Abstract
The complete genome sequence of cultivated rice (Oryza sativa) provides an unprecedented opportunity to understand the biology of this model cereal. An essential and necessary step in this effort is the determination of the coding information and expression patterns of each sequenced chromosome. Here, we report an analysis of the transcriptional activity of rice chromosome 4 using a tiling path microarray based on PCR-generated genomic DNA fragments. Six representative rice organ types were examined using this microarray to catalog the transcribed regions of rice chromosome 4 and to reveal organ- and developmental stage–specific transcription patterns. This analysis provided expression support for 82% of the gene models in the chromosome. Transcriptional activities in 1643 nonannotated regions were also detected. Comparison with cytologically defined chromatin features indicated that in juvenile-stage rice the euchromatic region is more actively transcribed than is the transposon-rich heterochromatic portion of the chromosome. Interestingly, increased transcription of transposon-related gene models in certain heterochromatic regions was observed in mature-stage rice organs and in suspension-cultured cells. These results suggest a close correlation between transcriptional activity and chromosome organization and the developmental regulation of transcription activity at the chromosome level.
INTRODUCTION
Rice (Oryza sativa) is the principle staple food for more than half of the world's population. With a compact genome spanning 430 megabase (Mb) pairs, an extensive genetic map (Harushima et al., 1998), and established synteny with other cereal crops (Ahn and Tanksley, 1993; Chen et al., 1997; Gale and Devos, 1998), cultivated rice represents a model for cereal as well as monocot plants (Shimamoto and Kyozuka, 2002). Furthermore, the rice genome is nearly completely sequenced (Feng et al., 2002; Sasaki et al., 2002; Rice Chromosome 10 Sequencing Consortium, 2003; Rensink and Buell, 2004). One of the next essential steps in deciphering the sequenced genome is to develop complete and accurate maps of actively transcribed regions during rice development. This will facilitate the identification of all genes and proteins encoded in the DNA sequence. Such information will allow further analysis of their function, regulation, and how they cooperate in complex biological processes in a systematic manner.
As a first attempt to decipher the rice genome, computational annotation has been successful, although improvements are needed (Yuan et al., 2003). Recent efforts to verify experimentally the gene model structure by sequencing cDNAs and ESTs have provided valuable information toward our understanding of gene structure and genome-coding capacity (Wu et al., 2002; Rice Full-Length cDNA Consortium, 2003; Rensink and Buell, 2004). However, to date, only approximately half of the predicted genome-coding capacity has had any cDNA or EST expression support. Massively parallel signature sequencing provides another tool to analyze the transcriptional activity of complex genomes (Meyers et al., 2004) and is just being applied to rice. Recently, tiling path DNA microarrays have made it possible to detect expression within almost any desired portion of the genome in an unbiased and high-throughput manner. Studies in several model genomes using tiling path microarray analysis revealed an abundance of previously unpredicted transcribed regions in each of the chromosomes or genomes investigated (Shoemaker et al., 2001; Kapranov et al., 2002; Rinn et al., 2003; Yamada et al., 2003). Clearly, experimental approaches complementary to computation-based genome annotation are essential for an understanding of genome structures. Because of the presence of large amounts of unfinished sequence data, unusual compositional gradients in genes, and the large size of the rice genome (Wong et al., 2002; Rensink and Buell, 2004), there is even greater need for experimental approaches in rice genome annotation.
A chromosome-scale transcriptional analysis will also expand our knowledge of possible chromosome-level transcriptional regulation. One prominent feature of eukaryotic chromosomes is their organization into heterochromatic and euchromatic regions. Heterochromatin was first distinguished from euchromatin cytologically as more intensely staining nuclear material throughout the cell cycle in Bryophyta (Heitz, 1928). For a long time, heterochromatin was considered a junkyard composed of only noncoding DNA and silent transposons. The cytological appearance of heterochromatin actually reflects a specific chromatin-packaging condition frequently associated with transcriptional dormancy (Hennig, 1999). In most well-studied eukaryotes, heterochromatin is found near centromeres and telomeres. Sequencing of heterochromatic regions revealed the existence of tandem long repeats and transposons (Hennig, 1999). On the other hand, the density of nonredundant protein-coding gene models and the recombination rate are low in heterochromatic regions (CSHL/WUGSC/PEB Arabidopsis Sequencing Consortium, 2000).
The idea that heterochromatin influences the regulation of nearby genes began with the early observation of position effect variegation in Drosophila (Müller 1930). It was further noted that a euchromatic site could become a heterochromatic site in nature under certain conditions (Henikoff and Comai, 1998). Recent genetic studies in plants provide supporting evidence for the chromatin-level regulation of gene expression (Hoekenga et al., 2000; Stam et al., 2002; Scheid et al., 2003). RNA interference has been suggested to be an underlying mechanism that acts by directing DNA and histone modifications to control gene expression (Bender, 2004; Lippman and Martienssen, 2004; Lippman et al., 2004; Matzke and Birchler, 2005). In fact, heterochromatin has emerged as a key regulator in the epigenetic control of gene expression, chromosome behavior, and evolution.
Heterochromatin was systematically investigated in maize (Zea mays) by chromosome staining (McClintock, 1929). Recent cytological studies have revealed dynamic cytological characteristics of plant chromosomes, especially in heterochromatic regions in Arabidopsis thaliana and rice (Fransz et al., 1998; Cheng et al., 2001). However, the functions of these patterns are not fully understood (Avramova, 2002). Different from Arabidopsis, cytologically defined heterochromatic regions of rice cover a significantly larger portion of the pericentric region in the majority of rice chromosomes. For example, approximately half of rice chromosome 4 is characterized as heterochromatic based on its dense staining pattern, including the entire short arm and another ∼9-Mb extension beyond the centromere into the long arm (Cheng et al., 2001). Little if any information is available regarding chromosome-level transcriptional activity regulation in distinct chromatic regions or the functional significance of rice heterochromatin (Cheng et al., 2001). Using tiling path microarray analysis as a tool, it is now possible to perform high-throughput profiling of the transcriptional activities along an entire sequenced chromosome to examine potential connections between transcription and cytologically defined chromatin organization.
In this study, we developed a tiling path DNA microarray consisting of overlapping PCR-amplified genomic fragments covering >33 Mb (95.5%) of japonica rice chromosome 4. Using this array, we analyzed the transcriptional activity of chromosome 4 in six representative organs or tissues. Chromosome-scale transcription patterns were analyzed and compared with cytologically observed chromatin organization and the distribution of transposon-related and various other gene model groups.
RESULTS
Construction of the Rice Chromosome 4 Tiling Microarray
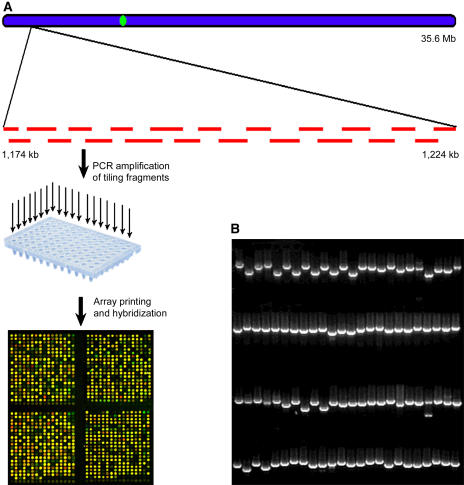
We constructed a minimal tiling path DNA microarray covering essentially the entire rice chromosome 4 (Figure 1A) using the very same DNA subclone fragments from which the finished sequence of this chromosome was obtained (Feng et al., 2002). The selected subclones have some overlaps at the junctions (Figure 1A). This degree of redundancy in coverage has proven beneficial for analytical purposes to increase resolution and to provide repetition (Sun et al., 2003). Each subclone was amplified by PCR using universal primers annealing to the flanking vector sequences, followed by agarose gel analysis to assess DNA fragment purity and abundance (Figure 1B). Importantly, all of the amplified fragments were sequenced from both ends to ensure accuracy. All quality-controlled fragments, together with both negative and positive controls, were printed on an aminosilane-coated glass slide to produce the tiling microarray (see Methods).
Figure 1.
Construction of the Rice Chromosome 4 Tiling DNA Microarray.
(A) A total of 14,742 PCR-amplified overlapping fragments, which were selected to cover the entire chromosome, were printed onto glass slides with negative and positive controls. An image of four subarrays of a sample microarray hybridized with probes originating from seedling shoots labeled with Cy3 and cultured cells labeled with Cy5 is presented. The bottom row of each subarray contains negative control spots.
(B) Quality-control gel image of 96 PCR-amplified fragments from one randomly chosen 96-well plate.
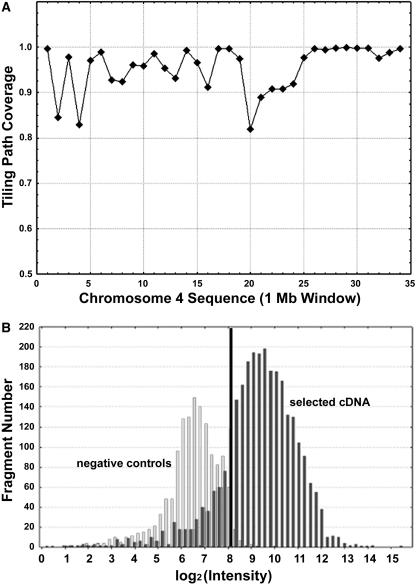
All subclone sequences contained on the microarray were mapped against the updated chromosome 4 sequence (The Institute for Genomic Research [TIGR] release version 2.0, April 2004). Subclones that were either too large or potentially chimeric in nature were flagged and excluded from further analysis (see Methods). The final tiling path consists of 14,742 subclone fragments covering >33 Mb or 95.5% of the chromosome 4 sequence. The average size of the subclone fragments is 3.08 kb, with an average overlap of 718 bp between two neighboring fragments. The average resolution of this microarray is 1.6 kb, considering subclone overlapping. Because of unfinished gaps in the sequence and the absence of suitable subclones, 910 gaps remained in the tiling path that were estimated to represent <4.5% of the chromosome. The average array coverage in 1-Mb windows along the length of the chromosome (ranging from 82 to 100%) is shown in Figure 2A.
Figure 2.
Tiling Array Coverage and Expression Threshold Determination.
(A) Coverage of the tiling path microarray. The coverage was calculated by dividing the tiling path covered region (overlaps removed) by the entire region in 1-Mb windows across rice chromosome 4.
(B) Expression threshold determination. The histogram in light gray shows the distribution of 257 negative control spots in a representative experiment. We selected a cutoff, shown by a black line, at which only 1% of the negative control spots scored as false positives. The distribution in dark gray represents the intensities of 515 selected fragments with cDNA support.
Tiling Array Analysis Provides Expression Support for Most of the Annotated Gene Models of Rice Chromosome 4
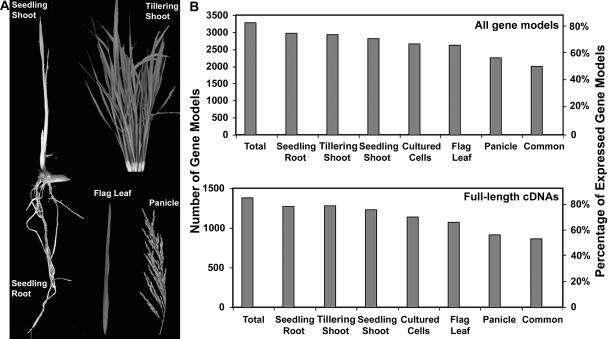
To map the transcribed regions of rice chromosome 4, we collected six representative organs or tissues at different developmental stages, including seedling shoot, seedling root, tillering-stage shoot, heading-stage flag leaf, and heading-stage panicle (Figure 3A), as well as suspension-cultured cells. Such a selection covered key representative organ types from both vegetative and reproductive developmental phases under normal growth conditions. Poly(A)+ RNA from each organ or tissue type was selectively transcribed using an oligodeoxythymidine primer, labeled with cyanine fluorescent dye, and hybridized to the array. Each organ type was represented by at least six experimental repeats using cDNA prepared from three independent biological samples.
Figure 3.
Expression of Chromosome 4 Gene Models in Representative Organs and Cultured Cells.
(A) Photographs of five representative rice organs selected for experimental analysis. Note that these images are not at the same magnification and thus do not reflect relative sizes in real samples.
(B) Number of annotated gene models and full-length cDNA genes for which expression was detected. The number of gene models whose expression was detected in at least one sample set is labeled Total, whereas the number of gene models transcribed in all six sample sets examined is labeled Common.
To objectively identify fragments with detectable expression, we performed a multiple-step statistical analysis. Initial normalization was performed on replicates. This accounts for and lessens the effect of artifacts caused by technical variation (Quackenbush, 2002). An expression threshold with a 1% false-positive rate was determined based on the distribution of the normalized intensities of the negative control spots on each array. All of our positive controls of genomic DNA had signals well above this threshold. We next explored the intensity distributions of 515 selected fragments covering available cDNA sequences. We found that usually >80% of them had expression above this empirical threshold (Figure 2B). Recent studies in Arabidopsis suggest that the detection rate of cDNA in an organ sample ranges from 77 to 84% (Yamada et al., 2003; Redman et al., 2004).
We mapped all currently annotated gene models along chromosome 4 to genomic DNA fragments in the minimal tiling path collection. In total, our tiling path array represents 5464 (96.2%) of the 5682 gene models (including transposon-related models). Expression for 82% (3296) of all 4025 nonredundant protein-coding gene models (excluding transposon-related models) was observed in at least one of the six organs or tissues (Figure 3B; see Supplemental Table 1 online). The expression detection rate in each individual organ or tissue ranged from 56% (panicle) to 73% (seedling root). Expression of ∼50% (2013) of nonredundant protein-coding gene models was detected in all six organs or tissues.
For the 1640 gene models matching available full-length cDNAs (Rice Full-Length cDNA Consortium, 2003), we detected expression of 1383 (86%) gene models in at least one organ or tissue sample (Figure 3B). The panicle sample had the lowest detection rate (57%) of cDNA-supported gene models, whereas seedling shoot and root had the highest rates (80 and 79%, respectively). Again, ∼54% of the cDNA-supported gene models were commonly expressed in all organs or tissues.
The rice genome has a long history of duplication (Blanc and Wolfe, 2004; Paterson et al., 2004; Yu et al., 2005). We used our expression data to estimate and compare the expression rates of duplicated genes and unique genes on chromosome 4. Approximately 19% of all gene models in this chromosome have >80% of their full length similar to another gene and were considered potential duplicated genes. The expression of 76% of those potential duplicated genes was detected in at least one sample. One the other hand, the expression detection rate for genes without any evidence of duplication, which account for 68% of all gene models, was 80%. In agreement with a recent classification of duplicated full-length cDNAs (Yu et al., 2005), we found that 91% of tandem duplicated, 87% of segmental duplicated, and 75% of background duplicated full-length cDNAs had expression detected.
Detection of Transcriptional Activity in the Nonannotated Regions of Rice Chromosome 4
Differing from genomic DNA amplicon microarrays that only represent annotated genes (Jiao et al., 2003; Kim et al., 2003), tiling path microarrays are also useful for detecting novel transcription in nonannotated regions. To this end, we applied the same expression threshold to examine fragments within nonannotated regions in each sample set. These surveys resulted in the detection of transcriptional activity in 1643 nonannotated regions (see Supplemental Table 2 online). Among these 1643 nonannotated regions, 1076 (65%) were commonly detected in all organs or cultured cells. This is similar to the 61% (2013) of the 3296 expressed gene models that were commonly expressed in all six organs and cultured cells.
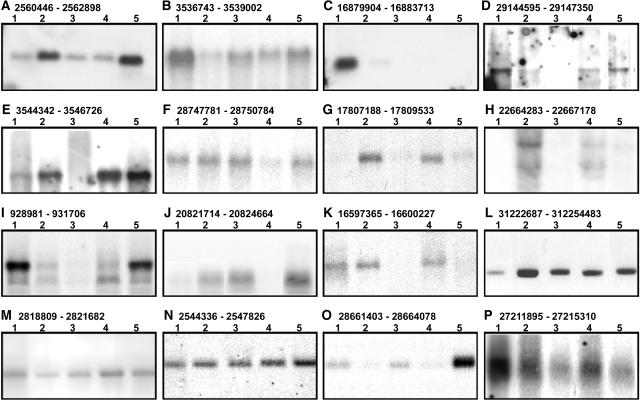
To provide independent experimental support for the transcriptional activity detected in these nonannotated regions, we randomly selected 21 such expressed nonannotated regions for RNA gel blot analysis using the same RNA samples used for microarray analysis. Specific RNA species in 16 of these 21 nonannotated regions were clearly detected in this RNA gel blot analysis (Figure 4). The RNA gel blot signal strengths for most bands were usually consistent with microarray hybridization intensities (data not shown).
Figure 4.
RNA Gel Blot Analysis of 16 Representative Nonannotated DNA Fragments.
The transcripts were detected in 16 nonannotated regions with the chromosome coordinates of each fragment shown at the top of each panel. In total, 21 nonannotated fragments were analyzed, with 16 (76%) showing hybridization signals. The remaining five nonannotated DNA fragments did not present hybridization signals under our conditions. The RNA samples from cultured cells or specific organs subjected to microarray analysis were used for RNA gel blot analysis with the following order, from lane 1 to lane 5: cultured cells, seedling shoot, seedling root, flag leaf, and panicle.
Organ-Specific Transcriptional Profiles Reflect Their Developmental and Physiological Characteristics
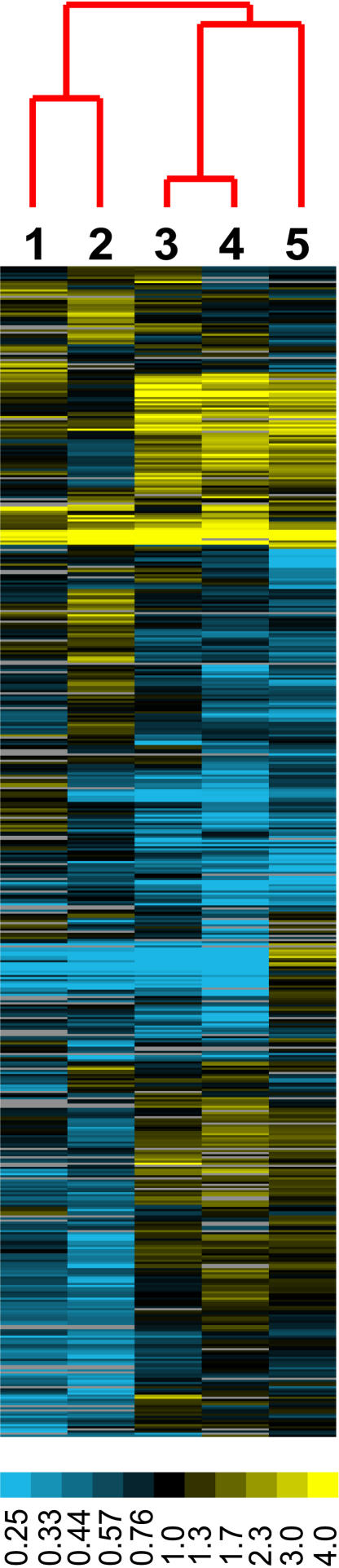
To examine the organ-specific transcriptional activities of chromosome 4, we compared the transcriptional profiles of five selected organs using cultured cells as a common control (Figure 3A). Unsupervised hierarchical clustering of all of the fragments with differential expression revealed the transcriptional similarities among different organs (Figure 5). This analysis indicated that seedling and tillering-stage shoots were closer to each other. Flag leaves and panicles, both at the heading stage, shared more similar transcription profiles. Roots exhibited a transcriptional profile quite distinct from those of other organ types. Each organ also contains a unique set of specifically enriched transcripts located on chromosome 4, as shown in Figure 5. These results suggest that it is plausible that a relatively small number of specifically expressed or enriched genes in each organ define its developmental and physiological characteristics.
Figure 5.
Cluster Display of Differentially Expressed Genomic Fragments in Distinct Organs.
Centroid linkage hierarchical clustering was performed on transcription ratios of each organ versus cultured cells. Lane 1, panicle; lane 2, flag leaf; lane 3, tillering shoot; lane 4, seedling shoot; lane 5, seedling root. Only those gene models that exhibited differential expression among five organ samples were included. Differential expression was determined by the analysis of variance F test with P < 0.001. Each fragment must have expression detected in at least one organ sample. A total of 1915 fragments were included in this cluster analysis. Yellow indicates high levels of expression in a specific organ relative to cultured cells; blue indicates low levels compared with cultured cells; and gray indicates missing data. The dendrogram shows the relationship among organs based on expression.
Possible Developmental Regulation of Transcription at the Chromosomal Scale
To examine transcriptional activity along the chromosome, we calculated the average intensity of all fragments in each 100-kb window for each tissue type (Figure 6). For this analysis, fragments covering transposon-related gene models or highly repetitive sequences were excluded. From Figure 6A, it is evident that the transcriptional activity along the chromosome is uneven and subject to developmental regulation. All samples from juvenile stages had stronger expression in the distal portion of the long arm.
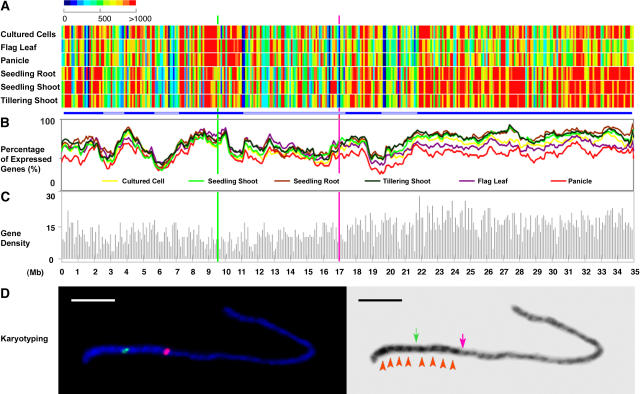
Figure 6.
Chromosomal Transcriptional Analysis of the Nonredundant Protein-Coding Gene Models of Rice Chromosome 4.
(A) Average expression intensities of transcribed fragments in each 100-kb window along the chromosome in each organ or cultured cells. Bars at bottom highlight chromosomal domains based on expression.
(B) Percentage of expressed gene models in a 100-kb window along the chromosome in each organ or cultured cells.
(C) Annotated nonredundant protein-coding gene model density along the chromosome in a 100-kb window.
In (A) to (C), the position of the centromere is represented by a green line, whereas the last major knob of the heterochromatin half of chromosome 4 (as defined in [D] below) is shown by a pink line.
(D) Karyotyping of rice pachytene chromosome 4 and positioning of the euchromatin and heterochromatin border region. Left, chromosome fluorescence in situ hybridization using the centromeric probe CentO (green signal) and a BAC clone, OSJNBa0034E24 (pink signal), located at 16.9 Mb. This BAC clone is located within the last major knob of the heterochromatin half of chromosome 4 and thus is close to the border of the heterochromatin and euchromatin domains. Right, the 4′6-diamidino-2-phenylindole dihydrochloride–stained chromosome 4 at left was converted to a black-and–white image to enhance the visualization of the distribution of euchromatin and heterochromatin. The centromere and euchromatin/heterochromatin border region are indicated by green and pink arrows, respectively. Eight heterochromatin knobs are highlighted by red arrowheads. Bar = 5 μm.
To correlate chromosome transcriptional activity with chromosome organization, a fluorescence in situ hybridization analysis of chromosome 4 was performed using the centromere-specific repeat sequence and a BAC clone located at position 16.9 Mb (Figure 6D). The fluorescence in situ hybridization image shown in Figure 6D clearly indicates that this BAC clone is located within the last of the eight major heterochromatin knobs, thus ∼0.5 to 2 Mb away from the bordering region of the heterochromatin and euchromatin of chromosome 4.
Interestingly, this defined separation of heterochromatin and euchromatin correlates with a general stronger transcriptional activity of juvenile-stage rice tissues in the euchromatin half (Figure 6A). Although the transcriptional profiles for root were quite distinct from those for shoot (Figure 5), they both had strong transcription in euchromatin. On the other hand, samples from reproductive-stage flag leaf and panicle had relatively weaker transcription in euchromatin but greater transcription in parts of the heterochromatic regions. These two reproductive-stage organs had strong transcription near the centromere region, which is located at ∼9.6 Mb (Zhang et al., 2004) (Figure 6D). Compared with the other samples, cultured cells had more uniform transcriptional activities along the chromosome.
We also examined the percentage of transcribed fragments among fragments with annotated gene models in 100-kb windows along the chromosome (Figure 6B). We noted that reproductive-stage panicle and flag leaf had fewer gene models transcribed in euchromatic regions from 18 Mb to the distal end (Figure 6D). In heterochromatic regions, there was generally no obvious difference in the percentage of transcribed fragments among different organs or cultured cells. One distinct exception was the first 2-Mb region at the distal end of the short arm, where juvenile samples, together with cultured cells, had stronger transcription than reproductive samples. In fact, the transcription properties of this region were quite similar to those of euchromatic regions in the long arm. Several other domains within heterochromatin show similar transcriptional profiles that are distinct from those of their neighboring regions. These short domains are usually 1 to several Mb in size, as emphasized by the bars in Figure 6A. In the region flanking the centromere, slightly stronger average transcriptional activity was detected in the two reproductive-stage samples, whereas the percentages of transcribed gene models were close among all samples. These patterns suggest slightly higher transcriptional activity of gene models in the 1-Mb region toward the centromere in reproductive flag leaf and panicle. Chromosomal domains with similar transcriptional profiles between different organs and cultured cells are highlighted by bars in Figure 6A. It is interesting that this discontinuous pattern of transcriptional activity in the heterochromatin half of chromosome 4 is somewhat reflected in its uneven cytological staining pattern. Both previous studies (Cheng et al., 2001) and the data in Figure 6D suggest that the heterochromatin half of chromosome 4 is composed of eight intensely stained knobs (four on each side of the centromere) with relatively weakly staining gaps.
We further examined the annotated gene model density along the chromosome and found that euchromatic regions generally have more nonredundant protein-coding gene models (excluding transposon-related models) than heterochromatic regions (Figure 6C). However, no general correlation between gene model density and transcription activity seems to exist in all samples examined.
Rice Chromosome 4 Gene Models with Significant Homology with Arabidopsis Genes Are Enriched in Euchromatic Regions
It has been reported that a large fraction of rice gene models are not significantly homologous at the sequence level with Arabidopsis genes (Rice Chromosome 10 Sequencing Consortium, 2003; Rice Full-Length cDNA Consortium, 2003). In principle, those rice gene models lacking significant homology with Arabidopsis genes may represent fast-evolving genes. Alternatively, the majority of this group of less conserved gene models may represent highly diverged transposon-related sequences and may not be real genes (Bennetzen et al., 2004). Therefore, grouping rice gene models based on their homology with the Arabidopsis genome may also separate gene models with high confidence from potentially misannotated models.
To this end, we compared all gene models of rice chromosome 4 against the Arabidopsis genome. An expectation value cutoff of 10−7 was used for the homology search (see Methods for details). Based on these criteria, 3166 rice chromosome 4 gene models exhibited significant sequence homology with Arabidopsis genes and were named high-homology (HH) gene models. The rest of the 2516 gene models lacked significant homologous counterparts in the Arabidopsis genome and were defined as low- (or no-) homology (LH) gene models.
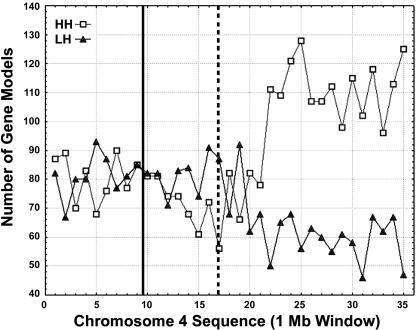
Figure 7 illustrates the chromosomal distribution of nonredundant HH and LH gene models along the chromosome by the density of each group. In the first half of the chromosome, which is mostly cytologically defined as heterochromatin, the densities of HH and LH gene models are quite similar. This part includes the entire short arm and the proximate portion of the long arm, for a total length of ∼19 Mb. In the rest of this chromosome (from 19 Mb toward the distal end of the chromosome), we found a conspicuous enrichment of HH gene models, with twice as many HH gene models present. The HH gene model density shows a dramatic increase from ∼80 gene models/Mb to ∼110 gene models/Mb, whereas the density for LH gene models exhibits a clear reduction of ∼25 gene models/Mb in euchromatic regions compared with heterochromatic regions.
Figure 7.
Density Distribution of HH and LH Gene Models Along Rice Chromosome 4 in a 1-Mb Window.
HH and LH gene models were defined by homology search with all Arabidopsis gene models using tBLASTN (for details, see Methods). Solid line, centromere; dotted line, the last major knob of the chromosome 4 heterochromatin half.
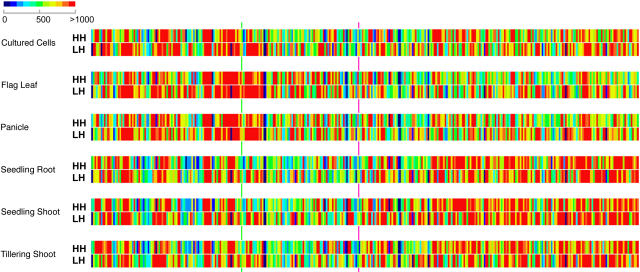
The expression of both nonredundant HH gene models and nonredundant LH gene models followed a similar pattern for all gene models as a whole. Both groups of gene models exhibited higher expression in euchromatin from the juvenile stages and increased expression in some domains in heterochromatin at the reproductive stage (Figure 8). In general, we did not find an obvious pattern specific for HH or LH gene models in expression at 100-kb resolution along the chromosome. A few short regions (<1 Mb) in the heterochromatic portion of the chromosome had stronger average transcription for LH gene models than for HH gene models. This observation in heterochromatin suggests that a fraction of LH gene models may in fact correspond to transposon-related elements, which were misannotated as nonredundant gene models. The differences in transcriptional strength are more distinct for reproductive-stage samples and cultured cells than for juvenile samples.
Figure 8.
Average Expression Levels of HH and LH Gene Models Along the Chromosome in Each Organ or Cultured Cells.
Average expression intensities of fragments with HH gene models and fragments with LH gene models were calculated separately. Fragments with annotated transposon-related gene models were excluded. All features were calculated from 100-kb windows across the chromosome. Green line, centromere; pink line, the last major knob of the chromosome 4 heterochromatin half.
General Increase of Transcriptional Activity of Transposon-Related Gene Models in the Reproductive Stage
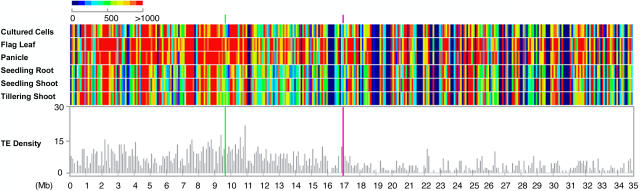
There are 1501 gene models on chromosome 4 annotated by TIGR as transposon-related. By comparing the average transcriptional activity of only those fragments covering transposon-related gene models in each 100-kb window along the chromosome, we found much weaker average transcription for them than for those fragments containing nonredundant protein-coding gene models (cf. Figures 6 and 9). On the other hand, a conspicuous increase in transposon-related gene model expression in reproductive-stage organs was detected (Figure 9). The transcriptional activity increase of transposon-related elements in flag leaf and panicle was dramatic in heterochromatin but was also observed in limited regions in euchromatin. Cultured cells also showed a stronger transcription of transposon-related models than juvenile-stage samples, although not as strong as those in the reproductive-stage flag leaf and panicle. This increased transcription in cultured cells was restricted to heterochromatin only. Both shoots and roots from juvenile stages showed low transcription of transposon-related models, which can be visualized by the dominant cold colors in Figure 9.
Figure 9.
Average Expression Levels of Transposon-Related Gene Model–Containing Fragments Along the Chromosome.
Average expression intensities in each sample as well as transposon (TE)-related gene model densities were calculated from 100-kb windows across the chromosome. Green line, centromere; pink line, the last major knob of the chromosome 4 heterochromatin half.
The stronger transcription of transposon-related gene models in heterochromatin than in euchromatin was also noticeable in all samples examined. Even though our measurement of the average transcriptional activity was independent of probe density, we found that regions with higher transposon-related element density, which are heterochromatic regions, generally had a higher average transcription of transposon-related models. Such higher transcription of more densely distributed transposable elements in heterochromatin was consistent in all of the samples profiled (Figure 9).
To further dissect transposon-related gene models on chromosome 4, we separated out retrotransposon gene models and DNA transposon gene models (Mao et al., 2000; Turcotte et al., 2001). A total of 1147 transposon-related gene models were classified as retrotransposons, and 344 were classified as DNA transposons. Among the retrotransposons, the gypsy-like subclass was the dominant group, with 505 members. Another 124 retrotransposon-related models matched the copia-like subclass. A total of 285 DNA transposon-related models matched the En/Spm superfamily.
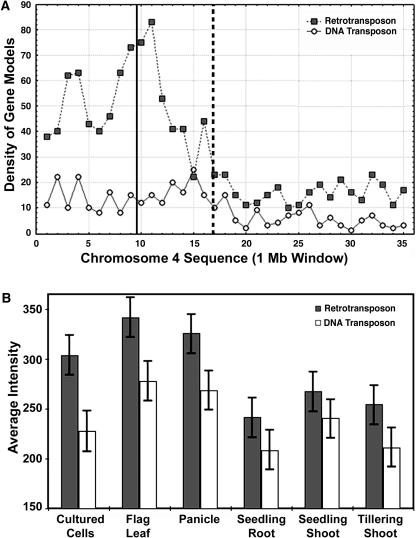
Although there are clearly more retrotransposon-related gene models in heterochromatin than in euchromatin, the DNA transposon gene models show less difference in distribution along the chromosome (Figure 10A). In euchromatic regions, retrotransposon gene models have a density of ∼15 gene models/Mb. The density of retrotransposon models has a threefold increase to ∼50 gene models/Mb in heterochromatin. The average density of DNA transposons increases from <10 gene models/Mb in euchromatin to ∼15 gene models/Mb in heterochromatin. Our result is consistent with the previously reported active transcription of retrotransposons in grass genomes by EST analysis (Vicient et al., 2001).
Figure 10.
Chromosomal Distribution of Two General Types of Transposon-Related Gene Models and Their Average Expression among Distinct Organs.
(A) Density distribution of transposon-related gene models. Transposon-related gene model classification was based on gene annotation from TIGR. The density was calculated from 1-Mb windows across the chromosome. Solid line, centromere; dotted line, the last major knob of the chromosome 4 heterochromatin half.
(B) Average expression intensities of retrotransposon-related gene models and DNA transposon-related gene models in each organ or cultured cells.
Comparison of the transcription of DNA transposon gene models and retrotransposon gene models suggests that retrotransposon models generally have stronger transcriptional activity than DNA transposon models (Figure 10B). This difference is more evident in the reproductive-stage flag leaf and panicle. Cultured cells show the most distinct difference in transcription between retrotransposon models and DNA transposon models. No obvious difference was found among the above-mentioned major DNA transposon subclasses or among retrotransposon subclasses.
DISCUSSION
In this study, we used a genomic DNA fragment-based tiling path microarray to provide transcriptional activity profiles of rice chromosome 4 in representative rice organs or tissues. As chromosome 4 was one of the first three completely sequenced rice chromosomes, experimental analysis of its annotation could provide a valuable resource for improvements to in silico gene annotation in rice. The chromosome-wide transcriptional analysis presented here also provides insights into the relationship between transcriptional activity and chromosome organization.
A Minimal Tiling Path Microarray to Represent the Entire Chromosome
As an extension of such PCR-based genomic DNA amplicon microarrays (Kim et al., 2003), which cover only annotated coding regions, a minimal tiling path microarray was used to survey global transcriptional activity. A minimal tiling path microarray has several advantages. First, its essentially full-chromosome coverage has the potential to detect novel sites of transcription, irrespective of available annotation information. Second, by using the same DNA fragments used for sequencing, it is possible to amplify the entire chromosome in an efficient and economical way using only universal primer pairs with high throughput and reproducibility. This tiling path microarray construction strategy was recently implemented to study transcription of the human genome at a small scale in a 1.7-Mb region (Li et al., 2004). Third, tiling path microarray analysis also has the potential to be used in other applications on a genome scale. For example, global protein–DNA binding and DNA modification sites can be mapped by a tiling path microarray (Sun et al., 2003).
However, there are also limitations to the minimal tiling path microarray. The resolution of this array is not adequate to examine the structure of a single gene model. Indeed, 19% of the gene models studied were represented by subclones that covered more than one gene model, although many of these were within clusters of short gene models. Cross-hybridization is a common and inherent problem of microarray analysis and for other DNA hybridization-based methods (Held et al., 2003). Both of these situations clearly affected the accuracy of our assessment to some extent. However, comparisons of average transcriptional activity for each 100-kb window overcame this weakness to some extent (Figure 6). To test the potential cross-hybridization caused by such short repetitive sequences with a high copy number in the genome as miniature inverted repeat transposon-related elements (MITEs) (Feng et al., 2002), we examined all nonannotated fragments used for RNA gel blot analysis and found nine of those fragments with MITEs. None of them showed a smear on the RNA gel blot, regardless of whether the main bands were detected (Figure 4). Among our 21 randomly selected nonannotated fragments, 7 of them also included simple repeats, such as (CAG)n. Some of these simple repeats have hundreds of copies in the genome. Again, RNA gel blot results did not suggest a cross-hybridization problem, because no smear was detected on RNA gel blots. Therefore, cross-hybridization may not be a major problem. However, considering the technical difference between RNA gel blot and microarray analyses, it is not feasible to estimate the exact extent of cross-hybridization in our microarray analysis.
Repetitive sequences without coding capacity did not significantly affect the hybridization of reverse-transcribed probes to the microarray or even to RNA directly (Figure 4). Although many transposons exist in copy numbers of only a few per genome and exhibit homology only to a low degree (Bennetzen et al., 2004), we cannot rule out the potential cross-hybridization among certain transposon-related families, specifically transcribed retrotransposons, and DNA transposons. The separation of transposon-related gene model–containing fragments from all other fragments largely isolated this problem to only transposon-related gene models. A sequence similarity search suggests that ∼60% of the transposon-related gene models have at least one other such gene model with high similarity (>80% at the nucleotide level) in the entire rice genome. A similar problem could also happen to duplicated genes and to nonannotated regions. Close to 19% of all gene models are considered potential duplicated genes. Interestingly, these potential duplicated genes do not exhibit stronger transcription than unique genes. We believe that portions of these duplicates, especially background duplicates, are actually pseudogenes. It has been shown that DNA sequences with identity above 80% exhibit detectable cross-hybridization (Hughes et al., 2001); therefore, when interpreting the hybridization results for transposon-related gene models, we should be cautious about this cross-hybridization effect.
Microarray Analysis Provides an Important Complement to Computational Annotation
We demonstrated that >81% of computationally annotated gene models have transcriptional activity. As a control, our detection rate of full-length cDNA-supported gene models was 86%. Recently, it was suggested that a significant portion of annotated rice-specific gene models might be transposon-related, and some of them may even be expressed (Bennetzen et al., 2004; Jabbari et al., 2004; Jiang et al., 2004). If this is the case, our tiling microarray analysis will be able to detect the expression of these transposon-related gene models (Bennetzen et al., 2004), although detection of expression would not directly imply a functional role of those gene models.
In addition to the annotated regions, we also identified transcriptional activities in 1643 nonannotated regions. Among them, 1076 had transcriptional activities in all samples. Using RNA gel blot analysis, we detected 16 of 21 hybridizing fragments. Those regions not detected by RNA gel blot analysis usually had a low abundance of transcripts based on microarray data. Importantly, in no case did we detect any smear or other minor bands, which would indicate cross-hybridization caused by possible repetitive sequences. However, we may still not be able to rule out cross-hybridization in the detected nonannotated transcription activities.
These novel nonannotated transcription activities may arise from a variety of situations. The first possibility is that some of the transcripts come from missed genes in nonannotated regions. These transcripts can include protein-coding genes and non-protein-coding RNAs with structural, catalytic, or regulatory capacity. These regions may also encode transposons that are transcriptionally active but lack homology with other transposons (Bennetzen et al., 2004). Alternatively, some of these novel transcripts might be missing parts of a neighboring annotated gene model. It has been reported that although computational annotation algorithms have high reliability for locating a gene model in the chromosome region, they are less reliable in predicting the precise structure of that gene (Zhang, 2002). For example, an exon can be missed or the 3′ or 5′ noncoding regions not as accurately identified as the coding regions. The resolution limitation of this microarray, however, prohibited us from further distinguishing these possibilities.
Chromosome-Level Regulation of Transcriptional Activity
The eukaryotic chromosome is organized into two forms, heterochromatin and euchromatin, which are defined by their cytological staining patterns. Recent sequencing of heterochromatin in Arabidopsis and rice successfully integrated cytological features and sequence data (CSHL/WUGSC/PEB Arabidopsis Sequencing Consortium, 2000; Feng et al., 2002; Sasaki et al., 2002; Rice Chromosome 10 Sequencing Consortium, 2003), providing new insights into this cytological structure. It has been suggested that Arabidopsis heterochromatic regions are determined by transposon-related elements and that their assembly is associated with chromatin remodeling, including DNA methylation, histone methylation, small interfering RNAs, and DNA replication (Soppe et al., 2002; Lippman et al., 2004).
Kim et al. (2003) have observed an association between chromosome organization and expression using microarrays in Arabidopsis chromosome 2. However, limited cytologically defined heterochromatin in the Arabidopsis chromosome makes it difficult to extend the learned relationship between chromatin features and transcription in Arabidopsis to rice, because the latter is dramatically different from Arabidopsis in its chromosome organization.
Using rice chromosome 4 as a case study, we systematically explored the molecular features and transcriptional activities of heterochromatin in rice. Integration of the cytological map with sequence data and the physical map (Chen et al., 2002; Feng et al., 2002; Zhao et al., 2002) suggests that an ∼17-Mb region starting from the telomere of the short arm is heterochromatic (Figure 6D). We found a high density of retrotransposon gene models in this heterochromatic region (Figure 10A). The density of DNA transposon gene models was also increased slightly in heterochromatin but not in euchromatin.
Profiling of the transcriptional activities of nonredundant protein-coding gene models showed that rice heterochromatin also has many transcriptionally active gene models embedded within it, although at a lower density. In spite of the fact that half of rice chromosome 4 is cytologically defined heterochromatin (Cheng et al., 2001), it is quite possible that the transcriptionally repressed regions are scattered among transcriptionally active gene models (Lamond and Earnshaw, 1998). Therefore, it is likely that the microscopically scattered, rather than continuous, heterochromatin regions on the short arm, and also part of the neighboring long arm region, form the cytological heterochromatin. Lippman et al. (2004) recently showed in Arabidopsis that transcriptionally active regions can indeed lie within small islands of transcriptionally repressed domains in heterochromatin. Furthermore, the heterochromatic region has a similar density of HH and LH gene models, whereas euchromatin has a much higher density of HH gene models than LH gene models (Figure 7). However, the causal relationship between HH/LH gene model distribution and heterochromatin and euchromatin division is not clear. We speculate that chromatin structure may play a global regulatory role in the transcription of nonredundant protein-coding gene models during development (Figures 6 and 8). Euchromatin showed active transcription in all juvenile-stage samples but a generally weaker transcription in all reproductive-stage samples. Heterochromatin showed relatively weak transcription in juvenile samples, but the transcription of a domain in heterochromatin is noticeably stronger at the reproductive stage. In reproductive-stage samples, the transcriptional activity in most of the heterochromatin is similar to that in euchromatin. Cultured cells as undifferentiated cell types show relatively more uniform transcriptional activity along the entire chromosome (Figure 6). Previous studies have shown the effect of chromatin structures on gene transcription in a wide range of plants, including monocots and dicots (Mlynárová et al., 1994; Tikhonov et al., 2000; Rudd et al., 2004), but our data extended this possible regulation to the chromosomal scale and tentatively associated this regulation with the heterochromatin and euchromatin structures. The underlying mechanisms of those regulations are not known, but modification at the DNA or histone level or RNA interference are possible routes (Henikoff and Comai, 1998; Pandey et al., 2002; Reyes et al., 2002; Lippman and Martienssen, 2004). Contrary to the nontransposon gene models, an increase in transcriptional activities of transposon-related elements in reproductive-stage organs, especially those located in heterochromatic regions, was observed (Figure 9). Cultured cells showed medium transcriptional activity between reproductive-stage and juvenile-stage organs. A very recent study has suggested that certain retrotransposon RNA may interact with chromatin structure to form complexes with the potential to maintain the chromatin functions in maize (Topp et al., 2004). In Arabidopsis, Lippman and colleagues (2004) demonstrated that DNA methylation and histone modification correlate with the activation of transposon-related gene models using a tiling path microarray for a heterochromatic region. It is quite possible that certain rice retrotransposon RNA species may also perform some functions to maintain the structure of chromatin, and rice also uses DNA methylation and histone modification to control the activities of transposon-related elements.
Thus, it is reasonable to speculate that the organization of the chromosome into heterochromatic and euchromatic regions may enable rice to have another level of regulation at the chromosome level. This regulation could be applied to both nonredundant protein-coding gene models and to transposon-related gene models. For both groups of gene models, it seems that the developmental phase, rather than the organ identity, may be the signal for the chromosome-level regulation of transcription. This type of developmental regulation at the chromosome level would be consistent with the reported developmental regulation of heterochromatin assembly and activity (Preuss, 1999; Meyer, 2000; Ahmad and Henikoff, 2001).
An increasing body of evidence has shown that transposon-related elements contribute to the evolution of a genome (Feschotte et al., 2002). Studies of the current genome sequence structure in maize have suggested that transposon-related elements could rapidly restructure a genome (SanMiguel et al., 1998). Our observation of the active expression of transposon-related gene models in reproductive stages suggests that this process may occur more frequently at the mature stage, or the end of the life cycle, in a rice plant. Although the movement of transposon-related elements can provide resources for selection during evolution, they are more likely to cause malfunction of useful genes. Activation of transposon-related elements, especially retrotransposons, only at the mature stage seems to be a solution for balancing both survival and the need for new gene creation.
METHODS
Collection of the Minimal Tiling Path Fragments of Rice Chromosome 4
The minimal tiling path was selected from sequenced subclones of chromosome 4 of rice (Oryza sativa ssp japonica cv Nipponbare). We selected subclones using the assembled sequences of BACs or P1-derived artificial chromosomes (PACs). All subclones were derived from libraries constructed from sheared BAC or PAC DNA and ligated into pBluescript vectors (Stratagene, La Jolla, CA). To reach a fine tiling coverage of the chromosome, we selected a single set of sequenced clones based on minimal overlaps between fragments. To this end, we initially determined the positions of the shotgun subclones within the regions that were defined by BACs and PACs (Zhao et al., 2002). Subclones at the overlapping regions between BACs and PACs were further streamlined to minimize redundancy. A minimal tiling path was then calculated to minimize the overlapped subclones (Figure 1A). The selection also tried to keep a uniform size and to have a minimal presence of repetitive sequences.
A chromosome 4 pseudomolecule, computational gene models, and their predicted transcripts (version 2.0, April, 2004) were downloaded from the TIGR rice genome database (Yuan et al., 2003; http://www.tigr.org/tdb/e2k1/osa1/). Full-length cDNA sequences were downloaded from the Knowledge-Based Oryza Molecular Biology Encyclopedia database (Rice Full-Length cDNA Consortium, 2003; http://cdna01.dna.affrc.go.jp/cDNA/). Subclone fragments and gene model transcripts were remapped to the pseudomolecule using BLAT (Kent, 2002). Fragments longer than 5 kb or containing sequence gaps were removed. In the final count, 14,742 fragments were successfully amplified and used in the tiling path microarray. The expression of each gene model was represented by the expression of the fragment with the longest overlap. Overlap calculation was based on BLAT results of gene models and fragment locations on the chromosome pseudomolecule. In general, fragments covering more than one gene model were not used unless they occurred in the following situations: (1) one gene model occupied the predominant portion of the sequence and it did not have any unique genomic fragment by itself; and (2) the other gene model took up only a small region of the fragment and its expression, as judged from the other overlapping fragment, was at a much lower level. In total, 845 fragments fit these criteria and were used for the calculation of expression.
Construction of the Tiling Path Microarray
Selected subclone fragments were amplified by PCR using subclone plasmid DNA as a template. We performed PCR using TaKaRa LA Taq and TaKaRa Ex Taq kits (TaKaRa, Dalian, China) with 0.02 nM of one of the following common primer pairs (LAS2, 5′-CCCAGTCACGACGTTGTAAAACGACGGCCAGTGCC-3′, and LAS4, 5′-GAATTGTGAGCGGATAACAATTTCACACAGGAAAC-3′; LAF, 5′-CCCTCGAGGTCGACGGTATCGATAAGCTTGATATC-3′, and LARb, 5′-GTAATACGACTCACTATAGGGCGAATTGGAGCTCC-3′; S2, 5′-CGTTGTAAAACGACGGCCAG-3′, and S4, 5′-CGGATAACAATTTCACACAG-3′; and F, 5′-CCTCGAGGTCGACGGTATCG-3′, and Rb, 5′-AATACGACTCACTATAGGGC-3′) and 5 ng of plasmid DNA. PCR amplicons were purified by ethanol precipitation. We resuspended purified PCR products in water and ran an equal amount of sample from each fragment on an agarose gel for quality control. Fragments that migrated as a single band of the predicted size and also had a DNA concentration >100 ng/μL were used for microarray printing.
Arabidopsis Functional Genomics Consortium microarray control sets were used as negative controls. These 18 controls were selected as showing no cross-hybridization with Arabidopsis thaliana RNA (http://www.arabidopsis.org/links/microarrays.jsp). After sequence comparison with rice and pilot experiments, we removed two additional controls with potential cross-hybridization. Each of the remaining 16 distinct controls was repeated 16 times for array printing.
For microarray printing, we combined the resuspended PCR fragments with DMSO (1:1) and transferred 8 μL of each sample to 384-well printing source plates (Whatman, Clifton, NJ). All 256 negative control DNA fragments and 16 rice genomic DNA samples were also resuspended in printing solution and transferred onto printing plates. All DNA fragments were arrayed onto Corning (Corning, NY) GAPS slides using a VersArray ChipWriter Pro system (Bio-Rad, Hercules, CA). Printed slides were allowed to dry at room temperature and cross-linked at 150 mJ in a Stratalinker (Stratagene). Print quality was confirmed by staining for total DNA with POPO-3 (Molecular Probes, Eugene, OR).
Plant Materials
The rice strain used for all experiments was japonica cv Nipponbare. Seeds were baked at 42°C for 3 d to break seed dormancy and then spread in water at 26 to 28°C. We collected 7-d-old seedling shoots and roots (Figure 5A) from normal light-grown plants. Two-week-old seedlings were transferred into soil in controlled-environment chambers (26 to 28°C, 13-h-light/11-h-dark light cycle). Tillering-stage shoots (with four tillers), heading-stage flag leaves, and heading-stage panicles were collected from those plants with normal growth. Suspension cell cultures were also derived from the same rice strain using previously described protocols (Yu et al., 1991).
Preparation of Labeled cDNA Probes and Microarray Hybridization Conditions
Plant materials were frozen in liquid nitrogen and ground to powder using a chilled mortar and pestle. Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) and purified using the RNeasy kit (Qiagen, Valencia, CA). Oligo(dT) was used to selectively synthesize and label cDNA from poly(A)+ mRNA. The probe-labeling protocols used for this study were modified from those used for EST microarrays (Ma et al., 2001). Total RNA (100 μg) was labeled by direct incorporation of amino-allyl–modified dUTP (Sigma-Aldrich, St. Louis, MO) during reverse transcription. After reverse transcription, template RNA was degraded. The amino-allyl–modified dUTP–labeled cDNAs were purified using a Microcon YM-30 filter (Millipore, Billerica, MA) and resuspended in 0.1 M NaHCO3. The cDNA probe was further fluorescently labeled by conjugating the monofunctional Cy3 or Cy5 dye (Amersham, Piscataway, NJ) to the amino-allyl functional groups. After coupling at room temperature for 45 to 60 min, the labeling reaction was stopped by ethanolamine. The fluorescent dye–labeled probe was separated from unincorporated monofunctional dye and concentrated to a final volume of 7 μL for hybridization using a Microcon YM-30 filter. Microarray hybridization, microarray slide washing, and array scanning were performed as described previously (Ma et al., 2001).
Microarray Experimental Design and Dye-Effect Assessment
We followed the reference design for microarray experiments (Clarke and Kempson, 1997; Wu et al., 2003). A suspension-cultured cell RNA sample was used as the reference sample, and all comparisons were made between an organ sample and the reference sample with the same direction of dye labeling. Each organ sample was collected from multiple plants, and three independent biological replicates were collected. Amplified cDNA was prepared twice from each RNA sample. Thus, six quality data sets from three independent RNA samples were obtained for each organ type. Pooled suspension-cultured cell RNA was used as a reference sample in all hybridizations. We selected suspension-cultured cells as the reference because it is an undifferentiated cell type and expresses a relatively high percentage of its genome.
We started with a set of pilot experiments to check the dye effect on our microarray data quality using seedling shoot and cultured cell RNA samples on this tiling array. Each RNA sample was labeled with Cy3 or Cy5. Seedling shoot RNA and cultured cell RNA samples labeled with different dyes were paired together and hybridized to two identical slides to obtain two repeats, with the only difference being the labeling direction. This step was repeated three times (with the three independent RNA samples) to provide three repeats labeled in one direction and three in the reverse direction. We calculated the correlation coefficient of logarithm-transformed raw intensities between each pair of intersample repeats before any correction or normalization. As shown in Supplemental Figure 1 online, we found that the correlations between repeats labeled with the same dye did not exhibit significant differences in data reproducibility compared with repeats labeled with different dyes. This relatively low dye effect is likely attributable to the fact that we used the amino-allyl dye-coupling method for labeling to reduce the incorporation bias of the two dyes (Lee et al., 2004). Based on this initial study, we chose to use the Cy5 dye for all organ sample labeling, whereas the reference sample was labeled with Cy3 dye.
Microarray Data Collection and Initial Normalization
Hybridized microarray slides were scanned with a GenePix 4000B scanner (Axon, Union City, CA), and TIFF images were initially processed using GenePix Pro 3.0 software. Spots with unusual morphology or high background were manually removed from further analysis. All spots with foreground intensity less than two times the background sd in both channels were also removed. Median foreground minus median background intensities were used for subsequent steps.
To identify and remove systematic sources of variation, including dye effects and spatial effects, we used the lowess normalization method for each print-tip group on each slide with the MAANOVA package for R (Yang et al., 2002; Wu et al., 2003). Normalized log2-transformed ratios of signal intensities had a median of zero.
Differential Expression Analysis
To identify differentially expressed spots among five organs, we fitted normalized replicate intensities of all organs together with cultured cell controls into an analysis of variance model. For each data set, the spot intensities of both organ and reference cultured cells (Cy3 and Cy5 channels) were used. The model is given by yijkl = μ + Ai + Dj + ADij + Sl + VSkl + DSjl + ASil + ɛijkl, where yijkl denotes the logarithm-transformed signal for spot l on slide i labeled with dye j of sample k. The overall mean effect was represented by μ; A, D, and S represent main effects from array, dye, and spot, respectively. The interaction terms AD, VS, DS, and AS represent array by dye, sample (variation) by spot, dye by spot, and array by spot, respectively. The random error is denoted by ɛijkl. We were interested in the term VS. The analysis of variance described above was performed on each spot using MAANOVA for R with F statistics computed on the James-Stein shrinkage estimates of the error variance (Kerr et al., 2000; Wu et al., 2003). The false-discovery rate controlling method (Benjamini and Hochberg, 1995) was used to control for multiple testing errors through a function included in MAANOVA. We selected spots with P < 0.001 in the F test after false-discovery rate adjustment as differentially expressed. To further examine these selected differentially expressed spots, the log2-transformed expression ratios between each organ sample and cultured cells were clustered using the CLUSTER program with an unsupervised centroid linkage hierarchical clustering algorithm (Eisen et al., 1998).
Determination of the Expression Threshold
The threshold for the detection of expression was determined for each hybridization repeat by constructing a distribution of normalized intensities obtained by considering the negative control spots. We found an intensity cutoff with a false-positive rate of 1% using Student's t test on log10-transferred intensities. All positive control spots of genomic DNA on each slide had expression above this cutoff. We also selected 515 fragments containing cDNA to further estimate type II errors. We found ∼80% of them had intensities above this cutoff in most samples (Figure 2B). Recent studies in Arabidopsis suggest that the detection rate of cDNA in an organ sample ranges from 77 to 84% (Yamada et al., 2003; Redman et al., 2004). Thus, such a 1% false-positive rate will also give a reasonable false-rejection (type II error) rate base to the experimental information described above. Spots with an expression signal above the cutoff in four of six data sets for the same organ were considered expressed in that organ type.
Estimation of Average Intensities for Each Fragment (Spot)
For the display of transcriptional activities along the chromosome, intensities of individual replicates for all samples were further normalized via quantile normalization (Bolstad et al., 2003). After this normalization, intensities of all replicates representing different organ or cultured cell samples reached a common median. All normalized intensities for each expressed spot were then averaged among all replicates of the same sample to obtain a single statistic, which was used to perform subsequent analyses of expression patterns along the chromosome.
Gene Model Classification
All annotated rice genes were divided into HH and LH genes based on their homology with Arabidopsis genes. Homology was based on a BLAST search of both annotated rice and Arabidopsis gene model sequences by means of tBLASTN (Altschul et al., 1997). We used the TIGR version 4.0 Arabidopsis gene model annotation downloaded from The Arabidopsis Information Resource (ftp://ftp.arabidopsis.org). Specifically, all rice gene models were compared against each Arabidopsis gene model. For genes encoding proteins with 200 residues or more, an expectation cutoff value of 10−7 for sequences not <100 residues was used. For genes encoding proteins with fewer than 200 residues, the expectation cutoff value of 10−7 for >50% of the full length was used. A total of 3166 rice genes were identified with significant homologs in the Arabidopsis genome (HH gene models). The rest of the 2516 rice genes were classified as LH gene models.
Transposon-related gene identification followed the TIGR version 2.0 annotation. In total, 1501 such gene models on chromosome 4 were covered by this tiling path microarray. Further classification of retrotransposons and DNA transposons also followed the annotation from TIGR. Retrotransposons included 124 from the Ty1/copis subclass, 505 from the Ty3/gypsy subclass, 10 from the LINE subclass, 8 gag proteins, and 500 unclassified proteins. DNA transposons included 285 from the CACTA En/Spm subclass, 47 from the mariner subclass, 7 from the ping/pong/SNOOPY subclass, 2 from the Ac/Ds subclass, and 3 unclassified transposons. MITEs and simple repeats were identified directly from the chromosome 4 pseudomolecule using RepeatMasker (http://www.repeatmasker.org).
RNA Gel Blot Analysis
Total RNA from the same organ type was used for RNA gel blot analysis. Subcloned DNA fragments were labeled with [α-32P]dCTP using a random primers DNA labeling system (Invitrogen). RNA gel blot analysis was performed as described by Li et al. (2002).
Microarray data from this article have been deposited with the NCBI Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE2358.
Supplementary Material
Acknowledgments
The authors thank Thomas Royce and Paul Harrison for advice on microarray data analysis, Kenneth Nelson for technical assistance with microarray printing, and Jessica Habashi, Elizabeth Strickland, and two anonymous reviewers for commenting on the manuscript. This work was supported by a special University-CAS cooperation grant from the Chinese Academy of Sciences, the 863 Rice Functional Genomics Program from the Ministry of Science and Technology of China, and grants from the National Institutes of Health (GM-47850) and the National Science Foundation (DBI-0421675) to X.W.D. Y.J. was the recipient of a Yale University Joseph F. Cullman, Jr. fellowship. L.M. was a long-term postdoctoral fellow of the Human Frontier Science Program.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Bin Han (bhan@ncgr.ac.cn) and Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031575.
References
- Ahmad, K., and Henikoff, S. (2001). Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104, 839–847. [DOI] [PubMed] [Google Scholar]
- Ahn, S., and Tanksley, S.D. (1993). Comparative linkage maps of rice and maize genomes. Proc. Natl. Acad. Sci. USA 90, 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramova, Z.V. (2002). Heterochromatin in animals and plants: Similarities and differences. Plant Physiol. 129, 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, J. (2004). Chromatin-based silencing mechanisms. Curr. Opin. Plant Biol. 7, 521–526. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bennetzen, J.L., Coleman, C., Liu, R., Ma, J., and Ramakrishna, W. (2004). Consistent over-estimation of gene number in complex plant genomes. Curr. Opin. Plant Biol. 7, 732–736. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad, B.M., Irizarry, R.A., Astrand, M., and Speed, T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Chen, M., et al. (2002). An integrated physical and genetic map of the rice genome. Plant Cell 14, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., SanMiguel, P., de Oliveria, A.C., Woo, S.-S., Zhang, H., Wing, R.A., and Bennetzen, J.L. (1997). Microcolinearity in the sh2-homologous regions of maize, rice, and sorghum genomes. Proc. Natl. Acad. Sci. USA 94, 3431–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Buell, C.R., Wing, R.A., Gu, M., and Jiang, J. (2001). Toward a cytological characterization of the rice genome. Genome Res. 11, 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, G.M., and Kempson, R.E. (1997). Introduction to the Design and Analysis of Experiments. (London: Arnold).
- CSHL/WUGSC/PEB Arabidopsis Sequencing Consortium. (2000). The complete sequence of a heterochromatic island from a higher eukaryote. Cell 100, 377–386. [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420, 316–320. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., Jiang, N., and Wessler, S.R. (2002). Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 3, 329–341. [DOI] [PubMed] [Google Scholar]
- Fransz, P., Armstrong, S., Alonso-Blanco, C., Fischer, T.C., Torres-Ruiz, R.A., and Jones, G. (1998). Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13, 867–876. [DOI] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, E. (1928). Das heterochromatin der Moose. Jehrb. Wiss. Botanik 69, 762–818. [Google Scholar]
- Held, G.A., Grinstein, G., and Tu, Y. (2003). Modeling of DNA microarray data by using physical properties of hybridization. Proc. Natl. Acad. Sci. USA 100, 7575–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Comai, L. (1998). Trans-sensing effects: The ups and downs of being together. Cell 93, 329–332. [DOI] [PubMed] [Google Scholar]
- Hennig, W. (1999). Heterochromatin. Chromosoma 108, 1–9. [DOI] [PubMed] [Google Scholar]
- Hoekenga, O.A., Muszynski, M.G., and Cone, K.C. (2000). Developmental patterns of chromatin structure and DNA methylation responsible for epigenetic expression of a maize regulatory gene. Genetics 155, 1889–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.R., et al. (2001). Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19, 342–347. [DOI] [PubMed] [Google Scholar]
- Jabbari, K., Cruveiller, S., Clay, O., Le Saux, J., and Bernardi, G. (2004). The new genes of rice: A closer look. Trends Plant Sci. 9, 281–285. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Bao, Z., Zhang, X., Eddy, S., and Wessler, S.R. (2004). Pack-MULES: Transposon-mediated gene evolution in plants. Nature 431, 569–573. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., et al. (2003). A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol. 133, 1480–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. (2002). Large-scale transcriptional activity in chromosomes 21 and 22. Science 296, 916–919. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. (2002). BLAT: The BLAST-like alignment tool. Genome Res. 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, M.K., Martin, M., and Churchill, G.A. (2000). Analysis of variance for gene expression microarray data. J. Comput. Biol. 7, 819–837. [DOI] [PubMed] [Google Scholar]
- Kim, H., Snesrud, E.C., Haas, B., Cheung, F., Town, C.D., and Quackenbush, J. (2003). Gene expression analyses of Arabidopsis chromosome 2 using a genomic DNA amplicon microarray. Genome Res. 13, 327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I., and Earnshaw, W.C. (1998). Structure and function in the nucleus. Science 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., et al. (2004). Sensitivity of 70-mer oligonucleotides and cDNAs for microarray analysis of gene expression in Arabidopsis and its related species. Plant Biotechnol. J. 2, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Li, C., Lee, G.I., and Howe, G.A. (2002). Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L.-H., Li, J.-C., Lin, Y.-F., Lin, C.-Y., Chen, C.-Y., and Tsai, S.-F. (2004). Genomic shotgun array: A procedure linking large-scale DNA sequencing with regional transcript mapping. Nucleic Acids Res. 32, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z., and Martienssen, R. (2004). The role of RNA interference in heterochromatic silencing. Nature 431, 364–370. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476. [DOI] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., Wood, T.C., Yu, Y., Budiman, M.A., Tomkins, J., Woo, S., Sasinowski, M., Presting, G., Frisch, D., Goff, S., Dean, R.A., and Wing, R.A. (2000). Rice transposable elements: A survey of 73,000 sequence-tagged-connectors. Genome Res. 10, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A., and Birchler, J.A. (2005). RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6, 24–35. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1929). Chromosome morphology in Zea mays. Science 69, 629. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Vu, T.H., Tej, S.S., Ghazal, H., Matvienko, M., Agrawal, V., Ning, J., and Haudenschild, C.D. (2004). Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 22, 1006–1011. [DOI] [PubMed] [Google Scholar]
- Meyer, P. (2000). Transcriptional transgene silencing and chromatin components. Plant Mol. Biol. 43, 221–234. [DOI] [PubMed] [Google Scholar]
- Mlynárová, L., Loonen, A., Heldens, J., Jansen, R.C., Keizer, P., Stiekema, W.J., and Nap, J.-P. (1994). Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix-associated region. Plant Cell 6, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H.J. (1930). Types of visible variations induced by X-rays in Drosophila. J. Genet. 22, 299–334. [Google Scholar]
- Pandey, R., Muller, A., Napoli, C.A., Selinger, D.A., Pikaard, C.S., Richards, E.J., Bender, J., Mount, D.W., and Jorgensen, R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., and Chapman, B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101, 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D. (1999). Chromatin silencing and Arabidopsis development: A role for polycomb protein. Plant Cell 11, 765–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J. (2002). Microarray data normalization and transformation. Nat. Genet. 32, 496–501. [DOI] [PubMed] [Google Scholar]
- Redman, J.C., Haas, B.J., Tanimoto, G., and Town, C.D. (2004). Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 38, 545–561. [DOI] [PubMed] [Google Scholar]
- Rensink, W.A., and Buell, C.R. (2004). Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 135, 622–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, J.C., Hennig, L., and Gruissem, W. (2002). Chromatin-remodeling and memory factors: New regulators of plant development. Plant Physiol. 130, 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium. (2003). In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300, 1566–1569. [DOI] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379. [DOI] [PubMed] [Google Scholar]
- Rinn, J.L., et al. (2003). The transcriptional activity of human chromosome 22. Genes Dev. 17, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, S., Frisch, M., Grote, K., Meyers, B.C., Mayer, K., and Werner, T. (2004). Genome-wide in silico mapping of scaffold/matrix attachment regions in Arabidopsis suggests correlation of intragenic scaffold/matrix attachment regions with gene expression. Plant Physiol. 135, 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., et al. (2002). The genome sequence and structure of rice chromosome 1. Nature 420, 312–316. [DOI] [PubMed] [Google Scholar]
- Scheid, O.M., Afsar, K., and Paszkowski, J. (2003). Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34, 450–454. [DOI] [PubMed] [Google Scholar]
- Shimamoto, K., and Kyozuka, J. (2002). Rice as a model for comparative genomics of plants. Annu. Rev. Plant Biol. 53, 399–419. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D.D., et al. (2001). Experimental annotation of the human genome using microarray technology. Nature 409, 922–927. [DOI] [PubMed] [Google Scholar]
- Soppe, W.J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M.S., Jacobsen, S.E., Schubert, I., and Fransz, P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21, 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., Belele, C., Dorweiler, J.E., and Chandler, V.L. (2002). Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16, 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L.V., Chen, L., Greil, F., Negre, N., Li, T.-R., Cavalli, G., Zhao, H., van Steensel, B., and White, K.P. (2003). Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. USA 100, 9428–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., Bennetzen, J.L., and Avramova, Z.V. (2000). Structural domains and matrix attachment regions along colinear chromosomal segments of maize and sorghum. Plant Cell 12, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp, C.N., Zhong, C.X., and Dawe, R.K. (2004). Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101, 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte, K., Srinivasan, S., and Bureau, T. (2001). Survey of transposable elements from rice genomic sequences. Plant J. 25, 169–179. [DOI] [PubMed] [Google Scholar]
- Vicient, C.M., Jaaskelainen, M.J., Kalendar, R., and Schulman, A.H. (2001). Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125, 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, G.K.-S., Wang, J., Tao, L., Tan, J., Zhang, J., Passey, D.A., and Yu, J. (2002). Compositional gradients in Gramineae genes. Genome Res. 12, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Kerr, K., Cui, X., and Churchill, G.A. (2003). MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. In The Analysis of Gene Expression Data: Methods and Software, G. Parmigiani, E.S. Garett, R.A. Irizarry, and S.L. Zeger, eds (Heidelberg: Springer), pp. 313–341.
- Wu, J., et al. (2002). A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2005). The genomes of Oryza sativa: A history of duplications. PLoS Biol. 3, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S.-M., Kuo, Y.-H., Sheu, G., Sheu, T.-J., and Liu, L.-F. (1991). Metabolic depression in suspension-cultured cells of rice. J. Biol. Chem. 266, 21131–21137. [PubMed] [Google Scholar]
- Yuan, Q., Ouyang, S., Liu, J., Suh, B., Cheung, F., Sultana, R., Lee, D., Quackenbush, J., and Buell, C.R. (2003). The TIGR rice genome annotation resource: Annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 31, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.Q. (2002). Computational prediction of eukaryotic protein-coding genes. Nat. Rev. Genet. 3, 698–709. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., et al. (2004). Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32, 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q., et al. (2002). A fine physical map of the rice chromosome 4. Genome Res. 12, 817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.