Abstract
JC virus (JCV), the causative agent of progressive multifocal leukoencephalopathy (PML), has a hypervariable regulatory region (JCV RR). A conserved archetype form is found in the urines of healthy and immunocompromised individuals, whereas forms with tandem repeats and deletions are found in the brains of PML patients. Type I JCV RR, seen in MAD-1, the first sequenced strain of JCV, contains two 98-bp tandem repeats each containing a TATA box. Type II JCV RR has additional 23-bp and 66-bp inserts or fragments thereof and only one TATA box. We cloned and sequenced JCV RR from different anatomic compartments of PML patients and controls and correlated our findings with the patients' clinical outcome. Twenty-three different sequences were defined in 198 clones obtained from 16 patients. All 104 clones with tandem repeats were type II JCV RR. Patients with poor clinical outcome had high proportions of JCV RR clones with both tandem repeats in plasma (54%) and brain or cerebrospinal fluid (85%). In those who became survivors of PML, archetype sequences predominated in these anatomic compartments (75 and 100%, respectively). In patients with advanced human immunodeficiency virus infection without PML, only 8% of JCV RR clones obtained in the plasma contained tandem repeats. These data suggest that the presence of tandem repeats in plasma and CNS JCV RR clones is associated with poor clinical outcome in patients with PML.
The human polyomavirus JC (JCV) is the etiologic agent of progressive multifocal leukoencephalopathy (PML). Eighty to ninety percent of the adult population is seropositive for JCV (29). After an asymptomatic primary infection during childhood, the virus remains quiescent in the kidneys and lymphoid tissue (1, 11, 21). In the setting of immunosuppression, JCV can reactivate and cause a lytic infection of oligodendrocytes. The JCV regulatory region (JCV RR) of urine isolates is highly conserved in healthy and immunocompromised individuals and has been called the archetype (1, 20, 32). The archetype has no duplications and contains a 23-bp insert and a 66-bp insert localized at nucleotides (nt) 36 and 92 compared to the first sequenced brain isolate of JCV, MAD-1. A hypervariable form of JCV RR is found in the brain and cerebrospinal fluid (CSF) of PML patients. In the MAD-1 isolate, the RR contains two identical 98-bp tandem repeats.
It has been hypothesized that rearrangements of JCV RR occur in the setting of immunosupression, leading to JC viremia, hematogenous spread of the virus to the central nervous system (CNS), and the development of PML. Indeed, JCV DNA is rarely found in the blood of healthy individuals, but it becomes more readily detectable in the blood of human immunodeficiency virus (HIV)-infected people who have <200 CD4+ T cells/μl of blood (16). Analyses of RR from JCV isolates obtained from blood samples have been limited and, for the most part, restricted to direct sequencing of PCR products (2, 5, 26, 27). In addition, the relationship between the types of JCV RR in the blood and disease progression has not been investigated.
The clinical course of PML is variable. Some patients have a fulminant evolution and die within 1 to 6 months of their diagnosis, whereas others have a protracted course and become PML survivors. We sought to determine the variability of JCV RR in the blood and CNS and if patterns of JCV RR found in the CNS arise in the blood. Moreover, we sought to correlate our findings with the patients' clinical outcome.
Urine, plasma, CSF, or brain samples were obtained from 16 patients. Of these patients, eight who were HIV positive (HIV+) and two who were HIV negative (HIV−) died from PML. One HIV+ patient and one HIV− patient were PML survivors and were still alive more than 2 years after the diagnosis of their neurologic disease. Four patients were HIV+ with other neurological diseases (HIV+/OND) including HIV encephalopathy, cytomegalovirus polyradiculopathy, and other non-PML leukoencephalopathies. These patients were also long-term survivors of their neurologic diseases.
DNA extraction from CSF, plasma, urine, and brain was performed as previously described (16). PCR amplification was performed using the external primers JCRS (5′-ATTAGTGCAAAAAAGGGAAAAACAAGGG 3′) (nt 5035 to 5062) and JCRAS (5′-CTCGGATCCAGCTGGTGACAAGCCAAAACAG-3′) (nt 272 to 242), which amplify a 368-bp fragment of JCV RR, followed by a semi-nested PCR using the internal primers JCRSN (5′-CTACTTCTGAGTAAGCTTGGAGG-3′) (nt 5100 to 5122) and JCRAS, which amplify a 303-bp fragment of the regulatory region. The first PCR was performed in a PE 9600 thermal cycler using 1 μg of brain or DNA extracted from 200 μl of CSF or 6 ml of plasma, 25 pmol of each primer, 1.5 mM MgCl, and 2.5 U of Ampli Taq Gold DNA polymerase in a final volume of 50 μl. The cycles of amplification consisted of 94°C for 10 min followed by 40 cycles of 94°C for 30 s, 58°C for 1 min, 72°C for 1 min, and 7 min of elongation at 72°C. For the semi-nested PCR, we used 10 μl of the PCR product of the first reaction and the following cycles: 94°C for 10 min followed by 30 cycles of 94°C for 30 s, 60°C for 1 min, 72°C for 1 min, and an elongation of 7 min at 72°C.
The PCR products were analyzed by UV transillumination after electrophoresis in a 2% agarose gel. Using this seminested PCR technique, we were able to detect 10 copies of MAD-1 plasmid used as a positive control. Bands of expected size were cut and gel purified with a gel extraction kit (Qiagen, Valencia, Calif.) and cloned with a PCR-Script Amp cloning kit (Stratagene, La Jolla, Calif.). Up to 12 clones of each PCR product were sequenced. The cycle-sequencing reaction was performed according to the manufacturer's instructions using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Kit (Applied Biosystems, Weiterstadt, Germany) with 400 ng of double-stranded DNA and 3.2 pmol of sequencing primer. Fluorescence-based DNA sequence analyses were obtained on an ABI 377 DNA Sequencer (Applied Biosystems). Sequence analysis was performed with Lasergene Software for Macintosh and Power PC, Megalign 3.12 (DNASTAR Inc., Madison, Wis.) comparing the sequences obtained to those of the prototype MAD-1 (nucleotide numbering system according to Frisque et al. [9]).
Since previous reports had shown archetype JCV RR to be present in the urine samples of most individuals regardless of their degree of immunosuppression (15, 21), we concentrated our efforts on DNA samples obtained from blood, CSF, and brain specimens. JCV exists in the blood mainly in a latent, nonreplicative stage (6). The JCV DNA load in the peripheral blood mononuclear cells (PBMC) is very low, between 10 and 90 copies of JCV/μg of PBMC DNA (16), and becomes detectable only after PCR amplification and the application of hybridization techniques. JCV has also been detected in cell-free plasma (7, 16, 17). A recent report has demonstrated that JCV sticks to the surfaces of blood cells rather than productively infecting them (30). In our experience, a stronger PCR signal is generated by extracting viral DNA from plasma. This also facilitates cloning of the amplified products. We therefore analyzed JCV RR from plasma samples rather than from PBMC.
A total of 23 different sequences were found in 198 clones derived from these 16 patients. Each patient had unique JCV RR sequences except for those who had an archetype JCV RR. In a few cases the differences between JCV RR sequences obtained from different patients were only single mutations, but these were consistent in all clones found in a particular anatomic compartment. These findings rule out the possibility of sample contamination.
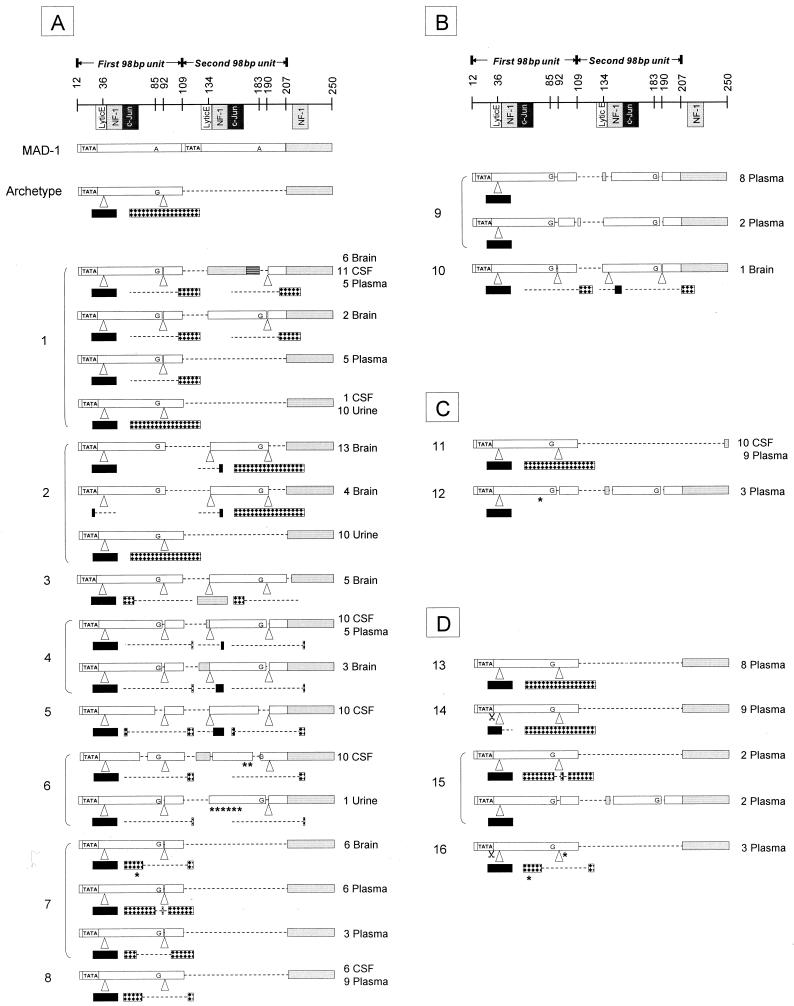
We first characterized JCV RR in various anatomic compartments in PML progressors, PML survivors, and HIV+/OND patients. Analysis of CSF and/or brain samples from the eight HIV+ patients who died of PML (Fig. 1A) showed that six of them had a JCV RR with two 98-bp tandem repeats (patients no. 1 through 6). All contained 23- and/or 66-bp inserts (at positions nt 36 and 134 and positions nt 92 and 190, respectively) which had variable deletions. In contrast, two patients had only archetype-like JCV RR with one single 98-bp unit (patients no. 7 and 8). Surprisingly, one patient (no. 1) also had an archetype form of JCV RR in 1 of 12 CSF clones sequenced.
FIG. 1.
Sequencing results of the JCV RR. On the top is a representation of the MAD-1 and archetype JCV RR. The nucleotide numbers are based on the prototype MAD-1 sequence. The known transcription regulation factor binding sites are indicated and include the lytic control element (Lytic E), nuclear factor 1 (NF-1), and c-Jun. Each 98-bp unit is represented by an open box. The TATA box is represented by TATA. The archetype contains only one 98-bp unit with two inserts. Black box, 23-bp insert; checkered box, 66 bp-insert; dotted lines, deletions in the 98-bp units or in the 23- or 66-bp inserts; grey box, region downstream of the 98-bp units. Some clones contain fragments of this region inserted in the second 98-bp unit. MAD-1 contains an adenine at positions 85 and 183, compared with all other sequences that contain guanine at these positions. Asterisks, single mutations; scissors, single nucleotide deletions in the TATA box. (A) Sequencing results of HIV+ PML progressor patients 1 to 8. At the right of each sequence is the number of clones obtained per anatomic compartment. The first sequence of patient no. 1 has a large deletion in the second 98-bp unit that is replaced by a fragment identical to nucleotides 208 to 247 (striped box) containing an NF-1 predicted binding site and next to it a c-Jun binding site. Patient 3, 4, and 6 sequences have smaller fragments of the region nt 208 to 250 inserted in the second 98-bp unit; only the patient 3 sequence contains the full length of the NF-1 binding site. (B) HIV-negative PML progressors. (C) PML survivors. No. 11 is HIV+ and no. 12 is HIV−. (D) HIV+ patients with low CD4+ T-cell counts and other, non-PML neurologic disorders.
In four of these HIV+ PML progressors, the JCV RR could be amplified and cloned from plasma samples. Clones were obtained from one of these individuals (no. 1) that had tandem repeats identical to his most prominent CSF JCV RR, as well as a distinct archetype-like JCV RR. Patient no. 4 had clones with only a tandem repeat JCV RR, identical to the one found in his CSF. Patients no. 7 and 8 had archetype-like JCV RR. Patient no. 7 had two distinct plasma JCV RR that were different from the one found in his brain, whereas the single pattern found in the plasma of patient no. 8 was identical to the CSF sequence. Urine specimens were analyzed for three of these patients. Two had archetype (no. 1 and 2) and one had tandem repeat (no. 6) JCV RR.
For the two HIV− PML progressor patients (Fig. 1B), plasma could be analyzed from one and brain from the other. We obtained two distinct sequences from the plasma of patient no. 9 and one type of sequence from the brain of patient no. 10. In all of them, tandem repeats were seen in the JCV RR.
CSF and plasma were analyzed from one HIV+ PML survivor (no. 11, Fig. 1C). In both samples, we found an archetype JCV RR with a deletion of 40 bp (nt 208 to 247) downstream of the 98-bp unit. This deleted region encodes a nuclear factor 1 (NF-1) binding site. The analysis of plasma from one HIV− PML survivor (no. 12) showed JCV RR with tandem repeats (Fig. 1C).
Plasma samples were analyzed from four HIV+ individuals with neurologic disorders other than PML (Fig. 1D). One of them, no. 13, had an archetype JCV RR. Two others, no. 14 and no. 16, had an archetype-like JCV RR, and no. 15 had two different sequences, an archetype-like JCV RR and a JCV RR with tandem repeats. These patients had undetectable JCV DNA in their CSF. Interestingly, the sequence found in the plasma of patient no. 14, which has a 10-bp deletion at the 3′ end of the 23-bp insert, is identical to that of a strain (PNG-1A) recently identified in the urine sample from a healthy individual from Papua New Guinea. Similar 10-bp deletions were also found in the same region in a few Central and East African strains (24). Patient no. 14 is originally from Haiti.
We then correlated the patterns of JCV RR in different anatomic compartments with the patients' clinical outcomes. Patients with poor clinical outcomes had a high proportion of tandem repeats in the JCV RR in the plasma (54%) and CNS (85%) (Table 1). In contrast, PML survivors had a high proportion of archetype JCV RR in the plasma (75%) and CNS (100%). Finally, HIV+ individuals with low CD4+ T-cell counts without PML had a low proportion of tandem repeats in JCV RR in the plasma (8%).
TABLE 1.
Types of JCV RRa found in plasma and CNS (brain and CSF) of groups with different clinical outcomes
| Clinical outcome and/or patient characteristic | Plasma
|
CNS
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients | No. of clones | No. (%) of indicated RR typea
|
No. of patients | No. of clones | No. (%) of indicated RR typea
|
|||||
| A | AL | TR | A | AL | TR | |||||
| PML progressor, HIV+/HIV− | 5 | 43 | 0 | 20 (46) | 23 (54) | 9 | 88 | 1 (1) | 12 (14) | 75 (85) |
| PML survivor, HIV+/HIV− | 2 | 12 | 9 (75) | 0 | 3 (25) | 1 | 10 | 10 (100) | 0 | 0 |
| HIV+/OND | 4 | 24 | 8 (33) | 14 (58) | 2 (8) | |||||
| Total | 11 | 79 | 10 | 98 | ||||||
A, archetype; AL, archetype like; TR, tandem repeats.
These sequence data are in agreement with previous reports (2, 23, 31) demonstrating that JCV RR is hypervariable in the body. We isolated two to four distinct JCV RR in 6 of 12 PML patients and in 1 of 4 patients without PML. These six PML patients were progressors. The presence of dual infection with different JCV strains was also detected in 6 of 21 (28.6%) PML patients in a recent study of JCV genotype, and this was found to be an additional risk factor for the development of PML (8). The precise anatomic compartment in which tandem repeats first appear has not been unequivocally determined, since they can be found both in blood and in CNS samples. It is also unclear if these tandem repeats are the cause or the consequence of JCV neurotropism. Patient no. 1 was the only one for whom it was possible to amplify, clone, and sequence JCV RR from four different anatomic compartments, (brain, CSF, plasma, and urine). We obtained four different JCV RR for this particular patient and saw an evolutionary gradient from archetype to tandem repeat going from the urine to the blood and then to the CNS. For the four HIV+ PML progressors whose plasma and CSF or brain samples could be analyzed (patients 1, 4, 7, and 8), JCV RR in the CNS samples had deletions of the 23- and 66-bp inserts in numbers either similar to or greater than those of the plasma (Fig. 1A). These data suggest that tandem repeats first originate in the plasma and that further rearrangements may occur once the virus enters the CNS.
Archetype JCV RR is usually not found in the CSF of PML patients. However, 1 of the 12 clones obtained from the CSF of HIV+ PML progressor patient no. 1 showed an archetype JCV RR while 10 of 10 clones from HIV+ PML survivor patient no. 11 had this sequence. In addition, HIV+ PML progressors no. 7 and no. 8 had archetype-like JCV RR in the brain or CSF. To our knowledge, this is the first report of fatal PML cases in which no tandem repeats were found in the CNS. Finally, one HIV+ PML progressor had a tandem repeat JCV RR isolated from the urine. Therefore, the dogma suggesting that archetype JCV RR is present only in the urine and that tandem repeats are restricted to the CNS is not absolute.
We also aimed to determine if additions or deletions of known protein binding sites in JCV RR correlated with clinical outcomes. Complex rearrangements of the JCV RR with additional inserts containing new predicted NF-1 or c-Jun binding sites were found only in patients with fatal outcomes (no. 1, 3, and 6). In contrast, deletions of predicted NF-1 or c-Jun binding sites were found only in patients with good outcomes (no. 11 and 14). NF-1 is a family of proteins, and the one that specifically binds to the JCV promoter is the NF-1 class D. This protein is predominantly expressed in glial cells (25). The productive infection and restricted lysis of oligodendrocytes by JCV reflect the fact that the JC virus early gene promoter is more active in glial cells than in cells of nonglial lineage (14). This observation is similar to that of studies done with murine polyomavirus, where mutations in the regulatory region were shown to confer a tissue-specific cis-advantage in viral replication (3, 4).
Most of the studies of JCV promoter activity have used the prototype MAD-1 regulatory region, which was the first JCV that was sequenced and has no 23-bp or 66-bp inserts. This JCV RR pattern was called type I. Subsequent studies have shown that the vast majority of JCV strains isolated in vivo contain these inserts or fragments thereof, as well as a deletion of the second TATA box. Such JCV RR were called type II (12, 22). The 23-bp insert contains a binding site for the transcription factor SP-1 (13). The promoters of the oligodendrocyte-specific cellular genes, myelin basic protein and proteolipid protein, contain similar binding sites. Together with the paired NF-1 and c-Jun binding sites, the SP-1 binding site in the 23-bp insert is part of a motif that is conserved between several glial-specific promoters (13). These findings are in agreement with in vitro studies demonstrating that the number of NF-1 binding sites is directly proportional to the level of viral transcription in glial cell lines (18, 19).
The function of the 66-bp insert has not yet been elucidated. No predicted binding sites of transcription regulators have been found in this region. Such sites have also not been shown in the region where it is inserted. Interestingly, deletions of ≥85% of the 66-bp insert correlated in our study with a fatal outcome. In contrast, the complete deletion of the 66-bp insert did not correlate with the clinical outcome.
The present study shows that PML progressors have a higher frequency of archetype-like and tandem repeat JCV RR in the plasma and CNS than do PML survivors. In these latter patients, archetype JCV RR predominates. In the present series of patients, the number of PML progressors studied was larger than the number of PML survivors. This is due to the fact that PML survivors are quite rare and account only for approximately 10% of all patients with this disease. They also usually have a lower JCV viral load in the CSF and undetectable JCV DNA in the blood. CSF JCV DNA often becomes undetectable following initiation of antiretroviral treatment. These changes parallel their higher CD4+ T-lymphocyte counts. Therefore, analysis of JCV RR from these patients' samples is often impossible. Attempts to amplify JCV RR from the plasma of two additional PML survivors were unsuccessful. However, analyses of plasma samples from four HIV+ patients without PML also showed only 8% tandem repeats.
Since MAD-1 was the first JCV strain to be isolated, the type I MAD-1 JCV RR has been employed in the study of JCV expression and replication in vitro. In addition, type II strains containing a 23-bp insert grow poorly in primary human glial cells (10). However, the present study indicated that in 103 clones from plasma and CNS with a tandem repeat pattern only type II JCV RR were detected. These results are in agreement with a recent study that showed type II JCV RR in the CSF of 11 of 14 (79%) of probable PML cases, whereas type I JCV RR was found only in three possible PML cases (28). Therefore, more emphasis should be directed to the study of type II JCV RR strains and the factors restricting their replication in available tissue culture systems.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences determined in this study are as follows, in order of appearance in Fig. 1: patient no. 1, AF354562, AF354565, AF354566, and AF354567; patient no. 2, AF354569, AF354570, and AF354571; patient no. 3, AF354572; patient no. 4, AF354573, and AF354574; patient no. 5, AF354575; patient no. 6, AF 354576 and AF354577; patient no. 7, AF354578, AF354579, and AF354580; patient no. 8, AF354581; patient no. 9, AF354582 and AF354583; patient no. 10, AF354584; patient no. 11, AF354585; patient no. 12, AF354587; patient no. 13, AF354588; patient no. 14, AF354589; patient no. 15, AF354590 and AF354591; patient no. 16, AF354592.
Acknowledgments
This study was supported by a grant from the Swiss National Science Foundation to L.-A.P. and by NIH grants NS01919 (I.J.K.) and AI-20729 (N.L.L.).
REFERENCES
- 1.Agostini H T, Ryschkewitsch C F, Stoner G L. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J Clin Microbiol. 1996;34:159–164. doi: 10.1128/jcm.34.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciappi S, Azzi A, De Santis R, Leoncini F, Sterrantino G, Mazzotta F, Mecocci L. Archetypal and rearranged sequences of human polyomavirus JC transcription control region in peripheral blood leukocytes and in cerebrospinal fluid. J Gen Virol. 1999;80:1017–1023. doi: 10.1099/0022-1317-80-4-1017. [DOI] [PubMed] [Google Scholar]
- 3.De Simone V, Amati P. Replicative cis-advantage of polyomavirus regulatory region mutants in different murine cell lines. J Virol. 1987;61:1615–1620. doi: 10.1128/jvi.61.5.1615-1620.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Simone V, La Mantia G, Lania L, Amati P. Polyomavirus mutation that confers a cell-specific cis advantage for viral DNA replication. Mol Cell Biol. 1985;5:2142–2146. doi: 10.1128/mcb.5.8.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorries K, Vogel E, Gunther S, Czub S. Infection of human polyomaviruses JC and BK in peripheral blood leukocytes from immunocompetent individuals. Virology. 1994;198:59–70. doi: 10.1006/viro.1994.1008. [DOI] [PubMed] [Google Scholar]
- 6.Dubois V, Dutronc H, Lafon M E, Poinsot V, Pellegrin J L, Ragnaud J M, Ferrer A M, Fleury H J. Latency and reactivation of JC virus in peripheral blood of human immunodeficiency virus type 1-infected patients. J Clin Microbiol. 1997;35:2288–2292. doi: 10.1128/jcm.35.9.2288-2292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubois V, Moret H, Lafon M E, Janvresse C B, Dussaix E, Icart J, Karaterki A, Ruffault A, Taoufik Y, Vignoli C, Ingrand D. Prevalence of JC virus viraemia in HIV-infected patients with or without neurological disorders: a prospective study. J Neurovirol. 1998;4:539–544. doi: 10.3109/13550289809113498. [DOI] [PubMed] [Google Scholar]
- 8.Ferrante, P., M. Mediati, R. Caldarelli-Stefano, L. Losciale, R. Mancuso, A. E. Cagni, and R. Maserati. Increased frequency of JC virus type 2 and of dual infection with JC virus type 1 and 2 in Italian progressive multifocal leukoencephalopathy patients. J. Neurovirol., in press. [DOI] [PubMed]
- 9.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frisque R J, White F A., III . The molecular biology of JC virus, causative agent of progressive multifocal leukoencephalopathy. In: Roos R P, editor. Molecular neurovirology. Totowa, N.J: Humana Press Inc.; 1992. pp. 69–72. [Google Scholar]
- 11.Gallia G L, Houff S A, Major E O, Khalili K. Review: JC virus infection of lymphocytes—revisited. J Infect Dis. 1997;176:1603–1609. doi: 10.1086/514161. [DOI] [PubMed] [Google Scholar]
- 12.Henson J, Saffer J, Furneaux H. The transcription factor Sp1 binds to the JC virus promoter and is selectively expressed in glial cells in human brain. Ann Neurol. 1992;32:72–77. doi: 10.1002/ana.410320112. [DOI] [PubMed] [Google Scholar]
- 13.Henson J W. Regulation of the glial-specific JC virus early promoter by the transcription factor Sp1. J Biol Chem. 1994;269:1046–1050. [PubMed] [Google Scholar]
- 14.Kenney S, Natarajan V, Strike D, Khoury G, Salzman N P. JC virus enhancer-promoter active in human brain cells. Science. 1984;226:1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura T, Satoh K, Tominaga T, Taguchi F, Tajima A, Suzuki K, Aso Y, Yogo Y. Alteration in the JC polyomavirus genome is enhanced in immunosuppressed renal transplant patients. Virology. 1994;198:341–345. doi: 10.1006/viro.1994.1038. [DOI] [PubMed] [Google Scholar]
- 16.Koralnik I J, Boden D, Mai V X, Lord C I, Letvin N L. JC virus DNA load in patients with and without progressive multifocal leukoencephalopathy. Neurology. 1999;52:253–260. doi: 10.1212/wnl.52.2.253. [DOI] [PubMed] [Google Scholar]
- 17.Koralnik I J, Schmitz J E, Lifton M A, Forman M A, Letvin N L. Detection of JC virus DNA in peripheral blood cell subpopulations of HIV-1-infected individuals. J Neurovirol. 1999;5:430–435. doi: 10.3109/13550289909029484. [DOI] [PubMed] [Google Scholar]
- 18.Kumar K U, Pater A, Pater M M. Human JC virus perfect palindromic nuclear factor 1-binding sequences important for glial cell-specific expression in differentiating embryonal carcinoma cells. J Virol. 1993;67:572–576. doi: 10.1128/jvi.67.1.572-576.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar K U, Tang S C, Pater M M, Pater A. Glial and muscle embryonal carcinoma cell-specific independent regulation of expression of human JC virus early promoter by cyclic AMP response elements and adjacent nuclear factor 1 binding sites. J Med Virol. 1996;49:199–204. doi: 10.1002/(SICI)1096-9071(199607)49:3<199::AID-JMV7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 20.Major E O, Amemiya K, Tornatore C S, Houff S A, Berger J R. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markowitz R B, Thompson H C, Mueller J F, Cohen J A, Dynan W S. Incidence of BK virus and JC virus viruria in human immunodeficiency virus-infected and -uninfected subjects. J Infect Dis. 1993;167:13–20. doi: 10.1093/infdis/167.1.13. [DOI] [PubMed] [Google Scholar]
- 22.Martin J D, King D M, Slauch J M, Frisque R J. Differences in regulatory sequences of naturally occurring JC virus variants. J Virol. 1985;53:306–311. doi: 10.1128/jvi.53.1.306-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman J T, Frisque R J. Identification of JC virus variants in multiple tissues of pediatric and adult PML patients. J Med Virol. 1999;58:79–86. [PubMed] [Google Scholar]
- 24.Ryschkewitsch C F, Friedlander J S, Mgone C S, Jobes D V, Agostini H T, Chima S C, Alpers M P, Koki G, Yanagihara R, Stoner G L. Human polyomavirus JC variants in Papua New Guinea and Guam reflect ancient population settlement and viral evolution. Microbes Infect. 2000;2:987–996. doi: 10.1016/s1286-4579(00)01252-1. [DOI] [PubMed] [Google Scholar]
- 25.Sumner C, Shinohara T, Durham L, Traub R, Major E O, Amemiya K. Expression of multiple classes of the nuclear factor-1 family in the developing human brain: differential expression of two classes of NF-1 genes. J Neurovirol. 1996;2:87–100. doi: 10.3109/13550289609146542. [DOI] [PubMed] [Google Scholar]
- 26.Sundsfjord A, Flaegstad T, Flo R, Spein A R, Pedersen M, Permin H, Julsrud J, Traavik T. BK and JC viruses in human immunodeficiency virus type 1-infected persons: prevalence, excretion, viremia, and viral regulatory regions. J Infect Dis. 1994;169:485–490. doi: 10.1093/infdis/169.3.485. [DOI] [PubMed] [Google Scholar]
- 27.Tornatore C, Berger J R, Houff S A, Curfman B, Meyers K, Winfield D, Major E O. Detection of JC virus DNA in peripheral lymphocytes from patients with and without progressive multifocal leukoencephalopathy. Ann Neurol. 1992;31:454–462. doi: 10.1002/ana.410310426. [DOI] [PubMed] [Google Scholar]
- 28.Vaz B, Pickhardt M, Weber T. Analysis of the transcription control region in progressive multifocal leukoencephalopathy. J Neurovirol. 2000;6:398–409. doi: 10.3109/13550280009018304. [DOI] [PubMed] [Google Scholar]
- 29.Walker D L, Padgett B L. The epidemiology of human polyomaviruses. New York, N.Y: Alan R. Liss; 1983. [PubMed] [Google Scholar]
- 30.Wei G, Liu C K, Atwood W J. JC virus binds to primary human glial cells, tonsillar stromal cells, and B-lymphocytes, but not to T lymphocytes. J Neurovirol. 2000;6:127–136. doi: 10.3109/13550280009013156. [DOI] [PubMed] [Google Scholar]
- 31.Yogo Y, Guo J, lida T, Satoh K, Taguchi F, Takahashi H, Hall W W, Nagashima K. Occurrence of multiple JC virus variants with distinctive regulatory sequences in the brain of a single patient with progressive multifocal leukoencephalopathy. Virus Genes. 1994;8:99–105. doi: 10.1007/BF01703608. [DOI] [PubMed] [Google Scholar]
- 32.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]