Abstract
Acinetobacter baumannii can cause prolonged infections that disproportionately affect immunocompromised populations. Our understanding of A. baumannii respiratory pathogenesis relies on an acute murine infection model with limited clinical relevance that employs an unnaturally high number of bacteria and requires the assessment of bacterial load at 24–36 hours post-infection. Here, we demonstrate that low intranasal inoculums in immunocompromised mice with a tlr4 mutation leads to reduced inflammation, allowing for persistent infections lasting at least 3 weeks. Using this “chronic infection model,” we determined the adhesin InvL is an imperative virulence factor required during later stages of infection, despite being dispensable in the early phase. We also demonstrate that the chronic model enables the distinction between antibiotics that, although initially reduce bacterial burden, either lead to complete clearance or result in the formation of bacterial persisters. To illustrate how our model can be applied to study polymicrobial infections, we inoculated mice with an active A. baumannii infection with Staphylococcus aureus or Klebsiella pneumoniae. We found that S. aureus exacerbates the infection, while K. pneumoniae enhances A. baumannii clearance. In all, the chronic model overcomes some limitations of the acute pulmonary model, expanding our capabilities to study of A. baumannii pathogenesis and lays the groundwork for the development of similar models for other important opportunistic pathogens.
Keywords: Acinetobacter baumannii, Murine model, Virulence factors, Antibiotic treatments, Polymicrobial infections, Pathogenesis
Introduction
Acinetobacter baumannii is a Gram-negative opportunistic pathogen that causes diverse infections including pneumonia, urinary tract infection (UTI), bone and soft tissue infection, and septicemia (1–5). While becoming an increasingly more common cause of community-acquired infections, A. baumannii still primarily causes hospital-acquired infections in critically ill and immunocompromised patients, ~25% of which are polymicrobial (6–11). These infections are associated with an alarming mortality rate, up to 80% in some populations, largely owing to extremely high rates of multi-drug resistance (8, 12, 13). Notably, A. baumannii isolates exhibit the highest rates of multi-drug resistance of all Gram-negative pathogens, leading the World Health Organization to classify the bacterium at its highest priority for research and development of new treatments (13, 14). There is consequently an urgent need to better understand the virulence mechanisms employed by A. baumannii to guide the development of novel therapeutic approaches to combat infections.
While A. baumannii can cause a variety of infections, it is most commonly associated with pneumonia (4, 15). In fact, A. baumannii causes up to 10% of all hospital-acquired pneumonia (HAP) cases in the United States, highlighting its importance in clinical settings (16, 17). Despite this, little is known regarding the pathogenesis of this bacterium in the respiratory tract (18). A major hindrance in the ability to investigate A. baumannii pneumonia is the lack of available clinically-relevant murine infection models. This is, in large part, due to the low virulence of most strains in immunocompetent mice. This is a shared feature among many pathogens that commonly cause HAP, including Pseudomonas aeruginosa and Staphylococcus aureus, for which animal models closely mimicking human infection are not available (19, 20). An acute infection model requiring a very high, and rather artificial, inoculum of 108-109 bacteria introduced intranasally or intratracheally is most often used to investigate these pathogens (10, 20, 21). Wild type (WT) mice will typically either succumb to infection or clear the organism by 72 h, thus requiring early readouts of infection such as bacterial pulmonary titers at 24–36 h. While this model may serve as a useful tool to study pathogenesis early during infection, the quick bacterial clearance does not allow for the study of bacterial virulence mechanisms at later timepoints. Importantly, A. baumannii respiratory infection in humans results in an average length of hospital stay of ~30 days, and this number is much higher in cases caused by multi-drug resistant strains, highlighting the need for a long-term infection model (22, 23). In this pursuit, some laboratories have used antibody or cyclophosphamide treatments to render mice neutropenic (24–30). These treatments initially make mice more susceptible to A. baumannii infection, enabling the study of bacterial pathogenesis up to 7 d post-infection (dpi) using lower inoculums (~107 bacteria). However, these models do not achieve stable neutropenia in mice which leads to clearance of infection. To maintain neutropenia over longer periods, multiple injections would be necessary, which can lead to fluctuating neutrophil levels, thereby altering the overall course of disease. A notable caveat to many reports using these currently available immunocompetent and immunocompromised infection models is that older, lab-domesticated strains and non-lung isolates, such as Ab19606 and Ab17978, are employed, despite the extensive literature demonstrating numerous genotypic and phenotypic differences between these and modern respiratory isolates (31–35). In all, there is an urgent need for alternative infection models to study bacterial pathogenesis during long-term infection by relevant clinical isolates.
Previous reports have used genetically immunocompromised mice to study the role of the host immune response to A. baumannii infection. One example is mice carrying a mutation in toll-like receptor 4 (TLR4). TLR4 recognizes the lipid A moiety of bacterial lipopolysaccharide (LPS) and lipooligosaccharide (LOS), the main component of the outer membrane of most Gram-negative bacteria (36–38). The recognition of lipid A by TLR4 triggers a signaling cascade through MyD88- or TRIF-dependent pathways, resulting in increased inflammatory cytokine and type 1 interferon production, respectively (39). The role of TLR4 during A. baumannii infection has been examined in murine septicemia, acute pneumonia, UTI, and catheter-associated UTI (CAUTI) models (40–42). In the acute pneumonia model, Knapp et al. showed that tlr4 mutant mice had increased A. baumannii CFU in the lungs with reduced inflammatory cytokines compared to WT mice (41, 43). Using a bloodstream infection model, Lin et al. demonstrated that WT C3H/FeJ and tlr4 mutant C3H/HeJ mice had similar bacterial burdens (40, 44). However, all WT mice succumbed to infection by day 4, whereas all tlr4 mutant mice survived. This could be attributed to WT mice experiencing septic shock associated with increased inflammatory cytokines. Finally, in a UTI model, our laboratory found that tlr4 mutant C3H/HeJ mice were more susceptible to infection than WT C3H/HeN mice (42). Moreover, we found that C3H/HeJ mice in the UTI model formed small intracellular populations in urothelial cells referred to as Acinetobacter baumannii intracellular reservoirs (ABIRs), which could seed a recurrent infection upon catheterization at higher rates relative to WT mice. In addition to playing a significant role during murine infection, TLR4 is relevant in clinical settings as well. In fact, numerous studies have identified links between tlr4 polymorphisms and infection outcomes from A. baumannii pneumonia (45–47). In all, these studies demonstrate the key role of TLR4 in controlling A. baumannii infection and disease progression and highlight the clinical relevance of the associated signaling cascade.
In this work we describe a novel murine model of A. baumannii pneumonia that employs tlr4 mutant mice and low bacterial inoculums (105 bacteria). Using this model, we show that clinically-relevant A. baumannii strains can establish chronic infection. We additionally demonstrate that our model enables the discovery of virulence factors not detectable in the acute infection model. Finally, we illustrate how our model can be employed to assess the efficacy of antibiotics over the course of infection and investigate polymicrobial infections.
Results
tlr4 mutant mice are susceptible to chronic infection at low inoculums.
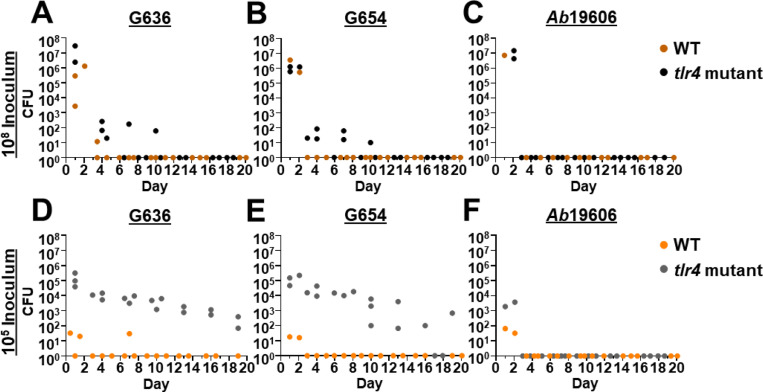
To assess if tlr4 mutant mice could serve as permissive hosts for long-term respiratory infection, we performed intranasal inoculations of WT (C3H/HeN) and tlr4 mutant (C3H/HeJ) mice with high (108) and low (105) inoculums of a modern A. baumannii respiratory isolate, G636, and sacrificed groups of mice every 3 days starting at 24 hours post-infection (hpi). At the higher inoculum, WT mice cleared infection by day 4, consistent with previously published results using the acute pulmonary infection model (Fig. 1A) (21). tlr4 mutant mice infected with the higher inoculum also cleared infection relatively early after inoculation, with most mice having no detectable bacteria in the lungs by ~7 dpi. Strikingly, while WT mice infected with the lower dose of 105 bacteria cleared infection after 1 day, tlr4 mutant mice maintained detectable bacteria in the lungs out to the latest timepoint tested, 19 dpi (Fig. 1D). Despite this long infection course, dissemination to distal organs was rarely detected. This is consistent with the clinical manifestations of non-ventilator A. baumannii pneumonia, as less than 20% of patients will develop subsequent bacteremia (48). Notably, by employing confocal microscopy, we were able to visualize bacteria in tlr4 mutant mice with the low inoculum; at early timepoints (4 hpi and 2 dpi), bacteria were identified inside cells in the bronchoalveolar lavage fluid (BALF), as well as extracellularly, consistent with our previously published results in the acute infection model (Fig. S1) (31).
Figure 1.
Low inoculums of modern respiratory A. baumannii clinical isolates result in chronic lung infection in tlr4 mutant mice. Groups of C3H/HeN (WT) or C3H/HeJ (tlr4 mutant) mice were intranasally inoculated with 108 G636 (A), 108 G654 (B), 108 Ab19606 (C), 105 G636 (D), 105 G654 (E), or 105 Ab19606 (F). Beginning at 24 hpi, groups of mice were sacrificed every three days, and bacteria in the lungs were quantified. Each data point indicates and individual mouse.
We then assessed if a second modern A. baumannii respiratory isolate, G654, behaves similarly to G636 (Fig. 1E). Again, detectable levels of bacteria were present in the lungs out to 19 dpi with the low inoculum in tlr4 mutant mice, while WT mice cleared this inoculum within 1 day. At the higher inoculum, G654 was cleared soon after inoculation regardless of TLR4 functionality (Fig. 1B). On the contrary, when we tested an older, lab-domesticated urinary isolate that is commonly used to study A. baumannii respiratory pathogenesis, Ab19606, we found that WT and tlr4 mutant mice cleared infection after 1 day regardless of inoculum size or mouse background (Fig. 1C and 1F). In all, these results indicate that modern A. baumannii respiratory isolates can cause infection out to nearly 3 weeks at lower and likely more clinically-relevant inoculums than previously used in the literature. The finding that Ab19606 was unable to establish long-term infection further highlights the differences between modern, infection site-specific isolates and lab-domesticated strains (31–35). Importantly, this infection duration with low inoculums of modern respiratory isolates in tlr4 mutant mice is the longest reported for A. baumannii in any animal model to date. We therefore chose to further characterize these conditions as a model to study pulmonary pathogenesis, referred to hereafter as the “chronic respiratory infection model.”
Lower A. baumannii inoculums result in a decreased immune response in tlr4 mutant mice.
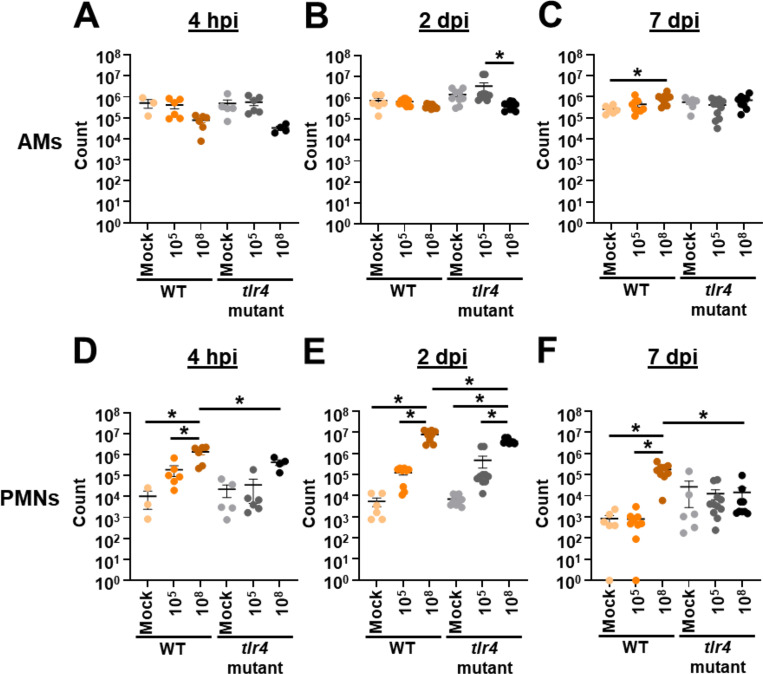
Given the unexpected result that tlr4 mutant mice exhibit chronic infection at lower inoculums, while WT and tlr4 mutant mice clear infection at higher inoculums, we sought to characterize the host immune response in these different conditions. We intranasally infected groups of WT and tlr4 mutant mice with 105 or 108 bacteria or mock infected them with phosphate-buffered saline (PBS). Then, at early timepoints of 4 hpi and 2 dpi and a later timepoint of 7 dpi, BALF was collected for immune cell quantification (Fig. 2). Regardless of timepoint, inoculum, or mouse background, few significant changes were observed in number of alveolar macrophages (AMs) (Fig. 2A–2C). In WT mice, the number of polymorphonuclear leukocytes (PMNs) was increased with the higher inoculum relative to lower and mock inoculums at every timepoint (Fig. 2D–2F). Additionally, at the higher inoculum, WT mice had increased PMNs relative to tlr4 mutant mice at every time point in line with previous results (41). Interestingly, at the lower inoculum, while PMN counts trended higher at 4 hpi for WT mice relative to tlr4 mutant mice, no significant differences were noted between these groups at any timepoint. This result suggests that, despite neutrophil influx being a predominant mechanism of A. baumannii clearance, PMN numbers alone may not account for differences in clearance between WT and tlr4 mutant mice at the lower inoculum (49–55).
Figure 2.
Lower intranasal A. baumannii inoculums result in reduced lung neutrophil influx. Groups of C3H/HeN (WT) or C3H/HeJ (tlr4 mutant) mice were intranasally inoculated with 105 G636, 108 G636, or mock inoculated with PBS. At 4 h (A and D), 2 d (B and E), and 7 d (C and F) pi, alveolar macrophages (AMs) (A-C) and polymorphonuclear leukocytes (PMNs) (D-F) in the BALF were enumerated by flow cytometry. Shown are pooled results from at least two independent experiments, and each data point represents an individual mouse. The horizontal line represents the mean, and the standard error of the mean (SEM) is indicated by error bars. *P < 0.05; two-way analysis of variance (ANOVA), Tukey’s test for multiple comparisons.
To further evaluate the host response, we quantified 13 common inflammatory cytokines in the BALF (Table S1). At the higher inoculum, WT and tlr4 mutant mice exhibited significantly increased levels of IL-1α, IFN-γ, TNF-α, MCP-1, IL-1β, IL-6, and IL-17A early during infection relative to the lower inoculum while levels dissipated by 7 dpi, consistent with bacterial clearance (See Fig. 1A). WT mice infected with the high inoculum had significantly increased levels of IFN-β relative to tlr4 mutant mice at 4 hpi and increased levels of IL-1α, IFN-γ, TNF-α, MCP-1, IL12-p70, IL-1β, IL-6, IL-27, and IL17A at 2 dpi, likely leading to the earlier clearance observed. At the lower inoculum, although WT mice clear infection within nearly 24 h and tlr4 mutant mice maintain infection out to at least three weeks, minimal significant differences in inflammatory cytokines were observed (See Fig. 1D). In fact, the only significant difference noted was the increased levels of GM-CSF at 4 hpi in WT mice relative to tlr4 mutant mice. Other inflammatory cytokines that trended higher at early timepoints in WT mice at the lower inoculum include the inflammasome-associated cytokines IL-1α and IL-1β, as well as TNF-α and IL-6. These elevated levels of inflammatory cytokines early during infection in WT mice could possibly account for the earlier clearance. Later during infection, however, tlr4 mutant mice had elevated, albeit not significantly higher, amounts of TNF-α, IL-1α, and IL-6 relative to WT mice, consistent with persistent infection.
The chronic respiratory infection model results in lung pathology.
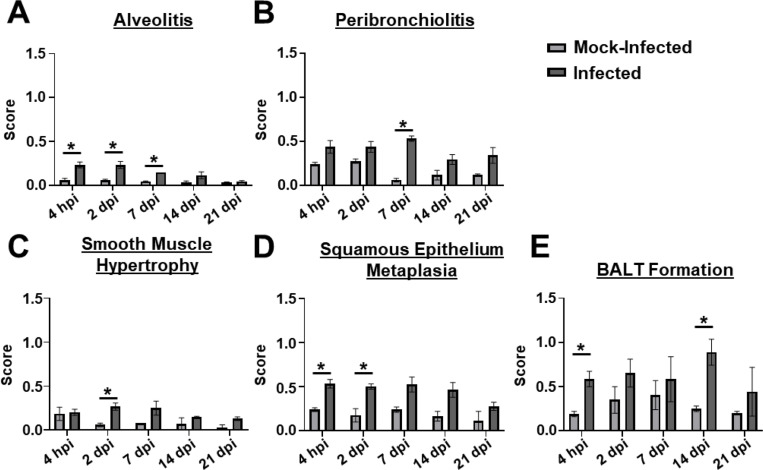
We next assessed if the chronic respiratory infection model is associated with lung pathology. tlr4 mutant mice were infected with 105 G636 or mock infected with PBS, and mice were sacrificed at 4 hpi, 2 dpi, 7 dpi, 14 dpi, and 21 dpi. Lungs were then sectioned, stained with hematoxylin and eosin (H&E), and scored for pathological changes as previously described (Fig. 3, Fig. S2, and Fig. S3) (56). The chronic model resulted in significant increases in alveolitis, peribronchiolitis, smooth muscle hypertrophy, squamous epithelium metaplasia, and formation of bronchus-associated lymphoid tissue (BALT) relative to mock-treated mice (Fig. 3 and Fig S2). Significant changes in goblet cell hyperplasia and fibrosis were not detected (Fig. S3). Of note, significant signs of disease were detected out to 14 dpi, and, even at 21 dpi, infected mice showed trends toward increased lung damage relative to mock-infected mice. In all, histopathological analyses revealed that the chronic respiratory infection model results in sustained lung damage, indicative of chronic infection.
Figure 3.
The chronic respiratory infection model results in lung pathology. Groups of C3H/HeJ (tlr4 mutant) mice were inoculated with 105 G636 or mock-inoculated with PBS, and at 4 hpi, 2 dpi, 7 dpi, 14 dpi, and 21 dpi, lungs slices were prepared, H&E stained, and scored for alveolitis (A), peribronchiolitis (B), smooth muscle hypertrophy (C), squamous epithelium metaplasia (D), and BALT formation (E). The mean is shown on the graph, and the SEM is indicated by error bars. *P < 0.05, Unpaired Student’s t-test.
InvL is a critical virulence factor for long-term infection.
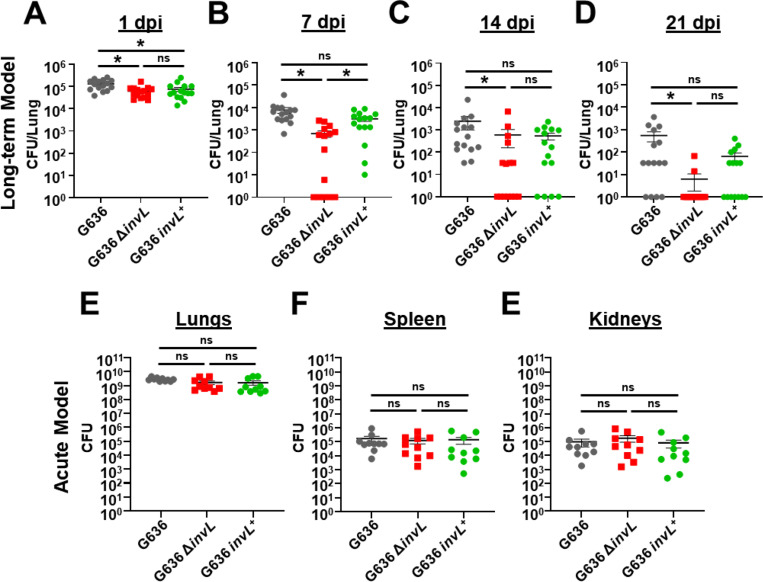
The acute pulmonary infection model has been widely used to characterize A. baumannii virulence factors (21). While the acute model is valuable for identifying bacterial proteins required at early timepoints, these mice clear infection within 3–4 dpi, not allowing for the identification of factors required for prolonged infection. As a proof of principle, we sought to determine if the chronic respiratory infection model could identify proteins required for bacterial persistence in the lungs. We hypothesized that prolonged adherence to respiratory epithelium would be required for persistence, so we first tested individual mutants lacking previously identified A. baumannii adhesins (Bap, Ata, FhaBC, and InvL) for attenuation in the chronic infection model (Fig. S4) (57–66). tlr4 mutant mice were infected with 105 G636 WT or mutant bacteria, and mice were sacrificed at 1 or 14 dpi for lung colony-forming units (CFU) quantification. This experiment indicated a possible role for InvL in long-term infection as mice began to clear bacteria in the lungs by 14 dpi. To further confirm the importance of InvL for bacterial persistence, we performed more extensive analyses with G636 WT, invL mutant (ΔinvL), and complemented invL mutant (invL+) strains in the chronic infection model, sacrificing mice at 1, 7, 14, and 21 dpi to quantify CFU in the lungs (Fig. 4A-D). Early during infection, the ΔinvL mutant exhibited only a modest defect. However, at later timepoints the infection defect became more pronounced, as some mice cleared the bacteria as early as 7 dpi. By 21 dpi, all but two mice had cleared the ΔinvL mutant, while the majority of mice infected with the WT strain still had detectable bacteria in their lungs. Genetic complementation partially rescued this defect at these later timepoints, as no significant difference was detected between WT and complemented strains.
Figure 4.
InvL is a critical virulence factor for long-term respiratory infection, but dispensable in the acute infection model. C3H/HeJ (tlr4 mutant) mice were infected with 105 G636, G636 ΔinvL, or G636 invL+. Groups of mice were then sacrificed at 1 dpi (A), 7 dpi (B), 14 dpi (C), and 21 dpi (D), and CFU in the lungs were quantified. Shown are the results from 3 independent experiments. For the acute infection model, groups of C57BL/6 mice were infected with 109 G636, G636 ΔinvL, or G636 invL+. 24 hpi, mice were sacrificed, and CFU in the lungs (A), spleen (B), and kidneys (C) were enumerated. Each data point represents an individual mouse, the horizontal line represents the mean, and the SEM is indicated by error bars. Shown are the results of 2–3 independent experiments. *P < 0.05; Kruskal-Wallis H test with Dunn’s test for multiple comparisons; ns = not significant.
We next compared results from the chronic respiratory infection model to the acute infection model. We infected C57BL/6 mice with 109 G636 WT, ΔinvL, or invL+ strains. 24 hpi, mice were sacrificed, and CFU in the lungs, spleens, and kidneys were quantified (Fig. 4E-F). As opposed to results seen in the chronic infection model, the ΔinvL mutant had no significant defect in bacterial load in the lungs. Additionally, no defect was noted in dissemination to the spleen and kidneys, indicating that InvL is dispensable in the acute infection model. In all, these results highlight the differences in required bacterial genes between these disparate pulmonary infection models and show the importance of continuing to explore models that can better approximate clinical disease. Additionally, these experiments establish InvL as the first known A. baumannii virulence factor required for long-term infection.
The chronic infection model can be used to study the outcome of antibiotic treatment.
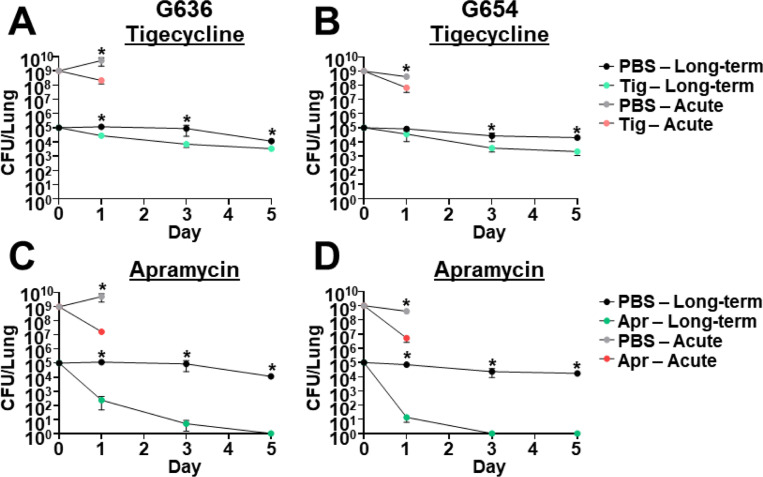
The acute pulmonary infection model has been employed extensively to assess effects of antibiotic treatment (21). However, this model only allows us to estimate the efficacy of antibiotics by measuring the initial reduction in the bacterial burden at 24–36 hpi due to rapid bacterial clearance by the host. A clear limitation of this model is that it does not inform if bacterial infection is cleared, or if persistent bacteria remain in the lung. The chronic respiratory infection model therefore represents a novel platform that could be used to track the kinetics of A. baumannii clearance due to antibiotic treatment. As a proof of principle, we assessed the effect of tigecycline, colistin, and imipenem in the chronic model with strains G636 and G654 at antibiotic concentrations similar to those previously used to determine treatment efficacy in mice (Fig. 5A-B and Fig. S5A-D) (67–73). We additionally assessed the effect of apramycin, a drug with demonstrated efficacy and safety in mice that is currently in Phase I clinical trials for use in humans (Fig. 5C-D) (74–76). Minimum inhibitory concentrations (MICs) for G636 and G654 for these and other commonly used antibiotics are listed in Table S2. Colistin was ineffective for both strains in the acute infection model, while initial reductions in CFU were noted in the chronic infection model (Fig. S5A-B). However, bacterial numbers appeared to stabilize over time in the chronic infection model, consistent with the development of bacterial persisters (discussed below). Imipenem showed limited efficacy against both strains in both models (Fig. S5C-D), as expected given the strains’ resistance in vitro (Table S2). At 24 hpi, tigecycline and apramycin treatment resulted in initial reductions in CFUs in both the chronic and the acute infection models relative to PBS-treated mice (Fig. 5A-D). However, the chronic model enabled us to differentiate the efficacy of both antibiotics at later times. Apramycin treatment ultimately led to clearance after 3–5 days, demonstrating the efficacy of this antibiotic. However, with tigecycline treatment, although there were initial reductions in CFU, bacterial numbers leveled out over time indicative of treatment failure. The behavior of bacteria in presence of tigecycline over time is consistent with the development of persisters. Notably, the efficacy of tigecycline and apramycin against A. baumannii cannot be distinguished at 24 hpi. These results indicate that the chronic model can be used to determine outcome of infection with therapeutic intervention, a significant advantage over the currently employed acute infection model.
Figure 5.
The chronic respiratory infection model can be used to study outcomes of antibiotic treatment. Groups of C3H/HeJ (tlr4 mutant) mice were infected with 105 G636 (A, C) or 105 G654 (B, D) and sacrificed at 1, 3, and 5 dpi (long-term). Additionally, groups of C57Bl/6 mice were infected with 109 G636 (A, C) or 109 G654 (B, D) and sacrificed at 24 hpi (acute). Mice in both infection models were treated intraperitoneally treated with PBS or 100 mg/kg tigecycline (tig) every 12 h (A, B) or PBS or 500 mg/kg apramycin (apr) every 12 h (C, D) with all treatments beginning 4 hpi. At each timepoint, CFU were quantified in the lungs. Shown are the results from at least two independent experiments, each data point represents an individual mouse, the horizontal line represents the mean, and the SEM is represented by error bars. *P < 0.05; Mann-Whitney U test.
Use of the chronic infection model to study bacterial co-infections reveals that Staphylococcus aureus exacerbates ongoing A. baumannii infection while Klebsiella pneumoniae leads to earlier clearance.
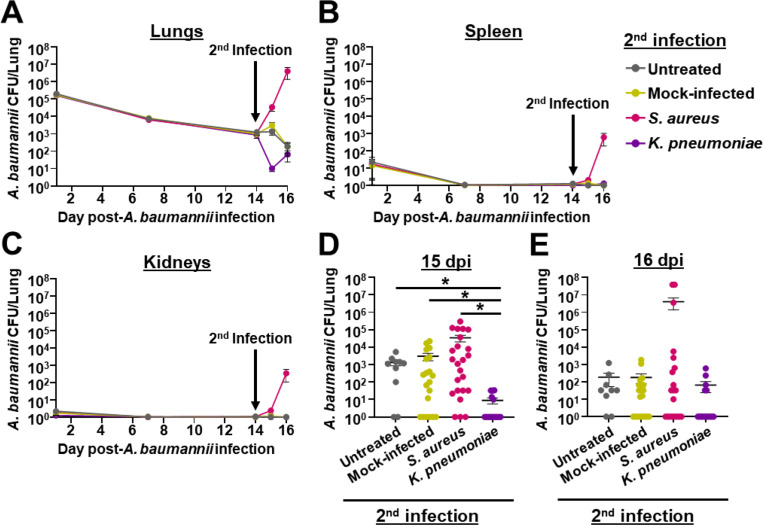
Approximately 25% of A. baumannii pulmonary infections are polymicrobial, and two of the most commonly co-infecting pathogens are Staphylococcus aureus and Klebsiella pneumoniae (9). We thus sought to assess the impact of secondary infections with these two bacteria on the outcome of A. baumannii infection in the context of the chronic respiratory infection model. For these experiments, we first established a primary A. baumannii infection by inoculating tlr4 mutant mice with 105 CFU of strain G636. Following 14 days of A. baumannii infection, we inoculated mice with 5 × 107 CFU of S. aureus strain Newman or K. pneumoniae strain TOP52, mock-treated mice with PBS, or left mice untreated. One and two days post-secondary infection, mice were sacrificed, and bacterial CFU were quantified in the lungs, spleens, and kidneys (Fig. 6, Fig. S6, and Fig. S7). Secondary infection with S. aureus led to a resurgence of A. baumannii CFU in the lungs of many mice, though the overall mean CFU in these mice were not significantly different from mock-infected and untreated groups (Fig. 6A and Fig. 6D-E). A. baumannii were also identified in the spleens and kidneys of some mice that received the secondary S. aureus infection, even though A. baumannii bacteremia rarely occurs in the context of this chronic respiratory infection model (Fig. S6). Additionally, S. aureus trended toward increased numbers in the lungs, spleens, and kidneys in the context of polymicrobial infection with A. baumannii relative to monomicrobial infection (Fig. S7A-C). Notably, two mice succumbed to A. baumannii-S. aureus polymicrobial infection ~24 hpi following the secondary inoculation, an outcome that did not occur with monomicrobial infection with either bacterium. Contrarily, secondary infection with K. pneumoniae significantly decreased A. baumannii CFU in the lungs relative to mock-infected and untreated groups (Fig. 6A and Fig. 6D-E). Additionally, polymicrobial infection with A. baumannii and K. pneumoniae resulted in significantly reduced K. pneumoniae CFU recovered in the lungs, spleens, and kidneys of mice relative to K. pneumoniae monomicrobial infection (Fig. S7D-E). Although understanding the interactions between these bacteria is beyond the scope of this work, these experiments indicate that S. aureus exacerbates A. baumannii infection, while K. pneumoniae attenuates infection in the context of the chronic infection model. Additionally, these results demonstrate the ability of the model to be used to study longer-term aspects of polymicrobial interactions that were not previously able to be done with the acute infection model.
Figure 6.
Bacterial secondary infection alters the course of chronic A. baumannii pneumonia. C3H/HeJ (tlr4 mutant) mice were intranasally inoculated with 105 G636, and groups of mice were sacrificed at 1, 7, and 14 dpi. At 14 days post-A. baumannii infection, groups of mice were either not inoculated (untreated), inoculated with PBS (mock-infected), infected with S. aureus, or infected on K. pneumoniae. Subsequently, on days 15 and 16 post-A. baumannii infection (1 and 2 days post-secondary infection), mice were sacrificed, and A. baumannii CFU were quantified in the lungs (A, D, E), spleen (B), and kidneys (C). In panels A, B, and C, each data point represents the mean, the SEM is represented by error bars, and the limit of detection is indicated by the dashed line. In panels D and E, each data point represents an individual mouse, the horizontal line represents the mean, and the SEM is indicated by error bars. Shown are results from at least 2 independent experiments. *P < 0.05; Kruskal-Wallis H test with Dunn’s test for multiple comparisons.
Discussion
A. baumannii has emerged as a significant cause of nosocomial pneumonia and is of major clinical importance due to its extremely high rates of multidrug resistance (8, 12, 13). Despite this, our understanding of A. baumannii respiratory pathogenesis is hindered by a shortage of clinically relevant infection models. Here, we aimed to address this significant gap in the field by developing a novel respiratory infection model. In this pursuit, we found that, at likely more clinically-relevant inoculums, tlr4 mutant mice maintain long-term respiratory infections by A. baumannii. We then demonstrate the versatility of this model which enabled i) the identification of a bacterial virulence factor required for long-term respiratory infection, which is not required in acute models, ii) the study of kinetics of bacterial clearance upon treatment with clinically-relevant antibiotics, and iii) the exploration of the impact of secondary infections with two commonly co-isolated respiratory pathogens.
In this study, we found that InvL is required for chronic infection, and, more importantly, at the later stages of infection. However, InvL was dispensable in the context of the acute infection model. There are multiple possible reasons for this discrepancy. First, the massive bacterial dose required for the acute infection model may mask potential defects that can now be detected with a smaller, more clinically-relevant inoculum. This is unlikely, as WT and invL mutant bacteria behave similarly at early time points in our model. An alternative reason could be that adhesins required early during infection/interaction with the healthy airway differ from those required during persistent interaction with a more inflamed or damaged airway. It is well-established that the airway extracellular matrix (ECM) is altered by bacterial infection, lung damage, and/or inflammation (91, 92). Long-term lung damage and inflammation results in increased fibronectin, collagen, laminin, and fibrinogen in the ECM (93–98). Moreover, specific pathogens elicit different inflammatory responses, resulting in distinct changes to the lung ECM. For example, in an acute mouse model of pneumonia, Pseudomonas aeruginosa induces versican deposition in the lungs, while Escherichia coli induces robust versican and hyaluronan deposition (99, 100). We previously showed that InvL can bind α5β1 integrin, collagen V, and fibrinogen (66). However, whether A. baumannii infection or the associated inflammation induces production of these protein(s) during pulmonary infection is unknown. Future work will investigate this possibility, as well as assess which InvL-host protein interactions are essential for chronic infection.
Herein, we demonstrate the potential to use the chronic respiratory infection model to study the efficacy of antibiotic treatments over time. One intriguing finding from these experiments is that with antibiotics such as colistin and tigecycline, an initial decrease in CFU (~10–100 fold) recovered from the lungs at 1 dpi was observed. However, following this decrease, the number of bacteria in the lungs appeared to stabilize over time. It is tempting to speculate that this is the result of the formation of bacterial persisters, defined as bacterial cells that become tolerant to antibiotics despite undergoing no genetic changes (101–103). Importantly, the commonly used acute infection model does not allow for the study of bacterial persisters due to the short time course of the model. Given that persister cells represent a major cause of treatment failure and chronic infection, the chronic infection model presented here represents a unique platform that is desperately needed to understand this aspect of A. baumannii pathogenesis. Furthermore, our model offers new possibilities to study efficacy of novel antibiotics in murine models before committing to expensive clinical trials.
While a significant portion of A. baumannii infections are polymicrobial, the acute infection model has limitations for use with polymicrobial infections. First the quick clearance of the bacteria usually only allows inoculation at a single timepoint, thus not enabling investigation of secondary infections. Second, the high required infectious dose often means that typical inoculums for bacteria used in these experiments must be adjusted, so mice do not succumb to infections at early timepoints. Here, we applied the chronic respiratory infection model to assess the result of secondary infection with two pathogens commonly co-isolated with A. baumannii, S. aureus and K. pneumoniae. We found opposite results with these different bacteria; S. aureus secondary infection trended toward exacerbation of A. baumannii infection, while K. pneumoniae secondary infection led to reduced A. baumannii numbers. The potential synergism of A. baumannii and S. aureus in the chronic infection model aligns with previous reports. For example, using a Tn-Seq-based approach, Li et al. demonstrated that the 49% of genes required by S. aureus for monomicrobial infection in a murine systemic infection model became non-essential upon A. baumannii co-infection (104). Another recent report showed that S. aureus can support A. baumannii growth in vitro by providing acetoin as a carbon source (105).
Although we found that K. pneumoniae secondary infection led to reduced A. baumannii numbers in the lungs in the chronic infection model, one study has shown that K. pneumoniae could cross-feed A. baumannii through products of sugar fermentation in vitro and demonstrated that co-infection led to reduced survival of Galleria mellonella relative to monomicrobial infection with either pathogen (106). This, in part, shows that these two bacteria can have beneficial interactions. There are two potential reasons however for the reduction of CFU for both bacteria in the context of the chronic infection model reported here; i) bacterial competition or ii) the host response to the secondary infection. Regarding bacterial competition, there have been several lines of evidence pointing to direct bacterial killing between diverse A. baumannii and K. pneumoniae strains mediated by the type VI secretion system (107–110). In addition to direct killing, this bacterial competition could be indirect as well, as both A. baumannii and K. pneumoniae may be competing for similar nutrients in the lung microenvironment. With respect to the immune response, a difference between this work and the above study is that the microenvironment encountered in the mammalian lung is not perfectly modeled by the wax moth (111). Our results may therefore be the result of TLR4-independent host response elicited by the combination of both bacteria that is not recapitulated by a G. mellonella model. While understanding the precise mechanism behind the in vivo interactions between A. baumannii and commonly co-isolated pathogens is outside the scope of the current study, these results highlight the practicality of applying the chronic respiratory infection model to better understand polymicrobial infections.
In this study, we have validated several different uses for the chronic respiratory infection model. However, there are also other potential uses for this model that were not previously investigable. For example, we can now perform experiments differentiating between virulence factors required for establishment of infection and factors required for maintenance of infection, assessing bacterial evolution during long-term infection, investigating changes in the pulmonary microbiome due to infection over time, and analyzing the long-term outcomes of novel therapies such as newly developed phage cocktails. Additionally, while this model was initially developed to study Acinetobacter respiratory infections, it has the potential to be applied to research with other respiratory pathogens in cases where suitable animal models are lacking. In all, this work describes the longest-term infection model available to investigate A. baumannii host-pathogen interactions to date, which will ultimately aid in the development of novel therapeutics to combat infection by this increasingly multidrug-resistant bacterium.
Materials and Methods
Bacterial plasmids, strains, and growth conditions.
Plasmids and strains used in this study are detailed in Table S3. Bacterial cultures were grown at 37°C in Lennox broth/agar supplemented with 10 μg/mL chloramphenicol, 50 μg/mL apramycin, 100 μg/mL ampicillin, 50 μg/mL kanamycin, 10 μg/mL tetracycline, or 10% sucrose when appropriate.
Murine pneumonia models.
All animal experiments were approved by the Washington University Animal Care and Use Committee, and we have complied with all relevant ethical regulations. The acute pneumonia model was performed similar to previously described experiments (21, 112). Briefly, overnight cultures were subcultured at a 1:200 dilution and grown shaking at 37°C for 3 h to mid-exponential growth phase. Six- to eight-week-old female C57BL/6 mice (Charles River Laboratories, Wilmington, MA) anesthetized with 4% isoflurane were intranasally inoculated with 109 CFU that were twice-washed in PBS. At 24 hpi, mice were sacrificed, and CFU in the lungs, spleen, and kidneys were quantified by serial dilution plating the homogenized organs. For experiments with C3H/HeN (Envigo International Holdings, Indianapolis, IN) and C3H/HeJ (Jackson Laboratory, Bar Harbor, ME) mice, A. baumannii, S. aureus, and K. pneumoniae inoculums were prepared and mice were intranasally inoculated as described above, with the exception that inoculums of 105 and 108 CFU were used for A. baumannii, and 5 × 107 CFU was used for S. aureus and K. pneumoniae. Following, at the indicated timepoints, mice were sacrificed and bacteria in the lungs, spleen, and kidneys were quantified as described above. For co-infections, A. baumannii was distinguished from S. aureus and K. pneumoniae by plating on LB agar supplemented with 10 μg/mL chloramphenicol. For antibiotic treatment experiments the indicated mice were treated intraperitoneally with PBS or 100 mg/kg tigecycline every 12 h, PBS or 5 mg/kg colistin every 8 h, PBS or 500 mg/kg apramycin every 12 h, or PBS or 100 mg/kg imipenem every 12 h with all treatments beginning 4 hpi. Antibiotics for intraperitoneal treatments were dissolved in PBS, and the injection volume was 100 μl.
Flow cytometry.
Flow cytometry was performed similarly to previously described methods (31). Briefly, BALF samples were collected in PBS supplemented with 1 mM EDTA, and cells were collected by centrifugation at 300 x g for 5 min. Cells were then resuspended in Pharm Lyse Buffer (BD Biosciences, Franklin Lakes, NJ) and incubated for 3 min at room temperature to lyse red blood cells. Cells were subsequently washed in fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 1% heat inactivated fetal bovine serum and 0.1% sodium azide) and blocked with TruStain FcX PLUS (BioLegend, San Diego, CA) for 15 min at 4°C. Samples were then stained with anti-CD45-BV605 (BioLegend), anti-CD11c-APC (BioLegend), anti-SiglecF-PerCP5.5 (BioLegend), and anti-Ly6G-BV421 (Biolegend) for 30 min at 4°C. Following, cells were washed in FACS buffer and fixed in 2% paraformaldehyde (PFA). Samples were read on a LSR II Fortessa cytometer (BD Biosciences) or an Aurora cytometer (Cytek Biosciences, Fremont, CA). Total cell counts in the BALF were calculated using Precision Count Beads (BioLegend) according to the manufacturer’s instructions.
Antibiotic protection assays.
Antibiotic protection assays were performed as previously described (31). To determine the number of total and intracellular bacteria present in BALF from A. baumannii infected mice, two 500 μl aliquots of lavage fluid were centrifuged at 4100 x g for 5 min. Pelleted cells were resuspended in warm Dulbecco’s Modified Eagle Medium (DMEM) (total bacteria) or DMEM with colistin (50 μg/mL) (intracellular bacteria) and incubated for 1h at 37°C. Samples were then washed three times with PBS and lysed with 500 μL of Triton X-100 (0.05%). CFUs were determined by serial dilutions of the bacterial suspensions. The remaining lungs following BALF collection were also homogenized, and CFUs were quantified by serial dilution plating.
Cytospin of BALF cells.
Cytospin of BALF cells was performed similar to previously described work (31). BALF samples were centrifugated at 300 x g for 5 min, and the pellets were resuspended in 1 mL Pharm Lyse Buffer (BD Biosciences) and incubated for 5 min on ice to lyse red blood cells. 9 mL of PBS was added to stop the lysis, viability was determined using Trypan Blue solution (Sigma-Aldrich, St. Louis, MO), and cells were counted using the TC20 Automated Cell Counter (Bio-Rad Laboratories, Hercules, CA). Samples were centrifugated at 300 x g for 6 min onto CytoPro Poly-L-Lysine Coated Microscope Slides (ELITechGroup Inc., Logan, UT) using a Cytospin Cytocentrifugue (Fisher Scientific, Hampton, NH). The slides were air-dried overnight at 4°C and fixed in 4% PFA for 30 min at room temperature. Samples were incubated with permeabilizing and blocking solution (PBS supplemented with 0.1% saponin, 0.5% bovine serum albumin, and 10% heat inactivated fetal bovine serum). Cells were stained with Alexa Fluor 555 Phalloidin (Cell Signaling Technology, Danvers, MA) and 4’,6-Diamidino-2-phenylindole dihydrochloride (DAPI) solution (Invitrogen) for 1 h at 37°C. After staining, the samples were rinsed with washing solution [PBS supplemented with 0.1% saponin and 0.5% bovine serum albumin (BSA)], and then rinsed with water and mounted on a coverslip in ProLong Gold Antifade Mountant (Invitrogen).
Confocal microscopy.
Confocal microscopy was performed as previously described (31, 66). Microscopy slides were analyzed with a Zeiss LSM880 laser scanning confocal microscope (Carl Zeiss AG, Oberkochen, Germany) equipped with 405nm diode, 488nm Argon, 543nm HeNe, and 633nm HeNe lasers. A Plan-Apochromat 63X DIC objective and ZEN black 2.1 SP3 software were used for image acquisition. Images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD) (113).
Cytokine analysis.
BALF was collected and centrifuged at 300 x g for 5 min. Supernatant containing cytokines was then collected and frozen at −20°C until the analysis was performed. Cytokine levels were determined using the LEGENDplex Mouse Inflammation Panel (13-plex) with V-bottom Plate (BioLegend) according to the manufacturer’s instructions. Samples were read using an Aurora cytometer (Cytek Biosciences).
Histopathology of lung slices.
Lung slices were prepared, stained, and scored as previously described by Castro et al. (71). Briefly, lungs were perfused with PBS, inflated with optimal cutting temperature (OCT) compound (Fisher Scientific) diluted in 4% PFA at a 1:1 ratio, snap-frozen, and stored at −80°C until sectioning. For histology imaging, 4 μm tissue sections were stained with H&E and imaged with a ZEISS Axioscan 7 Microscope Slide Scanner (Carl Zeiss AG). Lung tissues were blindly scored on a scale of 0 to 3 for alveolitis, peribronchiolitis, smooth muscle hypertrophy, squamous epithelium metaplasia, BALT formation, goblet cell hyperplasia, and fibrosis. Area affected was quantified, multiplied by previously defined intensity scores, and the resulting weighted scores are reported.
Generation of constructs and strains used in this study.
Primers used in this study are listed in Table S4. DNA fragments were assembled using either the In-Fusion HD EcoDry Cloning Kit (TaKaRa Bio, Mountain View, CA) or NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA). To generate the vector for generation of the invL mutational construct, pEX18Tc was amplified without the tetracycline resistance cassette (primers: 5’ pEX18 marker swap and 3’ pEX18 marker swap), the apramycin resistance cassette was amplified from pKD4-Apr (primers: 5’ Apr for pEX18Ap and 3’ Apr for pEX18Ap), and the amplicons were assembled, generating pEX18Ap (114, 115). The pEX18Ap mutational constructs were then made by amplifying the pEX18Ap vector (primers: 5’ pEX18Tc and 3’ pEX18Tc), a ~1000 bp region upstream of the genes of interest (invLKO primers: 5’ F1 G636 invLKO and 3’ F1 G636 invLKO; bapKO primers: 5’ F1 G636 bapKO and 3’ F1 G636 bapKO; ataKO primers: 5’ F1 G636 ataKO and 3’ F1 G636 ataKO; fhaBCKO primers: 5’ F1 G636 fhaBCKO and 3’ F1 G636 fhaBCKO), and a ~1000 bp region downstream of the genes of interest (invLKO primers: 5’ F2 G636 invLKO and 3’ F2 G636 invLKO; bapKO primers: 5’ F2 G636 bapKO and 3’ F2 G636 bapKO; ataKO primers: 5’ F2 G636 ataKO and 3’ F2 G636 ataKO; fhaBCKO primers: 5’ F2 G636 fhaBCKO and 3’ F2 G636 fhaBCKO), followed by assembly of these amplicons. Mutational constructs were then transformed into G636, and strains with the integrated plasmid were selected for by apramycin treatment. Counterselection for double crossover was performed by plating these strains on LB agar without NaCl supplemented with 10% sucrose. Mutants were then confirmed by PCR analyses and whole-genome sequencing.
The invL complementation construct was generated by amplifying the putative promoter region (~300 bp upstream) along with the invL open reading frame (primers: 5’ G636 fdeCKO Comp and 3’ G636 fdeCKO Comp-His6 v2) and the pUC18T-miniTn7T-Apr vector (primers: Tn7 linear Fwd-His6 and Tn7 liner Rev) (116). These amplicons were then assembled, generating pUC18T-miniTn7T-Apr::G636 invLKO comp. To generate the gfp integration construct, the gfp cassette was amplified from PB-FLuc+GFPd2 (primers: 5’ d2EGFP for pUC18T-mTn7 and 3’ d2EGFP for pUC18T-mTn7) and pUC18T-mTn7-Apr was amplified (primers: 5’ pUC18T-mTn7 for d2EGFP and 3’ pUC18T-mTn7 for d2EGFP). These fragments were then assembled, generating pUC18T-miniTn7T-Apr::gfpd2. pUC18T-miniTn7T-Apr::G636 invLKO comp and pUC18T-miniTn7T-Apr::gfpd2 were introduced into G636 ΔinvL and G636, respectively, using a four-parental conjugation technique, as previously described (116–119). Selection was achieved using LB supplemented with apramycin and chloramphenicol, and insertion of the respective fragments at the mTn7 site in the resulting G636 invL+ and G636-gfp strains was confirmed by PCR analyses.
Antibiotic susceptibility assays.
MIC analyses were performed using a two-fold broth dilution microtiter assay similar to previously described protocols (112, 120, 121). Briefly, overnight cultures were sub-cultured at 0.05 Abs600 and grown for 3 h shaking at 37 hpi. Mid-exponential growth phase cultures were then inoculated at 0.01 Abs600 into a 96-well microtiter plate (Corning Inc, Corning, NY) containing two-fold decreasing dilutions of the indicated antibiotics. Plates were then incubated at 37°C with shaking for 24 h. The MIC was defined as less than 10% of the Abs600 of an untreated control.
Statistical methods.
All statistical analyses were performed using GraphPad Prism version 9, and P values of <0.05 were considered statistically significant. When normally distributed, data sets were analyzed with Unpaired Student’s t-tests (comparing two samples), one-way analysis of variance (ANOVA) with Tukey’s test for multiple comparisons (comparing more than two samples), or two-way ANOVA with Tukey’s test for multiple comparisons (comparing more than two samples with two independent variables). For non-normally distributed data sets, the Mann-Whitney U test (comparing two samples) or the Kruskal Wallis H test with Dunn’s test for multiple comparisons (comparing more than two samples) was used.
Supplementary Material
Acknowledgements
This work was supported by funding to M.F.F. (R01AI166359 - CHECK), C.J.L. (T32AI007172), and C.B.L (R01AI137062) through the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. JM was supported through The American Association of Immunologists Careers in Immunology Fellowship Program and The Pediatric Cardiovascular and Pulmonary Research Training Program (5T32HL125241-07). The modern respiratory isolates used in this study, G636 (strain 3689) and G654 (strain 6919), were collected by the CDC-funded Georgia Emerging Infections Program’s (EIP) Multi-site Gram-Negative Surveillance Initiative (MuGSI) and kindly provided by Sarah Satola. We also acknowledge Jennifer Philips, Jacco Boon, and Gayan Bamunuarachchi for thoughtful discussion about the manuscript. We thank Wandy Beatty and the Washington University School of Medicine Molecular Microbiology Imaging Facility for microscopy assistance, Alma Johnson of the Washington University Center for Reproductive Health Sciences Histocore for lung tissue slide mounting and staining assistance, and De Chen of the Washington University Center for Cellular Imaging for assistance with the Zeiss AxioScan Z1. Finally, we thank Dakota Hall for technical assistance with experiments.
References
- 1.Vijayakumar S, Biswas I, Veeraraghavan B. 2019. Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci OA 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerqueira GM, Peleg AY. 2011. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life 63:1055–1060. [DOI] [PubMed] [Google Scholar]
- 3.Sarshar M, Behzadi P, Scribano D, Palamara AT, Ambrosi C. 2021. Acinetobacter baumannii: An Ancient Commensal with Weapons of a Pathogen. Pathogens 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Venanzio G, Flores-Mireles AL, Calix JJ, Haurat MF, Scott NE, Palmer LD, Potter RF, Hibbing ME, Friedman L, Wang B, Dantas G, Skaar EP, Hultgren SJ, Feldman MF. 2019. Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, Dramowski A, Dunachie SJ, Eckmanns T, Eibach D, Emami A, Feasey N, Fisher-Pearson N, Forrest K, Garrett D, Gastmeier P, Giref AZ, Greer RC, Gupta V, Haller S, Haselbeck A, Hay SI, Holm M, Hopkins S, Iregbu KC, Jacobs J, Jarovsky D, Javanmardi F, Khorana M, Kissoon N, Kobeissi E, Kostyanev T, Krapp F, Krumkamp R, Kumar A, Kyu HH, Lim C, Limmathurotsakul D, Loftus MJ, Lunn M, Ma J, Mturi N, Munera-Huertas T, Musicha P, Mussi-Pinhata MM, Nakamura T, Nanavati R, Nangia S, Newton P, Ngoun C, Novotney A, Nwakanma D, Obiero CW, Olivas-Martinez A, Olliaro P, Ooko E, Ortiz-Brizuela E, Peleg AY, Perrone C, Plakkal N, Ponce-de-Leon A, Raad M, Ramdin T, Riddell A, Roberts T, Robotham JV, Roca A, Rudd KE, Russell N, Schnall J, Scott JAG, Shivamallappa M, Sifuentes-Osornio J, Steenkeste N, Stewardson AJ, Stoeva T, Tasak N, Thaiprakong A, Thwaites G, Turner C, Turner P, van Doorn HR, Velaphi S, Vongpradith A, Vu H, Walsh T, Waner S, Wangrangsimakul T, Wozniak T, Zheng P, Sartorius B, Lopez AD, Stergachis A, Moore C, Dolecek C, Naghavi M. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dexter C, Murray GL, Paulsen IT, Peleg AY. 2015. Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther 13:567–573. [DOI] [PubMed] [Google Scholar]
- 7.Cisneros JM, Rodriguez-Baño J. 2002. Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features and treatment. Clin Microbiol Infect 8:687–693. [DOI] [PubMed] [Google Scholar]
- 8.Ibrahim S, Al-Saryi N, Al-Kadmy IMS, Aziz SN. 2021. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol Biol Rep 48:6987–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakonstantis S, Kritsotakis EI. 2021. Systematic review and meta-analysis of the proportion and associated mortality of polymicrobial (vs monomicrobial) pulmonary and bloodstream infections by Acinetobacter baumannii complex. Infection 49:1149–1161. [DOI] [PubMed] [Google Scholar]
- 10.Xiao D, Wang L, Zhang D, Xiang D, Liu Q, Xing X. 2017. Prognosis of patients with Acinetobacter baumannii infection in the intensive care unit: A retrospective analysis. Exp Ther Med 13:1630–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengstock DM, Thyagarajan R, Apalara J, Mira A, Chopra T, Kaye KS. 2010. Multidrug-resistant Acinetobacter baumannii: an emerging pathogen among older adults in community hospitals and nursing homes. Clin Infect Dis 50:1611–1616. [DOI] [PubMed] [Google Scholar]
- 12.Inchai J, Pothirat C, Bumroongkit C, Limsukon A, Khositsakulchai W, Liwsrisakun C. 2015. Prognostic factors associated with mortality of drug-resistant Acinetobacter baumannii ventilator-associated pneumonia. J Intensive Care 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giammanco A, Calà C, Fasciana T, Dowzicky MJ. 2017. Global Assessment of the Activity of Tigecycline against Multidrug-Resistant Gram-Negative Pathogens between 2004 and 2014 as Part of the Tigecycline Evaluation and Surveillance Trial. mSphere 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tacconelli E, Carrara E, Savoldi A, Kattula D, Burkert F. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. [DOI] [PubMed] [Google Scholar]
- 15.Kollef MH, Torres A, Shorr AF, Martin-Loeches I, Micek ST. 2021. Nosocomial Infection. Crit Care Med 49:169–187. [DOI] [PubMed] [Google Scholar]
- 16.Jones RN. 2010. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 51 Suppl 1. [DOI] [PubMed] [Google Scholar]
- 17.Sader HS, Castanheira M, Mendes RE, Flamm RK. 2018. Frequency and antimicrobial susceptibility of Gram-negative bacteria isolated from patients with pneumonia hospitalized in ICUs of US medical centres (2015–17). J Antimicrob Chemother 73:3053–3059. [DOI] [PubMed] [Google Scholar]
- 18.Weber BS, Harding CM, Feldman MF. 2015. Pathogenic Acinetobacter: from the Cell Surface to Infinity and Beyond. J Bacteriol 198:880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, Jania C, Doerschuk CM, Tilley SL, Duncan JA. 2012. Staphylococcus aureus α-Hemolysin Mediates Virulence in a Murine Model of Severe Pneumonia Through Activation of the NLRP3 Inflammasome. J Infect Dis 205:807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann N, Rasmussen TB, Jensen PØ, Stub C, Hentzer M, Molin S, Ciofu O, Givskov M, Johansen HK, Høiby N. 2005. Novel Mouse Model of Chronic Pseudomonas aeruginosa Lung Infection Mimicking Cystic Fibrosis. Infect Immun 73:2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmer LD, Green ER, Sheldon JR, Skaar EP. 2019. Assessing Acinetobacter baumannii Virulence and Persistence in a Murine Model of Lung Infection. Methods Mol Biol 1946:289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pogue JM, Zhou Y, Kanakamedala H, Cai B. 2022. Burden of illness in carbapenem-resistant Acinetobacter baumannii infections in US hospitals between 2014 and 2019. BMC Infect Dis 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alotaibi T, Abuhaimed A, Alshahrani M, Albdelhady A, Almubarak Y, Almasari O. 2021. Prevalence of multidrug-resistant Acinetobacter baumannii in a critical care setting: A tertiary teaching hospital experience. SAGE Open Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koomanachai P, Kim A, Nicolau DP. 2009. Pharmacodynamic evaluation of tigecycline against Acinetobacter baumannii in a murine pneumonia model. Journal of Antimicrobial Chemotherapy 63:982–987. [DOI] [PubMed] [Google Scholar]
- 25.Koomanachai P, Kim A, Nicolau DP. 2009. Pharmacodynamic evaluation of tigecycline against Acinetobacter baumannii in a murine pneumonia model. Journal of Antimicrobial Chemotherapy 63:982–987. [DOI] [PubMed] [Google Scholar]
- 26.Crandon JL, Kim A, Nicolau DP. 2009. Comparison of tigecycline penetration into the epithelial lining fluid of infected and uninfected murine lungs. Journal of Antimicrobial Chemotherapy 64:837–839. [DOI] [PubMed] [Google Scholar]
- 27.Braunstein A, Papo N, Shai Y. 2004. In Vitro Activity and Potency of an Intravenously Injected Antimicrobial Peptide and Its dl Amino Acid Analog in Mice Infected with Bacteria. Antimicrob Agents Chemother 48:3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joly-Guillou ML, Wolff M, Pocidalo JJ, Walker F, Carbon C. 1997. Use of a new mouse model of Acinetobacter baumannii pneumonia to evaluate the postantibiotic effect of imipenem. Antimicrob Agents Chemother 41:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song JY, Cheong HJ, Lee J, Sung AK, Kim WJ. 2009. Efficacy of monotherapy and combined antibiotic therapy for carbapenem-resistant Acinetobacter baumannii pneumonia in an immunosuppressed mouse model. Int J Antimicrob Agents 33:33–39. [DOI] [PubMed] [Google Scholar]
- 30.Manepalli S, Gandhi JA, Ekhar V V., Asplund MB, Coelho C, Martinez LR. 2013. Characterization of a cyclophosphamide-induced murine model of immunosuppression to study Acinetobacter baumannii pathogenesis. J Med Microbiol 62:1747–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Distel JS, Di Venanzio G, Mackel JJ, Rosen DA, Feldman MF. 2023. Replicative Acinetobacter baumannii strains interfere with phagosomal maturation by modulating the vacuolar pH. PLoS Pathog 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sycz G, Di Venanzio G, Distel JS, Sartorio MG, Le NH, Scott NE, Beatty WL, Feldman MF. 2021. Modern Acinetobacter baumannii clinical isolates replicate inside spacious vacuoles and egress from macrophages. PLoS Pathog 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valcek A, Philippe C, Whiteway C, Robino E, Nesporova K, Bové M, Coenye T, De Pooter T, De Coster W, Strazisar M, Van der Henst C. 2023. Phenotypic Characterization and Heterogeneity among Modern Clinical Isolates of Acinetobacter baumannii. Microbiol Spectr 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubio T, Gagné S, Debruyne C, Dias C, Cluzel C, Mongellaz D, Rousselle P, Göttig S, Seifert H, Higgins PG, Salcedo SP. 2022. Incidence of an Intracellular Multiplication Niche among Acinetobacter baumannii Clinical Isolates. mSystems 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato Y, Unno Y, Miyazaki C, Ubagai T, Ono Y. 2019. Multidrug-resistant Acinetobacter baumannii resists reactive oxygen species and survives in macrophages. Sci Rep 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cochet F, Peri F. 2017. The Role of Carbohydrates in the Lipopolysaccharide (LPS)/Toll-Like Receptor 4 (TLR4) Signalling. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akira S, Yamamoto M. 2010. Lipid A receptor TLR4-mediated signaling pathways. Adv Exp Med Biol 667:59–68. [DOI] [PubMed] [Google Scholar]
- 38.Homma JY, Matsuura M, Kanegasaki S, Kawakubo Y, Kojima Y, Shibukawa N, Kumazawa Y, Yamamoto A, Tanamoto K ichi, Yasuda T, Imoto M, Yoshimura H, Kusumoto S, Shiba T. 1985. Structural requirements of lipid A responsible for the functions: a study with chemically synthesized lipid A and its analogues. J Biochem 98:395–406. [DOI] [PubMed] [Google Scholar]
- 39.Kawasaki T, Kawai T. 2014. Toll-like receptor signaling pathways. Front Immunol 5:112681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, Montgomery JI, Reilly U, Barbacci EG, Hujer K, Bonomo RA, Fernandez L, Hancock REW, Adams MD, French SW, Buslon VS, Spellberg B. 2012. Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. mBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knapp S, Wieland CW, Florquin S, Pantophlet R, Dijkshoorn L, Tshimbalanga N, Akira S, Van Der Poll T. 2006. Differential roles of CD14 and toll-like receptors 4 and 2 in murine Acinetobacter pneumonia. Am J Respir Crit Care Med 173:122–129. [DOI] [PubMed] [Google Scholar]
- 42.Hazen JE, Di Venanzio G, Hultgren SJ, Feldman MF. 2023. Catheterization triggers resurgent infection seeded by host Acinetobacter baumannii reservoirs. Sci Transl Med 15:eabn8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dijkshoorn L, Aucken HM, Gerner-Smidt P, Kaufmann ME, Ursing J, Pitt TL. 1993. Correlation of typing methods for Acinetobacter isolates from hospital outbreaks. J Clin Microbiol 31:702–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo G, Lin L, Ibrahim AS, Baquir B, Pantapalangkoor P, Bonomo RA, Doi Y, Adams MD, Russo TA, Spellberg B. 2012. Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behairy MY, Abdelrahman AA, Toraih EA, Ibrahim EEDA, Azab MM, Sayed AA, Hashem HR. 2022. Investigation of TLR2 and TLR4 Polymorphisms and Sepsis Susceptibility: Computational and Experimental Approaches. Int J Mol Sci 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang WH, Nie LH, Zhang LJ, Jing LP, Dong F, Wang M, Zhang N, Liu Y, Zhang BH, Chen C, Lin HS, Wei XC, Yang G, Jing CX. 2015. Association of TLR2 and TLR4 non-missense single nucleotide polymorphisms with type 2 diabetes risk in a Southern Chinese population: A case-control study. Genetics and Molecular Research 14:8694–8705. [DOI] [PubMed] [Google Scholar]
- 47.Chatzi M, Papanikolaou J, Makris D, Papathanasiou I, Tsezou A, Karvouniaris M, Zakynthinos E. 2018. Toll-like receptor 2, 4 and 9 polymorphisms and their association with ICU-acquired infections in Central Greece. J Crit Care 47:1–8. [DOI] [PubMed] [Google Scholar]
- 48.Wang S-H, Teng C-K, Chan M-C, Yang K-Y, Sheu C-C, Liang S-J, Huang W-H, Feng J-Y, Chen C-M, Weng Z-X, Peng C-K. 2024. The impact and risk factors for developing pneumogenic bacteremia in carbapenem-resistant Acinetobacter baumannii nosocomial pneumonia in the intensive care unit: A multicenter retrospective study. Int J Infect Dis 146:107128. [DOI] [PubMed] [Google Scholar]
- 49.Van Faassen H, KuoLee R, Harris G, Zhao X, Conlan JW, Chen W. 2007. Neutrophils play an important role in host resistance to respiratory infection with Acinetobacter baumannii in mice. Infect Immun 75:5597–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Xu W. 2022. Neutrophil and Macrophage Response in Acinetobacter Baumannii Infection and Their Relationship to Lung Injury. Front Cell Infect Microbiol 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grguric-Smith LM, Lee HH, Gandhi JA, Brennan MB, DeLeon-Rodriguez CM, Coelho C, Han G, Martinez LR. 2015. Neutropenia exacerbates infection by Acinetobacter baumannii clinical isolates in a murine wound model. Front Microbiol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruhn KW, Pantapalangkoor P, Nielsen T, Tan B, Junus J, Hujer KM, Wright MS, Bonomo RA, Adams MD, Chen W, Spellberg B. 2015. Host fate is rapidly determined by innate effector-microbial interactions during Acinetobacter baumannii bacteremia. J Infect Dis 211:1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchiya T, Nakao N, Yamamoto S, Hirai Y, Miyamoto K, Tsujibo H. 2012. NK1.1(+) cells regulate neutrophil migration in mice with Acinetobacter baumannii pneumonia. Microbiol Immunol 56:107–116. [DOI] [PubMed] [Google Scholar]
- 54.Qiu H, KuoLee R, Harris G, Chen W. 2009. High susceptibility to respiratory Acinetobacter baumannii infection in A/J mice is associated with a delay in early pulmonary recruitment of neutrophils. Microbes Infect 11:946–955. [DOI] [PubMed] [Google Scholar]
- 55.Breslow JM, Meissler J, Hartzell RR, Spence PB, Truant A, Gaughan J, Eisenstein TK. 2011. Innate immune responses to systemic acinetobacter baumannii infection in mice: Neutrophils, but not interleukin-17, mediate host resistance. Infect Immun 79:3317–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castro ÍA, Yang Y, Gnazzo V, Kim D-H, Van Dyken SJ, López CB. 2023. Murine Parainfluenza Virus Persists in Lung Innate Immune Cells Sustaining Chronic Lung Pathology. bioRxiv 10.1101/2023.11.07.566103. [DOI] [PubMed]
- 57.Brossard KA, Campagnari AA. 2012. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun 80:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pérez A, Merino M, Rumbo-Feal S, Álvarez-Fraga L, Vallejo JA, Beceiro A, Ohneck EJ, Mateos J, Fernández-Puente P, Actis LA, Poza M, Bou G. 2017. The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence 8:959–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Astaneh SDA, Rasooli I, Gargari SLM. 2017. Filamentous hemagglutinin adhesin FhaB limits A.baumannii biofilm formation. Front Biosci (Elite Ed) 9:266–275. [DOI] [PubMed] [Google Scholar]
- 60.Darvish Alipour Astaneh S, Rasooli I, Mousavi Gargari SL. 2014. The role of filamentous hemagglutinin adhesin in adherence and biofilm formation in Acinetobacter baumannii ATCC19606(T). Microb Pathog 74:42–49. [DOI] [PubMed] [Google Scholar]
- 61.Bentancor L V., Camacho-Peiro A, C Bozkurt-Guzel, Pier GB, Maira-Litrán T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weidensdorfer M, Ishikawa M, Hori K, Linke D, Djahanschiri B, Iruegas R, Ebersberger I, Riedel-Christ S, Enders G, Leukert L, Kraiczy P, Rothweiler F, Cinatl J, Berger J, Hipp K, Kempf VAJ, Göttig S. 2019. The Acinetobacter trimeric autotransporter adhesin Ata controls key virulence traits of Acinetobacter baumannii. Virulence 10:68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatefi Oskuei R, Darvish Alipour Astaneh S, Rasooli I. 2021. A conserved region of Acinetobacter trimeric autotransporter adhesion, Ata, provokes suppression of Acinetobacter baumannii virulence. Arch Microbiol 203:3483–3493. [DOI] [PubMed] [Google Scholar]
- 64.Tram G, Poole J, Adams FG, Jennings MP, Eijkelkamp BA, Atack JM. 2021. The Acinetobacter baumannii Autotransporter Adhesin Ata Recognizes Host Glycans as High-Affinity Receptors. ACS Infect Dis 7:2352–2361. [DOI] [PubMed] [Google Scholar]
- 65.Ishikawa M, Nakatani H, Hori K. 2012. AtaA, a new member of the trimeric autotransporter adhesins from Acinetobacter sp. Tol 5 mediating high adhesiveness to various abiotic surfaces. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jackson-Litteken CD, Di Venanzio G, Le NH, Scott NE, Djahanschiri B, Distel JS, Pardue EJ, Ebersberger I, Feldman MF. 2022. InvL, an Invasin-Like Adhesin, Is a Type II Secretion System Substrate Required for Acinetobacter baumannii Uropathogenesis. mBio 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yesil C, Yalcin AN, Ogunc D, Ongut G, Ozhak B, Colak D, Er H, Sarltas ZE. 2022. Use of colistin with rifampicin, trimethoprim-sulfamethoxazole and teicoplanin in Acinetobacter mouse infection model. Future Microbiol 17:665–671. [DOI] [PubMed] [Google Scholar]
- 68.İzci F, Ture Z, Dinc G, Yay AH, Eren EE, Bolat D, Gönen ZB, Ünüvar GK, Yıldız O, Aygen B. 2023. The efficacy of mesenchymal stem cell treatment and colistin-fosfomycin combination on colistin-resistant Acinetobacter baumannii sepsis model. European Journal of Clinical Microbiology and Infectious Diseases 42:1365–1372. [DOI] [PubMed] [Google Scholar]
- 69.Dinc G, Demiraslan H, Elmali F, Ahmed SS, Metan G, Alp E, Doganay M. 2014. Efficacy of Sulbactam and Its Combination with Imipenem, Colistin and Tigecycline in an Experimental Model of Carbapenem-Resistant Acinetobacter baumannii Sepsis. Chemotherapy 59:325–329. [DOI] [PubMed] [Google Scholar]
- 70.Nicasio AM, Crandon JL, Nicolau DP. 2009. In Vivo Pharmacodynamic Profile of Tigecycline against Phenotypically Diverse Escherichia coli and Klebsiella pneumoniae Isolates. Antimicrob Agents Chemother 53:2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Joly-Guillou ML, Wolff M, Farinotti R, Bryskier A, Carbon C. 2000. In vivo activity of levofloxacin alone or in combination with imipenem or amikacin in a mouse model of Acinetobacter baumannii pneumonia. Journal of Antimicrobial Chemotherapy 46:827–830. [DOI] [PubMed] [Google Scholar]
- 72.Queenan AM, Davies TA, He W, Lynch AS. 2013. Assessment of the combination of doripenem plus a fluoroquinolone against non-susceptible Acinetobacter baumannii isolates from nosocomial pneumonia patients. Journal of Chemotherapy 25:141–147. [DOI] [PubMed] [Google Scholar]
- 73.Al-Madboly LA. 2022. A Novel Triple Combination To Combat Serious Infections with Carbapenem-Resistant Acinetobacter baumannii in a Mouse Pneumonia Model. Microbiol Spectr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang AD, Smith KP, Berg AH, Truelson KA, Eliopoulos GM, McCoy C, Kirby JE. 2018. Efficacy of apramycin against multidrug-resistant Acinetobacter baumannii in the murine neutropenic thigh model. Antimicrob Agents Chemother 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Becker K, Aranzana-Climent V, Cao S, Nilsson A, Shariatgorji R, Haldimann K, Platzack B, Hughes D, Andrén PE, Böttger EC, Friberg LE, Hobbie SN. 2021. Efficacy of EBL-1003 (apramycin) against Acinetobacter baumannii lung infections in mice. Clinical Microbiology and Infection 27:1315–1321. [DOI] [PubMed] [Google Scholar]
- 76.Zhao C, Chirkova A, Rosenborg S, Palma Villar R, Lindberg J, Hobbie SN, Friberg LE. 2022. Population pharmacokinetics of apramycin from first-in-human plasma and urine data to support prediction of efficacious dose. Journal of Antimicrobial Chemotherapy 77:2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ambrosi C, Scribano D, Sarshar M, Zagaglia C, Singer BB, Palamara AT. 2020. Acinetobacter baumannii Targets Human Carcinoembryonic Antigen-Related Cell Adhesion Molecules (CEACAMs) for Invasion of Pneumocytes. mSystems 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.An Z, Huang X, Zheng C, Ding W. 2019. Acinetobacter baumannii outer membrane protein A induces HeLa cell autophagy via MAPK/JNK signaling pathway. Int J Med Microbiol 309:97–107. [DOI] [PubMed] [Google Scholar]
- 79.Wang Y, Zhang K, Shi X, Wang C, Wang F, Fan J, Shen F, Xu J, Bao W, Liu M, Yu L. 2016. Critical role of bacterial isochorismatase in the autophagic process induced by Acinetobacter baumannii in mammalian cells. FASEB J 30:3563–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bist P, Dikshit N, Koh TH, Mortellaro A, Tan TT, Sukumaran B. 2014. The Nod1, Nod2, and Rip2 axis contributes to host immune defense against intracellular Acinetobacter baumannii infection. Infect Immun 82:1112–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parra-Millán R, Guerrero-Gómez D, Ayerbe-Algaba R, Pachón-Ibáñez ME, Miranda-Vizuete A, Pachón J, Smani Y. 2018. Intracellular Trafficking and Persistence of Acinetobacter baumannii Requires Transcription Factor EB. mSphere 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maure A, Robino E, Van der Henst C. 2023. The intracellular life of Acinetobacter baumannii. Trends Microbiol 31:1238–1250. [DOI] [PubMed] [Google Scholar]
- 83.Asensio NC, Rendón JM, Burgas MT. 2021. Time-Resolved Transcriptional Profiling of Epithelial Cells Infected by Intracellular Acinetobacter baumannii. Microorganisms 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rumbo C, Tomás M, Moreira EF, Soares NC, Carvajal M, Santillana E, Beceiro A, Romero A, Bou G. 2014. The Acinetobacter baumannii Omp33–36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect Immun 82:4666–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.An Z, Ding W. 2021. Acinetobacter baumannii up-regulates LncRNA-GAS5 and promotes the degradation of STX17 by blocking the activation of YY1. Virulence 12:1965–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kho ZY, Azad MAK, Han ML, Zhu Y, Huang C, Schittenhelm RB, Naderer T, Velkov T, Selkrig J, Zhou Q, Li J. 2022. Correlative proteomics identify the key roles of stress tolerance strategies in Acinetobacter baumannii in response to polymyxin and human macrophages. PLoS Pathog 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smani Y, Docobo-Peŕez F, Loṕez-Rojas R, Domińguez-Herrera J, Ibáñez-Martínez J, Pachón J. 2012. Platelet-activating factor receptor initiates contact of Acinetobacter baumannii expressing phosphorylcholine with host cells. J Biol Chem 287:26901–26910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 78:1952–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao D, Li Y, Peng C, Lin J, Yu F, Zhao Y, Zhang X, Zhao D. 2021. Outer membrane protein a in Acinetobacter baumannii induces autophagy through mTOR signalling pathways in the lung of SD rats. Biomedicine & Pharmacotherapy 135:111034. [DOI] [PubMed] [Google Scholar]
- 90.Stahl J, Bergmann H, Göttig S, Ebersberger I, Averhoff B. 2015. Acinetobacter baumannii Virulence Is Mediated by the Concerted Action of Three Phospholipases D. PLoS One 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tomlin H, Piccinini AM. 2018. A complex interplay between the extracellular matrix and the innate immune response to microbial pathogens. Immunology 155:186–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wight TN, Frevert CW, Debley JS, Reeves SR, Parks WC, Ziegler SF. 2017. Interplay of Extracellular Matrix and Leukocytes in Lung Inflammation. Cell Immunol 312:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Onishi Y, Kawamura T, Higashino T, Mimura R, Tsukamoto H, Sasaki S. 2021. Clinical features of acute fibrinous and organizing pneumonia: An early histologic pattern of various acute inflammatory lung diseases. PLoS One 16:e0249300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chapman HA. 2012. Epithelial Responses to Lung Injury. 10.1513/pats201112-053AW 9:89–95. [DOI] [PMC free article] [PubMed]
- 95.Åhrman E, Hallgren O, Malmström L, Hedström U, Malmström A, Bjermer L, Zhou XH, Westergren-Thorsson G, Malmström J. 2018. Quantitative proteomic characterization of the lung extracellular matrix in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. J Proteomics 189:23–33. [DOI] [PubMed] [Google Scholar]
- 96.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, Moore BB, Martinez FJ, Niklason LE, White ES. 2012. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med 186:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Annoni R, Lancas T, Tanigawa RY, Matsushita MDM, Fernezlian SDM, Bruno A, Da Silva LFF, Roughley PJ, Battaglia S, Dolhnikoff M, Hiemstra PS, Sterk PJ, Rabe KF, Mauad T. 2012. Extracellular matrix composition in COPD. Eur Respir J 40:1362–1373. [DOI] [PubMed] [Google Scholar]
- 98.White ES. 2015. Lung extracellular matrix and fibroblast function. Ann Am Thorac Soc 12:S30–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang MY, Tanino Y, Vidova V, Kinsella MG, Chan CK, Johnson PY, Wight TN, Frevert CW. 2014. A rapid increase in macrophage-derived versican and hyaluronan in infectious lung disease. Matrix Biol 34:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Snyder JM, Washington IM, Birkland T, Chang MY, Frevert CW. 2015. Correlation of Versican Expression, Accumulation, and Degradation during Embryonic Development by Quantitative Immunohistochemistry. J Histochem Cytochem 63:952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Niu H, Gu J, Zhang Y. 2024. Bacterial persisters: molecular mechanisms and therapeutic development. Signal Transduction and Targeted Therapy 2024 9:1 9:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial Persister Cell Formation and Dormancy. Appl Environ Microbiol 79:7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kunnath AP, Suodha Suoodh M, Chellappan DK, Chellian J, Palaniveloo K. 2024. Bacterial Persister Cells and Development of Antibiotic Resistance in Chronic Infections: An Update. Br J Biomed Sci 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li G, Shen W, Gong Y, Li M, Rao X, Liu Q, Yu Y, Zhou J, Zhu K, Yuan M, Shang W, Yang Y, Lu S, Wang J, Zhao Y. 2022. Essential Fitness Repertoire of Staphylococcus aureus during Co-infection with Acinetobacter baumannii In Vivo. mSystems 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Timme S, Wendler S, Klassert TE, Saraiva JP, da Rocha UN, Wittchen M, Schramm S, Ehricht R, Monecke S, Edel B, Rödel J, Löffler B, Ramirez MS, Slevogt H, Figge MT, Tuchscherr L. 2024. Competitive inhibition and mutualistic growth in co-infections: deciphering Staphylococcus aureus-Acinetobacter baumannii interaction dynamics. ISME communications 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Semenec L, Cain AK, Dawson CJ, Liu Q, Dinh H, Lott H, Penesyan A, Maharjan R, Short FL, Hassan KA, Paulsen IT. 2023. Cross-protection and cross-feeding between Klebsiella pneumoniae and Acinetobacter baumannii promotes their co-existence. Nat Commun 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krasauskas R, Skerniškytė J, Armalytė J, Sužiedėlienė E. 2019. The role of Acinetobacter baumannii response regulator BfmR in pellicle formation and competitiveness via contact-dependent inhibition system. BMC Microbiol 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Repizo GD, Gagné S, Foucault-Grunenwald ML, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, Salcedo SP. 2015. Differential Role of the T6SS in Acinetobacter baumannii Virulence. PLoS One 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Storey D, McNally A, Åstrand M, Santos JPG, Rodriguez-Escudero I, Elmore B, Palacios L, Marshall H, Hobley L, Molina M, Cid VJ, Salminen TA, Bengoechea JA. 2020. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog 16:e1007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsai CJY, Loh JMS, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McGuffey JC, Jackson-Litteken CD, Di Venanzio G, Zimmer AA, Lewis JM, Distel JS, Kim KQ, Zaher HS, Alfonzo J, Scott NE, Feldman MF. 2023. The tRNA methyltransferase TrmB is critical for Acinetobacter baumannii stress responses and pulmonary infection. mBio 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. [DOI] [PubMed] [Google Scholar]
- 115.Pontes MH, Groisman EA. 2019. Slow growth dictates non-heritable antibiotic resistance in Salmonella enterica. Sci Signal 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ducas-Mowchun K, De Silva PM, Crisostomo L, Fernando DM, Chao TC, Pelka P, Schweizer HP, Kumar A. 2019. Next Generation of Tn 7-Based Single-Copy Insertion Elements for Use in Multi- and Pan-Drug-Resistant Strains of Acinetobacter baumannii. Appl Environ Microbiol 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carruthers MD, Nicholson PA, Tracy EN, Munson RS. 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kumar A, Dalton C, Cortez-Cordova J, Schweizer HP. 2010. Mini-Tn7 vectors as genetic tools for single copy gene cloning in Acinetobacter baumannii. J Microbiol Methods 82:296–300. [DOI] [PubMed] [Google Scholar]
- 120.Leus I V., Adamiak J, Trinh AN, Smith RD, Smith L, Richardson S, Ernst RK, Zgurskaya HI. 2020. Inactivation of AdeABC and AdeIJK efflux pumps elicits specific nonoverlapping transcriptional and phenotypic responses in Acinetobacter baumannii. Mol Microbiol 114:1049–1065. [DOI] [PubMed] [Google Scholar]
- 121.Leus I V., Weeks JW, Bonifay V, Smith L, Richardson S, Zgurskaya HI. 2018. Substrate Specificities and Efflux Efficiencies of RND Efflux Pumps of Acinetobacter baumannii. J Bacteriol 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.M100 Performance Standards for Antimicrobial Susceptibility Testing A CLSI supplement for global application.
- 123.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RAR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. [DOI] [PubMed] [Google Scholar]
- 125.HUGH R, REESE R. 1968. A comparison of 120 strains of Bacterium anitratum Schaub and Hauber with the type strain of this species. Int J Syst Evol Microbiol 18:207–229. [Google Scholar]
- 126.DUTHIE ES, LORENZ LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol 6:95–107. [DOI] [PubMed] [Google Scholar]
- 127.Rosen DA, Hooton TM, Stamm WE, Humphrey PA, Hultgren SJ. 2007. Detection of Intracellular Bacterial Communities in Human Urinary Tract Infection. PLoS Med 4:1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.