Abstract
Purpose
To evaluate nnU-Net–based segmentation models for automated delineation of medulloblastoma tumors on multi-institutional MRI scans.
Materials and Methods
This retrospective study included 78 pediatric patients (52 male, 26 female), with ages ranging from 2 to 18 years, with medulloblastomas, from three different sites (28 from hospital A, 18 from hospital B, and 32 from hospital C), who had data available from three clinical MRI protocols (gadolinium-enhanced T1-weighted, T2-weighted, and fluid-attenuated inversion recovery). The scans were retrospectively collected from the year 2000 until May 2019. Reference standard annotations of the tumor habitat, including enhancing tumor, edema, and cystic core plus nonenhancing tumor subcompartments, were performed by two experienced neuroradiologists. Preprocessing included registration to age-appropriate atlases, skull stripping, bias correction, and intensity matching. The two models were trained as follows: (a) the transfer learning nnU-Net model was pretrained on an adult glioma cohort (n = 484) and fine-tuned on medulloblastoma studies using Models Genesis and (b) the direct deep learning nnU-Net model was trained directly on the medulloblastoma datasets, across fivefold cross-validation. Model robustness was evaluated on the three datasets when using different combinations of training and test sets, with data from two sites at a time used for training and data from the third site used for testing.
Results
Analysis on the three test sites yielded Dice scores of 0.81, 0.86, and 0.86 and 0.80, 0.86, and 0.85 for tumor habitat; 0.68, 0.84, and 0.77 and 0.67, 0.83, and 0.76 for enhancing tumor; 0.56, 0.71, and 0.69 and 0.56, 0.71, and 0.70 for edema; and 0.32, 0.48, and 0.43 and 0.29, 0.44, and 0.41 for cystic core plus nonenhancing tumor for the transfer learning and direct nnU-Net models, respectively. The models were largely robust to site-specific variations.
Conclusion
nnU-Net segmentation models hold promise for accurate, robust automated delineation of medulloblastoma tumor subcompartments, potentially leading to more effective radiation therapy planning in pediatric medulloblastoma.
Keywords: Pediatrics, MR Imaging, Segmentation, Transfer Learning, Medulloblastoma, nnU-Net, MRI
Supplemental material is available for this article.
© RSNA, 2024
See also the commentary by Rudie and Correia de Verdier in this issue.
Keywords: Pediatrics, MR Imaging, Segmentation, Transfer Learning, Medulloblastoma, nnU-Net, MRI
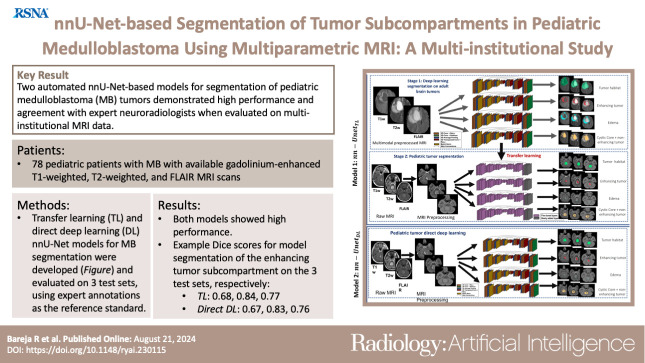
Summary
Two automated nnU-Net–based models for segmentation of pediatric medulloblastomas demonstrated high performance and agreement with expert neuroradiologists when evaluated on multi-institutional MRI data.
Key Points
■ Transfer learning and direct deep learning nnU-Net models exhibited high performance metrics scores for segmentation of pediatric medulloblastomas, with Dice scores of 0.68, 0.84, and 0.77 for the transfer learning model and Dice scores of 0.67, 0.83, and 0.76 for the direct deep learning model for enhancing tumor segmentation on three MRI test datasets.
■ Both models demonstrated robust segmentation performance across site-specific variations in multi-institutional studies (eg, Dice scores ranged from 0.81 to 0.88 for tumor habitat segmentation).
Introduction
Medulloblastoma is the most common malignant brain tumor in children, accounting for 20% of pediatric brain tumors (1). Currently, radiation treatment planning in medulloblastoma requires careful tumor delineation. Similarly, treatment response assessment in pediatric tumors (including medulloblastoma) requires tumor delineation to reliably compute measurements in two perpendicular planes (bidirectional or two-dimensional [2D]) based on consensus recommendations by the Response Assessment in Pediatric Neuro-Oncology (ie, RAPNO) working group (2–4). However, 2D measurements may not be sufficient to characterize phenotypically heterogeneous medulloblastomas, composed of enhancing tumor (ET), cystic core (CC) and nonenhancing tumor (NET), and peritumoral edema (ED), often manifested in varying proportions. Further, manual delineation of the tumor boundaries at MRI is time-consuming, hard to perform in real time during surgery, and is prone to interrater variability (5,6). There is thus an opportunity to develop automated tools for comprehensive and accurate segmentation of the entire tumor habitat (ie, ET, CC and NET, and peritumoral ED subcompartments) to enable more detailed volumetric, as well as downstream, computational analysis at MRI (7). This could, in turn, also benefit the development of reliable prognostic and predictive markers of treatment response in medulloblastomas.
This decade, deep learning approaches have emerged as powerful tools by training neural networks to learn higher-level to minute image features for semantic segmentation tasks (8). Of note, nnU-Net has emerged as a powerful approach in segmenting regions of interest on clinical scans, given its ability to offer standardized and self-configuring processes for preprocessing, network design, and architecture for a variety of segmentation tasks (9). In the context of adult brain tumors, many works have used publicly available datasets, such as the Brain Tumor Segmentation (BraTS) challenge (10,11), to employ deep learning architectures (including nnU-Net [12]) for brain tumor subcompartment segmentation. Unfortunately, most of these approaches have been developed in the context of adult and not pediatric studies, perhaps on account of there being fewer numbers of children with brain tumors than adults (13).
In this work, we explore nnU-Net–based segmentation approaches to segment the entire medulloblastoma tumor habitat, comprising the ET, peritumoral ED, and NET plus CC, and the specific subcompartments, on conventional MRI scans (T1-weighted [T1w], T2-weighted [T2w], and fluid-attenuated inversion recovery [FLAIR]).
Specifically, we evaluate two different segmentation models using the nnU-Net architecture. First, we use an nnU-Net–based transfer learning model to transfer knowledge from a related domain with a larger dataset (eg, adult brain tumor cohort in our case), as previous studies have demonstrated its value and suitability for brain tumor segmentation tasks (12). Our rationale for training the transfer learning model using adult tumor cases is that previous studies have demonstrated that domain-specific tasks, especially in medical imaging, perform better when trained on domain-specific images, as compared with ImageNet data (which mostly consists of images from nature) (14–16). Thus, in the context of our study, we hypothesize that the high-level image features can be learned from a large adult glioma cohort (17) and can be transferred over following fine-tuning on a smaller cohort of pediatric medulloblastoma cases to learn low-level domain-specific features within an nnU-Net model, thereby improving the segmentation of medulloblastoma tumor subcompartments on routine MRI scans. Second, we evaluate the efficacy of an nnU-Net model trained directly on the pediatric medulloblastomas (nnU-Net–based direct training model). Finally, we evaluate the robustness of both models to image variations across multi-institutional studies by using different combinations of the three datasets, with data from two sites at a time used for training and data from the third site used for testing. Our work presents one of the first attempts at automatic and systematic segmentation of medulloblastoma tumor subcompartments on routine MRI scans, via a multi-institutional dataset.
Materials and Methods
This retrospective, Health Insurance Portability and Accountability Act–compliant study was approved by the institutional review board of each site (identification no. 2022–1683). The need for informed consent was waived due to the study’s retrospective nature.
Notation
We define an image scene I as I = (C, f), where I is a spatial grid C of voxels c ∈ C in a three-dimensional (3D) space R3. Each voxel, c ∈ C, is associated with an intensity value f(c). IET, IED, and INET + CC correspond to the ET, ED, and NET plus CC subcompartments within every I, respectively, such that IET, IED, INET+CC ∈ I.ITH is the tumor habitat that comprises all the tumor subcompartments, representing IET + IED + INET + CC.
Workflow
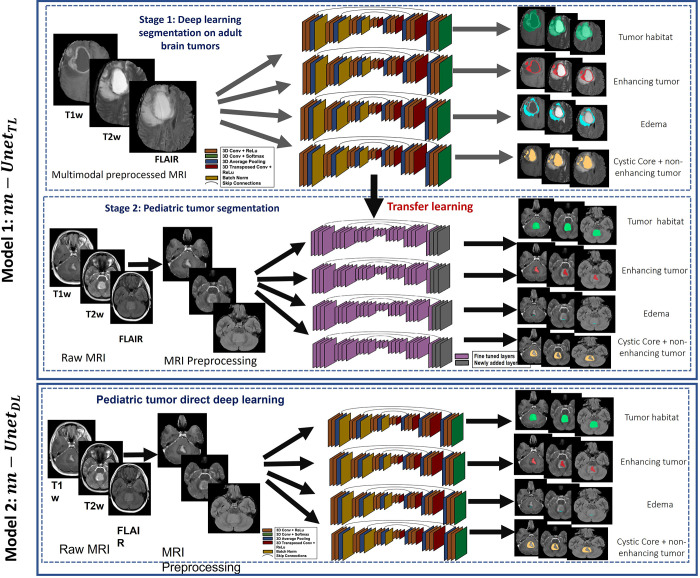
Figure 1 shows the pipeline for the two nnU-Net–based segmentation models evaluated in this work. First, for the nnU-Net transfer learning approach (denoted as nnU-NetTL), the three MRI protocols (gadolinium-enhanced T1w [Gd-T1w], T2w, and FLAIR) were employed on the BraTS glioma dataset to obtain three tumor subcompartments IET, IED, and INET + CC separately, as well as ITH, using the nnU-Net framework (9), accounting for a total of four pretrained segmentation models. Preprocessing was then conducted on our pediatric medulloblastoma datasets, including registration to age-appropriate atlases, skull stripping, bias correction, and intensity matching. Then, we used each of the four pretrained segmentation models trained in stage 1 to apply transfer learning on the preprocessed pediatric medulloblastoma MRI scans (Gd-T1w, T2w, and FLAIR) to obtain medulloblastoma tumor subcompartment segmentations. Second, for the nnU-Net deep learning approach (nnU-NetDL), after employing the preprocessing scheme on the pediatric medulloblastoma scans, the nnU-Net framework was directly applied on those scans to obtain the segmentations for the medulloblastoma tumor subcompartments.
Figure 1:
Workflow of the two proposed nnU-Net–based segmentation models. For the nnU-Net transfer learning model (nnU-NetTL), we illustrate the deep learning approach conducted on adult brain tumors in stage 1; then, in stage 2, we show the transfer learning approach employed on the pediatric medulloblastoma cases, using stage 1 results. We also illustrate the nnU-Net deep learning model (nnU-NetDL) trained directly on the pediatric medulloblastoma studies. Conv = convolutional, FLAIR = fluid-attenuated inversion recovery, ReLu = rectified linear unit, T1w = T1-weighted, T2w = T2-weighted, 3D = three-dimensional.
Data Curation
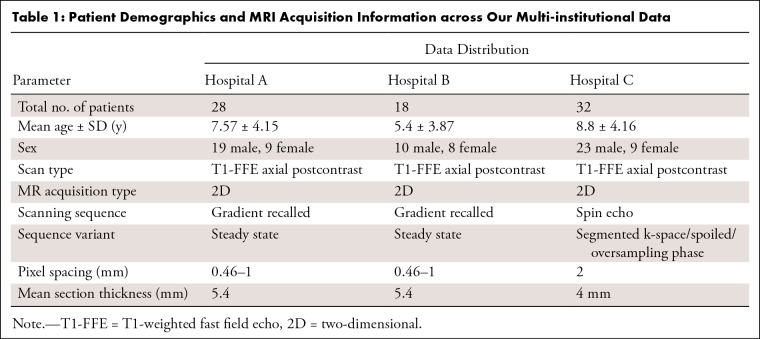
Our datasets consisted of 484 adult glioma studies (high grade and low grade) from the BraTS dataset (17), as well as 78 medulloblastoma studies that were retrospectively collected from patients between 2 and 18 years of age and performed from the year 2000 up to the date of institutional review board–approved data (May 16, 2019). The MRI scans of the medulloblastoma cases were obtained from three different institutions: Cincinnati Children’s Hospital Medical Center (hospital A), with 28 cases; Children’s Hospital Los Angeles (hospital B), with 18 cases; and Children’s Hospital of Philadelphia (hospital C), with 32 cases. The following inclusion criteria were used for our data: (a) availability of Gd-T1w, T2w, and FLAIR axial view MRI scans; (b) patients with only medulloblastoma tumors; and (c) acceptable diagnostic quality of the MRI scans, as identified by the collaborating radiologists. Following these inclusion criteria, we excluded 31 of the total 48 patients available for hospital B because of unavailability of all three sequences; similarly, hospital A had 42 cases, but only 28 had all three sequences for analysis. The 78 medulloblastoma studies included in our analysis were acquired using 1.5-T and 3-T Philips (Ingenia, Achieva) and Siemens MRI scanners. To account for variability in imaging scans across the multi-institutional data, we performed quality control of all MRI scans using an in-house, open-source tool called MRQy (18). Briefly, MRQy extracts a series of quality measures (eg, noise ratios, variation metrics) and MR image metadata (eg, voxel resolution and image dimensions) and identifies outliers (including poor-quality cases) using these measures and metadata from across the multi-institutional datasets. Patients with poor-quality MRI protocols, as identified by MRQy, were excluded from the analysis (a total of six patients from hospital C). Additional details regarding the available tumor characteristics (including tumor location, presence of metastasis, and extent of resection) for every patient included in our study are provided in Table S1.
Careful Reference Standard Annotations of Medulloblastoma Tumor Habitat and Individual Tumor Subcompartments
Reference standard labels were carefully and rigorously generated via consensus by two experienced board-certified neuroradiologists (expert 1 [A.N.], with 9 years of experience, and expert 2 [D.M.], with 8 years of radiology experience), on a per-section basis, using 3D Slicer (version 5.6.1; www.slicer.org) (19). IET was defined as the hyperintense region appearing on the Gd-T1w image, while IED was defined as signal hyperintensity on T2w and FLAIR scans. INET + CC was identified as gray or dark on Gd-T1w and FLAIR scans, with the only difference on T2w scans being the hyperintensity of the CC subcompartment (ICC). Finally, ITH was defined as the union of the three tumor subcompartments, IET + IED + INET + CC.
Preprocessing
The first step involved performing registration of our pediatric medulloblastoma scans to age-specific atlases to account for the anatomic differences across the different age groups because of brain development in pediatric patients. A total of four age-specific atlases (0–2, >2 to 5, >5 to 10, and >10 to 18 years) were used (20). We first registered the Gd-T1w images to the age-specific atlases (19) and then registered the corresponding T2w and FLAIR scans to the Gd-T1w atlas-registered scans using 3D Slicer (19). This was done for the purpose of aligning all MRI protocols (Gd-T1w, T2w, and FLAIR) to the same reference space. This process was followed by skull stripping using the Brain Extraction Tool in the FMRIB Software Library (21). Finally, correction for intensity inhomogeneities was conducted using N4ITK bias correction in 3D Slicer (19), followed by applying an intensity-matching approach (22).
Segmentation of Medulloblastoma Using nnU-Net–based Approaches
Direct-learned nnU-NetDL model.— Following preprocessing, we trained the nnU-Net model directly on the medulloblastomas. Specifically, the nnU-Net model flow was incorporated, which generates three different U-Net configurations: a 2D U-Net, a 3D U-Net, and a 3D U-Net cascade in which the first U-Net operates on downsampled images while the second is trained to refine the segmentation maps created by the former. Following cross-validation, nnU-Net then empirically chooses the best performing configuration or ensemble (9).
For training of both segmentation models (nnU-NetDL and nnU-NetTL), an initial learning rate of 0.01, stochastic gradient descent as optimizer, and a combination of Dice and cross-entropy as the loss function were used. For nnU-NetTL, separate nnU-Net architectures were trained for individual subcompartments of the tumor, IET, IED, and INET + CC, as well as for ITH, using the BraTS dataset with three MRI modalities (Gd-T1w, T2w, and FLAIR). All the training experiments were conducted in a fivefold cross-validation scheme.
Transfer-learned nnU-NetTL model.— We first trained a source nnU-Net model on 387 patients from the adult glioma dataset (BraTS 2018, known as the source dataset) with three MRI modalities (Gd-T1w, T2, and FLAIR) (17) and then validated the model on 97 patients from the dataset; separate nnU-Net models were trained for IET, IED, and INET (lumped with necrotic core subcompartment) subcomponents, as well as ITH. Following pretraining on adult glioma cases, transfer learning was incorporated within the nnU-Net framework to develop nnU-NetTL using Models Genesis (23) to fine-tune every layer of the model on our pediatric medulloblastoma training data. Specifically, this involved our target model copying all the model designs and their parameters from the source model containing the knowledge learned from the source dataset (BraTS) and applying it to the target dataset (pediatric medulloblastomas). An output layer for the target model was added, where the number of outputs was the number of class labels in our target dataset (medulloblastoma cases). The model parameters of the target model’s output layer were then randomly initialized. Finally, we trained the target model using the target dataset (ie, the pediatric brain tumor cases), where the output layer was trained from scratch but the parameters for all other layers were fine-tuned based on the pretrained weights of the source model.
Evaluating Robustness of nnU-Net Models across Multi-institutional Studies
To ensure the robustness of our segmentation models and their generalizability across multi-institutional data, we evaluated our models (nnU-NetDL and nnU-NetTL) using different combinations of training and test sets, with the data from two sites at a time used for training, while using the third site for testing. Specifically, we employed the multi-institutional data across the two segmentation approaches as follows: (a) hospital A and hospital B as training, hospital C (n = 32) as testing; (b) hospital A and hospital C as training, hospital B (n = 18) as testing; and (c) hospital B and hospital C as training, hospital A (n = 28) as testing.
Statistical Analysis
We evaluated the efficacy of both segmentation models (nnU-NetDL and nnU-NetTL) using multiple performance metrics (Tables S2–S6), namely Dice coefficient (24), Hausdorff distance (25), Fréchet distance (26), precision (27), recall (27), and Jaccard index (24).
Additionally, to statistically compare the performance of both segmentation approaches, we conducted a paired t test to identify any significant differences between the results of the two models. Specifically, for the test set used in each of the three data combinations, Dice scores obtained for each tumor subcompartment using nnU-NetDL and nnU-NetTL were input to a paired t test, followed by false discovery rate correction, to compute the two-tailed P value (with .05 determined as the level of significance). Twelve total experiments (four for the tumor subcompartments and the lesion habitat for each data combination of the three) were thus conducted using R software package (version 4.3.1; the R Foundation for Statistical Computing).
Data and Code Availability
The MRI scans obtained from hospitals A and B are protected through institutional compliance at the local institutions. The clinical repository of these patient scans can be shared per specific institutional review board requirements. Upon reasonable request, a data sharing agreement can be initiated between the interested parties and the clinical institution following institution-specific guidelines. Data from hospital C was obtained from the Children’s Brain Tumor Network (CBTN), based on an established agreement between the senior author and the CBTN. We will release the segmentations obtained from the CBTN studies into the CBTN network for future research purposes. CBTN membership can be obtained following the guidelines provided on their website to obtain access to the scans of the associated segmentations.
Results
Patient Characteristics
Our datasets included a total of 78 medulloblastoma studies that were retrospectively collected from patients between 2 and 18 years of age. The MRI scans of the medulloblastoma cases were obtained from three different institutions: 28 cases from hospital A (19 male, nine female; mean age = 3.85 years), 18 cases from hospital B (10 male, eight female; mean age = 5.4 years), and 32 cases from hospital C (23 male, nine female; mean age = 8.8 years). Table 1 shows the demographics of all study patients.
Table 1:
Patient Demographics and MRI Acquisition Information across Our Multi-institutional Data
Segmenting Medulloblastoma Tumor Habitat and Tumor Subcompartments Using nnU-NetTL Model
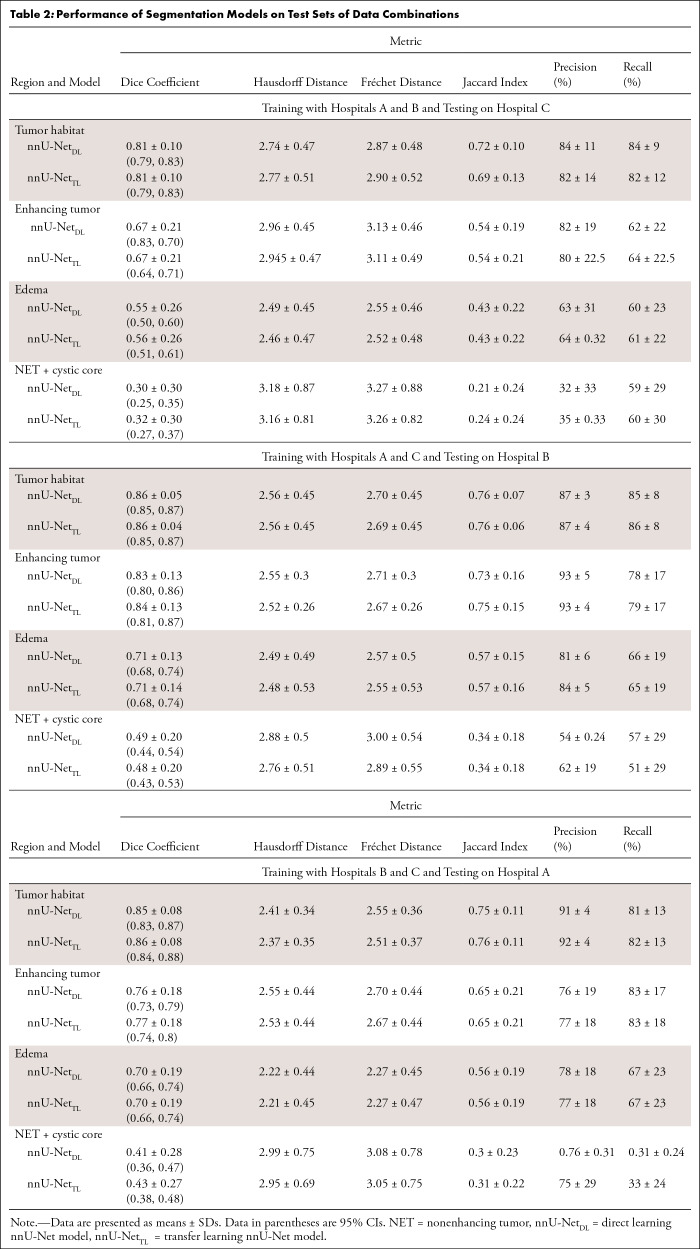
When training the source nnU-Net model on adult patients with glioma from the BraTS dataset, we obtained Dice scores of 0.90 ± 0.005 [SD], 0.78 ± 0.02, 0.81 ± 0.1, and 0.62 ± 0.007 for ITH, IET, IED, and INET (with necrotic core), respectively. Next, when applying the nnU-NetTL model on pediatric medulloblastoma cases using the three data schemes for training and testing (presented in the Evaluating Robustness of nnU-Net Models across Multi-institutional Studies section), our results demonstrated comparable accuracies and performance metrics scores in segmenting the tumor subcompartments (and the tumor habitat) across all experiments. For instance, Dice scores of 0.81, 0.86, and 0.86 were obtained for ITH segmentation for each of the three test datasets (hospital C, hospital B, and hospital A), respectively. Similarly, Dice scores of 0.68, 0.84, and 0.77 for IET segmentation, scores of 0.56, 0.71, and 0.69 for IED segmentation, and scores of 0.32, 0.48, and 0.43 in INET + CC segmentation were obtained for each of the three test datasets, respectively.
Segmenting Medulloblastoma Tumor Habitat and Tumor Subcompartments Using nnU-NetDL Model
When applying the nnU-NetDL model, our results similarly demonstrated comparable accuracies and performance metrics scores in segmenting the tumor subcompartments (and the tumor habitat) when using different combinations of training and test sets. For instance, Dice scores of 0.80, 0.86, and 0.85 were obtained for ITH segmentation for each of the three test datasets (hospital C, hospital B, and hospital A), respectively. Similarly, Dice scores of 0.67, 0.83, and 0.76 were obtained for IET segmentation for each of the three test datasets, respectively.
Comparison between nnU-NetTL and nnU-NetDL Model Performance
Detailed results of the performance metrics, including Dice coefficient, Hausdorff distance, Fréchet distance, precision, recall, and Jaccard index, that were used to assess the performance of the two segmentation models across different training and testing sets are provided in Tables S3–S5. Table 2 summarizes the performance of the two segmentation models on the test set of each of the three data combinations we employed in our work.
Table 2:
Performance of Segmentation Models on Test Sets of Data Combinations
When conducting the paired t tests, followed by the false discovery rate procedure, to compare the performance of the two segmentation models on the test sets, results showed that no evidence of differences were observed across the two segmentation approaches for the 12 conducted experiments, indicating that the two segmentation approaches exhibited a similar performance. In Table S6, we report the P values and the 95% CIs for the 12 conducted experiments.
Qualitative Examples of Good and Poor Segmentations across nnU-NetTL and nnU-NetDL Models
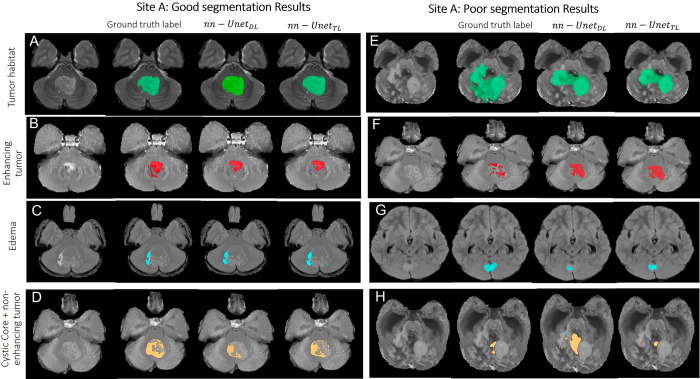
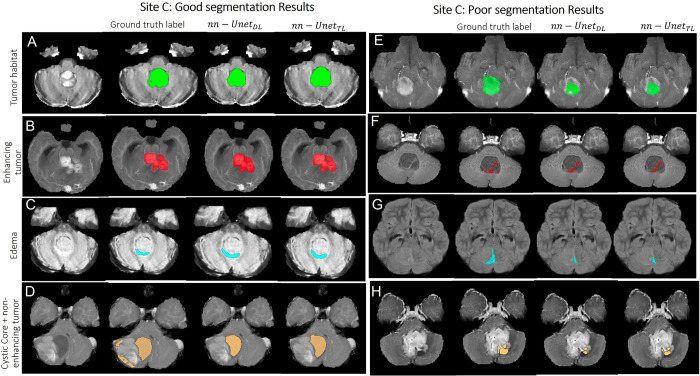
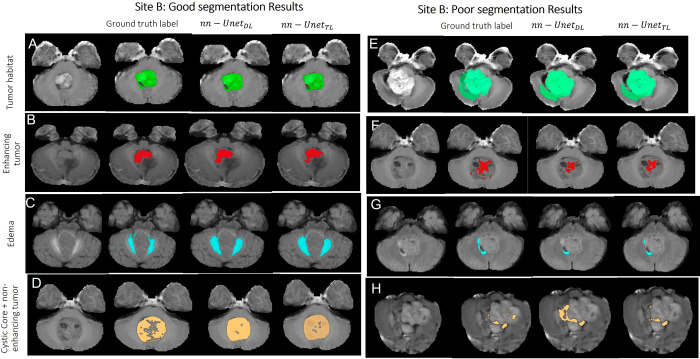
Figures 2–4 showcase some qualitative examples comparing performances of both models (nnU-NetTL and nnU-NetDL) on cases from hospitals A (Fig 2), B (Fig 3), and C (Fig 4). Specifically, Figure 2 presents both good (rows A–D) and poor (rows E–H) segmentation examples on cases obtained from hospital A using both nnU-NetTL and nnU-NetDL models. Based on our visual assessment, it was challenging to identify which model performed better, which is in line with the results of our statistical tests conducted for performance comparison not yielding significant differences across the two models. Figure 2 shows a qualitative example where nnU-NetTL undersegmented ITH, whereas nnU-NetDL undersegmented IED. Further, while nnU-NetDL oversegmented INET + CC, nnU-NetTL yielded an undersegmented result. Similarly, good (rows A–D) and poor (rows E–H) segmentation examples from hospital B, as shown in Figure 3, suggested that in these specific instances, the two models undersegmented IET, IED, and ITH, with nnU-NetDL notably undersegmenting IED more than nnU-NetTL did. Further, the nnU-NetDL model oversegmented the discontinued INET + CC label, while nnU-NetTL undersegmented it. Finally, in Figure 4, good (rows A–D) and poor (rows E–H) segmentation examples on cases obtained from hospital C suggested that both models undersegmented IED, ITH, and INET + CC. The good segmentation results in Figures 2–4 generally yielded similar performances across the two segmentation models.
Figure 2:
Representative examples of (A–D) good and (E–H) poor segmentation results obtained on patients from site A using both segmentation models. Each row shows, from left to right, the original MRI scan, the reference standard label for the subcompartment of interest, segmentation result from the nnU-NetDL model, and segmentation result from nnU-NetTL. Tumor habitat example segmentation results are shown in A (prediction accuracies of 0.9 for nnU-NetDL and 0.9 for nnU-NetTL) and E (accuracies of 0.54 for nnU-NetDL and 0.53 for nnU-NetTL). The poor results in E are perhaps on account of the subtle differences between enhancing and nonenhancing tumor and the highly heterogeneous tumor case. Enhancing tumor example segmentation results are shown in B (prediction accuracies of 0.73 for nnU-NetDL and 0.73 for nnU-NetTL) and F (accuracies of 0.44 for nnU-NetDL and 0.49 for nnU-NetTL). The poor results in F are perhaps on account of subtle differences between enhancing and nonenhancing tumor in this case. Edema example segmentation results are shown in C (prediction accuracies of 0.72 for nnU-NetDL and 0.714 for nnU-NetTL) and G (accuracies of 0.55 for nnU-NetDL and 0.58 for nnU-NetTL). Finally, nonenhancing tumor plus cystic core example segmentation results are shown in D (prediction accuracies of 0.65 for nnU-NetDL and 0.69 for nnU-NetTL) and H (accuracies of 0.15 for nnU-NetDL and 0.29 for nnU-NetTL). The poor results in H are perhaps on account of subtle differences between enhancing and nonenhancing tumor in this case. nnU-NetDL = direct learning nnU-Net model, nnU-NetTL = transfer learning deep learning model.
Figure 4:
Representative examples of (A–D) good and (E–H) poor segmentation results obtained on patients from site C using both segmentation models. Each row shows, from left to right, the original MRI scan, the reference standard label for the subcompartment of interest, segmentation result from the nnU-NetDL model, and segmentation result from nnU-NetTL. Tumor habitat example segmentation results are shown in A (prediction accuracies of 0.89 for nnU-NetDL and 0.89 for nnU-NetTL) and E (accuracies of 0.51 for nnU-NetDL and 0.53 for nnU-NetTL). Enhancing tumor example segmentation results are shown in B (prediction accuracies of 0.9 for nnU-NetDL and 0.93 for nnU-NetTL) and F (accuracies of 0.49 for nnU-NetDL and 0.51 for nnU-NetTL). Edema example segmentation results are shown in C (prediction accuracies of 0.78 for nnU-NetDL and 0.78 for nnU-NetTL) and G (accuracies of 0.42 for nnU-NetDL and 0.42 for nnU-NetTL). Finally, nonenhancing tumor plus cystic core example segmentation results are shown in D (prediction accuracies of 0.81 for nnU-NetDL and 0.78 for nnU-NetTL) and H (accuracies of 0.48 for nnU-NetDL and 0.51 for nnU-NetTL). nnU-NetDL = direct learning nnU-Net model, nnU-NetTL = transfer learning deep learning model.
Figure 3:
Representative examples of (A–D) good and (E–H) poor segmentation results obtained on patients from site B using both segmentation models. Each row shows, from left to right, the original MRI scan, the reference standard label for the subcompartment of interest, segmentation result from the nnU-NetDL model, and segmentation result from nnU-NetTL. Tumor habitat example segmentation results are shown in A (prediction accuracies of 0.85 for nnU-NetDL and 0.86 for nnU-NetTL) and E (accuracies of 0.8 for nnU-NetDL and 0.8 for nnU-NetTL). Enhancing tumor example segmentation results are shown in B (prediction accuracies of 0.897 for nnU-NetDL and 0.899 for nnU-NetTL) and F (accuracies of 0.38 for nnU-NetDL and 0.4 for nnU-NetTL). Edema example segmentation results are shown in C (prediction accuracies of 0.772 for nnU-NetDL and 0.773 for nnU-NetTL) and G (accuracies of 0.46 for nnU-NetDL and 0.52 for nnU-NetTL). Finally, nonenhancing tumor plus cystic core example segmentation results are shown in D (prediction accuracies of 0.8 for nnU-NetDL and 0.81 for nnU-NetTL) and H (accuracies of 0.28 for nnU-NetDL and 0.33 for nnU-NetTL). nnU-NetDL = direct learning nnU-Net model, nnU-NetTL = transfer learning deep learning model.
Discussion
Accurate segmentation of pediatric brain tumors plays a major role in radiation therapy planning yet is understudied. In this work, we presented two automated nnU-Net–based segmentation approaches for segmenting medulloblastomas using multi-institutional clinical MRI scans. The first model utilized transfer learning and Models Genesis on a pretrained nnU-Net model on a large adult brain tumor dataset to segment medulloblastomas, while the second method trained the nnU-Net model directly on patients with medulloblastoma. Further, to ensure the generalizability and robustness of our approaches, we employed data from three institutions by performing three experiments for each model (using data from two sites for training and data from the third site for testing). Our results demonstrated high performance metrics scores from both models for the segmentation task with no evidence of differences observed between the Dice scores obtained using each model across the different tumor subcompartments. Further, our experiments that employed different training and test sets per model exhibited relatively high and consistent Dice scores across the different training and test sets (with similar segmentation results for the two experiments when using hospitals B and C as the test sets), suggesting that the nnU-Net segmentation models may be generalizable and robust to site-specific variations.
A few approaches have been developed this decade in the context of pediatric brain tumors (28), yet with a focus on low- and high-grade gliomas and none being specifically targeted toward medulloblastomas (29–32). These studies have largely focused on deep learning–based or Bayesian approaches (30,31), but, to our knowledge, none have explicitly explored transfer learning or nnU-Net–based approaches for pediatric tumors. For the previous pediatric brain tumor segmentation approaches that considered medulloblastoma cases, the reported Dice scores from NET, CC, and ED subcompartments have been suboptimal (ranging from 0.25 to 0.6), underlining the challenges to segmenting the tumor subcompartments in a heterogeneous tumor such as medulloblastoma. For instance, Peng et al (32) developed a 3D U-Net neural network architecture model to automatically segment the tumors of high-grade gliomas, medulloblastomas, and other leptomeningeal diseases in pediatric patients on T1 contrast-enhanced and T2/FLAIR images. Similarly, the work in the study by Madhogarhia et al (31) employed a convolutional neural network–based model to segment the subcompartments of multiple pediatric brain tumors, primarily gliomas, and included a limited dataset of medulloblastoma cases (n = 24). The model processed images at multiple scales simultaneously, using a dual pathway. The first pathway kept the images at their normal resolution, while the second pathway downsampled them. While the model was able to differentiate between the enhancing and nonenhancing tumor compartments of medulloblastomas, the reported Dice scores were relatively low (0.62 for ET, 0.18 for ED, and 0.26 for NET), indicating that the model had undersegmented the tumor subcompartments. In contrast, our nnU-Net–based segmentation models (transfer learning and deep learning based) consistently yielded high values for most subcompartments, both on the training and test sets (eg, Dice scores when testing on hospital B were 0.84 and 0.83 for ET, 0.71 and 0.71 for ED, and 0.48 and 0.44 for CC plus NET for the transfer-learned and direct-learned nnU-Net models, respectively). We attribute our improved segmentation performance (compared with existing approaches) to our deliberate attempt to account for certain challenges that are pertinent to pediatric medulloblastoma studies during the training of our models.
There are some merits to our approach. First, we accounted for differences in brain anatomy by employing age-specific atlases during preprocessing before our model construction. Second, we employed data from three institutions, using different combinations of training and test sets, which yielded relatively high and consistent Dice scores across training and testing. This suggests that our segmentation models, when validated on larger studies, may be reproducible and robust to multi-institutional studies.
Our study also had limitations. First, a major challenge that we had to address was the difficulties in creating precise reference standard labels of the tumor subcompartments in retrospective pediatric brain tumor studies that are not typically encountered with adult brain tumor segmentations (33,34). This is due to reasons including the rapidly developing brains of children, the need for dedicated imaging protocols to acquire high-quality scans that show the tumor details (eg, compressed sensitivity encoding MRI) (35), which we did not have in our retrospective datasets, and the lack of availability of high-quality scans following patient sedation to control motion artifacts, which is usually conducted in the presence of anesthesia experts to mitigate the dangerous effects of sedating pediatric patients (34). In this work, we attempted to mitigate the issue of retrospective scans with relatively poor resolution by conducting intensity standardization (22), as well as excluding the scans that were identified as poor quality, using a quality control tool, MRQy (18). Another issue we faced was that some of the tumors were found to be heterogeneous (eg, mix of ET, NET, and CC), which made reference standard labeling arduous. Specifically, in our data, all three tumor subcompartments were present in only 70% of the patients. In the remaining 30% of the datasets, some of the subcompartments were missing. For instance, we found the ED subcompartment to be rarely present in medulloblastoma around the tumor core, as reported in literature (36). To increase class representation of ED to improve the model’s performance, we labeled ED around the ventricles, as ventricular ED has similar features as peritumoral ED. Additionally, the CC subcompartment has been reported in previous works to be in 40%–50% of pediatric medulloblastomas (37), whereas it was found scarcely in our datasets and had similar visual appearance to the NET on Gd-T1w and FLAIR scans. For this reason, we combined both these classes, resulting in improved pixel representation, which has been previously employed in the BraTS dataset as well (17). We also had a limitation with the availability of T1 precontrast scans, which were available for about 20% of the cases only, but were used when available to identify hemorrhage while creating reference standard labels.
Second, in some instances, distinguishing between ET and NET in our medulloblastoma datasets was difficult (unlike in adult brain tumors) when the intensity features looked similar. To account for this challenge, in consultation with collaborating radiologists who served as expert readers, we decided to make use of the center of the caudate nucleus region in the Gd-T1w postcontrast MRI scans as an intensity threshold to determine whether the subcompartment was ET or NET. The intensity of the tumor core region above the defined threshold was labeled as an ET region, while intensity below the threshold was labeled as a nonenhancing region. Third, we found that scans that yielded poor Dice scores were the ones where the tumor was small (corresponding to a few pixels) for a specific class. However, despite the small labels, our nnU-Net segmentation models, while not perfect, did manage to reasonably localize the tumor region. One approach that we will consider in the future to overcome the issue of segmenting small tumor regions is to develop a human-in-loop model (38) to improve the training labels and, in turn, the segmentation accuracy of our models. Last, a concern was that in some cases, portions of the brain tissue were removed during skull stripping. This could be due to the subtle intensity differences between the skull and the brain tissues in pediatric brain scans, which makes it challenging to accurately remove the skull from the images using automated approaches. While the skull stripping approach was carefully chosen following comparison across different skull stripping approaches (including FreeSurfer and Swiss Skull Strip), in the future, we will consider evaluating more recent approaches, such as HD-BET (39).
In conclusion, this work presented one of the first approaches to segment pediatric medulloblastoma cases via nnU-Net–based models. Our results, obtained on multi-institutional MRI scans, suggest that our nnU-Net–based automated segmentation models hold promise for improved tumor delineation, which in the future may yield development of robust diagnostic and prognostic markers for improved patient outcomes in medulloblastomas. Our future work will involve extending our segmentation models to larger multi-institutional data, with emphasis on curating high-resolution MRI scans while also accounting for the different histologic and molecular subgroups of medulloblastomas. We also plan to include subjective evaluations conducted by independent radiologists to complementarily evaluate the performance of our segmentation approaches and eventually help fine-tune our models. Another interesting future direction for our nnU-Net–based approaches would be pretraining the models on adult hemangioblastoma cases, which share similar characteristics with pediatric medulloblastomas, with both tumors arising from the posterior fossa (40), as well as on pediatric brain tumor cases, such as the ones recently released by the BraTS challenge (41).
We also plan to extend our segmentation analysis to other types of pediatric brain tumors, including high-grade and low-grade gliomas.
Acknowledgments
Acknowledgment
This research was conducted using data and/or samples made available by the Children’s Brain Tumor Network (CBTN).
M.I. supported by a Musella Foundation for Brain Tumor Research and Information grant, R&D Pilot Award, Departments of Radiology and Medical Physics, University of Wisconsin-Madison, which includes support for attending meetings and/or travel. P.T. supported by the National Cancer Institute (grant nos. 1U01CA248226, 1R01CA264017, 3U01CA248226-S1, 1R01CA277728, and P30CA014520), Department of Defense Peer-Reviewed Cancer Research Program (grant no. W81XWH-18-1-0404), Department of Veterans Affairs (grant no. 1 I01 BX005842), Dana Foundation David Mahoney Neuroimaging Program, V Foundation Translational Research Award, Johnson & Johnson WiSTEM2D Award, National Center for Advancing Clinical and Translational Science Clinical and Translational Science Award/UW Institute for Clinical and Translational Research Advancing Translational Research and Science Pilot Award (grant no. UL1TR002373), Wisconsin Alumni Research Foundation WARF Accelerator Oncology Diagnostics Grant (grant no. MSN281757), and an R&D Pilot Award, Departments of Radiology and Medical Physics, University of Wisconsin-Madison.
Data sharing: Data generated or analyzed during the study are available from the corresponding author by request.
Disclosures of conflicts of interest: R.B. Patent planned, issued, or pending for “Automatic segmentation of tumor sub-compartments in pediatric medulloblastoma using multiparametric MRI” (2022). M.I. Support for the present article from a Musella Foundation grant, R&D Pilot Award, Departments of Radiology and Medical Physics, University of Wisconsin-Madison, which includes support for attending meetings and/or travel. D.M. No relevant relationships. A.N. No relevant relationships. I.Y. Basic stipend provided by P.T. M.L. No relevant relationships. P.D. No relevant relationships. S.G. No relevant relationships. B.T. No relevant relationships. R.S. No relevant relationships. A.M. No relevant relationships. A.J. No relevant relationships. S.I. No relevant relationships. P.d.B. No relevant relationships. P.T. Support for the present work from the National Cancer Institute (grant nos. 1U01CA248226-01, 1R01CA264017-01A1, and 3U01CA248226-03S1), Department of Defense Peer-Reviewed Cancer Research Program (grant no. W81XWH-18-1-0404), Dana Foundation David Mahoney Neuroimaging Program, V Foundation Translational Research Award, and Johnson & Johnson WiSTEM2D Award; patent planned, issued, or pending for “Automatic segmentation of tumor sub-compartments in pediatric medulloblastoma using multiparametric MRI” (2022); co-founder of, equity holder in, and serves on the board of LivAi; serves as scientific consultant for Johnson & Johnson.
Abbreviations:
- BraTS
- Brain Tumor Segmentation
- CBTN
- Children’s Brain Tumor Network
- CC
- cystic core
- ED
- edema
- ET
- enhancing tumor
- FLAIR
- fluid-attenuated inversion recovery
- Gd-T1w
- gadolinium-enhanced T1w
- NET
- nonenhancing tumor
- T1w
- T1-weighted
- T2w
- T2-weighted
- 3D
- three-dimensional
- 2D
- two-dimensional
References
- 1. McKean-Cowdin R , Razavi P , Barrington-Trimis J , et al . Trends in childhood brain tumor incidence, 1973-2009 . J Neurooncol 2013. ; 115 ( 2 ): 153 – 160 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Taran SJ , Taran R , Malipatil N , Haridas K . Paediatric Medulloblastoma: An Updated Review . West Indian Med J 2016. ; 65 ( 2 ): 363 – 368 . [DOI] [PubMed] [Google Scholar]
- 3. DeNunzio NJ , Yock TI . Modern Radiotherapy for Pediatric Brain Tumors . Cancers (Basel) 2020. ; 12 ( 6 ): 1533 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wisoff JH , Sanford RA , Heier LA , et al . Primary neurosurgery for pediatric low-grade gliomas: a prospective multi-institutional study from the Children’s Oncology Group . Neurosurgery 2011. ; 68 ( 6 ): 1548 – 1554 ; discussion 1554–1555 . [DOI] [PubMed] [Google Scholar]
- 5. Gillies RJ , Kinahan PE , Hricak H . Radiomics: Images Are More than Pictures, They Are Data . Radiology 2016. ; 278 ( 2 ): 563 – 577 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quon JL , Chen LC , Kim L , et al . Deep Learning for Automated Delineation of Pediatric Cerebral Arteries on Pre-operative Brain Magnetic Resonance Imaging . Front Surg 2020. ; 7 : 517375 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stevens SP , Main C , Bailey S , et al . The utility of routine surveillance screening with magnetic resonance imaging (MRI) to detect tumour recurrence in children with low-grade central nervous system (CNS) tumours: a systematic review . J Neurooncol 2018. ; 139 ( 3 ): 507 – 522 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rizwan I , Haque I , Neubert J . Deep learning approaches to biomedical image segmentation . Inform Med Unlocked 2020. ; 18 : 100297 . [Google Scholar]
- 9. Isensee F , Jaeger PF , Kohl SAA , Petersen J , Maier-Hein KH . nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation . Nat Methods 2021. ; 18 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]
- 10. Bakas S , Reyes M , Jakab A , et al . Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge . arXiv 1811.02629 [preprint] https://arxiv.org/abs/1811.02629. Posted November 5, 2018. Updated April 23, 2019. Accessed February 23, 2023 . [Google Scholar]
- 11. Bakas S , Akbari H , Sotiras A , et al . Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features . Sci Data 2017. ; 4 ( 1 ): 170117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Isensee F , Jäger PF , Full PM , Vollmuth P , Maier-Hein KH . nnU-Net for Brain Tumor Segmentation . arXiv 2011.00848 [preprint] https://arxiv.org/abs/2011.00848. Posted November 2, 2020. Accessed February 23, 2023 . [Google Scholar]
- 13. Ostrom QT , Patil N , Cioffi G , Waite K , Kruchko C , Barnholtz-Sloan JS . CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017 . Neuro-oncol 2020. ; 22 ( 12 Suppl 2 ): iv1 – iv96 . [Published correction appears in Neuro Oncol 2022;24(7):1214.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alzubaidi L , Al-Amidie M , Al-Asadi A , et al . Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data . Cancers (Basel) 2021. ; 13 ( 7 ): 1590 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zoetmulder R , Gavves E , Caan M , Marquering H . Domain- and task-specific transfer learning for medical segmentation tasks . Comput Methods Programs Biomed 2022. ; 214 : 106539 . [DOI] [PubMed] [Google Scholar]
- 16. Ke A , Ellsworth W , Banerjee O , Ng AY , Rajpurkar P . CheXtransfer: Performance and Parameter Efficiency of ImageNet Models for Chest X-Ray Interpretation . arXiv 2101.06871 [preprint] https://arxiv.org/abs/2101.06871. Posted January 18, 2021. Updated February 21, 2021. Accessed February 23, 2023 . [Google Scholar]
- 17. Menze BH , Jakab A , Bauer S , et al . The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) . IEEE Trans Med Imaging 2015. ; 34 ( 10 ): 1993 – 2024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sadri AR , Janowczyk A , Zhou R , et al . Technical Note: MRQy - An open-source tool for quality control of MR imaging data . Med Phys 2020. ; 47 ( 12 ): 6029 – 6038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fedorov A , Beichel R , Kalpathy-Cramer J , et al . 3D Slicer as an image computing platform for the Quantitative Imaging Network . Magn Reson Imaging 2012. ; 30 ( 9 ): 1323 – 1341 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards JE , Xie W . Brains for all the ages: structural neurodevelopment in infants and children from a life-span perspective . Adv Child Dev Behav 2015. ; 48 : 1 – 52 . [DOI] [PubMed] [Google Scholar]
- 21. Smith SM . Fast robust automated brain extraction . Hum Brain Mapp 2002. ; 17 ( 3 ): 143 – 155 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madabhushi A , Udupa JK . Interplay between intensity standardization and inhomogeneity correction in MR image processing . IEEE Trans Med Imaging 2005. ; 24 ( 5 ): 561 – 576 . [DOI] [PubMed] [Google Scholar]
- 23. Zhou Z , Sodha V , Pang J , Gotway MB , Liang J . Models Genesis . Med Image Anal 2021. ; 67 : 101840 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taha AA , Hanbury A . Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool . BMC Med Imaging 2015. ; 15 ( 1 ): 29 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huttenlocher DP , Klanderman GA , Rucklidge WJ . Comparing images using the Hausdorff distance . IEEE Trans Pattern Anal Mach Intell 1993. ; 15 ( 9 ): 850 – 863 . [Google Scholar]
- 26. Alt H , Godau M . Computing the Fréchet distance between two polygonal curveS . Int J Comput Geom Appl 1995. ; 05 ( 01n02 ): 75 – 91 . [Google Scholar]
- 27. Russo DP , Zorn KM , Clark AM , Zhu H , Ekins S . Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction . Mol Pharm 2018. ; 15 ( 10 ): 4361 – 4370 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaari H , Kevrić J , Jukić S , et al . Deep Learning-Based Studies on Pediatric Brain Tumors Imaging: Narrative Review of Techniques and Challenges . Brain Sci 2021. ; 11 ( 6 ): 716 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Artzi M , Gershov S , Ben-Sira L , et al . Automatic segmentation, classification, and follow-up of optic pathway gliomas using deep learning and fuzzy c-means clustering based on MRI . Med Phys 2020. ; 47 ( 11 ): 5693 – 5701 . [DOI] [PubMed] [Google Scholar]
- 30. Zhang S , Edwards A , Wang S , Patay Z , Bag A , Scoggins MA . A Prior Knowledge Based Tumor and Tumoral Subregion Segmentation Tool for Pediatric Brain Tumors . arXiv 2109.14775 [preprint] https://arxiv.org/abs/2109.14775. Posted September 30, 2021. Accessed February 23, 2023 . [Google Scholar]
- 31.Madhogarhia R, Fathi Kazerooni A, Arif S, et al. Automated segmentation of pediatric brain tumors based on multi-parametric MRI and deep learning. In: Iftekharuddin KM, Drukker K, Mazurowski MA, Lu H, Muramatsu C, Samala RK, eds. Medical Imaging 2022: Computer-Aided Diagnosis. SPIE; 2022:124. [Google Scholar]
- 32. Peng J , Kim DD , Patel JB , et al . Deep learning-based automatic tumor burden assessment of pediatric high-grade gliomas, medulloblastomas, and other leptomeningeal seeding tumors . Neuro-oncol 2022. ; 24 ( 2 ): 289 – 299 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thukral BB . Problems and preferences in pediatric imaging . Indian J Radiol Imaging 2015. ; 25 ( 4 ): 359 – 364 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barkovich MJ , Li Y , Desikan RS , Barkovich AJ , Xu D . Challenges in pediatric neuroimaging . Neuroimage 2019. ; 185 : 793 – 801 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meister RL , Groth M , Jürgens JHW , Zhang S , Buhk JH , Herrmann J . Compressed SENSE in Pediatric Brain Tumor MR Imaging : Assessment of Image Quality, Examination Time and Energy Release . Clin Neuroradiol 2022. ; 32 ( 3 ): 725 – 733 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawamura A , Nagashima T , Fujita K , Tamaki N . Peritumoral brain edema associated with pediatric brain tumors: characteristics of peritumoral edema in developing brain . Acta Neurochir Suppl (Wien) 1994. ; 60 : 381 – 383 . [DOI] [PubMed] [Google Scholar]
- 37. Jaju A , Yeom KW , Ryan ME . MR Imaging of Pediatric Brain Tumors . Diagnostics (Basel) 2022. ; 12 ( 4 ): 961 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mosqueira-Rey E , Hernández-Pereira E , Alonso-Ríos D , Bobes-Bascarán J , Fernández-Leal Á . Human-in-the-loop machine learning: A state of the art . Artif Intell Rev 2023. ; 56 ( 4 ): 3005 – 3054 . [Google Scholar]
- 39. Isensee F , Schell M , Pflueger I , et al . Automated brain extraction of multisequence MRI using artificial neural networks . Hum Brain Mapp 2019. ; 40 ( 17 ): 4952 – 4964 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y , Chen C , Tian Z , Feng R , Cheng Y , Xu J . The diagnostic value of MRI-based texture analysis in discrimination of tumors located in posterior fossa: a preliminary study . Front Neurosci 2019. ; 13 : 1113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kazerooni AF , Khalili N , Liu X , et al . The Brain Tumor Segmentation (BraTS) Challenge 2023: Focus on Pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs) . arXiv 2305.17033 [preprint] https://arxiv.org/abs/2305.17033. Posted May 26, 2023. Updated May 23, 2024. Accessed February 23, 2023 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The MRI scans obtained from hospitals A and B are protected through institutional compliance at the local institutions. The clinical repository of these patient scans can be shared per specific institutional review board requirements. Upon reasonable request, a data sharing agreement can be initiated between the interested parties and the clinical institution following institution-specific guidelines. Data from hospital C was obtained from the Children’s Brain Tumor Network (CBTN), based on an established agreement between the senior author and the CBTN. We will release the segmentations obtained from the CBTN studies into the CBTN network for future research purposes. CBTN membership can be obtained following the guidelines provided on their website to obtain access to the scans of the associated segmentations.