relationships in RT and their clinical relevance. Here, we describe a remarkable in vivo case of insertions as long as 33 nucleotides into the β3-β4 loop coding region of the HIV-1 subtype E RT gene, demonstrate striking augmentation of MNR in the context of particular substitutions in the subtype E genome, and discuss the mechanisms responsible based on molecular modeling and mutagenesis of the subtype E RT. Our data illustrate a hitherto unappreciated mechanism, a long peptide insertion into RT, for HIV-1 adaptive evolution.
MATERIALS AND METHODS
Clinical history of the patient.
NH3 is a member of the NH family, in which a single HIV-1 subtype E (CRF01_AE) strain of Thai origin infected the father (NH1), the mother (NH2), and their child (NH3) (35). The plasma HIV-1 RNA level and CD4+ T-cell count in November 1997 were 7.5 × 105 copies/ml and 456 × 103 cells/ml, respectively. NH3 had initially been subjected to therapy with AZT and 2′,3′-dideoxyinosine (ddI) between January and December 1998. Subsequently, AZT, β-l-2′,3′-dideoxy-3′-thiacytidine (3TC), and a single protease inhibitor (nelfinavir or indinavir) had been administrated between December 1998 and November 1999. The plasma viral RNA level was monitored every 1 to 2 months between November 1997 and December 1999 by the AMPLICOR HIV-1 monitor test with an add-in primer (Roche Diagnostics), showing a temporary reduction to 2.8 × 103 copies/ml in February 1998. However, the viral RNA level rebounded soon thereafter and has stayed above 3.0 × 105 copies/ml since June 1999, with a continuous decrease in CD4+ T-cell numbers to 122 × 103/ml in October 1999.
Cells.
MAGIC-5 cells, a HeLa cell line that expresses HIV-1 receptors and contains an integrated copy of a β-galactosidase gene under the control of the HIV-1 long terminal repeat (LTR), was cultured as described previously (10). Peripheral blood mononuclear cells (PBMCs) were prepared from whole blood by Ficoll-Hypaque (Pharmacia LKB) density centrifugation.
HIV-1.
HIV-1 was isolated from an NH3 blood specimen collected in December 1999 (99JP-NH3-II) with MAGIC-5 cells (10). Briefly, MAGIC-5 cells were subjected to incubation with centrifugation-concentrated plasma, followed by collection of the culture supernatant at the peak of syncytium formation by the cells. The virus stock (99JP-NH3-IIvm) was kept at −152°C until use.
HIV-1 RT inhibitors.
AZT, ddI, 2′,3′-dideoxycytidine (ddC), and 2′,3′-didehydro-3′-deoxythymidine (d4T) were purchased from Sigma. 3TC and nevirapine (NVP) were provided by Glaxo Wellcome and Boehringer Ingelheim, respectively.
Drug susceptibility assay.
The susceptibility of HIV-1 to RT and protease inhibitors was determined with MAGIC-5 cells (10). Briefly, the number of blue-cell-forming units (BFU) of the virus stocks on MAGIC-5 cells was determined by endpoint dilution. Subsequently, MAGIC-5 cells were infected with a diluted virus stock (300 BFU) in increasing concentrations of inhibitors, cultured for 48 h, fixed, and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. The stained cells were counted under a light microscope, and drug concentrations inhibiting 50% of the stained cells of the drug-free control (IC50) were determined on the basis of the dose-response curve.
Phylogenetic analysis of the HIV-1 pol gene.
Viral RNA was extracted from the virus isolate stock or patient plasma with a High Pure Viral RNA kit (Roche Diagnostics), followed by RT-PCR with a TaKaRa One Step RNA PCR kit (Takara Shuzo, Otsu, Japan) and primers EPR350A (5′-CAA CAA GGG AAG GCC GGG AAA TT-3′) and ERT326B (5′-CTG TAC TTC TGC TAC TAA GTC TTT TGA TGG G-3′). The products were subjected to a second PCR with primers EPR351A (5′-GAA AGA CAA GGA ACA TCC TCA TCC-3′) and ERT328B (5′-CTG CCA ACT CTA ATT CTG CTT C-3′), purified with Centricon-100 (Amicon), and sequenced on an ABI PRISM310 automated DNA sequencer (Perkin-Elmer). Nucleotide sequences encoding the protease and the amino-terminal half of RT (1,035 or 1,068 bp) were aligned with HIV-1 subtype reference sequences (23) by CLUSTAL W, version 1.74, and a neighbor-joining tree with bootstrap values of 100 resamplings was constructed with the PHYLIP package as described previously (35).
Population-based sequence analysis.
Proviral DNA was extracted from PBMCs with a QIAamp DNA Blood Kit (QIAGEN, Hilden, Germany) and subjected to PCR by Pfu DNA polymerase (Promega, Madison, Wis.) with primers EPR350A and ERT326B. Viral RNA was extracted from plasma and subjected to RT-PCR with primers EPR350A and ERT326B. In both cases, the first PCR products were subjected to a second round of PCR by Pfu DNA polymerase with primers NH3-IIRT391A (5′-AAA GCA TTA ACA GAA ATT TGT GA-3′) and NH3-IIRT392B (5′-AGG AAT GGA GGT TCC TTC TGA TGC-3′). The PCR products (543 to 576 bp) were cloned into pPCR-Script Amp SK (+) (Stratagene), and 26 to 28 clones were sequenced for each blood sample with an automated DNA sequencer.
Molecular cloning of a full-length HIV-1 subtype E proviral genome.
Molecular cloning of a full-length HIV-1 subtype E proviral genome was carried out as described previously for HIV-1 subtype B (37). Briefly, a single restriction enzyme site (SpeI) in the HIV-1 subtype E 93JP-NH1 (35, 36) genome was identified by Southern blot analysis of the unintegrated circular DNAs prepared by the Hirt method (13) from 93JP-NH1-infected MT2 cells. Subsequently, the circular DNAs were digested with SpeI, purified on the basis of their sizes (9 to 12 kb), ligated with SpeI-digested arms of the ZAP Express lambda phage vector (Stratagene), and packaged in vitro with Gigapack III Gold packaging extract (Stratagene). The lambda phage library was screened by plaque hybridization with [32P]dCTP-labeled 93JP-NH1 gag and env DNA probes, and six positive clones out of 7.5 × 105 plaques were identified. The positive phages were purified and subjected to in vivo excision of the pBK-CMV phagemid vector (Stratagene) to generate plasmid DNA containing a circularly permuted HIV-1 genome (pBK-NH1).
A biologically active clone was screened from the six clones with MAGIC-5 cells. Briefly, pBK-NH1 was digested with SpeI and ligated to generate concatemers containing the two-LTR linear form of HIV-1 DNA. The DNAs were transfected into HeLa cells engineered to express a green fluorescent protein under the control of the HIV-1 LTR. Culture supernatants of the green fluorescent protein-positive cells were used for de novo infection of MAGIC-5 cells, and a clone that was able to initiate productive infection was identified (pBK-NH1–1).
A plasmid carrying the two-LTR linear form of HIV-1 93JP-NH1 (p93JP-NH1) was reconstructed from pBK-NH1-1 as follows. (i) The KpnI-SpeI fragment (1.6 kb) of pBK-NH1-1 (containing the 3′-end portion of the nef gene, the LTR, and the 5′ half of the gag gene) was subcloned into pLGKSC between the KpnI and SpeI sites to generate pLGNH1-5′. (ii) The SpeI-NarI fragment (8.2 kb) of pBK-NH1-1 (containing the 3′ half of the gag and pol-vif-vpr-tat-rev-vpu-env-nef genes and the LTR) was cloned into pLGNH1-5′ between the SpeI and ClaI sites to generate a plasmid (pLGNH1) containing the two-LTR linear form of the HIV-1 provirus. (iii) The BssHII-BssHII fragment (10 kb) of pLGNH1 (containing the full length HIV-1 93JP-NH1 DNA and multiple cloning sites of pLGKSC) was cloned into pBRKS between the BssHII and BssHII sites, resulting in p93JP-NH1 containing the two-LTR linear form of the 93JP-NH1 proviral genome (9,721 bp).
In vitro mutagenesis.
Site-directed mutagenesis (ERT-mt1 to -mt6) was carried out by the overlap extension method using PCR (14). Primers ΔBcl-374A (5′-GGC AAT AGG ATC AGA TAC TTA TAG-3′), HindIII-375B (5′-TAC TTT CTA AAG CTT TCA TCT AAA GG-3′), PR-376A (5′-GGA ATT GGA GGT TTT ATC AAG G-3′),and M41L-377B (5′-CTT CCA GCT CCT TAC AAA TTT CTG-3′) were used to generate a DNA fragment with the M41L mutation. ΔHind-378A (5′-TGA GAG CTT TAG AAA GTA TAC TGC-3′), Bsu-379B (5′-TTA GCT CCC CTG AGG AGT TTA CAC AG-3′), RT-380A (5′-AGT ACT AGA TGT GGG AGA TGC-3′), and L210W/T215Y-395B (5′-TGG TGT ATA AAA TCC CCA GCT CCA TAG ATG AGC-3′) were used for the L210W and T215Y mutations. Bcl-382A (5′-AAG GCA ATA TGA TCA GAT ACT TAT AG-3′), Ins1-383B (5′-GGC CGG GCC CTG GTC CCT TCC TCC GTG AAT GTT GTC CTT TTT CTT TAT AGC AAA TAC TGG-3′), Ins2-384A (5′-GGA GGA AGG GAC CAG GGC CCG GCC AGC ATT AAA TGG AGG AAA TTA GTA GAT TTC AGA GAG-3′), and HindIII-375B were used for the 33-nucleotide insertion and the T69I mutation. Bcl-382A, Ins1-383B, Ins3-396A (5′-GGA GGA AGG GAC CAG GGC CCG GCC AGC ACC AGA TGG AGG AAA TTA GTA GAT-3′), and HindIII-375B were used for the 33-nucleotide insertion. The PCR products with mutations were cloned between the BclI and HindIII sites or the HindIII and Bsu36I sites of pUC-NH1SpBm, a plasmid containing the SphI-to-BamHI fragment (2.8 kb) of p93JP-NH1. Subsequently, SphI-PmaCI fragments of pUC-NH1SpBm were cloned back into p93JP-NH1 between the SphI and PmaCI sites to generate full-length HIV-1 DNA clones.
ERT-mt7 and -mt8 carrying cloned RT segments of 99JP-NH3-II plasma viruses were constructed as follows. A pol DNA segment (1,191 bp) was amplified by RT-PCR from 99JP-NH3-II plasma RNAs with two sets of primers (outer, EPR350A and ERT326B; inner, EPR351A and ERT328B), digested with BclI and Bsu36I, and cloned between the BclI and Bsu36I sites of pUC-NH1SpBm; this was followed by construction of full-length clones as described for ERT-mt1 to -mt6. The nucleotide sequences of the PCR-amplified fragments and the sequences around the cloning sites of the RT mutants were verified with an automated sequencer.
Preparation of cell-free virus stocks of RT mutants by transfection.
HeLa cells (6 × 105) were grown in Dulbecco modified Eagle medium with 10% (vol/vol) fetal bovine serum in a T25 flask for 1 day and transfected with 3 μg of HIV-1 plasmid DNA using FuGENE 6 transfection reagent (Roche Diagnostics). The culture supernatants were collected at 48 and 72 h after transfection, filtered (0.45-μm pore size), analyzed for RT activity (42), and kept at −152°C until use.
Assay of viability of pJP93-NH1 and its RT mutants.
The viability of cell-free virus stocks of pJP93-NH1 and its RT mutants was assessed with MAGIC-5 cells (10). Briefly, MAGIC-5 cells (1 × 104) in a 96-well plate were infected in duplicate with a serially diluted (fivefold dilution) virus stock, cultured for 2 days, fixed, and stained as described previously (10). Stained cells (>100/well) were counted, and the infectious titer (BFU per microliter) of an undiluted virus sample was expressed as the mean value of duplicate samples.
Molecular modeling.
The three-dimensional (3-D) structure of a complex of the RT p66 subunit, the template-primer complex, and dTTP (16) was obtained from the Brookhaven Protein Data Bank (4) (identification code, 1RTD), and the A chain was used as a template structure for modeling. The 3-D structural models of 93JP-NH1 RT and ERT-mt6 RT were constructed and refined by energy minimization using the modeling program FAMS (28). In the modeling of ERT-mt6 RT with an insertion, to search for and construct a stable conformation of the β3-β4 loop, eight kinds of alignment conditions were set in the FAMS program and calculations were carried out three times for each condition, generating a total of 24 most likely loop positions. The stereochemical qualities of the 24 models were further assessed on the basis of a Ramachandran plot generated with the program PROCHECK (26), showing that all of the 24 structures are equally favorable (residue rates in most-favored regions ranged from 90.9 to 94.9%). The most favorable model was used as the backbone structure on which the positions of the 24 β3-β4 loops, primer-template complex, and dTTP were superimposed. For ERT-mt6 RT, the distance between the β3-β4 loop and an amino-acid residue or a protein region of RT was calculated with each of the 24 models and expressed as an average.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported here have been submitted to the DDBJ database under accession numbers AB052995 through AB053087.
RESULTS
Isolation of an HIV-1 MNR variant with a long insertion mutation in RT.
Clinical data on patient NH3 suggested that HIV-1 variants with multiple-drug resistance had emerged after chemotherapy (see Materials and Methods). Peripheral blood was taken from the patient in December 1999 (99JP-NH3-II), and the pol gene segment (1,035 bp) of the plasma viral RNA (99JP-NH3-IIp) and the RNA of the viruses isolated in culture (99JP-NH3-IIvm) were sequenced by direct sequencing. The deduced amino acid sequences were aligned with a subtype E consensus derived from early 1990s samples in Thailand (23), along with the sequence obtained in 1993 from NH3 (36).
Several remarkable changes were noticed that were unique to the 99JP-NH3-II RTs. First, they had an 11-amino-acid insertion between codons 67 and 68 in the β3-β4 loop coding region of the RT gene (Fig. 1A). Second, they had combinations of three or four amino acid substitutions (M41L, D67N, L210W, and T215Y) that can confer a high level of AZT resistance on HIV-1 subtype B (11, 15, 20, 25). Third, they carried five to seven other substitutions (6DE, K39E, E43K, T69I, G196E, T200R, and L228R) whose contribution to drug resistance has not been described. These viruses further possessed four substitutions (G16E, K20I, M89I, and Q92K) in the protease genes. However, their roles in drug resistance have not been reported either.
FIG. 1.
Isolation of an HIV-1 RT insertion mutant with MNR. (A) A 33-nucleotide insertion sequence. Cons E, HIV-1 subtype E consensus (23); 93JP-NH3, a virus isolated from NH3 before chemotherapy; 99JP-NH3-IIp and 99JP-NH3-IIvm, plasma virus and a virus isolated from NH3 after chemotherapy. The nucleotide sequence shown is in the mRNA sense. The protein sequence is shown in the single-letter amino acid code. Dots indicate identity with Cons E. (B) Susceptibilities of virus isolates to various RT inhibitors. IC50 of the indicated RT inhibitors were determined with MAGIC-5 cells (10), and fold increases in IC50 compared to those for the HIV-1 NL43 strain are shown.
The susceptibilities of 93JP-NH3 and 99JP-NH3-IIvm to various RT inhibitors were determined with MAGIC-5 cells (10). As expected from the previous clinical, genetic, and phenotypic data (35, 36), 93JP-NH3 was as sensitive to the inhibitors tested as reference HIV-1 strain NL43 was (Fig. 1B, 93JP-NH3; the fold increases in IC50 ranged from 1.0 to 1.8). In marked contrast, 99JP-NH3-IIvm exhibited an extremely high level of resistance to a broad range of NRTIs, as evidenced by a strikingly increased IC50 (Fig. 1B, 99JP-NH3-IIvm). However, 99JP-NH3-II was fully sensitive to nevirapine, a non-nucleoside RT inhibitor (Fig. 1B), and to the protease inhibitors so far tested (nelfinavir, indinavir, ritonavir, saquinavir, and amprenavir) (data not shown).
Evolutionary processes of the 99JP-NH3-II pol gene.
A neighbor-joining tree of the pol sequences (Fig. 2) revealed that the 99JP-NH3-II samples were within a monophyletic group of their AZT-sensitive predecessors of the NH family (36) (shaded box; bootstrap value, 65/100). This family cluster was most closely related to subtype E sequences from Thailand (CM240 and 93TH253), which is consistent with the evolutionary relationship of the gag and env genes of the NH family viruses (35). These data indicated that the evolutionary origin of the 99JP-NH3-II pol gene be placed with their drug-sensitive predecessors of the intrafamilial infection.
FIG. 2.
Neighbor-joining tree showing that the 99JP-NH3-II pol gene originated from the swarm of drug-sensitive predecessors in the NH family. A neighbor-joining tree of the HIV-1 pol gene sequences (1,035 bp) was constructed with the PHYLIP package and rooted with an HIV-1 group O strain (ANT70). Bootstrap values above 60/100 are indicated at the nodes of the tree. ∗, 99JP-NH3-II sequences; #, 93JP-NH3 sequence; shaded box, sequences collected between 1993 and 1997 from drug-sensitive subtype E strains of the intrafamilial infection case (NH1, NH2, and NH3) (35, 36); ¶, full-length subtype E molecular clone from the 93JP-NH1 virus isolate (see Fig. 4). Other sequences outside the NH family cluster represent HIV-1 group M (subtypes A to E) references (23).
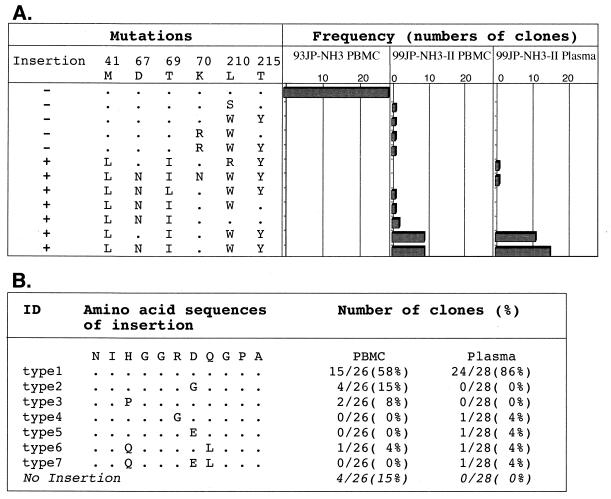
To assess the frequency and genetic background of the insertion mutants in the patient, a total of 82 independently cloned RT segments were obtained by PCR from NH3 blood specimens collected before and after chemotherapy and sequenced. Neither an insertion mutation, AZT resistance mutations, nor the T69I substitution located downstream of the insertion was detected in the 28 RT clones before drug administration (Fig. 3A, 93JP-NH3 PBMC). In contrast, the insertion mutants predominated in the peripheral blood after therapy, occurring in 85% (22 of 26) of the provirus clones (99JP-NH3-II PBMC) and 100% (28 of 28) of the plasma virus clones (99JP-NH3-II plasma). Virtually all of the insertion mutants in the blood (49 of 50) simultaneously carried two to four AZT resistance mutations and a T69I substitution (Fig. 3A). Some variations in the insertion sequence were found, while length polymorphism was not detected (Fig. 3B). A neighbor-joining tree of the cloned RT sequences revealed that the sequences of insertion mutants had much longer branch lengths than those without the insertion (data not shown). These data suggest that insertion mutants with a particular set of substitutions had a selective growth advantage during chemotherapy over variants without the insertion.
FIG. 3.
Data indicating that insertion mutants with three or four AZT resistance mutations and a T69I mutation became a dominant population in the patient's blood following chemotherapy. RT gene segments (543 to 576 bp) were amplified by PCR from PBMC-derived proviral DNAs and plasma-derived virus RNAs and cloned into pPCR-Script Amp SK (+), and 26 to 28 independent RT clones were sequenced for each blood sample. (A) Numbers of RT clones possessing the indicated set of mutations (insertion, AZT resistance mutations [M41L, D67N, K70R, L210W, and T215Y], and a T69I substitution) are shown. (B) Variations and frequency of the insertion amino acid sequences in RT clones from NH3 PBMC and plasma. ID, identification.
Roles of the various RT mutations in drug resistance.
To assess the roles of the RT mutations described above in actual MNR, a subtype E molecular clone (Fig. 2 and 4A, p93JP-NH1) was constructed from the 93JP-NH1 virus isolate, a drug-sensitive NH virus predecessor (36). The mutations were systematically introduced into its RT gene to generate a series of subtype E RT mutants (Fig. 4A). ERT-mt1 to -mt6 were made by site-directed mutagenesis of p93JP-NH1 RT and used to distinguish the roles of the AZT resistance mutations (M41L, L210W, and T215Y), the insertion, and the T69I substitution. ERT-mt7 and -mt8 carried the cloned RT segments from the 99JP-NH3-II plasma viruses in the backbone of p93JP-NH1. They were used to assess the roles of sporadic mutations in the context of the genetic backbone of ERT-mt6.
FIG. 4.
Data showing that the insertion mutation by itself does not have deleterious effects on RT activity and viral replication capability. (A) Construction of a full-length HIV-1 subtype E molecular clone (p93JP-NH1) and its RT mutants. The diagram at the top shows the approximate positions of the known open reading frames of p93JP-NH1. pUC-NH1SpBm is the p93JP-NH1-derived subclone in which site-directed mutagenesis (ERT-mt1 to -mt6) or replacement of the cloned 99JP-NH3-II RT fragments (ERT-mt7 and -mt8) was carried out. (B) Effects of RT mutations on RT activity and viral replication capability. Each HIV-1 DNA (3 μg) was transfected into HeLa cells (6 × 105) with FuGENE 6 (Roche Diagnostics). Culture supernatants were collected 2 and 3 days after transfection, and supernatant RT activity (42) and infectious titers on MAGIC-5 cells (10) were measured. d, day.
After transfection of equal amounts of the mutant DNAs into HeLa cells, the RT activities of the cell-free-virions released into the culture supernatants (42) were measured (Fig. 4B, middle). The mutants (ERT-mt1 to -mt8) had a level of RT activity that was approximately 2- to 10-fold lower than that of parental virus p93JP-NH1. Most of the mutants had a replication capacity comparable to that of the parental virus in MAGIC-5 cells (right panel, ERT-mt1 to -mt7). No evidence of replication was obtained for ERT-mt8. Thus, the insertion mutation by itself did not have any deleterious effect on RT activity and viral replication. Rather, it provided a basis for the generation of a panel of replication-competent variants in combination with substitutions.
The IC50 of each RT inhibitor was determined for each RT mutant by using MAGIC-5 cells (10) (Table 1). As expected, parental molecular clone p93JP-NH1 was as sensitive to the RT inhibitors as was NL43. Introduction of AZT resistance mutations alone (ERT-mt1) resulted in a reasonable increase (about 36-fold) in the IC50 of only AZT. An insertion mutation alone (ERT-mt2) and an insertion mutation with a T69I substitution (ERT-mt3) caused a slight increase in the IC50 of 3TC and AZT, respectively. However, the fold increases (5.2 and 8.8) were just above the variations (range, 0.1 to 5) seen in viruses from patients never treated with antiretroviral drugs (10).
TABLE 1.
Susceptibilities of RT mutants to various RT inhibitors
| Virus | IC50 (μM) (fold increase)a of:
|
|||||
|---|---|---|---|---|---|---|
| AZT | 3TC | d4T | ddI | ddC | NVP | |
| p93JP-NH1 | 0.038 (1.5) | 0.36 (0.7) | 1.9 (1.6) | 2.0 (1.3) | 0.48 (0.9) | 0.044 (0.4) |
| ERT-mt1 | 0.90 (36) | 0.69 (1.4) | 0.82 (0.7) | 1.7 (1.1) | 0.25 (0.4) | 0.029 (0.3) |
| ERT-mt2 | 0.027 (1.1) | 2.6 (5.2) | 1.7 (1.4) | 2.2 (1.5) | 0.54 (1.0) | 0.042 (0.4) |
| ERT-mt3 | 0.22 (8.8) | 1.2 (2.4) | 2.4 (2.0) | 6.4 (4.3) | 0.29 (0.5) | <0.01 (<0.1) |
| ERT-mt4 | >10 (>400) | 15.0 (30) | 14.0 (12) | 13.0 (8.7) | 0.25 (0.4) | 0.02 (0.2) |
| ERT-mt5 | >10 (>400) | 8.0 (16) | 5.2 (4.3) | 14.0 (9.3) | 1.3 (2.3) | 0.016 (0.1) |
| ERT-mt6 | >10 (>400) | 96 (192) | 30 (25) | 45 (30) | 2 (3.6) | 0.021 (0.2) |
| ERT-mt7 | >10 (>400) | >100 (>200) | 12 (10) | 39 (26) | 2.9 (5.2) | 0.023 (0.2) |
Fold increase compared to the IC50 for HIV-1 strain NL43.
Of note were the combinations of the insertion mutation, AZT resistance mutations, and the T69I mutation (Table 1, ERT-mt4 to -mt7). Introduction of the insertion with AZT resistance mutations (ERT-mt4 and -mt5) resulted in a marked increase in the IC50 of AZT (more than 400-fold) and a moderate increase in those of 3TC, d4T, and ddI (16- to 30-fold, 4.3- to 12-fold, and 8.7- to 9.3-fold, respectively). The addition of a T69I mutation to these mutations (ERT-mt6) enhanced the levels of 3TC, d4T, and ddI resistance of ERT-mt5 by approximately 12-, 6-, and 3-fold, respectively. Addition of other, sporadic substitutions to these mutations (ERT-mt7) resulted in increased 3TC resistance.
Superimposition of the mutations on the RT 3-D structure.
To obtain structural insight into the roles of mutations in MNR, 3-D structural models were generated for p93JP-NH1 RT and its MNR mutant with an insertion (ERT-mt6 RT) by using the FAMS program (28). The X-ray crystal RT structure (16) used as a template for this modeling assumes a hand-like structure in which the finger, palm, and thumb subdomains form the template-binding cleft and the deoxynucleoside triphosphate (dNTP)-binding pocket. The FAMS program generated a single most likely model of 93JP-NH1 RT that has only amino acid substitutions for the respective template residues (Fig. 5A), predicting that the overall 3-D structure, including the substrate-binding cleft, is indistinguishable from the X-ray structure.
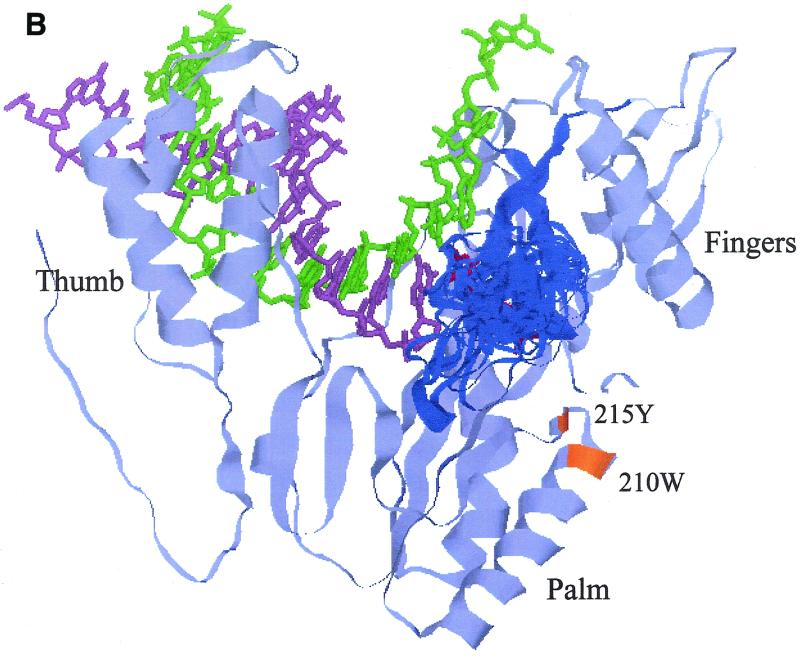
FIG. 5.
Models showing how the β3-β4 loop, extended by an insertion, in a highly flexible manner causes a reduction in the distance across the gap between the loop and the other RT subdomains in the polymerase active center. These 3-D structural models of 93JP-NH1 RT (A) and ERT-mt6 RT (B) were constructed by the FAMS program (28) using the X-ray crystal structure of HIV-1 subtype B RT (16). Only the finger, thumb, and palm subdomains of the RT p66 subunit (amino acid residues 1 to 334) are shown to highlight the polymerase active center. Backbone residues of the p66 models are pale blue. β3-β4 loops are dark blue. The template, primer, and dTTP backbones superimposed on the p66 models are green, light purple, and red, respectively. For the ERT-mt6 RT model, 24 β3-β4 loops with equally low-energy statuses were superimposed on the most favorable (26) model. Substitutions minimally required for development of MNR in combination with the insertion are orange.
The β3-β4 loop of ERT-mt6 RT accommodated an 11-amino-acid insertion without gross changes in the overall secondary structure of the finger subdomain, while a total of 24 loop models with equally low energy states could be generated (Fig. 5B). The results indicated that the β3-β4 loop of ERT-mt6 RT could be highly flexible. The ERT-mt6 RT models predicted the following marked structural changes along the polymerase active center. First, the various conformations of the β3-β4 loops extended to the 3′ end of the primer. Second, the loops extended over the nucleoside triphosphate-binding site (41). Third, the loops extended closer to the template in the cleft.
Table 2 compares the shortest distances across the gap between the β3-β4 loop and other protein regions or residues between the p93JP-NH1 and ERT-mt6 RT models. The average shortest distances in the main chain between the loops and the other subdomains were reduced by as much as 2.37 Å (from 10.63 to 12.63 Å in p93JP-NH1 RT to 9.30 to 11.69 Å in ERT-mt6) (Table 2, subdomains, main chain, average). The minimum shortest distance in the main chain was reduced to 5.47 to 7.44 Å in ERT-mt6 RT, i.e., by as much as by 6.20 Å (Table 2, subdomains, main chain, minimum). An essentially similar extent of reduction was found when the side-chain distances were compared (Table 2, subdomains, side chain) or when the shortest distances between the β3-β4 loop and particular residues involved in dNTP binding (16, 41) were compared (Table 2, residues involved in dNTP binding). Taken together, the molecular modeling demonstrated not only visually but also quantitatively a reduction in the distances between the β3-β4 loop and the other subdomains in the mutant RT, compared with the wild-type RT.
TABLE 2.
Shortest distances between β3-β4 loop and protein regions critical for polymerase activity
| Residue(s) | Location | Shortest distance to β3-β4 loop (Å)
|
|||||
|---|---|---|---|---|---|---|---|
| Main chain
|
Side chain
|
||||||
| 93JP-NH1 | ERT-mt6a
|
93JP-NH1 | ERT-mt6a
|
||||
| Avg | Minimum | Avg | Minimum | ||||
| Subdomains | |||||||
| Ile94–AspI13 | Palm | 10.63 | 9.84 (−0.79)b | 5.59 (−5.04) | 6.30 | 6.33 (+0.03) | 3.53 (−2.77) |
| Ile159–Thr240 | Palm | 11.67 | 9.30 (−2.37) | 5.47 (−6.20) | 4.17 | 3.24 (−0.93) | 1.42 (−2.75) |
| VaI241–Tyr319 | Thumb | 12.63 | 11.69 (−0.94) | 7.44 (−5.19) | 6.76 | 6.27 (−0.49) | 2.36 (−4.40) |
| Residues involved in dNTP binding | |||||||
| Tyr183–Asp186 | Palm | 14.72 | 13.50 (−1.22) | 10.47 (−4.25) | 6.40 | 6.48 (+0.08) | 5.69 (−0.71) |
| Asp110–Phe 116 | Palm-finger | 8.60 | 8.32 (−0.28) | 5.11 (−3.49) | 4.33 | 4.23 (−0.10) | 2.48 (−1.85) |
| Gln151 | Finger | 3.10 | 3.93 (+0.83) | 3.68 (+0.58) | 2.85 | 2.94 (+0.09) | 2.76 (−0.09) |
| Lys219 | Finger | 11.67 | 9.83 (−1.84) | 5.46 (−6.21) | 4.17 | 3.38 (−0.79) | 1.42 (−2.75) |
| His221 | Finger | 14.97 | 12.96 (−2.01) | 9.44 (−5.53) | 8.43 | 7.92 (−0.51) | 5.30 (−3.13) |
Average and minimum shortest distance of the 24 most likely models of ERT-mt6 RT are shown.
Difference in distance compared to 93JP-NH1.
Search for viral and cellular homologues to the insert.
To determine the origin of the 33-nucleotide insert, viral or cellular homologues to the seven insert sequences observed in December 1999 in patient NH3 (Fig. 3B) were screened as follows. First, a computer search for the p93JP-NH1 or other subtype E (23) sequences failed to identify HIV-1 genomic sequences with homology to the insert higher than 55%. Second, a computer search by the BLAST program (1) of the currently available DNA databases identified no viral and eucaryotic regions identical to the entire 33-nucleotide insert (highest score, 19-of-19-nucleotide identity with an E value of 0.14 on March 2001). Third, screening of a human leukocyte cDNA library (SuperScript Human Leukocyte cDNA Library; GIBCO BRL) identified four clones with only partial homology (20-to-25-nucleotide identity). Thus, while several sequences with partial homology were identified, no sequence with complete identity to the entire insert was found by these approaches (data not shown).
DISCUSSION
We have shown here that (i) the previously noted flexibility of the β3-β4 loop of HIV-1 subtype B in accepting extra amino acids is also maintained for the subtype E strain and is, notably, high enough to accommodate a foreign sequence as long as 11 amino acids (Fig. 1 and 2), (ii) variants with the insertion become virtually a single dominant population under selective pressure due to the presence of multiple NRTIs (Fig. 3), and (iii) this insertion mutation alone does not directly cause MNR but greatly augments the MNR on the basis of preexisting drug resistance mutations without compromising replication capacity (Fig. 4 and Table 1). These data demonstrate that incorporation of the long foreign fragment into the RT was a key event for HIV-1 to adapt to and survive the strong pressures in this particular patient. At the same time, our data illustrate a hitherto unappreciated mechanism, a long peptide insertion into RT, for HIV-1 adaptive change.
Of particular relevance to our present work is the biochemical study conducted by Kew et al. (21). Kew et al. successfully inserted a purely in vitro-designed 15-amino-acid sequence into the β3-β4 loop of subtype B RT without impairing enzyme activity. However, that study was conducted by using purified RT molecules in test tubes and thus was unable to address any biological and medical consequences of this insertion in the context of either viral replication in cells or drug sensitivity in vivo. That study also did not always predict that a long peptide insertion into the β3-β4 loop could take place during the evolutionary process of MNR variants in vivo.
How, then, does the 11-amino-acid insertion result in augmentation of MNR? Structure modeling suggested that the insertion makes the dNTP-binding pocket smaller (Fig. 5 and Table 2), as proposed for the Ser-Ser two-amino-acid insertion mutant of subtype B RT (24). This pocket size change may, in turn, provide a basis for a reduced probability of incoming NRTI binding to the enzyme active center. In addition, the insertion element possesses two charged residues (arginine and aspartic acid) (Fig. 1) and would therefore change the charged status of the surface area of the pocket, which may also affect substrate selectivity. Structural modeling further suggested that the insertion makes the template-primer cleft between the β3-β4 loop and the other subdomains narrower (Fig. 5 and Table 2), as noted previously with the 15-amino-acid insertion mutant of subtype B (21). This structural change may lead to more intimate interactions of the template-primer complex with the mutant RT, compared with the wild-type RT (21), leading to better sensing of DNA with the wrong geometry. This, in turn, may provide a basis for more efficient DNA repair at the chain-terminated position.
However, the profound structural changes induced by the insertion in the RT active center should be optimized for MNR by additional substitutions because the insertion alone was not sufficient to develop MNR (Table 1, ERT-mt2). Such a synergistic effect of an insertion mutation along with other substitutions on MNR has also been reported for the Ser-Ser insertion mutation in the subtype B β3-β4 loop (24, 27). The two AZT resistance mutations in the palm subdomain (L210W and T215Y) should play a key role in the fine modification of the RT conformation for MNR because the combination of the insertion and these mutations was the minimum requirement for MNR development (Table 1, ERT-mt4), as seen in the Ser-Ser mutation (24). These two mutations have been suggested to mediate biochemical changes in template-primer interaction, inducing higher processivity of DNA polymerization (2), which may be a prerequisite for MNR. A T69I mutation located downstream of the insertion enhanced the level of MNR (Table 1, ERT-mt6) and thus should also be critical to the optimization processes, possibly by affecting the orientation of the extended β3-β4 loop. Detailed biochemical comparisons of the wild-type and mutant RTs are necessary to address these hypothesized structure-function relationships of the subtype E RT.
Such an evolutionary survival strategy, the use of a long foreign peptide insertion, may not be very commonly utilized by HIV. However, there are other remarkable examples in adaptive changes of structural genes of RNA viruses, such as avian influenza A virus (22) and poliovirus (7). Furthermore, taking into account the characteristics of the HIV RT, which often undergoes template-primer misalignment (3) and recombination (18) during DNA polymerization, the high replication capacity of HIV in vivo (29) suggests that generation of insertion mutants occurs frequently in patients. Most of the insertion mutations probably have deleterious effects on infectivity or reduce relative fitness in a quasispecies under standard replication conditions because they are rarely detected in vivo. On the other hand, an insertion mutation generally causes much more profound changes in protein structure and function, compared with a single-point mutation, which, if not deleterious, may confer a substantial advantage when strong selective forces are encountered. In fact, the present study suggests that HIV tolerates long insertion mutations even in the most conserved retroviral gene under the pressure of multiple HIV inhibitors. Thus, insertion mutation appears to deserve more attention, particularly in the adaptive evolution of HIV, as well as the other RNA viruses.
ACKNOWLEDGMENTS
We thank M. A. Martin for critical reading of the manuscript.
This work was supported by grants from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arion D, Kaushik N, McCormick S, Borkow G, Parniak M A. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 3.Bebenek K, Abbotts J, Wilson S H, Kunkel T A. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 4.Bernstein F C, Koetzle T F, Williams G J, Meyer E E, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 5.Briones C, Mas A, Gomez-Mariano G, Altisent C, Menendez-Arias L, Soriano V, Domingo E. Dynamics of dominance of a dipeptide insertion in reverse transcriptase of HIV-1 from patients subjected to prolonged therapy. Virus Res. 2000;66:13–26. doi: 10.1016/s0168-1702(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 6.Briones C, Soriano V. Different outcome in the first two patients with an HIV-1 multinucleoside drug-resistant T69SSS insertion in Spain. Antivir Ther. 1999;4:125–127. [PubMed] [Google Scholar]
- 7.Charini W A, Todd S, Gutman G A, Semler B L. Transduction of a human RNA sequence by poliovirus. J Virol. 1994;68:6547–6552. doi: 10.1128/jvi.68.10.6547-6552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Antoni A, Foli A, Lisziewicz J, Lori F. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J Infect Dis. 1997;176:899–903. doi: 10.1086/516511. [DOI] [PubMed] [Google Scholar]
- 9.de Jong J J, Goudsmit J, Lukashov V V, Hillebrand M E, Baan E, Huismans R, Danner S A, ten Veen J H, de Wolf F, Jurriaans S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 10.Hachiya A, Aizawa-Matsuoka S, Tanaka M, Takahashi Y, Ida S, Gatanaga H, Hirabayashi Y, Kojima A, Tatsumi M, Oka S. Rapid and simple phenotypic assay for drug susceptibility of human immunodeficiency virus type 1 by using CCR5-expressing HeLa/CD4+ cell clone 1-10 (MAGIC-5) Antimicrob Agents Chemother. 2001;45:495–501. doi: 10.1128/AAC.45.2.495-501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrigan P R, Kinghorn I, Bloor S, Kemp S D, Najera I, Kohli A, Larder B A. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J Virol. 1996;70:5930–5934. doi: 10.1128/jvi.70.9.5930-5934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch M S, Brun-Vezinet F, D'Aquila R T, Hammer S M, Johnson V A, Kuritzkes D R, Loveday C, Mellors J W, Clotet B, Conway B, Demeter L M, Vella S, Jacobsen D M, Richman D D. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. J Am Med Assoc. 2000;283:2417–2426. doi: 10.1001/jama.283.18.2417. [DOI] [PubMed] [Google Scholar]
- 13.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Hooker D J, Tachedjian G, Solomon A E, Gurusinghe A D, Land S, Birch C, Anderson J L, Roy B M, Arnold E, Deacon N J. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 17.Imamichi T, Sinha T, Imamichi H, Zhang Y M, Metcalf J A, Falloon J, Lane H C. High-level resistance to 3′-azido-3′-deoxythimidine due to a deletion in the reverse transcriptase gene of human immunodeficiency virus type 1. J Virol. 2000;74:1023–1028. doi: 10.1128/jvi.74.2.1023-1028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jetzt A E, Yu H, Klarmann G J, Ron Y, Preston B D, Dougherty J P. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J Virol. 2000;74:1234–1240. doi: 10.1128/jvi.74.3.1234-1240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaliki V, Day N K, Dinglasan E, James-Yarish M, Hitchcock R, Skapura D, Chinta A, Johnson L, Andreopoulos A, Rey A, Good R A, Haraguchi S. Emergence of HIV-1 variants containing codon insertions and deletions in the beta3-beta4 hairpin loop domain of reverse transcriptase. Immunol Lett. 2000;74:173–175. doi: 10.1016/s0165-2478(00)00209-1. [DOI] [PubMed] [Google Scholar]
- 20.Kellam P, Boucher C A, Larder B A. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kew Y, Olsen L R, Japour A J, Prasad V R. Insertions into the beta3-beta4 hairpin loop of HIV-1 reverse transcriptase reveal a role for fingers subdomain in processive polymerization. J Biol Chem. 1998;273:7529–7537. doi: 10.1074/jbc.273.13.7529. [DOI] [PubMed] [Google Scholar]
- 22.Khatchikian D, Orlich M, Rott R. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature. 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 23.Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Mullins J, Wolinsky S, Korber B. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1999. [Google Scholar]
- 24.Larder B A, Bloor S, Kemp S D, Hertogs K, Desmet R L, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D K, Stuart D I, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski R, McArthur M, Moss D, Thornton J. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 27.Mas A, Parera M, Briones C, Soriano V, Martinez M A, Domingo E, Menendez-Arias L. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 2000;19:5752–5761. doi: 10.1093/emboj/19.21.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogata K, Umeyama H. An automatic homology modeling method consisting of database searches and simulated annealing. J Mol Graph Model. 2000;18:258–272. doi: 10.1016/s1093-3263(00)00037-1. , 305–306. [DOI] [PubMed] [Google Scholar]
- 29.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 30.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 31.Rakik A, Ait-Khaled M, Griffin P, Thomas T A, Tisdale M, Kleim J P. A novel genotype encoding a single amino acid insertion and five other substitutions between residues 64 and 74 of the HIV-1 reverse transcriptase confers high-level cross-resistance to nucleoside reverse transcriptase inhibitors. Abacavir CNA2007 International Study Group. J Acquir Immune Defic Syndr. 1999;22:139–145. doi: 10.1097/00126334-199910010-00005. [DOI] [PubMed] [Google Scholar]
- 32.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 33.Ross L, Johnson M, Ferris R G, Short S A, Boone L R, Melby T E, Lanier R, Shaefer M, St. Clair M. Deletions in the beta3-beta4 hairpin loop of HIV-1 reverse transcriptase are observed in HIV-1 isolated from subjects during long-term antiretroviral therapy. J Hum Virol. 2000;3:144–149. [PubMed] [Google Scholar]
- 34.Ross L, Johnson M, Graham N, Shaefer M, St. Clair M. The reverse transcriptase codon 69 insertion is observed in nucleoside reverse transcriptase inhibitor-experienced HIV-1-infected individuals, including those without prior or concurrent zidovudine therapy. J Hum Virol. 1999;2:290–295. [PubMed] [Google Scholar]
- 35.Sato H, Shiino T, Kodaka N, Taniguchi K, Tomita Y, Kato K, Miyakuni T, Takebe Y. Evolution and biological characterization of human immunodeficiency virus type 1 subtype E gp120 V3 sequences following horizontal and vertical virus transmission in a single family. J Virol. 1999;73:3551–3559. doi: 10.1128/jvi.73.5.3551-3559.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato H, Tomita Y, Shibamura K, Shiino T, Miyakuni T, Takebe Y. Convergent evolution of reverse transcriptase (RT) genes of human immunodeficiency virus type 1 subtypes E and B following nucleoside analogue RT inhibitor therapies. J Virol. 2000;74:5357–5362. doi: 10.1128/jvi.74.11.5357-5362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiura W, Matsuda M, Matsuda Z, Abumi H, Okano A, Oishi T, Moriya K, Yamamoto Y, Fukutake K, Mimaya J, Ajisawa A, Taki M, Yamada K, Nagai Y. Identification of insertion mutations in HIV-1 reverse transcriptase causing multiple drug resistance to nucleoside analogue reverse transcriptase inhibitors. J Hum Virol. 1999;2:146–153. [PubMed] [Google Scholar]
- 39.Tamalet C, Izopet J, Koch N, Fantini J, Yahi N. Stable rearrangements of the beta3-beta4 hairpin loop of HIV-1 reverse transcriptase in plasma viruses from patients receiving combination therapy. AIDS. 1998;12:F161–F166. doi: 10.1097/00002030-199814000-00003. [DOI] [PubMed] [Google Scholar]
- 40.Tamalet C, Yahi N, Tourres C, Colson P, Quinson A M, Poizot-Martin I, Dhiver C, Fantini J. Multidrug resistance genotypes (insertions in the beta3-beta4 finger subdomain and MDR mutations) of HIV-1 reverse transcriptase from extensively treated patients: incidence and association with other resistance mutations. Virology. 2000;270:310–316. doi: 10.1006/viro.2000.0261. [DOI] [PubMed] [Google Scholar]
- 41.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 42.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winters M A, Coolley K L, Girard Y A, Levee D J, Hamdan H, Shafer R W, Katzenstein D A, Merigan T C. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J Clin Investig. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]