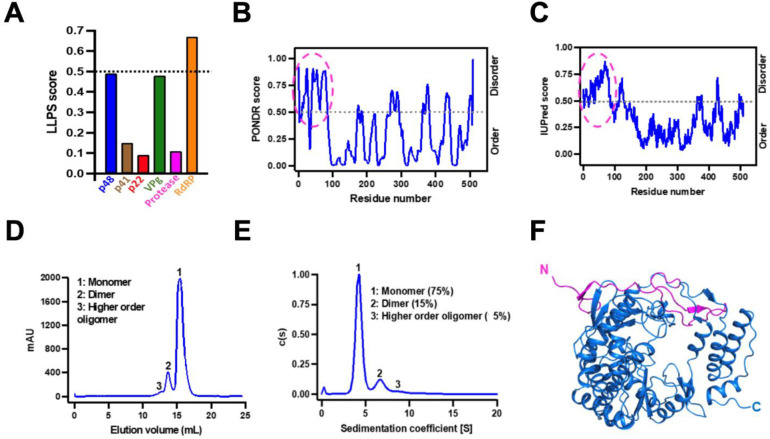
Figure 1. GII.4 RdRp has the required properties to undergo LLPS.
(A) Bioinformatics analysis of the primary amino acid sequences of all nonstructural proteins of HuNoV GII.4 to predict LLPS propensity using DeePhase. The dotted black line indicates the threshold LLPS score of proteins to undergo LLPS. (B-C) The disorder prediction of GII.4 RdRp primary amino acid sequence using bioinformatics tools (B) PONDR (C) IUPred2. The dotted black line in both panels indicates the threshold disorder score and the dotted pink circle shows the predicted disordered N-terminal region. (D-E) The oligomeric state of GII.4 RdRp analyzed using (D) size exclusion chromatography and (E) sedimentation velocity analytical ultracentrifugation. (F) A cartoon representation of the crystal structure of GII.4 RdRp showing the disordered/flexible N-terminal region colored in pink.