Abstract
Venezuelan equine encephalitis viruses (VEEV) belonging to subtype IC have caused three (1962–1964, 1992–1993 and 1995) major equine epizootics and epidemics. Previous sequence analyses of a portion of the envelope glycoprotein gene demonstrated a high degree of conservation among isolates from the 1962–1964 and the 1995 outbreaks, as well as a 1983 interepizootic mosquito isolate from Panaquire, Venezuela. However, unlike subtype IAB VEEV that were used to prepare inactivated vaccines that probably initiated several outbreaks, subtype IC viruses have not been used for vaccine production and their conservation cannot be explained in this way. To characterize further subtype IC VEEV conservation and to evaluate potential sources of the 1995 outbreak, we sequenced the complete genomes of three isolates from the 1962–1964 outbreak, the 1983 Panaquire interepizootic isolate, and two isolates from 1995. The sequence of the Panaquire isolate, and that of virus isolated from a mouse brain antigen prepared from subtype IC strain P676 and used in the same laboratory, suggested that the Panaquire isolate represents a laboratory contaminant. Some authentic epizootic IC strains isolated 32 years apart showed a greater degree of sequence identity than did isolates from the same (1962–1964 or 1995) outbreak. If these viruses were circulating and replicating between 1964 and 1995, their rate of sequence evolution was at least 10-fold lower than that estimated during outbreaks or that of closely related enzootic VEEV strains that circulate continuously. Current understanding of alphavirus evolution is inconsistent with this conservation. This subtype IC VEEV conservation, combined with phylogenetic relationships, suggests the possibility that the 1995 outbreak was initiated by a laboratory strain.
Venezuelan equine encephalitis viruses (VEEV) are single-stranded, message-sense RNA alphaviruses (Togaviridae) with a genome of approximately 11 kb (32, 35). The genome encodes four nonstructural proteins (nsP1 to nsP4) comprising the 5′ two-thirds of the genome (nucleotide positions 1 to 7514), and three structural proteins (capsid, E2, and E1 envelope glycoproteins) are encoded by a subgenomic message identical to the 3′ one-third of the genome. VEEV are transmitted among vertebrate hosts by mosquitoes and are serologically classified into six antigenic subtypes (I to VI). These viruses can also be classified into two distinct epidemiological types. Only epizootic/epidemic viruses in antigenic subtypes IAB and IC have been responsible for major, sporadic outbreaks of human and equine disease (44). In contrast, enzootic VEEV in subtypes ID, IE, IF, and II-VI are transmitted continuously in sylvatic foci among small mammalian reservoir hosts by mosquito vectors in the Culex (Melanoconion) subgenus (44). Although these enzootic viruses can cause severe human illness like epizootic strains (12, 46), they are not efficiently amplified by equines and have therefore not been associated with major epidemics. Epizootic VEEV (subtypes IAB and IC) cause major epidemics by exploiting equines as highly efficient amplification hosts and are transmitted by a wide range of non-Melanoconion mosquito vectors (37, 38, 44). With the exception of a 1983 VEEV subtype IC mosquito isolate from Panaquire in Miranda State, north-central Venezuela (Fig. 1) (22), no epizootic subtype IAB or IC VEEV has ever been isolated during interepizootic periods of up to 19 years (44, 47).
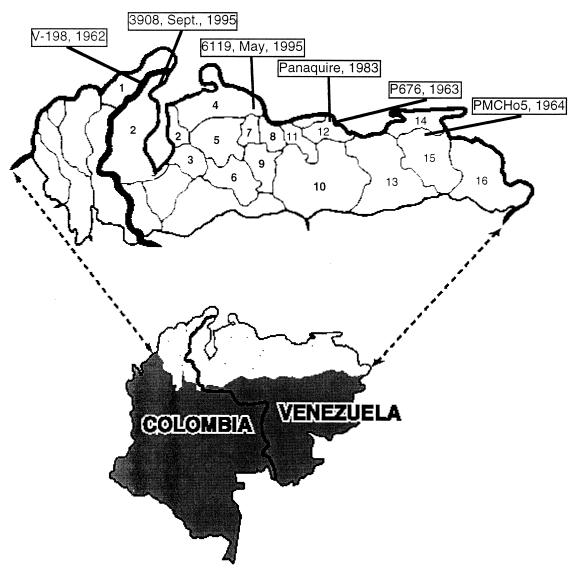
FIG. 1.
Map of the regions of Venezuela and Colombia affected by the 1962–1964 and 1995 outbreaks. Sites of isolation of the VEEV subtype IC isolates that we studied are identified by boxed strain names. The following bold numbers show the states involved in both the 1962–1964 and 1995 outbreaks: 1, Guajira (Colombia); 2, Zulia; 3, Trujillo; 4, Falcon; 5, Lara; 6, Portuguesa; 7, Yaracuy; 8, Carabobo; 9, Cojedes; and 10, Guarico. The following numbers show states involved only in the 1962–1964 outbreak: 11, Aragua; 12, Miranda; 13, Anzoategui; 14, Sucre; 15, Monagas; and 16, Delta Amacuro.
The first documented epidemic caused by a subtype IC VEEV began in October 1962, when clinical cases consistent with VEE were documented in the northwestern Venezuelan state of Zulia on the western shore of Lake Maracaibo (Fig. 1). Equine encephalitis outbreaks were noted in the same region during the preceding months. The outbreak moved both eastward and westward, and by the end of 1963 it had infected an estimated 14,000 people in Venezuela and also caused focal outbreaks within the Guajira Department of neighboring Colombia. The outbreak persisted into 1964, when the final isolate was made from a viremic human in eastern Venezuela (2). However, additional human cases from eastern Colombia in 1966 may represent an extension of this outbreak (36).
In 1995, an extensive epidemic and equine epizootic occurred in the same regions of western Venezuela and eastern Colombia (25, 53) (Fig. 1). In late April, similar to the findings in 1962, veterinarians with the Venezuelan Ministry of Agriculture reported focal outbreaks of disease in equines suggestive of VEE. Shortly thereafter, cases of febrile illness among humans living in the same areas were documented. However, unlike the 1962–1964 outbreak, which began on the western side of Lake Maracaibo, initial VEE cases in 1995 occurred in the eastern part of Falcon State, followed by similar reports from Carabobo, Yaracuy, and Lara States. By mid-July, the epidemic moved westward across Falcon State and into northeastern Zulia State. At the end of August, the disease appeared in western Zulia State and a major epidemic ensued in rural areas north of Maracaibo. Between August 27 and October 7, a total of 10,058 suspected human cases of VEE with 11 deaths were reported from Zulia State. During September and October, a large number of human and equine cases of VEE occurred in neighboring La Guajira Department of Colombia (25, 53). Estimates of total human cases during 1995 approached 100,000, with an estimated number of fatalities exceeding 300 (25, 53).
The viral source of VEE epizootics has remained an enigma for many years. Beginning in the 1930s, early subtype IAB epizootic VEEV isolates were used for the preparation of inactivated vaccines. Incompletely inactivated preparations may have initiated some VEE outbreaks until the establishment of the live, attenuated TC-83 IAB strain in the early 1970s as the vaccine of choice. However, subtype IC epizootic strains were not used for vaccine preparation, and so IC outbreaks cannot be explained by improperly prepared vaccines.
Previous investigations of a 1992–1993 VEEV subtype IC outbreak provided compelling evidence for the emergence of epizootic viruses via mutation of sympatric, enzootic subtype ID progenitors (15, 21, 24). However, the IC viruses isolated during the 1992–1993 outbreak are genetically unrelated to those isolated during 1962–1964 and 1995. Unlike the 1992–1993 subtype IC outbreak, no closely related enzootic ID viruses were isolated in the region where the 1995 outbreak began (45). Furthermore, the only enzootic VEEV strain ever isolated from this region in northern Venezuela was genetically unrelated to the 1995 emergence (29). Partial nucleotide sequences of the PE2 glycoprotein precursor gene of some isolates from both the 1962–1964 and 1995 outbreaks were identical, suggesting reemergence from a common source (53). In addition, a 1983 mosquito isolate from Panaquire in Miranda State had a partial PE2 sequence identical to that of a 1963 subtype IC isolate (P676) from the same region of north-central Venezuela, suggesting continuous interepizootic transmission of epizootic subtype IC viruses in northern Venezuela.
To examine more conclusively the plausibility of a cryptic epizootic transmission cycle in northern Venezuela, we sequenced the complete genome of three VEEV isolates from the 1962–1964 outbreak, the interepizootic Panaquire isolate from 1983, and two viruses from the 1995 outbreak. Rates of nucleotide substitution between 1964 and 1995 were compared to those during the outbreaks and to those of closely related, subtype ID viruses from Venezuela and Colombia. These estimates indicated that if the epizootic VEEV were circulating and replicating between 1964 and 1995, their rate of nucleotide substitution was at least 10-fold lower than that estimated to occur during enzootic or epizootic alphavirus transmission cycles. This conservation, as well as phylogenetic relationships, suggested a possible laboratory source for the 1995 outbreak.
MATERIALS AND METHODS
Virus preparation and PCR.
The VEEV isolates analyzed in this study are listed in Table 1. All virus strains were originally isolated either from human serum or pools of field-collected mosquitoes. For sequencing, virus stocks were prepared in C6/36 mosquito, baby hamster kidney (BHK-21), or African green monkey kidney (Vero76) cells at 37°C at a multiplicity of infection of less than 0.1. All viruses were passaged on separate days to avoid the possibility of cross-contamination. Virus strain P676-ag was isolated from an aliquot of sucrose-acetone-extracted mouse brain antigen that was prepared in 1982 from the P676 strain and used for serology at the National Institute of Hygiene in Caracas during 1983.
TABLE 1.
VEEV subtype IC strains used in genetic analyses
| Virus strain | Source | Location | Date | Cell type-passage levela | GenBank accession no. |
|---|---|---|---|---|---|
| V-198 | Human | La Guajira Department, Colombia | 1962 | sm-1, V-1 | U55342 |
| P676-gbb | Aedes triannulatus | Miranda State, Venezuela | Aug. 1963 | sm-1, V-7 | L04653 |
| P676-ag | A. triannulatus | Miranda State, Venezuela | Aug. 1963 | sm-4, BHK-1 | AF375051 |
| Panaquire | Mosquito | Panaquire, Miranda State, Venezuela | 1983 | sm-2, V-2, BHK-2 | |
| PMCHo5 | Human | Monagas State, Venezuela | 1964 | sm-1, V-1, BHK-1 | U55345 |
| 6119 | Human | Falcon State, Venezuela | May 24, 1995 | V-1, BHK-1 | U55347 |
| 3908 | Human | Sinamaica, Zulia State, Venezuela | Sept. 16, 1995 | C6/36-1 | U55350 |
BHK, BHK-21 cells; V, Vero-76. cells; sm, suckling mouse brain; C6/36, larval Aedes albopictus mosquito cells.
Strain sequenced previously (15).
Cytopathic effects (CPE) were observed in all cultures within 48 h of inoculation. After CPE was evident, RNA was extracted from 250 μl of culture supernatant by using TRIZOL LS (Gibco-BRL, Gaithersburg, Md.) as described previously (4). cDNA was synthesized by using the reverse-sense primer T25V-Mlu (−) or V-7651 (−) (Table 2) and Superscript reverse transcriptase (BRL) according to the manufacturer's recommendations.
TABLE 2.
Primers used for amplification and sequencing of VEEV subtype IC strains
| Primer namea | Sequence (5′-3′)b | Predicted melting temp (°C) |
|---|---|---|
| 5′ Xba (+) | GTGCCAAGCTTCTAGAAATTTAATACGACTC | 73.9 |
| ACTATAATGGGCGGCGCATGAGAGAAG | ||
| V-478 (+) | GGCAAGTCGCTGTTTACC | 60.9 |
| V-796 (−) | TTCTCGTGGTAGATGGTCGAGC | 67.5 |
| V-952 (+) | ATGCTGCTACGATGCACCG | 67.3 |
| V-1245 (−) | GCCTCTCATCTTCTTGATC | 56.3 |
| V-1526 (+) | CGATGAGGCTAAGGAAGTGCG | 67.9 |
| V-1714 (−) | GAAAGCACAGCGTAAGAGC | 60.3 |
| V-2010 (+) | AAAACTGTCAAGCCCAGCGAGCAC | 73.0 |
| V-2193 (−) | TGGTTGGTACTTGGTAAGGAG | 60.4 |
| V-2519 (+) | CCTGAAAGTGCATTTTAACC | 58.7 |
| V-3130 (+) | CCACTGAACAATGGAACAC | 58.9 |
| V-3419 (+) | GCGCAATTATGATCCGCG | 67.4 |
| V-3800 (+) | CGTCAGCATAGGTTATGGTTAC | 59.9 |
| V-4039 (−) | CCCTCGCACCACATGATATG | 66.3 |
| V-4669 (+) | AGGCGGCCAAGGATATAG | 61.5 |
| V-4738 (−) | TCACCGAGGATATACATGC | 58.4 |
| V-4883 (−) | GAGGCTTTTAGGCGTTGTAC | 60.3 |
| V-5065 (+) | AGAACCAATCCACAGAGGGGAC | 67.4 |
| V-5251 (−) | AGGAATGGACCAGGATGAGCTGG | 71.3 |
| V-5649 (+) | GAGGAGTTTGAGGCGTTC | 59.9 |
| V-6208 (+) | GCAGCTTTCCAAAGAAAC | 57.5 |
| V-6894 (+) | GAGCTGTTGACGCTGATTGA | 64.2 |
| V-6972 (−) | GGATTTCATCATGGCTCC | 60.4 |
| V-7514 (+) | ACYCTCTACGGCTRACCTRA | 59.2–61.6 |
| V-7651 (−) | CATCGCCAGAAAAGGGTCG | 67.8 |
| V-7886 (+) | AACAAGAAACCAGGCAAGAG | 60.8 |
| V-8659 (+) | AATTGAGGCAGTGAAGAGCGAC | 66.6 |
| V-8953 (−) | CTGCCTACAGGATTAAAT | 51.4 |
| V-9747 (+) | CGTGCCTAACTCCTTACC | 57.2 |
| V-9847 (−) | TGTTCCATAGGTGATCCAAGGAC | 65.9 |
| V-10247A (+) | TACCCNTTYATGTGGGG | 59.7–60.8 |
| V-10566 (+) | TTCCTGAGTATGGGGCAG | 61.2 |
| V-10664 (−) | TCGCTCCTGCTTTGGGTCTCTG | 71.8 |
| V-10830 (+) | CATTAGCCTTTGACATTCCC | 60.9 |
| T25V-Mlu (−) | TTACGAATTCACGCGTTTTTTTTTTTTTTTTTTTTTTTTTV | 50.4–51.4 |
Each numbers represents the 5′ genomic sequence position to which the primer anneals; the RNA strand orientation of each primer is given in parentheses.
R = A or G; Y = C or T; V = A, C, or G; N = A, C, G, or T.
The VEEV genomes were amplified in six overlapping amplicons by PCR. The primer sequences and their genome positions are listed in Table 2. The amplicons were (i) the 5′ end of the genome to V-1714; (ii) V-1526 to V-4738; (iii) V-3800 to V-5251; (iv) V-5065 to V-7651 for the nonstructural region of the genomes; (v) V-7514 to V-9847; and (vi) V-9747 to the poly(A) tail of the genome for the 26S structural region. Amplicons were synthesized using Pfu Turbo high-fidelity DNA polymerase (Stratagene, La Jolla, Calif.) with 300 ng of each antisense and sense primer. Amplification was conducted for 30 cycles with an annealing temperature 5°C below the lowest predicted melting temperature (Table 1) of the primer pair. A 2-min extension time at 72°C was allowed for each kilobase amplified.
Sequencing and sequence alignments.
Amplicons were sequenced directly, using a Big Dye Ready Reaction DNA sequencing kit (Perkin-Elmer, Foster City, Calif.) on an Applied Biosystems Prism 377 automated sequencer according to the manufacturer's protocol. Sequencing of the six individual amplicons was performed with the terminal PCR primers, and internal sequences were obtained by use of sequencing primers listed in Table 2. Sequences were aligned using the PILEUP program in the Wisconsin Package (Genetics Computer Group). Phylogenetic analysis was performed using the PAUP 4.0 (40) maximum parsimony and maximum likelihood programs. Parsimony analysis was performed using the heuristic algorithm with unordered characters. Bootstrapping (9) was performed using 1,000 replicates to estimate confidence indices of groupings within the phylogenetic trees. An outgroup comprised of subtype 1E strains MenaII (14) and 68U201 (18) was used to root the tree, and additional subtype I viruses described previously (13–15, 33) were included in the analysis.
Evolution rate estimations.
Evolutionary rates for enzootic VEEV lineages were estimated from maximum parsimony trees generated from partial PE2 gene sequences of a large number of subtype ID viruses sampled in the same regions of Colombia and Venezuela from the 1970s through 1997 (17). Rates were calculated using two methods: (i) a regression analysis in which each sequence within a geographically restricted clade was compared to the predicted sequence of its hypothetical ancestor based on maximum parsimony analysis, and the percent difference was plotted versus the year of isolation, and (ii) identification of sister sequences that were closely related and isolated at least 5 years apart in the same geographical region. This sister sequence method was also applied to trees generated from complete genomic sequences (45). The differences in nucleotide changes depicted in branch lengths separating each sister sequence from the predicted common ancestor were divided by the number of years between isolations to yield rates expressed as synonymous changes per year, and several estimates were compared to provide an estimated mean and standard deviation.
Serial virus passages.
To ascertain the role of cell culture passage history in some sequence differences, selected isolates were passaged serially in either BHK-21 or Vero76 cells at a multiplicity of infection of less than 0.1. Passages were conducted every 24 h at 37°C. Aliquots of cell culture medium were removed, and reverse transcription-PCR was performed as described above for the PE2 protein using primers V-8369 and V-9207 described previously (53).
Nucleotide sequence accession numbers.
The nucleotide sequences were submitted to the GenBank library. Accession numbers are listed in Table 1.
RESULTS
Virus isolation.
We obtained sucrose-acetone-extracted mouse brain antigen prepared from strain P676 and used in the Caracas laboratory where the Panaquire strain was isolated. This antigen preparation was resuspended and inoculated into Vero cells, and CPE appeared on the second day. The virus was characterized antigenically using monoclonal antibodies (25) and determined to belong to VEEV subtype IC.
Sequence analyses.
The complete genomic sequences excluding the 5′-terminal primer template sequences (11,391 total nucleotides) were determined for strains 6119 and 3908 from the 1995 outbreak, the 1983 interepizootic Panaquire isolate, and strains V-198, P676-ag, and VE64-PMCHo5 from the 1962–1964 outbreak. A complete list of sequence differences is presented in Table 3. When these sequences were compared, there were 35 variable nucleotides and 10 variable deduced amino acids. The majority of the nucleotide (31 of 35) and amino acid (9 of 10) sequence differences were found within the nonstructural protein genes, especially nsP3; however, sequence differences were found in every gene except the E3 and 6K protein genes.
TABLE 3.
Nucleotide and amino acid sequence differences between VEEV subtype IC isolates
| Genomic nucleotide positiona | Protein geneb | Nucleotide (amino acid)e
|
||||||
|---|---|---|---|---|---|---|---|---|
| P676-gb (1963c) | V-198 (1962) | P676-ag (1963) | PMCHo5 (1964) | Panaquire (1983) | 6119 (1995) | 3908 (1995) | ||
| 422 | nsP1 | T | A | A | A | A | A | A |
| 476 | nsP1 | G | A | |||||
| 1247 | nsP1 | T | C | |||||
| 1442 | nsP1 | G | A | |||||
| 1571 | nsP1 | T | A | A | A | A | A | A |
| 2051 | nsP2 | C | T | T | T | T | T | T |
| 2579 | nsP2 | C | T | T | T | T | T | T |
| 2668d | nsP2 (339) | T (Val) | A (Glu) | A (Glu) | A (Glu) | A (Glu) | A (Glu) | A (Glu) |
| 2947d | nsP2 (432) | T (Ile) | C (Thr) | C (Thr) | C (Thr) | C (Thr) | C (Thr) | C (Thr) |
| 3407 | nsP2 | C | T | T | ||||
| 3941 | nsP2 | C | T | |||||
| 4049 | nsP3 | G | A | |||||
| 4100 | nsP3 | T | C | C | C | C | C | C |
| 4187 | nsP3 | A | G | G | G | G | G | G |
| 4213d | nsP3 (60) | C (Ala) | T (Val) | T (Val) | T (Val) | T (Val) | T (Val) | T (Val) |
| 4215d | nsP3 (61) | G (Ala) | A (Thr) | A (Thr) | A (Thr) | A (Thr) | A (Thr) | A (Thr) |
| 4226 | nsP3 | C | T | T | T | T | T | T |
| 4241 | nsP3 | A | G | G | G | G | G | G |
| 4244 | nsP3 | A | C | C | C | C | C | C |
| 4974 | nsP3 | C | T | |||||
| 5291 | nsP3 | T | C | |||||
| 5344d | nsP3 (437) | G (Gly) | A (Glu) | A (Glu) | A (Glu) | A (Glu) | A (Glu) | A (Glu) |
| 5350d | nsP3 (439) | T (Val) | C (Ala) | C (Ala) | ||||
| 5453 | nsP3 | G | A | |||||
| 5469d | nsP3 (479) | C (Pro) | T (Ser) | |||||
| 5474 | nsP3 | T | C | |||||
| 6182 | nsP4 | T | G | |||||
| 6497 | nsP4 | T | A | |||||
| 6663d | nsP4 (320) | G (Asp) | T (Tyr) | T (Tyr) | T (Tyr) | T (Tyr) | T (Tyr) | T (Tyr) |
| 7229 | nsP4 | C | T | T | ||||
| 7312d | nsP4 (536) | T (Val) | C (Ala) | |||||
| 9158d | E2 (199) | A (Lys) | G (Glu) | G (Glu) | G (Glu) | G (Glu) | G (Glu) | |
| 9622 | E2 (199) | T | C | |||||
| 10306 | E1 | G | A | |||||
| 10912 | E1 | C | T | |||||
Bold text denotes nucleotide differences that are not unique to the P676-gb sequence.
Numbers in parentheses refer to amino acid positions within each protein.
Year of isolation.
Encodes a deduced amino acid change.
Blank spaces indicate the same nucleotide as in strain P676-gb.
Consistent with previous analyses (45), the IC VEEV strains from the 1962–1964 and 1995 outbreaks were relatively distantly related to those of the 1992–1993 outbreak, with ca. 2% nucleotide divergence. As expected, the GenBank sequence with the greatest identity to the subtype IC 1962–1964 and 1995 isolates that we sequenced was that of the 1963 P676-gb isolate described previously (15). When this sequence was included in the comparisons among the new subtype IC sequences, it was the most divergent. The P676-gb sequence had 16 nucleotide differences (seven deduced amino acids) versus the P676-ag sequence from the same strain but with a lower passage history (Table 1). The P676-gb virus had undergone seven Vero cell culture passages prior to being sequenced from subclones (15), while the P676-ag has only one cell culture passage.
The only amino acid (amino acid 199 of the E2 envelope glycoprotein) within the 26S subgenomic portion of the genome that varied among the 1962–1964 and 1995 VEEV subtype IC strains examined was shared by the Panaquire and the P676-gb sequences (Table 3). These two viruses had the longest cell culture passage histories (four and seven Vero cell culture passages, respectively). When strains from within the same 1962–1964 outbreak were compared, isolates V-198 (1962), P676-ag (1963), and VE64-PMCHo5 (1964) varied at 11 nucleotide positions (genome positions 476, 3941, 4049, 5350, 5453, 5469, 6182, 7229, 7312, 9622, and 10306). Two of these nucleotide differences (positions 5350 and 5469) resulted in deduced amino acid differences within the poorly conserved carboxy-terminal half of the nsP3 gene at amino acid positions 439 and 479. The last isolate available from the 1962–1964 outbreak, PMCHo5, had two unique nucleotide differences, at positions 3941 and 4049. The 1995 VEEV subtype IC isolates, 6119 and 3908, differed at a total of seven synonymous nucleotide positions (1247, 1442, 4974, 5291, 5474, 6497, and 10912) (Table 3).
When sequences from the 1962–1964 and 1995 outbreaks were compared, the V-198 and 3908 isolates, spanning the entire 33-year range examined, were different at a total of 14 nucleotide positions, with three nonsynonymous differences. Remarkably, only four nucleotide differences were identified between the 1963 P676-ag strain and the earliest isolate from the 1995 outbreak, strain 6119 (Table 4). Furthermore, the sequence of the P676-ag strain was identical to that of the 1983 Panaquire strain, with the exception of the E2-199 site implicated in cell culture adaptation (see below).
TABLE 4.
Percent nucleotide and amino acid identity between VEEV subtype IC isolates analyzeda
| Isolate | P676-gb | V-198 | PMCHo5 | Panaquire | P676-ag | 6119 | 3908 |
|---|---|---|---|---|---|---|---|
| P676-gb | 99.76 (9) | 99.79 (8) | 99.84 (6) | 99.81 (7) | 99.81 (7) | 99.81 (7) | |
| V198 | 99.78 (25) | 99.95 (2) | 99.89 (4) | 99.92 (3) | 99.92 (3) | 99.92 (3) | |
| PMCHo5 | 99.84 (18) | 99.92 (9) | 99.95 (2) | 99.97 (1) | 99.97 (1) | 99.97 (1) | |
| Panaquire | 99.88 (14) | 99.91 (10) | 99.96 (5) | 99.97 (1) | 99.97 (1) | 99.97 (1) | |
| P676-ag | 99.86 (16) | 99.92 (9) | 99.96 (4) | 99.99 (1) | 100 (0) | 100 (0) | |
| 6119 | 99.82 (20) | 99.89 (13) | 99.93 (8) | 99.96 (5) | 99.96 (4) | 100 (0) | |
| 3908 | 99.82 (21) | 99.88 (14) | 99.93 (8) | 99.95 (6) | 99.96 (5) | 99.94 (7) |
Numbers above the diagonal represent percent amino acid identity, and numbers below the diagonal represent percent nucleotide identity. Numbers in parentheses represent the total nucleotide (of 11,391 nucleotides) or amino acid (of 3,750 amino acids) sequence differences in each pairwise comparison.
Cell culture adaptation changes.
The P676-gb and Panaquire strains shared a unique, nonsynonymous mutation encoding a lysine at E2 amino acid position 199 (E2-199 Lys). A possible explanation is that seven Vero cell culture passages (Table 1) done before the P676 strain was sequenced by Kinney et al. (15) and the four vertebrate cell passages (two Vero and two BHK-21 [Table 1]) of the Panaquire strain may have caused the independent generation of this mutation via cell culture selection. To test this hypothesis, we passaged the P676-ag strain, which had the E2-199 Glu as its consensus sequence, 10 times in Vero cells, using two parallel but independent passage series; 24-h incubation times were used to represent typical laboratory passage times. Both passage series resulted in a mixed viral population at E2-199 (Lys and Glu) within two passages and complete replacement by the lysine residue within five passages. This suggested that the Lys mutation conferred higher fitness for replication in Vero cells. This Lys mutation alters the charge (Glu→Lys = +2) on the surface of the E2 glycoprotein within the E2g epitope that has been linked to the selection of monoclonal antibody-resistant variants in cell culture experimentation (10, 27, 28). Similar charge alterations have been linked to heparin sulfate receptor adaptation by Sindbis virus passaged in cell culture (3, 16). These results indicated that the E2-199 Lys found in the P676-gb and Panaquire isolates was probably the result of independent, convergent adaptation to Vero cell replication. This result is also consistent with a common source (the original P676 mosquito isolate, represented most accurately by the low passage P676-ag) for both the Panaquire and P676-gb strains, as indicated by the history of these strains.
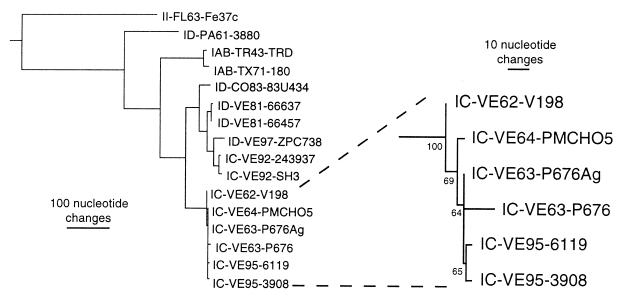
Phylogenetic analyses.
Phylogenetic analyses using maximum parsimony and maximum likelihood methods generated trees with the topology shown in Fig. 2. All analyses demonstrated that these VEEV subtype IC isolates diverged sometime before the isolation of strain V-198 in 1962. All methods yielded a bootstrap value of 100 for the ancestral node of this IC lineage, demonstrating strong support for the 1962–1964 and 1995 outbreak viruses sharing a common ancestor (Fig. 2). Confidence indices for the remainder of the nodes demonstrated limited resolution within the subtype IC analysis. The level of support for the 1995 and 1963–1964 clades was 63 to 74%, determined by the different phylogenetic methods. This low confidence index is related to the extremely close genetic identity (four or five nucleotide differences) of the IC viruses analyzed. However, the topology of all trees indicated that the P676-ag strain had a genomic sequence predicted to be identical to that of the direct ancestor of the 1995 outbreak clade. This topology also suggests that the P676-ag sequence is the authentic, wild-type sequence, and the long branch leading from the P676-ag to the P676-gb sequence probably reflects mutations associated with cell culture passage of the P676-gb strain.
FIG. 2.
Maximum parsimony phylogenetic analysis of VEEV subtype IC isolates from the 1962–1964 and 1995 outbreaks. An outgroup comprised of subtype IE strains MenaII (14) and 68U201 (18) was used to root the tree, and additional subtype I viruses described previously (13–15, 33) were included in the analysis. All virus strains are denoted by VEEV subtype, followed by country abbreviation and year of isolation, followed by strain name. Numbers at the nodes represent bootstrap confidence indices based on 1,000 replicates. The bar represents the number of nucleotide changes in each horizontal branch.
Evolutionary rates of VEEV subtype IC isolates.
To estimate the evolutionary rates of VEEV subtype IC isolates that were isolated over 30 years apart, the complete genomic nucleotide sequences of the earliest IC isolates from the 1962–1964 outbreak were compared to those from the 1995 outbreak. The results demonstrated an unusually high degree of conservation, as indicated previously from a portion of the PE2 glycoprotein precursor gene (53). A sister branch length comparison of IC isolates and their time between isolations estimated a maximum nucleotide substitution rate between the end of the 1962–1964 outbreak and the beginning of the 1995 outbreak (PMCHo5 versus 6119) of 2.3 × 10−5 substitutions per nucleotide per year. Using the same method, an even lower substitution maximum rate between the P676-ag and 6119 strains, 1.1 × 10−5 substitutions per nucleotide per year, was estimated. Direct comparison of sequences from these strains yielded the same maximum estimate. Conversely, the rates of substitution calculated from strains isolated during the same outbreak were 4.0 × 10−4 substitutions per nucleotide per year within the 1962–1964 outbreak and 1.84 × 10−3 within the 1995 outbreak. These rates estimates from within the two IC outbreaks were approximately 20- to 50-fold higher than the maximum rate of substitution between outbreaks.
To estimate the evolutionary rates of enzootic VEE viruses that circulate continuously in Venezuela and Colombia, subtype ID lineages sampled in the same regions of Colombia and Venezuela from the 1970s through 1997 were examined. Regression analysis of a monophyletic lineage of viruses from the Magdalena Valley of Colombia and the Catatumbo region of western Venezuela (17) yielded an estimate of 0.035% nucleotide sequence change per year, or 3.5 × 10−4 substitutions/nt/year (r2 = 0.63) (Fig. 3). Pairwise comparisons of branch lengths from sister sequences representing early and recent ID isolates from Venezuela and Colombia yielded an estimate of 2.9 × 10−4 (standard deviation = 2.2 × 10−4). Analysis of genomic sequences from two 1981 enzootic, subtype ID isolates from Zulia State in western Venezuela (strains 66637 and 66457) and a 1997 isolate from the Zulia State (ZPC738) (45) yielded estimates ranging from 2.9 × 10−4 to 3.2 × 10−4. These estimates were all 20 to 30 times greater than those described above for the maximum possible rate of sequence evolution of subtype IC viruses between 1963 and 1995 (Fig. 2). These data indicated that during the interepizootic period of 1964–1995, the IC viruses were not undergoing molecular evolution comparable to that observed during IC outbreaks or that exhibited by continuously circulating enzootic VEEV.
FIG. 3.
Regression analyses of sequence evolution rates of enzootic subtype ID (dashed line) and epizootic subtype IC (solid line) VEEV. Using maximum parsimony analyses, nucleotide changes from a node representing the common ancestor of a clade were plotted versus the year of isolation. The linear regression represents the rate of nucleotide substitution over time.
DISCUSSION
A central question in arbovirology for many decades has been the viral origin of VEE epidemics and epizootics. Several hypotheses have been proposed to explain the periodic emergence of VEE outbreaks separated by interepizootic periods approaching 20 years: (i) the periodic generation of epizootic viruses from enzootic progenitors, via mutations that enhance equine viremia; (ii) maintenance of epizootic virus subpopulations within enzootic virus populations; (iii) the presence of a cryptic epizootic virus transmission cycle that periodically emerges to cause human and/or equine disease; (iv) the recrudescence of latent VEEV infections; or (v) the administration of inadequately inactivated vaccines (11). The evidence for and against these hypotheses, in light of the present findings, is summarized below.
(i) Evolution of epizootic strains from enzootic subtype ID progenitors.
Previous sequencing studies described an extremely close genetic relationship between enzootic ID strains from western Venezuela and epizootic IC viruses from a sympatric outbreak of human and equine VEE that occurred in 1992–1993. Phylogenetic analysis of the amino terminus of the PE2 glycoprotein precursor gene indicated that the evolution of VEE subtype IC and IAB epizootics from enzootic ID progenitors has occurred on at least three different occasions (21, 24, 45, 50), with the 1992 emergence representing a recent evolution of the epizootic phenotype. However, while the 1962–1964 epidemic/epizootic began on the Guajira peninsula near sites where enzootic ID isolates have been isolated (43), the 1995 outbreak began in Falcon State, a region in which no closely related enzootic ID viruses have been isolated (29).
We cannot rule out the presence of an as yet undetected enzootic ID lineage in northern and/or western Venezuela that may have given rise to both the 1962–1964 and 1995 IC outbreaks. However, the only enzootic VEEV isolated to date in northern Venezuela is genetically unrelated to viruses from the 1962–1964 and 1995 outbreaks (29). Furthermore, repeated emergence of the IC phenotype via identical sets of mutations in a ID progenitor would also have to be accompanied by nearly complete genetic stasis of the enzootic progenitor lineage. Our estimates of enzootic VEEV evolution rates are inconsistent with the genetic conservation between 1962 and 1995, indicating that this scenario is an unlikely explanation for the 1962 and 1995 VEE emergences. Finally, even if a progenitor enzootic lineage underwent complete genetic stasis between 1962 and 1995, the quasispecies nature of alphavirus populations (48) suggests that the chance of sampling two progenitors with nearly identical sequences would be slim. For example, the 1981 enzootic VEEV subtype ID strains 66637 and 66457 were isolated within 8 days and only a few km apart near Sinamaica in northwestern Venezuela (43), yet their genomes differ by 10 nucleotides (45), 6 more than the 1963 strain P676-ag versus 1995 strain 6119 (Table 4).
(ii) Circulation of epizootic viruses as subpopulations within enzootic strains.
Extensive hydroxylapatite chromatography and plaque size analyses of enzootic virus isolates have been used to search for epizootic subpopulations (30, 31, 34). Stanick et al. (34) examined 23 different ID virus strains but were unsuccessful in detecting any epizootic phenotype variants. More sensitive methods and the analysis of unpassaged isolates are needed to further evaluate this hypothesis. However, even if subpopulations of epizootic viruses are present within enzootic strains, it is difficult to explain why they would undergo 20-fold-slower evolution than the enzootic majority populations when replicating continuously in nature.
(iii) Continuous circulation of epizootic viruses in cryptic cycles.
Field investigations before and after the 1995 outbreak failed to provide direct evidence for the maintenance of epizootic IAB or IC VEEV in continuous transmission cycles (5, 29, 41–43). The 1983 interepizootic subtype IC isolate from Panaquire provided the first possible evidence for a continuous epizootic virus transmission cycle. However, recent interepizootic investigations in the region where the Panaquire strain was isolated identified only enzootic, subtype ID VEEV circulation and provided no evidence for the circulation of subtype IC strains (29). Furthermore, the enzootic ID strains from this region of northern Venezuela are distantly related to all epizootic strains.
Our results cannot completely rule out the possibility that the P676 virus strain was maintained, without any nucleotide substitutions, in Panaquire within a cryptic transmission cycle from 1963 to 1983 and even beyond until 1995. However, considering the high mutation frequencies exhibited by all RNA viruses (6), including alphaviruses (48), this scenario would require complete genomic nucleotide sequence conservation via strong purifying selection at the nucleotide sequence level, combined with a complete absence of genetic drift. Our current IC sequence results, compared to those published previously for several other alphaviruses, indicate evolutionary rates at least 10 times lower than those for even eastern equine encephalitis virus and Highlands J viruses (1, 4, 49), which circulate only seasonally in temperate regions of North America. Furthermore, previous sequence analyses of several alphavirus groups have provided evidence for strong purifying selection at the amino acid but not the nucleotide level. Although the amount of VEEV normally transmitted in nature has not been estimated directly, at least occasional bottleneck transmission events probably occur when reservoir host viremia is just at the threshold level for vector infection, or when vector saliva titers decline after about 1 week of infection (54) such that transmission of small virus quantities occurs. These kind of transmission events probably result in genetic bottlenecks that have been shown to be accompanied by genetic change (drift and fitness declines) in equine encephalitis virus (51).
A far simpler explanation for the complete genomic sequence identity between the P676-ag and Panaquire strains is that the Panaquire isolate resulted from the laboratory contamination of the mosquito pool processed in 1983. Records indicate that the P676 antigen was in regular use in the Caracas laboratory at that time, and the P676-ag strain that we sequenced was isolated from a vial of this same antigen preparation. It is extremely unlikely that our isolation of live virus from this antigen preparation represented cross-contamination because no epizootic VEEV had been used in the laboratory for at least 3 days, and the sequence that we obtained from this isolate is unique. These P676-ag and Panaquire strains have only one nucleotide difference, which can be attributed to Vero cell passage of the Panaquire strain before sequencing. These results question the legitimacy of the Panaquire isolate.
(iv) Reemergence of epizootic strains from latent infections.
Although persistent or latent infections have not been described for VEEV, experimental infection of birds with western equine encephalitis virus yielded persistence of infectious virus for up to 10 months (23). However, persistent infection of animal cells with RNA viruses generally leads to rapid genetic change and diversification (7, 8) rather than stasis, as we have observed in subtype IC VEEV. Furthermore, the 31-year time period between the two major IC outbreaks exceeds the life expectancy of small mammalian reservoir hosts, equines, and mosquitoes. If recrudescence of a latent infection that was accompanied by complete genetic stasis was responsible for reemergence of the subtype IC virus in 1995, a longer-lived host such as a human would be a likely source. Although there is no evidence to support a role for latent infections in VEEV emergence, persistent alphavirus infections deserve further study as a possible mechanism of viral persistence in nature.
(v) Reemergence of epizootic strains from incompletely inactivated vaccines.
Beginning in the late 1930s after VEEV was first isolated, formalin-inactivated VEE vaccines were produced from epizootic, subtype IAB strains for equine immunization. The isolation of live virus from some human vaccinees (39) and the fear that incomplete inactivation might initiate outbreaks led to the development of the live, attenuated TC-83 vaccine in 1961 (44). Following the 1969–72 Central and North American outbreak, this vaccine largely replaced inactivated forms for equine vaccination. Laboratory workers and others at risk for occupational infection, which has occurred repeatedly following exposure to aerosols, are the primary human recipients of TC-83 (20).
Extensive sequencing studies of subtype IAB isolates isolated from 1938 to 1973 demonstrated almost complete genetic stasis, consistent with the hypothesis that many or all later IAB outbreaks resulted from the use of incompletely inactivated vaccine preparations (52). Following the introduction and widespread use of the attenuated TC-83 vaccine, there have been no reported outbreaks of subtype IAB viruses. This provides further support for a vaccine origin of the later IAB outbreaks rather than a natural source (52). However, in contrast to the IAB subtype, there is no record that VEEV subtype IC strains were ever used to produce either an attenuated or formalin-inactivated vaccine. This indicates that a vaccine source is a highly unlikely explanation for initiation of either the 1962–1964 or 1995 outbreak.
Alternative 1995 emergence scenarios.
Our genetic data, combined with other evidence reviewed previously (47), do not provide clear or direct support for any of the above hypotheses regarding the viral source of the 1995 VEE epizootic. Another possible explanation is that the 1995 outbreak was initiated by a laboratory virus strain. The P676-ag virus that we sequenced and used in our analysis was isolated from a 1982 antigen preparation used for diagnostic purposes for many years in Venezuela. The P676-ag sequence is identical to a predicted ancestral node at the base of the 1995 epizootic virus clade in all phylogenetic analyses that we performed (Fig. 2). This relationship suggests that the P676 strain initiated the 1995 outbreak, and this scenario would explain the extreme genetic stasis observed between the 1962–1964 and 1995 outbreaks. Possible scenarios for such a laboratory origin include a human infection or escape of an experimentally infected animal or mosquito. Laboratory infection via aerosol has been documented in over 150 human cases of VEE (19). Furthermore, viremias in humans infected with subtype IC strains are similar to those of equines (53) and are sufficient to infect mosquito vectors to initiate transmission (36). However, unless direct evidence of a 1995 laboratory infection and transmission to a natural epizootic cycle is found, this explanation for the origin of the 1995 outbreak will remain speculative.
ACKNOWLEDGMENTS
We thank Robert Tesh, Robert Shope, and Hilda Guzman for providing some of the VEEV isolates used in our analyses.
Aaron C. Brault was supported by a James L. McLaughlin Infection and Immunity Fellowship and NIH Emerging Tropical Diseases T32 training grant AI-107526. Ann M. Powers was supported by the James W. McLaughlin Fellowship Fund and NIH T32 training grant on emerging and reemerging infectious diseases AI-07536. This work was supported by Public Health Service grants AI-10984 and AI-39800 from the National Institutes of Health and by the National Aeronautics and Space Administration.
REFERENCES
- 1.Brault A C, Powers A M, Chavez C L, Lopez R N, Cachon M F, Gutierrez L F, Kang W, Tesh R B, Shope R E, Weaver S C. Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. Am J Trop Med Hyg. 1999;61:579–586. doi: 10.4269/ajtmh.1999.61.579. [DOI] [PubMed] [Google Scholar]
- 2.Briceno Rossi A L. Rural epidemic encephalitis in Venezuela caused by a group A arbovirus (VEE) Prog Med Virol. 1967;9:176–203. [PubMed] [Google Scholar]
- 3.Byrnes A P, Griffin D E. Binding of Sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–56. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cilnis M J, Kang W, Weaver S C. Genetic conservation of Highlands J viruses. Virology. 1996;218:343–351. doi: 10.1006/viro.1996.0203. [DOI] [PubMed] [Google Scholar]
- 5.Dickerman R W, Cupp E W, Groot H, Alarcon A M, Cura E, Dickerman A W, Ibagos A L, Ricco-Hesse R, Taylor C A, Weaver S C. Venezuelan equine encephalitis virus activity in northern Colombia during April and May 1983. Bull Pan Am Health Org. 1986;20:276–283. [PubMed] [Google Scholar]
- 6.Domingo E. Quasispecies and the implications for virus persistence and escape. Clin Diagn Virol. 1998;10:97–101. doi: 10.1016/S0928-0197(98)00032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo E, Baranowski E, Ruiz-Jarabo C M, Martin-Hernandez A M, Saiz J C, Escarmis C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521–527. doi: 10.3201/eid0404.980402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo E, Escarmis C, Sevilla N, Baranowski E. Population dynamics in the evolution of RNA viruses. Adv Exp Med Biol. 1998;440:721–727. doi: 10.1007/978-1-4615-5331-1_93. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 10.Johnson B J, Brubaker J R, Roehrig J T, Trent D W. Variants of Venezuelan equine encephalitis virus that resist neutralization define a domain of the E2 glycoprotein. Virology. 1990;177:676–683. doi: 10.1016/0042-6822(90)90533-w. [DOI] [PubMed] [Google Scholar]
- 11.Johnson K M, Martin D H. Venezuelan equine encephalitis. Adv Vet Sci Comp Med. 1974;18:79–116. [PubMed] [Google Scholar]
- 12.Johnson K M, Shelokov A, Peralta P H, Dammin G J, Young N A. Recovery of Venezuelan equine encephalomyelitis virus in Panama. A fatal case in man. Am J Trop Med Hyg. 1968;17:432–440. doi: 10.4269/ajtmh.1968.17.432. [DOI] [PubMed] [Google Scholar]
- 13.Kinney R M, Johnson B J B, Welch J B, Tsuchiya K R, Trent D W. Full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated derivative, strain TC83. Virology. 1989;170:19–31. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- 14.Kinney R M, Pfeffer M, Tsuchiya K R, Chang G J, Roehrig J T. Nucleotide sequences of the 26S mRNAs of the viruses defining the Venezuelan equine encephalitis antigenic complex. Am J Trop Med Hyg. 1998;59:952–964. doi: 10.4269/ajtmh.1998.59.952. [DOI] [PubMed] [Google Scholar]
- 15.Kinney R M, Tsuchiya K R, Sneider J M, Trent D W. Genetic evidence that epizootic Venezuelan equine encephalitis (VEE) viruses may have evolved from enzootic VEE subtype I-D virus. Virology. 1992;191:569–580. doi: 10.1016/0042-6822(92)90232-e. [DOI] [PubMed] [Google Scholar]
- 16.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moncayo, A. C., G. M. Medina, Z. Kalvatchev, R. Barrera, J. Boshell, C. Ferro, J. E. Freier, J. C. Navarro, R. Salas, J. de Siger, C. Vasquez, R. Walder, and S. C. Weaver. Genetic diversity and relationships among enzootic Venezuelan equine encephalitis virus field isolates from Colombia and Venezuela. Am. J. Trop. Med. Hyg. in press. [DOI] [PubMed]
- 18.Oberste M S, Parker M D, Smith J F. Complete sequence of Venezuelan equine encephalitis virus subtype IE reveals conserved and hypervariable domains within the C terminus of nsP3. Virology. 1996;219:314–320. doi: 10.1006/viro.1996.0254. [DOI] [PubMed] [Google Scholar]
- 19.Office of Health and Safety, Centers for Disease Control and Prevention. Biosafety in microbiological and biomedical laboratories. 4th ed. Washington, D.C.: U.S. Government Printing Office; 1999. [Google Scholar]
- 20.Pittman P R, Makuch R S, Mangiafico J A, Cannon T L, Gibbs P H, Peters C J. Long-term duration of detectable neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine. 1996;14:337–343. doi: 10.1016/0264-410x(95)00168-z. [DOI] [PubMed] [Google Scholar]
- 21.Powers A M, Oberste M S, Brault A C, Rico-Hesse R, Schmura S M, Smith J F, Kang W, Sweeney W P, Weaver S C. Repeated emergence of epidemic/epizootic Venezuelan equine encephalitis from a single genotype of enzootic subtype ID virus. J Virol. 1997;71:6697–6705. doi: 10.1128/jvi.71.9.6697-6705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Querales J, Plaz J, Fernandez H, Acive F. Estudio seroepidemiologico de encefalitis por virus Venezolano en la zona de Barlovento. Rev Inst Nac Hyg “Rafael Rangel”. 1985;18:43–49. [Google Scholar]
- 23.Reeves W C, Hutson G A, Bellamy R E, Serivani R P. Chronic latent infections of birds with western equine encephalomyelitis virus. Proc Soc Exp Biol Med. 1958;97:733–736. doi: 10.3181/00379727-97-23862. [DOI] [PubMed] [Google Scholar]
- 24.Rico-Hesse R, Weaver S C, de Siger J, Medina G, Salas R A. Emergence of a new epidemic/epizootic Venezuelan equine encephalitis virus in South America. Proc Natl Acad Sci USA. 1995;92:5278–5281. doi: 10.1073/pnas.92.12.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas F, Diaz L A, Cardenas V M, Daza E, Bruzon L, Alcala A, De la Hoz O, Caceres F M, Aristizabal G, Martinez J W, Revelo D, De la Hoz F, Boshell J, Camacho T, Calderon L, Olano V A, Villarreal L I, Roselli D, Alvarez G, Ludwig G, Tsai T. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J Infect Dis. 1997;175:828–832. doi: 10.1086/513978. [DOI] [PubMed] [Google Scholar]
- 26.Roehrig J T, Bolin R A. Monoclonal antibodies capable of distinguishing epizootic from enzootic varieties of subtype I Venezuelan equine encephalitis viruses in a rapid indirect immunofluorescence assay. J Clin Microbiol. 1997;35:1887–1890. doi: 10.1128/jcm.35.7.1887-1890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roehrig J T, Day J W, Kinney R M. Antigenic analysis of the surface glycoproteins of a Venezuelan equine encephalomyelitis virus (TC-83) using monoclonal antibodies. Virology. 1982;118:269–278. doi: 10.1016/0042-6822(82)90346-4. [DOI] [PubMed] [Google Scholar]
- 28.Roehrig J T, Mathews J H. The neutralization site on the E2 glycoprotein of Venezuelan equine encephalomyelitis (TC-83) virus is composed of multiple conformationally stable epitopes. Virology. 1985;142:347–356. doi: 10.1016/0042-6822(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 29.Salas, R. A., C. Z. Garcia, J. Liria, R. Barrera, J. C. Navarro, G. Medina, C. Vasquez, Z. Fernandez, and S. C. Weaver. Ecological studies of enzootic Venezuelan equine encephalitis in north-central Venezuela, 1997–1998. Am. J. Trop. Med. Hyg., in press. [DOI] [PubMed]
- 30.Scherer W F, Anderson K, Pancake B A, Dickerman R W, Ordonez J V. Search for epizootic-like Venezuelan encephalitis virus at enzootic habitats in Guatemala during 1969–1971. Am J Epidemiol. 1976;103:576–588. doi: 10.1093/oxfordjournals.aje.a112262. [DOI] [PubMed] [Google Scholar]
- 31.Scherer W F, Ordonez J V, Dickerman R W, Navarro J E. Search for persistent epizootic Venezuelan encephalitis virus in Guatemala, El Salvador and Nicaragua during 1970–1975. Am J Epidemiol. 1976;104:60–73. doi: 10.1093/oxfordjournals.aje.a112274. [DOI] [PubMed] [Google Scholar]
- 32.Schlesinger S, Schlesinger M J. Togaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 825–842. [Google Scholar]
- 33.Sneider J, Kinney R, Tsuchiya K, Trent D. Molecular evidence that epizootic Venezuelan equine encephalitis (VEE) I-AB viruses are not evolutionary derivatives of enzootic VEE subtype I-E or II viruses. J Gen Virol. 1993;74:519–523. doi: 10.1099/0022-1317-74-3-519. [DOI] [PubMed] [Google Scholar]
- 34.Stanick D R, Wiebe M E, Scherer W F. Markers of Venezuelan encephalitis virus which distinguish enzootic strains of subtype I-D from those of I-E. Am J Epidemiol. 1985;122:234–244. doi: 10.1093/oxfordjournals.aje.a114094. [DOI] [PubMed] [Google Scholar]
- 35.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suarez O M, Bergold G H. Investigations of an outbreak of Venezuelan equine encephalitis in towns of eastern Venezuela. Am J Trop Med Hyg. 1968;17:875–880. doi: 10.4269/ajtmh.1968.17.875. [DOI] [PubMed] [Google Scholar]
- 37.Sudia W D, Newhouse V F, Beadle I D, Miller D L, Johnston J G, Jr, Young R, Calisher C H, Maness K. Epidemic Venezuelan equine encephalitis in North America in 1971: vector studies. Am J Epidemiol. 1975;101:17–35. doi: 10.1093/oxfordjournals.aje.a112068. [DOI] [PubMed] [Google Scholar]
- 38.Sudia W D, Newhouse V F, Henderson B E. Experimental infection of horses with three strains of Venezuelan equine encephalomyelitis virus. II. Experimental vector studies. Am J Epidemiol. 1971;93:206–211. doi: 10.1093/oxfordjournals.aje.a121247. [DOI] [PubMed] [Google Scholar]
- 39.Sutton L S, Brooke C C. Venezuelan equine encephalomyelitis due to vaccination in man. J Am Med Assoc. 1954;155:1473–1476. doi: 10.1001/jama.1954.03690350015005. [DOI] [PubMed] [Google Scholar]
- 40.Swofford D L. PAUP∗. Phylogenetic analysis using parsimony (∗and other methods), version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 41.Walder R, Suarez O M. Studies of arboviruses in southwestern Venezuela. I. Isolations of Venezuelan and eastern equine encephalitis viruses from sentinel hamsters in the Catatumbo region. Int J Epidemiol. 1976;5:375–384. doi: 10.1093/ije/5.4.375. [DOI] [PubMed] [Google Scholar]
- 42.Walder R, Suarez O M, Calisher C H. Arbovirus studies in southwestern Venezuela during 1973–1981. II. Isolations and further studies of Venezuelan and eastern equine encephalitis, Una, Itaqui, and Moju viruses. Am J Trop Med Hyg. 1984;33:483–491. [PubMed] [Google Scholar]
- 43.Walder R, Suarez O M, Calisher C H. Arbovirus studies in the Guajira region of Venezuela: activities of eastern equine encephalitis and Venezuelan equine encephalitis viruses during an interepizootic period. Am J Trop Med Hyg. 1984;33:699–707. doi: 10.4269/ajtmh.1984.33.699. [DOI] [PubMed] [Google Scholar]
- 44.Walton T E, Grayson M A. Venezuelan equine encephalomyelitis. In: Monath T P, editor. The arboviruses: epidemiology and ecology. IV. Boca Raton, Fla: CRC Press; 1988. pp. 203–231. [Google Scholar]
- 45.Wang E, Barrera R, Boshell J, Ferro C, Freier J E, Navarro J C, Salas R, Vasquez C, Weaver S C. Genetic and phenotypic changes accompanying the emergence of epizootic subtype IC Venezuelan equine encephalitis viruses from an enzootic subtype ID progenitor. J Virol. 1999;73:4266–4271. doi: 10.1128/jvi.73.5.4266-4271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watts D M, Lavera V, Callahan J, Rossi C, Oberste M S, Roehrig J T, Cropp C B, Karabatsos N, Smith J F, Gubler D J, Wooster M T, Nelson W M, Hayes C G. Venezuelan equine encephalitis and Oropouche virus infections among Peruvian army troops in the Amazon region of Peru. Am J Trop Med Hyg. 1997;56:661–667. doi: 10.4269/ajtmh.1997.56.661. [DOI] [PubMed] [Google Scholar]
- 47.Weaver S C. Recurrent emergence of Venezuelan equine encephalomyelitis. In: Scheld W M, Armstrong D, Hughes J M, editors. Emerging infections 1. Washington, D.C.: ASM Press; 1998. pp. 27–42. [Google Scholar]
- 48.Weaver S C, Bellew L A, Gousset L A, Repik P A, Scott T W, Holland J J. Diversity within natural populations of eastern equine encephalomyelitis virus. Virology. 1993;195:700–709. doi: 10.1006/viro.1993.1421. [DOI] [PubMed] [Google Scholar]
- 49.Weaver S C, Bellew L A, Hagenbaugh A, Mallampalli V, Holland J J, Scott T W. Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol. 1994;68:158–169. doi: 10.1128/jvi.68.1.158-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver S C, Bellew L A, Rico-Hesse R. Phylogenetic analysis of alphaviruses in the Venezuelan equine encephalitis complex and identification of the source of epizootic viruses. Virology. 1992;191:282–290. doi: 10.1016/0042-6822(92)90190-z. [DOI] [PubMed] [Google Scholar]
- 51.Weaver S C, Brault A C, Kang W, Holland J J. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73:4316–4326. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weaver S C, Pfeffer M, Marriott K, Kang W, Kinney R M. Genetic evidence for the origins of Venezuelan equine encephalitis virus subtype IAB outbreaks. Am J Trop Med Hyg. 1999;60:441–448. doi: 10.4269/ajtmh.1999.60.441. [DOI] [PubMed] [Google Scholar]
- 53.Weaver S C, Salas R, Rico-Hesse R, Ludwig G V, Oberste M S, Boshell J, Tesh R B. Re-emergence of epidemic Venezuelan equine encephalomyelitis in South America. Lancet. 1996;348:436–440. doi: 10.1016/s0140-6736(96)02275-1. [DOI] [PubMed] [Google Scholar]
- 54.Weaver S C, Scott T W, Lorenz L H. Patterns of eastern equine encephalomyelitis virus infection in Culiseta melanura. J Med Entomol. 1990;27:878–891. doi: 10.1093/jmedent/27.5.878. [DOI] [PubMed] [Google Scholar]