Abstract
Signal peptides (SP) are key determinants for targeting glycoproteins to the secretory pathway. Here we describe the involvement in particle maturation as an additional function of a viral glycoprotein SP. The SP of foamy virus (FV) envelope glycoprotein is predicted to be unusually long. Using an SP-specific antiserum, we demonstrate that its proteolytic removal occurs posttranslationally by a cellular protease and that the major N-terminal cleavage product, gp18, is found in purified viral particles. Analysis of mutants in proposed signal peptidase cleavage positions and N-glycosylation sites revealed an SP about 148 amino acids (aa) in length. FV particle release from infected cells requires the presence of cognate envelope protein and cleavage of its SP sequence. An N-terminal 15-aa SP domain with two conserved tryptophan residues was found to be essential for the egress of FV particles. While the SP N terminus was found to mediate the specificity of FV Env to interact with FV capsids, it was dispensable for Env targeting to the secretory pathway and FV envelope-mediated infectivity of murine leukemia virus pseudotypes.
Signal peptides (SP) are key determinants for targeting and membrane insertion of secretory and membrane proteins (reviewed in reference 25). They can be removed co- or posttranslationally by the cellular membrane-bound signal peptidase or may, if not cleaved, serve as membrane anchors for proteins with distinct membrane orientations. In general, SP are composed of three domains, of which a central 6- to 15-amino-acid (aa)-long hydrophobic domain (h-domain) is the most essential. An N-terminal polar domain (n-domain) usually of net positive charge shows high variability in overall length, ranging from 15 to more than 50 aa. The composition and structure of the n-domain influences protein orientation in the membrane. The polar C-terminal domain (c-domain) often contains helix-breaking as well as small uncharged residues in positions -3 and -1 which determine the site of SP cleavage. In most cases, SP cleavage is thought to occur cotranslationally; however, for some proteins, e.g., the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein gp160, SP cleavage occurs inefficiently and very late after translocation (21). A basic amino acid stretch in the n-domain of gp160 is responsible for this phenomenon and believed to influence folding and exit of HIV-1 Env from the endoplasmic reticulum (ER) (21). Recent studies revealed that SP bear specific information accounting for distinct functions in targeting and membrane insertion or even for defined metabolic pathways after their cleavage from the parent protein (reviewed in reference 25). The HIV-1 SPEnv, for example, is further processed by the signal peptidase, leading to the release of an SP fragment into the cytosol, where it binds to calmodulin (26). The function of this process in viral replication is not known.
Foamy viruses (FV), as studied with the prototype member human foamy virus (HFV), follow a replication cycle which is characterized by several unique features setting them apart from the family of retroviruses. These are the independent expression of the Pol protein from a spliced mRNA, efficient reverse transcription prior to particle release, and intracellular retrotransposition (14, 24). The essential functions of retroviral glycoproteins are binding of the viral particle to cellular receptors and subsequent fusion of viral and cellular lipid membranes to release the viral capsid into the cytoplasm (reviewed in reference 19). The FV Env protein is unique among all retroviral glycoproteins since its expression is essential for the FV particle budding and release process (3, 7). Similar to B- or D-type retroviruses, FV particles assemble in the cytoplasm of infected cells. However, unlike the case for all other retroviruses, FV capsids do not bud across cellular membranes in the absence of FV Env, and heterologous viral glycoproteins cannot complement FV Env to enable particle release (3, 7, 28). The particle-associated FV Env glycoprotein is synthesized as a 130-kDa precursor. Analogous to other retroviral Env proteins, FV Env is cleaved during its transport to the cell surface by a cellular protease, yielding a 80- to 90-kDa surface (SU) and a 48-kDa transmembrane (TM) subunit (11, 23). However, the cytoplasmic domain (CyD) of the TM subunit contains an ER retrieval signal, leading to accumulation of FV Env in the ER when other FV structural proteins are absent (10, 11, 29). Thus, the export of FV capsids requires the coexpression of cognate Env protein, and vice versa, the surface localization of Env depends on the presence of cognate capsids. This implies inherent specific interactions between the two partners.
We have shown previously that the membrane-spanning domain (MSD) but not the CyD of Env TM is essential for the particle release process (28, 29). Since the C terminus of Env does not appear to mediate the interaction with Gag, we investigated whether the N-terminal SP sequence, besides targeting the Env protein to the secretory pathway, might have additional functions in the particle release process.
MATERIALS AND METHODS
Expression constructs.
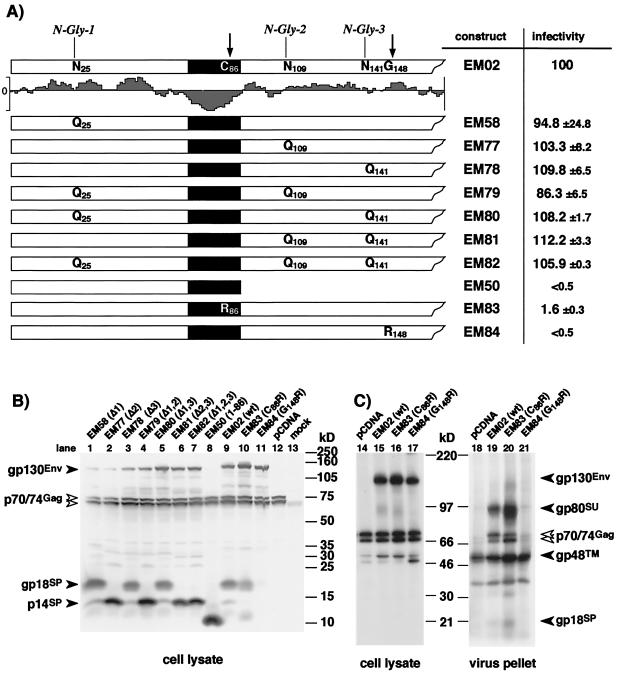
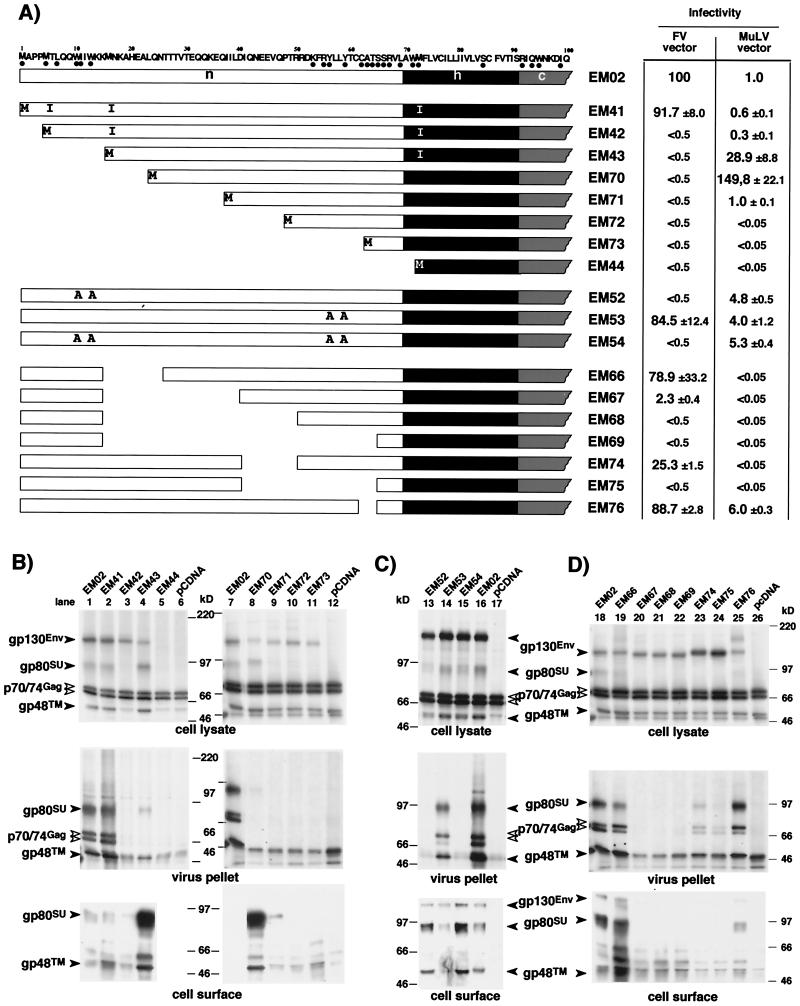
The eukaryotic expression constructs for various FV envelope mutants depicted in Fig. 4 and 5 are based on a previously described plasmid, pcHFE-wt (see Fig. 2A), which expresses only gp130 due to inactivation of the internal splice donor and splice acceptor pair within the FV Env coding region (EM02 mutation) (23, 28). All deletion and point mutants within the N region of the SP (pcHFVenv EM41 to 44 and EM66 to 76, described below) were generated by recombinant PCR techniques (18) with pcHFE-wt as the template and primers introducing the desired codon changes or deletions. The amplimers were cloned into the NheI/EcoRI sites of pcDNA3.1+zeo (Invitrogen) as XbaI/Kpn2I fragments together with a Kpn2I/EcoRI fragment of pcHFE-wt. For the N-glycosylation and signal peptidase complex (SPC) cleavage site mutants (EM58 and EM78 to EM84), a PpuMI/Kpn2I fragment of pcHFE-wt was replaced by the corresponding mutated PCR fragments. All PCR-derived inserts were completely sequenced to verify the presence of only the desired mutations. The resultant constructs contain the mutations and deletions in parentheses: N-terminal deletions, EM42 (Δ2–4, M16I, M72I), EM43 (Δ2–16, M72I), EM70 (Δ2–25), EM71 (Δ2–40), EM72 (Δ2–50), EM73 (Δ2–66), and EM44 (Δ2–72); internal deletions, EM66 (Δ16–25), EM67 (Δ16–40), EM68 (Δ16–50), EM69 (16–66), EM74 (Δ41–50), EM75 (Δ41–66), and EM76 (Δ63–66); C-terminal deletion, EM50 (Δ87–988); and point mutations EM41 (M4I, M16I, M72I), EM52 (W10A, W13A), EM53 (Y56A, Y59A), EM54 (W10A, W13A, Y56A, Y59A), EM58 (N25Q), EM77 (N109Q), EM78 (N141Q), EM79 (N25Q, N109Q), EM80 (N25Q, N141Q), EM81 (N109Q, N141Q), EM82 (N25Q, N109Q, N141Q), EM83 (C86R), EM84 (G148R).
FIG. 4.
Analysis of infectivity, SPEnv cleavage, and particle release of FV capside pseudotyped with different Env point mutants. 293T cells were cotransfected with the FV Gag/Pol-expressing pMH62 and with different FV Env expression constructs as indicated. (A) The N termini of the different FV Env proteins analyzed are schematically illustrated. The original sequences of the residues mutated that are found in the wild-type expression construct (EM02) are summarized at the top; below, amino acid sequences for the individual mutant expression constructs resulting from the introduced point mutations are shown. Relative infectivities of the respective 293T supernatants are given. (B) Western blot analysis (SDS-PAGE [10% gel]) of cellular lysates using polyclonal antisera specific for FV Gag and FV SPEnv. (C) RIPA (SDS-PAGE [7.5% gel]) of cellular lysates with antisera specific for Gag and SP/SU and metabolically labeled purified FV particles. The gp48TM protein often comigrates with an immunoreactive cellular protein present in negative controls.
FIG. 5.
Analysis of infectivity and FV particle release of FV capsids pseudotyped with different FV SPEnv deletion and point mutants. 293T cells were cotransfected with the FV Gag/Pol-expressing pMH62 or the MuLV Gag/Pol-expressing vector pHIT60 and the MuLV retroviral vector pczCFG2 fEGN together with different FV Env expression constructs as indicated. (A) Schematic illustration of the N termini of the different mutants used. The amino acid sequence of wild-type envelope construct (EM02) is shown at the top; amino acids conserved in Env proteins of different FV species are marked with black dots below the sequence. The relative infectivities of the respective supernatants measured by flow cytometry analysis are given. (B to D) RIPA of cellular lysates using polyclonal antisera specific for Gag and SP/SU (top) and metabolically labeled purified FV particles (middle), shown both after cotransfection of the indicated Env expression constructs together with the FV Gag/Pol-expressing vector plasmid pMH62 into 293T cells and metabolic labeling. Cell surface biotinylation detected by ECL after transfection of 293T cells with the indicated Env expression constructs in the absence of FV Gag/Pol-expressing vector is shown at the bottom. All SDS-PAGE were with 7.5% gels. The identity of each FV protein is indicated. The gp48TM protein often comigrates with an immunoreactive cellular protein present in negative controls.
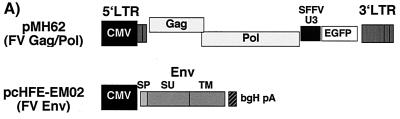
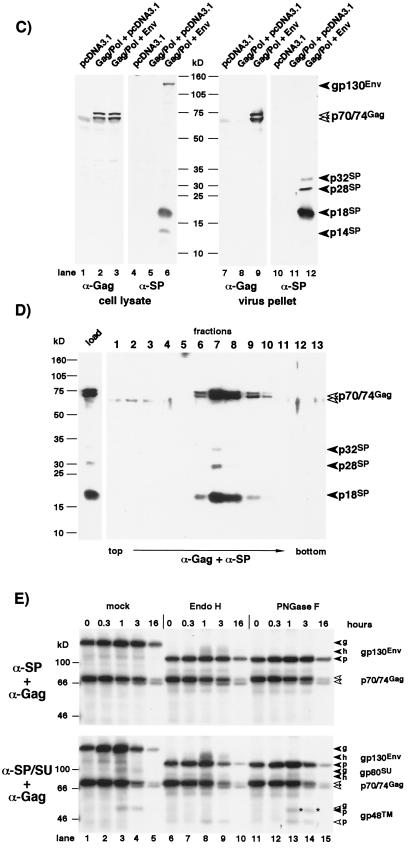
FIG. 2.
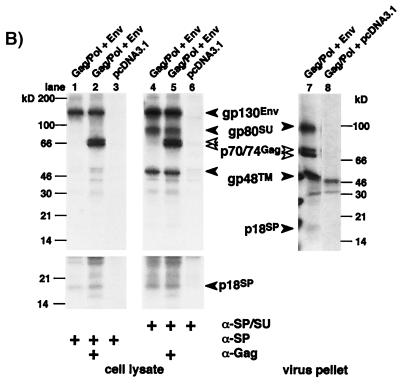
Identification of FV Env subdomains by different antisera. Cell or virus lysates of 293T cells transfected with the Gag/Pol-expressing FV vector pMH62 (Gag/Pol), wild-type FV Env expression construct pcHFE EM02 (Env), or empty expression vector (pCDNA3.1) as indicated were analyzed by RIPA or Western blotting. The identity of each FV protein is indicated. The gp48TM protein often comigrates with an immunoreactive cellular protein present in negative controls. (A) Schematic outline of the transfected expression constructs. SFFV, spleen focus-forming virus; bgH pA, bovine growth hormone polyadenylation site. (B) RIPA of cells labeled for 20 h using polyclonal rabbit sera specific for Gag (α-Gag), SPEnv/SU (α-SP/SU), or SPEnv (α-SP). A longer exposure of the lower part of the SDS–10% polyacrylamide gel with the cellular lysates is shown separately, and an autoradiogram of metabolically labeled purified FV particles separated by SDS-PAGE is shown to the right. (C) Western blot analysis (SDS-PAGE [12% gel]) of cellular lysates and purified FV particles using polyclonal rabbit sera specific for Gag (α-Gag) or SPEnv (α-SP). (D) Western blot analysis (SDS-PAGE [12% gel]) of equilibrium sedimentation gradient fractions using polyclonal rabbit sera specific for Gag (α-Gag) and SPEnv (α-SP). (E) Pulse-chase analysis of FV Env maturation. Transfected 293T cells were pulse-labeled for 30 min and then chased for different time periods as indicated at the top with fresh growth medium containing an excess of cold methionine and cysteine. Equal cell lysate samples were immunoprecipitated with FV-specific antisera as indicated to the left. Equivalent aliquots of the protein A eluates were incubated with glycosidases as indicated at the top prior to separation by SDS-PAGE (7.5% gel). Identities of the different forms (g [fully glycosylated], h [endo H resistant] and p [N-deglycosylated]) of gp130Env (solid arrows), gp80SU (shaded arrows), and gp48TM (open arrows) are indicated. The bands marked with asterisks at the 1- and 3-h time point after immunoprecipitation with anti-SP/SU antiserum and PNGase F treatment represent the fully N-deglycosylated form of SU running only slightly faster than the fully glycosylated form of TM.
The replication-deficient pMH62 vector (see Fig. 2A) was described previously (28). It expresses the FV Gag/Pol proteins and contains an internal spleen focus forming virus U3 promoter-directed enhanced green fluorescent protein (EGFP) marker gene expression cassette.
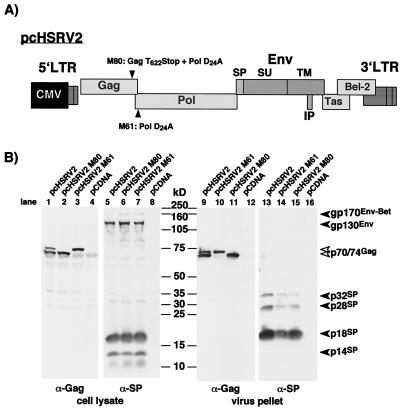
The parental human cytomegalovirus (CMV) immediate-early promoter-driven infectious proviral clone pcHSRV2 (see Fig. 3A) has been described previously (27). The pcHSRV2 mutant clones (M61 and M80) were generated by replacing a 1.83-kb SwaI/PacI fragment with respective PCR amplicons (18).
FIG. 3.
Analysis of FV SPEnv cleavage in FV protease-deficient proviral expression clones. (A) Schematic illustration of the mutant proviral constructs. Both constructs contain an active-site FV protease mutation, and M80 harbors in addition a stop codon introduced at the FV protease cleavage site in the FV Gag protein. IP, internal promoter. (B) 293T cells were transfected with the proviral expression clones or control expression vector as indicated at the top. Cellular lysates and purified viral particles were analyzed by Western blotting (SDS-PAGE [10% gel]) with polyclonal rabbit antisera specific for Gag (α-Gag) or SPEnv (α-SP). The identity of each FV protein is indicated.
The replication-deficient murine leukemia virus (MuLV) vector pczCFG2 fEGN is based on SFG GFPS65T (22). In pczCFG2 fEGN, the CMV enhancer/promoter replaces the U3 region of the 5′ long terminal repeat (LTR) and drives transcription in the producer cell; however, expression of the fEGN marker protein is driven by the reconstituted wild-type MuLV LTR upon reverse transcription and integration into the genome of the target cell. The fEGN marker gene was generated by recombinant PCR and contains the neomycin resistance gene fused in frame to the C terminus of the EGFP gene.
FV SPEnv-specific polyclonal antiserum.
The prokaryotic expression construct for the fusion protein of maltose binding protein (MBP) and HFV SPEnv was generated by inserting a Klenow enzyme-blunted BanI/EcoRI fragment of pcHFVenv EM50 into the XmnI/EcoRI sites of pMAL-C2 (New England Biolabs). The soluble fusion protein was generated in Escherichia coli TB1 cultures after induction with 0.5 mM isopropylthiogalactopyranoside (IPTG) for 3 to 6 h and affinity purified according to the manufacturer's instructions.
Cells.
The human kidney cell line 293T (4) and the fibrosarcoma cell line HT1080 were cultivated in Eagle's minimal essential medium and Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal calf serum and antibiotics.
Transfections and analysis of vector transduction.
Supernatants containing recombinant FV or MuLV retroviral particles were generated essentially as described earlier (22, 23, 32). FV supernatants were generated by cotransfection of 293T cells with the Gag/Pol-expressing FV vector pMH62 and an Env expression plasmid as indicated or by transfection of cells with the proviral expression construct pcHSRV2 and variants thereof. MuLV particles were obtained by cotransfection of 293T cells with the MuLV Gag/Pol expression vector pHIT60 (32), the MuLV retroviral vector pczCFG2 fEGN, and an Env expression vector as indicated.
The ability of each Env mutant to mediate infectivity was analyzed by transduction of HT1080 human fibrosarcoma cells with cell-free supernatant as described previously (15, 22). Briefly, 5 × 104 HT1080 cells were exposed to 1 ml of cell-free supernatant of 293T cells harvested 48 h posttransfection, and transduction efficiencies were determined by flow cytometry of the recipient cells 48 h later. Absolute percentages of EGFP-positive cells ranged from 30 to 60% and 2 to 4% for wild-type FV Env (EM02) with FV and MuLV vectors, respectively. Mock-transduced cells gave values of maximal 0.1% positive cells. All transduction experiments were performed at least three times; in each independent experiment, the values obtained with wild-type FV Env (EM02) were arbitrarily set to 100 in the case of FV capsids and to 1 in the case of MuLV capsids.
In some experiments involving FV vectors, intracellular viral particles were artificially released by a freeze-thawing of the transfected 293T cells and subsequent centrifugation and filtration of the supernatant through 0.45-μm-pore-size filters to remove cellular debris. The resulting supernatants were then assayed as described above.
Metabolic labeling and analysis of particle release.
For radioimmunoprecipitation analysis (RIPA), transiently transfected 293T cells were metabolically labeled with [35S]methionine and [35S]cysteine for approximately 20 h. Alternatively, cells were pulse-labeled for 30 min and chased for various time periods in fresh growth medium containing a 10-fold excess of cold methionine and cysteine. Subsequently the cells were lysed in RIPA buffer (20 mM Tris [pH 7.4], 0.3 M NaCl, 1% Triton X-100, 0.1% [wt/vol] sodium dodecyl sulfate [SDS]) containing protease inhibitors. Viral proteins were precipitated as described earlier (7, 23), using rabbit antisera directed against recombinant FV proteins and specific for Env (23) and Gag (13). For glycosidase treatment, protein A-Sepharose eluates were denatured by boiling in 0.5% SDS–1% β-mercaptoethanol and subsequently incubated with endoglycosidase H (endo H) peptide N-glycosidase or (PNGase F) in the appropriate incubation buffer as suggested by the manufacturer (New England Biolabs) prior to loading on gels for SDS-polyacrylamide gel electrophoresis (PAGE). Particle-associated proteins were analyzed after centrifugation through a 20% sucrose cushion as described previously (7, 23).
For Western blot analysis, transiently transfected 293T cells were lysed in RIPA buffer, and purified viral particles were obtained by ultracentrifugation as described above. Further purification by equilibrium sedimentation centrifugation using a 8.5 to 40% iodixonal (Optiprep; Gibco BRL) step gradient was essentially performed as described elsewhere (2). The protein samples were subjected to SDS-PAGE and semidry blotted onto nitrocellulose membranes (Amersham). The blots were incubated with rabbit antisera raised against recombinant HFV Gag (13) or the MBP-HFV SPEnv fusion protein described above and were developed with the Amersham ECL (enhanced chemiluminescence) detection system.
Cell surface biotinylation of 293T cells transiently transfected with the individual Env expression constructs was carried out essentially as described recently (29). Briefly, 293T cells were transiently transfected and metabolically labeled with [35S]methionine and [35S]cysteine. At 36 h after addition of the DNA, cell surface protein was labeled with N-hydroxysuccinimide–biotin (Calbiochem) at 1 mg/ml in phosphate-buffered saline for 30 min. Subsequently, the biotinylation reaction was stopped by adding phosphate-buffered saline containing 100 mM glycine prior to cell lysis in RIPA buffer. Lysates were precipitated with a FV-positive chimpanzee serum as described earlier (7, 23), separated by SDS-PAGE, and blotted onto nitrocellulose membranes (Hybond ECL; Amersham). Envelope protein expression at the cell surface was analyzed using streptavidin conjugated to horseradish peroxidase (Pierce), followed by detection with ECL (Amersham). The chemiluminescent biotin signal was allowed to fade overnight. Thereafter, the blot was exposed to X-ray film, and total cellular envelope expression was detected by autoradiography.
RESULTS
The FV SPEnv is cleaved posttranslationally, and cleavage products are viral particle associated.
A special structural feature of the FV Env protein is an unusually long N-terminal SP sequence with an SPC cleavage site predicted to be located after aa 86 (Fig. 1) (8, 35). The majority of secretory SP are cotranslationally cleaved; however, for some retroviral glycoproteins, e.g., HIV-1 and feline immunodeficiency virus (FIV), cleavage takes place late during intracellular transport (21, 34). To determine whether and when FV SP cleavage occurs we generated a FV SPEnv-specific polyclonal antiserum by immunizing rabbits with a protein containing the N-terminal 86 aa of HFV Env fused to MBP. The FV Env precursor glycoprotein gp130 was efficiently detected by this antiserum in immunoprecipitation analysis experiments using lysates of 293T cells cotransfected with the Gag/Pol-expressing FV vector pMH62 (28) and an expression vector for wild-type FV Env (Fig. 2A), indicating that gp130 still contains the SP (Fig. 2B, lane 1 to 3). Furthermore, upon longer exposure a specific faint band about 18 kDa in size was detected, but not the 80 kDa SU subunit (Fig. 2B, lanes 1 and 2).
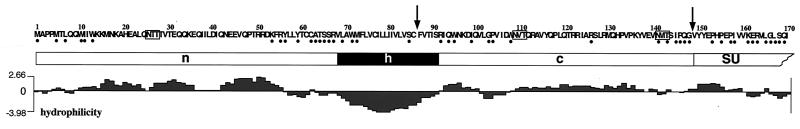
FIG. 1.
Schematic illustration of the N-terminal 170 aa of the HFV Env protein. The amino acid sequence starting from the first translation initiation codon in the Env ORF of HFV is given at the top. Amino acids conserved in Env proteins of different FV species are marked with black dots below the sequence. The SPC cleavage sites at position 86 as suggested by Flügel et al. (8) and Wang and Mulligan (35) and at position 148 as suggested in this study are indicated by arrows. Potential N-glycosylation sites are boxed. The predicted structural organization of this region of FV Env is schematically illustrated below, with the SP subdomains (n [N terminal], h [hydrophobic], and c [C terminal]) and the SU subunit indicated. At the bottom is a hydrophilicity plot of the region generated by the DNAstar Protean software.
In contrast, an anti-SP/SU serum raised against the N-terminal 571 aa (23) precipitated the precursor gp130 and, in addition, the p18 protein, gp80SU, and gp48TM (Fig. 2B, lanes 4 and 5). By Western blot analysis using the SP-specific antiserum, both the gp130 and the 18-kDa protein, and an additional 14-kDa protein were detected (Fig. 2C, lane 6). On autoradiograms of metabolically labeled FV particles derived from the supernatant of infected cell cultures and purified through a sucrose cushion, a faint band of 18 kD was detectable (Fig. 2B, lane 7). Western blot analysis of virus particle lysates using the SP-specific antiserum confirmed this protein to be the 18-kDa SP cleavage product (Fig. 2C, lane 12). Besides the 18-kDa protein, two additional minor cleavage products approximately 28 and 32 kDa in size were recognized by the SP-specific antiserum in viral particles (Fig. 2C, lane 12). To ensure the authenticity of these protein bands as integral parts of virus particles rather than inadvertently enriched cellular proteins, FV particles were further purified by equilibrium sedimentation gradient centrifugation. All three SP cleavage products described above were detected in fractions that contained the Gag proteins, demonstrating their particle association (Fig. 2D, fractions 6 to 10). Neither the gp130 precursor nor the p14 cleavage product seen in cell lysates was found in viral particle preparations (Fig. 2C, lane 12; Fig. 2D, fractions 6 to 10). Similar to what was observed with cell lysates, the gp80 SU subunit did not react with the anti-SP antiserum (Fig. 2C, lane 12; Fig. 2D).
For further analysis of the kinetics of FV SPEnv cleavage, transfected 293T cells were pulse-labeled for 30 min and then lysed immediately or chased for various time periods. Subsequently, maturation of the FV Env protein was analyzed by radioimmunoprecipitation using anti-SP or anti-SP/SU antisera combined with glycosidase treatment. As shown in Fig. 2E, gp130 was efficiently recovered with both anti-SP/SU and anti-SP antisera. Partially endo H-resistant forms of gp130 could be detected with both antisera at the 1- and 3-h time points, although somewhat more efficiently with the anti-SP/SU antiserum (Fig. 2E, lanes 8 and 9). Furthermore SU and TM subunit cleavage products were detectable only with the anti-SP/SU antiserum at the same time points. The TM subunit remained endo H sensitive during the complete chase period, indicating that the oligosaccharide chain are of high-mannose or hybrid but not of complex type.
Taken together, these data indicate that as described for the HIV-1 Env protein, no efficient cotranslational FV SPEnv processing occurs. However, in FV the SPEnv appears to be part of the mature virion. Since the SU subunit found in cells and viral particles no longer contains the SP, its removal probably occurs before or at the same time as SU/TM subunit processing.
The FV SPEnv is cleaved by a cellular protease.
Some retroviral glycoproteins are processed by the viral protease. The TM subunit p15E of MuLV Env, for example, is cleaved by the MuLV protease during or shortly after capsid budding to remove a 16-aa inhibitory peptide, thereby activating the fusogenic capacity of the protein (12, 16, 30, 31). Since the n-domain of the SP is located in the cytoplasm and therefore is theoretically accessible to the viral protease, we intended to determine whether the FV protease might be involved in SP cleavage. The FV protease removes a small 27-aa peptide from the C terminus of Gag as an essential step to retain viral infectivity (6, 36). Two protease active-site mutants in the context of an infectious molecular clone were used to address this question. Mutant pcHSRV2-M61 (D24A) inactivates the protease (20), whereas pcHSRV2-M80 has, in addition, a translational stop codon introduced at the protease cleavage site in the Gag open reading frame (ORF), terminating RAVN and thereby mimicking Gag cleavage (Fig. 3A). 293T cells were transfected with wild-type pcHSRV2 or the mutants, and FV Gag and Env processing was analyzed. As shown in Fig. 3B, efficient SP cleavage could be observed for both protease mutants, as indicated by the appearance of the cellular p18 and p14 (lanes 5 to 7) or viral p18, p28, and p32 cleavage products (lanes 13 to 15). Clearly, a cellular protease rather than the viral protease is responsible for FV SPEnv cleavage.
The FV SPEnv is glycosylated and is cleaved beyond aa 86.
The size of the major SP cleavage product of 18 kDa was larger than expected for an N-terminal peptide of 86 aa. Indeed, eukaryotic expression of the predicted 86-aa SP (EM50) yielded a protein of about 10 kDa (Fig. 4B, lane 8). To determine whether posttranslational modifications such as N-glycosylation and/or the use of an alternate cleavage site might account for this size discrepancy, several point mutants were analyzed. To test for N-glycosylation, the first three potential N-glycosylation sites in Env at N25, N109, and N141 (Fig. 1) were inactivated by N-to-Q changes either individually or in combinations (Fig. 4A). All mutants were able to generate infectious FV vector particles with similar infectivities as the wild-type Env (Fig. 4A). 293T cells were cotransfected with the individual mutants and the pMH62 vector, and cellular lysates were analyzed by Western blotting with SP- and Gag-specific antisera. Only inactivation of the second potential N-glycosylation site at N109 resulted in a change of the major cleavage product migration pattern (Fig. 4B, lane 2). Interestingly, this change resulted in comigration with the 14-kDa cleavage product (Fig. 4B, lane 1). Furthermore, glycosidase treatment of cell lysates prior to Western blot analysis had the same effect (data not shown). These data show that the FV SP is removed C terminally of N109 and, therefore, substantially larger than previously thought (8, 35). In addition, they imply that the 14-kDa cleavage product represents a form of gp18 lacking glycosylation at N109. As far as studied, however, SP glycosylation, does not appear to be essential for viral infectivity.
A computer-assisted comparison of all known FV Env sequences revealed a conserved IPQG motif at aa 148 of the HFV sequence. Interestingly, this conserved motif contains an PXG sequence as seen in C termini of many cleavable SP (25). To test whether this sequence motif might be involved in FV SPEnv cleavage, we analyzed the two mutants EM83 and EM84, bearing C86R and G148R mutation (Fig. 4A), respectively, in the putative −1 position of SPC cleavage sites. Similar mutations have been shown previously to inhibit SP cleavage (9). The EM83 mutation had no effect on SP cleavage (Fig. 4B, lane 10); however, infectivity was significantly reduced (Fig. 4A), indicating that C86 is structurally important for Env function but not for SP cleavage. In contrast, SP cleavage of the EM84 mutant was almost completely abolished (Fig. 4B, lane 11). In addition, no infectious FV vector particles were detected in the supernatant of EM84-transfected cells (Fig. 4A) because FV particle release by the EM84 mutant was heavily impaired, at least 10-fold compared to wild type (Fig. 4C, lane 21). These results show that the SP is not cleaved after C86 but support a role for SP cleavage of the conserved motif around aa 148. Furthermore, they indicate that SP cleavage is essential for efficient FV particle release and infectivity.
The n-region of the FV SP is required for FV membrane envelopment but not for targeting to the secretory pathway or envelope function.
The FV SPEnv contains four in-frame translation initiation codons upstream of the predicted SP cleavage site (Fig. 5A). To determine the actual translation initiation site and to examine whether certain domains of the FV SPEnv might be involved in FV particle maturation, we analyzed several N-terminal truncation and point mutants with respect to their cell surface transport and ability to support FV particle release (Fig. 5A). After cotransfection of 293T cells with the FV Gag/Pol-expressing vector pMH62 and the individual mutants, infectious FV vector particles were detectable only for the EM41 mutant, which utilizes the first translation initiation codon of the FV Env ORF, thereby containing a full-length SP (Fig. 5A). In agreement with the infectivity data, FV particle release was observed only with EM41 (Fig. 5B, lane 2). For the EM44 mutant, with the first 71 aa of the SP removed, no protein expression was detected (Fig. 5B, lane 5). For all other mutants, protein expression could be detected by RIPA in cell lysates (Fig. 5B). However, some of the mutants (EM42, EM71, EM72, and EM73) showed no detectable SU/TM processing (Fig. 5B, lanes 3 and 9 to 11). The reason for this is unclear. In addition, we analyzed cell-associated virus of the mutants in this study, because FV buds predominantly intracellularly and we previously identified a mutant FV Env protein that was deficient in particle release into the supernatant but still showed budding into intracellular compartments (28). Results obtained with the supernatants of freeze-thaw lysates of transfected cells indicated that potentially infectious FV particles were not intracellularly trapped in cases of those Env mutants that did not support release of infectious virus into the supernatant (data not shown). Electron microscopy analysis of deletion mutants revealed the presence of naked capsids in the cytoplasm of cells transfected with mutants not releasing infectious particles, while for EM41, which behaved like wild-type Env, particles associated with and budding through cellular membranes were observed (data not shown).
Interestingly, mutants with N-terminal deletions ranging from 5 up to 40 aa (EM42, EM43, EM70, and EM71) that were no longer able to support FV particle egress expressed protein at the cell surface (Fig. 5B, lanes 3, 4, 8, and 9) and gave rise to infectious pseudotyped MuLV capsids (Fig. 5A). Some of these mutants dramatically increased MuLV titers, one up to 150-fold (Fig. 5A). This observation correlated quite well with an enhanced cell surface biotinylation of the various mutants, although the EM43 and EM70 mutants, displaying similar cell surface expression levels (Fig. 5B, lane 4 and 8), showed a fivefold difference in their MuLV pseudotype titers. This indicated that the N-terminal truncations of the mutant Env proteins neither induced inherent defects in targeting the glycoprotein to the secretory pathway nor affected receptor binding and fusion capacities. The importance of N-terminal amino acid of the FV SP n-region for particle budding was further supported by analysis of point mutants in evolutionary conserved residues. Mutant EM52 had the N-terminal W10W13 motif replaced by alanines, EM53 had the C-terminal Y56Y59 motif replaced by alanines, and EM54 was a combination of both (Fig. 5A). The EM52 mutant as well as the EM54 double mutant no longer supported FV particle release, whereas results with EM53 were similar to those for the wild-type protein (Fig. 5A and C, lanes 13 to 15).
To further delineate the region of the SP required for FV particle release and infectivity, several mutants with internal deletions in the SP n-region were analyzed. Smaller deletions of 3 to 24 aa between aa 16 and 66, such as EM66, EM67, EM74, and EM76, were tolerated, resulting at the most in 45-fold-reduced infectivity (Fig. 5A and D, lanes 19, 20, 23, and 25). However, larger deletions of 25 to 50 aa, such as in EM68, EM69, and EM75, abolished infectivity, although for EM75 particle release into the supernatant could be detected (Fig. 5A and D, lanes 21, 22, and 24). 293T cells cotransfected with the EM66 mutant showed very strong syncytium formation, indicating that it had a highly increased fusogenic activity compared to the wild-type FV Env protein (data not shown). In contrast, all other internal deletion mutants showed no obvious difference in their fusogenic activity (data not shown). This might explain why no infectious MuLV vectors pseudotyped by EM66 could be detected.
Taken together, these data show that the region comprising the N-terminal 15 aa of the FV SPEnv, while being dispensable for targeting to the secretory pathway and proper envelope function, is specifically involved in the FV particle budding process. Furthermore, they point to a critical role of two conserved N-terminal tryptophan residues in this process. The central part of the SP n-region on the other side does not seem to play a crucial role, as it tolerates smaller deletions. However, there are some constraints regarding spacing of the N-terminal budding domain in respect to the h-region, as larger deletions negatively influence particle release and infectivity.
DISCUSSION
Recently it has become clear that SP can have several additional functions apart from being responsible and essential to target glycoproteins to the secretory pathway (reviewed in reference 25). For some retroviral envelope glycoproteins, a posttranslational cleavage of the SP sequence has been reported (21, 34). Similarly, we observed posttranslational cleavage of the FV SPENV sequence, as full-length FV gp130 was detected with an SP sequence-specific antiserum. The exact time point and cellular location of SP processing during the intracellular transport of FV Env cannot be deduced from our current analysis. However, we were able to detect neither an SU/TM Env precursor protein lacking the SP sequence nor an SP/SU intermediate. This could mean that the SP-SU and SU-TM cleavage events occur simultaneously or within a very short time. A more detailed analysis of FV Env subunit processing and intracellular transport is needed to resolve this question. Interestingly, cleavage products containing the SP sequence were found not only in cell lysates but also in purified FV particles, which, to our knowledge, has not been reported for any other viral glycoprotein.
Positive charges in the n-region of the SP sequence of the HIV-1 Env protein have been shown to negatively influence intracellular transport and secretion (21). Similarly FV Env mutants with deletions in the n-region resulting in a reduction of the overall net positive charge yielded in increased cell surface expression. Noteworthy, all of these mutants still contained a wild-type ER retrieval signal in the CyD of the TM subunit. This domain influences intracellular distribution of FV Env but is dispensable for particle release and infectivity (10, 11, 28, 29). This could mean that the ER retrieval signal requires the interaction with some sequences of the cytoplasmic SP n-region for proper function or, alternatively, that the n-region of the SP is the major determinant for cell surface expression of the FV Env protein. Further experiments are necessary to clarify these phenomena.
The most intriguing finding of our analysis, however, is that the n-region of the FV SPEnv, being dispensable for targeting to the secretory pathway and proper envelope function, is essential for budding of FV capsids across cellular membranes and their release into the supernatant. To our knowledge, this is the first report that the SP domain of a retroviral glycoprotein is involved in particle maturation. The analysis of deletion and point mutations shows that this budding domain of the FV SPEnv sequence comprises the N-terminal 15 aa and that two evolutionarily conserved tryptophan residues located within are critical for FV particle release and Env incorporation. Furthermore, the spacing of this domain relative to the cellular membrane seems to influence its function. The results point to an interaction of the FV SPEnv n-domain with the FV capsid. Obviously, further experiments are necessary to determine whether a direct Env-Gag interaction takes place and cellular proteins are involved. From our previous work it is clear that other FV Env domains, such as the MSD of the TM subunit, are also involved in FV budding (28). This is shown by the fact that FV Env mutants with wild-type SP and deleted MSD and CyD failed to support FV particle egress when alternatively attached to the cell surface through a phosphoglycolipid membrane anchor (28). Based on preliminary results with heterologous Env SP chimeras (unpublished observations), we currently think that the FV SPEnv budding domain mediates the primary interaction with the FV capsid and an interaction with MSD plays a role later during the budding process.
Interestingly, FV Gag proteins, unlike other retroviral Gag proteins, are not processed into matrix, capsid, and nucleocapsid subunits, and as mentioned earlier, wild-type FV capsids are not found to be associated with cellular membranes in the absence of FV Env expression. Therefore, it may be conceivable that the FV SPEnv performs functions in viral assembly and budding, such as membrane targeting of the capsid, analogously to the matrix subunit of other retroviruses. Furthermore, it is likely that the particle-associated SP cleavage products have additional functions in the FV replication cycle. Even a role of SP in binding to the cellular receptor or fusion of viral and cellular membranes should be addressed in further studies.
Surprisingly, deletion of the N-terminal budding domain of the FV Env protein but not internal SP n-region deletions dramatically increase pseudotype titers of MuLV capsids. This shows that the budding domain is physically separable from those SP domains required for targeting to the secretory pathway and normal envelope function, namely, receptor binding and membrane fusion. Furthermore, these result suggest an inhibitory role of this region for FV Env incorporation into heterologous retroviral particles, which may be a cause for the poor pseudotyping capacity observed for the wild-type FV Env protein (22). However, to determine if this is an active exclusion of FV Env proteins containing this domain from heterologous budding particles or simply a result of the different levels of cell surface expression observed for most of these mutants, a more detailed analysis is required.
Based on our findings, it will be interesting to analyze functions of other retroviral glycoproteins SP for Env incorporation and particle maturation. The FIV Env (33, 34) and the mouse mammary tumor virus Env (1, 17), for example, bear also unusually long SP sequences. FIV Env can tolerate extensive deletions in the SP sequence with no effect on membrane targeting and intracellular transport; however, the effects of such deletions on envelope particle incorporation and infectivity have not been investigated (33). Similarly, SP mutations and chimeras of HIV-1 Env have also been examined only with respect to their intracellular transport (5, 21).
ACKNOWLEDGMENTS
We thank Jörg Enssle for constructing pcHSRV2-M61 and -M80, Ralf Bartenschlager for helpful discussion, and Ottmar Herchenröder for critical reading of the manuscript.
This work was supported by grants from the Bayerische Forschungsstiftung, DFG (Li621/2-1, Li621/2–3, SFB479, Re627/6-1, and Europäisches Graduiertenkolleg “Gene regulation in and by microbial pathogens”), and EU (BMH4-CT97-2010), Bayerische Forschungsstiftung (FORBEN).
REFERENCES
- 1.Arthur L O, Copeland T D, Oroszlan S, Schochetman G. Processing and amino acid sequence analysis of the mouse mammary tumor virus env gene product. J Virol. 1982;41:414–422. doi: 10.1128/jvi.41.2.414-422.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin D N, Linial M L. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J Virol. 1999;73:6387–6393. doi: 10.1128/jvi.73.8.6387-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin D N, Linial M L. The roles of Pol and Env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Bridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellerbrok H, D'Auriol L, Vaquero C, Sitbon M. Functional tolerance of the human immunodeficiency virus type 1 envelope signal peptide to mutations in the amino-terminal and hydrophobic regions. J Virol. 1992;66:5114–5118. doi: 10.1128/jvi.66.8.5114-5118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J Virol. 1997;71:7312–7317. doi: 10.1128/jvi.71.10.7312-7317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Muller J G, Rethwilm A. Foamy virus particle formation. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flügel R M, Rethwilm A, Maurer B, Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987;6:2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folz R J, Nothwehr S F, Gordon J I. Substrate specificity of eukaryotic signal peptidase. Site-saturation mutagenesis at position −1 regulates cleavage between multiple sites in human pre (delta pro) apolipoprotein A-II. J Biol Chem. 1988;263:2070–2078. [PubMed] [Google Scholar]
- 10.Goepfert P A, Shaw K, Wang G, Bansal A, Edwards B H, Mulligan M J. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J Virol. 1999;73:7210–7217. doi: 10.1128/jvi.73.9.7210-7217.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goepfert P A, Shaw K L, Ritter G D, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn H, Baunach G, Brautigam S, Mergia A, Neumann Haefelin D, Daniel M D, McClure M O, Rethwilm A. Reactivity of primate sera to foamy virus Gag and Bet proteins. J Gen Virol. 1994;75:2635–2644. doi: 10.1099/0022-1317-75-10-2635. [DOI] [PubMed] [Google Scholar]
- 14.Heinkelein M, Pietschmann T, Jarmy G, Dressler M, Imrich H, Thurow J, Lindemann D, Bock M, Moebes A, Roy J, Herchenroder O, Rethwilm A. Efficient intracellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 2000;19:3436–3445. doi: 10.1093/emboj/19.13.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the Pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson L E, Sowder R, Copeland T D, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson L E, Sowder R, Smythers G, Oroszlan S. Terminal amino acid sequences and proteolytic cleavage sites of mouse mammary tumor virus env gene products. J Virol. 1983;48:314–319. doi: 10.1128/jvi.48.1.314-319.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, White T J, editors. PCR protocols; a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 177–183. [Google Scholar]
- 19.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 20.Konvalinka J, Löchelt M, Zentgraf H, Flügel R M, Kräusslich H G. Active foamy virus proteinase is essential for virus infectivity but not for formation of a Pol polyprotein. J Virol. 1995;69:7264–7268. doi: 10.1128/jvi.69.11.7264-7268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Luo L, Thomas D Y, Kang C Y. Control of expression, glycosylation, and secretion of HIV-1 gp120 by homologous and heterologous signal sequences. Virology. 1994;204:266–278. doi: 10.1006/viro.1994.1531. [DOI] [PubMed] [Google Scholar]
- 22.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindemann D, Rethwilm A. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J Virol. 1998;72:4088–4094. doi: 10.1128/jvi.72.5.4088-4094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 26.Martoglio B, Graf R, Dobberstein B. Signal peptide fragments of preprolactin and HIV-1 p-gp160 interact with calmodulin. EMBO J. 1997;16:6636–6645. doi: 10.1093/emboj/16.22.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moebes A, Enssle J, Bieniasz P D, Heinkelein M, Lindemann D, Bock M, McClure M O, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietschmann T, Heinkelein M, Heldmann M, Zentgraf H, Rethwilm A, Lindemann D. Foamy virus capsids require the cognate envelope protein for particle export. J Virol. 1999;73:2613–2621. doi: 10.1128/jvi.73.4.2613-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietschmann T, Zentgraf H, Rethwilm A, Lindemann D. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J Virol. 2000;74:4474–4482. doi: 10.1128/jvi.74.10.4474-4482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rein A, Mirro J, Gordon Haynes J, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephens E B, Butfiloski E J, Monck E. Analysis of the amino terminal presequence of the feline immunodeficiency virus glycoprotein: effect of deletions on the intracellular transport of gp95. Virology. 1992;190:569–578. doi: 10.1016/0042-6822(92)90894-u. [DOI] [PubMed] [Google Scholar]
- 34.Verschoor E J, Hulskotte E G, Ederveen J, Koolen M J, Horzinek M C, Rottier P J. Post-translational processing of the feline immunodeficiency virus envelope precursor protein. Virology. 1993;193:433–438. doi: 10.1006/viro.1993.1140. [DOI] [PubMed] [Google Scholar]
- 35.Wang G, Mulligan M J. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J Gen Virol. 1999;80:245–254. doi: 10.1099/0022-1317-80-1-245. [DOI] [PubMed] [Google Scholar]
- 36.Zemba M, Wilk T, Rutten T, Wagner A, Flügel R M, Löchelt M. The carboxy-terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology. 1998;247:7–13. doi: 10.1006/viro.1998.9234. [DOI] [PubMed] [Google Scholar]