Abstract
Herpesviridae infect nearly all humans for life, causing diseases that range from painful to life-threatening1. These viruses penetrate cells by employing a complex apparatus composed of separate receptor-binding, signal-transmitting, and membrane-fusing components2. But how these components coordinate their functions is unknown. Here, we determined the 4.19-angstrom cryoEM reconstruction of the central signal-transmitting component from herpes simplex virus 2, the gH/gL complex, in its elusive pre-activation state. Analysis of the continuum of conformational ensembles observed in cryoEM data revealed a series of structural rearrangements in gH/gL that allosterically transmit the fusion-triggering signal from the receptor-binding glycoprotein gD to the membrane fusogen gB. Furthermore, we identified a structural “switch” element in gH/gL that refolds and flips 180 degrees during the transition from pre-activation to activated form. Conservation of this “switch” in gH/gL homologs suggests that the proposed fusion triggering mechanism may apply to all Herpesviridae and points to a new target for subunit-based vaccines and treatment efforts.
Keywords: herpesvirus, herpes simplex virus, HSV-2, gB, gH/gL, viral entry, membrane fusion, glycoprotein, viral fusogen, cryoEM, antibody, complex, activation, conformation, ensembles, regulation, conformational rearrangement, refolding
INTRODUCTION
Herpesviridae, commonly known as Herpesviruses, are ancient DNA viruses that infect mammals, birds, and reptiles, and establish lifelong infections in >90% of people worldwide3. While often asymptomatic, herpesvirus infections can cause diseases ranging from painful mucocutaneous lesions to loss of vision, encephalitis, cancer, developmental abnormalities, and disseminated disease3,4. Current therapeutic and prophylactic options are very limited5,6.
Herpesviruses penetrate target host cells by binding an entry receptor on the cell surface and fusing their lipid envelopes with the plasma membrane or membrane of an endocytic vesicle7. Unusually, herpesviruses distribute entry functions – host receptor engagement, fusion regulation, and membrane fusion – across several viral glycoproteins8,9. Entry of the prototypical herpes simplex viruses 1 and 2 (HSV-1 and HSV-2) requires four viral glycoproteins: gD, gB, gH, and gL2,9,10. gD is a receptor-binding protein that engages one of its three cognate receptors: nectin-111,12, a cell adhesion molecule; herpes virus entry mediator (HVEM)12–14, an immune modulator; or an O-sulfonated heparan sulfate10,15,16. gB is a class III viral fusogen that catalyzes the merger of the viral and host membranes17,18. gH and gL form a uniquely folded heterodimeric complex, gH/gL, that somehow “senses” the receptor-binding status of gD and, in response, activates the fusogenic activity of gB10,19–21. Indeed, gH/gL can interact with both gD and gB22.
Previously, we reported the crystal structure of the N-terminally truncated gH/gL ectodomain from HSV-2, lacking residues 19–48 immediately following the signal sequence (gHectoΔ48/gL)19. This structure is thought to represent an activated state of gH/gL because the gHΔ48/gL variant supports low-level membrane fusion in the absence of gD, which suggests that the N terminus of gH could be autoinhibitory23. Its removal may activate gH/gL by mimicking conformational changes induced by gD/receptor interactions. But how gH/gL transmits the activating signal from gD to gB is unknown.
Here, we report the cryogenic electron microscopy (cryoEM) structure of the HSV-2 gHecto/gL containing intact gH N terminus in its pre-activation state, bound to a fragment of antibody binding (Fab) of a neutralizing antibody CHL2721. Comparison with the crystal structure of the gHectoΔ48/gL, representing the activated state, revealed two major structural differences, in addition to changes in domain orientations. First, the N termini of gH and gL, which were unresolved in the activated state, form an X-shaped feature near the gD-binding site that appears to stabilize the pre-activation state. Second, a structural “switch” element near the C terminus of gH refolds and flips towards the gB-binding face of gH/gL. Analysis of the conformational ensembles observed by cryoEM suggests a conformational pathway for refolding of gH/gL from the pre-activation to the activated state, in which interaction of receptor-bound gD (or CHL27 Fab) with the N terminus of gH/gL sets off a series of rearrangements that culminate in the refolding of the C-terminal switch to transmit the activating signal to gB. The proposed allosteric activation mechanism may be conserved across Herpesviridae.
RESULTS
Structure of the pre-activation state of gHecto/gL
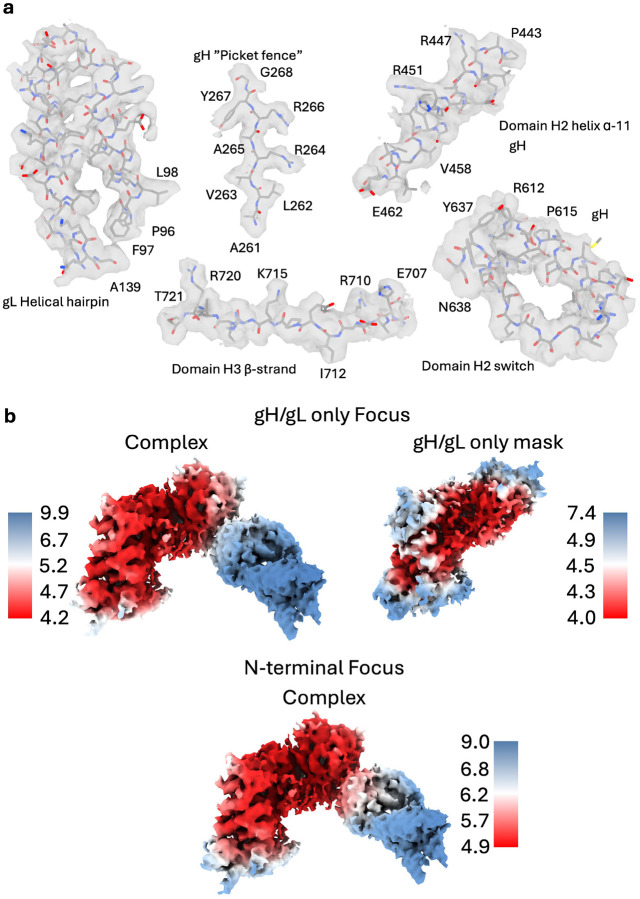
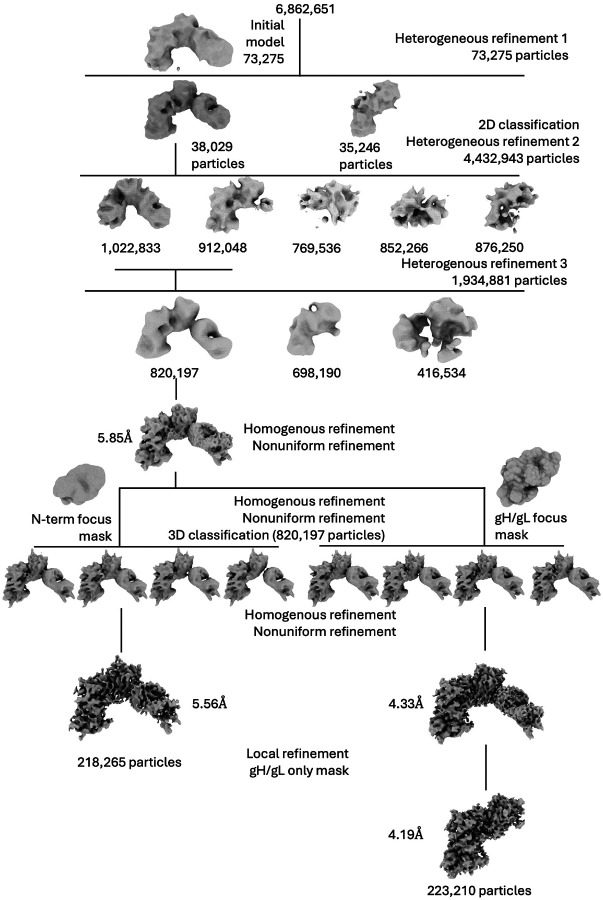
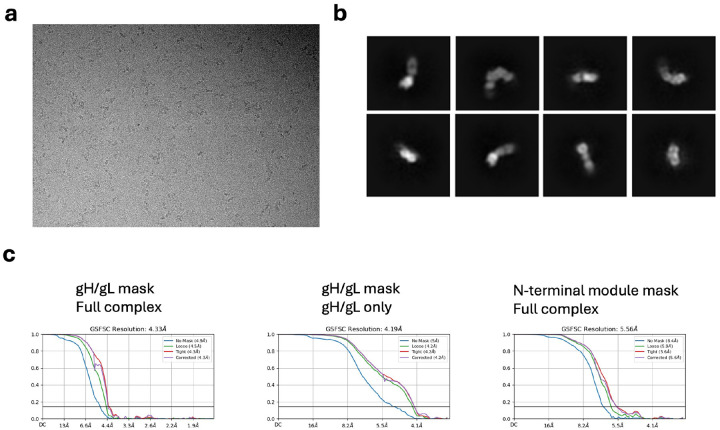
We co-expressed HSV-2 gHecto with HSV-2 gL in Sf9 cells and purified the heterodimeric complex (Extended Data Figure 1A and B). The gHecto/gL retains the gH N terminus, residues 19–48 (immediately following the signal sequence). The N terminus is autoinhibitory23. We therefore hypothesized that the gH N terminus may stabilize the pre-activation conformation of gH/gL. The N terminus also contains epitopes of several neutralizing antibodies21,23,24. To immobilize this unstructured region, we incubated gHecto/gL with a Fab of CHL27, a mouse neutralizing antibody that binds residues 37–47 and blocks gD binding21, and briefly cross-linked the complex with glutaraldehyde. After initial rounds of 3D classification and refinement in cryoSPARC, we obtained an 820,197-particle cryoEM data set. Focused 3D classification and local refinement using a mask focused on the gH/gL portion of the complex yielded a 4.19-Å reconstruction of gHecto/gL from a subset of 223,210 particles that allowed for placement of side chains with reasonable confidence (Extended Data Figures 2A and B).
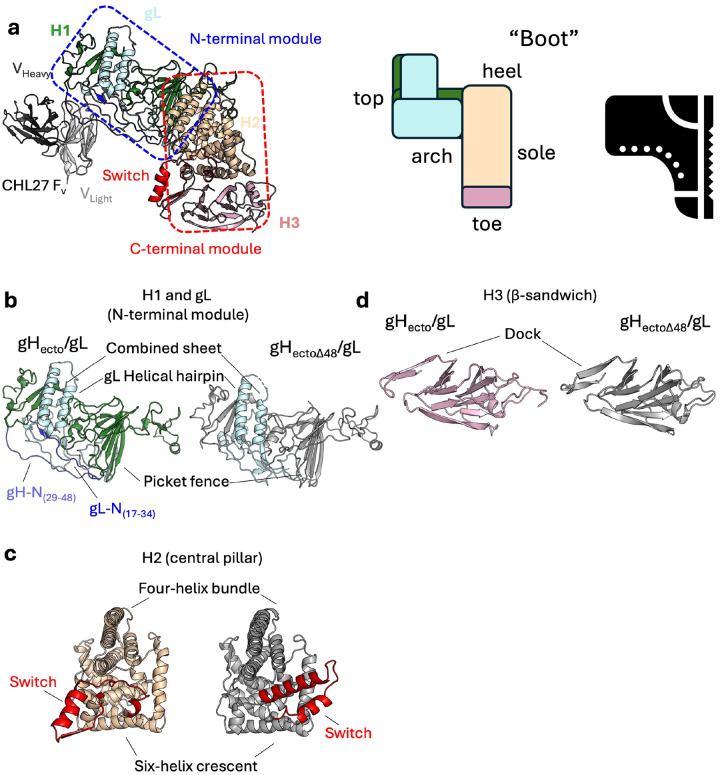
Overall, the structure of the gHecto/gL complex resembles the boot-like structure of the gHectoΔ48/gL (Figure 1A and Extended Data Figure 3A). The gHecto is composed of three domains, H1, H2, and H3 (Extended Data Figure 3A). Domain H1 (gH residues 29–332), located in the upper part of the boot, is composed of two subdomains that sandwich the gL core. This N-terminal H1/gL module of gH/gL features a β-sheet, composed of strands from both components, that packs against the helical hairpin of gL (Extended Data Figure 3B). The five-β-strand “picket fence” in H1 separates the N-terminal H1/gL module at the top of the boot from the C-terminal module in the lower part of the boot, composed of domains H2 and H3 (Figure 1A, Extended Data Figure 3A). The helix-rich domain H2 (gH residues 333–596) in the central part of the boot is composed of a four-helix bundle packed against a six-helix crescent (Extended Data Figure 3C). Domain H3, in the toe part of the boot, is a rigid, disc-like β-sandwich surrounded by loops and a helix (Extended Data Figure 3D). While the individual domains remain mostly unchanged, the bulk of the heterodimer (H1-gL-H2) is rotated and tilted 8° relative to domain H3 such that the gHecto/gL structure appears to stand more upright (Figure 1A).
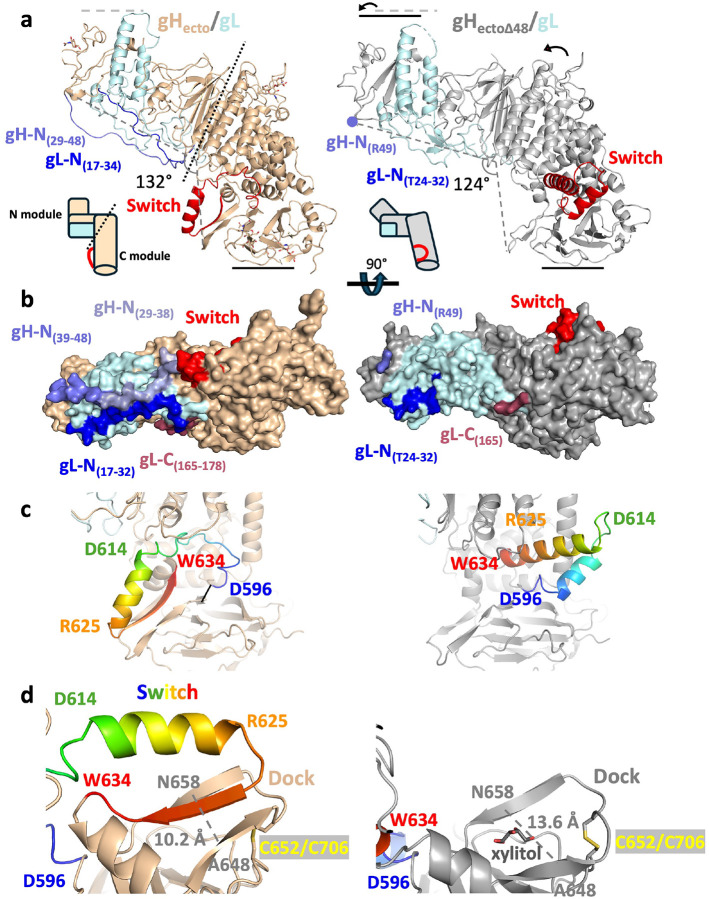
Figure 1. Comparison of the structures of the HSV-2 gHecto/gL (pre-activation state) and the gHectoΔ48/gL (activated state).
a) Ribbon diagrams of the two structures. gH is shown in wheat and grey; gL is shown in light blue. The N termini of gH and gL resolved in the gHecto/gL structure are shown in slate and blue, respectively. The switch region of gH is shown in red. The global differences in the overall conformations are shown as differences in the height and the angle. The approximate boundary between the N- and the C-terminal modules of gH/gL is shown as a dashed line. Insets depict the two states schematically. b) Surface diagrams, colored as in A. The N terminus of gH is colored according to the resolution of the cryoEM maps used in tracing, aa 39–48 (4.19 Å, slate) and aa 29–38 (5.56 Å, light blue). The C terminus of gL resolved in the gHecto/gL structure is shown in maroon. c) The switch motif (residues 596–636, color ramped from blue to red) adopts a loop-strand-helix conformation in the gHecto/gL structure and a helix-loop-helix conformation in the gHectoΔ48/gL structure. The loop is made up of residues 599–610, the helix is composed of residues 611–626, and the strand is made up of residues 628–636. Select residues are shown in colors matching their location. d) In the gHecto/gL structure, the β-strand of the switch is positioned between two β-strands of the dock (residues 644–659) completing a three-strand β-sheet. In the gHectoΔ48/gL structure, a xylitol molecule occupies the larger space between the two β-strands of the dock. The distance between the two β-strands of the dock is shown as a dashed line. PDB ID for activated gH/gL, 3M1C.
In addition to distinct domain orientations, the gHecto/gL structure has two major conformational differences from the gHectoΔ48/gL structure. First, the N-terminal regions of gH and gL that were missing or unresolved in the gHectoΔ48/gL structure, form an X-shaped feature covering one face of the N-terminal H1/gL module (Figure 1B). gH residues 29–48, missing from the gHectoΔ48/gL structure, form an extended polypeptide that runs from the picket fence along the surface composed of gL loops to the tip of the gH/gL heterodimer. gH residues 39–47 were resolved in the 4.19-Å map whereas residues 29–38 were only resolved in the 5.56-Å N-terminal focus map (described below) at lower map threshold values (Figure 1B). gL residues 17–23, unresolved in the gHectoΔ48/gL structure, also form an extended polypeptide that interacts with the N terminus of gH and with the C terminus of gL, residues 165–178, also unresolved in the gHectoΔ48/gL structure (Figure 1B).
Second, residues 596–636, located at the C terminus of domain H2 in the arch of the boot, adopt a very different fold. In the gHectoΔ48/gL structure, these residues adopt a helix-loop-helix conformation (Figure 1C). However, in the gHecto/gL structure, they instead adopt a loop-helix-strand conformation (Figure 1C). The entire loop-helix-strand element is flipped ~180 degrees relative to the gHectoΔ48/gL structure, akin to a light switch (Figure 1A, 1C, Supplemental Movie 1). We refer to this region as a “switch.” In the gHecto/gL structure, the β-strand (residues 628–636) of the switch is docked between two β-strands, residues 644–649 and 655–659, completing a three-strand β-sheet (Figure 1C and 1D). We refer to this region as a “dock”. In the gHectoΔ48/gL structure, the two strands are spaced farther apart, with the shortest distance CαA648-CαN658 of 13.2 Å (compared to 10.6 Å in the gHecto/gL structure) (Figure 1D). A xylitol molecule, used as a crystal cryoprotectant, occupies this larger interstrand space (Figure 1D, right). In both structures, the dock is anchored to the H3 disc by a disulfide between residues C652 and C706 (Figure 1D). Mutagenesis targeting disulfide-forming cysteines in HSV-1 and HSV-2 gH/gL showed that these cysteines are important for fusion25.
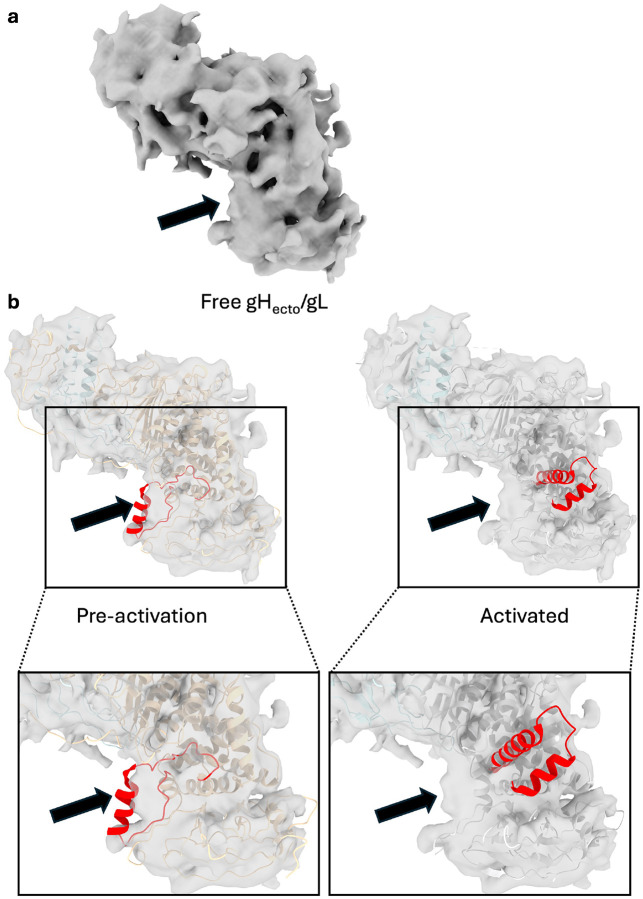
The presence of the intact gH N terminus along with the observed structural differences suggests that the cryoEM structure of gHecto/gL represents the pre-activation state of the gH/gL ectodomain. A 6.11-Å reconstruction of the unbound gHecto/gL heterodimer, which resembles that of the CHL27-bound gHecto/gL, further supports this notion (Extended Data Figure 4A and B).
CHL27 Fab primarily interacts with the N terminus of gH
To visualize the gHecto/gL-CHL27 interface, we used a focused classification and refinement of the complete particle set by imposing an N-terminal module mask (Extended Data Figure 5). The epitope-binding surface was resolved well enough in the 4.19 Å map to permit tracing of the N-terminal gH binding epitope (residues 38–47, slate), and N-terminal gH residues 29–37 (light blue) were placed into lower-confidence density in the 5.56 Å map (Figures 1B, and 2A).
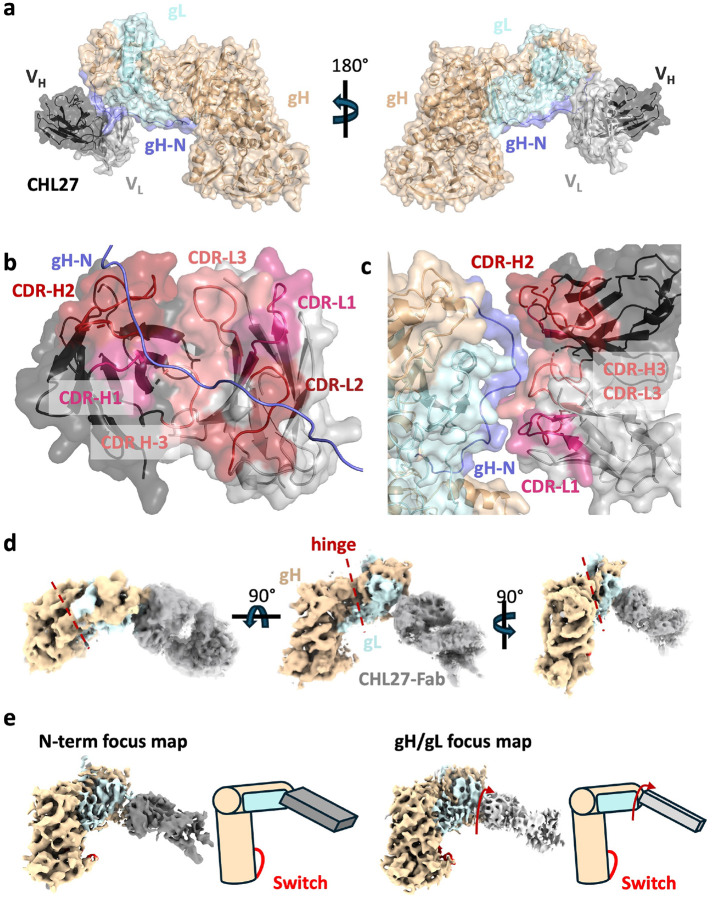
Figure 2. CHL27 Fab dynamically binds a membrane-distal surface of gH/gL.
a) The CHL27 Fab light (VL) and heavy (VH) chains bind the gH/gL N terminus at residues 43–47 of gH. b) Complementarity-determining regions (CDRs) of both the heavy and light chains of CHL27 interact with the gH N terminus. Most contacts with gH residues 43–47 occur with VH. c) VL is in proximity to gH and gL residues. CDR-L3 is proximal to gL residues 27–34, yet rigid-body fitting and refinement did not give conclusive evidence of amino-acid level interactions. d) CHL27-Fab is dynamically bound to the gH/gL heterodimer. e) Focused classification and refinement of the gH/gL-Fab complex using N-terminal or a full gH/gL mask reveals a mobile Fab that rotates and twists with respect to the gH/gL heterodimer. gH N terminus: slate, CHL27 VL: light gray, CHL27 VH: dark gray, CDR-H1: pink, CDR-H2: brick, CDR-H3: light salmon, CDR-L1: pink CDR-L2: brick, CDR-L3: light salmon.
Both VH and VL of CHL27 Fab contribute to interactions with gH and gL (Figure 2B, 2C). Residues 43–47 of gH fit into a vertical groove made up of the complementarity determining regions (CDRs) 1 and 3 of the VH chain, CDR-H1 and CDR-H3 (Figure 2B). The CDR-L3 completes the binding site for residues 43–47 of gH at the junction between the variable regions. Additionally, CDR-L1 and CDR-L2 are close enough to gL residues 27–34 and, possibly, gH residues 39–42 to interact (Figure 2C), but these interactions are not sufficiently resolved to discern side chain orientations. The gH/CHL27 interactions observed in our structure are consistent with prior epitope mapping studies using a series of overlapping peptides, which mapped the CHL27 epitope to gH residues 37–4721.
The low resolution of CHL27 and its epitope hinted at a highly dynamic binding mode. To identify possible global conformational heterogeneity, we reconstructed the entire 820,197-particle dataset into a 5.85-Å consensus map and performed a 3D Flexibility analysis (3DFlex) in cryoSPARC26. This analysis generated a volume series describing the motion of CHL27 Fab and its effects on the heterodimer. As the Fab moves relative to gH/gL, the N-terminal module also flexes about a hinge located between the gL helical hairpin and the gH picket fence (Figure 2D, Supplemental Movies 2 and 3). Comparison of reconstructions from the two different focused classifications recapitulated the motions suggested from 3DFlex analysis (Figure 2E). The highly mobile nature of the gHecto/gL-CHL27 interaction likely contributed to the low local resolution of the CHL27 Fab relative to gHecto/gL (Extended Data Figure 2B).
The gH/gL-CHL27 complex exists as an ensemble of conformational states spanning a pre-activation-to-activated continuum
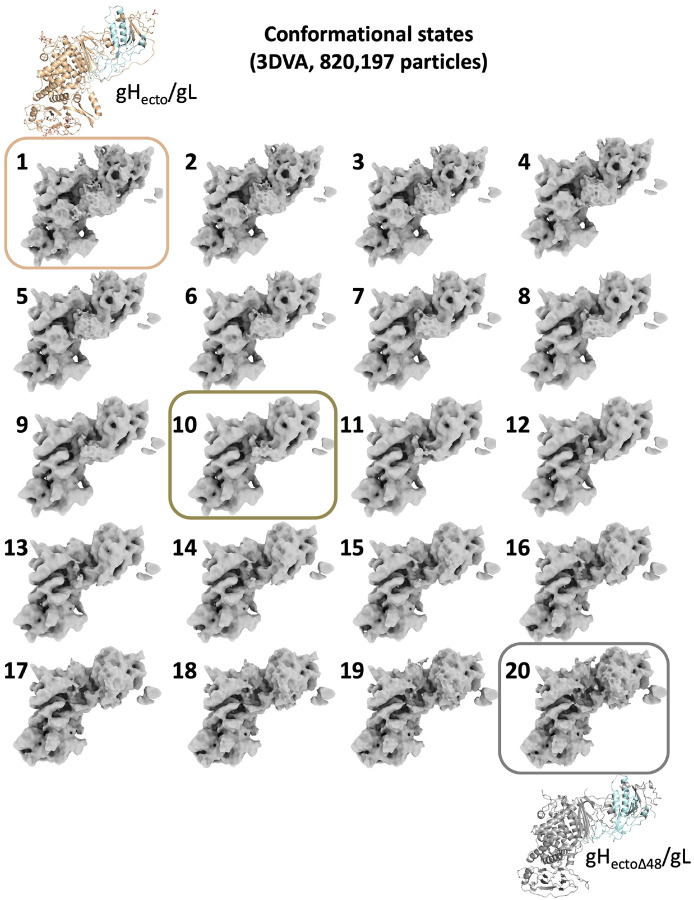
To uncover conformational differences within the gHecto/gL heterodimer that might have otherwise been overshadowed by the large-scale movements of the Fab, we generated a masked gH/gL-only cryoEM map and examined it for heterogeneity using 3D Variability analysis (3DVA)27. 3DVA resolved a series of 20 volumes representing continuous conformational heterogeneity in the gHecto/gL ectodomain (Extended Data Figure 6, Supplemental Movies 4 and 5). Thus, the gHecto/gL-CHL27 complex exists as a conformational ensemble in solution. The conformational states identified by this analysis were independently validated using an orthogonal machine-learning algorithm as implemented in CryoDRGN28 (Supplemental Movie 6). Conformational state 1 on one end of the continuum matches the 4.19-Å cryoEM reconstruction of the pre-activation state of gHecto/gL (Figure 3A, left and Extended Data Figure 6, top left). Conversely, conformational state 20 on the opposite end of the continuum resembles the activated state captured in the crystal structure of gHectoΔ48/gL (Figure 3A, right and Extended Data Figure 6, bottom right). Finally, one of the intermediate conformational states matches the 5.56-Å cryoEM map generated by imposing an N-terminal module mask (Figure 3A, middle and Extended Data Figure 6, middle).
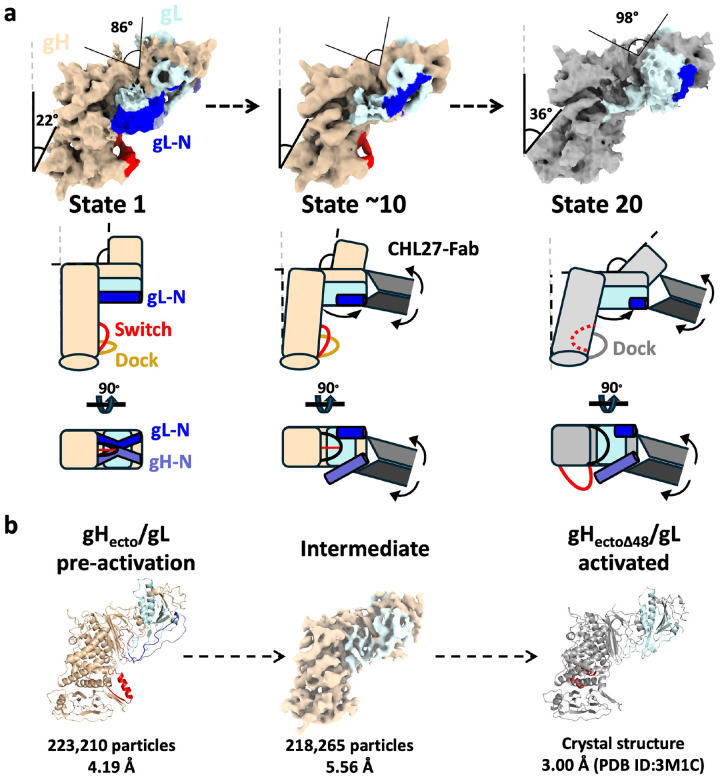
Figure 3. gH/gL-CHL27 complex exists in a conformational equilibrium.
a) Local and global conformational rearrangements as revealed by 3DVA. The conformational continuum is bookended by states that correspond to pre-activation (left) and post-activation gH/gL states. Locally, N-terminal gL density translates across the N-terminal module (top asterisk) as the helical density corresponding to the switch disappears (bottom asterisk). Globally, the angle between the H2 central pillar and the vertical increases, as does the angle between the gD-binding region and the rest of the N-terminal module. Electron densities are represented diagrammatically below the continuum, as are the structures determined for gHecto/gL and gHectoΔ48/gL. Model for the sequential activation of gH/gL by CHL27: (Left) gH/gL in its pre-activation state stands tall, with gH and gL N termini making contact with the picket fence. (Center) As the Fab binds and applies force, gH and gL disengage from their binding surfaces. Angles from the vertical and between the N- and C-terminal modules subsequently increase, leading to a “tilt” as seen in the gHectoΔ48/gL structure. (Right) The distance between the two strands of the dock increases. Density corresponding to the switch motif disappears from below the N-terminal module, indicating its refolding and repositioning into the helix-loop-helix configuration captured by the gHectoΔ48/gL crystal structure. b) Structures solved for the pre-activation and activated states of the gH/gL ectodomain. PDB ID: pre-activation gHecto/gL (9CZG), activated gHectoΔ48/gL (3M1C).
Both local and global conformational rearrangements occur during the transition from state 1 to state 20 along the continuum (Figure 3B and Extended Data Figure 6). Locally, the cryoEM density corresponding to the gL N terminus, residues 17–30, shifts across the N-terminal module. Concurrently, the density corresponding to the switch, residues 596–636, disappears from its position below the N-terminal module (Figure 3B, Supplemental movies 4 and 5). Globally, the angle between the gD-binding region and the rest of the N-terminal module increases from 86° to 98° (Figure 3). The entire gH/gL heterodimer tilts away from the vertical, evident from the angle between the sole of the gH/gL boot and the vertical increasing from 22° to 36°.
The resemblance between the terminal states 1 and 20 and the structures of the pre-activation and activated states of gH/gL, respectively, suggests that the observed continuum captures conformational transitions in gH/gL as it refolds from the pre-activation to activated state. The presence of the activated state, previously captured in crystals, in our cryoEM data further supports this notion.
DISCUSSION
gH/gL is a conserved component of the herpesviral entry machinery that transmits the receptor engagement signal from gD to gB thereby coupling host-receptor binding to membrane fusion during host entry. To achieve this, it must undergo conformational changes that somehow activate the fusogen, gB. The nature of these conformational changes has remained elusive, however. Here, we determined the structure of the gHecto/gL complex in its pre-activation state, prior to interactions with the receptor-bound gD. To achieve this, we preserved the N terminus of gH and used a neutralizing Fab CHL27 to stabilize its conformation and enable particle alignment, a strategy that previously succeeded with other macromolecules29. The other known conformational state of gH/gL, captured in the crystal structure of the gHectoΔ48/gL construct lacking the gH N terminus19, has been proposed to represent the activated state of the heterodimer23. Indeed, the match between this structure and one of the terminal states in the conformational ensemble observed in the cryoEM data supports this idea.
The N termini of gH and gL stabilize the pre-activation state
Our cryo-EM reconstruction of the gHecto/gL resolved the N termini of gH and gL that were either missing or disordered, respectively, in the gHectoΔ48/gL crystal structure19. In the gHecto/gL structure, these segments interact to form an X-shaped feature covering one face of the N-terminal module (Figure 1B). The structure clarifies why antibodies CΔ48L3 and CHL18, which bind epitopes within the C terminus of gL (173 to 183 and 208 to 219, respectively), interact with gHΔ48/gL variants but not with intact gH/gL21,23,24. This was puzzling because the C terminus of gL, which was unresolved in the gHectoΔ48/gL crystal structure, was expected to be surface exposed. The gHecto/gL structure shows that the C terminus of gL is covered by the N terminus of gL, which interacts with the N terminus of gH (Figure 1B). Thus, the epitopes of CΔ48L3 and CHL18 should be hidden in intact gH/gL, which also explains why these antibodies are non-neutralizing21,23,24. The lack of the N terminus of gH in the gHectoΔ48/gL destabilizes interactions between the N and C termini of gL, exposing these epitopes. Indeed, despite being non-neutralizing, CΔ48L3 and CHL18 block membrane fusion in the presence of gHΔ48/gL21,23,24.
Our analysis suggests that the N termini of gH and gL both help stabilize gH/gL in its high-energy pre-activation state (Figure 1B). This explains why the gHΔ48/gL variant lacking the gH N terminus can support fusion in a cell-cell fusion assay when co-transfected with gB, be it in the presence or absence of gD23.In the absence of the gH N terminus, the pre-activation state of gHectoΔ48/gL is stabilized by the N terminus of gL and can be activated by gD similarly to the intact gH/gL. However, lacking the stabilizing interactions provided by the gH N terminus, the pre-activation state of gHΔ48/gL is less stable and can be triggered even in the absence of gD, albeit less efficiently.
The gD-binding site in gH/gL and the role of CHL27
Activation of gH/gL requires binding to a receptor-bound gD. A panel of antibodies, including CHL27, that bind the N terminus of gH block gD-gH/gL binding. Thus, the gD-binding site in gH/gL at least partially overlaps the epitope of CHL27, residues 43–47 within the N terminus of gH. A more precise identification of the gD-binding site awaits the structure of the gD/receptor-gH/gL complex. Such a structure would also clarify the triggering of conformational rearrangements in gH/gL.
Initially, we hypothesized that binding of CHL27 to the N terminus of gH would help stabilize the pre-activation state of gH/gL. Serendipitously, this complex yielded an ensemble of conformational states spanning the range from pre-activation to intermediate and activated. Partial stabilization of the pre-activation state by CHL27 could be due to experimental conditions – brief crosslinking at room temperature after a short incubation of the two components of the complex on ice – that failed to trap all gH/gL in pre-activation state. However, we cannot yet rule out the possibility that CHL27, instead, triggered gH/gL to refold from its pre-activation to activated state, mimicking receptor-bound gD. In this latter case, neutralization by CHL27 could be due to non-productive conversion of gH/gL to its activated form in the absence of receptor. Regardless, the gH/gL-CHL27 complex allowed us to capture and characterize the conformational continuum of gH/gL states that lead to activation of gB.
Flipping of the switch in gH activates gB
The most striking difference between the gHecto/gL and the gHectoΔ48/gL structures, which represent the pre-activation and activated states, respectively, is the refolding and flipping of the switch element in domain H2. The switch adopts a loop-helix-strand conformation in the pre-activation state but refolds into a helix-loop-helix conformation in the activated state (Figure 1C, 1D). This refolding is accompanied by a ~180-degree flipping.
What is the role of switch? In the gHecto/gL structure, it is docked in the middle of a β-sheet and tucked under the N-terminal module. By contrast, in the gHectoΔ48/gL structure, the switch protrudes out from the wide surface near the putative epitope of the antibody LP11 that blocks gH/gL-gB interactions19. We hypothesize that in its flipped conformation, the switch would preclude gH/gL-gB interactions. This raises the question of how gH/gL and gB interact at different stages of fusion.
gH/gL-gB interactions
Our prior work using split luciferase reporters showed that gH/gL and gB interact before and during fusion22. We hypothesize that in its pre-activation state, gH/gL ectodomain interacts with the gB ectodomain and that this interaction helps stabilize the metastable prefusion form of gB. Conversely, interactions with the prefusion gB could help stabilize the pre-activation state gH/gL, locking both glycoproteins in a mutually inhibited complex on the surface of a virion or an infected cell. We also showed that in the absence of gL, not only all gH but also a large portion of gB is retained in the ER, suggesting that gH/gL and gB are likely transported as a complex to the cell surface22. Flipping of the switch during transition of gH/gL to its activated state would disrupt interactions with the prefusion gB ectodomain, providing a mechanism for releasing gB and allowing it to refold into the postfusion conformation. This model implies that rather than being activated by interactions with gH/gL, gB is released from an autoinhibited complex with gH/gL. In support of this idea, complexes of prefusion gB bound to gH/gL have been observed on the surface of HCMV virions by cryoelectron tomography and immunoprecipitation30,31. Thus, we posit that gH/gL serves as a “lynchpin,” constraining gB in its metastable prefusion state prior to host-receptor binding by gD.
How do gH/gL and gB maintain interactions during fusion, i.e., post-triggering? Previously, we found that the intraviral (cytoplasmic) regions of gB and gH regulate the membrane fusion process and proposed the “clamp-and-wedge” model32. The cytoplasmic domain (CTD) of gB forms a membrane-interacting inhibitory clamp that helps restrain the gB ectodomain in its prefusion conformation, presumably, through interactions with the transmembrane and membrane-proximal regions on the opposing side of the membrane32. The cytoplasmic tail (CT) of gH disrupts the inhibitory gB CTD clamp by acting as a wedge, which helps trigger the fusogenic refolding of the gB ectodomain. Using mutagenesis and modeling, we narrowed down the interacting regions to a hydrophobic pocket at the interprotomeric interface in the gB CTD trimer and residue V831 of gH CT33.
We hypothesize that the loss of interactions between the ectodomains of gH/gL and gB coincides with the disruption of the gB CTD clamp by the insertion of the gH CT wedge (Figure 4). This model helps explain how gB and gH/gL can maintain interactions before and during fusion despite undergoing conformational changes. We propose that prior to fusion, gB and gH/gL primarily interact through their ectodomains whereas during fusion, the interactions are primarily mediated by the cytoplasmic domains. Thus, the activation process involves coordinated actions across both the ectodomains and the cytoplasmic domains of gH/gL and gB. Future studies will unveil the precise order of these events and their coordination. Indirect evidence of gB-gH/gL complexes before and during fusion would be greatly bolstered by determining the structure of that complex.
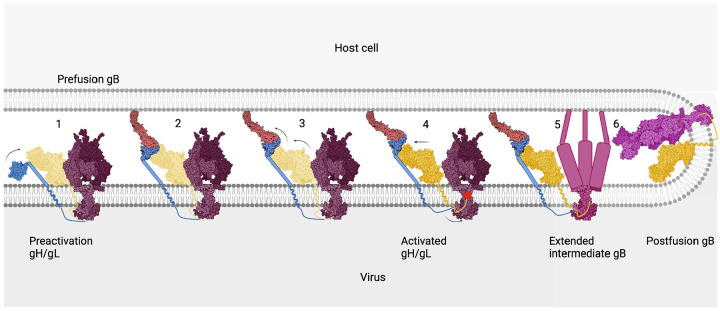
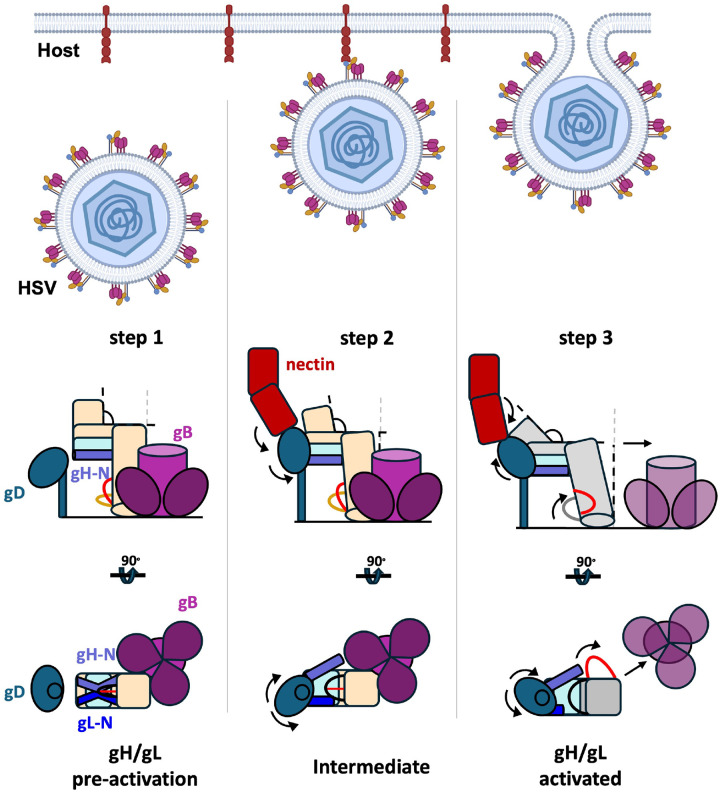
Figure 4. Stepwise model for gH/gL-mediated activation of gB.
1) Pre-activation gH/gL interacts with prefusion gB through their ectodomains. 2) Binding to receptor causes gD to bind the N terminus of gH/gL. 3) Binding of the gD/receptor complex to the N terminus of gH/gL causes a series of conformational changes in gH/gL that propagate throughout the heterodimer, culminating in the release and refolding of the switch. 4) In its activated state, the ectodomain of gH/gL no longer interacts with the prefusion gB. On the opposing side of the membrane, the cytoplasmic tail of gH engages the cytoplasmic domain of gB. This interaction destabilizes the cytoplasmic “clamp” that releases its grip onto the ectodomain. 5) Prefusion gB refolds into its extended intermediate conformation, exposing the fusion loops that interact with the membrane of the host cell. 6) As gB continues to refold into its final postfusion conformation, the fusion loops and the transmembrane spans are brought into proximity. This action brings apposing membranes close enough to merge, completing the fusion process. gD (blue), nectin-1 (brick), pre-activation gH/gL (pale yellow), prefusion gB (bordeaux), activated gH/gL (goldenrod), intermediate gB (dark rose), and postfusion gB (magenta). PDB IDs: gD (2C36), nectin-1 (3U83), activated gH/gL (3M1C), prefusion gB (6Z9M and 5V2S), and postfusion gB (5V2S).
Flip-switch transmission of the receptor-binding signal from gD to gB through gH/gL
We hypothesize that the continuum of conformational states observed by cryoEM here recapitulates the steps that transmit the activating signal from receptor-bound gD to prefusion gB (Extended Data Figure 7). We envision that gH/gL folds into a high-energy, pre-activation state stabilized by the interactions between the gH and gL N termini and, potentially, prefusion gB (Extended Data Figure 7, step 1). When receptor-bound gD binds to the N terminus of gH, this releases it from its contacts with the N terminus of gL and the picket fence (Extended Data Figure 7 step 2, Supplemental Movies 4 and 5). This action also releases the N terminus of gL, which we observe as a large shift in cryoEM density toward the top of the heterodimer (Figures 1B and 3, Supplemental Movies 4 and 5). The release of the N termini coincides with the tilting of domain H2 relative to domain H3 (Figures 1C and 3). The switch is then released by weakened inter-strand interactions when the distance between the two outer strands of the dock (residues 644–662) is widened by ~3 angstroms, allowing it to flip and refold into the helix-loop-helix conformation and captured in the gHectoΔ48/gL structure (Figure 1C, D, Extended Data Figure 7, step 3, and Supplemental Movie 1). In this manner, the conformational rearrangements described above propagate across the gH/gL heterodimer from the N terminus of domain H1 to the C terminus of domain H2 (Figure 3, Supplemental Movie 1). In this conformation, the switch juts out from the gB-binding surface of gH/gL.
The ”lynchpin” model for fusion regulation by gH/gL
These structural studies lead us to propose a lynchpin model for how the gH/gL ectodomain regulates gB fusion activity (Figure 4). Prior to gD binding of a host receptor, pre-activation gH/gL and prefusion gB interact through their ectodomains, helping stabilize each other (step 1). Binding to receptor causes gD to bind the N-terminus of gH/gL (step 2). Binding of gD/receptor complex to gH/gL initiates a series of conformational changes that propagate throughout the heterodimer, culminating in the release, refolding, and flipping of the switch (step 3). Once flipped, the switch disrupts the interactions between the ectodomains of gH/gL and gB. In its activated state, the ectodomain of gH/gL no longer binds the prefusion gB ectodomain. Meanwhile, on the opposing side of the membrane, the gH CT wedges into the gB CTD, which destabilizes the clamp and causes it to release its grip on the gB ectodomain (step 4). The loss of the stabilizing interactions with the gH/gL ectodomain allows the prefusion gB to refold into the extended intermediate (step 5) and, ultimately, (6) to its stable, low-energy postfusion conformation, completing the fusion process. We hypothesize that refolding of the gH switch from loop-helix-strand to helix-loop-helix is thermodynamically favorable. Thus, gH/gL is a “one-shot” machine that irreversibly refolds from pre-activation to activated form, similarly to gB, which irreversibly refolds from prefusion to the postfusion form18.
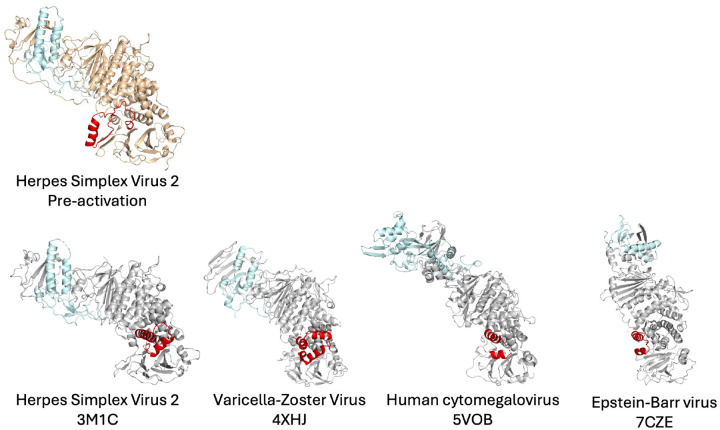
There are several implications to our model that potentially apply across all Herpesviridae subfamilies. First, the switch element, in its activated helix-loop-helix conformation, is found across all known structures of gH/gL homologs (Extended Data Figure 8). Therefore, we hypothesize that this element has a conserved role in the gB activation mechanism by gH/gL. Thus, we suspect that our proposed model for how gH/gL regulates the activity of gB could be generally applicable across the entire family Herpesviridae. Finally, the identification of the switch mechanism for gB activation by gH/gL points to a target for subunit-based vaccines and treatment efforts, possibly applicable to all herpesviral infections.
MATERIALS AND METHODS
Expression and purification of the recombinant HSV-2 gH/gL
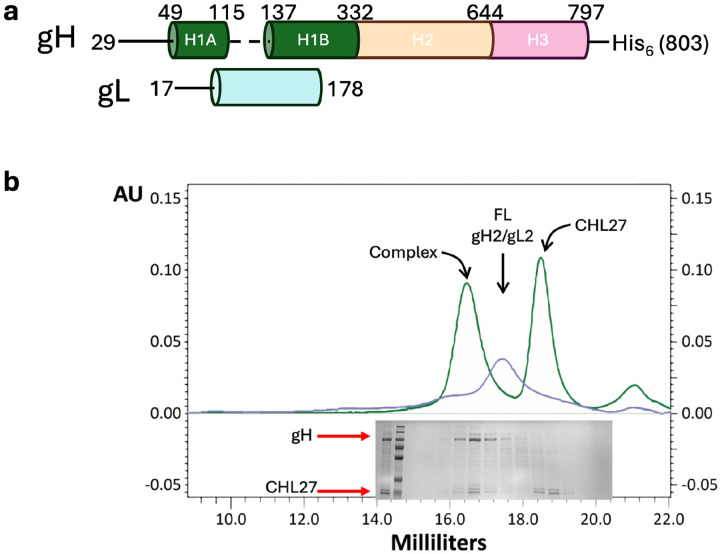
Recombinant baculovirus (pTC605) encoding HSV-2 gH ectodomain, residues 1–803, with a C-terminal His6 tag and full-length HSV-2 gL, residues 1–224, was generated using the pFastBac Dual system as previously described (Extended Data Figure 8A)34. Virus titer was increased by two rounds of serial passaging in Sf9 cells to passage 3 (P3). For protein expression, 4.5–6-L cultures of Sf9 cells were infected with 8 mL of virus per 1 L of cells at a density of 2×106 cells/mL. Supernatants containing the secreted gHecto/gL were collected 72–80 hours post-infection and centrifuged at 4,000 g for 40 minutes in a Sorvall Lynx 4000 floor centrifuge (ThermoFisher) using a swinging bucket rotor. The supernatant was decanted and filtered using 0.2 μm polyethylenesulfonate (PES) bottle top vacuum filters. 100 mM phenylmethylsulfonylfluoride (PMSF) was added to the supernatant at a 1:1000 dilution for a working concentration of 100 μM.
The supernatant was concentrated from 4.5 or 6 L to 1.2 L using a tangential flow filtration (TFF) system as previously described35. Supernatant was incubated with 3–4 mL Ni-NTA Excel resin (GE/Cytiva) pre-equilibrated in phosphate buffered saline (PBS) for 45 minutes at 4 °C in 50-mL conical tubes under nutation. The resin was pelleted by centrifugation at 1,000 g for 5 minutes in a tabletop centrifuge using a swinging bucket rotor, and the supernatant containing the flow-through fraction was carefully removed. The resin was washed using gravity flow with 60–80 mL wash buffer 1 (25 mM Tris, pH 7.9, 150 mM NaCl, 10 mM imidazole); 60–80 mL wash buffer 2 (25 mM Tris, pH 7.9, 500 mM NaCl, 20 mM imidazole); and 60–80 mL of wash buffer 3 (25 mM Tris, pH 7.9, 150 mM NaCl, 20 mM imidazole) prior to elution with 25 mL elution buffer (25 mM Tris, pH 7.9, 150 mM NaCl, 140 mM imidazole) into a 50-mL conical tube containing 50 μL of 0.5 M ethylenediaminetetraacetic acid (EDTA) for a final concentration of 1 mM. The eluate fraction was concentrated using 100 kDa MWCO 15 mL spin concentrators (Amicon/Millipore) to 500 μL, centrifuged at 21,000 g at 4 °C for 10 minutes to remove aggregates, and injected onto a 10/300 Superose 6 column (GE/Cytiva) pre-equilibrated with size exclusion/storage buffer (25 mM HEPES, pH 7.4, 150 mM NaCl) using a Biorad NGC system. Chromatographic peaks were analyzed by SDS-PAGE (Extended Data Figure 1B). Relevant fractions were pooled and concentrated to ~5 mg/mL in storage buffer with a 100 kDa MWCO 4 mL spin concentrator (Amicon/Millipore) for immediate use or snap-frozen in 25-μL aliquots for future use.
Purification of CHL27 Fab
CHL27 Fab was isolated using a Pierce Fab Preparation Kit (Thermo Scientific), according to the manufacturer’s protocol. Briefly, 5 mgs of murine IgG were incubated with 250 μl papain immobilized on agarose resin for 4h at 37°C. After digestion, Fab was purified using a Protein A spin column. Protein concentration was determined by absorbance at 280nm using an estimated extinction coefficient of 1.4.
CHL27 isotyping and hybridoma sequencing
Mouse CHL27 Mab was isotyped using Pierce Rapid Isotyping Kit following the manufacturer’s protocol. Hybridoma sequencing were performed by Genscript (Piscataway, NJ). Total RNA was isolated from 2×106 cells and reverse-transcribed into cDNA using isotype-specific (mouse IgG1 heavy chain, kappa light chain) anti-sense primers or universal primers following PrimeScriptTM 1st Strand cDNA Synthesis Kit’s manual. Antibody fragments of VH, VL, CH and CL were amplified according to the standard operating procedure of rapid amplification of cDNA ends of GenScript. The sequences of different clones were aligned, and the consensus sequence was provided (Extended Data Figure 8B).
Cryo-EM sample preparation and data collection
gHecto/gL/CHL27 Fab complex formation was verified by size exclusion chromatography (Extended Data Figure 1B). Complexes were assembled and crosslinked in situ during sample preparation. gHecto/gL was diluted to 1.2 mg/mL using 1.25x molar excess of CHL27 Fab in 30μL of storage buffer. After 30 minutes of incubation on ice, electron microscopy-grade 25% glutaraldehyde (Invitrogen) was added to a final v/v percentage of 0.025% and incubated for 5 or 10 minutes. The crosslinking reaction was quenched with 4X quenching buffer (36 mM Tris, pH 8.0 with 0.4% w/v 2-octylpyranoglucoside (2-OG) (Affymetrix)) for a final Tris and detergent concentration of 9 mM and 0.1%, respectively. Sample was immediately applied to glow-discharged holey C-flat grids (1.2/1.3, 4C 400 mesh Cu grids Mitegen Cat #:M-C413) and vitrified in nitrogen-cooled liquid ethane with a Gatan CP3 cryoplunger using blot setting 4.
Samples were screened on a Talos Arctica microscope (ThermoFisher) with a K3 camera (Gatan) operated at 200 kV at the Harvard Cryo-EM center and the highest quality grids were imaged on a Titan Krios Microscope (ThermoFisher) with a K3 camera (Gatan) at the New York Structural Biology Center (NYSBC) National Center for CryoEM Access and Training (NCCAT) operating at 300 kV acceleration voltage. 50–60 frame movies were collected in counting mode at 105,000x magnification with a total dose of ~65 e−/Å2 and a pixel size of 0.825 Å. In total 29,989 movies were collected with a defocus range of −1.2 – −2.2 μm. Movies were motion-corrected and dose-weighted via the NCCAT automated pipeline using MotionCor236, and contrast transfer function (CTF) parameters were estimated using CTFFIND437 (representative micrograph, Extended Data Figure 9A). Some of this work was performed at the National Center for CryoEM Access and Training (NCCAT) and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539,) and by grants from the Simons Foundation (SF349247) and NY State Assembly.
Image processing and 3D reconstructions
Data processing and analysis was performed chiefly using the cryoSPARC38 suite. Templates were generated from prior low-resolution density maps from images collected at the Harvard CryoEM Center (11,286 50-frame movies were collected on a Titan Krios at 300kV acceleration voltage in counting mode at 105,000x magnification with a total dose of ~75 e−/Å2, a pixel size of 0.825 Å, and a defocus range of −1.2– −2.2 μm). These data were processed using RELION’s39 processing pipeline. The templates generated by this initial analysis were used to pick 6,862,651 total particles (Extended Data Figure 5). After successive 2D classification analyses, an initial 3D model was generated using ab-initio reconstruction from 73,725 particles and was further improved by heterogeneous refinement. After further 2D classification of the full particle set (representative 2D classes, Extended Data Figure 9B), these initial models along with “junk” 3D density maps were used for heterogeneous refinement. A subsequent heterogeneous refinement step combined two 3D classes that corresponded to two distinct “views” of the gHecto/gL/CHL27 complex. The density map corresponding to the full complex (820,197 particles) was refined using homogenous refinement followed by nonuniform refinement. This density map (5.85 Å resolution) was used for cryoSPARC 3D Flexibility and 3D Variability analyses26,27 as well as cryoDRGN continuous heterogeneity analysis28. This map was then further improved by focused 3D classification using two different masks: an N-terminal-module-focused mask and a full gHecto/gL mask. Although the N-terminally focused 3D classification only marginally improved the overall resolution to 5.56 Å (GFSC curves, Extended Data Figure 9C) (218,265 particles), it enabled tracing of the gH and gL N termini. The gHecto/gL-focused 3D classification significantly improved the resolution to 4.33 Å for the gHecto/gL/CHL27 complex (223,210 particles). The resolution was further improved to 4.19 Å by masking away the Fab density and using local refinement of only gHecto/gL. Reference-based motion correction was attempted on these final electron density maps but did not result in a meaningful improvement of the density map quality. Data collection and refinement statistics are reported in Extended Data Table 1.
Model building
The crystal structure of the N-terminally truncated gH/gL (gHectoΔ48/gL, PDBID:3M1C) was used for initial model building. It was fit into the electron density in ChimeraX40, and the model was manually refined using COOT41 and ChimeraX ISOLDE module42. Models were real-space refined in PHENIX43 against the gHecto/gL-focused and refined map (4.33 Å, 223,210 particles) and against a combination of the N-terminally focused map (5.56 Å, 218,265 particles) and the gHecto/gL-only locally refined map (4.19 Å, 223,210 particles) using PHENIX’s combine_focused_maps module43. A homology model of CHL27 Fab was generated using the ROSIE2 server44 and fit into the N-terminally focused map (5.56 Å, 218,265 particles) in ChimeraX. The homology model of the CHL27 Fv region was further rigid-body fitted into the density in COOT. The Fab was then rigid-body refined against the combined map. Once the gHecto/gL and the CHL27 Fab models were merged into a single model, real-space refinement of the residues at the gHecto/gL/CHL27 interface was done to prevent clashes using COOT and ISOLDE. The final model was refined against the combined focused map using real-space refinement to generate the statistics report (Extended Data Table 1).
Extended Data
Extended Data Figure 1. Domain architecture and purification of gHecto/gL-CHL27 complex.
a) Domain architecture of gH/gLFL. Domain diagram shown here represents density visible in our EM reconstruction; dotted line indicates missing density. b) Chromatographic and SDS-PAGE analysis confirms that gHecto/gL and CHL27Fab form a complex. Green trace, complex; slate trace, gHecto/gL alone. gHecto/gL structure resembles large-scale features of the gHectoΔ48/gL structure.
Extended Data Figure 2. EM density quality and local resolution of gHecto/gL-CHL27 complex.
a) Density quality across gH/gL was high enough to confidently place side chains despite the relatively low overall resolution of 4.19Å. All volumes were contoured in ChimeraX to 0.86 except for the switch motif, which was contoured to 0.38 to visualize the mobile helix. b) Local resolution of full complex and masked gH/gL-only maps. Top: gH/gL focused maps. Left map is the full complex, right map generated by masking the Fab and performing local refinement after particle subtraction. Bottom: N-terminal module-focused map of the complex.
Extended Data Figure 3. Overall structure of a gHecto/gL-CHL27 Fab complex.
a) gHecto/gL structure resembles large-scale features of the gHectoΔ48/gL structure, retaining its boot-like appearance. b) The N-terminal module contains a gH-gL combined β-sheet, gL helical hairpin, and a five-strand β-sheet picket fence. c) The H2 central pillar is composed of a four-helix bundle packed against a six-helix crescent. The C-terminal switch is in both a different configuration and position. d) Domain H3 is made up of a β-sandwich lined by loops and a helix. The switch locks into the dock in the gHecto/gL structure.
Extended Data Figure 4. Unbound gHecto/gL more closely resembles CHL27-bound gHecto/gL than gHectoΔ48/gL.
a) 6.11 angstrom EM reconstruction of unbound gHecto/gL. Arrow points at putative switch density. b) ChimeraX fit of pre-activation gHecto/gL (left) and activated gHectoΔ48/gL (right) into free gHecto/gL EM map. Indicated density is empty when gHectoΔ48/gL is fit, whereas switch motif fits reasonably well in the density. Inset zoomed into switch region.
Extended Data Figure 5.
CryoEM analysis workflow.
Extended Data Figure 6. 3D Variability analysis-generated maps of gH/gL-focused masking reveal the conformational continuum of gHecto/gL when in complex with CHL27Fab.
3DVA resolved continuous heterogeneity across 20 volumes. State 1 (boxed in wheat) refers to the pre-activation conformation, state 10 (boxed in dark brown) refers to an intermediate conformation, and state 20 (boxed in gray) refers to the activated conformation.
Extended Data Figure 7. Stepwise “flip-switch” model for allosteric signal transduction through gH/gL.
Step 1: Prior to host-receptor binding, gH/gL (wheat and pale cyan, respectively) and gB (maroon/dark pink) are locked in a mutually inhibited complex on the virion surface. Step 2: host-receptor (nectin, brick) binds gD (sapphire) and leads to gD-gH/gL ectodomain interactions. Movements between nectin, gD, and gH/gL release gL N terminus from its contacts with the gH picket fence. Step 3: Release of the N termini leads to a relaxation and tilting of gH/gL that widens the distance between outer β-strands of the C-terminal dock, allowing the switch motif to flip out toward the gB-binding surface of the heterodimer. We posit this destabilizes gH/gL-gB interactions, freeing the labile prefusion gB to transition to its extended intermediate conformation.
Extended Data Figure 8. Switch motif is conserved across all known structures of Herpesviridae gH/gL homologs.
Structures of human-infecting herpesvirus gH/gL ectodomain (PDBIDs: HSV-2 3M1C, VZV 4XHJ, HCMV 5VOB, EBV 7CZE) all show conservation of the switch in its activated, helix-loop-helix configuration. Our structure of HSV-2 gHecto/gL is the only known structure with the switch in its pre-activation conformation. Models were aligned in Pymol by domains H3/D4.
Extended Data Figure 9.
gH/gL and CHL27 sequence and domain maps.
Extended Data Figure 10. CryoEM data collection and analysis.
a) Representative cryoelectron micrograph collected on Titan Krios “Caitlin” at the New York Structural Biology Center. b) 2D classes for the 820,197 particles before focused 3D classification. c) Gold-standard Fourier shell correlation curves for the three reconstructed maps used for model building and structural analysis.
Extended Data Table 1.
Cryo-EM data collection, refinement, and validation statistics.
| Data collection and processing | gHecto/gL2 only PDBID: 9CZG | gHecto/gL-CHL27 PDBID: 9CZR |
|---|---|---|
| Magnification | 105,000x | |
| Voltage (kV) | 300 | |
| Electron exposure (e−/ Å2) | ~64 | |
| Defocus range (μm) | −1.2 - −2.2 | |
| Pixel size (Å) | 0.825 | |
| Symmetry imposed | C1 | |
| Initial particle images (no.) | 6,862,651 | |
| Final particle images (no.) | 223,210 | 218,265 |
| Map resolution (Å) at 0.143 FSC threshold | 4.19 | 5.56 |
| Refinement | ||
| Initial model used (PDB code) | 3M1C | |
| Map sharpening B factor (Å2) | ||
| gH/gL local refinement C-term focus | −196 | −256 |
| Composition (no.) | ||
| Chains | 2 | 4 |
| Non-hydrogen atoms | 7113 | 8827 |
| Residues | 916 | 1138 |
| Ligands | 7 (BMA:2, NAG: 5) | 7 (BMA:2, NAG: 5) |
| Bonds (RMSD) | ||
| Length (Å) (# > 4σ) | 0.008 (14) | 0.103 (71) |
| Angles (°) (# > 4σ) | 1.46 (86) | 2.88 (205) |
| MolProbity score | 2.48 | 2.66 |
| Clash score | 19.5 | 27.4 |
| Ramachandran plot (%) | ||
| Outliers | 0.88 | 0.98 |
| Allowed | 3.96 | 4.02 |
| Favored | 95.2 | 95.0 |
| Rotamer outliers (%) | 2.94 | 3.22 |
| Model vs. Data | ||
| CC (mask) | 0.77 | 0.58 |
| CC (box) | 0.79 | 0.67 |
| CC (peaks) | 0.68 | 0.49 |
| CC (volume) | 0.77 | 0.59 |
| Mean CC for ligands | 0.72 | 0.70 |
Supplementary Material
Acknowledgments:
We thank Carmen Rivera for help with protein purification experiments. We thank Patricia Spear (Northwestern University) for the gift of plasmids. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We dedicate this publication to the memory of Roselyn J. Eisenberg (University of Pennsylvania).
Funding:
National Institutes of Health grants K12GM133314 (GLGDP), R01AI164698 (EEH) Howard Hughes Medical Institute grant 55108533 (EEH)
Funding Statement
National Institutes of Health grants K12GM133314 (GLGDP), R01AI164698 (EEH) Howard Hughes Medical Institute grant 55108533 (EEH)
Footnotes
Competing interests: Authors declare that they have no competing interests.
Supplementary Materials
Movies S1 to S6
Data and materials availability:
Structure model and maps have been deposited to EMDB using the following accession numbers: 9CZG – gH/gL only, 9CZR – gH/gL-CHL27 complex.
REFERENCES
- 1.Bernard N., F., 1950-, K., David M. (David Mahan), & Peter M., H.. Fields virology. (2013). [Google Scholar]
- 2.Atanasiu D., Saw W. T., Cohen G. H. & Eisenberg R. J. Cascade of Events Governing Cell-Cell Fusion Induced by Herpes Simplex Virus Glycoproteins gD, gH/gL, and gB▿. J Virol 84, 12292–12299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L. Fields Virology, 6th Edition. Clin Infect Dis 59, 613–613 (2014). [Google Scholar]
- 4.Jeffery-Smith A. & Riddell A. Herpesviruses. Medicine 49, 780–784 (2021). [Google Scholar]
- 5.Eisenberg R. J. et al. Herpes Virus Fusion and Entry: A Story with Many Characters. Viruses 4, 800–832 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poole C. L. & James S. H. Antiviral Therapies for Herpesviruses: Current Agents and New Directions. Clin. Ther. 40, 1282–1298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heldwein E. E. & Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci 65, 1653–1668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pino G. L. G.-D. & Heldwein E. E. Well Put Together-A Guide to Accessorizing with the Herpesvirus gH/gL Complexes. Viruses 14, 296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly S. A., Jardetzky T. S. & Longnecker R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 19, 110–121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns T. M. & Connolly S. A. Entry of Alphaherpesviruses. Curr. Issues Mol. Biol. 41, 63–124 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Lu G. et al. Crystal Structure of Herpes Simplex Virus 2 gD Bound to Nectin-1 Reveals a Conserved Mode of Receptor Recognition. J Virol 88, 13678–13688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giovine P. D. et al. Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1. Plos Pathog 7, e1002277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly S. A. et al. Structure-Based Analysis of the Herpes Simplex Virus Glycoprotein D Binding Site Present on Herpesvirus Entry Mediator HveA (HVEM). J Virol 76, 10894–10904 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stiles K. M. et al. Herpes Simplex Virus Glycoprotein D Interferes with Binding of Herpesvirus Entry Mediator to Its Ligands through Downregulation and Direct Competition▿. J Virol 84, 11646–11660 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shukla D. et al. A Novel Role for 3-O-Sulfated Heparan Sulfate in Herpes Simplex Virus 1 Entry. Cell 99, 13–22 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Tiwari V. et al. Soluble 3-O-sulfated heparan sulfate can trigger herpes simplex virus type 1 entry into resistant Chinese hamster ovary (CHO-K1) cells. J Gen Virol 88, 1075–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 17.White J. M., Ward A. E., Odongo L. & Tamm L. K. Viral Membrane Fusion: A Dance Between Proteins and Lipids. Annu. Rev. Virol. 10, 139–161 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison S. C. Viral membrane fusion. Virology 479–480C, 498–507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdary T. K. et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17, 882–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cairns T. M. et al. Capturing the Herpes Simplex Virus Core Fusion Complex (gB-gH/gL) in an Acidic Environment. J Virol 85, 6175–6184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairns T. M. et al. Localization of the Interaction Site of Herpes Simplex Virus Glycoprotein D (gD) on the Membrane Fusion Regulator, gH/gL. J Virol 94, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pataki Z., Viveros A. R. & Heldwein E. E. Herpes Simplex Virus 1 Entry Glycoproteins Form Complexes before and during Membrane Fusion. MBio 13, e0203922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atanasiu D. et al. Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. MBio 4, e00046–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atanasiu D., Saw W. T., Cairns T. M., Eisenberg R. J. & Cohen G. H. Using Split Luciferase Assay and Anti-Herpes Simplex Virus Glycoprotein Monoclonal Antibodies To Predict a Functional Binding Site between gD and gH/gL. J Virol 95, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cairns T. M., Landsburg D. J., Whitbeck J. C., Eisenberg R. J. & Cohen G. H. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 332, 550–562 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Punjani A. & Fleet D. J. 3DFlex: determining structure and motion of flexible proteins from cryo-EM. Nat. Methods 20, 860–870 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Punjani A. & Fleet D. J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Zhong E. D., Bepler T., Berger B. & Davis J. H. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods 18, 176–185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh R. M. et al. Structural principles of distinct assemblies of the human α4β2 nicotinic receptor. Nature 557, 261–265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Si Z. et al. Different functional states of fusion protein gB revealed on human cytomegalovirus by cryo electron tomography with Volta phase plate. Plos Pathog 14, e1007452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanarsdall A. L., Howard P. W., Wisner T. W. & Johnson D. C. Human Cytomegalovirus gH/gL Forms a Stable Complex with the Fusion Protein gB in Virions. Plos Pathog 12, e1005564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper R. S., Georgieva E. R., Borbat P. P., Freed J. H. & Heldwein E. E. Structural basis for membrane anchoring and fusion regulation of the Herpes Simplex Virus fusogen gB. Nat. Struct. Mol. Biol. 25, 416–424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pataki Z., Sanders E. K. & Heldwein E. E. A surface pocket in the cytoplasmic domain of the herpes simplex virus fusogen gB controls membrane fusion. PLOS Pathog. 18, e1010435 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannah B. P. et al. Herpes Simplex Virus Glycoprotein B Associates with Target Membranes via Its Fusion Loops. J. Virol. 83, 6825–6836 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White E. M., Stampfer S. D. & Heldwein E. E. Herpes Simplex Virus, Methods and Protocols. Methods Mol Biology 2060, 377–393 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohou A. & Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Punjani A., Rubinstein J. L., Fleet D. J. & Brubaker M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Fernandez-Leiro R. & Scheres S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. Sect. D 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goddard T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biological Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croll T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D: Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biological Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyskov S. et al. Serverification of Molecular Modeling Applications: The Rosetta Online Server That Includes Everyone (ROSIE). PLoS ONE 8, e63906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure model and maps have been deposited to EMDB using the following accession numbers: 9CZG – gH/gL only, 9CZR – gH/gL-CHL27 complex.