Abstract
A novel member of the nectin family, nectin1γ, was molecularly cloned. The cDNA has the same ectodomain as nectin1α and nectin1β, the two known transmembrane isoforms that serve as receptors for herpes simplex virus (HSV) entry into human cell lines (nectin1α and nectin1β, also called PRR1-HveC and HIgR, respectively). The 1.4-kb transcript, which originated by alternative splicing, is expressed in human cell lines, and appears to have a narrow distribution in human tissues. The sequence does not have a hydrophobic anchoring region, and the protein is secreted in the culture medium of cells transfected with the cDNA. Nectin1γ, purified from culture medium, can compete with membrane-bound nectin1β and reduce HSV infectivity. The expression of nectin1γ cDNA in cells resistant to HSV infection and lacking HSV receptors enables HSV to enter the cell, which implies that it is present at the cell surface. Thus, nectin1γ has the potential both to mediate and to reduce HSV entry into cells.
Nectin1 (recently assigned to CD111) is a member of a new family of human receptors that belongs to the immunoglobulin (Ig) superfamily (1, 2, 22). The prototype molecule of the family is the poliovirus receptor (PVR/CD155) (16). The family includes nectin1, also known as PRR1 (for poliovirus receptor related), nectin2/PRR2, and the recently described nectin3/PRR3 (5, 7, 8, 13, 19). All members of the cluster are structurally related, and their ectodomain is made of three Ig domains (one V-type domain and two C-type domains). For each member of the family, at least two transmembrane isoforms that originate by differential splicing are known. The ectodomain of the isoforms is the same, but the C-terminal regions are different (5, 7, 8, 13). Alternative designations for human nectin1α and nectin1β are HveC and HIgR, and nectin2/PRR2α is also called HveB (summarized in Table 1) (for recent reviews, see references 1, 2, and 22). Soluble isoforms for PVR/CD155 have also been described (9). Nectin1 and -2, but not nectin3, molecules are expressed in a broad range of human tissues and cell lines of different lineages (5, 7, 8, 13, 19). They are adhesion molecules localized at cell-to-cell junctions of endothelial and epithelial cells (11, 23). Homophilic adhesion (or trans interaction) of nectin2 is correlated to cis dimerization of the molecule at the cell surface and to tyrosine phosphorylation of the long δ isoform (11). The localization at the adherens junction in epithelial cells is mediated by the interaction of the C-terminal consensus region (A/ExYV) with the PDZ domain of afadin that anchors the nectin/PRR molecules to F-actin (23). The human and murine nectin1 and nectin2 molecules mediate entry into the cell and cell-to-cell spread of herpes simplex virus (HSV) and of animal alphaherpesviruses, porcine pseudorabies virus, and bovine herpesvirus type 1 (4, 5, 8, 12, 17, 21, 24). In particular, nectin1α and -β appear to be major HSV receptors due to their broad distribution in human cell lines and tissues targeted by HSV and to their ability to serve as receptors for all the HSV type 1 and 2 strains tested (5, 8).
TABLE 1.
Nomenclature of human nectin1 isoforms
In this report, we describe a novel isoform of human nectin1, nectin1γ, that lacks a transmembrane region, is secreted in culture medium, and therefore represents a natural soluble isoform of the receptor which originates by alternative splicing.
Molecular cloning of a novel isoform of human nectin1.
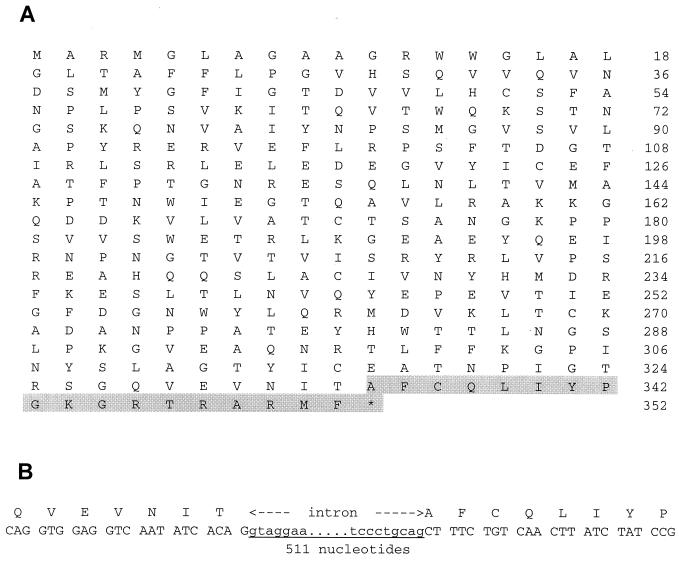
BLAST analysis of the human expressed sequence tag (EST) cDNA library with the sequence of the third C domain of nectin1 cDNA led to the identification of five different sequences that differ from nectin1α and nectin1β at a splicing site located 1,003 nucleotides from the ATG codon (see below). EST accession numbers were R73842, R66178, N59143, AI871188, and AW005044. Clone R73842 was entirely sequenced and comprised an entire open reading frame, which included the entire ectodomain of nectin1 and ended with a poly(A) tail, preceded by the polyadenylation AATAAA motif (Fig. 1A). The deduced sequence is 352 amino acids (aa) long (compared to 517 and 458 aa for α and β transmembrane isoforms, respectively) and does not carry a putative transmembrane hydrophobic region (Fig. 1A), suggesting that it encodes a natural soluble isoform of nectin1. The C-terminal region specific to the novel isoform is 18 aa long (Fig. 1A).
FIG. 1.
(A) Full-length amino acid sequence of the nectin1γ isoform. The sequence is 352 aa long, and the first 334 aa are identical to those of α and β isoforms. The nectin1γ-specific C-terminal sequence is shaded. (B) Mapping of the intron located between the exon of the third C domain and the C-terminal exon of the soluble sequence. The 511-bp intronic sequence was amplified by PCR using the R1S1 (CGCT CAGG CCAG GTGG AGGT C) and the R1S3 (GCCA TTTA TTGA CAGA CTGA TC) primers from human genomic placental DNA.
To characterize the intron located downstream of the splicing site (nucleotide 1003), we amplified a 511-bp DNA fragment from placental genomic DNA by using a 5′ primer located on the third C domain of the molecule and a 3′ primer located in the 3′ region. Boundary sequences are in accordance with intronic type I consensus splice sequence (Fig. 1B). This result shows that the exon encoding the C terminus of the novel isoform is located immediately downstream of the exon that encodes the third C domain. We have named the novel predicted isoform nectin1γ.
Expression of the nectin1γ isoform in human tissues and human cell lines.
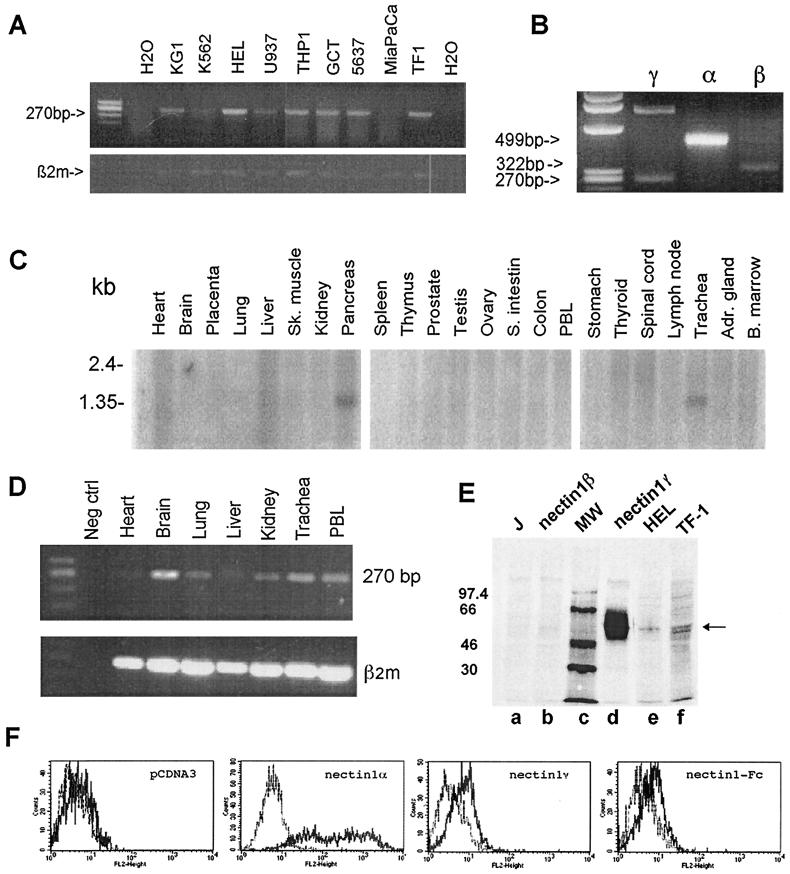
To determine that the EST-derived isoform of nectin1 was actually expressed, its expression was preliminarily assessed in a number of human cell lines and tissues by reverse transcription-PCR (RT-PCR). Total RNA (2 μg), extracted with TRIzol reagent (Gibco-BRL), or purchased from Clontech, was retrotranscribed with SuperScript II reverse transcriptase (Gibco-BRL). The three different isoforms of nectin1 were amplified with a common 5′-end primer and with 3′ primers specific to each isoform (see the legends to Fig. 1 and 2). A 270-bp amplification product corresponding to the new nectin1 cDNA isoform (nucleotides 973 to 1243) was detected in seven of nine human cell lines from different lineages (Fig. 2A). This isoform was coexpressed with nectin1α and nectin1β in the hematopoietic TF-1 cell line (Fig. 2B). The broad distribution among human cell lines is a property in common with the transmembrane isoforms nectin1α and nectin1β (5, 8). Expression in human tissues was first assessed by Northern blot analysis of multiple Northern blot membranes (Clontech). The probe consisted of the nectin1γ cDNA sequence (nucleotides 973 to 1243) and revealed a 1.4-kb band compatible with the 1,417-bp sequence of the nectin1γ cDNA. The nectin1γ isoform was detectable in two tissues, pancreas and trachea, and not in other tissues (Fig. 2C) with two independent series of membranes. With a third series of membranes, only pancreas resulted positive, probably reflecting the relatively low overall abundance. This narrow distribution contrasts with the broad distribution of the α and β transmembrane isoforms (5), confirmed here by means of a 429-bp probe that encompasses the V domain of nectin1. By hybridizing the same membranes with the latter probe, we detected three different transcripts after a long exposure time (data not shown). One of the transcripts found was a 5.9-kb transcript strongly expressed in numerous tissues, including the brain, spinal cord, peripheral blood mononuclear cells, and prostate, in addition to the pancreas and trachea. The second transcript had a similar broad pattern of expression, although at lower levels. This distribution is in accordance with results of previous studies, the only difference being the size of the second transcript, 2.5 kbp rather than 3.5 kbp, as estimated previously (5). The third transcript was the faint 1.4-kbp band. These transcripts likely correspond to the α, β, and γ isoforms of nectin1. When a more sensitive assay (RT-PCR) was employed to assess expression in human tissues, the soluble γ isoform was detectable in all tissues tested, namely, the brain, kidney, peripheral blood mononuclear cells, and trachea (Fig. 2D). The results suggest that expression of the γ isoform (at low levels) may be broader than inferred by Northern blot analysis and may reflect either expression in selected cell types or an overall low-level broad expression.
FIG. 2.
Nectin1γ distribution in human cell lines and tissues and its secretion in culture medium. (A) Distribution of nectin1γ in human cell lines assessed by RT-PCR. RT-PCR amplification experiments were performed on RNAs from the indicated cell lines using the common 5′ primer R1S1, located upstream of the splice site (in the third C domain), and one of the 3′ primers specific for each isoform (R1IC for nectin1α [GCTA CTGG TAGC CCAG AGTC CGG], HIIC for nectin1β [GCAG GGAC AGCT TCTG CAAA GTCC], and R1S3 for nectin1γ. PCR conditions were as follows: (i) 5 min at 95°C, (ii) 30 cycles, with 1 cycle consisting of 1 min at 95°C, 1 min at 60°C, and 1 min at 72°C, and (iii) 30 min at 72°C. The predicted size of the amplification product was 270 bp. Amplification of β2-microglobulin (β2m) cDNA is shown in the lower panel. H2O, water (negative control). (B) Simultaneous detection of the three isoforms of nectin1 (α, β, and γ) in the TF-1 cell line. The amplification products of the three isoforms were obtained by using the above primers. The expected sizes of the three amplification products were 499, 322, and 270 bp, respectively. The upper 782-bp band in lane γ results from contaminating genomic DNA. (C) Distribution of nectin1γ in tissues. The distribution of nectin1γ was assessed by Northern blot analysis of multiple tissue samples applied to membranes (Clontech) hybridized with a 270-bp fragment (nucleotides 973 to 1243) probe specific for nectin1γ, obtained by PCR, sequenced, and labeled with [32P]dCTP. The membranes were also probed with the β-actin probe to verify the hybridization conditions (not shown). Abbreviations: Sk. muscle, skeletal muscle; S. intestin, small intestine; PBL, peripheral blood lymphocytes; Adr. gland, adrenal gland; B. marrow, bone marrow. (D) RT-PCR analysis of nectin1γ expression in human tissues. RNA from the indicated tissues was retrotranscribed and amplified as described above for panel A. The lower panel shows amplification of β2-microglobulin (β2m). Neg ctrl, negative control. (E) Immunoprecipitation of nectin1γ from the culture medium of J1.1-2 (J) cells expressing nectin1γ constitutively. J cells (lane a), J cells constitutively expressing nectin1γ (lane d) or nectin1β (lane b), HEL (lane e), and TF-1 (lane f) cells were labeled overnight with [35S]methionine-cysteine (Trans-label; Radiochemical Center, Amersham). The media were concentrated by using Microcon Y3 centrifugal filters (Millipore). Immunoprecipitation was done with MAb R1.302. After electrophoresis, the gels were analyzed by means of a Bio-Rad phosphorimager. The numbers to the left of the gel are the migration positions of the molecular weight (MW) markers. The arrow points to nectin1γ. (F) FACS analysis of Cos1a cells expressing nectin1γ. Cos1a cells were transfected with the following vectors: pcDNA3, pLX1.12 (nectin1α), pCDSR1.D1 (nectin1γ), and CFLR1VCC (nectin1-Fc). Nectin1 at the cell surface was detected by MAb R1.302 conjugated to phycoerythrin (grey curve) and compared to that of control mouse isotypic IgG1 conjugated to phycoerythrin (black curve).
Nectin1γ is a natural soluble isoform of nectin1.
To identify and characterize the protein encoded by the nectin1γ cDNA, we cloned the entire cDNA (1,245 bp) under the control of the cytomegalovirus promoter at the BamHI and ApaI sites in the pCDNA3 vector, generating pCDSR1.D1. This was transfected into J1.1-2 cells, which do not express human nectin1 and are resistant to HSV infection (5). The derivative cell line stably expressing nectin1γ was labeled with a [35S]methionine-cysteine mixture (The Radiochemical Center, Amersham) overnight. The culture medium was concentrated 10 times, and nectin1 was immunoprecipitated from culture medium with monoclonal antibody (MAb) R1.302, known to react with the ectodomain of human nectin1 (3, 14). Radioimmunoprecipitation was performed in parallel from a cell line expressing the transmembrane isoform human nectin1β and from two cell lines, HEL and TF-1, shown above by RT-PCR to be positive for nectin1γ. The results in Fig. 2E show a prominent band in the culture medium (lane d) of J1.1-2 cells transformed with pCDSR1.D1, corresponding to a protein with an average apparent molecular mass of 56 kDa. A weak band with the same migration position was detectable in the culture medium of TF1 and HEL cells (lanes e and f). The culture medium of untransfected J1.1-2 (lane a) or J1.1-2 cells expressing nectin1β (lane b) was negative. The results indicate that (i) the novel nectin1 is indeed secreted in the culture medium of the cell line harboring nectin1γ cDNA and is weakly expressed in hematopoietic cell lines and (ii) a soluble form resulting from proteolytic cleavage of transmembrane nectin1 is not detected.
In the next series of experiments, we analyzed the level of cell surface expression of nectin1γ in transiently expressing Cos1a cells by fluorescence-activated cell sorting (FACS) analysis with MAb R1.302. We argued that it should be low and should be similar to that of nectin1-Fc, a recombinant soluble form of nectin1 in which the ectodomain is fused to the Fc portion of human IgG (3). For both proteins, the level of cell surface expression was indeed low compared to that of the transmembrane isoform nectin1α, indicating similar expression patterns for nectin1γ and recombinant soluble nectin1-Fc (Fig. 2F). The cell surface expression of nectin1γ may result from the endogenous protein that is transported to the plasma membrane and/or from reassociation of previously secreted protein to endogenous transmembrane isoforms of nectin1 or nectin3 (20). To investigate the latter possibility, we examined whether exogenous nectin1-Fc, when added to J1.1-2 cells, can associate with their plasma membrane. J1.1-2 cells are known to express an endogenous hamster homolog of nectin3/PRR3, but not an homolog of nectin1, at least not an homolog amplificable with the primers designed for the human sequence (reference 5 and our unpublished observation). FACS analysis reveals binding of exogenous nectin1-Fc (data not shown), consistent with the possibility that localization at the plasma membrane results from reassociation of the secreted molecules with the endogenous transmembrane nectin molecules.
The nectin1γ secreted in the culture medium can reduce HSV infectivity.
Previous studies have shown that the transmembrane nectin1α and -β isoforms mediate the entry of HSV into cells (5, 8) and that the chimeric nectin1-Fc can compete with cell-bound nectin1 and reduce HSV infectivity (3, 8). In this study, we wanted to ascertain whether nectin1γ displays an inhibitory activity on HSV infectivity.
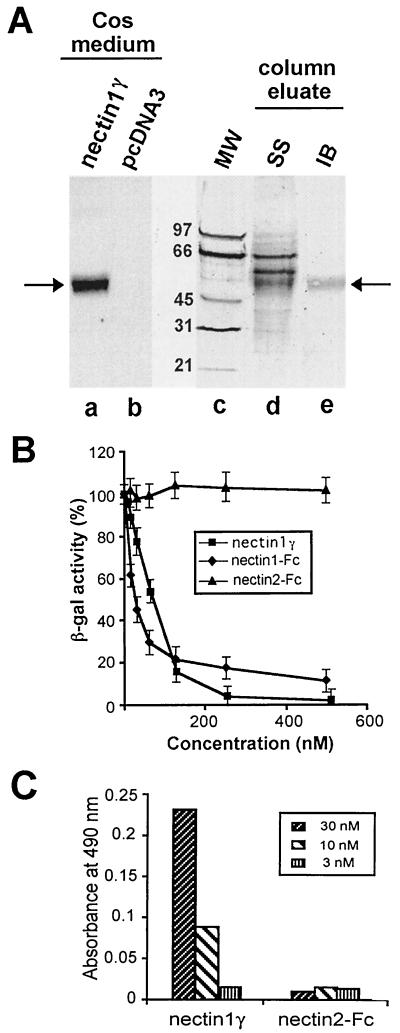
To this end, first we derived a rabbit antipeptide polyclonal serum specific for the C terminus of nectin1γ (QLIYPGKGRTRARMF). Its reactivity to nectin1γ secreted in the culture medium of transfected Cos1a cells is shown in Fig. 3A. A batch of nectin1γ was then produced by transient transfection in Cos1a cells. It was purified by affinity chromatography on a column of MAb R1.302 immobilized to Sepharose, and its purification was traced by reactivity with the C-terminal antipeptide serum. The electrophoretic profile of the proteins eluted from the affinity column shows the presence of a 56-kDa protein specifically recognized by the antinectin1γ serum (Fig. 3A, lane e), together with two additional bands detected by silver staining (lane d) but not recognized by the antiserum and therefore likely contaminants. By densitometric analysis, we estimated that nectin1γ accounts for about 30% of total protein content of the partially purified preparation.
FIG. 3.
(A) Purification of nectin1γ from culture medium of transfected Cos1a cells. To produce and affinity purify nectin1γ, R1.302 IgGs were precipitated overnight at 4°C from R1.302 ascitic fluid with ammonium sulfate (50% saturated) and then conjugated to CNBr-activated Sepharose 4B (Amersham Pharmacia Biotech AB) according to the manufacturer's instructions. The column was loaded with about 1 liter (concentrated using Microcon YM10 filters [Millipore]) of culture medium from Cos1a cells transfected with pCDSR1.D1 plasmid, harvested 6 days after transfection. The concentrated medium was allowed to adsorb overnight at 4°C and eluted with 3 M potassium thiocyanate in equilibration buffer (10 mM Tris-HCl [pH 7.5], 0.5 M NaCl). Pooled protein fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining (SS) (lane d) or by immunoblotting (IB) with an antipeptide serum directed against the C terminus of nectin1γ (lane e). Lanes a and b illustrate the specificity of the antipeptide immune serum. Shown is the immunoblot reactivity of the serum to the medium of Cos1a cells transfected with pCDSR1.D1, encoding nectin1γ, or with the pcDNA3 vector alone. The migration positions of the molecular weight markers (MW) are shown in lane c. Arrows point to the nectin1γ secreted in the culture medium of transfected Cos1a cells and in the affinity column eluate. (B) Inhibition of HSV-1 R8102 infectivity by nectin1γ and nectin1-Fc. R8102 virions were preincubated with increasing concentrations of the partially purified nectin1γ, nectin1-Fc, or nectin2-Fc for 1 h at 37°C. Virions were then allowed to infect cells expressing nectin1β, grown in 96-well plates, for 2 h at 4°C. Infection was quantified as β-Gal expression by ONPG staining. Each point represents the average of three values. The concentration of nectin1γ was estimated by multiplying the actual concentration by 0.3 to take the partial purification shown in panel A into consideration. (C) Binding of nectin1γ to virions. The wells in 96-well plates were coated with gradient-purified R7032 virions (10) and reacted with nectin1γ at the indicated concentrations, followed by the rabbit antipeptide serum (1:400), and peroxidase-conjugated anti-rabbit IgG (17). As a background control, R7032 virions were reacted with nectin2-Fc, a recombinant form of nectin2, in which the ectodomain is fused to the Fc portion of human IgGs (12). Binding was detected with peroxidase-conjugated anti-human IgG.
This preparation was employed in virus infectivity inhibition assays and compared with nectin1-Fc. Nectin1-Fc was purified by affinity chromatography to protein A, as described previously (11). R8102 virions were preincubated with increasing concentrations of nectin1γ or nectin1-Fc and allowed to infect a derivative of J1.1-2 cells stably expressing the human transmembrane isoform nectin1β (5). R8102 carries a lacZ reporter gene inserted between the UL3 and UL4 genes under the α27 promoter, and infection can be quantified as β-galactosidase (β-Gal) expression 16 h after infection, using o-nitrophenyl-β-d-galactopyranoside (ONPG), as previously described (5, 18). Figure 3B shows that R8102 infection was reduced in a dose-dependent manner by nectin1γ, with a curve similar to that of nectin1-Fc. To demonstrate that inhibition occurs because of the direct binding of nectin1γ to virions, binding was measured in an enzyme-linked immunosorbent assay (ELISA) in which purified virions, immobilized onto ELISA plates, were reacted with nectin1γ (17). R7032, a recombinant virus lacking the genes encoding gE and gI (15), two glycoproteins with Fc-binding activity, was employed in this assay to avoid the nonspecific binding of IgGs to virions. Nectin1γ binding to virions was revealed by using the antipeptide serum specific for nectin1γ, described above. As shown in Fig. 3C, a dose-dependent binding was readily detected. Altogether, these experiments provide evidence that nectin1γ can compete with cell-bound nectin1β and reduce HSV entry mediated by nectin1β.
Nectin1γ expressed in receptor-negative cells enables HSV-1 entry.
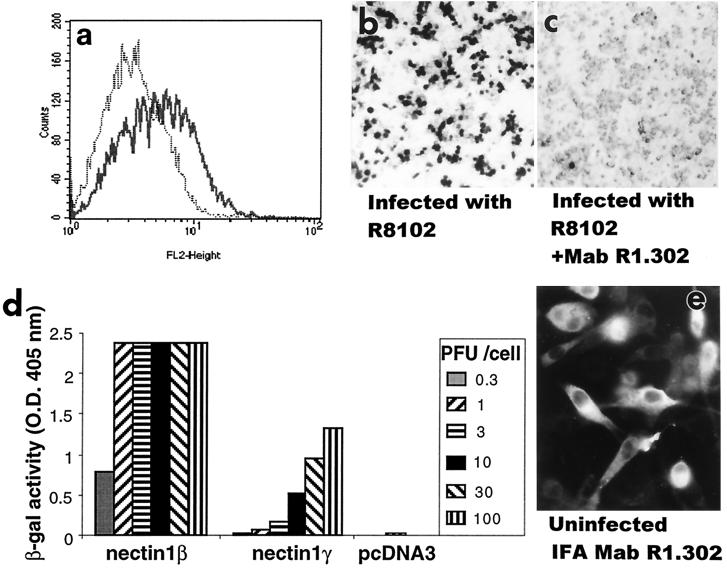
FACS analysis of Fig. 2 shows a small but significant amount of the nectin1γ transiently expressed in Cos1a cells is located at the cell surface. To investigate whether nectin1γ can mediate HSV entry, stable transformants of J1.1-2 cells expressing nectin1γ were exposed to increasing amounts of R8102, and infection was monitored as β-Gal activity. Cell surface expression of nectin1γ was determined in parallel by FACS. The results in Fig. 4a show that nectin1γ was expressed at detectable levels at the plasma membrane of stably transformed cells, in accordance with what was observed in the Cos1a transient-expression system reported above in Fig. 2. Figure 4b shows that expression of nectin1γ rendered J1.1-2 cells capable of being infected with HSV. Infection was abolished by MAb R1.302, indicating that it was indeed mediated by nectin1γ (Fig. 4c). Infection, however, required a higher multiplicity of infection than in cells expressing the β transmembrane isoform (Fig. 4d), probably as a consequence of the paucity of nectin1γ available on the plasma membrane. When the intracellular distribution of nectin1γ was analyzed by an indirect immunofluorescence assay, we noted that it differed from that of nectin1β (5); the former localized diffusely to the cytoplasm (Fig. 4e), whereas the latter localized predominantly to plasma membranes and vesicle-like structures (5).
FIG. 4.
Nectin1γ mediates HSV infectivity. A clonal transformant of J1.1-2 cells expressing nectin1γ was stained with MAb R1.302 (a) and infected with R8102 virus (b and c). (c) Prior to infection, cells were exposed to MAb R1.302 directed against nectin1 (1:25-diluted ascitic fluid) or with an irrelevant control antibody to human herpesvirus 6 (1:25-diluted ascitic fluid or purified IgGs [0.16 μg/μl] [not shown]) for 2 h at 4°C. Infection was monitored as β-Gal activity, by staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). (d) Stable cell lines expressing nectin1β, nectin1γ, or pcDNA3 were infected with R8102, at increasing multiplicities of infection ranging from 0.3 to 100 PFU/cell. Infection was monitored as β-Gal activity. O.D. 405 nm, optical density at 405 nm. (e) Intracellular distribution of nectin1γ detected by indirect immunofluorescence assay (IFA) with MAb R1.302. J1.1-2 cells expressing nectin1γ were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min.
Conclusions.
The results presented here show the following. (i) Nectin1γ, a novel isoform of the nectin family, is expressed in human cell lines and in specific human tissues. (ii) The amino acid sequence predicts a protein with a signal sequence but lacking a transmembrane-anchoring domain, and indeed, the protein is secreted in the culture medium of cells transfected with the cDNA. (iii) Nectin1γ obtained from culture medium behaves similarly to recombinant soluble nectin1, in that it competes with cell-bound nectin1 and reduces the ability of HSV to infect cells. (iv) Nectin1γ, expressed at low levels at the cell surface, is capable of mediating HSV entry. Thus, nectin1γ has the potential to modulate HSV infectivity positively and negatively.
Nectin1γ is a novel soluble isoform of the nectin1 subfamily. It shares the ectodomain made of three Ig-like sequences with the α and β isoforms; the remaining sequence diverges from that of the other nectin1 isoforms at the exact site where the sequences of the α and β isoforms begin to diverge (5, 13). This isoform originates by splicing, as we have identified the sequence of the intron located between the exon encoding the third C domain and the exon encoding the carboxyl terminus. The same donor site is therefore employed to generate the three isoforms of nectin1 known so far. We note that alternative splicing to generate transmembrane and soluble isoforms also occurs in the case of PVR/CD155, for which four isoforms are known; two of the four isoforms carry transmembrane domains (α and δ), and the other two (β and γ) do not, and the isoforms result from the excision of the exon encoding the transmembrane sequence (9). The intracellular distribution of the different isoforms of nectin1 is consistent with the presence of transmembrane sequences. Thus, the α and β isoforms of nectin1, which carry predicted transmembrane domains, localize to the plasma membrane, particularly to the cell-cell junctions, and intracellularly to vesicle-like structures (5). By contrast, nectin1γ, which does not carry a predicted transmembrane-anchoring region, shows a more diffuse cytoplasmic distribution, and the protein is found in the culture medium of cells expressing the cDNA. Nectin1γ is expressed in human cell lines derived from different hematopoietic lineages and from solid tumors. It was coexpressed with the transmembrane isoforms in at least in one cell line; the overall level of expression was rather low. The tissue distribution of nectin1γ appears to be narrow by Northern blot analysis, in contrast with that of the α and β transmembrane isoforms, which are expressed in a broader range of human tissues. However, a more sensitive assay showed a broader distribution. At present, the significance of the secreted isoforms of these receptors is unknown.
Nectin1γ can modulate HSV infectivity both positively and negatively. Thus, the nectin1γ present in the culture medium, when incubated with virions, is capable of preventing them from interacting with cell-bound nectin1β, reducing entry. By contrast, its expression in receptor-negative J1.1-2 cells conferred susceptibility to HSV infection. The latter property, although surprising, had been observed previously with the soluble isoform of PVR/CD155 (9) and also with other, totally unrelated, cellular receptor systems, such as avian sarcoma virus and leukosis retrovirus (6). This finding raises the issue as to how nectin1γ localizes to the plasma membrane to enable HSV entry. One plausible explanation rests on the observation that nectin1 can trans interact with itself (homophilic interaction) or trans interact with nectin3 (heterophilic interaction). These interactions are dependent on a cis dimerization of the molecule at the cell surface. Thus, it is possible that nectin1γ dimerizes, in cis or in trans, to endogenous transmembrane forms of nectins. The partners could be nectin1 or nectin3 or an as yet unidentified partner that by itself cannot mediate HSV entry. The finding that exogenously added recombinant nectin1-Fc binds to J1.1-2 cells is consistent with the possibility of trans interaction with endogenous cell surface partners.
The expression in human tissues of a receptor molecule identified in cell culture is at the moment one of the few means by which we can assess its role in rendering humans susceptible to infection or in modulating susceptibility. The dual enhancing and inhibitory activities of nectin1γ raise a hypothesis of wide-ranging significance that nectin1γ has the potential to modulate HSV infection in human tissues both positively and negatively. Thus, it is possible that tissues positive for nectin1γ expression may create a microenvironment with a reduced susceptibility to HSV infection. Conversely, tissues that do not express the α or β transmembrane isoforms might be susceptible to infection because of the interaction of nectin1γ with resident nectin3 molecules.
Nucleotide sequence accession numbers.
The nucleotide sequence of nectin1γ gene and the intronic type I consensus splice sequence have been deposited in the GenBank database under accession no. AY029539 and AY032612, respectively.
Acknowledgments
We thank Elisabetta Romagnoli and Eric Lecocq for invaluable assistance with cell cultures. We thank B. Roizman (Chicago, Ill.) for kindly providing MAb R8102.
The work done at the University of Bologna was supported by grants from Telethon grant A141, Target Project in Biotechnology/CNR, MURST 40%, University of Bologna 60%, and pluriannual plan. The work done at INSERM U.119 was supported by INSERM, the Association pour la Recherche contre le Cancer (ARC), and the Ligue Nationale Française Contre le Cancer (LNFCC) (Comité des Bouches du Rhône).
REFERENCES
- 1.Campadelli-Fiume G. Virus receptor arrays, CD46 and human herpesvirus 6. Trends Microbiol. 2000;8:436–438. doi: 10.1016/s0966-842x(00)01804-7. [DOI] [PubMed] [Google Scholar]
- 2.Campadelli-Fiume G, Cocchi F, Menotti L, Lopez M. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev Med Virol. 2000;10:305–319. doi: 10.1002/1099-1654(200009/10)10:5<305::aid-rmv286>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cocchi F, Menotti L, Dubreuil P, Lopez M, Campadelli-Fiume G. Cell-to-cell spread of wild-type herpes simplex virus type 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (PRR1/HveC/HIgR) and nectin2 (PRR2/HveB) J Virol. 2000;74:3909–3917. doi: 10.1128/jvi.74.8.3909-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damico R, Bates P. Soluble receptor-induced retroviral infection of receptor-deficient cells. J Virol. 2000;74:6469–6475. doi: 10.1128/jvi.74.14.6469-6475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberlé F, Dubreuil P, Mattei M G, Devilard E, Lopez M. The human PRR2 gene, related to the human poliovirus receptor gene (PVR), is the true homolog of the murine MPH gene. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 8.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 9.Koike S, Horie H, Ise I, Okitsu A, Yoshida M, Iizuka N, Takeuchi K, Takegami T, Nomoto A. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 1990;9:3217–3224. doi: 10.1002/j.1460-2075.1990.tb07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez M, Aoubala M, Jordier F, Isnardon D, Gomez S, Dubreuil P. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood. 1998;92:4602–4611. [PubMed] [Google Scholar]
- 12.Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Campadelli-Fiume G. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J Virol. 2000;74:1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez M, Eberlé F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Complementary DNA characterization and chromosomal localization of a human gene related to the poliovirus receptor-encoding gene. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 14.Lopez M, Jordier F, Bardin F, Coulombel L, Chabannon C, Dubreuil P. CD155 workshop: identification of a new class of IgG superfamily antigens expressed in hemopoiesis. In: Kishimoto T, et al., editors. Leukocyte typing VI: white cell differentiation antigens. New York, N.Y: Garland Publishing; 1997. pp. 1081–1083. [Google Scholar]
- 15.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 16.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 17.Menotti L, Lopez M, Avitabile E, Stefan A, Cocchi F, Adelaide J, Lecocq E, Dubreuil P, Campadelli-Fiume G. The murine homolog of human-nectin1δ serves as a species nonspecific mediator for entry of human and animal αherpesviruses in a pathway independent of a detectable binding to gD. Proc Natl Acad Sci USA. 2000;97:4867–4872. doi: 10.1073/pnas.97.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 19.Reymond N, Borg J, Lecocq E, Adelaide J, Campadelli-Fiume G, Dubreuil P, Lopez M. Human nectin3/PRR3: a novel member of the PVR/PRR/nectin family that interacts with afadin. Gene. 2000;255:347–355. doi: 10.1016/s0378-1119(00)00316-4. [DOI] [PubMed] [Google Scholar]
- 20.Satoh-Horikawa K, Nakanishi H, Takahashi K, Miyahara M, Nishimura M, Tachibana K, Mizoguchi A, Takai Y. Nectin-3, a new member of immunoglobulin-like cell adhesion molecules that shows homophilic and heterophilic cell-cell adhesion activities. J Biol Chem. 2000;275:10291–10299. doi: 10.1074/jbc.275.14.10291. [DOI] [PubMed] [Google Scholar]
- 21.Shukla D, Rowe C L, Dong Y, Racaniello V R, Spear P G. The murine homolog (Mph) of human herpesvirus entry protein B (HveB) mediates entry of pseudorabies virus but not herpes simplex virus types 1 and 2. J Virol. 1999;73:4493–4497. doi: 10.1128/jvi.73.5.4493-4497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spear P G, Eisenberg R J, Cohen G H. Three classes of cell surface receptors for alphaherpesvirus entry. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, Takai Y. Nectin/PRR: an immunoglobulin-like cell adhesion molecule recruited to cadherin-based adherens junctions through interaction with afadin, a PDZ domain-containing protein. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]