Abstract
There is increasing recognition that the sex chromosomes, X and Y, play an important role in health and disease that goes beyond the determination of biological sex. Loss of the Y chromosome (LOY) in blood, which occurs naturally in aging men, has been found to be a driver of cardiac fibrosis and heart failure mortality. LOY also occurs in most solid tumors in males and is often associated with worse survival, suggesting that LOY may give tumor cells a growth or survival advantage. We analyzed LOY in lung adenocarcinoma (LUAD) using both bulk and single-cell expression data and found evidence suggesting that LOY affects the tumor immune environment by altering cancer/testis antigen expression and consequently facilitating tumor immune evasion. Analyzing immunotherapy data, we show that LOY and changes in expression of particular cancer/testis antigens are associated with response to pembrolizumab treatment and outcome, providing a new and powerful biomarker for predicting immunotherapy response in LUAD tumors in males.
Keywords: loss of Y, lung adenocarcinoma, immune evasion, cancer
1. Introduction
Beyond its role in male sex determination, the role of the Y chromosome in health and disease has been greatly understudied. Mounting evidence suggests that the Y chromosome is a driver of men’s health (Maan et al, 2017) and that loss of the Y chromosome (LOY) profoundly affects both health and disease (Wang and Sano, 2024). For example, LOY in blood has been shown to lead to cardiac fibrosis and increased heart failure mortality in mouse models (Sano et al, 2022), to be associated with higher risk and shorter survival in human cancers (Wright et al, 2017; Thompson et al, 2019; Forsberg et al, 2014), and with shorter lifespans in human males (Loftfield et al, 2018, 2019). Others have reported that LOY in the blood occurs naturally in aging men (Forsberg et al, 2014) and that such LOY increases with smoking (Dumanski et al, 2015).
Although the functional consequences of LOY in blood have been reasonably well-studied, LOY in solid tissue is less well-understood. Several early studies reported LOY in various cancer types, including non-small-cell lung cancer (Center et al, 1993), bladder cancer (Sauter et al, 1995), and papillary renal cell carcinoma (Brunelli et al, 2003) but the mechanisms driving LOY and its consequences have remained elusive.
Recently, investigations of large, publicly available pan-tissue resources such as TCGA and GTEx have found that LOY is evident in most solid tumors but not corresponding healthy tissue and that LOY is often associated with reduced cancer survival (Qi et al, 2023; Müller et al, 2023). LOY has also been suggested as a potential driver event for cancer (Cáceres et al, 2020; Qi et al, 2023; Müller et al, 2023), but the connection has not been fully explored.
Cáceres et al (2020) reported an extreme down-regulation of Y-chromosome genes that they hypothesize as a functional mediator between LOY and cancer but did not establish a direct connection between down-regulation and loss of the Y-chromosome. Both Qi et al (2023) and Müller et al (2023) investigated LOY in solid tumors, including in lung adenocarcinoma (LUAD), using data from TCGA. Qi et al (2023) provided the first systematic catalog of LOY in cancer, showing that LOY is a frequent somatic event in primary tumors. Considering both gene-expression-based “functional” LOY as well as full and partial LOY estimated through CNVs, they further showed that TP53 mutations are enriched in particular LOY-affected cancers and that LOY is a driver event for uveal melanoma. Müller et al (2023) demonstrated that a binary LOY indicator derived from CNVs is correlated with tumor mutational burden across cancers and provided further evidence that TP53 mutations are associated with LOY in some cancers, including LUAD. A more recent study showed that LOY in mouse models of bladder cancer was associated with changes in the tumor immune microenvironment (TIME) and increased tumor growth (Abdel-Hafiz et al, 2023), possibly explaining the poor prognosis they found associated with LOY in human patients with bladder cancer.
Lung cancer is the leading cause of cancer death accounting for an estimated 1.8 million cancer deaths in 2020. Adenocarcinoma (LUAD) is the most common type of lung cancer, accounting for nearly 40% of all lung cancers. Although LOY has not been extensively studied in LUAD it has been estimated that approximately 32% lo LUAD tumors exhibit LOY (Müller et al, 2023); a result that is based on an assumed detection threshold. In this manuscript, we present an analysis of LOY in individuals with LUAD (Figure 1) using bulk and single-cell sequencing from The Cancer Genome Atlas (TCGA), the Genotype-Tissue Expression project (GTEx), and the Human Tumor Atlas Network (HTAN). We identified a characteristic pattern of LOY in TCGA LUAD tumor tissue that is absent in both TCGA adjacent normal tissue and in lung tissue from GTEx participants. We also provide evidence that LOY is selected for rather than being the result of a random chromosomal aberration, show that LOY does not occur in infiltrating immune cells, and that antigen presentation-related processes differ between cells with and without LOY.
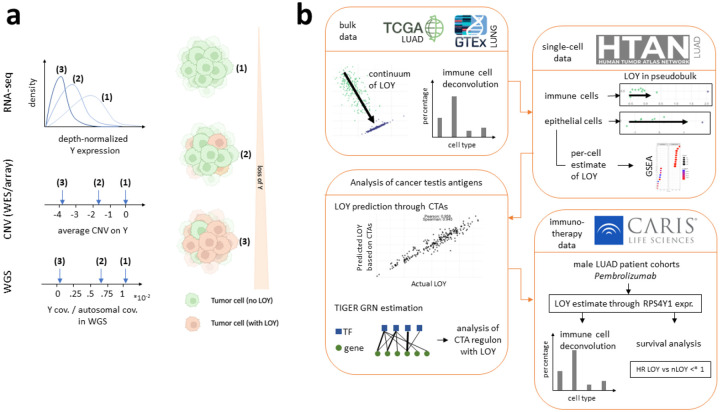
Fig. 1. Measures of LOY and study overview.
a Measures of LOY including RNA-seq, copy number variant (CNV) estimates, and ratios of Y coverage to autosome coverage from whole genome sequence depicted as they would manifest with three different ratios of LOY to non-LOY cells (primary tumor). b Overview of this study. Using lung adenocarcinoma as a model, we identify measures of LOY, demonstrate that this is driven by tumor and not immune cells, identify an association between LOY and cancer testis antigens and use gene regulatory network inference to analyze their expression, and then explore how immunotherapy response changes with LOY.
Cancer/testis antigens (CTAs) are proteins encoded by a group of autosomal genes that are primarily expressed in the testis, and occasionally in the placenta and ovary, all of which are immune-privileged sites. They are, however, aberrantly expressed in many tumor types where they present as antigens to the immune system, initiating changes in the Tumor Immune Microenvironment (TIME) (Kortleve et al, 2022) and altering immune response. In bladder cancer, a causal link between LOY and changes in the TIME has been established (Abdel-Hafiz et al, 2023), although the mechanism of action remains unclear. We find that autosomal CTAs are highly correlated with the degree of LOY and, using gene regulatory network modeling, demonstrate that differences in CTA regulation are linked to LOY. As such, this study provides the first description of a process in which tumor cells exhibiting LOY may be selected for through a mechanistic association between LOY, CTAs, and processes contributing to immune evasion (Figure 2).
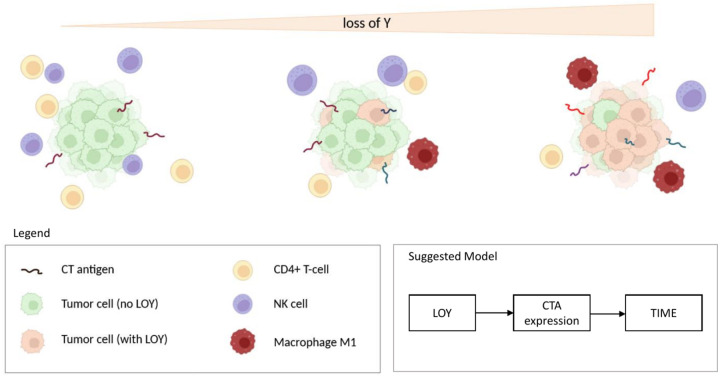
Fig. 2. Proposed model of loss of Y in LUAD.
With increasing LOY we observed a change in tumor immune environment (TIME) associated with alterations in the expression of various cancer testis antigens (CTAs). The amount of NK, and CD4+ T-cells decreases with LOY, while M1 macrophage proportions consistently increase, steady proportions of immune cells are not shown. Created in BioRender. Lopes Ramos, C. (2024) BioRender.com/v90i141. Bottom right: a graphical mechanistic model of LOY.
The alterations in immunogenicity associated with LOY led us to speculate whether increased LOY, coupled with changes in CTA expression and associated changes in the cancer’s immune environment, could affect the efficacy of immunotherapy. We explored the relationship between immunotherapy outcomes and differential expression of Y-genes and CTAs in male cohorts treated with pembrolizumab, a checkpoint inhibitor that targets the PD-1/PD-L1 pathway and one of the most commonly used immunooncology agents. We found that pembrolizumab treatment has a significantly better survival outcome in individuals with low RPS4Y1 expression (the best single-gene indicator for LOY we found in TCGA LUAD cases). Furthermore, several CTAs for which we found significant regulatory changes with LOY also show significant association with immunotherapy survival outcomes, providing further evidence of CTA-mediated effects of LOY on the TIME. Our results suggest that LOY measures might serve as a biomarker for the directed selection of immunotherapeutic treatments for individual patients and that CTA-targeting immunotherapy might complement those treatments to improve overall treatment outcomes.
2. Results
A continuum of loss of Y in lung adenocarcinoma
Although microdeletions of Y-chromosomal segments are known, we considered LOY in a single tumor cell as a binary quantity reflecting whether or not in genetically male individuals (those carrying both X and Y chromosomes), the Y chromosome is absent. In contrast, LOY in a collection of cells (such as a tumor biopsy) is a continuous measure reflecting the fraction of cells that have lost the Y chromosome. We defined the amount of LOY based on a linear mapping of principal components of Y gene expression in a given dataset (“pc-LOY”; Figure 3; Methods). pc-LOY reflects the continuous nature of LOY in tumors and is general enough to apply to a wide variety of data sources, as it is solely based on bulk gene expression.
Fig. 3. Loss of Y in LUAD.
a First two principal components of Y genes in TCGA primary tumor tissue of LUAD patients, providing evidence for LOY based on gene expression levels. b Visualization of the approach we used to estimate LOY in primary tumors, which uses the first three components of PCA applied to the expression of Y-chromosome genes, showing the first two principal components for patient i (only PC1 and PC2 are shown). c First two principal components of Y genes in TCGA adjacent normal tissue of LUAD patients. d First two principal components on Y genes of pseudobulked gene expression of annotated epithelial cells for all samples in HTAN LUAD data. e First two principal components on Y genes of pseudobulked gene expression of annotated immune cells for all samples in HTAN LUAD data. f Change of TF-regulated expression of CTAs along LOY as estimated by TIGER on TCGA LUAD. To account for individual-specific noise, we built seven equal-width bins across observed male pc-LOY, averaging networks across individuals within one bin. Visualized are CTAs that show a significant correlation with LOY after Benjamini-Hochberg correction.
In TCGA-LUAD, pc-LOY correlates well with Y-chromosome copy number variations (Pearson ρ = 0.74, p< 2.2 * 10−16; Supplementary Figure 1a) and with estimates of chromosomal loss based on whole genome sequencing (WGS) (Pearson ρ = 0.78, p= 1.491 * 10−12; Supplementary Figure 1b). We note that pc-LOY is a data-driven measurement that is specific to each dataset and is re-estimated for every new cohort. Examining pc-LOY in non-tumor tissue reveals no evidence of LOY in either TCGA LUAD adjacent “normal” lung tissue samples or GTEx lung samples (Figure 3c, Supplementary Figure 1c).
To rule out potential biases in the pc-LOY signal, we tested four potential drivers and confounders of LOY in TCGA LUAD tumors—age, smoking history (cigarettes per day and years of smoking), sample purity estimates (Aran et al, 2015), and sample batches (Supplementary Figure 1d–f, Supplementary Figure 2c, Supplementary Figure 2d)—and found no significant association between these variables and pc-LOY (Pearson correlation based t-test, p-value cutoff 0.05; Supplementary Tab. [TCGA_LUAD_covariates]).
We also tested whether the LOY could be a random aberration due to the small size of the Y chromosome or its relatively sparse gene landscape by comparing it to the loss of chromosome 21, which is comparably small and gene-poor (see Supplementary Figure 2b). We found no evidence of an association between chromosome 21 loss (defined as read coverage relative to genome-wide coverage) and LOY (Pearson correlation 0.05), providing further evidence that LOY is a systematic loss compared to autosomal genes.
LOY in tumor cells alters immune cell interactions
Although RNA-seq data has been instrumental in exploring the molecular mechanisms driving cancer, it does not capture the complex interplay between tumor and stromal cells. In males, LOY has been reported to be an age-associated phenomenon in white blood cells (Sano et al, 2022; Brown and Machiela, 2020; Cáceres et al, 2020; Qi et al, 2023; Müller et al, 2023), leading us to question whether the tumor-associated LOY we observed was driven by infiltrating immune cells. We analyzed single-cell RNA-seq data in LUAD tumors from the Human Tumor Atlas Network (HTAN) (Chan et al, 2021) separating the single-cell data into epithelial and immune cell groups based on the author-provided cell labeling. We constructed tumor and white blood cell pseudobulk datasets by pooling the respective scRNA-seq data and performed pc-LOY analysis on each separately. We found a clearly evident pc-LOY trajectory in epithelial cells, but not in immune cells within the tumor sample (Figure 3d,e), indicating that LOY in these cancers is a tumor cells phenomenon; these results were confirmed in independent LUAD single-cell data from Kim et al (2020) (Supplementary Figure 2e,f). The evidence that LOY is present only in tumor cells–not in healthy GTEx samples, in adjacent normal TCGA-LUAD tissue, or in immune cells within the tumor microenvironment–suggests that changes in the microenvironment may reflect selective pressure driving LOY in tumor cells.
Abdel-Hafiz and colleagues reported that in mouse models of bladder cancer, LOY induces a change of immune cell composition in the tumor microenvironment (Abdel-Hafiz et al, 2023). This led us to speculate that tumor immune invasion might select for LUAD cells that have lost their Y chromosome. Using TCGA LUAD data, we estimated the fractions of different immune cell types through computational deconvolution using quanTIseq (Finotello et al, 2019; Sturm et al, 2020). We examined the distribution of immune cell types with pc-LOY and found that immune cell makeup was predictive of LOY (least squares fit from multivariate regression of pc-LOY against all quanTIseq immune cell types, adj. R2 = .14, p = 3.143e − 06). Significant predictors of LOY (univariate p < .01) in this model include M1 macrophages increasing in quantity with LOY, and NK cells, Neutrophils, and CD4+ T-cells decreasing in quantity with LOY (see Supplementary Figure 4c). Both NK cells and CD4+ T-cells are of particular importance for tumor control and tumor immune response through mediation of cytotoxicity (Wu et al, 2020), with CD4+ T-cells explicitly recognizing antigens presented through HLA-II (Jhunjhunwala et al, 2021). In the HTAN LUAD single-cell data, we found a similar trend, with the percentage (as a fraction of the total immune cell population) of both NK cells and T-cells on decreasing with LOY. Annotation for macrophages was not available for the HTAN LUAD dataset.
Abdel-Hafiz and colleagues also reported that changes in bladder cancer TIME may be induced through cellular programs associated with LOY. We used HTAN LUAD single-cell data to compare gene expression in tumor cells that lost Y and those that did not and identified significant enrichment of several Gene Ontology (GO) biological process terms related to antigen presentation in tumor cells with LOY (Benjamini-Hochberg (FDR)-corrected enrichment p < .05, Supplementary Figure 3a). Analyzing gene expression in an independent LUAD single-cell dataset from Kim et al (2020), we also found enrichment of GO terms related to adaptive immune response (FDR-corrected enrichment p < .05, Supplementary Figure 3b). This supports the hypothesis that LOY may indeed induce changes in tumor cells that alter their interactions with immune cells.
Immune cells recognize other cells by identifying antigens on their surfaces. Cancer/testis antigens (CTAs) (Fratta et al, 2011) are a unique group of antigens generally expressed only in the human germ cells of the testis (and occasionally in the placenta and ovary, all “immunoprivileged” sites), not in other normal tissues, but they are also aberrantly expressed in various types of cancer (Maxfield et al, 2015; Jhunjhunwala et al, 2021). Several CTAs have been shown to have direct gene regulatory effects in cancer, such as PAGE4 through modulation of the MAPK pathway (Lv et al, 2019), CEP55 by regulating FOXO1 signaling (Zhang et al, 2022) and being linked with effects on cancer, with FBXO39 knockdowns slowing down tumor growth in squamous cell tumors (Yang et al, 2022). Another CTA, ADAM29, and its mutations significantly influence the proliferation, migration, and invasion of breast cancer cells (Zhao et al, 2016), and MAGEC3 promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma (Wu et al, 2021). CTAs have also been shown to have an explicit T-cell response in testicular cancer (Pearce et al, 2017) and have been identified as a potential key component in the mechanisms of cancer immune evasion (Kortleve et al, 2022).
We investigated whether changes in CTA expression might explain the TIME changes we observed; a simple linear fit shows a significant correlation between autosomal CTA expression and LOY (least squares fit adj. R2 = .43, p-value 0.0118). We then tested for a possible regulatory relationship between LOY and CTAs. We searched CTA promoter regions for binding sites of the Y chromosome-based transcription factors SRY and HSFY2; we also identified the Androgen Receptor (AR) binding sites as AR is regulated by the Y-encoded KDM5D. A complete list of CTA promoters containing binding sites for SRY, HSFY2, and AR can be found in Supplementary Tab. [YToTestis_TFBinding]) and indicate that, at the least, these CTAs may alter their expression in response to LOY.
For a more principled approach to exploring the regulatory changes induced by LOY, we computationally inferred gene regulatory networks using TIGER. TIGER uses Bayesian matrix factorization to simultaneously infer transcription factor (TF) regulomes and TF activities from RNA-seq data (Chen and Padi, 2024). We computed TIGER gene regulatory networks for male TCGA-LUAD subjects (see Methods 4) and selected the 1000 genes with the greatest changes in expression that were explained by LOY-associated TF regulatory disruption. We used gene set enrichment analysis and found significant (p < .05, FDR-corrected) enrichment for pathways related to cell cycle and repair, processes reflective of tumorigenesis. Considering all autosomal CTA genes in the TIGER model, we find 29 of 266 for which expression variation by TF regulation is significantly correlated with LOY (t-test on Pearson correlation coefficient; FDR-corrected p-values < 0.05; Figure 3f). Among these is XAGE1B, which has been reported to trigger a humoral immune response in non-small cell lung cancer patients (Nakagawa et al, 2005), linking LOY and CTA expression changes in the TIME.
We used the TIGER networks to analyze the relationship between LOY and the regulation of 37 known immune-related genes, including drivers of the immune response, pro-inflammatory cytokines, genes encoding components of the T-cell receptor, and known cytotoxic mediations (Davoli et al (2017), see Supplementary Tab. [immunegene_list]). In the TIGER models, one finding of note is that regulation of CCL19 is significantly negatively associated with LOY (Pearson correlation: −0.28; FDR-corrected p = 0.0038); CCL19 can chemoattract T- and B-cells (Kim et al, 1998) and has been shown to mediate potent immune responses against tumors in mouse models of lung cancer (Hillinger et al, 2006). This suggests a potential causative link between LOY and decreased immune infiltration in tumors. These regulatory network analyses further clarify the mechanism by which LOY may be selected for–that LOY may alter the regulation of CTAs and immune-response-related genes, altering the infiltration of white blood cells in the tumor and aiding tumor cell immune evasion.
Although a node-based analysis of gene regulatory network models can identify elements associated with phenotypic features, this approach does not fully exploit the patterns of interconnectivity represented in the network model. “Active” sub-network analysis integrates gene expression data with the network structure to pinpoint areas where there is a notable deviation in activity compared to a control or baseline condition. We used the approach described by Dittrich et al (2008) (see Methods 4) to identify active subnetworks in the TIGER LUAD GRN that are associated with LOY. We identified 337 nodes with positive scores resulting in a bipartite active subnetwork consisting of 210 nodes (10 transcription factors, 200 genes) and 233 TF-gene edges.
Key attributes of the ten TFs in the network are shown in Supplementary Table 2. The highest degree TF was NR3C2 (weighted absolute outdegree: 23.96), followed by KLF15 (16.85). Both NR3C2 and KLF15 have a strong negative association with LOY, indicating lower expression among individuals who have lost the Y chromosome. KLF15 has been shown to regulate pathways that intersect with immune responses, potentially contributing to immune evasion mechanisms by altering the tumor microenvironment and immune cell infiltration (Shu et al, 2024). NR3C2 is known primarily for its role in regulating electrolyte balance but also has been found to play a part in cancer biology, including immune evasion (Sun et al, 2024b). In lung adenocarcinoma, NR3C2 can influence immune response by affecting the expression of immune-regulatory genes. This modulation can help the tumor evade immune surveillance by altering the activation and infiltration of immune cells (Chen et al, 2023; Shu et al, 2024). In most cases, their targets also have a negative association with LOY, consistent with the network trend for NR3C2 and KLF15 as activators (Supplementary Figure 5a; Supplementary Figure 5b). The active subnetwork also contains the TF GATA3, which also decreased in expression with LOY (effect=−0.34, p=1.52e − 06, FDR=1.19e − 04). GATA3 plays a critical role in the development and function of T cells and other cell types, including playing a role in cell differentiation and proliferation Wan (2014). Aberrant expression of GATA3 can alter the regulation of cytokines and other signaling molecules that modulate the immune system’s ability to recognize and attack tumor cells (Anichini et al, 2020).
Only two TFs, ZC3H8 and ADNP, increased in expression with LOY. The association between ADNP and LOY is not statistically significant (effect = 0.03, p = 0.63) and the heuristic algorithm likely selected ADNP for inclusion in the active subnetwork because of the strong associations of several of its target genes with LOY. However, high levels of ADNP expression have been linked with poor prognosis and enhanced tumor progression, and it is thought to play some role in immune modulation (Wang et al, 2023). ZC3H8 exhibits a significant positive association with LOY (effect = 0.39, p=1.91 * 10−7, FDR=6.18 * 10−5) and serves as a repressor of six of the seven of its target genes in the active GRN subnetwork (Supplementary Figure 5c). ZC3H8 is a known repressor of GATA3 (Schmidt et al, 2018), consistent with the relationship we see between the expression of these genes. Thus, ZC3H8 likely influences immune regulation as we would expect; its increased expression has been linked to poor outcomes in LUAD (Sun et al, 2024a).
We also investigated the properties of the 200 targeted genes in the active subnetwork using an over-representation analysis. We used the Canonical Pathways (CP) subset of the MolSigDB gene set repository as our annotation source; this set includes the KEGG-Medicus, REACTOME, WikiPathways, and KEGG legacy gene sets. The background was set to all genes in the full TIGER network. Cytochrome p450 (CYP) pathways were consistently found to be significant when using all three annotation sources: the KEGG “drug metabolism cytochrome p450” pathway (FDR = 0.013), the WikiPathways “oxidation by cytochrome p450” pathway (FDR = 0.034), and the REACTOME “cytochrome p450 arranged by substrate type” pathway (FDR = 0.035). Other top pathways enriched among the targeted genes, consistent with the activation of CYP genes, included the REACTOME “phase 1 functionalization of compounds” (FDR = 8.6e-5) and “biological oxidations” (FDR = 0.010) and the KEGG “fatty acid metabolism” pathway (0.014). Full results of the over-representation analysis, including lists of overlap genes in each pathway, are available in Supplementary Table 1.
CYP enzymes play significant roles in the metabolism of both endogenous and exogenous compounds, influencing not only cancer progression and treatment outcomes but also the immune responses within the tumor microenvironment. CYP enzymes are known to play significant roles in xenobiotic and drug metabolism, and the regulation of CYP enzymes by pro-inflammatory cytokines has been implicated in cancer prognosis and therapeutic response (Stipp and Acco, 2021). In lung adenocarcinoma, sex differences in CYP expression have been associated with the increased risk of developing lung adenocarcinoma in females (Oyama et al, 2007).
LOY and immunotherapy
The LOY-driven changes we observed in tumor cell gene expression and TIME composition led us to speculate that LOY may affect response to treatment with cancer immunotherapy agents. We explored the association between LOY and response in a dataset of 832 individuals with LUAD (kindly provided by Caris Life Sciences) consisting of bulk gene expression data for cohorts treated with pembrolizumab (pembro, N=832), with response assessed through survival after treatment. Further available data for cohorts treated with nivolumab (N=125) and atezolizumab (N=55) were not considered for in-depth analysis due to low statistical power caused by an order of magnitude smaller sample size. We provide hazard ratio plots based on LOY and CTA expression across all treatments in Supplementary Figure 6. In compliance with HIPAA, no individual-level information was provided so the analyses presented here use summary statistics for data partitions such as high vs low LOY (see Methods).
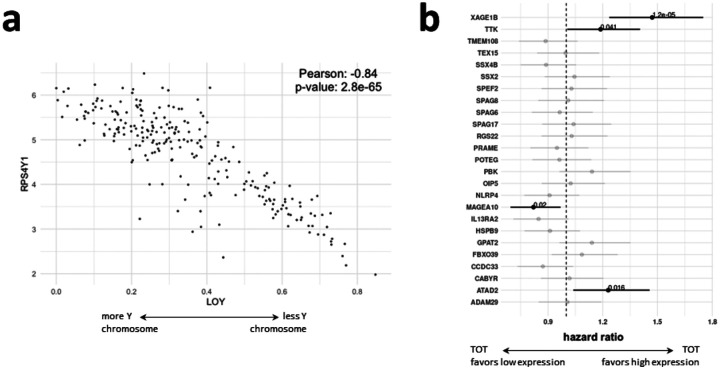
Because the Caris data did not have individual-level Y chromosome information, we could not calculate LOY directly and instead used the expression of RPS4Y1 as a proxy. We chose RPS4Y1 because it was the Y-chromosome gene we found to have the greatest weight in pc-LOY and had the highest single-gene correlation with LOY in bulk sequencing data (Pearson correlation 0.84 in male individuals in TCGA-LUAD; Figure 4a). Consequently, we used the median RPS4Y1 expression to define high and low LOY groups. We compared estimated TIME composition in those groups using quanTIseq; consistent with our analyses in other datasets, we found significant differences between high and low LOY for M1 and M2 macrophages, neutrophils, monocytes, B-cells, and CD8+ T-cells (FDR-adjusted p-value < 0.05; Supplementary Tab. [quantiseq caris]). In the Caris data, we found that M1 macrophages increase with LOY; previous work has shown M1 macrophage infiltration in tumors to be predictive of immunotherapy response in NSCLC (Wu et al, 2024).
Fig. 4. Immunotherapy response in context of LOY.
a Correlation of RPS4Y1 gene expression and pc-LOY (larger value means more LOY) in TCGA LUAD data. b Hazard ratio (y-axis) when comparing male LUAD patients treated with pembro split into two groups based on median expression value of CTA (x-axis, low expression group vs high expression group). HR values with confidence intervals and p-values in bold, all non-significant observed HRs are transparent for clarity of the results.
We compared treatment outcomes for individuals with and without LOY. For pembro-treated individuals, LOY was associated with better outcomes as measured by the hazard ratio between the two partitions (HR = 0.831, 95% CI = 0.722–0.956, p= 0.0095), which is consistent with Abdel-Hafiz et al (2023), which reported improved response to PD-1 antibodies in a mouse model of LOY.
Because we had seen that LOY was correlated with CTA expression and that the expression of 26 CTAs was correlated with survival in TCGA, we wanted to understand whether the expression of these CTAs was predictive of immunotherapy response. Data was available from Caris for 25 of the predictive CTAs, and for each, we partitioned each treatment group into low- and high-expression for that CTA according to cutpoints at the 75th percentile. The relationship between CTA expression and survival was generally treatment-specific, with four of the 25 measured CTAs showing a significant association with survival in the pembro treatment group (p < .05; Figure 4b). These provide evidence that LOY can affect immunotherapy outcomes, likely by modifying the TIME through CT antigen expression. Hence, an individual’s likely treatment response could be estimated using the amount of LOY or expression of specific CTAs, and treatment adjusted to increase the likelihood of a positive outcome. Our findings also suggest that immunotherapy treatments explicitly targeting CTAs such as XAGE1B (Saito et al, 2016) could further improve the treatment of lung cancer, particularly if those treatments are conditioned on LOY. This may be particularly important for sub-populations with poor responses to current treatments such as those showing high expression of MAGEA10, for whom a complementary treatment with CTA-targeting immunotherapy (Simister et al, 2022) could improve therapeutic response.
3. Discussion
Despite substantial evidence that there are clinical differences between biological males and females in disease risk, development, and response to therapy, the role of sex chromosomes and the molecular processes associated with them have only recently begun to be studied in earnest in the context of cancer (Lopes-Ramos et al, 2018, 2020; Arnold and Disteche, 2018). The role that the male Y chromosome plays in cancer risk, development, and response to therapy was also poorly understood despite growing evidence that LOY in solid tumors is associated with poor outcomes and that LOY may be a cancer driver (Brown and Machiela, 2020; Cáceres et al, 2020; Qi et al, 2023; Müller et al, 2023).
Our analyses provide support for the hypothesis that LOY is not just a passive driver but that it is selected for lung adenocarcinoma development in biological males. We first demonstrated that LOY is associated with alterations in patterns of expression of Y-chromosomal genes. While not surprising, this observation provides means of inferring LOY in datasets that do not include direct sequencing of the Y chromosome. Armed with this information, we tested whether the LOY we observed was due to chromosomal loss in tumor cells or infiltrating lymphocytes. We found that the LOY in tumors is confined to cancer cells, not the lymphocytes present in the tumor. This analysis was important because it is known that LOY occurs in the white blood cells of aging males, and so the LOY in the tumor could have been confounded by LOY in the infiltrating immune cells. Next, we examined the composition of white blood cells in tumors as a function of Y chromosome loss. We discovered that there were associated changes in immune cell composition including increased levels of M1 macrophages that are well correlated with LOY. We also tested for expression of cancer/testes antigens (CTAs) and found that CTA expression was correlated with both LOY and changes in the tumor immune microenvironment (TIME).
Collectively, this set of facts points to a simple model of selection for LOY in solid tumors in biological males. As cancer develops, immune cells recognize CTAs on tumor cells and attach to them. However, some tumor cells lose their Y chromosome, resulting in diminished expression of CTAs. This allows them to avoid immune attack, selecting for those cells that have lost the Y chromosome. As tumor cells that have experienced LOY begin to dominate the tumor cell population, the overall composition of the TIME begins to change, resulting in the process of tumor evolution that is selectively enriching for LOY. This model is, in fact, consistent with the report that LOY in bladder cancer is a driver of tumor growth and is accompanied by a change in the tumor immune environment that facilitates immune evasion (Abdel-Hafiz et al, 2023).
Our proposed mechanistic model not only sheds light on the role of the LOY in LUAD through its role in immune invasion but also suggests that LOY or CTA expression patterns may be relevant to understanding immunotherapy response. Indeed, CTA expression has been reported to have a strong association with survival in LOY-affected individuals, some have suggested using CTA expression as a biomarker for immunotherapy (Gjerstorff et al, 2015; Al-Khadairi and Decock, 2019; Meng et al, 2021; Seager et al, 2024), and CTAs themselves have been suggested as targets for immunotherapy (Saito et al, 2016; Simister et al, 2022). Although we were only able to draw strong conclusions for pembro, the results for nivo and atezo suggest further investigation of whether LOY can be used to determine optimal immunotherapy treatment.
A consistent observation across our studied datasets, including immunotherapy data, is that M1 macrophages increase with LOY. M1 macrophages have been reported to be associated with better immunotherapy treatment response in lung cancer (Wu et al, 2024), which may help explain why the changes in the TIME associated with LOY result in better treatment effects. There is also evidence that the presence of M1 macrophages improves treatment efficacy for multiple immunotherapeutic treatments Duan and Luo (2021); Huo et al (2023). As such, LOY could also serve as a biomarker for selecting therapies with responses linked to the presence of M1 macrophages.
What we have learned about the role that LOY plays in lung tumor development and in affecting changes in the immune environment provides a strong motivation for a more complete study of the role that sex chromosomes play in disease processes. The allosomes have long been neglected in the study of health and disease (Sun et al, 2023), and while LOY is gaining recognition as a marker of some cancers and male aging, its functional role is still relatively unknown. Our findings point to a simple but compelling model for why LOY is prevalent in male solid tumors: selective pressure. As tumor cells lose their Y chromosome, they alter the expression of CTAs and the composition of the TIME. These changes allow cells that have undergone LOY to better evade immune surveillance, giving them an advantage over tumor cells that retain the Y chromosome. This also strongly suggests that an exploration of the mosaic loss of X could further our understanding of sex differences in immune response (Klein and Flanagan, 2016) and may shed light on sex-biased processes that are active in the tumors of biological females.
As is true of all studies, this study is limited by the quantities and types of data that we have available or which we can practically generate. Lacking those data, we are confident in the model because of the large number of lines of evidence derived from multiple sources that support it. Cancer development and progression is fundamentally a process that is driven by selection in which tumor cells gain an advantage over normal cells and expand in numbers. As put quite eloquently by Martincorena et al (2017), “By definition, cancer genes are genes under positive selection in tumor cells.” It is simply logical to extend this concept to subpopulations of tumor cells defined by their genetic makeup—including whether or not they have a Y chromosome.
Similarly, the conclusions that we reached regarding the association between LOY and immunotherapy response are based on a relatively small number of individuals with LUAD receiving immunotherapy. Recall that for two of the drugs, nivo and atezo, there was insufficient data to draw significant conclusions. While the results for pembro were statistically significant, they suggest further independent validation in larger independent datasets. Nonetheless, the model here is also compelling—LOY clearly influences CTA expression, tumor progression, immune processes and the TIME, and M1 cell infiltration—providing a strong argument for the potential use of LOY as a marker for immunotherapy response in male LUAD tumors.
4. Methods
Bulk RNA-seq data retrieval and processing
We obtained publicly available bulk RNA-sequencing data from the Genotype-Tissue Expression (GTEx) project and The Cancer Genome Atlas (TCGA). In both cases, data were uniformly reprocessed using recount3 (Wilks et al, 2021), version 1.8.0. Raw counts were normalized by conversion to transcripts per million (TPM); a pseudocount was added, and data were log (base e) transformed as log(TPM + 1).
TCGA data were separated into adjacent normal and primary tumor tissue. Duplicates were removed, keeping only the sample with the greatest sequencing depth.
Estimating sample purity for TCGA tumor samples
Sample purity estimates for TCGA data were obtained from Aran et al. and are based on RNA-seq (Aran et al, 2015).
Whole-genome sequencing retrieval and processing
Whole-genome sequencing (WGS) alignment files of N=278 male LUAD patients in TCGA were accessed through dbGaP (IDs can be found in Supplementary Tab. [gdc_manifest_LUAD_WGS_bam_maleonly]). Coverage of Y and autosomal genes was computed using samtools (Li et al, 2009) version 1.18. To facilitate the comparison of WGS-based LOY estimates with gene expression-based LOY estimates, we considered only high-coverage WGS samples that had at least 50% of the maximum observed coverage across samples (N=57).
Calculating WGS-based loss of Y in GTEx and TCGA
We computed LOY from WGS based on the number of reads covering Y divided by the number of reads covering autosomes, which gives a unit-less relative measure of the loss of chromosome Y. Assuming that reads are distributed uniformly at random over all chromosomes, potential individual biases in read depth were normalized to provide a standardized measure of LOY.
Calculating copy number variation-based LOY in TCGA
To obtain copy number variation (CNV)-based LOY estimates we use the TCGAbiolinks package (Colaprico et al, 2016), version 2.25.3, to retrieve copy number segment information for CNVs. We then computed the mean CNV value across segments on the Y chromosome.
Single-cell expression data retrieval and processing
Single-cell RNA-seq data for 8 male and 16 female LUAD patients were obtained from the Human Tumor Atlas Network (HTAN) (Chan et al, 2021) through CELLx-GENE (Abdulla et al, 2023) on 2/24/23, which had previously been processed using Seurat (Stuart et al, 2019). Data were split into epithelial and immune cells according to the authors’ cell-type annotation and filtered for lung tumor tissue. To conduct patient-level analysis, we pseudobulked the single-cell data by adding up read counts across cells of the same patient, for each cell normalizing the count by the total number of overall reads in this cell (read depth). For each patient, we then further adjusted this value by the number of cells for this patient. To validate our findings, we downloaded single-cell LUAD expression data from Kim et al (2020) through GEO (GSE131907) and used the same procedure of filtering for lung tissue, partitioning cells into epithelial and immune groups, and pseudobulking.
Estimating loss of Y
To create a measure for LOY based on bulk gene expression in TCGA and GTEx, we began by computing the principal components (PCs) of the Y-chromosome RNA-seq data within each dataset. The PCs are computed on data containing both male and female samples; female samples serve as a reference point for gene expression in individuals without a Y chromosome. We excluded Y genes in pseudoautosomal regions (PAR1 and PAR2) (Ross et al, 2005), as they frequently contain mismapped reads.
In TCGA LUAD, the first three PCs account for 93.5% of the variance in the data (see Supplementary Figure 4b). Projecting the samples onto these PCs produces a clear separation of the data into two clusters corresponding to male and female samples (Supplementary Figure 4a).
To obtain a one-dimensional measure of LOY based on these PCs, we take the vector connecting the female centroid to the male centroid . We map each male individual’s PC coordinates to a univariate measure by orthogonal projection onto this vector(Figure 3b).
We then transform these PC-based values to a [0, 1] measure of LOY (“pc-LOY”) by scaling to the maximum observed LOY value.
Association of pc-LOY with covariates
We analyzed the association of pc-LOY with covariates available as annotated meta-data from recount3 (Wilks et al, 2021), version 1.8.0. Specifically, we considered age at diagnosis, batch effect as given by the annotated CGC case batch number, smoking exposure as indicated by the number of years smoked, and smoking exposure as indicated by cigarettes per day. We further considered sample purity as estimated by the ESTIMATE method (Aran et al, 2016).
Predicting LOY with cancer/testis antigen expression
Annotated cancer/testis antigens (CTAs) were retrieved from CTdatabase (Almeida et al, 2009) on 10/19/2022. Using the N=266 CTAs that are not located on the Y chromosome, we fitted a multivariate least-squares model predicting LOY for male samples based on the expression levels of these CTAs.
Investigating regulation in the promoter region of CTAs
Motif matches for binding sites of the two Y chromosome transcription factors SRY and HSFY2 and the androgen receptor in the promoter regions of CTAs were extracted from human motif prior in the GRAND database v1.5.0 (Ben Guebila et al, 2021), which are derived from motif scans using FIMO (Grant et al, 2011) against gene promoter regions defined as ±1kb from human reference genome hg38. They define a motif for each TF using position weight matrices from the CIS-BP 1.94d database (Weirauch et al, 2014). The motif call was done for an adjusted p-value cutoff of 10−4.
Adjudicating LOY in single-cell data
To adjudicate LOY on a single-cell level from scRNA-seq, we define cells with less than 0.04% of total reads coming from the Y chromosome to be affected by LOY. All other cells were considered as non-LOY. This threshold was determined based on a dip in the coverage distribution (see Supplementary Figure 4d).
Gene set enrichment analysis in single-cell LOY data
We conducted Gene Set Enrichment Analysis based on log-fold-change between mean observed gene expression in LOY vs non-LOY cells. We use the clusterProfiler v4.10.0 (Yu et al, 2012; Korotkevich et al, 2021) R package with minimum gene set size set to five and report pathways as enriched based on a p-value cutoff of 0.05 for Benjamini-Hochberg corrected p-values.
Predicting LOY through deconvoluted cell quantities
We predicted the pc-LOY as the response, taking all cell type quantities estimated through quanTIseq v3.16 (Finotello et al, 2019) as features in a multivariate least squares regression.
Estimating sample-specific gene regulatory networks
To estimate sample-specific gene regulatory networks for the TCGA LUAD data, we used TIGER (Chen and Padi, 2024) from NetZooR 1.5.4 (Ben Guebila et al, 2023), a Bayesian matrix factorization method for inferring regulatory networks that leverages prior knowledge of transcription factor (TF) binding to decompose a gene expression matrix into the product of an overall gene regulatory network and sample-specific TF activities. More specifically, TIGER decomposes a gene expression matrix of m genes and n samples into G = WZ, where is a gene regulatory matrix for the t TFs, specifying the regulatory effect of each TF on each gene, and specifies the “availability” of each TF in each sample.
TIGER incorporates prior knowledge about the binding of TFs to gene promoters. Here, we consider signed TF-gene interactions with high confidence levels according to the DoRothEA database v1.12.0 (Garcia-Alonso et al, 2019), excluding TFs with support from only one computational resource of DoRothEA level E, resulting in N=735 TFs in the prior. We considered the 16856 protein-coding genes in the input expression matrix that were expressed in more than 25% of the samples. Using these TFs and genes, including X and Y TFs and genes, we applied TIGER to the set of N=193 males in the TCGA LUAD data to estimate the regulatory network W and the individual TF activities Z.
To study individual-specific regulatory effects of TF i on gene j in a given sample k, we compute
As an aggregate measure of the regulation explained by all TFs on gene j in a given sample k, we compute
For assessment of the relationship between LOY and changes in gene regulation, we calculated the correlation between pc-LOY and for each gene j across all samples k. Based on the 1000 genes exhibiting the regulatory changes most strongly correlated (in absolute value) with pc-LOY, we carried out a gene set enrichment analysis in KEGG pathways. The set of all 16856 genes used in the gene regulatory network was used as a background set. We report significantly over-represented pathways based on a p-value cutoff of 0.05 for FDR-corrected p-values.
We also analyzed individual CTAs and immune-related genes using gene-level regulatory scores Rg. Here, we computed the Pearson correlation coefficient along with its t-test statistics between each , for CTA and immune-related genes j summing over all samples k, and the pc-LOY. FDR multiple test correction was applied and corrected p-values < .05 were reported as significant.
Active subnetwork detection
Active subnetwork detection uses graph search methods to identify subnetworks that are enriched for nodes associated with a particular outcome (Nguyen et al, 2019). To identify active subnetworks enriched for association with LOY in the TIGER LUAD GRN, we used a heuristic variation of the integer linear programming algorithm developed by Dittrich et al. (Dittrich et al, 2008) as implemented in the runFastHeinz function of the BioNet R package v 1.49.1 (Beisser et al, 2010). This algorithm searches subgraphs in a node-weighted graph by maximizing a subgraph score based on the sum of the node weights; in this scheme, positive node weights are desirable. Following the approach of Dittrich et al. (Dittrich et al, 2008), we used p-values previously calculated for the association of each node in the TIGER network with the LOY trajectory. We fit a beta-uniform mixture (BUM) model to the distribution using the fitBumModel function and converted p-values into a score using the scoreNodes function, which assigns higher scores to lower p-values and vice-versa. By design, the BUM model controls the FDR at a user-specified rate such that scores are positive for nodes below the FDR threshold and negative for nodes above it (Dittrich et al, 2008).
Immunotherapy data
Real-world clinical data were obtained from insurance claims, which encompass detailed records of health services, including prescribed medications, procedures performed, and established diagnoses. Time-on-treatment (TOT) was determined as the interval from the initiation to the conclusion of treatment. Kaplan-Meier survival estimates were generated for cohorts defined by molecular characteristics. Hazard ratios (HR) were computed utilizing the Cox proportional hazards model, and significant differences in survival times were assessed with the log-rank test, where P < 0.05 was considered significant.
For RNA expression measurement, FFPE specimens underwent pathology review to measure percent tumor content and tumor size; a minimum of 10% of tumor content in the area for microdissection was required to enable enrichment and extraction of tumor-specific RNA. Qiagen RNA FFPE tissue extraction kit was used for extraction, and the RNA quality and quantity were determined using the Agilent TapeStation. Biotinylated RNA baits were hybridized to the synthesized and purified cDNA targets, and the bait-target complexes were amplified in a post-capture PCR reaction. The Illumina NovaSeq 6500 was used to sequence the whole transcriptome from patients to an average of 60M reads. Raw data was demultiplexed by Illumina Dragen BioIT accelerator, trimmed, counted, PCR-duplicates removed, and reads were aligned to human reference genome hg19 using STAR aligner (Dobin et al, 2013). Transcripts per million (TPM) values were generated using the Salmon expression pipeline (Patro et al, 2017). RNA deconvolution was performed using quanTIseq method to characterize the tumor microenvironment (Finotello et al, 2019).
4.1. Reproducibility of Code
We provide code and documentation for generation of the presented analysis at https://github.com/QuackenbushLab/LOY_lung_cancer.
Supplementary Material
Acknowledgments
The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. This work was supported by grants from the National Institutes of Health: JF, KHS, CC, VF, ES, PM, MBG, CMLR and JQ were supported by R35CA220523; MBG and JQ were also supported by U24CA231846; JQ received additional support from P50CA127003; JQ and DLD were supported by R01HG011393; KHS and DLD were supported by P01HL114501; KHS was supported by T32HL007427; DLD was supported by K24HL171900; CMLR was supported by K01HL166376; CMLR and ES were also supported by the American Lung Association grant LCD-821824. Immunotherapy data were provided in aggregate by Caris Life Sciences; JQ serves on the Caris Scientific Advisory Board but was not compensated in any way for the analysis presented here.
References
- Abdel-Hafiz HA, Schafer JM, Chen X, et al. (2023) Y chromosome loss in cancer drives growth by evasion of adaptive immunity. Nature 619(7970):624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulla S, Aevermann B, Assis P, et al. (2023) Cz cell×gene discover: A single-cell data platform for scalable exploration, analysis and modeling of aggregated data. bioRxiv 10.1101/2023.10.30.563174, URL https://www.biorxiv.org/content/early/2023/11/02/2023.10.30.563174, https://www.biorxiv.org/content/early/2023/11/02/2023.10.30.563174.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khadairi G, Decock J (2019) Cancer Testis Antigens and Immunotherapy: Where Do We Stand in the Targeting of PRAME? Cancers (Basel) 11(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida LG, Sakabe NJ, deOliveira AR, et al. (2009) CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res 37(Database issue):D816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anichini A, Perotti VE, Sgambelluri F, et al. (2020) Immune escape mechanisms in non small cell lung cancer. Cancers 12(12). 10.3390/cancers12123605, URL https://www.mdpi.com/2072-6694/12/12/3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sirota M, Butte AJ (2015) Systematic pan-cancer analysis of tumour purity. Nat Commun 6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Sirota M, Butte AJ (2016) Corrigendum: Systematic pan-cancer analysis of tumour purity. Nat Commun 7:10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Disteche CM (2018) Sexual inequality in the cancer cell. Cancer Res 78(19):5504–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisser D, Klau GW, Dandekar T, et al. (2010) Bionet: an r-package for the functional analysis of biological networks. Bioinformatics 26(8):1129–1130 [DOI] [PubMed] [Google Scholar]
- Ben Guebila M, Wang T, Lopes-Ramos CM, et al. (2023) The network zoo: a multi-lingual package for the inference and analysis of gene regulatory networks. Genome Biology 24(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Guebila M, Lopes-Ramos CM, Weighill D, et al. (2021) GRAND: a database of gene regulatory network models across human conditions. Nucleic Acids Research 50(D1):D610–D621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Machiela MJ (2020) Why Y? Downregulation of Chromosome Y Genes Potentially Contributes to Elevated Cancer Risk. J Natl Cancer Inst 112(9):871–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli M, Eble JN, Zhang S, et al. (2003) Gains of chromosomes 7, 17, 12, 16, and 20 and loss of Y occur early in the evolution of papillary renal cell neoplasia: a fluorescent in situ hybridization study. Mod Pathol 16(10):1053–1059 [DOI] [PubMed] [Google Scholar]
- Cáceres A, Jene A, Esko T, et al. (2020) Extreme Downregulation of Chromosome Y and Cancer Risk in Men. J Natl Cancer Inst 112(9):913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center R, Lukeis R, Vrazas V, et al. (1993) Y chromosome loss and rearrangement in non-small-cell lung cancer. Int J Cancer 55(3):390–393 [DOI] [PubMed] [Google Scholar]
- Chan JM, Quintanal-Villalonga A, Gao VR, et al. (2021) Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 39(11):1479–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Padi M (2024) Flexible modeling of regulatory networks improves transcription factor activity estimation. NPJ Syst Biol Appl 10(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Yin Z, Chen Y, et al. (2023) Validation of prognostic signature and exploring the immune-related mechanisms for nr3c2 in clear cell renal cell carcinoma. Translational Cancer Research 12(10):2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Silva TC, Olsen C, et al. (2016) TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44(8):e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, et al. (2017) Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355(6322):eaaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich MT, Klau GW, Rosenwald A, et al. (2008) Identifying functional modules in protein–protein interaction networks: an integrated exact approach. Bioinformatics 24(13):i223–i231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, et al. (2013) Star: ultrafast universal rna-seq aligner. Bioinformatics 29(1):15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Luo Y (2021) Targeting macrophages in cancer immunotherapy. Signal transduction and targeted therapy 6(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanski JP, Rasi C, Lönn M, et al. (2015) Smoking is associated with mosaic loss of chromosome y. Science 347(6217):81–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotello F, Mayer C, Plattner C, et al. (2019) Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 11(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LA, Rasi C, Malmqvist N, et al. (2014) Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet 46(6):624–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta E, Coral S, Covre A, et al. (2011) The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol Oncol 5(2):164–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alonso L, Holland CH, Ibrahim MM, et al. (2019) Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 29(8):1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerstorff MF, Andersen MH, Ditzel HJ (2015) Oncogenic cancer/testis antigens: prime candidates for immunotherapy. Oncotarget 6(18):15772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7):1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillinger S, Yang S, Batra R, et al. (2006) Ccl19 reduces tumour burden in a model of advanced lung cancer. British journal of cancer 94(7):1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Zhang H, Sa L, et al. (2023) M1 polarization enhances the antitumor activity of chimeric antigen receptor macrophages in solid tumors. Journal of Translational Medicine 21(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, Hammer C, Delamarre L (2021) Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 21(5):298–312 [DOI] [PubMed] [Google Scholar]
- Kim CH, Pelus LM, White JR, et al. (1998) Ckβ−11/macrophage inflammatory protein-3β/ebi1-ligand chemokine is an efficacious chemoattractant for t and b cells. The Journal of Immunology 160(5):2418–2424 [PubMed] [Google Scholar]
- Kim N, Kim HK, Lee K, et al. (2020) Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 11(1):2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Flanagan KL (2016) Sex differences in immune responses. Nat Rev Immunol 16(10):626–638 [DOI] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, et al. (2021) Fast gene set enrichment analysis. bioRxiv [Google Scholar]
- Kortleve D, Coelho RML, Hammerl D, et al. (2022) Cancer germline antigens and tumor-agnostic CD8+T cell evasion. Trends Immunol 43(5):391–403 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield E, Zhou W, Graubard BI, et al. (2018) Predictors of mosaic chromosome y loss and associations with mortality in the uk biobank. Scientific Reports 8(1):12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield E, Zhou W, Yeager M, et al. (2019) Mosaic Y Loss Is Moderately Associated with Solid Tumor Risk. Cancer Res 79(3):461–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Ramos CM, Kuijjer ML, Ogino S, et al. (2018) Gene regulatory network analysis identifies sex-linked differences in colon cancer drug metabolism. Cancer Res 78(19):5538–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Ramos CM, Quackenbush J, DeMeo DL (2020) Genome-wide sex and gender differences in cancer. Front Oncol 10:597788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv C, Fu S, Dong Q, et al. (2019) PAGE4 promotes prostate cancer cells survive under oxidative stress through modulating MAPK/JNK/ERK pathway. J Exp Clin Cancer Res 38(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan AA, Eales J, Akbarov A, et al. (2017) The y chromosome: a blueprint for men’s health? European Journal of Human Genetics 25(11):1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, et al. (2017) Universal patterns of selection in cancer and somatic tissues. Cell 171(5):1029–1041.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield KE, Taus PJ, Corcoran K, et al. (2015) Comprehensive functional characterization of cancer-testis antigens defines obligate participation in multiple hallmarks of cancer. Nat Commun 6:8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Sun X, Liu Z, et al. (2021) A novel era of cancer/testis antigen in cancer immunotherapy. Int Immunopharmacol 98:107889. [DOI] [PubMed] [Google Scholar]
- Müller P, Velazquez Camacho O, Yazbeck AM, et al. (2023) Why loss of y? a pan-cancer genome analysis of tumors with loss of Y chromosome. Comput Struct Biotechnol J 21:1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Velazquez Camacho O, Yazbeck AM, et al. (2023) Why loss of Y? A pancancer genome analysis of tumors with loss of Y chromosome. Comput Struct Biotechnol J 21:1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Noguchi Y, Uenaka A, et al. (2005) XAGE-1 expression in non-small cell lung cancer and antibody response in patients. Clin Cancer Res 11(15):5496–5503 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Shrestha S, Tran D, et al. (2019) A comprehensive survey of tools and software for active subnetwork identification. Frontiers in genetics 10:426749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Sugio K, Uramoto H, et al. (2007) Cytochrome p450 expression (cyp) in non-small cell lung cancer. Front Biosci 12(2299–2308):10–2741 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, et al. (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nature methods 14(4):417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce H, Hutton P, Chaudhri S, et al. (2017) T-cell responses directed against cancer testis antigens are present in the peripheral blood of testicular cancer patients. Eur J Immunol 47(7):1232–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Pang J, Mitsiades I, et al. (2023) Loss of chromosome y in primary tumors. Cell 186(14):3125–3136.e11 [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, et al. (2005) The DNA sequence of the human X chromosome. Nature 434(7031):325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nakayama E, Valmori D (2016) Immune responses to the cancer testis antigen xage-1b in non small cell lung cancer caucasian patients. PloS one 11(3):e0150623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano S, Horitani K, Ogawa H, et al. (2022) Hematopoietic loss of y chromosome leads to cardiac fibrosis and heart failure mortality. Science 377(6603):292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter G, Moch H, Wagner U, et al. (1995) Y chromosome loss detected by FISH in bladder cancer. Cancer Genet Cytogenet 82(2):163–169 [DOI] [PubMed] [Google Scholar]
- Schmidt JA, Danielson KG, Duffner ER, et al. (2018) Regulation of the oncogenic phenotype by the nuclear body protein zc3h8. BMC cancer 18:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seager RJ, Senosain MF, Van Roey E, et al. (2024) Cancer testis antigen burden (CTAB): a novel biomarker of tumor-associated antigens in lung cancer. J Transl Med 22(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L, Tang J, Liu S, et al. (2024) Plasma cell signatures predict prognosis and treatment efficacy for lung adenocarcinoma. Cellular Oncology 47(2):555–571. 10.1007/s13402-023-00883-w, URL 10.1007/s13402-023-00883-w [DOI] [PubMed] [Google Scholar]
- Simister PC, Border EC, Vieira JF, et al. (2022) Structural insights into engineering a t-cell receptor targeting mage-a10 with higher affinity and specificity for cancer immunotherapy. Journal for ImmunoTherapy of Cancer 10(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp MC, Acco A (2021) Involvement of cytochrome p450 enzymes in inflammation and cancer: a review. Cancer chemotherapy and pharmacology 87(3):295–309 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, et al. (2019) Comprehensive integration of single-cell data. Cell 177:1888–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G, Finotello F, List M (2020) Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol Biol 2120:223–232 [DOI] [PubMed] [Google Scholar]
- Sun L, Wang Z, Lu T, et al. (2023) eXclusionarY: 10 years later, where are the sex chromosomes in GWASs? Am J Hum Genet 110(6):903–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Ma Q, Chen Y, et al. (2024a) Identification and analysis of prognostic immune cell homeostasis characteristics in lung adenocarcinoma. The Clinical Respiratory Journal 18(5):e13755. 10.1111/crj.13755, URL https://onlinelibrary.wiley.com/doi/abs/10.1111/crj.13755, https://onlinelibrary.wiley.com/doi/pdf/10.1111/crj.13755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YY, Gao HC, Guo P, et al. (2024b) Identification of NR3C2 as a functional diagnostic and prognostic biomarker and potential therapeutic target in non-small cell lung cancer. Cancer Innov 3(4):e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DJ, Genovese G, Halvardson J, et al. (2019) Genetic predisposition to mosaic y chromosome loss in blood. Nature 575(7784):652–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YY (2014) Gata3: a master of many trades in immune regulation. Trends in Immunology 35(6):233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Peng H, Zhang G, et al. (2023) Adnp is associated with immune infiltration and radiosensitivity in hepatocellular carcinoma for predicting the prognosis. BMC Medical Genomics 16(1):178. 10.1186/s12920-023-01592-x, URL 10.1186/s12920-023-01592-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sano S (2024) Why y matters? the implication of loss of y chromosome in blood and cancer. Cancer Science 115(3):706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158(6):1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks C, Zheng SC, Chen FY, et al. (2021) recount3: summaries and queries for large-scale RNA-seq expression and splicing. Genome Biol 22(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DJ, Day FR, Kerrison ND, et al. (2017) Genetic variants associated with mosaic Y chromosome loss highlight cell cycle genes and overlap with cancer susceptibility. Nat Genet 49(5):674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang W, Wang Y, et al. (2021) MAGE-C3 promotes cancer metastasis by inducing epithelial-mesenchymal transition and immunosuppression in esophageal squamous cell carcinoma. Cancer Commun (Lond) 41(12):1354–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Sheng Q, Liu P, et al. (2024) M1 macrophage-related gene model for NSCLC immunotherapy response prediction. Acta Biochim Biophys Sin (Shanghai) 56(3):379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Fu T, Jiang YZ, et al. (2020) Natural killer cells in cancer biology and therapy. Mol Cancer 19(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhao Y, Sun G, et al. (2022) FBXO39 predicts poor prognosis and correlates with tumor progression in cervical squamous cell carcinoma. Pathol Res Pract 238:154090. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, et al. (2012) clusterprofiler: an r package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 16(5):284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu Q, Li E, et al. (2022) CEP55 predicts the poor prognosis and promotes tumorigenesis in endometrial cancer by regulating the Foxo1 signaling. Mol Cell Biochem [DOI] [PubMed] [Google Scholar]
- Zhao M, Jia W, Jiang WG, et al. (2016) ADAM29 Expression in Human Breast Cancer and its Effects on Breast Cancer Cells In Vitro. Anticancer Res 36(3):1251–1258 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.