Abstract
Paddy fields are a major emission source of the greenhouse gas methane. In the present study, the addition of ferrihydrite to xylan-amended paddy soil microcosms suppressed methane emissions. PCR-based and metatranscriptomic analyses revealed that the addition of ferrihydrite suppressed methanogenesis by heterogeneous methanogens and simultaneously activated Geobacteraceae, the most abundant iron-reducing diazotrophs. Geobacteraceae may preferentially metabolize xylan and/or xylan-derived carbon compounds that are utilized by methanogens. Geomonas terrae R111 utilized xylan as a growth substrate under liquid culture conditions. This may constitute a novel mechanism for the mitigation of methane emissions previously observed in ferric iron oxide-applied paddy field soils.
Keywords: paddy field soil, methanogenesis, ferrihydrite, iron-reducing diazotrophs, metatranscriptome
Rice paddy field soils are major emission sources of methane, the global warming potential (GWP) of which is greater than that of carbon dioxide (Ehhalt and Schmidt, 1978; Dijkstra et al., 2012). The factors affecting methane emissions from paddy field soils have been extensively examined, and the application of ferric iron oxide was shown to delay decreases in the soil redox potential (Eh) and suppress methane generation in extremely low-Eh environments (Jäckel and Schnell, 2000; Ali et al., 2008). On the other hand, we previously reported that the application of ferric iron oxide to paddy soil enhanced the nitrogen-fixing activity of diazotrophic iron-reducing bacteria (Geobacteraceae and Anaeromyxobacter), the predominant diazotrophs in paddy soil (Masuda et al., 2021; Zhang et al., 2023). Acetate, a major metabolite of the decomposition of microbial rice straw (Glissmann and Conrad, 2000), is used as an electron donor and substrate for methanogenesis by acetate-utilizing methanogens (Falz et al., 1999; Conrad, 2007), and is also utilized as an electron donor by iron-reducing bacteria (Sanford et al., 2007; Masuda et al., 2020). Therefore, methanogens and iron-reducing diazotrophs compete for straw-derived carbon compounds, such as acetate. Based on these findings, we hypothesized that when methanogenesis is suppressed by the application of ferric iron oxide, the activation of iron-reducing diazotrophs simultaneously occurs in paddy soil due to the preferential utilization of straw-derived carbon compounds by diazotrophic iron-reducing bacteria. The purpose of the present study was to test this hypothesis using soil microcosms amended with xylan/cellulose, the main components of rice straw.
Paddy soil microcosms were constructed as follows. Paddy soil was collected at an experimental paddy field in the Niigata Agricultural Research Institute. Ten grams of soil was amended with cellulose (36.8 mg) or xylan (26.3 mg), the major component of rice straw, ferrihydrite (50 mg), a ferric iron oxide, was added, and the mixture was then placed in a vial (50 mL) (Supplementary document, Fig. S1). Each vial containing soil and air was sealed with a butyl rubber stopper and plastic screw cap and incubated at 30°C for 31 days. The amount of methane that accumulated in the gas phase was measured periodically by gas chromatography equipped with a flame ionization detector. Detailed information on the materials and methods used is provided in the supplementary document. Soils without carbon (cellulose or xylan) amendments (N, without ferrihydrite and NF, with ferrihydrite) did not emit methane during the incubation period (Fig. 1). Methane emission was observed from soils amended with cellulose and xylan. In cellulose-amended soils (C and CF), no significant differences were observed in the amount of methane that accumulated between ferrihydrite-amended (CF) and non-amended (C) soils during the incubation period. Meanwhile, in xylan-amended soils (X and XF), the amount of methane that accumulated was significantly lower in ferrihydrite-amended soil (XF) than in non-amended soil (X) after Day 9. These results suggest that the addition of ferrihydrite reduced the substrates available for methanogenesis (acetate and CO2/H2) that were derived from the microbial decomposition of xylan in soil.
Fig. 1.
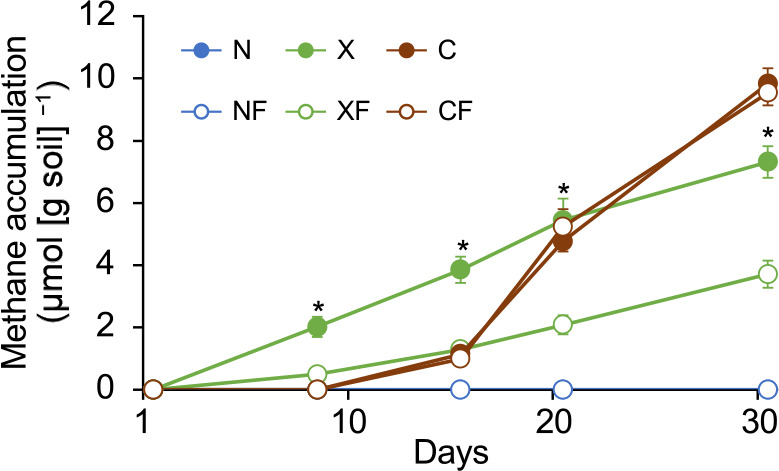
Methane accumulation in vials from control (N), ferrihydrite-amended (NF), xylan-amended (X), xylan and ferrihydrite-amended (XF), cellulose-amended (C), and cellulose and ferrihydrite-amended (CF) paddy soils (mean±SD). * indicates ferrihydrite-amended soils, in which the accumulation of methane was significantly lower than in the sample without ferrihydrite (the Mann–Whitney U test; P<0.05).
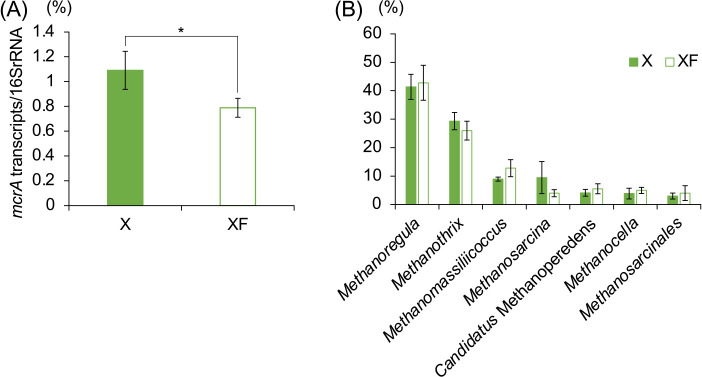
A metatranscriptomic analysis of X and XF soils (Day 16) targeting methanogenesis-related gene (mcrA) transcripts revealed that the detection frequency of mcrA transcripts was significantly lower in XF soil than in X soil (Fig. 2A), which is consistent with the results showing that methanogenesis was suppressed in XF soil. The taxonomic composition of mcrA-transcribing archaea, i.e., active methanogens, did not significantly differ between XF and X soils (Fig. 2B). Among microbes in the three most predominant genera, Methanoregula is a CO2/H2-utilizing methanogen, Methanothrix is an acetate-utilizing methanogen, and Methanosarcina utilizes both CO2/H2 and acetate (Huser et al., 1982; Jetten et al., 1992; Bräuer et al., 2011; Bai et al., 2020). These results suggest that the addition of ferrihydrite to xylan-amended paddy soils suppressed methanogenesis by heterogeneous methanogens, including that by CO2/H2-utilizing and acetate-utilizing methanogens, in a non-specific manner. The addition of ferrihydrite may have decreased both acetate and CO2/H2, which are substrates of methanogenesis derived from microbial xylan decomposition.
Fig. 2.
Ratio of the number of mcrA transcript reads to the number of rRNA transcript reads (A) and the taxonomic composition of mcrA-derived archaea (B) obtained by a metatranscriptomic analysis of xylan-amended (X) and xylan and ferrihydrite-amended (XF) paddy soils on Day 16 (mean±SD). * indicates ferrihydrite-amended soils, in which the mcrA ratio was significantly lower than in soils without ferrihydrite (the Mann–Whitney U test; P<0.05).
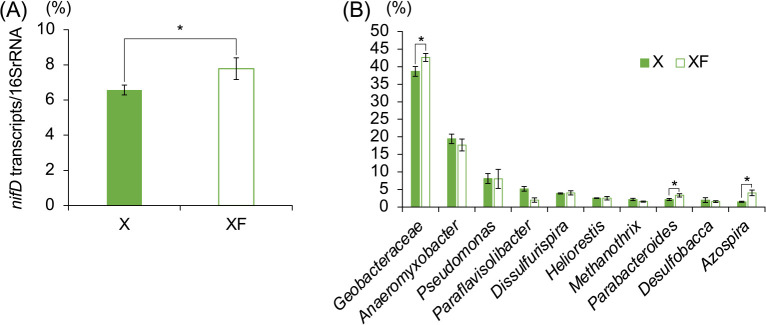
We also investigated whether the addition of ferrihydrite activated iron-reducing diazotrophs in xylan-amended soils (X and XF soils) and suppressed methanogenesis. In a qPCR-based analysis targeting the nitrogenase gene (nifD) of iron-reducing diazotrophs, the relative abundance of the nifD/16S rRNA gene increased during the first half of the incubation period and was significantly higher in XF soils than in X soils on Days 9 and 21 (Fig. S2). These results indicate that iron-reducing diazotrophs grew in soils by utilizing xylan-derived carbon compounds as carbon and energy sources and also that their growth was stimulated by the addition of ferrihydrite. In a metatranscriptomic analysis targeting nifD transcripts in X and XF soils (Day 16), the detection frequency of nifD transcripts was significantly higher in XF soil than in X soil (Fig. 3A), indicating that the addition of ferrihydrite activated nitrogen fixation in xylan-amended paddy soil. A taxonomic composition analysis revealed that the large majority of nifD-transcribing microbes belonged to Geobacteraceae, which are iron-reducing bacteria, and their abundance was higher in XF soils than in X soils (Fig. 3B), suggesting that these microbes significantly contributed to the activation of nitrogen fixation in xylan-amended paddy soils supplemented with ferrihydrite. These results supported our hypothesis that when methanogenesis was suppressed by the addition of ferrihydrite, the activation of iron-reducing diazotrophs, mostly Geobacteraceae, occurred in xylan-amended paddy soil.
Fig. 3.
Ratio of the number of nifD transcript reads to the number of rRNA transcript reads (A) and the taxonomic composition of nifD-transcribing microbes (B) obtained by a metatranscriptomic analysis of xylan-amended (X) and xylan and ferrihydrite-amended (XF) paddy soils on Day 16 (mean±SD). * indicates ferrihydrite-amended soils, in which the nifD ratio was significantly higher than in soils without ferrihydrite (the Mann–Whitney U test; P<0.05).
A possible mechanism for the simultaneous suppression of methanogenesis and activation of Geobacteraceae is that Geobacteraceae preferentially consumed acetate, the substrate of methanogenesis by acetate-utilizing methanogens. However, as described above, the addition of ferrihydrite suppressed methanogenesis by heterogeneous methanogens, including that by CO2/H2-utilizing and acetate-utilizing methanogens. Therefore, we hypothesized that members of Geobacteraceae may utilize carbon compounds other than acetate, i.e., xylan and/or carbon compounds generated from the decomposition of xylan. A liquid culture experiment using Geomonas terrae R111, a representative strain of Geobacteraceae (Xu et al., 2019), demonstrated that this strain grew by utilizing xylan as carbon and energy sources and ferric iron as an electron acceptor for respiration (Fig. S3A and B) and exhibited nitrogen-fixing activity (Fig. S3B). Consequently, the present results strongly suggest that in xylan-amended soil, the addition of ferrihydrite activated the consumption of xylan and nitrogen fixation by xylan-utilizing Geobacteraceae, thereby decreasing the generation of acetate and CO2/H2 from xylan decomposition and subsequently suppressing methanogenesis.
In terms of the taxonomic composition of nifD-transcribing microbes (Fig. 3B), Anaeromyxobacter, iron-reducing bacteria, were the second most dominant; however, their abundance did not significantly differ between X and XF soils. One possible reason for this is their inability to utilize xylan, unlike Geomonas. Anaeromyxobacter diazotrophicus Red267T, a representative strain of the diazotrophic genus Anaeromyxobacter (Itoh et al., 2022), did not utilize xylan as its growth substrate in our cultivation study (data not shown). Parabacteroides and Azospira were significantly more abundant in XF soil than in X soil; however, their contributions to soil nitrogen fixation were small because of their low abundance among nifD-transcribing microbes.
As shown in Fig. 1, the addition of ferrihydrite to cellulose-amended soil did not effectively suppress methanogenesis. Unlike xylan, cellulose may not be utilized by Geobacteraceae in soil. G. terrae Red111T did not utilize cellulose as a growth substrate in our culture experiment (data not shown). In addition, cellulose was not utilized by A. diazotrophicus Red267T (data not shown). Methanogens may compete with iron-reducing bacteria for substrates (acetate and CO2/H2) derived from cellulose even in ferrihydrite-amended soil.
In summary, in xylan-amended paddy soil microcosms, the addition of ferrihydrite activated nitrogen fixation by Geobacteraceae, the most abundant iron-reducing diazotrophs, and suppressed methanogenesis by heterogeneous methanogens. From a mechanistic perspective, xylan itself and/or carbon compounds generated from microbial xylan decomposition appeared to be preferentially consumed by Geobacteraceae, leading to decreases in available acetate and CO2/H2 as substrates of methanogenesis by methanogens. This may constitute a novel mechanism for mitigating methane emissions via the application of ferric iron oxide previously observed in paddy field soils. Further studies using 13C-labeled carbon compounds are needed to clarify this mechanism in more detail.
Citation
Masuda, Y., Chihara, M., and Senoo, K. (2024) Ferrihydrite Addition Activated Geobacteraceae, the Most Abundant Iron-reducing Diazotrophs, and Suppressed Methanogenesis by Heterogeneous Methanogens in Xylan-amended Paddy Soil Microcosms. Microbes Environ 39: ME24028.
https://doi.org/10.1264/jsme2.ME24028
Supplementary Material
Acknowledgements
We thank H. Ohba, Y. Shiratori, and the technical staff of the Niigata Agricultural Research Institute for their assistance with field management for providing the soil samples. The present study was supported by JSPS KAKENHI (Grant Numbers JP20H00409, JP20H05679, and JP20K15423), the Japan Science and Technology Agency (JST)-Mirai Program (Grant Number JPMJMI20E5), and Kubota Corporation.
References
- Ali, B.H., Blunden, G., Tanira, M.O., and Nemmar, A. (2008) Some phytochemical, pharmacological and toxicological properties of ginger (Zingiber officinale Roscoe): a review of recent research. Food Chem Toxicol 46: 409–420. [DOI] [PubMed] [Google Scholar]
- Bai, Y., Zhou, L., Irfan, M., Liang, T.T., Cheng, L., Liu, Y.F., et al. (2020) Bioelectrochemical methane production from CO2 by Methanosarcina barkeri via direct and H2-mediated indirect electron transfer. Energy (Oxford, U. K.) 210: 118445. [Google Scholar]
- Bräuer, S.L., Cadillo-Quiroz, H., Ward, R.J., Yavitt, J.B., and Zinder, S.H. (2011) Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol 61: 45–52. [DOI] [PubMed] [Google Scholar]
- Conrad, R. (2007) Microbial ecology of methanogens and methanotrophs. Adv Agron 96: 1–63. [Google Scholar]
- Dijkstra, F.A., Prior, S.A., Runion, G.B., Torbert, H.A., Tian, H., Lu, C., and Venterea, R.T. (2012) Effects of elevated carbon dioxide and increased temperature on methane and nitrous oxide fluxes: evidence from field experiments. Front Ecol Environ 10: 520–527. [Google Scholar]
- Ehhalt, D.H., and Schmidt, U. (1978) Sources and sinks of atmospheric methane. Pure Appl Geophys 116: 452–464. [Google Scholar]
- Falz, K.Z., Holliger, C., Großkopf, R., Liesack, W., Nozhevnikova, A.N., Müller, B., et al. (1999) Vertical distribution of methanogens in the anoxic sediment of Rotsee (Switzerland). Appl Environ Microbiol 65: 2402–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glissmann, K., and Conrad, R. (2000) Fermentation pattern of methanogenic degradation of rice straw in anoxic paddy soil. FEMS Microbiol Ecol 31: 117–126. [DOI] [PubMed] [Google Scholar]
- Huser, B.A., Wuhrmann, K., and Zehnder, A.J. (1982) Methanothrix soehngenii gen. nov. sp. nov., a new acetotrophic non-hydrogen-oxidizing methane bacterium. Arch Microbiol 132: 1–9. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Xu, Z., Mise, K., Masuda, Y., Ushijima, N., Hayakawa, C., et al. (2022) Anaeromyxobacter oryzae sp. nov., Anaeromyxobacter diazotrophicus sp. nov. and Anaeromyxobacter paludicola sp. nov., isolated from paddy soils. Int J Syst Evol Microbiol 72: 005546. [DOI] [PubMed] [Google Scholar]
- Jäckel, U., and Schnell, S. (2000) Suppression of methane emission from rice paddies by ferric iron fertilization. Soil Biol Biochem 32: 1811–1814. [Google Scholar]
- Jetten, M.S.M., Stams, A.J.M., and Zehnder, A.J.B. (1992) Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Rev 8: 181–197. [Google Scholar]
- Masuda, Y., Yamanaka, H., Xu, Z.X., Shiratori, Y., Aono, T., Amachi, S., et al. (2020) Diazotrophic Anaeromyxobacter isolates from soils. Appl Environ Microbiol 86: e00956-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, Y., Shiratori, Y., Ohba, H., Ishida, T., Takano, R., Satoh, S., et al. (2021) Enhancement of the nitrogen-fixing activity of paddy soils owing to iron application. Soil Sci Plant Nutr 67: 243–247. [Google Scholar]
- Sanford, R.A., Wu, Q., Sung, Y., Thomas, S.H., Amos, B.K., Prince, E.K., and Löffler, F.E. (2007) Hexavalent uranium supports growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower than predicted biomass yields. Environ Microbiol 9: 2885–2893. [DOI] [PubMed] [Google Scholar]
- Xu, Z., Masuda, Y., Itoh, H., Ushijima, N., Shiratori, Y., and Senoo, K. (2019) Geomonas oryzae gen. nov., sp. nov., Geomonas edaphica sp. nov., Geomonas ferrireducens sp. nov., Geomonas terrae sp. nov., four ferric-reducing bacteria isolated from paddy soil, and reclassification of three species of the genus Geobacter as Members of the genus Geomonas gen. nov. Front Microbiol 10: 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Masuda, Y., Xu, Z., Shiratori, Y., Ohba, H., and Senoo, K. (2023) Active nitrogen fixation by iron-reducing bacteria in rice paddy soil and its further enhancement by iron application. Appl Sci 13: 8156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.