Abstract
Chronic hepatitis C virus (HCV) infection is a major cause of liver disease. The HCV polyprotein contains a hypervariable region (HVR1) located at the N terminus of the second envelope glycoprotein E2. The strong variability of this 27-amino-acid region is due to its apparent tolerance of amino acid substitutions together with strong selection pressures exerted by anti-HCV immune responses. No specific function has so far been attributed to HVR1. However, its presence at the surface of the viral particle suggests that it might be involved in viral entry. This would imply that HVR1 is not randomly variable. We sequenced 460 HVR1 clones isolated at various times from six HCV-infected patients receiving alpha interferon therapy (which exerts strong pressure towards quasispecies genetic evolution) and analyzed their amino acid sequences together with those of 1,382 nonredundant HVR1 sequences collected from the EMBL database. We found that (i) despite strong amino acid sequence variability related to strong pressures towards change, the chemicophysical properties and conformation of HVR1 were highly conserved, and (ii) HVR1 is a globally basic stretch, with the basic residues located at specific sequence positions. This conservation of positively charged residues indicates that HVR1 is involved in interactions with negatively charged molecules such as lipids, proteins, or glycosaminoglycans (GAGs). As with many other viruses, possible interaction with GAGs probably plays a role in host cell recognition and attachment.
Hepatitis C virus (HCV) is a small enveloped RNA virus that belongs to the Flaviviridae family. It causes chronic liver disease, including chronic active hepatitis in up to 80% of infected individuals, as well as cirrhosis and hepatocellular carcinoma (3). The presently approved treatment is based on the combination of alpha interferon (IFN-α) and ribavirin, and sustained clearance of HCV replication is achieved in about 40% of patients (31, 39). HCV exists within its hosts as a pool of genetically distinct but closely related variants, referred to as quasispecies (29, 48). This confers a significant survival advantage, as the simultaneous presence of multiple variant genomes allows rapid selection of mutants better suited to new environmental conditions. The fittest infectious particles are continuously selected as a result of selective pressures exerted by their interactions with host cell proteins and host immune responses.
Sequence analysis of a large number of HCV isolates has revealed hypervariable genomic sequences. Hypervariable region 1 (HVR1) is a 27-amino-acid sequence located at the N terminus of the second envelope glycoprotein E2. This region is highly tolerant for amino acid substitutions. Being a target for anti-HCV neutralizing antibodies and, possibly, cytotoxic responses, it is also subjected to strong positive selection pressure (18, 24, 48). For these reasons, HVR1 has been widely used as a model to study HCV genome quasispecies distribution. It was recently shown that, in treated patients who did not clear HCV RNA, IFN-α therapy generates shifts in virus populations (35, 36). IFN-α therapy thus provides a good model to study constraints on HVR1 quasispecies sequences in drastically changing environmental conditions.
The biological role of HVR1 is unknown. It was recently proposed that HVR1 could serve as a decoy for the immune system during acute infection (40). Antibodies directed against HVR1 have been shown to be neutralizing in vitro, protecting chimpanzees against HCV infection after in vitro neutralization of the corresponding strain (18, 19). In addition, anti-HVR1 antibodies apparently inhibit viral adsorption to the surface of cultured cells of various types (44, 50, 51). It was recently reported that an HCV clone lacking HVR1 was infectious but attenuated in a chimpanzee (21). Thus, although HVR1 may not be essential for infection in chimpanzees, it probably plays a role in HCV strain infectivity. In addition, HVR1 is always present in strains infecting humans, suggesting that any virus containing the intact HVR1 has a significant survival advantage over emerging mutants lacking a part of or the full-length HVR1. Together, these findings suggest a role of HVR1 in viral entry. This would imply that HVR1 is not randomly variable and that its chemicophysical properties must be at least partly conserved. Previous studies have identified both invariant and variable positions within the HVR1 sequence (30, 42, 43). McAllister et al. (30), comparing HVR1 sequences in quasispecies variants isolated from individuals infected from a common source, found evidence that amino acid substitutions in HVR1 are not only due to random accumulation of mutations but are also driven by positive selection pressures and constrained by negative selection pressures. In addition, strong selection pressure to maintain the size of HVR1 has been reported (8).
Although HVR1 is reported to be structurally flexible and antigenically variable, little attention has been paid to its conformation. In this study we combined two complementary approaches to assess HVR1 variability and conservation, including a longitudinal study of HVR1 quasispecies evolution in six patients receiving IFN-α therapy, i.e., subjected to strong pressures towards change, and an analysis of the largest possible number of nonredundant HVR1 sequences of natural HCV variants collected from the EMBL database. The data presented here indicate that HVR1 conformation is well conserved and that HVR1 is a basic stretch likely involved in intermolecular interactions with negatively charged molecules such as lipids, proteins, or glycosaminoglycans (GAGs). HVR1 could thus be involved in viral attachment and possibly cell tropism.
HVR1 sequence collection and genetic characterization.
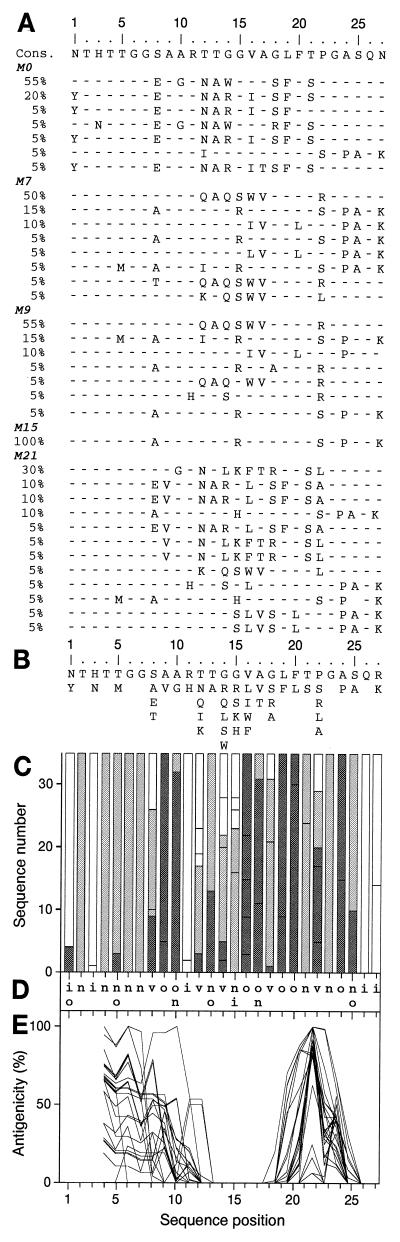
In the longitudinal study of HVR1 quasispecies evolution, six patients infected with HCV genotype 1 who did not clear HCV RNA after 6 months of treatment with 3 MU of IFN-α-2a three times per week (patients A to F) were randomly selected for extensive HVR1 quasispecies analysis (see reference 35 for further information). Patient E was re-treated with the same IFN-α regimen 3 months after the end of the first course (i.e., from month 9 to month 15) and was monitored until month 21, i.e., 6 months after IFN-α withdrawal (Fig. 1A); he did not clear HCV RNA after the second course of IFN-α. Blood samples were taken at various times before, during, and after IFN-α treatment, and a total of 460 HVR1 clones (20 clones per time point per patient) were generated from the six patients. Partial genetic analyses of these sequences have been reported elsewhere (35). Briefly, in all but one case (patient D) HVR1 genetic evolution during and after IFN-α therapy was characterized by successive shifts of the virus populations, as illustrated in Fig. 1A, which shows the HVR1 quasispecies sequences isolated from patient E at indicated times during follow-up. HVR1 quasispecies changes were evolutionary in all instances, and HVR1 genetic evolution appeared to be related to positive pressures towards change rather than to random accumulation of mutations during HCV replication (35). These results were in keeping with the view that HVR1 is highly tolerant of amino acid substitutions and is a target for anti-HCV immune responses.
FIG. 1.
Analysis of HVR1 quasispecies sequences from patient E (genotype 1b) based on sequences of 20 clones per time point at five time points, i.e., 100 distinct clones. (A) Alignment of HVR1 quasispecies sequences at month zero (M0, before IFN-α treatment), M7 (1 month after the end of the first IFN-α course), M9 (beginning of the second IFN-α course), M15 (end of the second IFN-α course), and M21 (6 months after the end of the second IFN-α course). The frequency of each sequence in the quasispecies is given as a percentage in the left-hand column. Cons, the derived consensus amino acid sequence. Amino acids identical to those in the consensus sequence are represented by a hyphen. (B) Repertoire of patient E's HVR1 amino acids per position from the analysis of the 36 nonredundant sequences observed at the five time points. Amino acids are listed in decreasing order of observed frequency, from top to bottom. (C) Histogram showing the hydropathic character of the residues at each position in HVR1. The height of the box in each bar indicates the number of sequences with a given residue at a given position. The boxes are presented in order of decreasing hydrophobicity, from bottom to top, according to the hydrophobicity scale of Black and Mould (4). Each box is colored according to the hydrophobic character of the residue: dark gray for hydrophobic (F, I, W, Y, L, V, M, P, C, A), light gray for neutral (G, T, S), and white for hydrophilic (K, Q, N, H, E, D, R). (D) Consensus hydropathic pattern of HVR1 quasispecies deduced from the latter. o, hydrophobic residue; n, neutral residue; i, hydrophilic residue; v, variable residue. (E) Antigenicity profiles of HVR1 sequences calculated according to the method of Parker et al. (34) with a window of 7 amino acids. Therefore, the antigenicity of the first three HVR1 positions could not be estimated.
To better characterize HVR1 tolerance of amino acid substitutions, together with its chemicophysical properties and the constraints on its sequence, we studied the HVR1 amino acid repertoires in the six patients' quasispecies at each time point. The hydropathic and chemicophysical features of the residues were characterized, and the antigenicity profiles of the variants were compared. To determine whether our findings on genotype 1 HVR1 quasispecies would also apply to other HVR1 sequences of the most frequent HCV genotypes, we extended the analysis to the 1,382 HVR1 sequences recovered from the EMBL database, including unambiguously genotyped HVR1 sequences.
For this, all HCV sequences in the EMBL database were downloaded to our HCV database website (HCVDB; http://hepatitis.ibcp.fr). The first 30 amino acid residues of envelope glycoprotein E2 taken from published HCV polyproteins representative of the most frequent HCV genotypes (9, 41) were used to select HVR1 sequences by means of the FASTA program (37). HVR1 sequences were aligned with CLUSTAL W (46) by using HCVDB Network Protein Sequence Analysis website facilities (http://pbil.ibcp.fr/NPSA) (5, 14). A final set of 1,382 nonredundant HVR1 sequences was analyzed. To select unambiguously genotyped HVR1 sequences, HVR1 sequences reported together with the flanking E1 sequence were sought with FASTA by using the sequence of a stretch spanning the C terminus of E1 and the N terminus of E2 (nucleotide positions 915 to 1,632 in the HCV-H prototype strain). The HCV clade and subtype were determined for each HVR1 sequence on the basis of the flanking E1 sequence using representative clade and subtype sequences (9), including clades 1 to 6 and subtypes 1a, 1b, 1c, 2a, 2b, 2c, 3a, 3b, 4a, 5a, 6a, 10a (classified into clade 3), and 11a (classified into clade 6). Among the unambiguously genotyped HVR1 sequences, by far the most numerous were subtypes 1a and 1b (85 and 119 HVR1 sequences, respectively).
HVR1 amino acid repertoire.
The amino acids observed at various time points at each of the 27 HVR1 positions in patient E's quasispecies are presented in Fig. 1B. Like in the other five patients (data not shown), residues were conserved at certain HVR1 positions, whereas most positions were variable. This was also the case when the repertoires of 119 unambiguously genotyped HCV genotype 1b sequences (Fig. 2A) and 85 HCV genotype 1a sequences (data not shown) were analyzed. Figure 3A presents the HVR1 amino acid repertoire for the 1,382 HVR1 sequences of 13 different genotypes from the EMBL database. Again, certain positions in HVR1 appeared to be far more variable than others.
FIG. 2.
Analysis of 119 unambiguously genotyped HCV genotype 1b HVR1 sequences from the EMBL database. Residues observed at a given position in only one sequence were not taken into consideration. (A) Repertoire of genotype 1b HVR1 residues per position. Residues are listed in decreasing order of observed frequency, from top to bottom. Residues within the box correspond to those observed in more than 10% of the sequences. (B) Histogram showing the hydropathic character of residues at each position (see the legend to Fig. 1C). (C) Consensus hydropathic pattern of genotype 1b sequences (see the legend to Fig. 1D). (D) Antigenicity profiles of the 10 most distantly related sequences of genotype 1b sequences (see the legend to Fig. 1E). Selection of these 10 among 119 sequences was based on the HVR1 phylogenetic tree of genotype 1b.
FIG. 3.
Analysis of 1,382 HVR1 sequences from the EMBL database. Amino acids observed at a given position in fewer than five distinct sequences (<0.3%) were not taken into consideration. (A) Repertoire of HVR1 residues per position in the 1,382 unrelated EMBL HVR1 sequences. Amino acids are listed in decreasing order of observed frequency, from top to bottom. Residues within the box correspond to those observed in more than 10% of the sequences. (B) Histogram showing the hydropathic character of residues at each position in HVR1. The height of the box in each bar indicates the frequency of sequences with a given residue at a given position. (C) Consensus hydropathic pattern from the 1,382 HVR1 sequences. (D) Comparison of the antigenicity profiles of HVR1 sequences representative of the principal HCV subtypes of clades 1 to 6 (the EMBL accession number of each sequence is indicated in parentheses): 1a (M67463), 1b (D90208), 1c (D14853), 2a (D00944), 2b (D10988), 2c (D50409), 3a (D28917), 3b (D49374), 4a (Y11604), 5a (Y13184), 6a (Y12083), 10a (D63821; classified in clade 3), and 11a (D63822; classified in clade 6).
Conservation of the hydropathic characters of HVR1 residues.
Comparisons of HVR1 sequences using the Kyte and Doolittle method (26) showed very similar hydrophobicity profiles among quasispecies variants and HCV isolates of various genotypes (data not shown). The small size of the HVR1 stretch allowed a more precise analysis of the hydropathic character of the residues at each HVR1 position. HVR1 hydropathic patterns are shown in Fig. 1C for patient E's quasispecies and in Fig. 2B for the 119 unambiguously genotyped HCV genotype 1b sequences. A letter-coded motif summarizing the corresponding HVR1 hydropathic patterns is shown in Fig. 1D and 2C, respectively. Typically, in patient E's quasispecies 12 positions bore exclusively one type of residue: hydrophobic, neutral, or hydrophilic; 13 positions were occupied by two classes of residues only; and all three classes of residues were observed at only two positions (positions 1 and 8), which were considered truly variable (Fig. 1D). Thus, despite HVR1 variability, the hydropathic characters of the residues were conserved at most positions in patient E's quasispecies variants. Findings for the five other patients were similar, and only minor differences were observed when compared to findings for patient E. These differences were related to the isolate rather than to the HCV genotype. Indeed, the hydropathic patterns of unambiguously genotyped 1a and 1b HVR1 sequences from EMBL differed from each other at only five positions (positions 5, 7, 9, 18, and 25; Fig. 2D and data not shown).
Regardless of the HCV genotype, common features were again found when this analysis was extended to the 1,382 EMBL HVR1 sequences (Fig. 3B and C). The consensus hydropathic pattern shown in Fig. 3C highlights the positions in HVR1 that are conserved or variable for the hydropathic character. In summary, a glycine residue is always found at position 23; three positions (positions 16, 19, and 20) are exclusively hydrophobic; two positions (positions 26 and 27) are exclusively hydrophilic; two positions (positions 2 and 6) are always neutral; six positions (positions 4, 5, 10, 13, 17, and 24) are either hydrophobic or neutral; and the remaining 13 positions can harbor any of the three classes of amino acids and constitute the truly hypervariable positions (positions 1, 3, 7, 8, 9, 11, 12, 14, 15, 18, 21, 22, and 25). Overall, as many as 12 HVR1 positions exclusively harbor hydrophobic or neutral residues (positions 2, 4, 5, 6, 10, 13, 16, 17, 19, 20, 23, and 24). Interestingly, the HVR1 consensus hydrophobic pattern presents some analogy with HLA binding motifs that contain both highly conserved anchor residues and variable ones. It is likely that conserved hydrophobic and neutral residues ensure HVR1 anchoring to the E2 glycoprotein. Typically, the fully conserved hydrophobic residues at positions 16, 19, and 20 probably interact with the hydrophobic core of E2. Overall, hydrophobic and neutral residue conservation at specific HVR1 positions indicates that HVR1 conformation is conserved and suggests that these residues play an important role in maintaining HVR1 conformation within the E2 glycoprotein.
Conservation of the predicted HVR1 antigenicity profile.
To further assess HVR1 conformation conservation, we compared the antigenicity profiles of HVR1 sequences. These profiles were calculated according to the guidelines of Parker et al. (34) using the ANTHEPROT package 5 program (http://antheprot-pbil.ibcp.fr) (16). Parker's method uses a combination of the best three parameters for hydrophilicity, accessibility, and flexibility to predict antigenic protein surface sites. No prediction was made for the first three HVR1 positions because this method uses a calculation window of seven residues. In patient E (Fig. 1E), although the position with the highest antigenic score differed from one quasispecies variant to the next, the antigenicity profiles defined two antigenic segments, between residues 1 and 13 and between residues 19 and 25. In contrast, segments 14 to 18 and 24 to 27 were never predicted to be antigenic. Similar profiles were obtained for the five other patients (data not shown).
Although individual profiles could differ markedly, the same two antigenic regions were predicted when we analyzed the 10 most distantly related HCV genotype 1b sequences (Fig. 2D), the 10 most distantly related HCV genotype 1a sequences (data not shown), and sequences representative of the principal HCV genotypes (Fig. 3D). In all instances the two antigenic regions were comprised between positions 1 and 13 and positions 19 and 24, whereas regions 14 to 18 and 25 to 27 were never predicted to be antigenic. These results strongly support HVR1 conformation conservation among HCV quasispecies variants and isolates of the most frequent HCV genotypes.
Prediction of HVR1 secondary structure.
The secondary structures of HVR1 sequences were predicted by using a large set of methods available at the NPSA website (http://npsa-pbil.ibcp.fr) (14). A secondary structure was consistently predicted for a segment comprised between positions 16 and 20, but the predicted conformation state (α-helix or β-strand) varied according to the HVR1 sequence and the method used (data not shown). The prediction scores for the two conformational states were close, suggesting that, as reported for glucagon, for instance (13), this HVR1 segment may be able to adopt either conformation. A turn was predicted in the region comprised between positions 21 and 24 in almost all the HVR1 sequences. Indeed, most of the residues observed at these positions were those frequently involved in β-turns, namely Pro, Gly, Asp, Asn, and Ser (12). This turn might explain Gly conservation at position 23. In contrast, the 1 to 15 segment appears to be rich in small and neutral residues and to be rather flexible. This flexibility, together with the variability of the predicted conformation state in segment 16 to 20 (α-helix or β-strand), suggests that conformational changes might occur in HVR1, possibly as a consequence of intermolecular interactions.
HVR1 amino acid composition.
The frequency of each of the 20 amino acids was calculated in the 1,382 HVR1 sequences and compared with their average frequency in protein sequences from the complete SWISS-PROT database (http://www.expasy.ch). Overall, with only 35% of hydrophobic residues versus 47% in the SWISS-PROT database, HVR1 tended to be hydrophilic. However, neutral residues were more frequent in HVR1 than in SWISS-PROT (42 and 20%, respectively), whereas hydrophilic residues were less frequent (23 and 33%, respectively).
Examination of amino acid repertoires (Fig. 1B, 2A, and 3A and data not shown) revealed that residues known to constrain polypeptide conformation were uncommon in HVR1 sequences. Indeed, Cys (which can form disulfide bridges) and Trp were almost never present, while Pro was only observed at positions 22 and 24, both of which are predicted to be involved in a turn (see above). Large hydrophobic residues were rare, except at specific positions (positions 16, 19, and 20). In contrast, small, flexible residues Ala, Gly, Thr, and Ser were found at most positions. As these residues are able to adopt any conformation, their presence might be required to compensate for structural constraints imposed by the large and poorly flexible residues present at certain positions.
As the HVR1 sequences contained a large number of Thr, Ser, and Asn residues, we checked the presence of putative N-glycosylation sites, characterized by the N-{P}-[S,T]-{P} PROSITE pattern (http://www.expasy.ch/prosite/), in the 1,382 EMBL HVR1 sequences. This analysis yielded only 13 hits, i.e., 0.9% of HVR1 variants, indicating a negative selection of variants with N-glycosylation sites in HVR1. Glycosylation is known to reduce protein accessibility by steric hindrance. Thus, negative selection of variants with N-glycosylation sites suggests that HVR1 should remain accessible at the surface of the E2 envelope glycoprotein to be functional.
HVR1 is a basic stretch.
Among the hydrophilic residues, acidic ones (Asp and Glu) were much less frequent in HVR1 sequences than in SWISS-PROT proteins (1.7 and 11.6%, respectively), whereas the difference in the frequency of basic residues was weak (11.2 and 13.3% for HVR1 and SWISS-PROT, respectively). This paucity of acidic residues has already been noted by McAllister et al. (30). As a result, the basic/acidic ratio was much higher in HVR1 than in common proteins (6.50 instead of 1.15). Detailed analysis of the 1,382 EMBL HVR1 sequences revealed that nearly all of them contained at least one basic residue (Table 1); about 87% of them contained two to four, including 41.2% that contained three, and 7% contained five or more. In contrast, only 41% of the examined HVR1 sequences contained acidic residues; the presence of two was rare (6.5%) and that of three was exceptional (0.4%) (Table 1). Among the 1,382 HVR1 sequences, only 31 (2.2%) were globally neutral (i.e., contained the same number of acidic and basic residues), and only three sequences (0.2%) were globally acidic. The remaining 1,348 HVR1 sequences (97.5%) were globally basic. Thus, HVR1 is intrinsically a basic stretch.
TABLE 1.
Number of acidic and basic residues among the 27 HVR1 amino acids in each of the 1,382 HVR1 sequences from the EMBL database
| Basic residuesa
|
Acidic residuesb
|
||
|---|---|---|---|
| No. of basic residues | Percentage of HVR1 sequences | No. of acidic residues | Percentage of HVR1 sequences |
| 0 | 0.2 | 0 | 59.1 |
| 1 | 5.2 | 1 | 34.0 |
| 2 | 24.4 | 2 | 6.5 |
| 3 | 41.2 | 3 | 0.4 |
| 4 | 21.7 | 4 | 0 |
| 5 | 6.2 | 5 | 0 |
| 6 | 1.0 | 6 | 0 |
| 7 | 0.07 | 7 | 0 |
Arginine, lysine, and histidine.
Aspartic acid and glutamic acid.
Basic residues are located at specific positions.
Figure 4A shows that acidic residues, when present, were mainly found at position 1. The remaining acidic residues were principally observed at positions 8, 12, and 27. Basic residues were most frequently observed at positions 1, 3, 11, 14, 15, 25, and 27 (Fig. 4B). Interestingly, positions 3, 11, 14, 15, and 25 were almost never occupied by acidic residues, whereas they correspond to variable positions of HVR1 (see above). Moreover, 51, 77, 31, 13, 38, and 46% of HVR1 sequences had a basic residue at positions 3, 11, 14, 15, 25, and 27, respectively. These positions are involved in the consensus basic patterns observed among the 1,382 EMBL HVR1 sequences (Fig. 4C). Although 311 different basic patterns were encountered in the 1,382 sequences, a small set of only 14 basic patterns accounted for 46% of sequences (Fig. 4C), and the five most frequent patterns accounted for 28%. In addition, most of the remaining basic patterns were close to the 14 most frequent and could thus be considered as variants of these patterns. Taken together, these results clearly indicate that both basic residues and basic patterns are conserved within the HVR1 sequence, arguing for positive selection of positively charged residues in HVR1 and their involvement in a biological function.
FIG. 4.
Frequency of acidic and basic residues and conserved basic patterns in the 1,382 HVR1 sequences from EMBL. The frequency of the residues at each position was calculated by dividing the number of observed acidic or basic residues by the total number of examined sequences (n = 1,382). (A) Frequency of acidic residues. (B) Frequency of basic residues. (C) Fourteen most common basic patterns among the 1,382 HVR1 sequences from EMBL. Basic residues (Arg, Lys, and His) are represented by a B, while nonbasic residues are represented by a hyphen. The observed frequency of each pattern is given in the left-hand column.
Role of HVR1 on the basis of conserved structural features.
Two complementary approaches–a prospective study of HVR1 quasispecies evolution under strong pressure towards change and a retrospective analysis of the largest available number of nonredundant HVR1 sequences–were used to assess the possible biological role of HVR1 on the basis of conserved structural features. Comparison of 460 quasispecies sequences and 1,382 unrelated HVR1 sequences from the EMBL database showed that amino acid changes in this region are strongly constrained by a well-ordered structure that tolerates amino acid substitutions as long as the chemicophysical properties of the residues are conserved at specific positions. In addition, although the frequency of basic residues in HVR1 was not statistically different from that of the rest of HCV E2 (30), HVR1 appears to be an intrinsic basic stretch with positively charged residues located at specific positions determining specific basic motifs despite the apparent hypervariability of residues. The principal structural characteristics of HVR1 are summarized in Fig. 5.
FIG. 5.
Summary of HVR1 structural analysis and predicted amino acid functions. The HVR1 consensus hydropathic pattern was established from the 1,382 HVR1 sequences from EMBL. o, n, i, and v, mean hydrophobic, neutral, hydrophilic, and variable positions, respectively; G is a fully conserved Gly residue. The black boxes indicate the main anchoring regions, and the gray box indicates a putative turn. The plus signs indicate positions often occupied by basic residues.
Experimental structural analysis of synthetic HVR1 peptides by circular dichroism and nuclear magnetic resonance showed no stable folding of HVR1 alone (R. Montserret and F. Penin, unpublished data). Therefore, the stabilization of HVR1 conformation depends on its interactions with the rest of the E2 glycoprotein. Conservation of the HVR1 conformation is supported by conservation of the hydropathic characters of amino acids at specific positions and the full conservation of predicted antigenicity profiles. We were able to define a consensus HVR1 hydropathic pattern distinguishing truly variable HVR1 positions from positions exhibiting conserved hydropathic characters despite amino acid variability. In folded proteins, hydrophobic residues are generally oriented within the hydrophobic core of the protein and participate in maintaining protein-folding stability (15). In contrast, hydrophilic residues are generally located at the protein surface and can be involved in molecular interactions. Our results suggest that HVR1 positions that bear hydrophobic and neutral residues likely play a role both in HVR1 anchorage to the E2 glycoprotein core and in maintaining the HVR1 conformation. In contrast, variable positions and purely hydrophilic positions are probably accessible at the E2 glycoprotein surface and are involved in molecular interactions (Fig. 5). This type of organization is similar to that of immunoglobulins and T-cell receptor variable domains. Indeed, these latter can have very different sequences but a highly conserved conformation, as shown by 3D structural analysis (11, 22). Furthermore, crystal structure analyses of immunoglobulins have shown that poorly variable regions are framework regions, whereas variable regions form the antigen binding sites (33). Thus, the amino acid residues located at HVR1 truly variable positions are very likely involved both in HVR1 antigenicity and in molecular interactions.
The two viral glycoproteins E1 and E2 are considered to be the major components of the viral envelope and form heterodimers (17). These heterodimers are probably involved in the interaction between HCV and molecules acting as receptors at the target cell surface. The molecules involved in HCV entry and the mechanisms of cell infection are unknown. It was recently shown that a truncated form of E2 can bind in vitro to the tetraspanin/CD81 molecule, a ubiquitous human cell surface molecule (20, 38). The CD81 molecule could therefore be involved in viral attachment, but productive infection has not yet been obtained through this interaction. It has also been shown that HCV associates with beta-lipoprotein (47) and has been proposed that the low-density lipoprotein (LDL) receptor could mediate internalization of HCV particles covered with LDL and very-low-density lipoprotein leading to an infectious cycle (2). Again, productive infection has not yet been obtained after internalization through this pathway. Finally, it was recently suggested that HCV could interact with GAGs at the cell surface (45, 49). GAGs are unbranched polysaccharides ubiquitously present at the cell surface, acquiring a net negative charge through N and O sulfatation (25). Numerous viruses have been shown to interact with GAGs (e.g., heparan sulfate) in an early step of virus-receptor interaction. This is the case of flaviviruses such as dengue virus (10) and pestiviruses such as classical swine fever virus (23), both of which are members of the Flaviviridae family. An early interaction between HCV and cell surface GAGs could therefore permit or facilitate viral attachment to target cells.
We found that basic residues are frequent in HVR1 sequences, whereas acidic residues are rare, indicating that positively charged HVR1 variants are positively selected. Moreover, not only the global HVR1 conformation but also the number of basic residues and their location throughout the HVR1 sequence are conserved, suggesting that basic residues play an important role in HVR1 function. It is possible that HVR1 basic residues could be involved in interactions with negatively charged groups borne by proteins or phospholipids present at the cell surface or in LDL. Alternatively, HVR1 basic residues could be involved in molecular interactions with negatively charged molecules, such as GAGs, present at the cell surface. This is in agreement with the fact that heparin-binding domains are located in protein regions rich in positively charged residues that can exhibit specific consensus structural motifs (7, 28). Other basic regions of the viral envelope proteins could also be involved in such interactions with GAGs in association with HVR1 (49). Further work is in progress to test HVR1-GAG interaction using isolated E1-E2 glycoproteins deleted or not for the HVR1 sequence.
The existence of extrahepatic sites of HCV replication remains controversial (27). It is, however, supported by the observed compartmentalization of HCV quasispecies variants in the liver, peripheral blood, and various peripheral blood mononuclear cell subsets in a given patient (1, 6, 27, 32). Typically, HVR1 sequences are not randomly distributed among the different cellular compartments, including the liver and various peripheral blood mononuclear cell subsets (1, 6, 27, 32), and the basic HVR1 motifs differ among different compartments (1). This suggests that HVR1 might be involved in selective cell recognition. At the cell surface, the specificity of the interaction cannot be ascribed to putative receptor molecules such as CD81 and the LDL receptor, which are ubiquitously distributed. In contrast, GAGs exhibit significant cellular specificity. Interaction of HVR1 basic residues with GAGs at the surface of target cells equipped with the appropriate receptor molecules might therefore play a role in the tropism of HCV variants in vivo. HVR1 hypervariability might thus not only result from host immune pressure but might also allow HCV to adapt to various cell phenotypes in a given host.
In conclusion, the present evolutionary and chemicophysical study clearly indicates that the conformation of hypervariable region 1 of HCV envelope glycoprotein E2 is well conserved, pointing to a biological role in the virus life cycle. Conservation of positively charged amino acid residues at specific positions further suggests that HVR1 likely interacts with negatively charged compounds, such as lipids, proteins, and GAGs, and might be involved in target cell recognition and virus attachment.
Acknowledgments
We thank Daniel Dhumeaux for providing patients' samples and Alexandre Soulier for excellent technical assistance in quasispecies studies. We are also grateful to Jean Dubuisson and Geneviève Inchauspé for their critical reviews of the manuscript.
This work was supported by the Centre National pour la Recherche Scientifique, EU grant QLK2-1999-00356, and grant 1178 from the Association pour la Recherche sur le Cancer (F.P., C.C., and G.D.), by grant AOM 96–136 from the Programme Hospitalier de Recherche Clinique (G.G., P.-O.F., and J.-M.P.), and the Réseau National Hépatite of the French Ministry for Education, Research, and Technology.
REFERENCES
- 1.Afonso A M, Jiang J, Penin F, Tareau C, Samuel D, Petit M A, Bismuth H, Dussaix E, Feray C. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J Virol. 1999;73:9213–9221. doi: 10.1128/jvi.73.11.9213-9221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agnello V, Abel G, Elfahal M, Knight G B, Zhang Q-X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alter H J. To C or not to C: these are the questions. Blood. 1995;85:1681–1695. [PubMed] [Google Scholar]
- 4.Black S D, Mould D R. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal Biochem. 1991;193:72–82. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- 5.Blanchet C, Combet C, Geourjon C, Deléage G. MPSA: integrated system for multiple protein sequence analysis with client/server capabilities. Bioinformatics. 2000;16:286–287. doi: 10.1093/bioinformatics/16.3.286. [DOI] [PubMed] [Google Scholar]
- 6.Cabot B, Esteban J I, Martell M, Genesca J, Vargas V, Esteban R, Guardia J, Gomez J. Structure of replicating hepatitis C virus (HCV) quasispecies in the liver may not be reflected by analysis of circulating HCV virions. J Virol. 1997;71:1732–1734. doi: 10.1128/jvi.71.2.1732-1734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardin A D, Weintraub H J R. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 8.Casino C, McAllister J, Davidson F, Power J, Lawlor E, Yap P L, Simmonds P, Smith D B. Variation of hepatitis C virus following serial transmission: multiple mechanisms of diversification of the hypervariable region and evidence for convergent genome evolution. J Gen Virol. 1999;80:717–725. doi: 10.1099/0022-1317-80-3-717. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain R W, Adams N J, Taylor L A, Simmonds P, Elliott R M. The complete coding sequence of hepatitis C virus genotype 5a, the predominant genotype in South Africa. Biochem Biophys Res Commun. 1997;236:44–49. doi: 10.1006/bbrc.1997.6902. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 11.Chothia C, Gelfand I, Kister A. Structural determinants in the sequences of immunoglobulin variable domain. J Mol Biol. 1998;278:457–479. doi: 10.1006/jmbi.1998.1653. [DOI] [PubMed] [Google Scholar]
- 12.Chou P Y, Fasman G D. Beta-turns in proteins. J Mol Biol. 1977;115:135–175. doi: 10.1016/0022-2836(77)90094-8. [DOI] [PubMed] [Google Scholar]
- 13.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 14.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 15.Darby N J, Creighton T E, editors. Protein structure. Oxford, United Kingdom: IRL Press; 1993. pp. 1–41. [Google Scholar]
- 16.Deleage G, Combet C, Blanchet C, Geourjon C. ANTHEPROT: an integrated protein sequence analysis software with client/server capabilities. Comput Biol Med. 2001;31:259–267. doi: 10.1016/s0010-4825(01)00008-7. [DOI] [PubMed] [Google Scholar]
- 17.Dubuisson J. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr Top Microbiol Immunol. 2000;242:135–148. doi: 10.1007/978-3-642-59605-6_7. [DOI] [PubMed] [Google Scholar]
- 18.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Eagle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint M, McKeating J A. The role of the hepatitis C virus glycoproteins in infection. Rev Med Virol. 2000;10:101–117. doi: 10.1002/(sici)1099-1654(200003/04)10:2<101::aid-rmv268>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 21.Forns X, Thimme R, Govindarajan S, Emerson S U, Purcell R H, Chisari F V, Bukh J. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc Natl Acad Sci USA. 2000;97:13318–13323. doi: 10.1073/pnas.230453597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia K C, Teyton L, Wilson I A. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 23.Hulst M M, van Gennip H G, Moormann R J. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein Erns. J Virol. 2000;74:9553–9561. doi: 10.1128/jvi.74.20.9553-9561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Lerat H, Rumin S, Habersetzer F, Berby F, Trabaud M A, Trepo C, Inchauspe G. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood. 1998;91:3841–3849. [PubMed] [Google Scholar]
- 28.Margalit S, Fischer N, Ben-Sasson S A. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J Biol Chem. 1993;268:19228–19231. [PubMed] [Google Scholar]
- 29.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAllister J, Casino C, Davidson F, Power J, Lawlor E, Yap P L, Simmonds P, Smith D B. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J Virol. 1998;72:4893–4905. doi: 10.1128/jvi.72.6.4893-4905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 32.Navas S, Martin J, Quiroga J A, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–1646. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padlan E A, Abergel C, Tipper J P. Identification of specificity-determining residues in antibodies. FASEB J. 1995;9:133–139. doi: 10.1096/fasebj.9.1.7821752. [DOI] [PubMed] [Google Scholar]
- 34.Parker J M, Guo D, Hodges R S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 35.Pawlotsky J M, Germanidis G, Frainais P O, Bouvier M, Soulier A, Pellerin M, Dhumeaux D. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J Virol. 1999;73:6490–6499. doi: 10.1128/jvi.73.8.6490-6499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pawlotsky J M, Roudot-Thoraval F, Bastie A, Darthuy F, Remire J, Metreau J M, Zafrani E S, Duval J, Dhumeaux D. Factors affecting treatment responses to interferon-alpha in chronic hepatitis C. J Infect Dis. 1996;174:1–7. doi: 10.1093/infdis/174.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 39.Poynard T, Marcellin P, Lee S S, Niederau C, Minuk G S, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, Albrecht J. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon α2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group. Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 40.Ray S C, Mao Q, Lanford R E, Bassett S, Laeyendecker O, Wang Y M, Thomas D L. Hypervariable region 1 sequence stability during hepatitis C virus replication in chimpanzees. J Virol. 2000;74:3058–3066. doi: 10.1128/jvi.74.7.3058-3066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, Gojobori T, Maertens G, Mizokami M, Nainan O, Netesov S, Nishioka K, Shini T, Simmonds P, Smith D, Stuyver L, Weiner A. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch Virol. 1998;143:2493–2503. doi: 10.1007/s007050050479. [DOI] [PubMed] [Google Scholar]
- 42.Sekiya H, Kato N, Ootsuyama Y, Nakazawa T, Yamauchi K, Shimotohno K. Genetic alterations of the putative envelope proteins encoding region of the hepatitis C virus in the progression to relapsed phase from acute hepatitis: humoral immune response to hypervariable region 1. Int J Cancer. 1994;57:664–670. doi: 10.1002/ijc.2910570509. [DOI] [PubMed] [Google Scholar]
- 43.Sherman K E, Andreatta C, O'Brien J, Gutierrez A, Harris R. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology. 1996;23:688–694. doi: 10.1002/hep.510230405. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H, Yoshikura H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 45.Takikawa S, Ishii K, Aizaki H, Suzuki T, Asakura H, Matsuura Y, Miyamura T. Cell fusion activity of hepatitis C virus envelope proteins. J Virol. 2000;74:5066–5074. doi: 10.1128/jvi.74.11.5066-5074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomssen R, Bonk S, Thiele A. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med Microbiol Immunol. 1993;182:329–334. doi: 10.1007/BF00191948. [DOI] [PubMed] [Google Scholar]
- 48.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- 49.Yagnik A T, Lahm A, Meola A, Roccasecca R M, Ercole B B, Nicosia A, Tramontano A. A model for the hepatitis C virus envelope glycoprotein E2. Proteins. 2000;40:355–366. doi: 10.1002/1097-0134(20000815)40:3<355::aid-prot20>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y H, Shimizu Y K, Esumi M. Monoclonal antibodies to the hypervariable region 1 of hepatitis C virus capture virus and inhibit virus adsorption to susceptible cells in vitro. Virology. 2000;269:276–283. doi: 10.1006/viro.2000.0227. [DOI] [PubMed] [Google Scholar]
- 51.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]