Abstract
The trivalent oral poliomyelitis vaccine (OPV) contains three different poliovirus serotypes. It use therefore creates particularly favorable conditions for mixed infection of gut cells, and indeed intertypic vaccine-derived recombinants (VdRec) have been frequently found in patients with vaccine-associated paralytic poliomyelitis. Nevertheless, there have not been extensive searches for VdRec in healthy vaccinees following immunization with OPV. To determine the incidence of VdRec and their excretion kinetics in primary vaccinees, and to establish the general genomic features of the corresponding recombinant genomes, we characterized poliovirus isolates excreted by vaccinees following primary immunization with OPV. Isolates were collected from 67 children 2 to 60 days following vaccination. Recombinant strains were identified by multiple restriction fragment length polymorphism assays. The localization of junction sites in recombinant genomes was also determined. VdRec excreted by vaccinees were first detected 2 to 4 days after vaccination. The highest rate of recombinants was on day 14. The frequency of VdRec depends strongly on the serotype of the analyzed isolates (2, 53, and 79% of recombinant strains in the last-excreted type 1, 2, and 3 isolates, respectively). Particular associations of genomic segments were preferred in the recombinant genomes, and recombination junctions were found in the genomic region encoding the nonstructural proteins. Recombination junctions generally clustered in particular subgenomic regions that were dependent on the serotype of the isolate and/or on the associations of genomic segments in recombinants. Thus, VdRec are frequently excreted by vaccinees, and the poliovirus replication machinery requirements or selection factors appear to act in vivo to shape the features of the recombinant genomes.
Genomic rearrangement by recombination during viral replication is a common mechanism of genetic variability and evolution of many RNA viruses (see references 1, 32, 41 and 54 for reviews). The first genetic evidence for recombination of RNA viruses was obtained with poliovirus (PV), the causative agent of poliomyelitis (22, 33, 48). Biochemical evidence of intermolecular recombination was reported subsequently from studies of PV and foot-and-mouth disease virus, another virus of the Picomaviridae family (27, 28, 46). Members of this family, and in particular PV, are still suitable models for studying genetic exchange between RNA viral genomes (2).
PV is a nonenveloped virus composed of an icosahedral capsid made of 60 copies of four proteins (VP1 to VP4) surrounding the single-stranded messenger-sense genomic RNA. This genomic RNA is polyadenylated at the 3′ terminus and covalently attached to a genome-linked protein (VPg) at the 5′ terminus. Two noncoding regions flank the single large open reading frame, which is translated in the cytoplasm of infected cells by a cap-independent mechanism. The resulting single polyprotein is subsequently cleaved to yield the structural and the nonstructural proteins including the RNA-dependent RNA polymerase (3Dpol). The 5′ and 3′ noncoding (5′NC and 3′NC) regions are involved in viral replication and translation (reviewed in references 5, 6 and 52).
PV replicates in the human digestive tract and can induce paralysis by infecting and destroying motor neurons. Attenuated strains of all three serotypes have been selected by numerous passages of wild-type strains in monkey tissues in vivo and in vitro (47). These strains (Sabin 1, 2, and 3), which replicate in the human gut and induce a strong immunity including a local intestinal immunity, have been used as an oral poliomyelitis vaccine (OPV). Because OPV acts against the fecal-oral transmission of PV strains in humans, it has been the tool of choice for the eradication of poliomyelitis. However, in rare cases (1 case per 2.5 to 0.2 million doses), OPV strains have been implicated in vaccine-associated paralytic poliomyelitis (VAPP). Phenotypic changes due to the genetic variability of the Sabin strains are probably one of the main causes of VAPP (38). This variability leads to the spread in the environment and possible circulation of vaccine-derived pathogenic strains. This could make the final steps of the eradication of the PV species more difficult to achieve. A poliomyelitis outbreak due to vaccine-derived PV strains was recently reported (9).
PV genomes with deletions in defective interfering particles, and deletions and insertions in the genomes of PV pseudorevertants, have been described (31, 44). Additionally, in vitro transduction of human rRNA by PV has also been described (10). Nevertheless, PV genome rearrangement frequently takes place through homologous RNA recombination involving accurate substitution of a similar genomic region without insertion, deletion, or mismatch such that the genetic organization is unchanged (1). It is generally accepted that RNA recombination in PV occurs by a copy-choice mechanism in which the viral RNA polymerase switches templates during negative-strand synthesis (29). However, other possible mechanisms have recently been proposed. Premature termination of transcription could generate RNA fragments of variable lengths which could subsequently be aligned to and extended on a different template (43). A nonreplicative RNA recombination model has also been proposed: recombining RNAs are cleaved, and the exposed termini are cross-ligated (20).
Although PV recombination has been evidenced and studied mostly in the laboratory, PV has been shown to recombine in nature. The ability of Sabin PV strains in vaccinees to exchange genetic material was first described by Kew and Nottay in 1984 (25). Since then, there have been extensive searches for similar PV intertypic vaccine-derived recombinants (VdRec), mostly in patients with VAPP. Such recombinants were found to appear very frequently (11, 13, 14, 18, 19, 34, 36). In some cases, recombinants between Sabin and wild strains have also been isolated from VAPP cases (13, 21, 45).
VdRec have also been reported in healthy vaccinees (7, 13, 36, 39). By introducing three different PV serotypes simultaneously, OPV creates particularly favorable conditions for mixed infection of the gut cells. Nevertheless, there have been no extensive searches for VdRec in vaccinees following immunization with trivalent OPV. In particular, neither the incidence of VdRec in primary vaccinees nor the general genomic features of the corresponding recombinant genomes have been established.
This report presents the characterization of the genomes of PV Sabin isolates excreted by 67 vaccinees following primary immunization with OPV during a mass campaign (23). Recombinant strains were identified by multiple restriction fragment length polymorphism (RFLP) assays (3, 14, 21). The localization of junction sites in recombinant genomes was also determined. VdRec appeared early after vaccination and were excreted by many of vaccinees. We found that there were both preferred associations of genomic segments and preferred recombination sites in the genomes. This indicates that mechanistic or selective factors acting in vaccinees and possibly in host cells determine the characteristics of VdRec genomes.
MATERIALS AND METHODS
Cells and viral reference strains.
HEp-2c and Vero cells were grown in monolayers in Dulbecco modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum.
The attenuated PV Sabin 1, 2, and 3, were obtained from the World Health Organization master seeds [Behringwerke (SO+1)] for OPV preparation. The second passage at 34°C in HEp-2c cells of the seed was used to prepare viral stocks.
Study group and specimen collecting.
The vaccine-derived PV isolates characterized in this study were isolated from a group of 67 healthy 2- to 5-month-old children and vaccinated with their first dose of OPV (106, 105, and 105.7 tissue culture infective dose units of Sabin 1, 2, and 3 strains, respectively; Sclavo, Siena, Italy). None of the children had received either OPV or inactivated poliomyelitis vaccine prior to immunization. The vaccine was administered as part of a mass campaign in spring 1993 in Bucharest, Romania (23).
Virus isolation, identification, and serotyping.
The primary isolation of enteroviruses from stool specimens was performed on Vero cells using standard procedures (53). Type-specific PV-neutralizing antisera produced at the Cantacuzino Institute were used for serotyping. Strains were isolated from mixtures of PV of different serotypes by neutralization tests with these sera. Viral stocks were obtained after a second passage on Hep-2c cells at 34°C to increase viral titers. RFLP assays were used for intratypic differentiation between wild-type and vaccine-derived viruses (see below) (3).
In some cases, to separate mixtures of viruses of the same serotype but different genotypes, viruses were plaque purified under agar overlay. Briefly, infected Hep-2c cells in six-well plates were maintained under 0.9% agarose in DMEM supplemented with 2% fetal calf serum and 50 mM MgCl2. After 72 h of incubation at 34°C, cells were stained with neutral red, and then individual plaques were picked and used to inoculate fresh cells.
Reverse transcription (RT) and PCR.
These techniques were used for various investigations: to confirm the presence of vaccine-derived viruses of a given serotype with a serotype-specific PCR assay, to synthesize amplicons for RFLP assays, and for sequencing.
Viral RNA was reverse transcribed directly from the cell-free supernatant of infected cells. For the synthesis of cDNA, a mixture of 1 μl of supernatant, 0.5 μl of RNasin (40 U/μl; Promega) 10 pmol of antisense oligonucleotide primer, and distilled water to a final volume of 14 μl was heated for 5 min at 80°C for denaturation and for 5 min at 42°C for annealing. Six microliters of a mixture containing 4 μl of 5× transcription buffer (Promega), 1 μl of deoxynucleoside triphosphate at 10 mM each, and 1 U of avian myeloblastosis virus reverse transcriptase (Promega) was added. RT was performed at 42°C for 30 min and stopped by heat inactivation at 95°C for 5 min, and the sample was placed immediately on ice. It was then mixed with 3 μl of 10× amplification buffer (Eurobio or Promega), 10 pmol of sense primer (or 100 pmol of primer UG17), 1.25 U of Taq polymerase (Eurobio or Promega), and distilled water to a final volume of 100 μl. The PCR was performed using 30 cycles of denaturation at 94°C for 1 min, annealing at 45°C (or at 50°C for RFLP-3D-3′) for 2 min, and extension at 70°C for 1 min; a final elongation step of 10 min at 70°C was also used. Aliquots (10 μl) were run on 1.5% agarose gel in the presence of ethidium bromide (0.5 μg/ml), and DNA amplicons were visualized under UV light.
Serotype- and strain-specific RT-PCR.
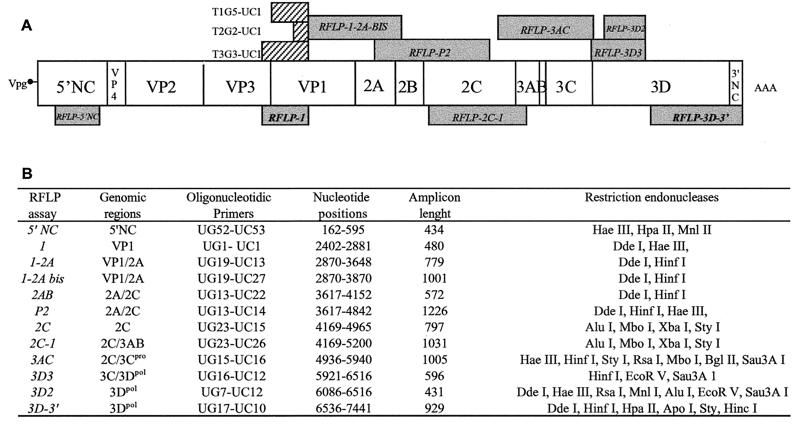
The serotype of viral isolates was confirmed by RT-PCR using strain serotype-specific oligonucleotide primers. A PV universal antisense primer, UC1, and specific sense primers T1G5 (5′-AGGTCAGATGCTTGAAAGCA-3′) T2G2 (5′-TGCTCCGACAAAGCGTGCCAG-3′) and T3G3 (5′-GTGACATACAGACAGACTACAC-3′) were used. These primers allow the amplification of genomic fragments of 396, 117, and 596 bp from the Sabin 1, 2, and 3 strains, respectively (Fig. 1A).
FIG. 1.
Molecular tools used for analyzing PV genomes. (A) Structure of the genome, showing the two noncoding regions (5′NC and 3′NC) covalently linked to the small viral protein VPg (3B) and to the poly(A) sequence (AAA), respectively. Genomic regions encoding viral proteins (VP4 to 3D) are indicated. Positions of the sequences used for strain-specific RT-PCR are shown as hatched boxes, and the corresponding pairs of primers are given. Names of RFLP assays and the corresponding genomic regions are indicated in grey boxes; the two assays used systematically are in bold. (B) Details of RFLP assays and genomic regions. Names of the primers used for RT-PCR are given (UG and UC denote genomic sense and complementary sense primers). Primer sequences are given in Materials and Methods. The nucleotide positions for amplicons are indicated according to Sabin 1 genome numbering.
RFLP assays.
RFLP assays involved the synthesis of genomic amplicons by RT-PCR amplification, followed by the comparative analysis of amplicon restriction profiles on agarose gel. These assays give strain-specific restriction profiles and were used to analyze different distant regions of the genome. Oligonucleotides and genomic regions analyzed by RFLP assays are shown in Fig. 1. Sequences of all of the oligonucleotides except UC22 (5′-TCAGTAAATTTCTTCAACCA-3′), UC26 (5′-GGAGTCAACTGCTTGGAGCA-3′), and UC27 (5′-AATGCAGGCCCGAGTGACTC-3′) are given in reference 21. Amplicons (20 to 40 μl) were digested for 2 h with 10 U of restriction endonuclease at the suitable temperature and in the appropriate buffer as indicated by the manufacturer. In most experiments, two or three different restriction endonucleases were used for each amplicon. Digested PCR products were then analyzed by electrophoresis in 3 to 4% agarose gels at 5 V/cm for 2 h using Tris-acetate-EDTA as an electrophoresis buffer and visualized by ethidium bromide staining. Restriction profiles of isolated PV strains were compared with those of the Sabin 1, 2, and 3 reference strains.
To check that there was no competition between the three Sabin strain genomes during RFLP assays, the sensitivity of some of the RFLP assays (RFLP-1 and RFLP-3D-3′) to detect each of the three vaccine serotypes was determined using two different restriction enzymes (DdeI-HaeIII and HinfI-DdeI, respectively). Different homotypic and heterotypic viral preparations were used: dilutions of viral stocks of each of the three vaccine serotypes, and dilutions of mixtures containing equal or unequal titers of each pair of the three serotypes. Pure virus solutions or 1:1 mixtures of Sabin 1, 2, and 3 strains were diluted 1:2 in DMEM, from 106 to 3.12 × 104 PFU/ml. Mixtures containing unequal titers of viruses were made up to ratios of 9:1, 8:2, etc., to 1:9. RFLP assays were performed as described above.
Localization of recombination junctions.
Recombination junctions were first mapped using RFLP assays and subsequently, in many cases, by sequencing RT-PCR products. Before sequencing, RT-PCR products were purified by using a Qiaquick spin column purification kit (Qiagen). Sequencing was performed using a BigDye Terminator Cycle Sequencing Ready Reaction kit according to the procedure recommended by Applied Biosystems Perkin-Elmer. Sequences were compared with those of the Sabin vaccine reference strains by using the Clustal W software (49). Recombination sites were thus mapped between two restriction sites or two nucleotides that differentiate the genomes of the parental Sabin strains. Nucleotide positions for the three Sabin strains are indicated according to the numbering of the Sabin 1 genome (51).
RESULTS
Detection of recombinant strains.
Stools were collected from 67 healthy children who had been vaccinated with their first dose of OPV. Stool specimens were collected from each child on day 0 and 2, 4, 7, 14, 21, 28, and 60 days following vaccination (23). PV strains were isolated from stool specimens on Vero cells, and strains of each serotype were identified and separated from mixture using type-specific antisera.
A set of 138 isolates, composed of isolates of each serotype that were excreted the latest by each child (maximum of three isolates per child), was analyzed in detail. The serotypes of these isolates and their vaccine origin were confirmed by using both vaccine serotype-specific oligonucleotides for RT-PCR and specific restriction sites present in the amplicon DNA obtained from a genomic region encoding part of the capsid protein VP1 (RFLP-1 assay [Fig. 1]) (3). Fifty five isolates belonging to type 1, 39 belonging to type 2, and 44 belonging to type 3 were identified.
We screened for recombinant strains by comparing the results of this RFLP-1 assay with those of the RFLP-3D-3′ assay, which analyzes a segment encompassing the region encoding the C-terminal part of the 3Dpol and the entire 3′NC region (Fig. 1) (21). In many cases, the results of the RFLP-1 assay were also compared with those of the RFLP-5′NC assay, analyzing a genomic segment located in the 5′NC region (42). Moreover, to detect multiple recombinant strains and to localize recombination junctions RFLP analysis was subsequently extended to other regions of the genome encoding nonstructural PV proteins. In particular, the RFLP-3D2 assay, analyzing the region encoding the N terminus of the 3Dpol, was used in most cases (Fig. 1).
In many cases (61 of 138 isolates), the segment corresponding to the 3′-terminal part of the genome (RFLP-3D-3′ assay) was derived from a vaccine strain serotype different from that expected from serotyping and from RFLP-1 assays (Fig. 2). These results indicated the presence of intertypic recombinant genomes. The 5′NC genomic region was analyzed (RFLP-5′NC assay) for 45 isolates (21, 11, and 13 isolates of serotypes 1, 2, and 3, respectively) and was found in all cases to be derived from the same vaccine strain serotype as that identified by serotyping. This indicated that genetic intertypic recombination involving large RNA segments occurred rarely in the genomic stretch between the analyzed 5′NC and capsid regions or did not occur at all; this stretch encodes major serotype antigenic determinants and includes the VP1 region analyzed by the RFLP-1 assay.
FIG. 2.
RFLP analysis of vaccine-derived PV strains. The 3′-terminal parts of the genomes of strains isolated from vaccinees were amplified by RT-PCR and digested with restriction enzymes HinfI and DdeI (RFLP-3D-3′ assay). The restriction profiles of the reference Sabin 1, 2 and 3 (S1, S2, and S3) strains and of some type 1 (lane 1), type 2 (lanes 2 to 4), and type 3 (lanes 5 to 7) strains isolated from vaccinees and identified by serotyping are shown. The profiles shown in lanes 1 to 7 correspond to recombinant genomes. Whereas most profiles correspond to a single genotype, lanes 1 and 4 show mixtures of two different genotypes. Lanes 3 (HinfI) and 5 (DdeI) show hybrid restriction fragments indicating the locations of recombinant sites in the amplified fragment. Faint minor bands (lane 1) corresponding to S3 genomic fragments are indicated by dashes. Other minor bands resulting from incomplete digestion were observed in a few lanes (lanes 1 and 2). MW, HaeIII-digested fragments of bacteriophage φX174 DNA serving as molecular weight markers; ND, not digested.
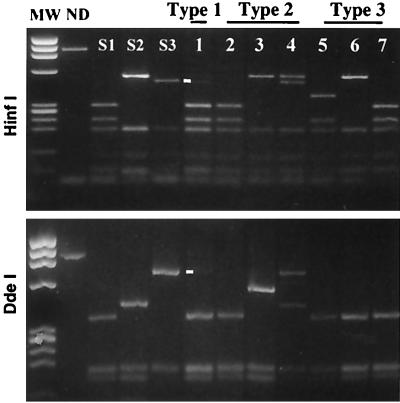
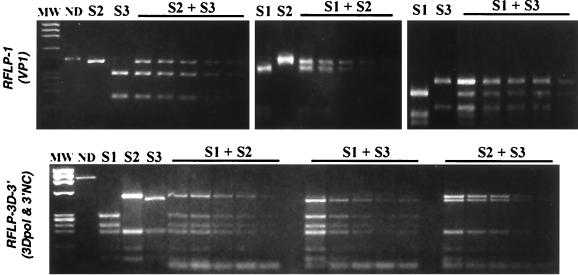
A single type of genomic segment per isolate was generally detected by the RFLP-3D-3′ assay; nevertheless, in several cases, two different types of segments were evidenced. This indicated the presence in the same isolate of mixture of different virus strains belonging to the same serotype (homotypic viruses) but with different genotypes (one recombinant plus one nonrecombinant genome, or two different recombinant genomes). For this reason, we hereafter distinguish between the term “isolate,” used for the product of the identification and purification of samples by seroneutralization, and the term “strain,” used for viruses identified by RFLP analysis of the genomes present in isolates. Isolates frequently contained mixtures of different homotypic strains, and therefore we checked that there was no competition during the RT-PCR amplification between homologous segments of different serotypes; the RT-PCR assay could have favored the detection of one segment and masked the presence of another. The sensitivity of the RFLP-1 and RFLP-3D-3′ assays to detect each of the three vaccine serotypes either in homotypic viral preparations or in heterotypic mixtures was thus determined (see Materials and Methods and Fig. 3). In homotypic preparations, the threshold of detection was 10 to 40 PFU per RT-PCR for the RFLP-1 assay and 40 to 100 PFU for the RFLP-3D-3′ assay. In heterotypic preparations containing each of the three different pairs of serotypes (1:1 ratio), the thresholds of detection were similar for all serotypes (around 30 PFU per RT-PCR for the RFLP-1 assay and from 30 to 60 PFU for the RFLP-3D-3′ assay). There was no evidence of preferential amplification or detection of homologous genomic segments for any pair of serotypes (Fig. 3). Both genotypes were also clearly detectable in heterotypic mixtures at ratios of 1:4 to 4:1 but not 1:9. The intensity of the restriction profiles was proportional to the virus concentrations or proportions in each analyzed mixture. Thus, these RFLP assays were similarly sensitive for the three vaccine serotypes both in homotypic preparations and in mixtures.
FIG. 3.
Sensitivity of RFLP assays for detection of the genome of various vaccine serotypes in a mixed population. Heterotypic mixtures containing pairs of the Sabin strains (S1, S2, and S3) combined in equal proportions were prepared. Mixtures were diluted (1:2) from 106 to 6.25 × 104 PFU/ml per strain. RFLP assays were performed using the restriction enzymes DdeI or HaeIII for RFLP-1 and HinfI for RFLP-3D-3′. The restriction profiles of the reference Sabin 1, 2, and 3 strains are shown. The two strains present in each mixture are indicated; restriction profiles for the first five dilutions are shown. MW, molecular weight markers (φX174 DNA HaeIII-diested fragments); ND, not digested.
To confirm that RFLP assays were able to detect various viral genotypes in the same isolate, several viral plaques from infected monolayers maintained under agarose overlays were picked and analyzed. Two isolates appearing as mixtures of recombinant and nonrecombinant type 1 strains (S1 plus S1/S3 [see above and Fig. 4]) and type 2 strains (S2 plus S2/S1) were tested. Analysis of the plaque-purified viruses confirmed the presence of the two viral populations in the same isolate (not shown). Moreover, the presence in a third isolate of two different type 2 recombinant genomes (S2/S1) with different recombination sites was also evidenced by this method. These results confirm that RFLP assays are suitable for detecting different viral populations present in a mixed isolate (mixtures of homotypic viruses with different genotypes).
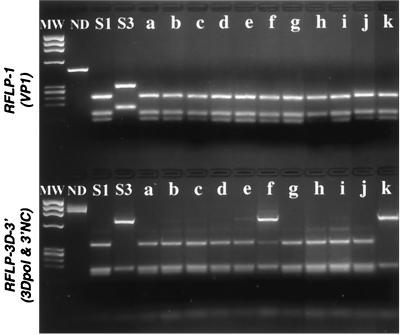
FIG. 4.
RFLP analysis of plaque-purified viruses isolated from a mixed isolate containing a nonrecombinant S1 population and a recombinant S1/S3 population. Plaque-purified viruses (lanes a to k) derived from the original isolate (Fig. 2, lane 1) were analyzed by RFLP-1 and RFLP-3D-3′ assays (restriction enzymes HaeIII and DdeI, respectively). Whereas most profiles correspond to a single genotype, some RFLP-3D-3′ profiles show traces of the second genotype (first round of purification) and minor bands due to incomplete digestion. The restriction profiles of the reference Sabin 1 and 3 strains are shown. MW, φX174 DNA HaeIII digested fragments used as molecular weight markers; ND, not digested.
In most isolates (83%), only a single viral recombinant or nonrecombinant genotype was detected (Table 1). Most recombinant strains had a bipartite genome produced by a single recombination event. However, tripartite genomes produced by two recombination events were also detected in six type 3 isolates. Most mixed isolates were mixtures of one recombinant and one nonrecombinant strain, although mixtures of two different recombinants were also detected. Four isolates gave complex RFLP patterns suggesting mixtures of three different genotypes or of bipartite and tripartite recombinant genomes (Table 1). These four isolates were not analyzed in detail.
TABLE 1.
Categories of recombinant genomes in isolates and localization of junctions by RFLP
| No. of isolates | Recombinant genotypea | Type of junctionb | Localization of junctions
|
|
|---|---|---|---|---|
| Region | Restriction site intervalsc | |||
| 1 | S2/S1 | 2C | 4911StyI-S1-5097AluI-S2 | |
| 1 | S2/S1 | 3B-3Cpro | 5432RsaI-S1-5537HaeIII-S2 | |
| 3 | S2/S1 | 3Dpol | 6302HaeIII-S2-6399Dde I-S1 | |
| 1 | S2/S1 | 3Dpol | 6416DdeI-S2-6626BglII-S2 | |
| 3 | S2/S1 | 3Dpol | 6650HpaII-S1-6711HpaII-S2 | |
| 2 | S2/S1 | 3Dpol | 6756HinfI-S1-7053PvuII-S1 | |
| 1 | S2/S3 | 3Dpol | 6856DdeI-S2-7052HpaII-S3 | |
| 1 | S3/S1 | 2C | 4728StyI-S1-5038StyI-S2 | |
| 2 | S3/S1 | 2C | 4886XbaI-S1-4911StyI-S1 | |
| 1 | S3/S1 | 2C | 4911ScyI-S1-5073MboI-S1 | |
| 1 | S3/S2 | 2A | 3571HinfI-S3-3722DdeI-S2 | |
| 5 | S3/S2 | 2C | 4422HinfI-S2-4563DdeI-S3 | |
| 2 | S3/S2 | 2C | 4686AluI-S3-5107StyI-S1 | |
| 2 | S3/S2 | 2C | 4728StyI-S1-4886XbaI-S1 | |
| 6 | S3/S2 | 2C | 4686AluI-S3-4801MboI-S2 | |
| 1 | S3/S2/S1 | S3/S21st | VP1-2A | 3363DdeI-S3-3502DdeI-S2 |
| S2/S12nd | 3Dpol | 6713HpaII-S2-6757HinfI-S2 | ||
| 1 | S3/S2/S3 | S3/S21st | VP1-2A | 3363DdeI-S3-3502DdeI-S2 |
| S2/S32nd | 3Dpol | 6713HpaII-S2-6858HpaII-S3 | ||
| 1 | S3/S2/S1 | S3/S21st | 2A | 3363DdeI-S3-3502DdeI-S2 |
| S2/S12nd | 3Dpol | 6756HinfI-S1-7215DdeI-S1 | ||
| 1 | S3/S2/S1 | S3/S21st | 2C | 4414HaeIII-S3-4563DdeI-S3 |
| S2/S12nd | 3Dpol | 6418DdeI-S2-6713HpaII-S2 | ||
| 1 | S3/S2/S3 | S3/S21st | 2C | 4414HaeIII-S3-4563DdeI-S3 |
| S2/S32nd | 3Dpol | 6588HinfI-S3-6894DdeI-S3 | ||
| 1 | S3/S2/S3 | S3/S21st | 2C | 4561AluI-S3-4678MbpI-S2 |
| S2/S32nd | 3Dpol | 6302HaeIII-S2-6586HinfI-S3 | ||
| 1 | S1+S1/S3 | 3Cpro-3Dpol | 5980HinfI-S1-6132HinfI-S3 | |
| 1 | S2+S2/S1 | 3Dpol | 6221Sau3A1-6371EcoRV-S1 | |
| 1 | S2+S2/S1 | 3Dpol | 6439Mnl-S1-6645ApoI-S1 | |
| 1 | S2+S2/S1 | 3Dpol | 6650HpaII-S1-6713HpaII-S2 | |
| 1 | S2/S1+S2/S1∗ | 2C-3A | 4732HaeIII-S2-5128BglII-S2 | |
| 3Dpol | 6474Sau3AI-S1-6650HpaII-S1 | |||
| 2 | S2/S1+S2/S1∗ | 3Dpol | 6221Sau3AI-S2-6399DdeI-S1 | |
| 3Dpol | 6474Sau3AI-S1-6650Hpa-II-S1 | |||
| 1 | S2+S2/S3 | 3Cpro | 5743Sau3AI-S2-5883HinfI-S3 | |
| 2 | S2+S2/S3 | 3Dpol | 6031Sau3AI-S2-6132HinfI-S3 | |
| 1 | S2+S2+S3 | 3Dpol | 6713HpaII-S2-6858HpaII-S3 | |
| 1 | S2+S2/S3 | 3Dpol | 7052HpaII-S3-7282DdeI-S2 | |
| 1 | S3+S3/S2 | 3Dpol | 6134DdeI-S2-6165DdeI-S3 | |
| 1 | S3+S3/S2 | 2C | 4801Sau3AI-S2-4845HinfI-S2 | |
| 1 | S3/S1+S3/S1∗ | 2C | 4414HaeIII-S3-4563DdeI-S3 | |
| 3Dpol | 6756HinfI-S1-7048HinfI-S3 | |||
| 3 | S3/S2+S3/S2 | 2C | 4423HinfI-S2-4563DdeI-S3 | |
| 2C | 4801Sau3AI-S2-4845HinfI-S2 | |||
| 1 | S3/S2+S3/S2 | None-3885DdeI-S3 | ||
| 2C | 4801Sau3AI-S2-4845HinfI-S2 | |||
| 1 | S2+S2/S1+S2/S1 | 3Dpol | 6414HinfI-S2-6474Sau3AI-S1 | |
| 1 | S3/S2+S3/S2/S3 | S3/S2 | 2C | 4686AluI-S3-4801MboI-S2 |
| S2/S32nd | 3Dpol | 6578HinfI-S2-6712HpaII-S2 | ||
| 1 | S3/S2+S3/S2/S3 | S3/S2 | 2C | 4414HaeIII-S3-4563DdeI-S3 |
| 1 | S3/S2+S3/S2/S1 | S3/S2 | 2C | 4801Sau3AI-S2-4845-HinfI-S2 |
| S3/S21st | 2C | 4423HinfI-S2-4563DdeI-S3 | ||
Asterisks indicate mixtures of recombinant strains of the same category but with different junctions; italics indicate complex mixtures of genotypes which were not accurately resolved.
Positions in tripartite recombinant genomes are indicated.
Restriction sites flanking the recombination junctions are given. Numbers indicate the first nucleotide of the restriction site and refer to Sabin 1 strain numbering. The restriction enzyme and the genome (serotype) cut specifically by this enzyme are given.
Incidence and characteristics of recombinant strains in vaccinees.
Among the 138 analyzed isolates, the percentage of isolates containing VdRec genomes varied considerably according to their serotype: 82% of type 3 isolates and 62% of type 2 isolates contained strains with recombinant genomes. Of the 55 type 1 isolates, only one was found to contain such an VdRec genome, and it was a mixture of recombinant and nonrecombinant genomes. For each serotype, the relative proportion of recombinant and nonrecombinant strains in isolates containing one genotype and that in isolates containing two different genotypes did not differ significantly (Fischer exact test; P = 0.25 for type 2 and 0.7 for type 3 isolates). Therefore, all well-defined strains present in isolates containing either one or two different genotypes (134 isolates) were considered for calculating the percentages of recombinant strains and for subsequent analysis. Two, 53, and 79% of strains belonging to serotypes 1, 2, and 3, respectively, were found to be VdRec.
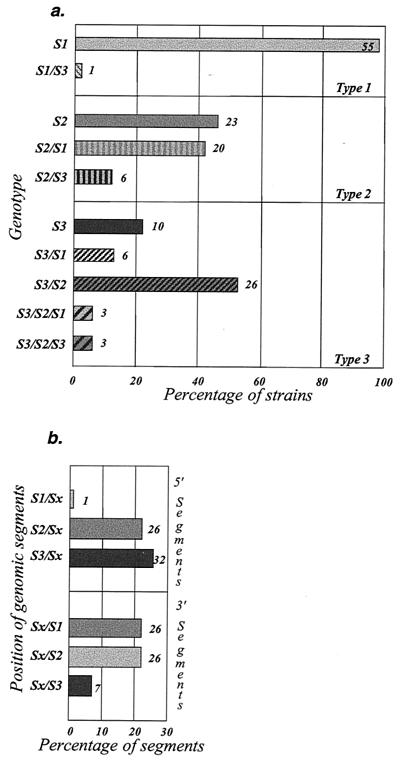
There was a preferential association of segments according to serotype origin (Fig. 5a). For example, type 2 VdRec (with the 5′ moiety of the genome including the capsid genomic region, and thus the antigenicity, of the Sabin 2 strain) were more frequently associated with Sabin 1-derived genomic segments in the 3′ moiety of the genome (20 S2/S1 versus 6 S2/S3 strains). Similarly, type 3 VdRec preferred Sabin 2-derived segments (26 S3/S2 versus 6 S3/S1). Sabin 1-derived genomic segments were more frequent in the 3′ moiety of the type 2 VdRec genomes than in that of the type 3 VdRec genomes (20 S2/S1 versus 6 S3/S1 strains). No difference was found in the association of segments between recombinant strains present in isolates containing one genotype and those in isolates containing two different genotypes (not shown). The six type 3 VdRec with tripartite genomes were not considered in this comparative analysis.
FIG. 5.
Association of genomic segments in recombinant strains. The genomes of strains present in isolates were analyzed by RFLP assays, and the genomic segments of recombinant genomes were identified and classified according to the vaccine strains from which the genomic segment are derived (see Table 1 for details) and according to the positions of these segments in the genome. (a) The different categories of nonrecombinant or recombinant genomes are classified according to the serotype of the isolate from which they are derived. The numbers of analyzed strains are shown. (b) The relative proportion of each of the three genomic segments in bipartite recombinant genomes is indicated according to position in the 5′ part (S1/Sx; includes S1/S2 and S1/S3, for example) or in the 3′ part (Sx/S1, for example) of the recombinant genomes. The number of different segments present in bipartite recombinant genomes described in panel a (118 segments) was considered 100%.
We determined the numbers of genomic segments of each serotype present in bipartite or tripartite recombinant genomes. All three serotypes were represented, but Sabin 1 genomic segments were less frequent than the others in recombinants. In bipartite recombinant genomes, the genomic segments were classified according to their position on the 5′ or 3′ side of the recombination site (Fig. 5b). Sabin 1 segments were almost never found on the 5′ side of the recombination junctions, and Sabin 3 segments were relatively rare on the 3′ side. Sabin 2 segments were found equally on both sides of the junctions. The molecular features of recombinant genomes thus indicated preferred well-ordered associations of genomic segments.
Localization of recombination junctions in recombinant genomes.
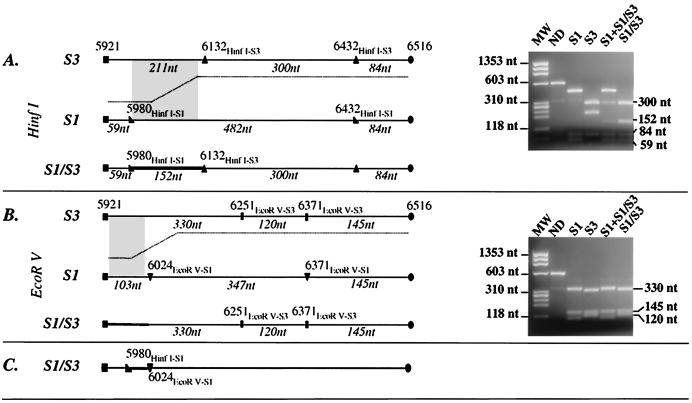
Recombination junctions were located using RFLP assays covering most of the genomic region encoding nonstructural viral proteins. These assays map the junction to intervals (restriction site intervals) flanked by two endonuclease restriction sites that differentiate the two parental genomes. A detailed analysis of the restriction patterns obtained from the type 1 recombinant strain (S1/S3) is presented as an example. The recombination junction of the S1/S3 strain was first mapped by combining RFLP-1, -3D3 and -3D-3′ assays using the initial type 1 mixed isolate S1+S1/S3. The assay of the VP1 capsid genomic region gave a clear single Sabin 1 pattern (not shown). The RFLP 3D-3′ assay (analyzing the 3′ extremity of the genome) gave a mixture of Sabin 1 and Sabin 3 patterns (Fig. 2, lane 1), clear evidence of the presence of both the original Sabin 1 strain and a recombinant genome S1/S3. The RFLP-3D3 assay (genomic region corresponding to the N-terminal half of the polymerase 3D) revealed an unusual restriction fragment of 153 nucleotides after HinfI endonuclease digestion. This fragment can be explained by the juxtaposition, after recombination, of the Sabin 1 HinfI restriction site at nucleotide position 5980 (HinfI-S1) and of the Sabin 3 HinfI restriction site at nucleotide position 6132 (HinfI-S3). The recombination site was thus mapped to the 5980HinfI-S1-6132 HinfI-S3 restriction site interval. Plaque-purified recombinant virus strains derived from this isolate were analyzed: some of these viruses gave Sabin 1 patterns in the RFLP-1, -1-2A-Bis, -P2, and -3AC assays and Sabin 3 patterns in the RFLP-3D-3′ assay (Fig. 4). As expected, the RFLP-3D3 HinfI restriction patterns were modified and presented a hybrid recombinant fragment (Fig. 6A). EcoRV restriction patterns were used to map the recombination junction to the 5980HinfI-S1-6024EcoRV-S1) restriction site interval (Fig. 6B and C).
FIG. 6.
RFLP pattern (RFLP-3D3) and restriction map of the amplicon carrying the recombination site of an S1/S3 recombinant genome. Restriction profiles obtained from the reference Sabin 1 and 3 strains (S1 and S3), from the original mixed isolate (S1+S1/S3), and from a plaque-purified recombinant virus (S1/S3) are shown. Nucleotide positions of the amplicon extremities and of the restriction sites on the PV genome are indicated according to Sabin 1 strain numbering. Genomic regions believed to include the recombination junction are indicated by grey zones and bold lines. Lengths of restriction fragments (in nucleotides [nt]) are indicated. Molecular weight markers (MW) are φX174 DNA HaeIII digested fragments. ND, nondigested. (A) HinfI restriction map and profiles on an agarose gel; (B) EcoRV maps and profiles; (C) localization of the recombination junction of the S1/S3 genome inferred from profiles in panels A and B.
Most of the recombination junctions of the bipartite type 2 recombinant S2/S1 and S2/S3 genomes were thereby shown to be in the P3 genomic region (encoding proteins 3A to 3Dpol) and, in particular, in the 3Dpol-coding region between nucleotides 6302 and 7053 (21 of 27 junctions). The junctions of only two S2/S1 genomes were found elsewhere, in the 2C- and 2C-3A-coding regions (Table 1 and Fig. 7).
FIG. 7.
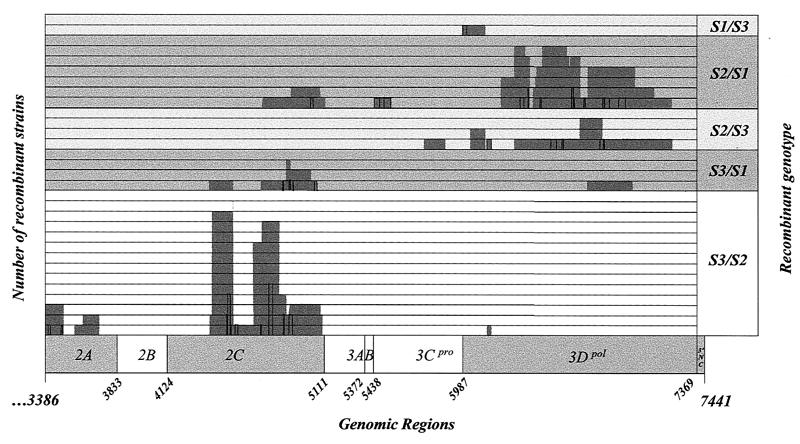
Localization of recombination junctions in PV genomes. The recombination junctions were localized by RFLP and by sequencing in homologous genomic segments flanked by two restriction sites and two nucleotides that differentiate the genomes of the parental Sabin strains. These genomic segments are indicated on the schematized genome as grey boxes (RFLP) and lines (sequencing). The data shown are from Tables 1 and 2. The numbers of recombination junctions located in the same genomic segment or in overlapping segments are given (one line per junction). The genomic regions of the PV genome are indicated as described for Fig. 1.
In most cases, the recombination junctions of type 3 recombinant genomes (S3/S1 and S3/S2) were located in the P2 region (encoding proteins 2A to 2C) and, in particular, in the 2C protein-coding region between nucleotides 4422 and 5107 (28 of 32 junctions). The junction of one S3/S2 genome was found in the 2A-coding region between nucleotides 3571 and 3722.
In the six tripartite genomes (S3/S2/S1 or S3/S2/S3), the upstream recombination junctions (S3/S21st) were found in the 2C-coding region (nucleotides 4414 to 4678) or in the VP1-2A-coding region (nucleotides 3363 to 3502; more precisely in 2A, as determined by sequencing). All downstream recombination junctions (S2/S12nd and S2/S32nd) of the tripartite genomes were in the 3Dpol-coding region (6302 to 7215). These results are in good agreement with the locations of the recombination junctions of bipartite S3/S2, S2/S1, and S2/S3 genomes (Table 1).
To confirm the results obtained by RFLP and to map the recombination junctions more precisely, amplicons obtained from RT-PCR were sequenced. Only bipartite or tripartite genomes present in isolates composed of a single genotype were considered. Recombination sites were localized in homologous nucleotide stretches flanked by two nucleotides that differentiate the genomes of the parental strains. All recombination sites were found in the restriction site intervals determined by RFLP above. No point mutations, deletions, or nucleotide insertions were observed. Recombination sites were located in genomic segments 5 to 26, 2 to 44, and 26 nucleotides long for the S3/Sx, S2/Sx, and S1/S3 junctions, respectively (Table 2 and Fig. 7).
TABLE 2.
Localization of recombination junctions by nucleotidic sequencing
| Type of junction | No. of junctions | Localizationa (nt) | Length (nt) | Nucleotide sequence (5′-3′) |
|---|---|---|---|---|
| S1/S3 | 1 | 5984–6009 | 26 | CAAGGTGAAATCCAGTGGATGAGACC |
| S2/S1 | 1 | 5036–5053 | 18 | CAGATCACTACAATGATT |
| S2/S1 | 1 | 5465–5493 | 29 | ATGGCTAAAAGAAACATTGTTACAGCAAC |
| S2/S1 | 1 | 6338–6363 | 26 | TACCCTTATGTAGCAATGGGAAAGAA |
| S2/S1 | 2 | 6389–6390 | 2 | AC |
| S2/S1 | 1 | 6464–6471 | 8 | GATGAACT |
| S2/S12nd | 1 | 6659–6672 | 14 | ATGGAAGAGAAGCT |
| S2/S1 | 2 | 6659–6672 | 14 | ATGGAAGAGAAGCT |
| S2/S1 | 1 | 6674–6678 | 5 | TTTGC |
| S2/S12nd | 1 | 6737–6744 | 8 | ATGGTGCT |
| S2/S12nd | 1 | 6854–6864 | 11 | TGCTCAGGCAC |
| S2/S1 | 1 | 6866–6894 | 29 | TACCCTTATGTAGCAATGGGAAAGAA |
| S2/S1 | 1 | 6953–6996 | 44 | ATGATTGCCTATGGTGATGATGTAATTGCTTCCTACCCCCATGA |
| S2/S32nd | 1 | 6530–6534 | 5 | AATGA |
| S2/S32nd | 1 | 6599–6608 | 8 | GTCACTGG |
| S2/S32nd | 1 | 6836–6858 | 23 | GGCGGCATGCCATCTGGCTGCTC |
| S2/S3 | 1 | 6860–6864 | 5 | GGCAC |
| S3/S1 | 1 | 4865–4869 | 5 | ATTCA |
| S3/S1 | 1 | 4904–4911 | 8 | AACATGGC |
| S3/S1 | 1 | 4927–4929 | 3 | GTG |
| S3/S1 | 1 | 5060–5076 | 17 | GAGAGAAACAGAAGATC |
| S3/S21st | 2 | 3404–3417 | 14 | AAAGCTGTGTACAC |
| S3/S21st | 1 | 3483–3492 | 10 | TCATGTGGAA |
| S3/S2 | 2 | 4514–4518 | 5 | CCAGG |
| S3/S2 | 2 | 4520–4536 | 17 | ACAGGAAAATCAGTTGC |
| S3/S21st | 2 | 4520–4536 | 17 | ACAGGAAAATCAGTTGC |
| S3/S21st | 1 | 4541–4542 | 2 | AA |
| S3/S21st | 1 | 4565–4584 | 20 | GAGAAAGAGAACACCTCCAC |
| S3/S2 | 1 | 4724–4728 | 5 | CCACC |
| S3/S2 | 5 | 4775–4800 | 26 | GTTTTAGCCTCCACCAACTCCAGTCG |
| S3/S2 | 2 | 4868–4872 | 5 | CAAGT |
| S3/S2 | 2 | 4901–4923 | 23 | CTCAACATGGCAATGGCTACTGA |
Recombination sites were localized (sequencing) in homologous segments flanked by two nucleotides that differentiate the genomes of the parental Sabin strains. Nucleotide (nt) numbering refers to Sabin 1 strain sequence.
Excretion kinetics of recombinant strains in vaccinees.
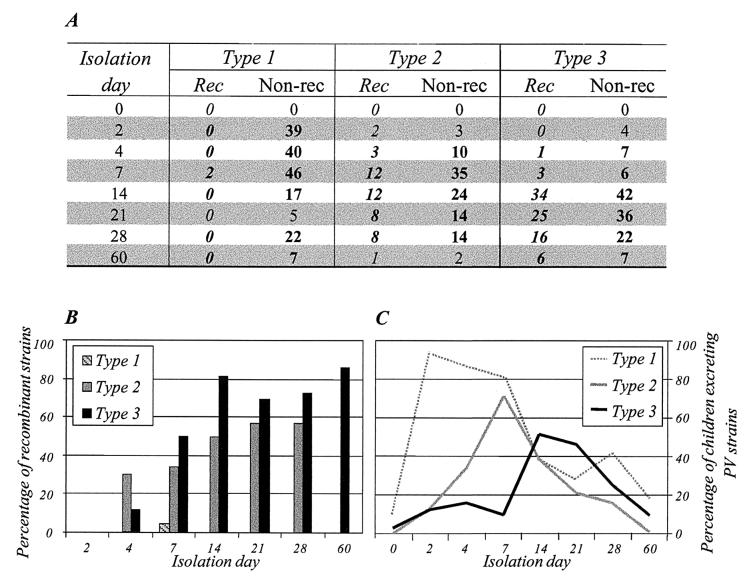
We studied excretion of recombinants from days 2 to 60 following primary vaccination on a total of 356 of 421 isolates, including the last-excreted isolates described above. Screening for recombinant genomes involved comparing the serotype origin of the genomic segments corresponding to the capsid protein VP1 and to the 3′-terminal part of the genome (RFLP-1 and RFLP-3D-3′ assays). Type 2 recombinants appeared as early as 2 days after vaccination, and type 3 recombinants were detected from day 4 (Fig. 8). Only one additional type 1 recombinant (S1/S3) was found (on day 7, coexisting with the parental Sabin 1 strain). The maximum relative abundances of recombinant strains (about 55 and 75% of all type 2 and type 3 strains, respectively) were on day 14 and had not increased substantially by day 21, 28, or 60. These relative abundances were not significantly different from those for the last-excreted isolates (53 and 79% being recombinants for the type 2 and type 3 strains, respectively). Moreover, the proportion of S2/S1 genomes among type 2 VdRec genomes (S2/S1 plus S2/S3) was stable throughout the excretion period (from 75 to 87%) and was similar to that for the last-excreted isolates (77%) (not shown). The various categories of type 3 recombinant genomes were not compared because the search for multiple recombinants was not completed.
FIG. 8.
Excretion kinetics of recombinants in vaccinees. PV strains were isolated from days 2 to 60 following primary OPV vaccination and analyzed for recombination using RFLP assays. (A) Numbers of recombinants (Rec) and nonrecombinant (Non-rec) strains, classified according to day of isolation and strain serotype. (B) Percentages of recombinant strains per day of isolation, given according to strain serotype. Only percentages including more than five strains are considered (indicated in bold in panel A). (C) Percentages of children excreting PV strains, given according to strain serotype and day of isolation (these results are adapted from reference 23 and described in Discussion).
DISCUSSION
To determine the incidence and genomic characteristics of intertypic recombinants in vaccinees, we studied PV isolates collected from children over a period of 60 days after feeding administration of their first dose of trivalent OPV. Strains with intertypic recombinant genomes were frequently found. Molecular analysis of recombinant genomes indicated preferred associations of genomic segments and preferred regions for junction sites.
A large proportion of strains isolated from patients with VAPP were found to have intertypic recombinant genomes: about 80% for type 2 and type 3 strains (21). We investigated only strains isolated from healthy primary vaccinees, and the percentages of type 2 and type 3 VdRec were again high (53 and 79% of strains, respectively, in the last-excreted isolates). Moreover, type 3 isolates presented a variety of recombinant genomes, including double recombinants (with tripartite genomes) and mixtures of different recombinants. Complex mixtures of different recombinant genotypes have been demonstrated in isolates from recipient VAPP cases (18, 19). Only two type 1 VdRec have been reported in previous studies of strains isolated from patients with VAPP (11, 15). However, the rarity of type 1 VdRec could not be clearly ascertained due to the small number of type 1 vaccine strains implicated in vaccine-associated disease (14, 16, 21). We analyzed 174 type 1 strains. Only two of them were found to be recombinant. Thus, type 1 recombinants are rare at least in primary vaccinees.
The excretion by vaccinees of such a high proportion of recombinant type 2 and 3 viruses is surprising and suggests that these viruses were not generated, or not generated solely, during viral replication in the digestive tract. However, it seems very unlikely that the OPV itself contains recombinants because the virus seeds used by OPV producers are derived from clonally purified virus (47). Moreover, monotypic virus stocks of all three serotypes are made and checked separately before mixing and therefore cannot coinfect cells during manufacture of the vaccine. It is also very unlikely that many recombinants were generated during the isolation process on coinfected cells inoculated with fecal suspensions containing different serotypes, given the low proportion of recombinants obtained by the mixed infection of cells with different PV serotypes (29, 52). Moreover, in this study, the second parental serotype strain was not isolated from most of the fecal suspensions (73%) containing a recombinant strain of a given serotype. Further evidence that the recombinants were generated during multiplication of the trivalent OPV in the gut of vaccinees is provided by the increase in the proportion of recombinants excreted over time (Fig. 8).
The percentage of type 2 recombinant strains in healthy vaccinees (about 55%) was lower than that in patients with VAPP (80%) (21). This suggests that there is a selection of type 2 recombinant strains (but not of type 3 recombinants) in VAPP patients and thus that these strains have an advantage over nonrecombinants to multiply and/or to induce the disease. Many other type 2 recombinant strains implicated in VAPP have been described (34). However, these previous data were obtained in different contexts and are therefore not strictly comparable with those obtained in our study. In particular, only strains excreted by children given a single dose of OPV were studied. Moreover, the doses of the vaccine strains present in the OPV used in this study differ slightly from the usual doses (ratio of 10:1:5 versus 10:1:3 for the type 1, 2, and 3 vaccine strains, respectively). This could also explain some differences between our results and those from previous studies indicating a lower frequency of type 2 VdRec and a higher frequency of complex type 3 VdRec in healthy vaccinees (7, 39).
Type 1 VdRec were rare in vaccinees. Nevertheless, Sabin 1-derived genomic segments encoding nonstructural proteins were frequently found in type 2 and type 3 recombinant strains, indicating that the Sabin 1 strain was not excluded from recombination events. All three strain serotypes were well represented among all genomic segments present in recombinants (Fig. 5). This suggests that replication machinery requirements prevent the inclusion of Sabin 1 segments encoding capsid and antigenic determinants in the 5′ moiety of recombinant vaccine-derived genomes. It is also possible that there is frequently selection against type 1 VdRec, when generated, in vaccinees. Similar factors (replication requirements or selective pressure) may favor some association of genomic segments in intertypic recombinants, as S2/S1 and S3/S2 genomes were significantly more frequent than S2/S3 and S3/S1 genomes. The analysis of type 2 and type 3 strains isolated from VAPP cases also provided evidence for similar nonrandom distributions (21). VdRec thus display preferred well-ordered association of genomic segments. To our knowledge, this is the first time that such a phenomenon has been described for recombinant RNA viruses isolated from infected organisms.
All recombination junctions were found in genomic regions P2 and P3 encoding nonstructural viral proteins, i.e., proteins implicated in proteolytic cleavage and in genomic replication. Recombination events in the 5′NC region and/or in the capsid region were not found in this study. Recombination in the 5′ moiety of the genome should be a rare event, since only one vaccine-derived strain that has the entire 5′NC genomic region and part of the capsid protein VP4 region from a nonvaccine strain has been described (18, 21). However, we cannot exclude that there are substitutions of small genomic fragments (double recombination events), too small to have been detected. Moreover, a possible explanation for the lack of detection of recombination event in the capsid region is the methodology used for the serotype identification and separation of viruses. This method uses type-specific PV neutralizing antisera allowing the separation of type 1, 2, and 3 PV present in isolates. However, hybrid antigenic viruses produced by recombination events may fail to be detected by the standard technique. All strains that we analyzed clearly reacted with serotype-specific antibodies and displayed at least part of the genomic VP1 region (according to the RFLP-1 assay), in agreement with the seroneutralization test. A methodology allowing the detection of viruses displaying the antigenicity of two different serotypes should be used to search for possible natural antigenic chimeras. Some antigenic chimeras have been produced by manipulation of infectious PV cDNA (37). However, many of them were shown to be nonviable or unstable when the recombination junctions were located in the capsid-coding region, indicating that the integrity of the capsid region of PV seems to be important (30). The integrity of the 5′NC region could also explain the absence of recombinants in the 5′ NCR, since this region is involved in replication and in the internal entry of ribosomes allowing the traduction of the viral genome (6, 52).
Another interesting finding was that most recombination junctions in the type 2 recombinants were in the P3 genomic region and in the type 3 recombinants in the P2 region (Fig. 7). If recombination can take place at every nucleotide with equal probability, the relative probability of finding a recombinant within any single region would be directly proportional to the size of the region. This was clearly not the case, and our results indicate that the recombination junctions are nonrandomly distributed. The nonrandom distribution appeared to apply equally to the rare S1/S3 recombinant sites: four S3/S1/S3 tripartite recombinant strains have been described, and their S1/S3 junctions (nucleotides 5672 to 6169) are in the same subgenomic region as that of the recombinant S1/S3 found in this study (nucleotides 5980 to 6024) (11, 17). These results indicate that some genomic regions (hot spots) are preferred for recombination during the replication process or that recombinant genomes with such recombination sites are selected in coinfected cells or in the infected host.
The association of genomic segments and locations of recombination sites appear to be interdependent in PV recombinants. The preferential regions for recombination were dependent on the serotype of the recombinant strains and/or of characteristics the 5′ genomic segment encoding the antigenic determinants, irrespective of the 3′ moiety. These results are in good agreement with published data describing the recombination sites of naturally occurring VdRec and with a recent analysis of VdRec isolated from patients with VAPP (J. Balanant, unpublished results; 7, 11, 17, 19, 26, 34). Nevertheless, cumulating all previous results indicates that there are differences in the location of recombination sites between type 2 S2/S1 and S2/S3 recombinants, suggesting that the 3′ moiety of recombinant genomes may also have an effect. In addition to the P3 genomic region, another region, at the 3′ end of the P2 region, frequently contains the recombination sites of S2/S1 recombinant genomes (two examples only in this study [Fig. 7]) but not those of S2/S3 recombinants.
Tolskaya et al. have described hot spots of recombination in type 3 recombinants (S3/S1) isolated after coinfection of cells and subsequently selected using genetic markers (50). Recombination junctions were nonrandomly distributed within the genomic region considered, which included most of the P2 region. Hot spots of recombination were found in the 2B and 2C regions. However, in this study, the genomic region available for recombination was artificially determined by the location of the genetic markers used for selecting recombinants (nucleotides 3386 to 4547). In our and previous works with natural recombinants isolated from vaccinees, only 3 of 13 recombination junctions of this category (S3/S1 junctions) were located inside the genomic region considered by Tolskaya et al. (7, 11, 36, 50).
Recombination hot spots have also been found in other RNA viruses, including brome mosaic virus, coronaviruses, and retroviruses (4, 35, 40). Such hot spots have been shown to be sequence-dependent or associated with RNA secondary structures (8, 35, 40). It was suggested that certain RNA structures favor RNA recombination mechanistically. However, studies of coronavirus recombination indicate that recombination events are random but that some types of recombinants have a selective advantage for multiplying in cultured cells (4).
Analysis of recombination sites of intertypic PV recombinants isolated from coinfected cells revealed no clear consensus sequence for recombination, although some features, in particular a high degree of homology between the parental genomes on the 3′ side of the sites, have been noted (24, 26, 29). Thus, the positions of recombination sites may be random, and some genotypes are then selected during subsequent multiplication. Nevertheless, the localization of recombination sites in recombinants selected in acellular systems, or in coinfected cells, has been found to be temperature dependent and not due to subsequent selection (12). RNA structures have also been described as being important for promoting PV recombination (50).
In humans, preferential association of genomic segments and recombination sites could result from numerous factors. Infected cell types allowing PV excretion in the digestive tract of infected individuals are not yet clearly known. The replication machinery and subsequently the association of genomic segments and the localization of recombination sites may depend on the viral replication complexes and/or the target cell's machinery. This could favor the synthesis and/or the encapsidation of particular genomic rearrangements. Alternatively, the fittest recombinants may be selected either in the in vivo-coinfected target cells and/or in the host. Indeed, the dominance of a recombinant population could be the result of a selective growth advantage in the gastrointestinal tract and/or of greater resistance against the various selective pressures of the infant intestine and immune system. However, the frequent presence of recombinants in the human gut could be the result of small numbers of target cells, of high concentrations of replicating viruses in rare permissive compartments, and of random sampling (15). We are currently investigating factors that could explain the appearance of particular VdRec genotypes.
The excretion kinetics of recombinant strains in vaccinees indicate that VdRec can be excreted early (during the first week) following primary vaccination and that the maximum relative abundance of recombinant strains (according to serotype) was on day 14. It has been reported that four children who excreted type 3 PV for a period of more than 12 days following primary vaccination produced VdRec (7, 39). More surprising is the fact that in our study, the relative abundance of recombinant strains did not vary (increase) significantly from days 14 to 60, suggesting that there is no subsequent selection or synthesis of recombinants in vaccinees. Possibly, during the first 14 days following primary vaccination there is simultaneous replication of vaccine strains in the gut, optimizing the chances of two viruses infecting the same cell and thereby exchanging genetic material by recombination. Thereafter, the acquired gut immunity of vaccinees or some unknown host factors may restrict the replication of some serotypes and thus limit the possibility of additional recombination events. Alternatively, the spread of the virus in the gut may be restricted to a few discrete compartments of susceptible cells, and these compartments may all be infected after 14 days. It is tempting to suggest that the level and characteristics of recombinants are determined by the kinetics of replication of each of the three serotype strains. Indeed, most vaccinees excreted first the type 1, then the type 2, and finally the type 3 viruses (Fig. 8C). This may explain why the type 2 viruses recombined frequently with type 1 (frequent S2/S1 genomes) and type 3 with type 2 (frequent S3/S2 genomes). However, this does not explain why the reciprocal genomic associations (S1/S2 and S2/S3 genomes, respectively) are not equally found in excreted recombinants. Therefore, beside the possible effect of the kinetics of replication of the different strains, the role of replication requirements or of selection factors should also be considered.
In conclusion, analysis of the genome structure of vaccine strains isolated from healthy vaccinees revealed a high frequency of PV intertypic recombinants, a nonrandom association of genomic fragments, preferential genomic regions for recombination, and linkage between these two phenomena. To our knowledge, this is the first time that such characteristics of viral RNA genetic recombination have been described. By increasing our knowledge of the factors (mechanistic requirements or otherwise) involved in enterovirus recombination, we may increase our understanding of RNA virus evolution. This may also provide greater insight into the genetic variability of live attenuated viral vaccine strains, which should make it possible to improve their safety.
ACKNOWLEDGMENTS
Adolfo Suarez is kindly acknowledged for constructive criticism, and F. Colbère-Garapin is thanked for critical reading of the manuscript.
This work was partly supported by grants to F.D. from the Délégation au Réseau des Instituts Pasteur et Instituts Associés (AC 98 Entérovirus and AC 99 Entérovirus) and from the Direction des Recherches sur I'Environnement (Recherche Environnement 6724) and by grants to R.C. from the World Health Organization (V26/181/107) and from the European Commission (Copemicus-CIPA CT94-0123 and Inco-Copernicus ERBIC 15 CT96-0912).
REFERENCES
- 1.Agol V I. Recombination and other rearrangements in picomaviruses. Semin Virol. 1997;8:77–84. [Google Scholar]
- 2.Agol V I, Paul A V, Wimmer E. Paradoxes of the replication of picornaviral genomes. Virus Res. 1999;62:129–147. doi: 10.1016/s0168-1702(99)00037-4. [DOI] [PubMed] [Google Scholar]
- 3.Balanant J, Guillot S, Candrea A, Delpeyroux F, Crainic R. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology. 1991;184:645–654. doi: 10.1016/0042-6822(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 4.Banner L R, Lai M M C. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology. 1991;185:441–445. doi: 10.1016/0042-6822(91)90795-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belsham G J, Sonemberg N. Picomavirus RNA translation: roles for cellular proteins. Trends Microbiol. 2000;8:330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 6.Blondel B, Duncan G, Couderc T, Delpeyroux F, Pavio N, Colbere-Garapin F. Molecular aspects of poliovirus biology with a special focus on the interactions with nerve cells. J Neurovirol. 1998;4:1–26. doi: 10.3109/13550289809113478. [DOI] [PubMed] [Google Scholar]
- 7.Cammack N, Phillips A, Dunn G, Patel V, Minor P D. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology. 1988;167:507–514. [PubMed] [Google Scholar]
- 8.Cascone P J, Haydar T F, Simon A E. Sequences and structures required for recombination between virus-associated RNAs. Science. 1993;260:801–805. doi: 10.1126/science.8484119. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Public health dispatch: outbreak of poliomyelitis—Dominican Republic and Haiti, 2000. Morb Mortal Wkly Rep. 2001;49:1094–1104. [Google Scholar]
- 10.Charini W A, Todd S, Gutman G A, Semler B L. Transduction of a human RNA sequence by poliovirus. J Virol. 1994;68:6547–6552. doi: 10.1128/jvi.68.10.6547-6552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driesel G, Diedrich S, Kunkel U, Schreier E. Vaccine-associated cases of poliomyelitis over a 30 year period in East Germany. Eur J Epidemiol. 1995;11:647–654. doi: 10.1007/BF01720298. [DOI] [PubMed] [Google Scholar]
- 12.Duggal R, Wimmer E. Genetic recombination of poliovirus in vitro and in vivo: temperature-dependent alteration of crossover sites. Virology. 1999;258:30–41. doi: 10.1006/viro.1999.9703. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich F, Da-Silva E F, Schatzmayr H G. Type 2 poliovirus recombinants isolated from vaccine-associated cases and from healthy contacts in Brazil. Acta Virol. 1996;40:27–33. [PubMed] [Google Scholar]
- 14.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology. 1993;196:199–208. doi: 10.1006/viro.1993.1468. [DOI] [PubMed] [Google Scholar]
- 15.Gavrilin G V, Cherkasova E A, Lipskaya G Y, Kew O M, Agol V I. Evolution of circulating wild poliovirus and of vaccine-derived poliovirus in an immunodeficient patient: a unifying model. J Virol. 2000;74:7381–7390. doi: 10.1128/jvi.74.16.7381-7390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgescu M M, Balanant J, Macadam A, Otelea D, Combiescu M, Aubert-Combiescu A, Crainic R, Delpeyroux F. Evolution of the Sabin type 1 poliovirus in humans: characterization of strains isolated from patients with vaccine-associated paralytic poliomyelitis. J Virol. 1997;71:7758–7768. doi: 10.1128/jvi.71.10.7758-7768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgescu M M, Balanant J, Ozden S, Crainic R. Random selection-a model for poliovirus infection of the central nervous system. J Gen Virol. 1997;78:1819–1828. doi: 10.1099/0022-1317-78-8-1819. [DOI] [PubMed] [Google Scholar]
- 18.Georgescu M M, Delpeyroux F, Crainic R. Tripartite genome organization of a natural type 2 vaccine nonvaccine recombinant poliovirus. J Gen Virol. 1995;76:2343–2348. doi: 10.1099/0022-1317-76-9-2343. [DOI] [PubMed] [Google Scholar]
- 19.Georgescu M M, Delpeyroux F, Tardy-panit M, Balanant J, Combiescu M, Guillot S, Crainic R. High diversity of poliovaccine strains isolated from the central nervous system of patients with vaccine-associated paralytic poliomyelitis. J Virol. 1994;68:8089–8101. doi: 10.1128/jvi.68.12.8089-8101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gmyl A P, Belousov E V, Maslova S V, Khitrina E V, Chetverin A B, Agol V I. Nonreplicative RNA recombination in poliovirus. J Virol. 1999;73:8958–8965. doi: 10.1128/jvi.73.11.8958-8965.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillot S, Caro V, Cuervo N, Korotkova E, Combiescu M, Persu A, Aubert-Combiescu A, Delpeyroux F, Crainic R. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J Virol. 2000;74:8434–8443. doi: 10.1128/jvi.74.18.8434-8443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst G K. Genetic recombination with Newcastle disease virus, poliovirus and influenza. Cold Spring Harbor Symp Quant Biol. 1962;27:303–308. doi: 10.1101/sqb.1962.027.001.028. [DOI] [PubMed] [Google Scholar]
- 23.Ion-Nedelcu N, Strebel P M, Toma F, Biberi-Moroeanu S, Combiescu M, Persu A, Aubert-Combiescu A, Plotkin S A, Sutter R W. Sequential and combined use of inactivated and oral poliovirus vaccines: Dolj District, Romania, 1992–1994. J Infect Dis. 1997;175:S241–S246. doi: 10.1093/infdis/175.supplement_1.s241. [DOI] [PubMed] [Google Scholar]
- 24.Jarvis T C, Kirkegaard K. Poliovirus RNA recombination—mechanistic studies in the absence of selection. EMBO J. 1992;11:3135–3145. doi: 10.1002/j.1460-2075.1992.tb05386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kew O M, Nottay B K. Evolution of the oral poliovaccine strains in humans occur by both mutation and intramolecular recombination. In: Chanock R, Lerner R, editors. Modern approaches to vaccines. N.Y: Cold Spring Harbor Press; 1984. pp. 357–367. [Google Scholar]
- 26.King A M. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 1988;16:11705–11723. doi: 10.1093/nar/16.24.11705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King A M, McCahon D, Slade W R, Newman J W. Biochemical evidence of recombination within the unsegmented RNA genome of aphthovirus. J Virol. 1982;41:66–77. doi: 10.1128/jvi.41.1.66-77.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King A M, McCahon D, Slade W R, Newman J W. Recombination in RNA. Cell. 1982;29:921–928. doi: 10.1016/0092-8674(82)90454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohara M, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. A recombinant virus between the Sabin 1 and Sabin 3 vaccine strains of poliovirus as a possible candidate for a new type 3 poliovirus live vaccine strain. J Virol. 1988;62:2828–2835. doi: 10.1128/jvi.62.8.2828-2835.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuge S, Saito I, Nomoto A. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J Mol Biol. 1986;192:473–487. doi: 10.1016/0022-2836(86)90270-6. [DOI] [PubMed] [Google Scholar]
- 32.Lai M M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ledinko N. Genetic recombination with poliovirus type 1. Virology. 1963;20:107–109. doi: 10.1016/0042-6822(63)90145-4. [DOI] [PubMed] [Google Scholar]
- 34.Lipskaya G Y, Muzychenko A R, Kutitova O K, Maslova S V, Equestre M, Drozdov S G, Bercoff R P, Agol V I. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J Med Virol. 1991;35:290–296. doi: 10.1002/jmv.1890350415. [DOI] [PubMed] [Google Scholar]
- 35.Lund A H, Mikkelsen J G, Schmidt J, Duch M, Pedersen F S. The kissing-loop motif is a preferred site of 5′ leader recombination during replication of SL3–3 murine leukemia viruses in mice. J Virol. 1999;73:9614–9618. doi: 10.1128/jvi.73.11.9614-9618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macadam A J, Arnold C, Howlett J, John A, Marsden S, Taffs F, Reeve P, Hamada N, Wareham K, Almond J, et al. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology. 1989;172:408–414. doi: 10.1016/0042-6822(89)90183-9. [DOI] [PubMed] [Google Scholar]
- 37.Minor P, Ferguson M, Katrak K, Wood D, John A, Howlet J, Dunn G, Burke K, Almond J W. Antigenic structure of chimeras of type 1 and type 3 poliovirus involving antigenic sites 2, 3, and 4. J Gen Virol. 1991;72:2475–2481. doi: 10.1099/0022-1317-72-10-2475. [DOI] [PubMed] [Google Scholar]
- 38.Minor P D. The molecular biology of poliovaccines. J Gen Virol. 1992;73:3065–3077. doi: 10.1099/0022-1317-73-12-3065. [DOI] [PubMed] [Google Scholar]
- 39.Minor P D, John A, Ferguson M, Icenogle J P. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J Gen Virol. 1986;67:693–706. doi: 10.1099/0022-1317-67-4-693. [DOI] [PubMed] [Google Scholar]
- 40.Nagy P D, Bujarski J J. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J Virol. 1997;71:3799–3810. doi: 10.1128/jvi.71.5.3799-3810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 42.Petitjean J, Quibriac M, Freymuth F, Fuchs F, Laconche N, Aymard M, Kopecka H. Specific detection of enteroviruses in clinical samples by molecular hybridization using poliovirus subgenomic riboprobes. J Clin Microbiol. 1990;28:307–311. doi: 10.1128/jcm.28.2.307-311.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierangeli A, Bucci M, Forzan M, Pagnotti P, Equestre M, Bercoff R P. ‘Primer alignment-and-extension’: a novel mechanism of viral RNA recombination responsible for the rescue of inactivated poliovirus cDNA clones. J Gen Virol. 1999;80:1889–1897. doi: 10.1099/0022-1317-80-8-1889. [DOI] [PubMed] [Google Scholar]
- 44.Pilipenko E V, Gmyl A P, Agol V I. A model for rearrangements in RNA genomes. Nucleic Acids Res. 1995;23:1870–1875. doi: 10.1093/nar/23.11.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rico-Hesse R, Pallansch M A, Nottay B K, Kew O M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987;160:311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- 46.Romanova L I, Tolskaya E A, Kolesnikova M S, Agol V I. Biochemical evidence for intertypic genetic recombination of polioviruses. FEBS Lett. 1980;118:109–112. doi: 10.1016/0014-5793(80)81229-4. [DOI] [PubMed] [Google Scholar]
- 47.Sabin A B, Boulger L R. History of Sabin attenuated poliovirus oral live vaccine strains. J Biol Stand. 1973;1:115–118. [Google Scholar]
- 48.Sergiescu D, Horodniceanu F, Klein R, Crainic R. Genetic transfer of guanidine resistance from type 2 to type 1 poliovirus. Arch Gesamte Virusforsch. 1966;18:231–235. doi: 10.1007/BF01241844. [DOI] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolskaya E A, Romanova L I, Blinov V M, Viktorova E G, Sinyakov A N, Kolesnikova M S, Agol V I. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology. 1987;161:54–61. doi: 10.1016/0042-6822(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 51.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura I, Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984;174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 52.Wimmer E, Hellen C U T, Cao X M. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 53.World Health Organization. Manual for poliovirus surveillance WHO/EPI/GEN/97.01. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 54.Worobey M, Holmes E C. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]