Abstract
Background
Changes in gene expression related to obesity are linked to microRNAs, such as miR‐141, which play a crucial role in metabolic homeostasis. Sirtuin 1 (SIRT1), an enzyme that plays a crucial role in regulating various cellular functions and metabolism, is implicated in obesity and the ensuing non‐alcoholic fatty liver disease (NAFLD). The aim of this research was to evaluate the levels of miR‐141 and its relationship with SIRT1 and NAFLD.
Methods
A group of 100 adults (50 with obesity and 50 with normal‐weight) were selected and underwent complete clinical evaluation and anthropometric measurements. Biochemical parameters were assessed in blood serum, and the levels of miR‐141 in plasma were measured by real‐time PCR. The expression of the SIRT1 gene was also evaluated in the peripheral blood mononuclear cells using Real‐time PCR. The ELISA technique was used to determine insulin levels. Liver steatosis was assessed by ultrasound.
Results
The results showed that levels of miR‐141 were significantly increased in participants with obesity compared with the control group. Conversely, the expression of the SIRT1 gene in individuals with obesity was lower than that in control participants. A strong negative correlation was observed between miR‐141 and SIRT1 and a strong positive association was observed between miR‐141 and metabolic parameters. Furthermore, participants with fatty liver had significantly elevated levels of miR‐141 gene expression and lower expression of SIRT1 gene, compared to those without fatty liver.
Conclusion
elevated levels of miR‐141 in individuals with obesity might be a contributing factor in the repression of SIRT1 in obesity and its consequences, including NAFLD. Therefore, miR‐141 might serve as a suitable diagnostic and therapeutic target in obesity and NAFLD.
Keywords: microRNA, miR‐141, non‐alcoholic fatty liver disease, obesity, sirtuin1
1. INTRODUCTION
The increasing prevalence of obesity has had a dramatic impact on the occurrence of obesity‐related illnesses, one of which is non‐alcoholic fatty liver disease (NAFLD), first described in 1980. 1 , 2 , 3 NAFLD is characterized by the excessive accumulation of lipids in liver cells, disrupting metabolic processes and leading to inflammation, insulin resistance, oxidative stress, liver fibrosis, and even hepatocellular carcinoma. 4 , 5 Although various metabolic factors involved in the progression of NAFLD have been identified, the underlying mechanisms are not fully understood. Recent research has linked certain microRNAs to obesity and its associated metabolic disturbances. 6
MicroRNAs (miRNAs) are naturally occurring, small non‐coding RNA molecules approximately 21–25 nucleotides in length. MicroRNAs regulate gene expression by binding to the 3′‐untranslated region (3′UTR) of mRNA molecules, thereby repressing translation and potentially causing mRNA cleavage and deacetylation. 7 , 8 miRNAs play a crucial role in lipid metabolism, particularly fatty acids and cholesterol, glucose homeostasis, insulin signaling, and the expression of key regulatory enzymes. 8 , 9 , 10 , 11 Furthermore, miRNAs are involved in both normal biological processes, such as organismal development, metabolism, and physiology, and the pathogenesis of diseases such as diabetes, cancer, cardiovascular disease, and inflammation. 12 Several miRNAs have been implicated in liver pathophysiology, including NAFLD, cirrhosis, and hepatocellular carcinoma. 13 Among these, miR‐141, a member of the miR‐200 family, has emerged as an miRNA with potential roles in liver physiology and pathology. 14 , 15 , 16 miR‐141 has been proposed as a biomarker for various liver disorders and as a marker for liver aging. 17
Sirtuins comprise a group of enzymes that play an important regulatory role in various cellular processes, especially carbohydrate and lipid metabolism. Sirtuin1 (SIRT1), an NAD‐dependent deacetylase enzyme, has been shown in recent years to be dysregulated in obesity and seems to play an important role in the pathophysiology of NAFLD. 18 , 19 Various mechanisms relate SIRT1 function to protection against NAFLD. Autophagy, which is an important parameter in many disease states including NAFLD, is modulated by SIRT1. For instance, SIRT1 augmentation by active components of ginseng root can ameliorate NAFLD by restoring autophagic flux. Additionally, SIRT1 affects metabolism in liver cells by increasing CPT1A level and decreasing triglyceride levels in HepG2 cells. SIRT1 can also attenuate oxidative stress, suppress mitochondrial fission, improve mitochondrial biogenesis, and decrease hepatocyte oxidative stress by deacetylating Nrf2. 20
Generally, the expression of SIRT1 in liver cells is high, and its inhibition leads to increased glucose and lipid levels. 21 Low levels of SIRT1 have been reported in animal models of obesity and patients with obesity, especially those with insulin resistance, diabetes, and NAFLD. 19 , 22
According to a bioinformatics study, miR‐141 aligns with the 3′UTR region of sirtuin1 (SIRT1). Our previous experimental study confirmed this binding and revealed that upregulation of miR‐141 could suppress SIRT1 expression and induce lipid accumulation in cultured hepatocytes, and that it can directly target and align with the 3′‐UTR of SIRT1 mRNA. 23 The aim of this study was to investigate the levels of miR‐141 together with SIRT1 gene expression in participants with obesity and their relationship with insulin resistance, liver function, and metabolic syndrome (MetS) parameters with the hypothesis that miR‐141 might be responsible for the suppression of SIRT1 expression in participants with obesity and subsequent NAFLD.
2. MATERIAL AND METHODS
2.1. Participants
This study involved 100 participants, an obesity group composed of with 50 participants with obesity (BMI over 30), while the remaining half consisted of healthy individuals with a normal weight who served as the control group. Prior to enrollment, written informed consent was obtained from each individual. Patients were carefully selected from those visiting the clinics of the Institute of Endocrinology and Metabolism or the Minimally Invasive Surgery Research Center, while healthy controls were chosen from volunteers following clinical evaluation. The control subjects did not have extensive recent weight loss, and none of them had undergone any process for weight loss, such as bariatric surgery. Individuals with various chronic and acute medical conditions, such as heart and kidney diseases, infections, malignancies, and inflammatory conditions, as well as those taking medications that could affect lipid and carbohydrate metabolism, were excluded from the study. Specific investigations were conducted to rule out abnormalities in adrenal and thyroid hormones, known endocrine causes of obesity, as well as diabetes. The participants were not following any special diet and had normal levels of physical activity and muscle mass. Pregnant and lactating individuals were not included. All participants underwent comprehensive examinations by a single clinician, which included documenting their height, weight, waist and hip measurements, blood pressure, and family medical history. Waist circumference was measured by a single clinican, using a non‐elastic measuring tape, with the patient in the standing position, at the midpoint between the lower margin of the last palpable rib and the top of the iliac crest.
To assess MetS based on IDF standards, 24 specific factors were taken into consideration, including abdominal obesity (waist circumference greater than 102 cm for males and 88 cm for females), serum triglyceride levels exceeding 150 mg/dl, HDL cholesterol levels below 40 mg/dl, systolic hypertension (over 130 mmHg), diastolic blood pressure above 85 mmHg, and fasting plasma glucose levels above 110 mg/dl. Individuals who met three or more of these characteristics were diagnosed with MetS. Ultrasonography was performed on the liver to evaluate the degree of hepatic steatosis.
2.2. Sample collection and preparation
Blood samples were collected from each individual following an overnight fast and immediately divided into two tubes. One tube contained EDTA (ethylenediaminetetraacetic acid) as an anticoagulant for the separation of peripheral blood mononuclear cells (PBMCs), while the other tube was plain for the separation of serum. After centrifugation, the separated serum samples were frozen and stored at −80°C for future analyses.
To isolate mononuclear cells from both patients and healthy donors, fresh samples were processed using a Ficoll‐Hypaque density gradient (Sigma‐Aldrich, USA). The isolated cells were stored in tubes that were free from RNase and DNase contamination. Briefly, 10 mL of peripheral blood samples diluted with phosphate buffer saline (PBS) were layered over 3 mL of Ficoll‐Hypaque. The tubes were then centrifuged for 20 min at 5000 RPM, allowing for the isolation of PBMCs from the interlayer. The PBMCs were subsequently washed twice with PBS.
2.3. Biochemical measurements
Biochemical parameters, including fasting blood glucose (FBS), triglyceride (TG), total cholesterol (TC), high‐density lipoprotein cholesterol (HDL‐C), low‐density lipoprotein cholesterol, aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase, total bilirubin, and direct bilirubin, were measured using commercially available enzymatic colorimetric kits (Pars Azmoon, Iran) and analyzed on a Hitachi autoanalyzer. Insulin levels were measured using the enzyme‐linked immunosorbent assay (ELISA) Kit (Monobind, USA).
2.4. Gene expression assessments
Separate RNA purification was conducted from both plasma and PBMCs. To extract total RNA from plasma, which contains small RNA molecules of less than 100 nucleotides, a sample volume of 200 μL was processed using the GeneAll Hybrid‐R Blood RNA isolation kit (GeneAll, Korea). For RNA purification from PBMCs, the Hybrid‐R miRNA isolation kit (GeneAll, Korea) was utilized. This kit is optimized for the extraction of small RNA molecules (<200 nucleotides) following cell lysis with RiboEx lysis reagent. To initiate cDNA synthesis from miRNAs, a poly(A) tail was added to elongate the miRNAs using Poly(A) Polymerase (New England Biolabs). The High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) was used for cDNA synthesis, employing a primer containing a complementary sequence for the poly(A) tail and an adapter sequence (refer to Table 1 for details).
TABLE 1.
The sequence of primers.
| Primers | Sequence |
|---|---|
| miR‐141‐3p | 5′‐CGTAACACTGTCTGGTAAAGATGG ‐3′ |
| miR‐16‐5p | 5′‐GGGTAGCAGCACGTAAATATTGG ‐3′ |
| Adapter | 5′‐CGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTAG ‐3′ |
| Universal | 5′‐GCGAGCACAGAATTAATACGACTC ‐3′ |
| SIRT1‐F | 5′‐TGCGGGAATCCAAAGGATAA ‐3′ |
| SIRT1‐R | 5′‐CAGGCAAGATGCTGTTGCA ‐3′ |
| GAPDH‐F | 5′‐ GGGAAGGTGAAGGTCGGAGT‐3′ |
| GAPDH‐R | 5′‐TCCACTTTACCAGAGTTAAAAGCAG ‐3′ |
Real‐time quantitative PCR was utilized to investigate the levels of miR‐141 and SIRT1 mRNA in plasma and PBMCs, respectively, using SYBR Premix EX Taq II (Takara, Japan). The reference genes used in this assay were miR‐16 and GAPDH, which served as normalization controls. A reverse primer, complementary to the adapter sequence, was employed for amplifying miR‐141. The primer sequences can be found in Table 1. Cel‐miR‐39‐1 was added to the samples as a Spike‐In control, using its primer for amplification, to ensure reproducibility. A standard curve containg 1 x 103 ‐ 1 x 106 copies of Cel‐miR‐39 cDNA was generated independent of a serum/plasma sample and RNA purification procedure, allowing estimation of the recovery of miRNeasy Serum/Plasma Spike‐In Control.
Gene expression quantification was performed using the ΔCt method, which involved subtracting the Ct value of the reference gene from the Ct value of the target gene.
2.5. Statistical analysis
The normal distribution of the data was assessed using the Kolmogorov‐Smirnov test. Variables that exhibited a normal distribution were analyzed using the independent sample t‐test and analysis of variance, followed by Tukey's post hoc test. For variables that did not follow a normal distribution, the Mann‐Whitney U test or Kruskal‐Wallis test was applied. The association between variables was examined using either Pearson or Spearman correlation analysis. Statistical analysis was conducted using IBM SPSS version 24 and MedCalc version 18.9.10 (Ostend, Belgium). A significance level of p < 0.05 was considered to indicate a statistically significant difference.
2.6. Institutional board approval
This study was approved by the Ethics Committee of Iran University of Medical Sciences (Number 94‐05‐30‐27493).
3. RESULTS
The anthropometric characteristics of the study groups are shown in Table 2. The participants were matched in terms of gender, age, and height, and there were no significant differences between the two groups. Individuals with obesity exhibited significant increases in various anthropometric indices, including weight, BMI, waist and hip circumference, as well as systolic and diastolic blood pressures. Among the biochemical factors, FBS and HOMA‐IR were significantly higher in the obesity group compared to the control participants, while other biochemical parameters did not differ significantly between the two groups (Table 2).
TABLE 2.
Anthropometric characteristics of the studied subjects.
| Variable | Obese | Control | p‐value |
|---|---|---|---|
| Gender (M/F) | 26/24 | 16/34 | 0.043 |
| Age (Year) | 43.1 ± 17.7 | 40.4 ± 10.7 | 0.22 |
| Height (cm) | 168.4 ± 8.6 | 172.1 ± 7.4 | 0.025 |
| Weight (kg) | 105.4 ± 21.7 | 71.6 ± 10.7 | <0.001 |
| BMI | 36.6 ± 7.3 | 23.7 ± 3.2 | <0.001 |
| WC (cm) | 111.5 ± 17.4 | 85.8 ± 8.9 | <0.001 |
| HC (cm) | 123.9 ± 19.9 | 96.2 ± 6.9 | <0.001 |
| WHR | 0.9 ± 0.03 | 0.89 ± 0.05 | 0.28 |
| SBP (mmHg) | 122.4 ± 5.9 | 119.4 ± 2.3 | <0.001 |
| DBP (mmHg) | 82.8 ± 6.8 | 78.8 ± 3.2 | <0.001 |
| FBS (mg/dl) | 101.4 ± 25.6 | 91.9 ± 17.8 | 0.031 |
| TG (mg/dl) | 146.1 ± 82.6 | 153.8 ± 103.9 | 0.68 |
| TC (mg/dl) | 175.4 ± 33.8 | 178.06 ± 32.3 | 0.69 |
| LDL‐C (mg/dl) | 98.08 ± 20.08 | 102.8 ± 18.9 | 0.31 |
| HDL‐C (mg/dl) | 41.1 ± 10.04 | 40.08 ± 7.9 | 0.85 |
| Insulin (mIU/L) | 11.3 ± 8.2 | 8.8 ± 6.3 | 0.098 |
| HOMA‐IR | 3.09 ± 3.1 | 2.08 ± 1.6 | 0.044 |
| AST (IU/L) | 31.5 ± 21.3 | 26.02 ± 8.7 | 0.095 |
| ALT (IU/L) | 31.08 ± 25.5 | 30.7 ± 17.9 | 0.44 |
| ALP (IU/L) | 206.5 ± 47.6 | 193.2 ± 28.6 | 0.095 |
| T bilirubin (mg/dl) | 0.71 ± 0.47 | 0.73 ± 0.35 | 0.83 |
| D bilirubin (mg/dl) | 0.24 ± 0.12 | 0.24 ± 0.08 | 0.92 |
Note: Values are expressed as mean ± SD.
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; D bilirubin, direct bilirubin; DBP, diastolic blood pressure; FBS, fasting blood glucose; HC, hip circumference; HDL‐C, high density lipoprotein cholesterol; HOMA‐IR, homeostatic model assessment of insulin resistance; LDL‐C, low‐density lipoprotein cholesterol; SBP, systolic blood pressure; T bilirubin, total bilirubin; TC, total cholesterol; TG, triglycerides; WC, waist circumference; WHR, waist‐to‐hip ratio.
In the obesity group, 21 (49%) patients were diagnosed with MetS, and 38 (76%) patients showed varying degrees of liver lipid accumulation. Among them, 36% had Grade 1, 46% had Grade 2%, and 18% had Grade 3 fatty liver. Table 3 presents a comparison of anthropometric indices and biochemical parameters in the studied participants based on the degree of hepatic steatosis.
TABLE 3.
Comparison of anthropometric indices between groups with different degrees of hepatic steatosis.
| Variable | Normal | Grade 1 | Grade 2 | Grade 3 |
|---|---|---|---|---|
| Number | 61 | 14 | 17 | 7 |
| Age | 40.68 ± 10.92 | 45.5 ± 15.3 | 43.72 ± 10.51 | 38.71 ± 6.99 |
| BMI | 25.13 ± 4.6 | 37.14 ± 6.97*** | 38.58 ± 7.6*** | 39.12 ± 8.45*** |
| WC (cm) | 88.31 ± 10.8 | 110.29 ± 13.37*** | 118.28 ± 18.94*** | 115.29 ± 21.3*** |
| HC (cm) | 99.02 ± 9.92 | 123.29 ± 17.5*** | 130.44 ± 23.14*** | 127.57 ± 20.67*** |
| WHR | 0.89 ± 0.05 | 0.89 ± 0.04 | 0.9 ± 0.04 | 0.89 ± 0.02 |
| SBP (mmHg) | 119.25 ± 2.18 | 122.86 ± 8.25 | 123.33 ± 5.94* | 122.83 ± 4.88 |
| DBP (mmHg) | 79.05 ± 3 | 82.86 ± 8.25 | 83.33 ± 8.4* | 82.86 ± 4.88 |
| FBS (mg/dl) | 91.6 ± 16.4 | 99.4 ± 18.2 | 104.05 ± 21.8 | 116.4 ± 49.4* |
| TG (mg/dl) | 154.3 ± 97.4 | 149.8 ± 121.3 | 147.3 ± 71.3 | 119 ± 41.3 |
| TC (mg/dl) | 173.8 ± 32.03 | 158.7 ± 26.9 | 189.7 ± 32.3 | 204.24 ± 29.4 |
| LDL‐C (mg/dl) | 99.6 ± 19 | 89.2 ± 18.7 | 107.1 ± 18.6 | 116.8 ± 14.7 |
| HDL‐C (mg/dl) | 39.7 ± 7.5 | 36.5 ± 7.3 | 44.8 ± 10.5 | 51.2 ± 10.02* |
| Insulin (mIU/L) | 8.6 ± 6 | 10.44 ± 6.31 | 12.8 ± 10.2 | 15.26 ± 9.13 |
| HOMA‐IR | 2.01 ± 1.52 | 2.72 ± 2.14 | 3.66 ± 4.13 | 4.58 ± 3.32* |
| AST (IU/L) | 25.72 ± 8.9 | 22.71 ± 5.59 | 27.88 ± 11 | 69.71 ± 33.46*** |
| ALT (IU/L) | 29 ± 17.14 | 24 ± 10.92 | 33.22 ± 17.94 | 79.14 ± 32.95*** |
| ALP (IU/L) | 189.33 ± 33.8 | 212.85 ± 53.36 | 212.38 ± 40.58 | 231.52 ± 24.42* |
| T bilirubin (mg/dl) | 0.71 ± 0.34 | 0.67 ± 0.31 | 0.73 ± 0.59 | 0.88 ± 0.65 |
| D bilirubin (mg/dl) | 0.23 ± 0.08 | 0.23 ± 0.04 | 0.23 ± 0.08 | 0.35 ± 0.26* |
Note: Values are expressed as mean ± SD.
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; D bilirubin, direct bilirubin; DBP, diastolic blood pressure; FBS, fasting blood glucose; HC, hip circumference; HDL‐C, high density lipoprotein cholesterol; HOMA‐IR, homeostatic model assessment of insulin resistance; LDL‐C, low‐density lipoprotein cholesterol; SBP, systolic blood pressure; T bilirubin, total bilirubin; TC, total cholesterol; TG, triglycerides; WC, waist circumference; WHR, waist‐to‐hip ratio.
***p < 0.001, **p < 0.01, *p < 0.1.
The expression of miR‐141 in plasma was compared between the two groups, and the results revealed that individuals with obesity exhibited increased levels of miR‐141 compared to normal‐weight individuals (p < 0.001) (Figure 1A). miR‐141 was also compared in participants diagnosed with and without MetS, and the results showed that miR‐141 was significantly increased in MetS (p < 0.05) (Figure 1B). The same comparison was performed for SIRT1, and its expression was found to be significantly decreased in participants with obesity (p < 0.001) (Fi 1c). However, there was only a slight decrease in SIRT1 expression among individuals with MetS, which did not yield a statistically significant difference (p = 0.3) (Figure 1D).
FIGURE 1.
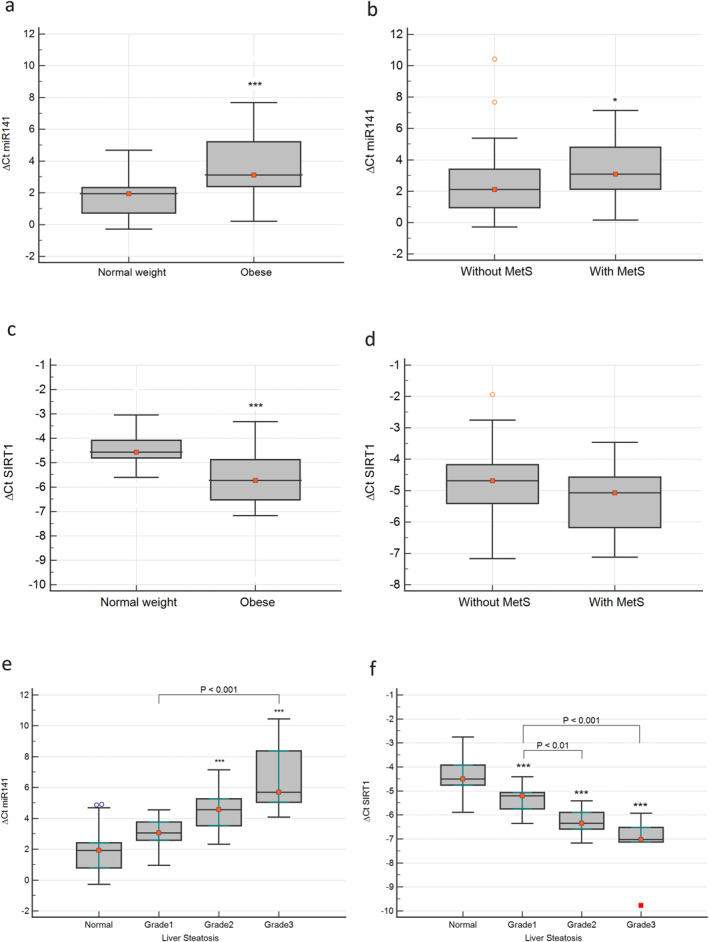
Comparison of miR‐141 between the obesity and control groups (A), and the participants with or without metabolic syndrome (MetS) (B). Comparison of SIRT1 gene expression between the obesity and control groups (C), and the participants with or without MetS (D). The comparison of miR‐141 (E) and SIRT1 (F) gene expression of the different groups categorized based on the grade of liver steatosis. *p < 0.05, **p < 0.01, ***p < 0.001.
The levels of circulatory miR‐141 were compared in the studied groups according to the degree of hepatic steatosis. miR‐141 levels showed an increasing trend with increased liver steatosis, and the expression of miR‐141 was significantly increased in patients with Grade 2 and 3 NAFLD (Figure 1E). On the contrary, the results of the expression of the SIRT1 gene displayed a significant decline along with increased lipid accumulation in the liver, and significantly reduced expression of SIRT1 was observed in the various grades of NAFLD compared to the control group (Figure 1F).
In the correlation analysis, a positive association was observed between miR‐141 and all parameters of metabolic dysregulation, including BMI, WC, HC, SBP, DBP, FBS, TC, HDL‐C, insulin, HOMA‐IR, and AST (Table 4). On the other hand, the relationship between SIRT1 and obesity parameters, glycemic indices, lipid profile, and liver function tests, including ALT and AST, was negative. When correlation analysis was performed only in the obesity group, the relationship between SIRT1 and miR‐141 with most parameters still remained significant, except with SBP, DBP, and FBS for SIRT1, and AST for miR‐141. The correlation analysis also revealed a significant reverse correlation between SIRT1 and miR‐141 (r = −0.801 in all participants and r = −0.813 in those with obesity), which remained significant after controlling for BMI (p < 0.001) (Figure 2).
TABLE 4.
Correlation coefficients indicating the relationship of Sirt1 and miR‐141 with biochemical indices.
| All subjects | Participants with obesity | |||
|---|---|---|---|---|
| Variable | SIRT1 | miR‐141 | SIRT1 | miR‐141 |
| Age | −0.009 | 0.099 | 0.027 | 0.133 |
| BMI | −0.509** | 0.533** | −0.314* | 0.445** |
| WC (cm) | −0.489** | 0.247** | −0.405** | 0.438** |
| HC (cm) | −0.468** | 0.489** | −0.294* | 0.406* |
| WHR | −0.139 | −0.058 | −0.178 | 0.411* |
| SBP (mmHg) | −0.238* | 031** | −0.235 | 0.289 |
| DBP (mmHg) | −0.214* | 0.294* | −0.117 | 0.289 |
| FBS (mg/dl) | −0.333** | 0.25* | −0.222 | 0.358* |
| TG (mg/dl) | −0.024 | 0.080 | 0.170 | −0.055 |
| TC (mg/dl) | −0.264** | 0.246* | −0.517** | 0.346* |
| LDL‐C (mg/dl) | −0.209* | 0.191 | −0.604** | 0.359* |
| HDL‐C (mg/dl) | −0.293** | 0.289* | −0.558** | 0.374* |
| Insulin (mIU/L) | −0.324** | 0.285* | −0.336* | 0.369* |
| HOMA‐IR | −0.346** | 0.338** | −0.354* | 0.432** |
| AST (IU/L) | −0.317** | 0.358** | −0.402** | 0.278 |
| ALT (IU/L) | −0.274** | 0.215 | −0.461** | 0.195 |
| ALP (IU/L) | −0.174 | 0.2 | −0.242 | 0.302 |
| T bilirubin (mg/dl) | −0.034 | −0.013 | −0.122 | 0.031 |
| D bilirubin (mg/dl) | −0.149 | 0.15 | −0.238 | 0.226 |
Note: Values are expressed as mean ± SD.
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; D bilirubin, direct bilirubin; DBP, diastolic blood pressure; FBS, fasting blood glucose; HC, hip circumference; HDL‐C, high density lipoprotein cholesterol; HOMA‐IR, homeostatic model assessment of insulin resistance; LDL‐C, low‐density lipoprotein cholesterol; SBP, systolic blood pressure; T bilirubin, total bilirubin; TC, total cholesterol; TG, triglycerides; WC, waist circumference; WHR, waist‐to‐hip ratio.
***p < 0.001, **p < 0.01, *p < 0.1.
FIGURE 2.
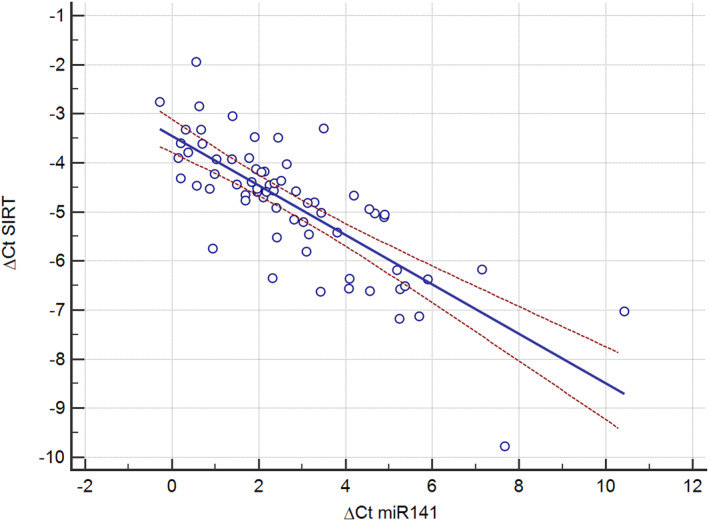
The correlation between the expression of SIRT1 gene and miR‐141.
4. DISCUSSION
This study found that overweight individuals had elevated levels of miR‐141 compared with those of average body weight. A significant correlation between miR‐141 and BMI and WC was also found. Although miR‐141 has not been previously investigated in patients with obesity, there is evidence showing that many other microRNAs are disrupted in obesity. These alterations have profound effects on the processes related to obesity, such as adipogenesis, lipid metabolism, and inflammation. 25 For example, miR‐141‐3p has been found to be significantly up‐regulated in mice fed a high‐fat diet, and it has been shown to modulate ATP production and induce oxidative stress. 26 In addition, miR‐141 has been found to be increased in pre‐eclampsia and is involved in inflammation associated with this disorder. 27
MicroRNAs have also been introduced to be responsible for various disorders associated with obesity, including endothelial dysfunction, cardiovascular diseases, insulin resistance, diabetes, MetS, and NAFLD. 25 In our study, we evaluated MetS and NAFLD in the participants and found that miR‐141 was significantly increased in both conditions. Furthermore, the level of miR‐141 showed a consistent increase with the severity of hepatic steatosis. We also observed significant correlations between miR‐141 and MetS components, such as insulin resistance indices and blood pressure. Consistently, Li et al. identified 40 upregulated and 10 downregulated miRNAs in patients with MetS. 28 In line with our results, miR‐141 has been associated with insulin resistance, a key component of MetS. Elevated levels of miR‐141 have been observed in diabetic mice and elderly individuals with diabetes, and treatment with pioglitazone, a widely used therapy for insulin resistance, has been shown to correct miR‐141 expression. The increased expression of miR‐141 has been linked to impaired insulin secretion and reduced proliferation of pancreatic β cells. Conversely, suppressing miR‐141 in β cells has been found to decrease the damage associated with type 2 diabetes. 29
In the case of fatty liver, the expression of certain microRNAs is reduced (e.g., miR‐22), while others are increased (e.g., miR‐34a and miR‐146b). These changes in expression levels ultimately affect the expression of genes involved in liver fat metabolism, contributing to the development of fatty liver. 30 In hepatocytes, miR‐141 has been extensively studied and its inhibition has been shown to suppress HSC activation via the AKT/mTOR pathway by targeting PTEN. This identified miR‐141 as a potential therapeutic target for the treatment of liver fibrosis. 31 In our previous research, we had demonstrated that inhibiting miR‐141 could reduce lipid accumulation in hepatocytes, while its upregulation resulted in increased intracellular lipid accumulation. 23 Tran et al. also showed that suppressing miR‐141 levels led to reduced hepatic steatosis and inflammation by reprogramming lipid and inflammation signaling pathways. 32
SIRT1 is the most extensively studied sirtuin and is implicated in both alcoholic and non‐alcoholic fatty liver diseases. Increasing SIRT1 activity can decrease fat accumulation in adipocytes, enhance mitochondrial energy production, and promote fatty acid oxidation. 33 Additionally, SIRT1 plays a beneficial role in regulating hepatic lipid metabolism, managing hepatic oxidative stress, and modulating hepatic inflammation. In patients with obesity presenting with severe fatty liver, SIRT1 levels are reduced in visceral fats, which can have a negative impact on mitochondrial function. 34 It has been demonstrated that SIRT1 is downregulated in children and adolescents with obesity, 22 although the exact mechanism of its suppression is not fully understood. Although we had limitations in extending the sample size, low levels of SIRT1 were observed in participants with obesity, which correlated well with the levels of miR‐141. Therefore, the decreased levels of SIRT1 in obesity may be attributed to the increased levels of miR‐141. This notion is supported by a previous in vitro study showing that miR‐141 repressed SIRT1 activity and expression, as well as a bioinformatics exploration indicating alignment of miR‐141 with the 3′‐UTR of SIRT1 mRNA. 23
5. CONCLUSION
The results of the current study demonstrated elevated levels of miR‐141 in individuals with obesity, which were associated with MetS and hepatic steatosis. There was a correlation between miR‐141 and SIRT1 expression, as well as metabolic parameters. Given the significant role of SIRT1 in obesity and its related conditions, such as NAFLD, miR‐141 could be regarded as a potential target for addressing disorders associated with obesity; however, further experiments are required to establish the translational value of miR‐141 in the context of obesity.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This study was financially supported by Iran University of Medical Sciences, grant number 94‐05‐30‐27493.
This research was presented at the 57th Annual meeting of the European Society for Pediatric Endocrinology, ESPE Abstracts (2018) 89 FC6.6, https://abstracts.eurospe.org/hrp/0089/hrp0089fc6.6.
Dadkhah Nikroo N, Jafarinejad H, Yousefi Z, et al. Elevated mir‐141 in obesity: Insights into the interplay with sirtuin 1 and non‐alcoholic fatty liver disease. Obes Sci Pract. 2024;e70007. 10.1002/osp4.70007
REFERENCES
- 1. Buzzetti E, Pinzani M, Tsochatzis EA. The multiple‐hit pathogenesis of non‐alcoholic fatty liver disease (NAFLD). Metabolism. 2016;65(8):1038‐1048. 10.1016/j.metabol.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 2. Kanuri G, Bergheim I. In vitro and in vivo models of non‐alcoholic fatty liver disease (NAFLD). Int J Mol Sci. 2013;14(6):11963‐11980. 10.3390/ijms140611963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohajan D, Mohajan HK. Obesity and its related diseases: a new escalating alarming in global health. Journal of Innovations in Medical Research. 2023;2(3):12‐23. 10.56397/jimr/2023.03.04 [DOI] [Google Scholar]
- 4. Kanwal F, Kramer JR, Mapakshi S, et al. Risk of hepatocellular cancer in patients with non‐alcoholic fatty liver disease. Gastroenterology. 2018;155(6):1828‐1837.e2. 10.1053/j.gastro.2018.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ziolkowska S, Binienda A, Jabłkowski M, Szemraj J, Czarny P. The interplay between insulin resistance, inflammation, oxidative stress, base excision repair and metabolic syndrome in nonalcoholic fatty liver disease. Int J Mol Sci. 2021;22(20):11128. 10.3390/ijms222011128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerhard GS, DiStefano JK. Micro RNAs in the development of non‐alcoholic fatty liver disease. World J Hepatol. 2015;7(2):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11(5):276‐288. 10.1038/nrendo.2015.25 [DOI] [PubMed] [Google Scholar]
- 8. Vienberg S, Geiger J, Madsen S, Dalgaard LT. Micro RNA s in metabolism. Acta Physiol. 2017;219(2):346‐361. 10.1111/apha.12681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23(1):175‐205. 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- 10. Wang J, Du Y, Deng J, Wang X, Long F, He J. MicroRNA‐506 is involved in regulation of the occurrence of lipopolysaccharides (LPS)‐induced pulpitis by sirtuin 1 (SIRT1). Med Sci Mon Int Med J Exp Clin Res: International Medical Journal of Experimental and Clinical Research. 2019;25:10008‐10015. 10.12659/msm.918172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Macvanin M, Obradovic M, Zafirovic S, Stanimirovic J, Isenovic ER. The role of miRNAs in metabolic diseases. Curr Med Chem. 2023;30(17):1922‐1944. 10.2174/0929867329666220801161536 [DOI] [PubMed] [Google Scholar]
- 12. Dexheimer PJ, Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020;8:409. 10.3389/fcell.2020.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gramantieri L, Fornari F, Callegari E, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12(6a):2189‐2204. 10.1111/j.1582-4934.2008.00533.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vrba L, Jensen TJ, Garbe JC, et al. Role for DNA methylation in the regulation of miR‐200c and miR‐141 expression in normal and cancer cells. PLoS One. 2010;5(1):e8697. 10.1371/journal.pone.0008697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yaman Agaoglu F, Kovancilar M, Dizdar Y, et al. Investigation of miR‐21, miR‐141, and miR‐221 in blood circulation of patients with prostate cancer. Tumor Biol. 2011;32(3):583‐588. 10.1007/s13277-011-0154-9 [DOI] [PubMed] [Google Scholar]
- 16. Lynch SM, O'Neill KM, McKenna MM, Walsh CP, McKenna DJ. Regulation of miR‐200c and miR‐141 by methylation in prostate cancer. Prostate. 2016;76(13):1146‐1159. 10.1002/pros.23201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Capri M, Olivieri F, Lanzarini C, et al. Identification of miR‐31‐5p, miR‐141‐3p, miR‐200c‐3p, and GLT 1 as human liver aging markers sensitive to donor–recipient age‐mismatch in transplants. Aging Cell. 2017;16(2):262‐272. 10.1111/acel.12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43(3):198‐211. 10.3109/07853890.2010.547211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deng XQ, Chen LL, Li NX. The expression of SIRT1 in nonalcoholic fatty liver disease induced by high‐fat diet in rats. Liver Int. 2007;27(5):708‐715. 10.1111/j.1478-3231.2007.01497.x [DOI] [PubMed] [Google Scholar]
- 20. Tian C, Huang R, Xiang M. SIRT1: harnessing multiple pathways to hinder NAFLD. Pharmacol Res. 2024;203:107155. 10.1016/j.phrs.2024.107155 [DOI] [PubMed] [Google Scholar]
- 21. Colak Y, Ozturk O, Senates E, et al. SIRT1 as a potential therapeutic target for treatment of nonalcoholic fatty liver disease. Med Sci Mon Int Med J Exp Clin Res: international medical journal of experimental and clinical research. 2011;17(5):HY5‐HY9. 10.12659/msm.881749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arab Sadeghabadi Z, Nourbakhsh M, Pasalar P, et al. Reduced gene expression of sirtuins and active AMPK levels in children and adolescents with obesity and insulin resistance. Obes Res Clin Pract. 2018;12(2):167‐173. 10.1016/j.orcp.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 23. Yousefi Z, Nourbakhsh M, Abdolvahabi Z, et al. microRNA‐141 is associated with hepatic steatosis by downregulating the sirtuin1/AMP‐activated protein kinase pathway in hepatocytes. J Cell Physiol. 2020;235(2):880‐890. 10.1002/jcp.29002 [DOI] [PubMed] [Google Scholar]
- 24. Alberti KG, Zimmet P, Shaw J. Metabolic syndrome‐‐a new world‐wide definition. A consensus statement from the international diabetes federation. Diabet Med. 2006;23(5):469‐480. 10.1111/j.1464-5491.2006.01858.x [DOI] [PubMed] [Google Scholar]
- 25. Nourbakhsh M. MicroRNAs as targets for the management of obesity. In: Rahman AU, Choudhary MI, eds. Anti Obesity Drug Discovery and Development. Bentham Science Publishers; 2018. [Google Scholar]
- 26. Ji J, Qin Y, Ren J, et al. Mitochondria‐related miR‐141‐3p contributes to mitochondrial dysfunction in HFD‐induced obesity by inhibiting PTEN. Sci Rep. 2015;5(1):16262. 10.1038/srep16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu D, Shi L, Chen F, Lin Q, Kong J. Methylation status of the miR‐141‐3p promoter regulates miR‐141‐3p expression, inflammasome formation, and the invasiveness of HTR‐8/SVneo cells. Cytogenet Genome Res. 2022;161(10‐11):501‐513. 10.1159/000519740 [DOI] [PubMed] [Google Scholar]
- 28. Li Y, Meng Y, Zhu X, Van Wijnen A, Eirin A, Lerman LO. Metabolic syndrome is associated with altered mRNA and miRNA content in human circulating extracellular vesicles. Front Endocrinol. 2021;12. 10.3389/fendo.2021.687586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu X, Zhong L. Pioglitazone/microRNA‐141/FOXA2: a novel axis in pancreatic β‐cells proliferation and insulin secretion. Mol Med Rep. 2018;17(6):7931‐7938. 10.3892/mmr.2018.8813 [DOI] [PubMed] [Google Scholar]
- 30. Szelenberger R, Kacprzak M, Saluk‐Bijak J, Zielinska M, Bijak M. Plasma MicroRNA as a novel diagnostic. Clin Chim Acta. 2019;499:98‐107. 10.1016/j.cca.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 31. Liang H, Wang X, Si C, et al. Downregulation of miR‐141 deactivates hepatic stellate cells by targeting the PTEN/AKT/mTOR pathway. Int J Mol Med. 2020;46(1):406‐414. 10.3892/ijmm.2020.4578 [DOI] [PubMed] [Google Scholar]
- 32. Tran M, Lee SM, Shin DJ, Wang L. Loss of miR‐141/200c ameliorates hepatic steatosis and inflammation by reprogramming multiple signaling pathways in NASH. JCI Insight. 2017;2(21). 10.1172/jci.insight.96094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Colak Y, Yesil A, Mutlu HH, et al. A potential treatment of non‐alcoholic fatty liver disease with SIRT1 activators. J Gastrointestin Liver Dis. 2014;23(3):311‐319. 10.15403/jgld.2014.1121.233.yck [DOI] [PubMed] [Google Scholar]
- 34. Alshehri AS, El‐kott AF, El‐Kenawy AE, Khalifa HS, AlRamlawy AM. Cadmium chloride induces non‐alcoholic fatty liver disease in rats by stimulating miR‐34a/SIRT1/FXR/p53 axis. Sci Total Environ. 2021;784:147182. 10.1016/j.scitotenv.2021.147182 [DOI] [PubMed] [Google Scholar]


