SUMMARY
Using halo-tagged PRC2 and “CLAP” methodology, Guo et al. recently came to the conclusion that PRC2 is not an RNA binding protein (RBP). They suggested that previous findings are CLIP artifacts and argue that RNA cannot play a direct role in PRC2 regulation. Here, we perform a re-analysis of the authors’ raw datasets and come to contrary conclusions. First, CLAP demonstrates significant PRC2 enrichment throughout the transcriptome, including in XIST’s Repeat A (RepA) motif. Second, our re-analysis of the authors’ CLAP and CLIP datasets demonstrates that the two methods yield similar outcomes, with both showing PRC2 enrichment in the transcriptome. Furthermore, PRC2 demonstrates more RNA binding peaks than SAF-A and PTBP1. Additionally, re-analysis of CLAP contradicts the authors’ conclusion that CTCF and YY1 are not RBP. The discrepancies may be attributable to the authors’ unconventional data normalization, methods of determining significance, and lack of minus-tag and input controls in some experiments.
INTRODUCTION
Many studies have shown that Polycomb repressive complex 2 (PRC2) is an RNA binding protein (RBP) that associates with a large family of transcripts in mammalian cells. Affinity capture methods such as native RNA immunoprecipitation (RIP)1–3, UV-crosslinked immunoprecipitation (CLIP)4–6, and denaturing CLIP7 have identified large RNA interactomes inclusive of thousands of transcripts in vivo2,5–7. In affinity capture studies that use an RNA bait to survey interacting proteins, PRC2 subunits have turned up as direct interactors for transcripts such as XIst and TERRA RNA8,9. Furthermore, RNA crosslinked proteomic surveys have identified PRC2 components as interacting partners10. Studies show that RNA contacts PRC2 along dispersed amino acid patches on its surface11–14 and binds the EZH2 subunit near its catalytic domain14. Subunits JARID214–16, AEBP2, and RBBP48 also contact RNA. The PRC2 subunits are now known to recognize RNA via several motifs, including CU-rich and G-rich sequences7,17,18. Some of the G-rich sequences fold into RNA G-quadruplexes (rG4) in vivo, bind PRC2 with 10–90 nM affinities19, and function in PRC2 recruitment20,21, regulation of EZH2’s catalytic activity (H3-K27 trimethylation)2 19 4,15–18, and eviction from chromatin17. They have also been implicated in POL-II transcriptional pausing at active genes7,22 and induction of B2/ALU ribozyme activity during the stress response23 24.
Given the existing body of work, the recent work of Guo et al. claiming that PRC2 is not a RBP25 is perplexing. The authors state that prior conclusions about PRC2’s RNA binding capacity1,6,7,14,15,17,26,27 stemmed from reassociation artifacts during RIP and CLIP procedures. RIP and CLIP depend on antibody-based precipitation of the protein of interest, which in turn would pull down interacting transcripts, if they are bound inside cells. The low nanomolar affinities of antibody-antigen binding are known to constrain the stringency of washes during the purification step and could, in theory, lead to reassociation artifacts — i.e., non-interacting RNAs binding nonspecifically to PRC2 during cell lysis, immunoprecipitation, and/or RNA purification. These nonspecific RNAs would then be captured during library preparation and lead to false positive results. Such limitations have spawned variants of the CLIP method, such as denaturing CLIP (dCLIP) and iCLIP7,28. In the latest iteration, Guo et al. developed CLAP (Covalent Linkage and Affinity Purification) and provided evidence for its superior performance relative to CLIP25. CLAP employs Halo-tags for crosslinking PRC2 components to substrate-coated beads and uses denaturing washes to remove non-crosslinked co-precipitants25, which would thereby minimize reassociation artifacts. Through the lens of the authors’ analytical approach and interpretation, CLIP replicated the large RNA interactome for PRC2 but CLAP failed to identify any enriched RNA, including Xist/XIST. The authors therefore urged a critical re-evaluation of RNA’s role in regulating PRC2 activity.
Here, in light of the contrary publication, we conducted a re-analysis of the raw datasets deposited in GEO by Guo et al. in order to reconcile differences and to exclude their analysis pipeline as a source of discrepancy. To our surprise, our independent analysis of the same raw data produced significantly different outcomes. Indeed, our analysis indicates that CLAP reaffirms the longstanding notion that PRC2 is an RBP. Thousands of transcripts — including the previously identified XIST RNA — are significantly enriched over their corresponding input levels for binding to various PRC2 subunits. In fact, XIST RNA has more enriched EZH2 and SUZ12 binding than SAF-A, PTBP1, and SPEN — three RBPs provided by the authors as positive controls. We also find that CLAP does not have superior performance relative to the CLIP. Thus, there is a major discrepancy in reported results despite the use of the same raw datasets. We provide potential explanations for the significant deviations.
RESULTS AND DISCUSSION
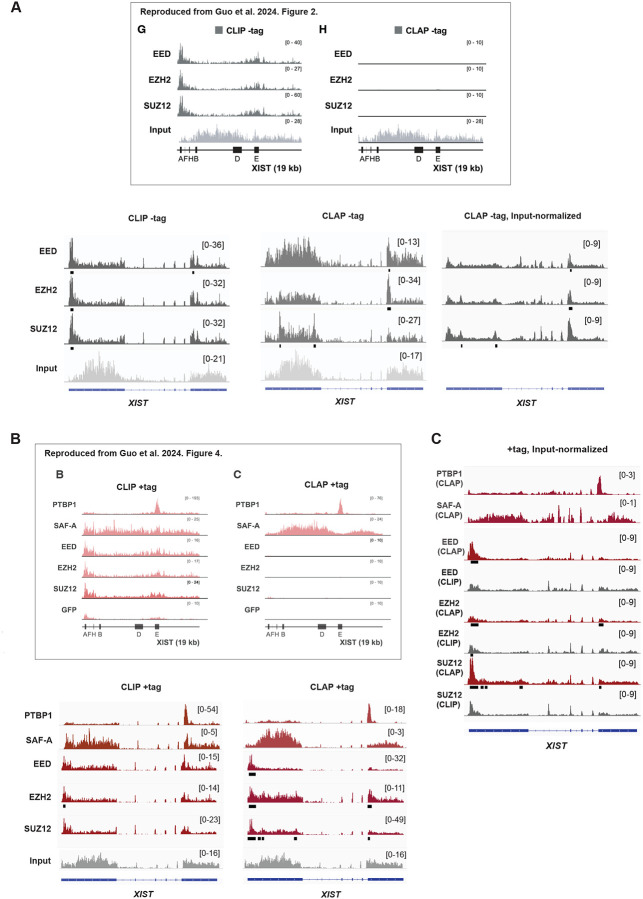
Guo et al25. employed cell mixing practices to determine the signal-to-noise ratio in CLIP and CLAP experiments. V5-Halo-tagged proteins of interest were expressed from the plasmid transfection in either mouse or human cells for affinity capture. Plus (+) tag cells harbor tagged proteins of interest, either in human or mouse cells. The +tag cells from one species are then mixed with wildtype cells from the other species prior to performing CLIP/CLAP. When the +tag is expressed in human cells, the admixed wildtype mouse cells provide a control for reassociation artifact in human CLIP/CLAP experiment. Reciprocally, when the +tag is expressed in mouse cells, the admixed -tag human cells provide a control for reassociation artifact in the mouse CLIP/CLAP. In the latter case, if human RNA reads are enriched in the mouse CLIP or CLAP, the human signals must result from reassociation artifacts during the procedure, as the human cells lack the Halo-tagged target protein. In Guo et al., CLIP analysis of PRC2 subunits with the -tag human cells accumulated significant human RNA reads (Fig. 1A, inset, left panel (“G”)), leading the authors to conclude that CLIP is prone to procedural artifacts25. In contrast, the authors reported no such artifacts when performing CLAP25 (Fig. 1A, inset, right panel (“H”)). From these results, the authors argue that CLAP is a superior method to CLIP. Furthermore, the authors’ CLAP showed no enrichment of PRC2 binding in human XIST (Fig. 1B, inset, right panel (“C”)), contrasting sharply with enrichment of SAF-A, PTBP1, and SPEN in various domains of XIST25. The authors therefore conclude that PRC2 is not an RBP.
Figure 1. CLAP affirms that PRC2 is an RBP.
(A) -tag experiments: CLIP and CLAP results for PTBP1, SAF-A, PRC2 (EED, EZH2, and SUZ12 subunits) -tag human samples. RPM normalized RNA reads are plotted across human XIST. Significant peaks are indicated by black bars below each track. Scale shown in brackets. Upper panel reproduced without alteration from Guo et al.25. And shows the spliced human XIST transcript. Bottom panel, re-analysis in this study displayed across the unspliced human XIST structural gene. Input normalized tracks are generated as indicated in the method section.
(B) +tag experiments: CLIP and CLAP results for PTBP1, SAF-A, PRC2 (EED, EZH2, and SUZ12 subunits) +tag human samples. RPM normalized RNA reads are plotted across human XIST. Significant peaks are indicated by black bars below each track. Scale shown in brackets. Upper panel reproduced without alteration from Guo et al.25. And shows the spliced human XIST transcript. Bottom panel, re-analysis in this study displayed across the unspliced human XIST structural gene
(C) +tag input-normalized tracks for human XIST. Enrichment profiles are generated as indicated in the method section. Significant peaks are indicated by black bars below each track. Scale shown in brackets.
Given this discrepancy, we re-analyzed the raw sequencing data to verify the validity of the reported findings. Our re-analysis of the raw sequencing files revealed strikingly different results. Notably, our re-alignment of the authors’ CLIP datasets showed that the profiles for PRC2 subunits, SAF-A, and PTBP1 in human XIST RNA were consistent with these proteins being robust RBPs — and were nearly identical to the CLIP profiles presented by Guo et al. (Fig. 1B, main section)25. The similar findings for SAF-A and PTBP1 indicate that our bioinformatic pipeline for read preprocessing and alignment is in accord with theirs. Furthermore, the input-normalized CLAP profiles for PTBP1 and SAF-A generated by our pipeline were also similar to those reported by Guo et al.25 (Fig. 1C), further confirming the consistency of our bioinformatics approach with the authors’ methodologies.
However, when we applied the same bioinformatic pipeline to the deposited raw data for PRC2 subunits — EZH2, SUZ12, and EED — both RPM-normalized and input--normalized CLAP profiles for these subunits showed a significant deviation from the results reported by Guo et al.25 (Fig. 1B, main section, CLAP +tag, and inset, right panel (“C”; Fig. 1C)). Notably, we observed substantial enrichment in XIST, in sharp contrast to the near-absence of signal reported by the authors25. Remarkably, the CLAP profiles generated by our pipeline closely matched the CLIP profiles for all three PRC2 subunits. This observation indicated that results from the CLIP and CLAP techniques, as applied by the authors, were in fact in close agreement with each other.
To identify significant PRC2 binding regions, we detected statistically significant peaks relative to the reference input for each protein of interest. In XIST RNA, significant binding was reproducibly observed at the 5’ end around Repeat A (RepA), a motif previously identified as a PRC2 binding site both in vitro and in vivo1,15,29. We note that the +tag enrichment for PRC2 was in excess of the -tag “background” (compare Fig. 1A to 1B). In the -tag CLAP negative controls, the 5’ enrichment was notably absent (Fig. 1A). When input-normalized, the enrichment peaks in XIST persisted in the CLAP experiment (Fig. 1C). Thus, our re-analysis of the deposited datasets confirms that RepA serves as a PRC2 binding domain.
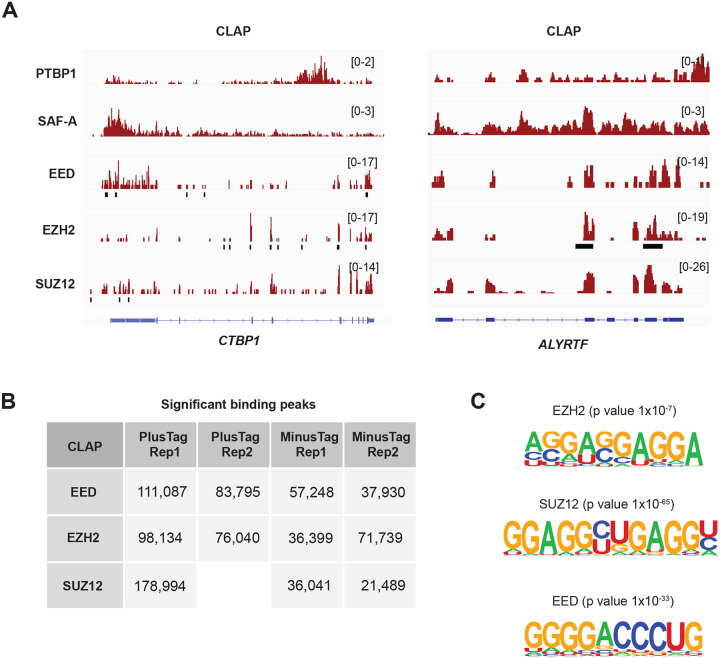
Significant binding peaks were detected across the transcriptome, with two notable examples including CTBP1 and ALYRTF (Fig. 2A). The PRC2 +tag CLAP samples consistently showed more prominent peaks compared to the -tag samples (Fig. 2B). Motif analysis of the significant binding sites for EZH2, SUZ12, and EED in the +tag samples identified G-rich motifs for all three subunits (Fig. 2C). This finding is consistent with prior biochemical and in vivo studies highlighting a preference for G-tracts and G-quadruplex-forming sequences1,7,17,18. These results reinforce the concept of sequence-specific in vivo binding of PRC2 in the mammalian transcriptome.
Figure 2. PRC2 binds significantly to selective sites throughout the transcriptome.
(A) Two representative PRC2 target transcripts are shown. Note the presence of significant peaks in both CTBP1 and ALYRTF.
(B) Table showing the numbers of significantly enriched CLIP and CLAP peaks in the indicated datasets. Blank spaces in the table indicate missing datasets in the study of Guo et al.
(C) Motif analysis from the +tag CLAP experiments for EZH2, SUZ12, and EED. Note G-rich motifs were significantly enriched.
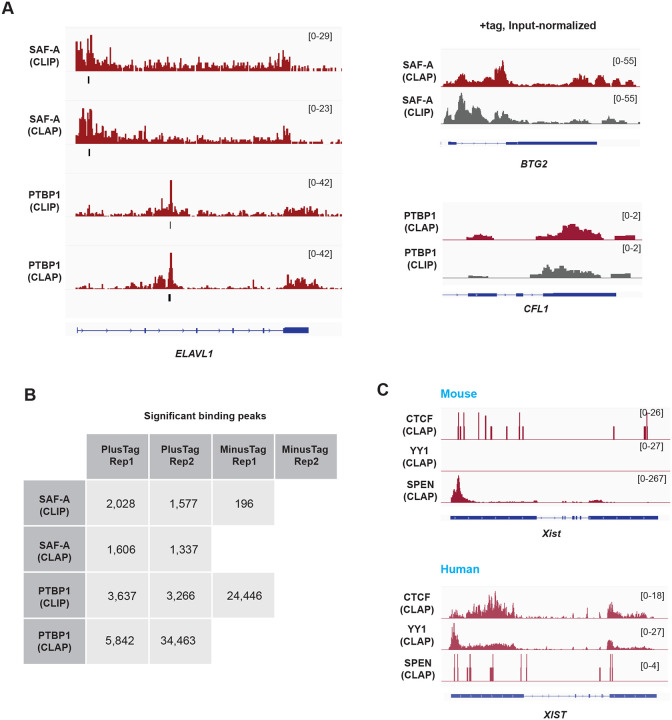
Guo et al. employed the known RBPs — SAF-A, PTBP1, and SPEN as positive controls to exclude PRC2 as an RBP25. To assess the performance of CLAP for these RNA-binding proteins, we applied the same informatics pipeline we used for analyzing PRC2 components. For both SAF-A and PTBP1, re-analysis of CLIP and CLAP datasets showed similar gene alignment patterns (Fig. 3A). Along ELAVL1, for example, CLIP and CLAP demonstrated significantly enriched peaks in identical RNA regions for SAF-A and PTBP1 (Fig. 3A, left panel). Input-normalized tracks also show similar alignment patterns between CLIP and CLAP (Fig. 3A, right panel). Global peak calling analysis showed comparable numbers of peaks in the CLIP and CLAP datasets, with the exception of PTBP1 replicate 2 (Fig. 3B). Notably, however, the authors’ replicate 2 lacked -tag controls (Fig. 3B). Thus, unlike for the PRC2 datasets, a determination of signal-to-noise levels for SAF-A and PTBP1 was not possible due to the absence of -tag controls. The missing data precludes a meaningful comparison of PRC2’s performance as an RBP relative to SAF-A and PTBP1.
Figure 3. CLAP and CLIP both confirm that SAF-A and PTBP1 are RBP.
(A) SAF-A and PTBP1 RNA binding profiles in the + tag samples. Left panel, alligned RNA reads are plotted across the human ENVIL1 for CLIP and CLAP. Significant peaks are indicated with the black lines below each track. Right panel, input normalized tracks of CLIP and CLAP. Note that CLAP was not superior to CLIP.
(B) Numbers of significantly enriched CLIP and CLAP peaks in the indicated datasets. Blank spaces in the table indicate missing datasets. -tag controls were missing for SAF-A CLAP. -tag controls also missing for replicate 2 (Rep2) in all cases.
(C) CTCF and YY1 show a large number of reads in the human +tag samples. Human XIST does not show enriched SPEN counts, while mouse Xist shows a enrichment at the 5’ end.
Similar criticisms can be applied to the analysis of SPEN as a positive control. There was only a single deposited mouse +tag SPEN CLAP sample. Because the study of Guo et al. was largely based on human +tag analysis, a +tag human sample should have been provided in order to compare RNA binding of SPEN to PRC2, PTBP1, and SAF-A. Moreover, a -tag sample should have been provided to assess the signal-to-noise difference. As for PTBP1 and SAF-A, the missing data precludes a comparison of SPEN to PRC2 in terms of their relative RNA binding potential. The authors therefore did not apply an appropriate level of rigor to the analysis of SAF-A, PTBP1, and SPEN.
A similar critique applies to the authors’ analysis of the chromosome architecture protein CTCF and transcription factor YY125. In these cases, the authors questioned the previously established RNA-binding status of CTCF and YY12,28,30–38. Nevertheless, we were able to observe significant raw RNA signals from the human XIST and other transcriptomic regions in both CTCF and YY1 datasets (Fig. 3C). On the other hand, the lack of input control libraries within CTCF and YY1 CLAP datasets prevented peak-calling and assessment of RNA binding significance. Additionally, the absence of -tag controls for both proteins precluded evaluation of the signal-to-noise ratio. Notably, the authors applied the -tag control to PRC2, PTBP1, and SAF-A to evaluate these proteins of interest as RBP. Consequently, we find that no meaningful conclusions can be drawn from the CTCF and YY1 CLAP analyses conducted by Guo et al.
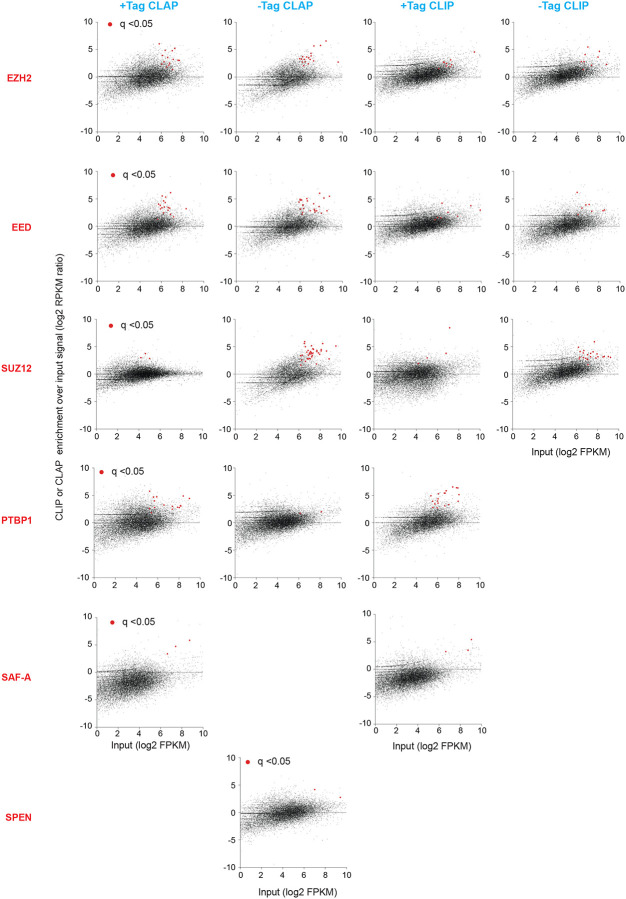
Given the lack of controls in the CLAP experiments, we questioned whether CLAP performed better than CLIP. The presence of a large number of peaks in the -tag PRC2 samples (Fig. 2B, 3B) indicates that CLAP may not be more effective at controlling noise than proposed. Guo et al. had called enrichment of RNA binding based on scatterplots of RNA expression versus CLAP enrichment, on the principle that true PRC2 binding must occur over the RNA’s representation in the transcriptome25. However, our re-analysis in scatterplot format showed only a modest enrichment of transcripts in the +tag versus -tag experiments for any protein of interest (noting that -tag controls were missing for several proteins)(Fig. 4). When compared to CLIP, CLAP did not demonstrate much improvement in enrichment (Fig. 4). Thus, based on the authors’ own criteria, these results suggest that CLAP does not deliver a superior method over CLIP.
Figure 4. The CLIP and CLAP of Guo et al. performed similarly.
Comparison of CLAP and CLIP pulldowns for various proteins of interest (red text) in relationship to the transcripts abundance. CLIP or CLAP enrichments over input were plotted on the y-axis, and then rearranged with the input abundance on the x-axis for visualization. Each dot represents a transcript. Red dots, significantly enriched CLIP or CLAP pulldowns relative to gene expression. A statistical test was performed using the Cufflinks differentially expressed gene calculation. False discovery rate (FDR) adjusted p-value (q-value) of 0.05 was utilized as the cutoff to determine significance.
While we did not attempt to recreate the detailed bioinformatic pipeline used by Guo et al., their publication indicates that they identified RNA binding enrichment by comparing the normalized ratio of reads in protein elution samples (IP) to their corresponding input samples across 100-nucleotide windows within annotated genes. They employed a binomial test to determine statistical significance with a threshold of < 10−⁶. In contrast, we avoided using a predefined window normalization approach, as we believe it may fail to capture the broader context of RNA-protein interactions, potentially leading to over-normalization, signal distortion, and the overlooking of significant binding events outside fixed window sizes. Rather, our approach involved merging biological replicates when available, preprocessing, and aligning reads, followed by separating them into Forward and Reverse strands. To ensure comparability, we randomly down-sampled the larger library (IP or Input) to match the smaller one, performing this separately for Forward and Reverse strands. We then analyzed the down-sampled IP and Input library pairs using MACS2, which employs Poisson distribution-based statistical analysis to identify enriched regions39,40. This analysis generated signal coverage tracks, which were then used to calculate a fold-enrichment track, normalizing each IP signal to its corresponding Input.
Acknowledging that these differing methodologies may lead to minor variations in results, we were puzzled by the substantial discrepancy between Guo et al.’s findings and ours. Guo et al. reported a near-complete absence of PRC2 RNA interactions by CLAP, suggesting that PRC2 subunits are not true RBPs. In contrast, our PRC2 CLAP results showed a significant RNA signal, particularly for EED and SUZ12, which were comparable to or slightly higher than those identified by the authors’ CLIP. This inconsistency in our respective CLAP analyses is perplexing and difficult to reconcile, especially since Guo’s pipeline was able to identify PRC2 binding signals over XIST in the corresponding CLIP datasets (Fig. 1C). Our data showed that CLAP signals were actually slightly stronger than the CLIP signals. Additionally, in our hands, the PRC2 CLAP results displayed significant RNA signals in both -tag and +tag samples, highlighting the high background in the CLAP method (Fig. 1A, 1B) and leading us to question whether CLAP performs better than CLIP.
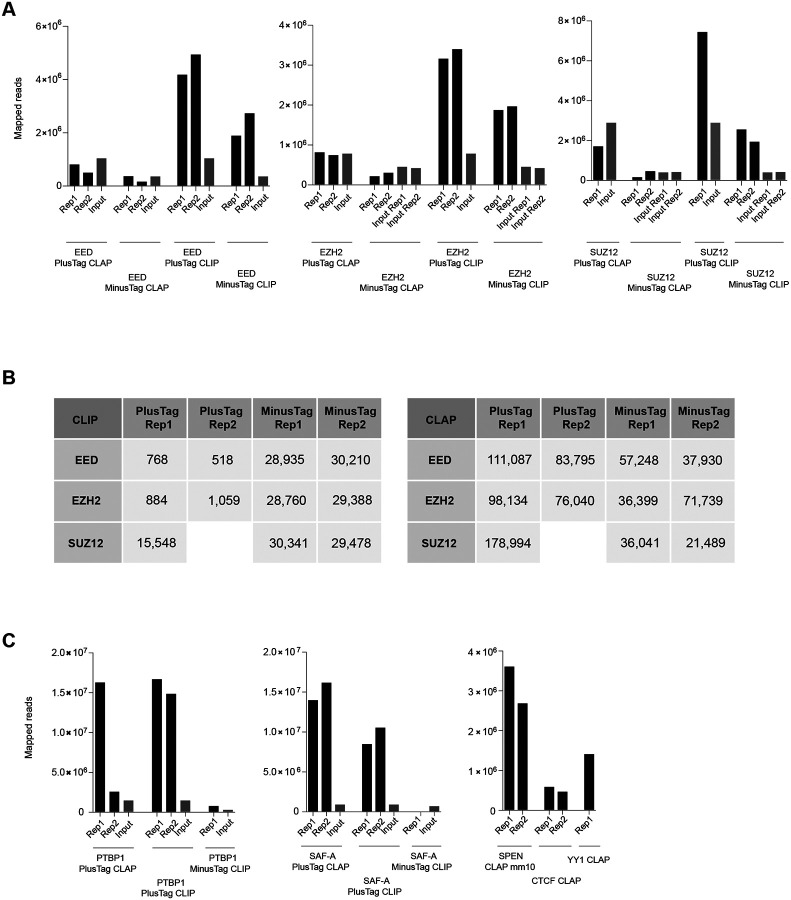
The CLIP and CLAP profiles for PTBP1 and SAF-A obtained from our pipeline are consistent with those reported by Guo et al. for both CLIP and CLAP. Similarly, the PRC2 CLIP tracks aligned well between the two methods. Given this, it is highly improbable that significant discrepancies would arise specifically in the CLAP analysis of PRC2 subunits between our re-analysis and the original results of Guo et al. This suggests that Guo et al. likely applied additional processing or normalization uniquely to PRC2. We considered the possibility that they scaled the CLAP datasets based on input to account for read depth. However, the total mapped reads were not substantially different between the -tag and +tag samples, yet the number of significant peaks was 2–3 folds greater in the +tag CLAP results compared to -tag (Fig. 5).
Figure 5. CLIP and CLAP library read depth comparison.
(A) Total mapped read of PRC2 CLIP and CLAP libraries. EED, EZH2, and SUZ12 PlusTag and MinusTag library reads aligned to the human reference genome were shown. Input read numbers are indicated with gray bars.
(B) Numbers of significant CLIP and CLAP peaks over input. Missing datasets are indicated with blanks.
(C) Total mapped read of CLIP and CLAP libraries for the other proteins.
Guo et al. opted to use a 100-nt window for normalization, an unconventional choice compared to the more straightforward bioinformatic method standardly employed by the field (and which we used here). No clear justification was provided for selecting such a limited window size. It would have been prudent to validate their findings across different window sizes and, in parallel, to avoid window-based normalization altogether. Consistency across multiple window sizes would then argue against an artifact of window-dependent normalization.
As for other proteins of interest, the numbers of mapped reads in the input files were substantially small in PTBP1 and SAF-A data, adding extra complication to the downstream applications (Fig. 5Fig. 4C). Also, as mentioned above, SPEN, YY1, and CTCF CLAP experiments were missing the corresponding input files altogether (Fig. 5Fig. 4C). We emphasize that, given that controversial nature of the main findings by Guo et al. – specifically, the claim that PRC2 is not a genuine RBP, which contradicts much of the existing literature – this conclusion should have been more rigorously tested and demonstrated. Similarly, the conclusion that YY1 and CTCF are not bona fide RBP could not have been reached without more rigorous demonstration and without the appropriate -tag and input controls.
In summary, our re-analysis of the CLIP and CLAP datasets deposited by the authors in GEO did not reproduce the conclusions of their publication. Our examination of the CLAP data revealed significant PRC2 enrichment across the transcriptome, including in XIST RNA. In fact, PRC2 demonstrates more substantial RNA binding peaks compared to other RBPs tested by the authors, such as SAF-A and PTBP1 (Fig. 2B, 3B). Additionally, our results challenge the authors’ claim that CTCF and YY1 are not RBPs. We observed the significant enrichment of RNA signal on human XIST compared to the control mouse Xist RNA with both CTCF and YY1 (Fig. 3C). While we do not discount the possibility of differences in informatics method, we attribute the discrepancies and the difficulty in deriving meaningful conclusions from the original publication to the lack of essential negative controls, including as -tag experiments for SAF-A, and both -tag experiments and input controls for CTCF and YY1. Our re-analysis also suggests that the authors applied additional processing or normalization specifically to PRC2, which therefore led to the misleading conclusion that PRC2 is not an RBP.
Finally, due to the absence of proper controls in the CLAP experiments and the high background in the -tag samples, it is inappropriate to claim that CLAP outperforms CLIP or to draw definitive statements about the RNA-binding properties of the proteins studied. The high number of peaks observed in the -tag PRC2 samples (Fig. 1A, 2B) suggests that CLAP may not be as effective at controlling noise as claimed25. We recommend that, whenever characterizing an RNA interactome is desired for proteins of interest, it is essential to perform perturbation studies and functional analyses to validate results. A large body of work currently supports functional PRC2-RNA interactions – such as in the fine-tuning of gene expression via transcriptional pausing7,22, targeting to specific chromatin sites1,5,26,41, control of a protein’s catalytic activity14,16,19, regulation of PRC2 eviction from RNA versus DNA1,4,17,19,42, and control of SINE B2/ALU ribozyme activity during the stress response23 24 . The CLAP analysis provided by Guo et al. does not invalidate the established biochemical and functional studies for PRC2-RNA interactions.
METHODS
Datasets
All CLIP and CLAP datasets were retrieved from GEO: GSE25347725.
Read sequence processing and alignment to the reference genome
FastQ files were preprocessed by performing adapter trimming using Trim Galore. To exclude reads mapping to non-target regions, the resulting trimmed reads were initially aligned using STAR aligner43 to a mouse (mm10) reference composite comprising genomic sequences from rDNAs, small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), 45S pre-rRNA, and tRNA. Unaligned reads were further filtered by aligning them to a human (hg38) reference composite containing the corresponding human-specific non-target genomic elements. Subsequently, the remaining reads were aligned to a composite reference including both mouse (mm10) and human (hg38) genomes. Duplicate reads were removed using sambamba_v0.6.644. BAM files were sorted, indexed, and split into forward and reverse strands. Strand-specific coverage tracks were generated by processing BAM files with the bamCoverage tool from deepTools45, using a bin size of 10 and normalizing with RPKM (Reads Per Kilobase Million) to account for differences in library size and gene length.
Normalization by fold-enrichment relative to input
FastQ files of IP libraries (CLIP or CLAP) and their corresponding Input samples were preprocessed and aligned as described earlier. When two replicates of the same type were available, they were merged, preprocessed, and aligned. Reads were then separated into Forward and Reverse strands. To ensure comparability between the IP and its corresponding Input, the larger library (either IP or Input) was randomly downsampled to match the size of the smaller library, with this process being performed separately for Forward and Reverse strands. The downsampled library pairs (IP and Input) were then used as input for MACS2 analysis39,40 with the parameter `--format BAM –nomodel –bdg –SPMR` to generate signal tracks (.bdg files) for both IP and Input libraries. MACS2’s `bdgcmp` utility was employed with the Fold Enrichment (FE) model to compare the IP track to its corresponding Input track. This comparison provided the fold enrichment signal of the IP over the Input, effectively normalizing the IP signal to the input control.
Quantification for CLIP and CLAP signal enrichment over genes
FDR-corrected q-values and enrichment over input metrics for each gene were calculated using Cufflinks v2.21, with the strand-specific library flag (-library-type fr-firststrand) applied. Sorted BAM files, prior to strand splitting, were provided to the cuffdiff module of the Cufflinks program. Enrichment and statistical analyses were conducted on the annotated genes from the GRCh38 (human) or GENCODE version M25 (mouse) reference gene assemblies. Scatter plots were generated using GraphPad Prism. The y-axis represents CLIP and CLAP signals on the genes normalized by the input signal, and then the dots were rearranged with the input abundance (log2 FPKM) on the x-axis for visualization.
Peak-Calling
Strand-separated BAM files were used for peak calling, and significantly enriched peaks over input reads were indicated by MACS239,40 program with the no-shifting model parameters (-p 0.01 -f BAM --nomodel -g hs).
Motif analysis
HOMER46 module (findMotifs.pl) analyzed enriched RNA motifs from the sequences of significant peak regions. Significant peak regions commonly observed in biological replicates that do not overlap with negative control (minus tag) peaks are used for the analysis.
ACKNOWLEDGMENTS
We thank Richard Jenner, Roberto Bonasio, Rodrigo Aguilar, and the Lee Lab for discussion and feedback. This work was supported by an NIH grant (R01-HD097665) to JTL.
Footnotes
STATEMENT OF FINANCIAL INTEREST
JTL is an Advisor to Skyhawk Therapeutics, a co-founder of Fulcrum Therapeutics, and a non-executive Director of the GSK. To the author’s knowledge, none of these entities work in the subject area covered by this study.
REFERENCES
- 1.Zhao J., Sun B.K., Erwin J.A., Song J.J. & Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J. et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40, 939–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khalil A.M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106, 11667–72 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beltran M. et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 26, 896–907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guil S. et al. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol 19, 664–70 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Kaneko S., Son J., Shen S.S., Reinberg D. & Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20, 1258–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg M. et al. Motif-driven interactions between RNA and PRC2 are rheostats that regulate transcription elongation. Nat Struct Mol Biol 28, 103–117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minajigi A. et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu H.P. et al. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 170, 86–101 e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C. et al. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol Cell 64, 416–430 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao L. & Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko S. et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24, 2615–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long Y. et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q. et al. RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat Struct Mol Biol 26, 237–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cifuentes-Rojas C., Hernandez A.J., Sarma K. & Lee J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55, 171–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko S., Son J., Bonasio R., Shen S.S. & Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28, 1983–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltran M. et al. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat Struct Mol Biol 26, 899–909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X. et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell 65, 1056–1067 e5 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Lee Y.W., Weissbein U., Blum R. & Lee J.T. G-quadruplex folding in Xist RNA antagonizes PRC2 activity for stepwise regulation of X chromosome inactivation. Mol Cell 84, 1870–1885 e9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colognori D., Sunwoo H., Kriz A.J., Wang C.Y. & Lee J.T. Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol Cell 74, 101–117 e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colognori D., Sunwoo H., Wang D., Wang C.Y. & Lee J.T. Xist Repeats A and B Account for Two Distinct Phases of X Inactivation Establishment. Dev Cell 54, 21–32 e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajit K., Alagia A., Burger K. & Gullerova M. Tyrosine 1-phosphorylated RNA polymerase II transcribes PROMPTs to facilitate proximal promoter pausing and induce global transcriptional repression in response to DNA damage. Genome Res 34, 201–216 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zovoilis A., Cifuentes-Rojas C., Chu H.P., Hernandez A.J. & Lee J.T. Destabilization of B2 RNA by EZH2 Activates the Stress Response. Cell 167, 1788–1802 e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez A.J. et al. B2 and ALU retrotransposons are self-cleaving ribozymes whose activity is enhanced by EZH2. Proc Natl Acad Sci U S A 117, 415–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J.K. et al. Denaturing purifications demonstrate that PRC2 and other widely reported chromatin proteins do not appear to bind directly to RNA in vivo. Mol Cell 84, 1271–1289 e12 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanhere A. et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38, 675–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J. et al. Structural basis for inactivation of PRC2 by G-quadruplex RNA. Science 381, 1331–1337 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konig J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17, 909–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidovich C. et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 57, 552–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao R. et al. Pervasive Chromatin-RNA Binding Protein Interactions Enable RNA-Based Regulation of Transcription. Cell 178, 107–121 e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigova A.A. et al. Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mumbach M.R. et al. HiChIRP reveals RNA-associated chromosome conformation. Nat Methods 16, 489–492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeon Y. & Lee J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saldana-Meyer R. et al. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev 28, 723–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kung J.T. et al. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell 57, 361–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen A.S. et al. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol Cell 76, 395–411 e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saldana-Meyer R. et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol Cell 76, 412–422 e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh H.J. et al. Jpx RNA regulates CTCF anchor site selection and formation of chromosome loops. Cell 184, 6157–6173 e24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J., Liu T., Qin B., Zhang Y. & Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7, 1728–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long Y. et al. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat Genet 52, 931–938 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemphill W.O., Fenske R., Gooding A.R. & Cech T.R. PRC2 direct transfer from G-quadruplex RNA to dsDNA has implications for RNA-binding chromatin modifiers. Proc Natl Acad Sci U S A 120, e2220528120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J. & Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinz S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–89 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]