Abstract
Background
Kidney biopsy is the standard of care for the diagnosis of various kidney diseases. In particular, chronic histopathologic lesions, such as interstitial fibrosis and tubular atrophy, can provide prognostic information regarding chronic kidney disease progression. In this study, we aimed to evaluate historadiological correlations between CT-based radiomic features and chronic histologic changes in native kidney biopsies and to construct and validate a radiomics-based prediction model for chronicity grade.
Methods
We included patients aged ≥ 18 years who underwent kidney biopsy and abdominal CT scan within a week before kidney biopsy. Left kidneys were three-dimensionally segmented using a deep learning model based on the 3D Swin UNEt Transformers architecture. We additionally defined isovolumic cortical regions of interest near the lower pole of the left kidneys. Shape, first-order, and high-order texture features were extracted after resampling and kernel normalization. Correlations and diagnostic metrics between extracted features and chronic histologic lesions were examined. A machine learning-based radiomic prediction model for moderate chronicity was developed and compared according to the segmented regions of interest (ROI).
Results
Overall, moderate correlations with statistical significance (P < 0.05) were found between chronic histopathologic grade and top-ranked radiomic features. Total parenchymal features were more strongly correlated than cortical ROI features, and texture features were more highly ranked. However, conventional imaging markers, including kidney length, were poorly correlated. Top-ranked individual radiomic features had areas under receiver operating characteristic curves (AUCs) of 0.65 to 0.74. Developed radiomics models for moderate-to-severe chronicity achieved AUCs of 0.89 (95% confidence interval [CI] 0.75–0.99) and 0.74 (95% CI 0.52–0.93) for total parenchymal and cortical ROI features, respectively.
Conclusion
Significant historadiological correlations were identified between CT-based radiomic features and chronic histologic changes in native kidney biopsies. Our findings underscore the potential of CT-based radiomic features and their prediction model for the non-invasive assessment of kidney fibrosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12880-024-01434-x.
Keywords: Radiomics, Kidney fibrosis, Histopathology, Chronic kidney disease
Introduction
Kidney biopsy is an essential tool for the diagnosis and etiological evaluation of diverse kidney diseases by detecting histological alterations. In particular, chronic histological changes such as interstitial fibrosis (IF), tubular atrophy (TA), glomerulosclerosis (GS), and arteriosclerosis are key findings for pathologic classification and severity grading of different kidney diseases, including IgA nephropathy, diabetic nephropathy, and lupus nephritis [1–3]. These chronic lesions are considered a common pathway of chronic kidney disease (CKD), and strongly predict kidney outcomes irrespective of etiology [4]. The semi-quantitative scoring of chronic lesions could provide additional prognostic information that complements glomerular filtration rate (GFR) and proteinuria [5]. Nonetheless, kidney biopsy is an invasive procedure associated with post-biopsy bleeding requiring transfusion or intervention [6]. Furthermore, patients with a solitary or small kidney are not eligible for kidney biopsy. Thus, kidney biopsy is performed selectively based on consideration of risk-benefit balance. On the other hand, non-invasive biomarkers are highly disease-specific and cannot be used as alternatives to kidney biopsy [7, 8]. Advanced imaging techniques, such as multi-parametric magnetic resonance imaging (MRI) and ultrasound (US) elastography, are particularly sensitive for IF [9, 10], but as yet, many of these techniques are unvalidated and lack standardization for the clinical use.
Recently, radiomics has been extensively studied due to its ability to quantify pathophysiological signals from medical images. In particular, radiomic analysis has been shown to have potential utility in the diagnosis and prognosis of various cancers [11–13]. A large proportion of radiomic features consist of high-order texture features that possibly reflect microstructural characteristics not completely detected or quantified by human eyes. Radiomics can bridge the gap between the macroscopic findings of medical images and micrometer-sized histological findings and further molecular phenotypes [14]. Indeed, close relationships have been widely reported between tumor histology and radiomic features [15–17]. Radiomic texture features are also highly related to fibrosis of other organs, including liver, lung, and intestine [18–20]. However, radiomic studies on kidney histopathologic findings are lacking. Relatively few studies have linked radiomic features of kidney imaging to pathologic findings, and most are based on kidney US [21]. Ultrasound-based images are quite operator-dependent, whereas CT images are more reproducible and appropriate for data-driven quantitative analysis. Furthermore, CT examinations are less costly than MRI and enable extensive data acquisition in routine practice. Given the increasing use of CT in clinical practice, historadiological correlations based on CT-derived radiomic features might be utilized to construct a non-invasive diagnostic tool for kidney diseases. In this study, we aimed to explore the association between CT-based radiomic features and chronic lesions in native kidney biopsies and assess the clinical implications of radiomic features and prediction models in kidney histopathological evaluation.
Methods
Study participants
We retrospectively recruited adult patients ≥ 18 years of age who underwent native kidney biopsy from October 2018 to December 2023 and underwent an abdomen CT scan at Keimyung University Hospital within the 7 days prior to kidney biopsy. Patients who underwent a CT scan immediately after kidney biopsy were not included because post-biopsy bleeding was present in some of these patients. Cases with poor image quality, artifacts, or fewer than 10 glomeruli in their biopsy specimen were excluded [22]. Image quality was assessed by two radiologists, and CT scans with a slice thickness of ≥ 5 mm were excluded. The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Keimyung University Hospital (IRB No. 2023-01-021). The requirement for informed consent was waived due to the retrospective nature of the study. A flow diagram of the study subjects is provided in Supplementary Fig. 1.
CT image acquisition and data preprocessing
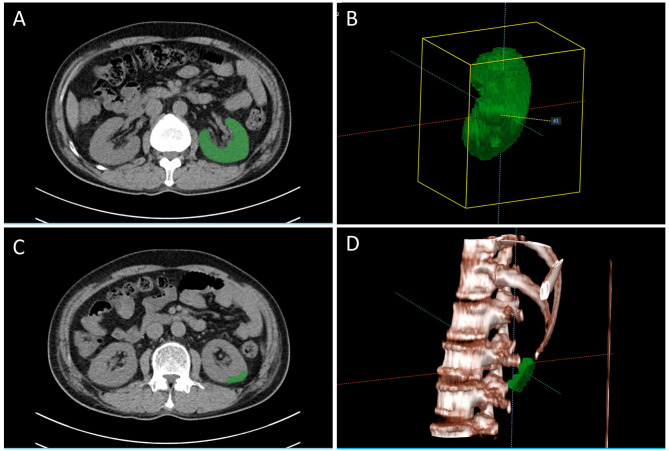
All the participants received non-contrast abdominal CT scans according to the institution’s standard protocol with different types of CT scanners (SOMATOM Definition Edge, SOMATOM Force, and SOMATOM Drive manufactured by Siemens). We only included CT scans with a slice thickness of 3 mm. Further information about CT image acquisition is provided in Supplementary Table 1. CT images were resampled to a spatial resolution of 1 mm x 1 mm x 1 mm and reconstruction kernels were normalized [23, 24]. The target labels used for segmentation models were manually generated by two radiologists with 10 and 11 years of experience in abdominal radiology, blinded to clinical information. Manual segmentation was performed using a semi-automated software (AVIEW Research; Coreline Soft, Seoul, South Korea) and modified and confirmed by consensus between the radiologists. In addition to the total kidney mask, we further defined isovolumic regions of interest (ROI) selected with a volume of 5,000 mm3 in the cortical region near the inferior pole of the left kidney (Fig. 1).
Fig. 1.
Three-dimensional masking processes. (A) A representative axial slice of a CT image and its corresponding mask. (B) 3D reconstruction image of a resampled mask. (C) An example of an isovolumic mask of a cortical ROI. (D) 3D reconstruction image of a cortical ROI mask
Histological assessment
All included participants underwent kidney biopsy for the histological diagnosis of kidney diseases in routine practice. Kidney biopsy was performed under ultrasound guidance using a 16-gauge core needle by experienced nephrologists. Biopsy specimens were obtained from the lower pole of the left kidney. Histological evaluations were done by light microscopy using hematoxylin and eosin, periodic acid-Schiff, Masson trichrome, and periodic acid methenamine silver staining, immunofluorescence microscopy, and electron microscopy. All biopsies were retrospectively reviewed and scored by two experienced kidney pathologists. Histological findings were graded by semi-quantitative scoring. IF, TA, and GS were graded as 0 (absence or minimal, < 10%), 1 (mild, 10–25%), 2 (moderate, 26–50%), or 3 (severe, > 50%), and arterial intimal thickening (IT) was graded as 0 (absence) and 1 (presence). Total chronicity score (CS) was calculated using histological scores for IF, TA, GS, and IT ranging from 0 to 10 and graded as minimal (0–1), mild (2–4), moderate (5–7), or severe (≥ 8) [4].
Segmentation model
A 3D-based transformer model termed Swin UNEt TRansformers (Swin UNETR) was employed for the segmentation task. SwinUNETR integrates the strengths of the Swin Transformer architecture with the UNet-like encoder-decoder structure, offering powerful context modeling and enhanced spatial understanding [25]. The hierarchical representation and shifted window mechanisms inherent in SwinUNETR enable effective capture of both global and local features, which are critical for precise organ segmentation in medical imaging. Three-dimensional segmentation of left kidneys was performed by fine-tuning a SwinUNETR model pre-trained on 5 publicly available datasets with a total of 5,050 CT images. This model was validated for different abdominal organs including both kidneys. Fine-tuning was done using the AdamW optimizer with an initial learning rate of 0.0005 and a weight decay of 0.01 for 15 epochs with a batch size of 2. We used 75 cases from the dataset to implement fine-tuning and split the remaining 20 into 10 each for the validation and test sets. The sub-volumes of 96 × 96 × 96 voxels cropped from each sample were fed into the model. Since only the left kidney was selected, flipping and rotation were not used in the transformation. Our final dice similarity coefficient for the hold-out test was 0.962.
Radiomic feature extraction
Several types of radiomic features, including shape, first-order, and high-order texture features, were extracted using PyRadiomics package (version 3.1.0; https://pyradiomics.readthedocs.io, accessed on 30 November 2023). Extracted radiomic feature categories were shape, first-order, gray level co-occurrence matrix (GLCM), gray level size zone matrix (GLSZM), gray level run length matrix (GLRLM), neighbouring gray tone difference matrix (NGTDM), and gray level dependence matrix (GLDM) features [26]. A detailed description of each radiomic feature is provided in Supplementary Table 2. Extracted radiomic features with interobserver intraclass correlation coefficients (ICCs) < 0.80 were excluded from the analysis (Supplementary Table 3). ICCs were calculated using a two-way random effects model with absolute agreement, using based on the radiomic features of kidney segmentations performed by the two radiologists on 30 randomly selected cases [27]. In addition, voxel-based feature maps were generated to visualize highly correlated radiomic features.
Prediction model construction
We constructed a prediction model for moderate to severe chronicity using extracted radiomic features. The entire dataset was split into 70% training and 30% test set in a stratified manner. To obtain an optimal feature subset, we first used the minimum redundancy maximum relevance (MRMR) algorithm [28], and then implemented recursive feature elimination (RFE) based on random forest with cross-validation. The light gradient boosting machine (light GBM) algorithm (29) was used to build the predictive model [29].Hyperparameter tuning was done using 5-fold cross-validation and a grid search. Feature importance plots were generated based on the SHapley Additive exPlanation (SHAP) [30].
Statistical analysis
Participant characteristics are presented as medians and interquartile ranges for continuous variables and as counts and percentages for categorical variables. Correlations between radiomic features and histopathologic grades were determined using Spearman’s correlation coefficients. Correlation matrix plots of radiomic features were also depicted based on hierarchical clustering. Box and whisker plots and the Mann-Whitney-Wilcoxon test were used to assess differences according to histopathologic grades for highly correlated radiomic features. Receiver operating characteristic curve (ROC) analysis for single radiomic features was performed to distinguish between moderate to severe lesions and absence to mild lesions (< 2 vs. ≥ 2 scores for IF, TA, GS, and IT, and < 8 vs. ≥8 scores for CS). Performance metrics for developed machine learning prediction models include sensitivity, specificity, F1 score, accuracy, and AUC. The 95% confidence intervals for accuracy, sensitivity, and specificity were calculated using Wilson’s method, while those for F1 score and AUC were calculated using the bootstrap method [31]. P-values of < 0.05 were considered statistically significant. All analyses were performed using Python software (Python Software Foundation, version 3.8.0) and R software version 4.1.2. (R Foundation for Statistical Computing, Vienna, Austria). For the training and evaluation of deep learning models for 3D segmentation, the Medical Open Networks for AI (MONAI) framework developed by NVIDIA was used. The deep learning experiments were conducted using CUDA 12.1 and PyTorch version 2.4.0. The hardware setup included a 13th Gen Intel Core i9-13900 H CPU at 2.60 GHz, 128 GB of RAM, and an RTX 3090 GPU.
Results
Characteristics of participants
A total of ninety-five patients were included in the analysis. Participant characteristics are provided in Tables 1; 55.8% were male, median age was 52 years, and 26.3% were diabetic patients. Mean estimated glomerular filtration rate (eGFR) was 67.8 ml/min/1.73m2, and 43 patients (45.2%) had an eGFR of < 60 ml/min/1.73m2. Median time between CT scans and kidney biopsy was 1 day (range: 0–6 days). Pathological diagnoses included IgA nephropathy, focal segmental glomerulosclerosis, diabetic nephropathy, ANCA-associated vasculitis, minimal change disease, membranous nephropathy, hypertensive nephrosclerosis, and tubulointerstitial nephritis.
Table 1.
Characteristics of the participants
| Variables (n = 95) | |
|---|---|
| Age (yr), median [IQR] | 52.0 [37.5–62.5] |
| Male sex (%) | 53 (55.8) |
| Comorbidities | |
| Hypertension | 55 (57.9) |
| Diabetes | 25 (26.3) |
| Estimated GFR (ml/min/1.73m2)†, median [IQR] | 67.8 [30.2–91.0] |
| 90–120 ml/min/1.73m2 | 25 (26.3) |
| 60–90 ml/min/1.73m2 | 27 (28.4) |
| 30–60 ml/min/1.73m2 | 19 (20.0) |
| < 30 ml/min/1.73m2 | 24 (25.2) |
| Time interval between CT scan and kidney biopsy (days), median [IQR] | 0 [0–1] |
| Pathological diagnosis | |
| IgA nephropathy | 29 (30.5) |
| Focal segmental glomerulosclerosis | 10 (10.5) |
| Diabetic nephropathy | 9 (9.5) |
| ANCA-associated vasculitis | 7 (7.4) |
| Minimal change disease | 5 (5.3) |
| Membranous nephropathy | 5 (5.3) |
| Hypertensive nephrosclerosis | 5 (5.3) |
| Tubulointerstitial nephritis | 2 (2.1) |
| Others | 23 (24.2) |
| Histological findings | |
| Glomerulosclerosis | |
| < 10% | 27 (28.4) |
| 10–25% | 24 (25.3) |
| 26–50% | 26 (27.4) |
| > 50% | 18 (18.9) |
| Interstitial fibrosis | |
| < 10% | 33 (34.7) |
| 10–25% | 44 (46.3) |
| 26–50% | 12 (12.6) |
| > 50% | 6 (6.3) |
| Tubular atrophy | |
| < 10% | 38 (40.0) |
| 10–25% | 38 (40.0) |
| 26–50% | 14 (14.7) |
| > 50% | 5 (5.3) |
| Chronicity index | |
| Minimal (0–1) | 24 (25.3) |
| Mild (2–4) | 34 (35.8) |
| Moderate (5–7) | 29 (30.5) |
| Severe (≥ 8) | 8 (8.4) |
†eGFR is based on serum creatinine measured on the day of kidney biopsy
Correlation analysis
A total of 1,226 radiomic features were included in the analysis (17 shape features, 236 first-order features, and 973 high-order texture features, which included NGTDM, GLDM, GLSZM, GLCM, and GLRLM). Extracted radiomic features displayed several cluster groups in a correlation matrix plot, which was more pronounced for total parenchymal features (Supplementary Fig. 2). The top 5 highly correlated radiomic features showed moderate correlations with chronic histopathologic scores (Table 2). Overall, radiomic features extracted from total parenchyma showed higher correlations with chronic histology than those extracted from cortical ROI: correlation coefficients ranged from 0.36 to 0.42 for total parenchyma and from 0.24 to 0.38 for cortical ROI. Of note, the majority of highly correlated feature types were higher-order texture features for total parenchyma but first-order features for cortical ROI. Correlations of total parenchymal radiomic features with CS were stronger than those with IF, TA, and GS. On the other hand, the correlation between chronic lesions and kidney length was relatively poor. The correlation coefficients for major axis length ranged from − 0.25 to -0.10 (data not shown). Also, kidney volume (mesh volume) was weakly correlated with IF, TA, and CS (correlation coefficient: -0.20, -0.17, and − 0.28, respectively). In addition, a relationship between the top 5 features and histopathologic grades was identified using box and whisker plots (Supplementary Figs. 3 and 4). Of the highly ranked features, several GLCM and GLRLM features showed significant increasing trends as histopathologic grades advanced. Voxel-based feature maps for selected features were produced to better understand the spatial distributions of these features (Supplementary Fig. 5). As the chronicity grade increased, more high-intensity voxels were distributed within parenchyma with particularly bright signals at outer kidney margins.
Table 2.
Top five radiomic features ranked according to correlations with chronic histopathologic grades
| Kidney parenchyme | Correlation coefficient (95% CI) | P-value | Cortical sample | Correlation coefficient (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| Interstitial fibrosis | GLCM MCC (original) | 0.38 (0.20 to 0.54) | < 0.001 | GLRLM SRLGLE (HHH-wavelet) | 0.38 (0.20 to 0.54) | < 0.001 |
| GLRLM SRE (LoG 1.0) | 0.38 (0.20 to 0.54) | < 0.001 | First-order RMAD (LoG 3.0) | -0.32 (-0.49 to -0.12) | < 0.001 | |
| GLCM SS (HLH-wavelet) | -0.37 (-0.53 to -0.18) | < 0.001 | GLDM DV (HHL-wavelet) | -0.31 (-0.48 to -0.12) | 0.002 | |
| GLCM lmc2 (original) | 0.37 (0.18 to 0.53) | < 0.001 | GLDM DE (HHL-wavelet) | -0.31 (-0.48 to -0.11) | 0.003 | |
| GLRLM SRE (HHH-wavelet) | 0.36 (0.17 to 0.53) | < 0.001 | First-order IQR (LoG 3.0) | -0.31 (-0.48 to -0.11) | 0.003 | |
| Tubular atrophy | GLCM MCC (original) | 0.39 (0.21 to 0.55) | < 0.001 | GLRLM SRLGLE (HHH-wavelet) | 0.33 (0.14 to 0.50) | 0.001 |
| GLCM lmc2 (original) | 0.37 (0.18 to 0.53) | < 0.001 | First-order 90 percentile (LLL-wavelet) | 0.31 (0.12 to 0.48) | 0.002 | |
| GLCM lmc1 (original) | -0.37 (-0.53 to -0.18) | < 0.001 | First-order 90 percentile (original) | 0.31 (0.12 to 0.48) | 0.002 | |
| GLCM Correlation (original) | 0.36 (0.18 to 0.53) | < 0.001 | GLCM Idmn (LLL-wavelet) | 0.30 (0.10 to 0.47) | 0.003 | |
| GLRLM SRE (LoG 1.0) | 0.36 (0.17 to 0.53) | < 0.001 | First-order median (original) | 0.28 (0.08 to 0.45) | 0.007 | |
| Glomerulosclerosis | GLSZM LALGLE (LHL-wavelet) | -0.39 (-0.55 to -0.21) | < 0.001 | First-order Kurtosis (LoG 4.0) | 0.31 (0.12 to 0.48) | 0.002 |
| GLDM DV (LoG 2.0) | 0.39 (0.20 to 0.55) | < 0.001 | GLRLM SRLGLE (LoG 2.0) | 0.29 (0.09 to 0.46) | 0.004 | |
| GLRLM SRE (LoG 2.0) | 0.38 (0.20 to 0.54) | < 0.001 | GLSZM SZNU (LHL-wavelet) | 0.26 (0.06 to 0.44) | 0.01 | |
| GLRLM RLNU (LLH-wavelet) | -0.38 (-0.54 to -0.19) | < 0.001 | First-order RMAD (LoG 3.0) | -0.24 (-0.42 to -0.04) | 0.02 | |
| NGTDM Coarseness (LHL-wavelet) | 0.37 (0.18 to 0.53) | < 0.001 | First-order MAD (LoG 3.0) | -0.24 (-0.42 to -0.04) | 0.02 | |
| Chronicity index | GLRLM SRE (LoG 1.0) | 0.42 (0.24 to 0.58) | < 0.001 | GLRLM SRLGLE (HHH-wavelet) | 0.33 (0.14 to 0.50) | 0.001 |
| GLRLM SRE (HHH-wavelet) | 0.41 (0.23 to 0.56) | < 0.001 | First-order RMAD (LoG 3.0) | -0.32 (-0.49 to -0.13) | 0.002 | |
| GLRLM SRLGLE (HHH-wavelet) | 0.41 (0.23 to 0.56) | < 0.001 | First-order Kurtosis (LoG 4.0) | 0.32 (0.13 to 0.49) | 0.002 | |
| GLRLM SRHGLE (HHH-wavelet) | 0.40 (0.22 to 0.56) | < 0.001 | First-order IQR (LoG 3.0) | -0.31 (-0.48 to -0.11) | 0.002 | |
| GLDM SDE (LoG 1.0) | 0.39 (0.20 to 0.55) | < 0.001 | GLDM DNUN (HHL-wavelet) | 0.28 (0.08 to 0.45) | 0.006 |
MCC, maximal correlation coefficient; SRE, short run emphasis; SS, LALGLE, large area low gray-level emphasis; DV, difference variance; RLNU, gray level nonuniformity; SRLGLE, short run low gray-level emphasis; SRHGLE, short run high gray-level emphasis; SDE, small dependence emphasis; RMAD, ; DE, difference entropy; IQR, interquartile range; SZNU, size zone nonuniformity; MAD, mean absolute deviation; DNUN, dependence nonuniformity normalized
Discriminability of radiomic features
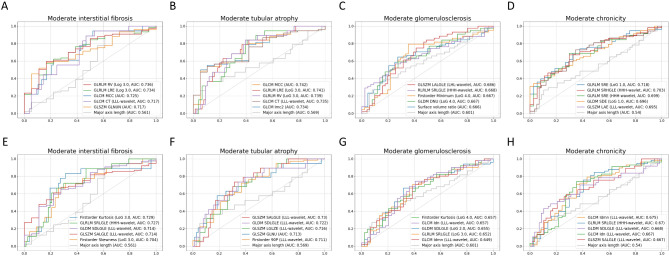
Highly discriminable radiomic features for chronic regions were quite different according to extracted ROI (Fig. 2 and Supplementary Tables 4 and 5). Overall, features extracted from total parenchymal ROI had higher AUCs than those extracted from cortical ROI. The top five total parenchymal features had AUCs of ≥ 0.7 for moderate-to-severe IF, TA, and chronicity grade, but not for GS, while the top 5 cortical ROI features had AUCs ≥ 0.7 only for moderate-to-severe IF and TA. Discriminability for GS was relatively low irrespective of ROI types. Most of the highly ranked total parenchymal features were high-order texture features, but one shape feature (surface-to-volume ratio; SVR) showed the fifth highest AUC for GS. GLRLM run variance and GLCM maximal correlation coefficient were the texture features with the highest AUCs for IF and TA, both achieving a value of 0.74. For cortical ROI features, some first-order features, such as skewness, kurtosis, and 90 percentile, had good discriminability for IF and TA. Several highly ranked features had significantly higher AUCs for moderate-to-severe chronicity grade than major axis length as determined by the Delong test.
Fig. 2.
Receiver operating characteristic curves and areas under the curves of the radiomic features for chronic histopathologic lesions. Total parenchymal features predicting (A) interstitial fibrosis, (B) tubular atrophy, (C) glomerulosclerosis, and (D) chronicity grade. Cortical ROI features predicting (E) interstitial fibrosis, (F) tubular atrophy, (G) glomerulosclerosis, and (H) chronicity grade. The top five features, ranked by AUC, were plotted alongside the conventional marker, major axis length
Prediction model using radiomic features
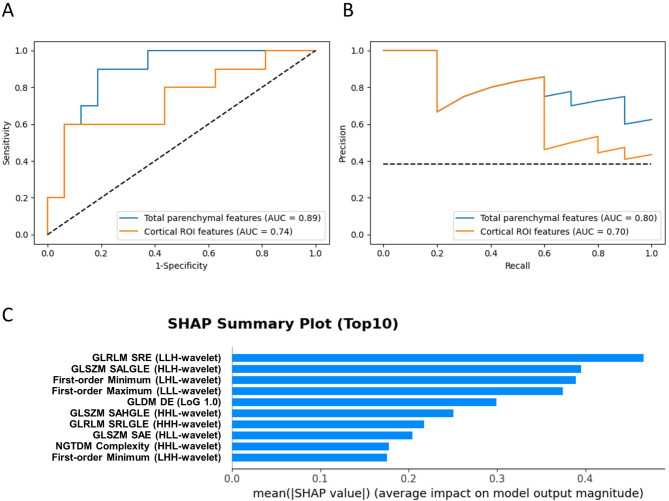
After dimension reduction using MRMR followed by RFE, 22 and 5 features were selected for model construction from total parenchymal and cortical ROIs, respectively. The Light GBM-based prediction model using total parenchymal features had better performance than the model using cortical ROI features (AUC: 0.89 versus 0.74, Table 3; Fig. 3). We evaluated model performance separately on multiple independent test sets and found the consistent metrics (Supplementary Fig. 6). Other diagnostic metrics, such as sensitivity, accuracy, and F1 score were also better in the total parenchymal feature model. Of the selected features, GLRLM and GLSZM features were highly ranked based on the feature importance using SHAP values (Fig. 3). Of note, GLRLM short run emphasis and short run low gray level emphasis identified by correlation analysis were also included in the highly ranked features.
Table 3.
Diagnostic metrics of radiomics models using total parenchymal or cortical ROI features
| Model | Sensitivity (95% CI) | Specificity (95% CI) | Accuracy (95% CI) | F1 score (95% CI) | AUC (95% CI) |
|---|---|---|---|---|---|
| Total parenchymal feature model | 0.90 (0.60–0.98) | 0.81 (0.57–0.93) | 0.85 (0.66–0.94) | 0.82 (0.55–0.95) | 0.89 (0.75–0.99) |
| Cortical ROI feature model | 0.60 (0.31–0.83) | 0.94 (0.72–0.99) | 0.81 (0.62–0.91) | 0.71 (0.33–0.90) | 0.74 (0.52–0.93) |
Fig. 3.
Evaluation metrics and feature importance of the developed radiomics model. (A) Receiver operating characteristic curves and (B) precision-recall curves of the model based on total parenchymal and cortical ROI features. (C) SHAP summary plot for the model developed using total parenchymal features
Discussion
In this study, we presented historadiological correlations between CT-based radiomic features and chronic lesions detected by native kidney biopsy. Several 3D radiomic features correlated well with fibrotic lesions such as IF and TA. Most of these radiomic features primarily consisted of high-order texture features and showed better discriminability for kidney fibrosis than conventional imaging markers. In addition, a machine learning prediction model was constructed for chronicity grade using selected radiomic features. The suggested model had relatively high performance for differentiating moderate-to-severe chronicity grade, which could be useful for diagnostic or prognostic purposes.
Regardless of the underlying disease etiologies, chronic histopathological lesions accompanied by kidney fibrosis serve as a prognostic indicator and guide treatment decision-making [5, 32]. However, semi-quantitative pathological grading is available only through invasive core needle biopsy. Despite the variety of kidney imaging modalities available, limited information is utilized with few quantitative measures. At the tissue level, kidney fibrosis disrupts kidney microstructures by excessive extracellular matrix accumulation [33], which can be linked to the macroscopic appearances of CT images. Our findings indicate that CT-based radiomic texture features possibly reflect histopathological changes related to kidney fibrosis based on the spatial interrelationship between neighboring pixels.
Previous radiomics studies using kidney images have primarily focused on predicting kidney function [21, 34, 35]. However, given kidney biopsy sampling bias and the highly organized structures of kidney compartments, predicting kidney pathology appears to be more complex than predicting kidney function. In earlier sonographic studies, decreased kidney length and cortical thickness have been considered typical quantifiable features of CKD. However, these parameters are only weakly correlated with chronic histologic lesions. Moghazi et al. reported correlation coefficients between kidney length and GS, IF, and TA of only − 0.26, -0.14, and − 0.20, respectively, and even lower correlations for cortical thickness [36]. Cortical echogenicity reportedly is slightly better correlated with IF and TA, but echogenicity is a qualitative measure and subject to operator variability [37, 38]. By contrast, we identified a set of CT-based radiomic texture features that were moderately correlated with chronic lesions. As was expected, the radiomic features best correlated were high-order texture features, such as GLCM, GLRLM, and GLDM. By definition, high-order texture features can effectively incorporate information about biopsy target areas and surrounding lesions. In addition, we identified texture features highly correlated with chronic lesions were intensely distributed around kidney margins in voxel-based radiomic feature maps. Of note, multiple wavelet-transformed radiomic features were included in the top-ranked correlated features. Theoretically, low-pass filtered features can offer coarse textures, while high-pass filtered features offer fine textures. Among cortical ROI features, a number of first-order features were highly correlated with chronic lesions. First-order features, which are calculated from histogram-based signal intensity, may reflect regional cortical intensity, similar to sonographic cortical echogenicity [36]. On the other hand, most shape features showed poor discriminability for chronic lesions; however, SVR had high AUC for moderate to severe GS. This finding is consistent with our previous study, in which SVR showed the highest AUC for eGFR [39].
In the differentiation of moderate-to-severe chronicity, highly ranked single radiomic features exhibited AUCs of ≥ 0.7, whereas sonographically measured kidney length and cortical thickness reportedly have AUCs of around 0.6 [40]. Several radiomic texture features were found to be significantly more diagnostic than major axis length, and constructed radiomics-based machine learning model performed better with an AUC as high as 0.89. We selected cortical ROI to surround lesions likely to undergo kidney biopsy, but the diagnostic value of cortical ROI features was inferior to that of total parenchymal features. These results could be due to the fact that the total parenchymal features can contain all imaging information from the entire kidney, including the medulla, without data loss, and can also convey regional differences of the kidney. Given that corticomedullary differentiation is a typical imaging feature of CKD, radiomic features extracted from total parenchyma can contrast differences in signal intensity or texture across different regions of the kidney.
In recent years, MRI-derived imaging parameters, such as apparent diffusion coefficient in diffusion-weighted imaging and T1 or T2 mapping, have attracted research interest as a promising measure of analyzing kidney textures [10], and significant correlations between these MRI parameters and interstitial fibrosis have been reported [41, 42]. However, most MRI studies were conducted experimentally using a few quantitative parameters from small samples due to high cost. With the exception of kidney tumors, MRI-based radiomics research remains in its infancy. Also, US-based radiomics studies have been conducted to evaluate kidney fibrosis [43, 44]. However, the performance of radiomics models remains insufficient, which suggests additional information, such as clinical information or elastography data, should be incorporated into models [44]. CT-based radiomics models have the advantage of utilizing 3D information such as volume, 3D shape, and 3D texture, which are not available from US-based radiomics data. Moreover, CT images offer quantitative data in Hounsfield units, enabling more reproducible analysis. Nonetheless, few CT-based radiomics studies have attempted to evaluate kidney histopathology. Thus, the current study provides valuable information regarding the characterization and predictive value of CT-based radiomic features in relation to chronic kidney histopathology. If validated, CT-based radiomics models could serve as non-invasive diagnostic and/or prognostic tools when biopsy is contraindicated. Moreover, given that repeat or follow-up kidney biopsy is rarely performed due to possible complications, CT-based radiomics could enable non-invasive histopathology monitoring.
Our study has several limitations that require consideration. We analyzed data with small sample size obtained from a single institution. Nevertheless, no thorough investigation has been previously conducted on the histopathological correlations of CT-based radiomics in native kidney biopsy. Notably, despite the retrospective nature of this study, the time intervals between CT scans and kidney biopsy were short (a median of 1 day), which allowed accurate capture of kidney histology. As iodinated contrast is not recommended for patients with kidney diseases, we used radiomics data from non-contrast CT scans, which may be advantageous in clinical applications. In addition, automated segmentation using the recent deep learning algorithm enabled more clinically applicable radiomics models. On the other hand, the histological findings of kidney biopsies may reflect pathological features specific to individual kidney diseases beyond chronicity; therefore, CT-based radiological features have the potential to predict individual glomerular diseases such as IgA nephropathy and diabetic nephropathy, which can be an important topic for future research. For further validation of our findings, a larger multicenter study and techniques that aggregate data from different institutions are warranted.
Conclusion
We identified significant correlations between CT-based radiomic features and chronic histopathology in native kidney biopsies and presented a machine-learning prediction model for chronicity grade with high discriminability. Our results could serve as a preliminary step towards further applicative research on non-invasive tools capable of assessing and grading kidney fibrosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
None.
Author contributions
YHC contributed to the methodology and implemented software. J-EK performed data preprocessing and formal analysis. RWL and BK participated in image preprocessing and segmentation. HCS and MC performed histological examination. YK, WYP, KJ, and SH contributed to data acquisition and curation. JHP contributed to the conception and supervision of the work, KK drafted the manuscript and designed the study. All authors read and approved the final version of the manuscript.
Funding
None.
Data availability
The data used and analyzed in the current study are available from the corresponding authors on reasonable request.
Declarations
Clinical trial number
Not applicable.
Ethical approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Keimyung University Hospital (IRB No. 2023-01-021). The requirement for informed consent was waived due to the retrospective nature of the study.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jin Hyuk Paek and Kipyo Kim contributed equally to this work as corresponding authors.
Contributor Information
Jin Hyuk Paek, Email: novawang@naver.com.
Kipyo Kim, Email: kpkidney@inha.ac.kr.
References
- 1.Herzenberg AM, Fogo AB, Reich HN, Troyanov S, Bavbek N, Massat AE, Hunley TE, Hladunewich MA, Julian BA, Fervenza FC, et al. Validation of the Oxford classification of IgA nephropathy. Kidney Int. 2011;80(3):310–7. [DOI] [PubMed] [Google Scholar]
- 2.Tervaert TW, Mooyaart AL, Amann K, Cohen AH, Cook HT, Drachenberg CB, Ferrario F, Fogo AB, Haas M, de Heer E, et al. Pathologic classification of diabetic nephropathy. J Am Soc Nephrol. 2010;21(4):556–63. [DOI] [PubMed] [Google Scholar]
- 3.Bajema IM, Wilhelmus S, Alpers CE, Bruijn JA, Colvin RB, Cook HT, D’Agati VD, Ferrario F, Haas M, Jennette JC, et al. Revision of the International Society of Nephrology/Renal Pathology Society classification for lupus nephritis: clarification of definitions, and modified National Institutes of Health activity and chronicity indices. Kidney Int. 2018;93(4):789–96. [DOI] [PubMed] [Google Scholar]
- 4.Sethi S, D’Agati VD, Nast CC, Fogo AB, De Vriese AS, Markowitz GS, Glassock RJ, Fervenza FC, Seshan SV, Rule A, et al. A proposal for standardized grading of chronic changes in native kidney biopsy specimens. Kidney Int. 2017;91(4):787–9. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava A, Palsson R, Kaze AD, Chen ME, Palacios P, Sabbisetti V, Betensky RA, Steinman TI, Thadhani RI, McMahon GM, et al. The Prognostic Value of Histopathologic Lesions in native kidney biopsy specimens: results from the Boston kidney biopsy cohort study. J Am Soc Nephrol. 2018;29(8):2213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poggio ED, McClelland RL, Blank KN, Hansen S, Bansal S, Bomback AS, Canetta PA, Khairallah P, Kiryluk K, Lecker SH, et al. Systematic review and Meta-analysis of native kidney biopsy complications. Clin J Am Soc Nephrol. 2020;15(11):1595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barinotti A, Radin M, Cecchi I, Foddai SG, Rubini E, Roccatello D, Sciascia S. Serum biomarkers of renal fibrosis: a systematic review. Int J Mol Sci 2022, 23(22). [DOI] [PMC free article] [PubMed]
- 8.Huang E, Mengel M, Clahsen-van Groningen MC, Jackson AM. Diagnostic potential of minimally invasive biomarkers: a biopsy-centered viewpoint from the Banff Minimally Invasive Diagnostics Working Group. Transplantation. 2023;107(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ce M, Felisaz PF, Ali M, Re Sarto GV, Cellina M. Ultrasound elastography in chronic kidney disease: a systematic review and meta-analysis. J Med Ultrason (2001) 2023, 50(3):381–415. [DOI] [PubMed]
- 10.Buchanan CE, Mahmoud H, Cox EF, McCulloch T, Prestwich BL, Taal MW, Selby NM, Francis ST. Quantitative assessment of renal structural and functional changes in chronic kidney disease using multi-parametric magnetic resonance imaging. Nephrol Dial Transpl. 2020;35(6):955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin J, Seo N, Baek SE, Son NH, Lim JS, Kim NK, Koom WS, Kim S. MRI Radiomics Model predicts pathologic complete response of rectal Cancer following Chemoradiotherapy. Radiology. 2022;303(2):351–8. [DOI] [PubMed] [Google Scholar]
- 12.Vicini S, Bortolotto C, Rengo M, Ballerini D, Bellini D, Carbone I, Preda L, Laghi A, Coppola F, Faggioni L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: focus on the three most common cancers. Radiol Med. 2022;127(8):819–36. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Lou X, Kong N, Xu M, Gao C. Can quantitative peritumoral CT radiomics features predict the prognosis of patients with non-small cell lung cancer? A systematic review. Eur Radiol. 2023;33(3):2105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomaszewski MR, Gillies RJ. The Biological meaning of Radiomic features. Radiology. 2021;298(3):505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Gao L, Ma H, Arefan D, He J, Wang J, Liu H. Radiomics-based features for prediction of histological subtypes in Central Lung Cancer. Front Oncol. 2021;11:658887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee P, Cintra M, Huang C, Zhou M, Zhu S, Colevas AD, Fischbein N, Gevaert O. CT-based Radiomic Signatures for Predicting Histopathologic Features in Head and Neck squamous cell carcinoma. Radiol Imaging Cancer. 2020;2(3):e190039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Perucho JAU, Hu Y, Choi MH, Han L, Wong EMF, Ho G, Zhang X, Ip P, Lee EYP. Computed Tomographic Radiomics in differentiating histologic subtypes of epithelial ovarian carcinoma. JAMA Netw Open. 2022;5(12):e2245141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park HJ, Lee SS, Park B, Yun J, Sung YS, Shim WH, Shin YM, Kim SY, Lee SJ, Lee MG. Radiomics Analysis of Gadoxetic Acid-enhanced MRI for staging liver fibrosis. Radiology. 2019;290(2):380–7. [DOI] [PubMed] [Google Scholar]
- 19.Meng J, Luo Z, Chen Z, Zhou J, Chen Z, Lu B, Zhang M, Wang Y, Yuan C, Shen X, et al. Intestinal fibrosis classification in patients with Crohn’s disease using CT enterography-based deep learning: comparisons with radiomics and radiologists. Eur Radiol. 2022;32(12):8692–705. [DOI] [PubMed] [Google Scholar]
- 20.Refaee T, Salahuddin Z, Frix AN, Yan C, Wu G, Woodruff HC, Gietema H, Meunier P, Louis R, Guiot J, et al. Diagnosis of idiopathic pulmonary fibrosis in high-resolution computed Tomography scans using a combination of handcrafted Radiomics and Deep Learning. Front Med (Lausanne). 2022;9:915243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandara MS, Gurunayaka B, Lakraj G, Pallewatte A, Siribaddana S, Wansapura J. Ultrasound Based Radiomics features of chronic kidney disease. Acad Radiol. 2022;29(2):229–35. [DOI] [PubMed] [Google Scholar]
- 22.Amann K, Haas CS. What you should know about the work-up of a renal biopsy. Nephrol Dial Transpl. 2006;21(5):1157–61. [DOI] [PubMed] [Google Scholar]
- 23.Park D, Oh D, Lee M, Lee SY, Shin KM, Jun JS, Hwang D. Importance of CT image normalization in radiomics analysis: prediction of 3-year recurrence-free survival in non-small cell lung cancer. Eur Radiol. 2022;32(12):8716–25. [DOI] [PubMed] [Google Scholar]
- 24.Gallardo-Estrella L, Lynch DA, Prokop M, Stinson D, Zach J, Judy PF, van Ginneken B, van Rikxoort EM. Normalizing computed tomography data reconstructed with different filter kernels: effect on emphysema quantification. Eur Radiol. 2016;26(2):478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y, Yang D, Li W, Roth HR, Landman B, Xu D, Nath V, Hatamizadeh A. Self-supervised pre-training of swin transformers for 3d medical image analysis. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition: 2022; 2022: 20730–20740.
- 26.Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, et al. The image Biomarker Standardization Initiative: standardized quantitative Radiomics for High-Throughput Image-based phenotyping. Radiology. 2020;295(2):328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo TK, Li MY. A Guideline of selecting and reporting Intraclass correlation coefficients for Reliability Research. J Chiropr Med. 2016;15(2):155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005;3(2):185–205. [DOI] [PubMed] [Google Scholar]
- 29.Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q, Liu T-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In: Neural Information Processing Systems: 2017; 2017.
- 30.Lundberg S. A unified approach to interpreting model predictions. arXiv Preprint arXiv:170507874 2017.
- 31.Rothman KJ. Epidemiology: an introduction. OUP USA; 2012.
- 32.Lv J, Shi S, Xu D, Zhang H, Troyanov S, Cattran DC, Wang H. Evaluation of the Oxford classification of IgA nephropathy: a systematic review and meta-analysis. Am J Kidney Dis. 2013;62(5):891–9. [DOI] [PubMed] [Google Scholar]
- 33.Humphreys BD. Mechanisms of Renal Fibrosis. Annu Rev Physiol. 2018;80:309–26. [DOI] [PubMed] [Google Scholar]
- 34.Li LP, Leidner AS, Wilt E, Mikheev A, Rusinek H, Sprague SM, Kohn OF, Srivastava A, Prasad PV. Radiomics-based image phenotyping of kidney apparent diffusion coefficient maps: preliminary feasibility & efficacy. J Clin Med 2022, 11(7). [DOI] [PMC free article] [PubMed]
- 35.Zhang G, Liu Y, Sun H, Xu L, Sun J, An J, Zhou H, Liu Y, Chen L, Jin Z. Texture analysis based on quantitative magnetic resonance imaging to assess kidney function: a preliminary study. Quant Imaging Med Surg. 2021;11(4):1256–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moghazi S, Jones E, Schroepple J, Arya K, McClellan W, Hennigar RA. O’Neill WC: correlation of renal histopathology with sonographic findings. Kidney Int. 2005;67(4):1515–20. [DOI] [PubMed] [Google Scholar]
- 37.Manley JA, O’Neill WC. How echogenic is echogenic? Quantitative acoustics of the renal cortex. Am J Kidney Dis. 2001;37(4):706–11. [DOI] [PubMed] [Google Scholar]
- 38.Hricak H, Cruz C, Romanski R, Uniewski MH, Levin NW, Madrazo BL, Sandler MA, Eyler WR. Renal parenchymal disease: sonographic-histologic correlation. Radiology. 1982;144(1):141–7. [DOI] [PubMed] [Google Scholar]
- 39.Choi YH, Jo S, Lee RW, Kim JE, Paek JH, Kim B, Shin SY, Hwang SD, Lee SW, Song JH et al. Changes in CT-Based morphological features of the kidney with declining glomerular filtration rate in chronic kidney disease. Diagnostics (Basel) 2023, 13(3). [DOI] [PMC free article] [PubMed]
- 40.Araujo NC, Rebelo MAP, da Silveira Rioja L, Suassuna JHR. Sonographically determined kidney measurements are better able to predict histological changes and a low CKD-EPI eGFR when weighted towards cortical echogenicity. BMC Nephrol. 2020;21(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beck-Tolly A, Eder M, Beitzke D, Eskandary F, Agibetov A, Lampichler K, Hambock M, Regele H, Klager J, Nackenhorst M, et al. Magnetic Resonance Imaging for Evaluation of Interstitial Fibrosis in kidney allografts. Transpl Direct. 2020;6(8):e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berchtold L, Crowe LA, Combescure C, Kassai M, Aslam I, Legouis D, Moll S, Martin PY, de Seigneux S, Vallee JP. Diffusion-magnetic resonance imaging predicts decline of kidney function in chronic kidney disease and in patients with a kidney allograft. Kidney Int. 2022;101(4):804–13. [DOI] [PubMed] [Google Scholar]
- 43.Chen Z, Ying MTC, Wang Y, Chen J, Wu C, Han X, Su Z. Ultrasound-based radiomics analysis in the assessment of renal fibrosis in patients with chronic kidney disease. Abdom Radiol (NY). 2023;48(8):2649–57. [DOI] [PubMed] [Google Scholar]
- 44.Ge XY, Lan ZK, Lan QQ, Lin HS, Wang GD, Chen J. Diagnostic accuracy of ultrasound-based multimodal radiomics modeling for fibrosis detection in chronic kidney disease. Eur Radiol. 2023;33(4):2386–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used and analyzed in the current study are available from the corresponding authors on reasonable request.