Abstract
MicroRNAs (miRNAs) affect the morphology of flowering plants by the posttranscriptional regulation of genes involved in critical developmental events. Understanding the spatial and temporal dynamics of miRNA activity during development is therefore central for understanding miRNA functions. We describe a microarray suitable for detection of plant miRNAs. Profiling of Arabidopsis thaliana miRNAs during normal development extends previous expression analyses, highlighting differential expression of miRNA families within specific organs and tissue types. Comparison of our miRNA expression data with existing mRNA microarray data provided a global intersection of plant miRNA and mRNA expression profiles and revealed that tissues in which a given miRNA is highly expressed are unlikely to also show high expression of the corresponding targets. Expression profiling was also used in a phylogenetic survey to test the depth of plant miRNA conservation. Of the 23 families of miRNAs tested, expression of 11 was detected in a gymnosperm and eight in a fern, directly demonstrating that many plant miRNAs have remained essentially unchanged since before the emergence of flowering plants. We also describe an empirical strategy for detecting miRNA target genes from unsequenced transcriptomes and show that targets in nonflowering plants as deeply branching as ferns and mosses are homologous to the targets in Arabidopsis. Therefore, several individual miRNA regulatory circuits have ancient origins and have remained intact throughout the evolution and diversification of plants.
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nucleotide regulatory RNAs that derive from stem-loop regions of endogenous precursor transcripts (Ambros, 2004; Bartel, 2004). Many miRNAs are known to pair to the messages of protein-coding genes to target these mRNAs for posttranscriptional repression. In plants, this repression is primarily through the mechanism of miRNA-directed cleavage of the mRNA (Llave et al., 2002; Tang et al., 2003; Schwab et al., 2005).
miRNAs are important controllers of development in flowering plants (Dugas and Bartel, 2004). The majority of known miRNA targets in Arabidopsis thaliana code for proteins with a known or suspected role in developmental control (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004). Dysfunction of individual miRNAs and/or their ability to properly regulate their targets has been shown to cause floral and leaf-patterning defects (miR159 and miR319; Palatnik et al., 2003; Achard et al., 2004; Millar and Gubler, 2005), floral development and timing defects (miR172; Aukerman and Sakai, 2003; Chen, 2004), loss of organ polarity and altered vascular development (miR165/166; McConnell et al., 2001; Emery et al., 2003; Juarez et al., 2004; Mallory et al., 2004b; McHale and Koning, 2004; Zhong and Ye, 2004; Kim et al., 2005), defective organ separations and aberrant numbers of floral organs (miR164; Laufs et al., 2004; Mallory et al., 2004a; Baker et al., 2005), aberrant phyllotaxis, reduced fertility, and abortion of the shoot apical meristem (miR168; Vaucheret et al., 2004), and cotyledon and rosette leaf shape and symmetry defects, reduced fertility, and misexpression of early auxin response genes (miR160; Mallory et al., 2005). A null mutation in the Dicer-Like 1 (DCL1) locus, which codes for an endonuclease critical for miRNA accumulation (Park et al., 2002; Reinhart et al., 2002), causes embryonic lethality (Schauer et al., 2002), further implicating plant miRNAs in the elaboration of the multicellular plant body plan. Given these clear roles in plant development, it has been proposed that precise regulation of miRNA activity during various stages of growth and in specific cell types is of central importance for normal plant development (Rhoades et al., 2002; Bartel, 2004).
Many Arabidopsis miRNAs are conserved among flowering plants. For most miRNAs cloned from Arabidopsis, exact or nearly exact matches can be found in the rice (Oryza sativa) genome, which if transcribed would be in a context predicted to fold into stem-loops characteristic of miRNA primary transcripts (Reinhart et al., 2002; Bonnet et al., 2004; Wang et al., 2004). Similarly, rice homologs of many Arabidopsis miRNA targets have conserved miRNA complementary sites, implying that these miRNA–target interactions have been functioning at least since the last common ancestor of monocots and eudicots (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). Additional evidence for conservation of plant miRNAs has come from EST sequence data from diverse flowering plants and occasional nonflowering plants, in which sequences containing miRNA hairpins as well as sequences homologous to the known or predicted Arabidopsis targets retaining miRNA complementary sites have been observed (Palatnik et al., 2003; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). Direct observations have shown that miRNAs in the miR165/166 family are expressed and functional in wheat (Triticum aestivum; Tang et al., 2003; Mallory et al., 2004b) and maize (Zea mays; Juarez et al., 2004) and are guiding cleavage of homologous target mRNAs in basal plants such as the lycopod Selaginella kraussiana (Floyd and Bowman, 2004), implying that the miR165/166 regulatory circuit has remained intact since the last common ancestor of vascular plants.
Several different approaches enabling multiplexed detection of miRNAs using microarray technologies have been reported (Krichevsky et al., 2003; Babak et al., 2004; Liu et al., 2004; Miska et al., 2004; Nelson et al., 2004; Sun et al., 2004; Thomson et al., 2004; Liang et al., 2005), including one from our laboratory (Baskerville and Bartel, 2005). In our approach, probes consist of Tm-normalized DNA oligonucleotides antisense to the given small RNA sequence. Sample preparation begins by selecting small RNAs with the characteristic features of miRNAs, followed by reverse transcription and PCR amplification with a fluorescently labeled primer. Single-stranded Cy3-labeled biological samples are then hybridized to the array along with a synthetic reference library containing a constant amount of Cy5-labeled single-stranded DNA sample, which allows internal normalization of the experiments. This technology has proven to be semiquantitative, sensitive, and highly reproducible in experiments with vertebrate miRNAs (Baskerville and Bartel, 2005).
In this study, a microarray suitable for the detection of Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis miRNAs is described. Using this platform, the overall miRNA expression profile within the major organs of Arabidopsis was determined, providing a useful baseline for understanding the developmental dynamics of plant miRNA expression. Comparison with existing mRNA expression data revealed a significant negative correlation between the levels of miRNAs and those of their target messages. The array was also used as a phylogenetic profiling tool to probe RNA samples derived from specimens representative of major clades of land plants. We detected members of 11 miRNA families in a gymnosperm, eight in a fern, three in a lycopod, and two in a moss, indicating that many plant miRNA families have been long conserved during land plant evolution. Using a strategy for identification and validation of miRNA-regulated transcripts in the absence of any genomic information, we identified targets for several of these conserved miRNAs in organisms as divergent as Arabidopsis and moss. The newly identified targets of miR160, miR167, miR170/171, and miR172 in nonflowering plants were all homologous to the known Arabidopsis targets, demonstrating that multiple miRNA–target interactions have remained unchanged over very long periods of plant evolution.
RESULTS
Design and Validation of a Plant miRNA Array
To enable the simultaneous detection of multiple miRNAs from three model organisms, we designed a microarray using a recently described technique (Baskerville and Bartel, 2005). The 5′ C6 amino-link DNA oligonucleotides antisense to 88 C. elegans miRNAs, 79 D. melanogaster miRNAs, and 63 plant small RNAs were designed and spotted in quadruplicate to glass slides. Subsequent to printing of the array, experimental evidence for some additional plant miRNAs has been reported (Sunkar and Zhu, 2004; Wang et al., 2004; Adai et al., 2005); most of these were not included in this array. A full list of probes and sequences for the array may be found in Supplemental Table 1 online.
Many plant miRNAs are members of closely related families that differ by only a few nucleotides in sequence. We arrayed separate spots for closely related family members if there were one or more nucleotide differences in the center portion of the sequence (more than four nucleotides from both the 5′ and 3′ termini). To test the discrimination between these closely related sequences, one-half of the synthetic reference library was labeled with Cy5 and the other with Cy3 and hybridized in duplicate to the array. The selection of Cy5- and Cy3-labeled oligonucleotides was such that nearly all members of closely related families were tested against each other. This experiment showed that 13 out of the 63 plant spots were cross-hybridizing (see Supplemental Table 2 online). Twelve of these cases could be sorted into seven families of small RNAs within which a closely related probe could be found that most likely accounts for the cross-hybridization. Thus, signals from spots within these seven families reflect the combination of the closely related family members. This experiment also revealed that for five other closely related plant small RNAs, discrimination between species differing by one or two nucleotides was achieved (see Supplemental Table 2 online). Because these experiments tested equimolar concentrations of all samples, cross-hybridization might still be a problem if a slightly mismatched miRNA was present in a biological sample at much higher concentrations than was the perfectly matched RNA. Nonetheless, because all cases of observed cross-hybridization among plant-specific probes, save that of miR158, could be accounted for by closely related probes, we conclude that the array is specifically reporting the abundance of the intended miRNA families.
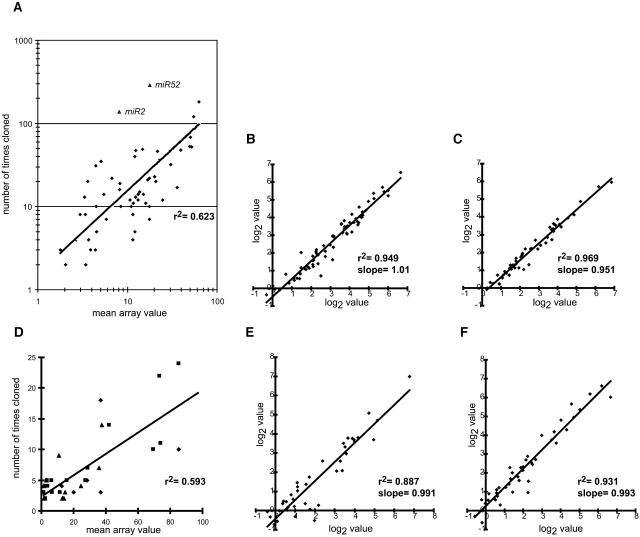
Previously, it has been demonstrated that the cloning frequency of C. elegans miRNAs from a mixed stage total RNA sample correlates with absolute molecular abundance, as determined by quantitative RNA gel blots (Lim et al., 2003). When C. elegans miRNAs were analyzed by hybridization to the array, a positive linear correlation was observed between cloning frequency and array value, indicating that array value is generally indicative of RNA abundance (Figure 1A). However, it is important to note that there was significant variation in this relationship between array value and RNA abundance and that, therefore, two miRNAs with a similar array value may in fact vary substantially in their absolute abundance within a given sample. Of course, the primary purpose of expression analysis is not to compare the abundance of different miRNAs within a sample, but to compare the relative abundance of individual miRNAs across several samples. To investigate the array's utility for this purpose, four small RNA hybridizations were performed from two samples of C. elegans total RNA [wild type, mixed stage worms, and glp-4(bn2) worms; see Supplemental Table 3 online]. Figures 1B and 1C show that the technical variation was very low; linear regressions gave r2 values of 0.949 and 0.969 [for the wild type and glp-4(bn2), respectively]. Therefore, we concluded that an individual array value was highly reproducible and that large variations in the array value for a given miRNA across an experimental panel were indicative of different steady state levels of that species within the different samples.
Figure 1.
The miRNA Array Is Semiquantitative and Highly Reproducible.
(A) Mean array values for each miRNA detected (average of two hybridizations) are plotted versus the number of times the corresponding RNAs were cloned and sequenced from wild-type, mixed stage C. elegans (Lim et al., 2003). The linear regression line shown has a correlation coefficient of 0.623; this regression excludes two outliers, miR-2 and miR5-2, which are noted as triangles.
(B) Technical replicate array hybridizations. Log2-transformed array values from two hybridizations of independently labeled C. elegans wild-type, mixed stage RNA from the same total RNA sample.
(C) Technical replicate array hybridizations. Log2-transformed array values from two hybridizations of independently labeled C. elegans glp-4(bn2) RNA from the same total RNA sample.
(D) Mean array values from Arabidopsis rosette leaves (diamonds), inflorescences (triangles), and seedlings (squares) are plotted versus the number of times the corresponding RNAs were cloned and sequenced from the corresponding Col-0 tissues. The linear regression line shown has a correlation coefficient of 0.593.
(E) Biological replicate array hybridizations. Log2-transformed array values from two hybridizations of independently labeled Arabidopsis inflorescence RNA from two RNA samples derived from different crops of plants grown under identical conditions.
(F) Biological replicate array hybridizations. Log2-transformed array values from two hybridizations of independently labeled Arabidopsis rosette leaf RNA from two RNA samples derived from different crops of plants grown under identical conditions.
Lines in (A) to (F) represent linear regressions with the given correlation coefficients (r2) and slopes.
Global Expression Profile of Arabidopsis Small RNAs
To obtain a global overview of miRNA levels in wild-type Arabidopsis tissues, two samples of total RNA from siliques, stems, cauline leaves, roots, short-day seedlings, long-day seedlings, and rosette leaves as well as four samples from inflorescences were analyzed. All duplicate samples were harvested from independent crops of plants grown under the same conditions. Array values were obtained and processed as described in Methods and reported as log2-transformed values (see Supplemental Table 4 online). A positive correlation between the cloning frequencies (Reinhart et al., 2002; R. Rajagopalan and D.P. Bartel, unpublished data) from three different organs and the mean array values from those organs was observed despite the low numbers of clonings for many Arabidopsis miRNAs (Figure 1D), indicating that the array values were representative of the abundance of cloned miRNAs in the experimental samples. Examples of the biological variation found in these samples are shown in Figures 1E and 1F, where linear regressions gave r2 values of 0.887 and 0.931 with slopes of 0.991 and 0.993 for inflorescences and rosette leaves, respectively. The mean r2 value for all of the biological replicates included in the organ map study was 0.888 ± 0.089 with a mean slope of 0.914 ± 0.078.
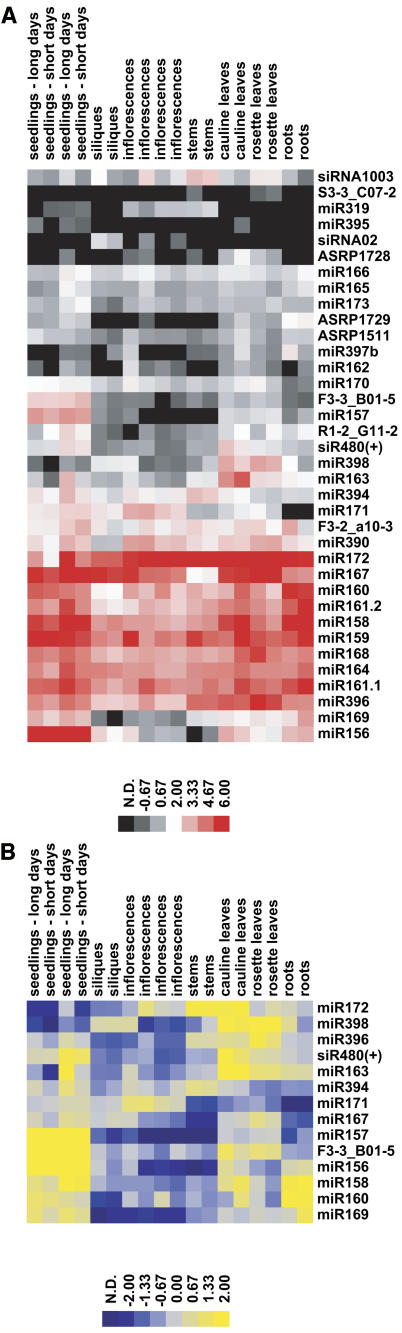
A graphical representation of array values organized by hierarchical clustering of both genes and experiments is shown in Figure 2A. For clarity, values derived from spots designed for predicted rice homologs as well as any spot that was not above the detection threshold in both replicates in at least one organ were omitted from display. Consistent with the high reproducibility of these biological replicates, clustering of the experiments placed all biological replicates as most closely related to each other, with the exception of the long- and short-day seedling replicates, which were intercalated.
Figure 2.
A Global Expression Profile of Arabidopsis Small RNAs.
(A) Log2-transformed array values for Arabidopsis spots that were above the detection threshold (see Methods) in both replicates for at least one organ were hierarchically clustered both by gene and by hybridization sample. Log2-transformed values are displayed as a color gradient from gray (low values) through white (intermediate values) to bright red (high values), with black indicating not detected (N.D.)
(B) Differentially expressed small RNAs in Arabidopsis tissues. Arabidopsis small RNAs whose array values in at least one organ were significantly different than those in other organs were F3-3_B01-5, miR157, miR172, miR156, miR396, miR398, miR160, and miR163 (single-factor analysis of variance with Bonferroni-Holm corrected P-values of < 0.05). The other differentially expressed small RNAs with lower confidence were miR167 (P = 0.056), miR169 (P = 0.069), miR394 (P = 0.064), miR158 (P = 0.073), siR480(+) (P = 0.118), and miR171 (P = 0.117). To highlight the relative differences between tissues, the expression values for each miRNA were normalized on a per-gene basis to their median level of expression and log2-transformed. Yellow indicates high relative expression, gray indicates moderate relative expression, and blue indicates low relative expression. N.D., not detected.
Figure 2B illustrates miRNAs that were differentially expressed between different organs and shows that there were several different patterns of miRNA accumulation. For example, miRNAs from the miR156/157 family were strongly expressed during seedling development and downregulated in mature tissues, whereas miR172 had a roughly opposite expression profile with its strongest expression detected in mature stems, cauline leaves, and inflorescences, with very weak expression in seedlings (Figure 2B). miR398, miR396, and miR163 shared a broadly similar expression profile with peak expression within rosette and/or cauline leaves and depressed expression within the stems.
The organ expression map showed that non-miRNA small RNAs can be developmentally regulated. For instance, the DCL1-independent and RNA-dependent RNA polymerase 2–dependent species siRNA02, which originates from an inverted repeat on chromosome V (Xie et al., 2004), was detected by the array only in siliques and in two of four inflorescence samples. It is also worthwhile to note that many miRNAs were found to exhibit relatively uniform accumulation across the panel of tissues assayed. This does not necessarily imply that in these cases precise tissue- or cell type–specific miRNA activities are not important; indeed, such high-resolution miRNA accumulation patterns would be lost when assaying RNA from entire organs. Higher resolution methods to determine spatio-temporal accumulation patterns of miRNAs, such as in situ blot analysis (Chen, 2004; Juarez et al., 2004; Kidner and Martienssen, 2004) or sensor transgenes (Brennecke et al., 2003; Parizotto et al., 2004), will be necessary to discover the precise locations of many of these plant miRNAs.
miRNA Expression Is Generally Anticorrelated with That of Targeted mRNAs
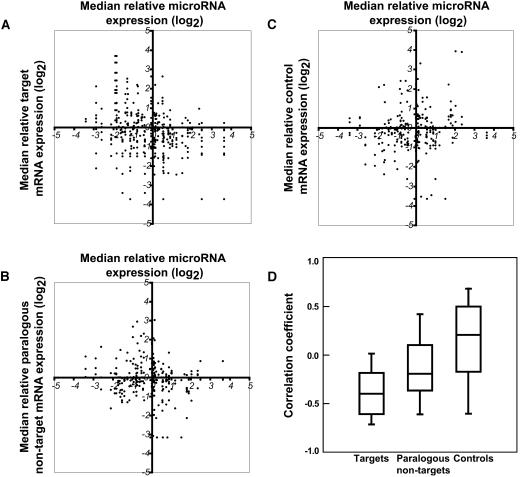
Plant miRNAs generally direct endonucleolytic cleavage of mRNAs (Llave et al., 2002; Tang et al., 2003; Schwab et al., 2005), consistent with the suggestion that plant miRNAs enable rapid clearance of target mRNAs at specific points during plant development (Rhoades et al., 2002; Bartel, 2004). This hypothesis predicts a negative correlation between the expression of a miRNA and its target mRNAs within a given tissue or organ. We tested this hypothesis by comparing the expression levels of the differentially expressed miRNAs shown in Figure 2B with the expression levels of their known and predicted targets (Figure 3). To do this, we made use of portions of the AtGenExpress expression atlas of wild-type Arabidopsis development (Schmid et al., 2005), available from The Arabidopsis Information Resource (www.arabidopsis.org). These expression data consist of triplicates of Affymetrix ATH1 array hybridizations using RNA derived from various organs and growth stages of Columbia-0 (Col-0) wild-type plants. For each of the seven organs sampled in Figure 2, the median relative expression level of each differentially expressed miRNA was plotted versus the median relative expression level of the corresponding targets (Figure 3A). As controls, we compared the expression levels of randomly selected WRKY and MADS box transcription factors (two large families of plant-specific transcription factors, neither of which have any Arabidopsis members known to be regulated by miRNAs) to miRNA levels (Figure 3C). As expected, there was no apparent correlation between the expression of these control genes and that of the differentially expressed miRNAs.
Figure 3.
Comparison of miRNA and Target mRNA Expression Levels.
(A) The median relative level of miRNA expression plotted versus the median relative level of target mRNA expression within seven Arabidopsis organs.
(B) Analysis as in (A), except the median expression levels of miRNA targets were replaced with the median expression levels of the closest paralogs of each target that do not have recognized miRNA complementary sites. Paralogous nontargets were chosen as the top nontarget Arabidopsis BLASTp hits at an e-value of < 1e-10 to a target that were called present in greater than half of the array experiments (four maximum per miRNA).
(C) Analysis as in (A), except the median expression levels of miRNA targets were replaced with median expression levels of nonparalogous mRNAs. These control RNAs were randomly selected members of the WRKY and MADS box families of transcription factors.
(D) Plots showing the distributions of linear correlation coefficients between relative miRNA expression and expression of target (n = 43), paralogous nontarget (n = 33), and control (n = 35) mRNAs within seven Arabidopsis organs. Central lines indicate median values, boxes bound the 25th to 75th percentiles, and the bars indicate the 10th and 90th percentiles. The median value of the targets was significantly lower than the medians of both the paralogous nontargets and the controls (P = 0.0038 and P < 0.0001, respectively; Mann-Whitney U-test). The median value of the paralogous nontargets was also significantly less then the median of the controls (P = 0.0079; Mann-Whitney U-test).
In tissues where the miRNA was relatively high, the targets of that miRNA were unlikely also to be high (Figure 3A). At lower miRNA expression levels, no such negative (or positive) correlation with target gene expression was observed. We also examined the relative expression levels of mRNAs that are paralogs of the miRNA targets but do not contain recognized miRNA complementary sites. Curiously, paralogous nontargets were also unlikely to be highly expressed in tissues where the corresponding miRNA was also highly expressed (Figure 3B). This reflected the fact that in many cases, the expression profiles of targets and their closely related nontargets are similar.
For each set of miRNA versus target, paralogous nontarget, and control mRNA expression data, a correlation coefficient was calculated (Figure 3D). The expression of the majority of miRNA targets was negatively correlated with expression of their corresponding miRNAs. The paralogous nontarget set was also somewhat negatively correlated with miRNA expression, whereas the control set had a large range of correlation coefficients, as expected from a random selection of genes. The median correlation coefficient of the targets was significantly lower than those of both the paralogous nontargets and the controls (P = 0.0038 and P < 0.0001, respectively; Mann-Whitney U-test), demonstrating that in plants, expression of miRNAs and that of their targets are generally negatively correlated.
Many Plant miRNA Families Are Ancient
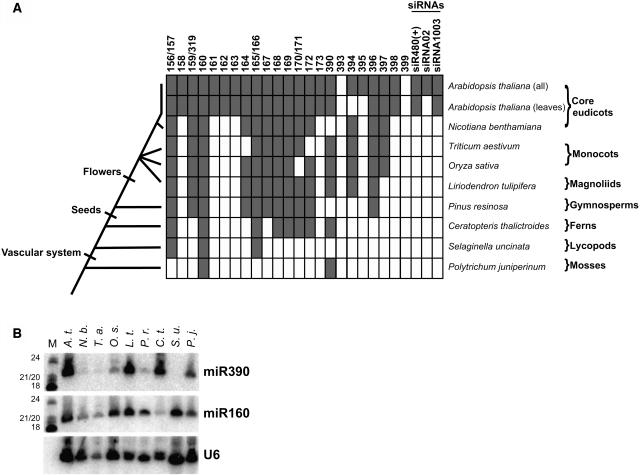
Cloning and computational analyses of Arabidopsis small RNAs suggest that many plant miRNAs and their predicted targets are conserved between monocots and eudicots, which are thought to have diverged >125 million years ago (Reinhart et al., 2002; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004; Adai et al., 2005). For the miR165/166 family, functional conservation between eudicots and monocots has been experimentally demonstrated (McConnell et al., 2001; Emery et al., 2003; Tang et al., 2003; Juarez et al., 2004; Mallory et al., 2004b; McHale and Koning, 2004; Zhong and Ye, 2004), and conservation of target mRNA cleavage at the canonical site has been shown to occur in the lycopod S. kraussiana and at an offset potential target site in the moss Physcomitrella patens (Floyd and Bowman, 2004). To directly assay multiple miRNA families for conservation between distantly related land plants, the plant miRNA array was used to analyze samples derived from the eudicot Nicotiana benthamiana, the monocots rice and wheat (T. aestivum), the magnoliid Liriodendron tulipifera, the gymnosperm Pinus resinosa (pine), the fern Ceratopteris thalictroides, the lycopod Selaginella uncinata, and the moss Polytrichum juniperinum. miRNAs, but not endogenous small interfering RNAs (siRNAs), trans-acting siRNAs, or any of the nine families of unclassified Arabidopsis small RNAs with probes present on the array were detected outside of Arabidopsis (see Supplemental Table 1 online; data not shown). miR161, miR163, and ASRP1729 (data not shown) were not detected outside of Arabidopsis, consistent with the hypothesis that these genes emerged recently (Allen et al., 2004). Out of the 23 families of Arabidopsis miRNAs analyzed, we detected expression of 21 in Arabidopsis (composite of all experiments), 19 in Arabidopsis rosette leaves, 13 in N. benthamiana leaves, 12 in wheat germ lysate, 13 in rice seedlings, 13 in magnoliid leaves, 11 in pine needles, eight in fern leaves and stems, three in lycopod leaves and stems, and two in moss leafy gametophytes (Figure 4A; see Supplemental Table 5 online). We noted that miR158 was detected in N. benthamiana, T. aestivum, and L. tulipifera (data not shown), but we suspect these detections were false positives because this probe was the one that uniquely gave unexplained cross-hybridization in our control experiments (see Supplemental Tables 2 and 5 online), miR158 homologs are not computationally evident in either the rice (Jones-Rhoades and Bartel, 2004) or poplar genome (M. Jones-Rhoades, personal communication), and RNA gel blot analysis could not detect miR158 in these RNA samples (data not shown). RNA gel blots were performed for the two miRNAs that were detected in the moss sample, miR160 and miR390, using DNA probes antisense to the Arabidopsis RNA sequences (Figure 4B). Both miRNAs were detected in the same samples as those indicated by the array analyses. For the samples in which the array analysis did not detect miR390, blot analysis showed very slight (N. benthamiana and pine [P. resinosa]) or no (lycopod [S. uncinata]) accumulation of the miRNA, demonstrating that, with the exception of the abnormally performing probe for miR158, the method used to determine the lower limit of detection in the array analyses was sufficiently stringent to prevent false positives, with sensitivity at least comparable to that of RNA gel blots.
Figure 4.
Detection of Ancient miRNAs.
(A) Duplicate array hybridizations were used to probe RNA samples from various specimens representative of major clades of land plants. RNAs detected in the species indicated at right for each row are shaded gray, whereas those not detected are left unshaded. Arabidopsis is represented by two rows, one for the composite of all organs (all), and the other for the analysis of rosette leaves only (leaves). Names of the major groups represented by each species are shown at the far right. An abbreviated cladogram displaying the evolutionary relationships of the sampled species is shown at left. Major evolutionary innovations are marked on the cladogram by perpendicular hash marks. Detection of three endogenous siRNAs is indicated. Unclassified small RNAs, of which none were detected outside of Arabidopsis, were omitted from this figure.
(B) RNA gel blot using probes specific for Arabidopsis miR390, miR160, and the U6 small nuclear RNA. The blot was sequentially probed and stripped. Markers (M) are 33P-labeled RNAs with sizes indicated at left. Initials are abbreviations of the species listed above.
This analysis almost certainly underestimated the true extent of miRNA conservation for several reasons: First, the array is relatively intolerant of nucleotide substitutions (see Supplemental Table 2 online) and would be unlikely to detect a homolog that differs from the probe sequence by more than two nucleotides and may not detect molecules differing by only one nucleotide. Second, with the exception of Arabidopsis, only one tissue type was sampled for each of the organisms reported in Figure 4A; therefore, it is possible that other miRNA families could accumulate in different organs or from specimens exposed to different environmental conditions. For instance, the accumulation of miR395 in Arabidopsis occurs under conditions of limiting sulfate (Jones-Rhoades and Bartel, 2004) and was not detectable in Arabidopsis leaves nor in any other species tested. Additionally, not all of the organs sampled from the various specimens are homologous: The moss sample was derived from gametophytic tissue, whereas all of the other samples were derived from sporophytic tissues. It is possible that conserved miRNAs that are specific to the sporophytic phase of the plant life cycle were not detected in the moss sample. Therefore, the number of miRNA families reported here as conserved should be considered a lower limit. These data show that at least eight miRNA families have remained essentially unchanged since before the emergence of seed plants and that at least two families (miR160 and miR390) have remained unchanged since the last common ancestor of mosses and flowering plants.
miRNA Targets in Nonflowering Plants Are Homologous to Those in Arabidopsis
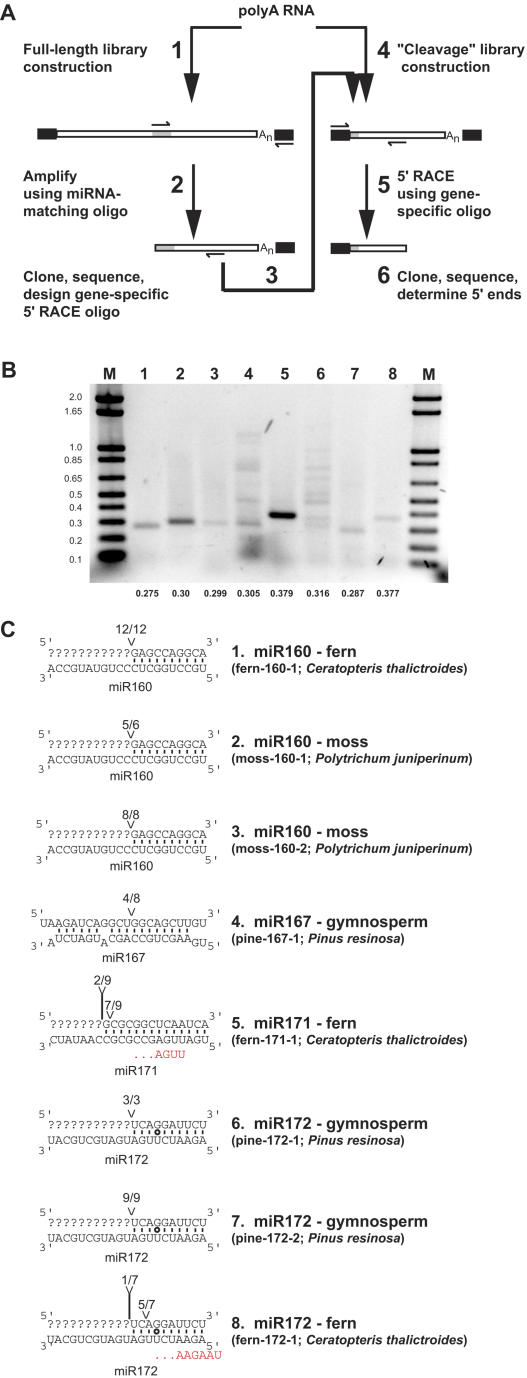
The extraordinary conservation of the plant miRNAs shown in Figure 4 raised the question of whether their target mRNAs, and by inference their biological functions, have also been conserved. All known plant miRNA:mRNA interactions are characterized by extensive base pairing, and disruption of this pairing has been shown to render the regulatory circuit dysfunctional in numerous independent studies (reviewed in Dugas and Bartel, 2004). In the absence of genomic sequence data, this functionally critical complementarity was exploited to probe the transcriptomes of pine (P. resinosa), fern (C. thalictroides), lycopod (S. uncinata), and moss (P. juniperinum) for mRNAs containing a putative miRNA binding site (Figure 5A). Degenerate oligonucleotides corresponding to the expected sequences of functional miRNA target sites were used as gene-specific primers, along with an oligo(dT) adapter primer, to amplify the 3′ regions of potential miRNA targets. The sequences from the 3′ regions of these candidate targets were then used to design gene-specific primers for subsequent 5′ rapid amplification of cDNA ends (5′-RACE) experiments. A plurality of 5′-RACE amplicons that terminate at the nucleotide that pairs to the tenth nucleotide of the miRNA is strong evidence that the mRNA in question is a miRNA target (Llave et al., 2002). Initial amplifications were attempted with oligonucleotides representing miRNA complementary sites for most of the miRNAs detected in pine, fern, lycopod, and moss, resulting in 29 candidate targets (see Supplemental Table 6 online). Subsequent 5′-RACE experiments using libraries enriched in uncapped messages as templates yielded single PCR products of the predicted size for six candidates (Figure 5B). For two other candidates, multiple bands were recovered, and in each case one of them corresponded to the predicted size for a cleavage product (Figure 5B, lanes 4 and 6). For the other 21 candidate sequences tested, either no 5′-RACE products could be obtained (n = 12) or no evidence for cleavage was observed (n = 9; see Supplemental Table 6 online). We suspect that most of these other 21 candidate targets do not contain miRNA complementary sites and instead represent artifacts obtained from using short, degenerate oligonucleotides during the initial PCR. Single bands from 5′-RACE reactions were gel-excised before cloning and sequencing, as were lanes with multiple bands (gel slices containing all visible species were excised in these two cases).
Figure 5.
Identification of miRNA Targets in Nonflowering Land Plants.
(A) Scheme of strategy for empirical discovery of miRNA targets. Black rectangles represent adapter sequences added before or during cDNA synthesis, and gray rectangles represent miRNA complementary sites.
(B) Ethidium bromide–stained gel showing 5′-RACE products from candidate miRNA targets. Pixel values were inverted for ease of viewing. Bands in lanes 1 to 8 are identified in (C). Numbers at bottom indicate the predicted sizes of cleavage products (in kilobases) assuming termination at position 10 relative to the alignment with the Arabidopsis miRNA. Cloning and sequencing showed that products with higher molecular weights in lanes 4 and 6 were PCR artifacts resulting from amplification of nontarget cDNAs. Marker lanes (M) contain DNA standards, with sizes (kilobases) noted at the left.
(C) Mapping 5′ ends of cleavage fragments of miRNA targets. In each duplex, mRNA sequence of the target is shown at top, aligned with the Arabidopsis miRNA sequence on the bottom. Base pairs are indicated by solid lines and G:U wobbles by circles. Predominant positions of 5′ target ends are indicated. Fractions refer to the number of independently cloned RACE products whose 5′ end terminated at the indicated position (numerator) over the total number of sequenced clones matching the target gene (denominator). In most cases, no 5′-RACE products were obtained upstream of the miRNA complementary site, precluding determination of the target sequence that pairs with the 3′ of the miRNA. Nucleotides in red indicate the 5′ ends of fern miR171 and fern miR172 as determined by PCR and sequencing.
In all eight cases, sequencing the 5′ ends revealed strong evidence for target cleavage (Figure 5C). Most 5′ ends terminated at the nucleotide that would pair with the tenth nucleotide of the Arabidopsis miRNA, as expected if a miRNA with similar sequence to the Arabidopsis miRNA was present in these nonflowering plants and directed the cleavage of the mRNA target. However, there were two exceptions. In the case of the fern miR170/171 target (fern-171-1), the 5′ ends mapped to position 13 or 14. Because this was clearly a predominant 5′ end of this transcript (Figure 5B, lane 5) and the transcript itself was highly homologous to the known Arabidopsis targets of miR170/171 (see below), we suspected that the fern miR170/171 homolog was offset from that of higher plants. The fern miR172 target (fern-172-1) also had an apparent cleavage site offset from that predicted by alignment with Arabidopsis miR172, which also could be due to an offset miR172 homolog in ferns.
To test directly whether the fern miR171 and miR172 are offset relative to their Arabidopsis homologs, we performed PCR from a fern small RNA library using oligonucleotides designed for miR171 and miR172 detection and 5′ end definition (Lim et al., 2003). In both cases, the experimentally determined 5′ ends were indeed offset relative to the Arabidopsis homologs: fern miR171 was shifted three nucleotides to the 3′ relative to Arabidopsis, whereas fern miR172 was shifted two nucleotides to the 5′ relative to Arabidopsis (Figure 5C). Using these offset miRNAs as guides, the fern-171-1 and fern-172-1 target cleavage sites, as mapped in Figure 5C, were precisely at the nucleotide expected for miRNA-directed cleavage. Interestingly, register-shifted miRNAs have been cloned at low frequencies from Arabidopsis as well, including a shifted miR171 that matches the miR171 version that appears to predominate in the fern C. thalictroides (miR171.2/ASRP444; Gustafson et al., 2005). An apparent offset target site in the moss (P. patens) homolog of a miR165/166 HD-ZIP target has also been suggested (Floyd and Bowman, 2004). Together, these examples indicate that deeply conserved plant miRNAs can diverge from each other by shifts in register. If these offsets are too large relative to the Arabidopsis or rice sequences for which probes were designed, the miRNA will not be detected by the array. The putative offset target site in the P. patens miR165/166 target proposed by Floyd and Bowman (2004) was shifted by 10 nucleotides, which could explain why we did not detect a miR165/166 ortholog in our moss (P. juniperinum) sample.
To assign putative functions to the newly discovered miRNA targets, deduced protein sequences were used to query the Arabidopsis protein database. In all cases, the best hit in the database was found to be either a confidently predicted or confirmed target of that miRNA in Arabidopsis (Table 1). For instance, fern-160-1, moss-160-1, and moss-160-2 are all most similar to the Arabidopsis gene Auxin Response Factor 16 (ARF16), a target of Arabidopsis miR160 (Mallory et al., 2005). Pine-172-1 and pine-172-2 are most similar to two Pinus genes annotated as coding for Apetala 2 (AP2)-like proteins (Shigyo and Ito, 2004), whereas the full-length fern-172-1 is most similar to Arabidopsis AP2; in Arabidopsis, miR172 is known to target AP2 and related mRNAs (Aukerman and Sakai, 2003; Chen, 2004). Pine-167-1 is most similar to the Arabidopsis ARF6 gene, which is a predicted target of Arabidopsis miR167 (Rhoades et al., 2002). Fern-171-1 is most homologous to the Arabidopsis Scarecrow-Like 6-III (SCL6-III) gene, which is a confirmed target of miR170/171 in Arabidopsis (Llave et al., 2002). In summary, our direct detection of miRNAs and empirical target discovery demonstrate that plant miRNA–target interactions are frequently conserved between mosses, ferns, gymnosperms, and flowering plants, implying that these regulatory circuits have long been critical components of land-plant development.
Table 1.
Validated miRNA Targets from Gymnosperms, Ferns, and Mosses
| miRNA | Confirmed or Predicted Arabidopsis Targetsa | Species | Target | Accession No. | Homolog/Species/e-Valueb | Description of Homolog |
|---|---|---|---|---|---|---|
| 160 | At1g77850(ARF17), At2g28350(ARF10), At4g30080(ARF16) | C. thalictroides | Fern-160-1 | AY974155 | At4g30080/Arabidopsis/1e-06 | ARF16 |
| P. juniperinum | Moss-160-1 | AY974165 | At4g30080/Arabidopsis/8e-19 | ARF16 | ||
| P. juniperinum | Moss-160-2 | AY974166 | At4g30080/Arabidopsis/7e-16 | ARF16 | ||
| 167 | At1g30330(ARF6), At5g37020(ARF8) | P. resinosa | Pine-167-1 | AY974146 | At1g30330/Arabidopsis/3e-29 | ARF6 |
| 171 | At2g45160(SCL6-II), At3g60630(SCL6-III), At4g00150(SCL6-IV) | C. thalictroides | Fern-171-1 | AY974159 | At3g60630/Arabidopsis/2e-45 | SCL6-III |
| 172 | At2g28550(TOE1), At2g39250, At4g36920(AP2), At5g60120(TOE2), At5g67180(TOE3) | P. resinosa | Pine-172-1 | AY974149 | BAD16604.1/P. thunbergii/1e-32 | APETALA2-like protein 2 |
| P. resinosa | Pine-172-2 | AY974150 | BAD16603.1/P. thunbergii/0.015 | APETALA2-like protein 1 | ||
| C. thalictroides | Fern-172-1c | AY974163 | At4g36920/Arabidopsis/3e-58 | APETALA2 |
Target predictions from Jones-Rhoades and Bartel (2004). Experimentally confirmed targets are underlined. Citations for individual target confirmations are found in Jones-Rhoades and Bartel (2004), with the exception of ARF16, which was confirmed recently by Mallory et al. (2005).
BLASTp was performed using the deduced protein sequence of the target against Arabidopsis proteins (AGI proteins); the protein scoring the highest is listed along with the e-value. In cases where no significant match was found in Arabidopsis, BLASTx was performed using the entire 3′ DNA sequence against the nonredundant protein database and the best hits listed, along with the e-value.
A full-length cDNA was isolated for fern-172-1 and used for the subsequent BLAST searches.
A Moss Small RNA Population Has Similarities with Those of Higher Plants
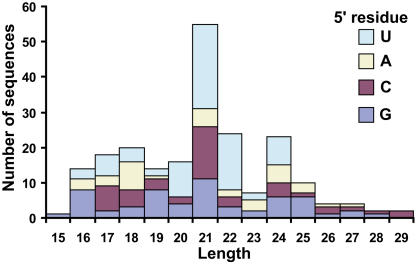
The observation of two deeply conserved miRNAs that were shifted in register compared with the flowering plant versions raised the possibility that basal land plants might contain many more conserved miRNAs with register shifts that would render them undetectable by the microarray. To begin to test this, we cloned 214 unique, non-tRNA or rRNA-derived small RNAs from the moss P. juniperinum and compared their sequences to the known set of plant miRNAs. As expected, we cloned miR160, which was identical in sequence to the Arabidopsis version. However, there were no other moss small RNAs that were recognizable homologs of known miRNAs in the set of 214 sequences (see Supplemental Table 7 online).
The moss small RNA population had some characteristics reminiscent of those previously observed for Arabidopsis small RNA populations (Tang et al., 2003): As seen in Arabidopsis, a strong peak was observed at 21 nucleotides in length, and uridine was the most frequent 5′ residue of these 21mers (Figure 6). However, the strong peak of 24 nucleotide species possessing a 5′ adenine residue that is seen in Arabidopsis small RNA populations was not apparent in moss.
Figure 6.
Moss Small RNAs.
The lengths and 5′ residues of 214 nonredundant small RNAs from the moss P. juniperinum. RNA fragments that were obvious tRNA or rRNA fragments were eliminated before this analysis, and sequences cloned multiple times are only represented once. Sequences are listed in Supplemental Table 7 online.
DISCUSSION
We present an expression profile of Arabidopsis miRNAs within the major organs of the wild-type plant, highlighting negative correlation between miRNA and target mRNA accumulation. Exploiting the plant miRNA array to analyze RNA samples derived from widely divergent specimens revealed the deep conservation of many plant miRNA families, with at least eight families conserved since before the emergence of seed plants. The targets of these deeply conserved miRNAs in nonflowering plants are homologous to the known targets in Arabidopsis. This shows that regulatory units defined by given miRNA–target pairs have been conserved throughout the evolution of plants.
Expression Profile of Arabidopsis miRNAs and Their Targets
The global expression profile shown in Figure 2 demonstrates the organ specificity of some miRNAs, while highlighting a fairly uniform level expression for many others. In instances where there are previous RNA gel blot data, the global expression profile corresponded well: For instance, miR156/157 expression is very strong in seedlings (Reinhart et al., 2002), miR398 is strongly expressed in leaves (Jones-Rhoades and Bartel, 2004), and miR171 is strongly expressed within inflorescences (Llave et al., 2002).
Accumulation of miRNA target mRNAs was frequently negatively correlated with that of the corresponding miRNAs (Figure 3D). There are several probable sources of noise in the comparison of the two expression profiling data sets: The experiments used different RNA samples from specimens grown under slightly different conditions and in both cases represented only a crude dissection of the organism where many cell types were combined in single samples. Additionally, some plant miRNA targets have very stable 3′ cleavage fragments that may raise the mRNA expression value as detected by the ATH1 microarray. Because of these sources of noise, it is possible that the true extent of the negative correlations is higher. Expression profiling in human cells has revealed a similar phenomenon: Probable target genes (defined by partial complementarity to the miRNA and by their repression upon ectopic expression of the miRNA) have their lowest expression values within tissues where the corresponding miRNA is maximally expressed (Lim et al., 2005).
The observation that miRNA targets rarely accumulate to high levels in the organs in which the corresponding miRNAs are most highly expressed (Figure 3A) is consistent with the hypothesis that plant miRNAs often act to clear target messages at certain stages of development. However, this observation and the more general anticorrelation between miRNAs and their targets is likely to have more complex causes, with other contributions to mRNA target expression in addition to miRNA-mediated clearance. In animals, the magnitude of the miRNA-induced downregulation of targets appears too small to fully explain the anticorrelation between miRNA and target expression, suggesting that the miRNAs are reinforcing regulation also occurring at the transcriptional and other levels (Lim et al., 2005). The same could be true in plants. Indeed, the paralogs of miRNA targets, which themselves have not been confidently predicted as targets, also rarely accumulate to high levels in the organs in which the corresponding miRNAs are most highly expressed (Figure 3B). In some instances, this could indicate functional miRNA complementary sites with more mispaired residues than have been allowed when reliably predicting plant miRNA targets. For instance, one of the paralogs of the miR172 targets, AINTEGUMENTA (At4g37750), has a site with six mismatches to miR172, none of which occur in the region complementary to the 5′ of the miRNA—a region that has been shown to be critical for plant miRNA function (Mallory et al., 2004b; Parizotto et al., 2004; Schwab et al., 2005). This pairing to miR172, together with its AP2-like domain, which is present in all of the more confidently predicted miR172 targets, suggests that AINTEGUMENTA may also be a direct target of miR172. Nonetheless, in the absence of any evidence to the contrary, it is possible that the similar expression profiles of miRNA targets and their closely related paralogs is not due to the direct action of miRNAs on highly mispaired complementary sites, but is instead due to alternative, non-miRNA–mediated control that enables the expression of the paralogs of the miRNA targets to mirror that of the targets. Such non-miRNA–mediated control processes could also be influencing expression of the miRNA targets. This being said, the observation that most miRNA targets are transcription factors (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004) brings up the possibility that in some cases expression of nontarget paralogs might be transcriptionally regulated by miRNA targets, such that miRNA-mediated repression of targets indirectly leads to repression of paralogous nontargets as well.
The Antiquity of Plant miRNAs: Evolutionary and Developmental Implications
The microarray platform allowed the direct detection of deeply conserved miRNAs from a gymnosperm, a fern, a lycopod, and a moss. The fact that these basal land plants with radically different lifestyles and morphologies share miRNAs in common with flowering plants indicates that these miRNAs have long been under selection pressure. Sequencing of a limited amount of moss small RNAs showed that there is a large and diverse population of 21-nucleotide species that predominantly possess a 5′ uridine residue. Arabidopsis miRNAs and trans-acting siRNAs are most often 21 nucleotides in length (Reinhart et al., 2002; Vazquez et al., 2004), and miRNAs have a strong bias toward uridine as the 5′ residue (Reinhart et al., 2002); thus, this initial sampling of small RNAs in moss suggests a wealth of small silencing RNAs in lower plants. The anticipated completion of the P. patens genome will soon enable the examination of potential miRNA families that have emerged specifically in the bryophyte lineage or have been lost in flowering plants.
Most of the 11 miRNA families detected in pine and all eight families detected in fern have targets in Arabidopsis that are developmentally implicated, either as DNA binding transcription factors or as a core component of the miRNA machinery itself (miR168; AGO1). The deeply conserved miR390, cloned from Arabidopsis by the Carrington group as ASRP754 (http://asrp.cgrb.oregonstate.edu/; Gustafson et al., 2005), from rice by Sunkar et al. (2005), and computationally predicted by several groups (Bonnet et al., 2004; Wang et al., 2004; Adai et al., 2005), does not have any confirmed targets in Arabidopsis. However, Sunkar et al. (2005) have recently demonstrated that rice miR390 targets an mRNA encoding a Leu-rich repeat containing receptor-like kinase (RLK). Pairing guidelines used to predict the targets of plant miRNAs (Jones-Rhoades and Bartel, 2004) suggest that miR390 could potentially regulate several Arabidopsis RLK mRNAs (At1g34110, At1g55610, At1g56130, At1g73070, At3g24660, At3g43740, At4g08850, At5g07180, At5g14210, At5g44700, At5g49660, and At5g62230), which are homologous to the confirmed miR390 target in rice. However, despite extensive attempts, we have been unable to detect 3′ cleavage fragments indicative of miR390-mediated cleavage for any of these possible RLK targets in Arabidopsis. There are 3 to 3.5 mismatches (counting G:U wobbles as 0.5 mismatches) between miR390 and each of these Arabidopsis RLK mRNAs, which is just at the cutoff for confident target prediction (Jones-Rhoades and Bartel, 2004). The Arabidopsis miRNAs whose targets are not obviously involved in developmental control (e.g., miR161, miR163, miR397, and miR398) were not detected outside of flowering plants. Because the predicted or confirmed Arabidopsis targets of all of the miRNA families detected in nonflowering plants have at least potential developmental connections, we propose that the deeply conserved miRNAs are primarily involved in ancient circuits of gene regulation whose outputs have been affecting the morphology of plants throughout their diversification.
The discovery of miRNA targets from basal plants (Figure 5) demonstrates unequivocally that several miRNA–target interactions have been constant throughout plant evolution. mir160, miR167, miR170/171, and miR172 all direct cleavage of targets in nonflowering plants whose closest known homologs are the very same targets that they are known or thought to cleave in Arabidopsis, as do miRNAs in the miR165/166 family (Floyd and Bowman, 2004).
Technical limitations of our target discovery strategy might have prevented identification of additional targets: Target discovery depends first upon the presence of full-length target mRNAs in the sample and is probably helped by having target sites close to the 3′ end of the transcript and by targets with short 3′ untranslated regions, all of which combine to make amplification of the 3′ portion of the message more robust (Figure 5A, step 2). Target validation depends on there also being a reasonably sized pool of somewhat stable 3′ cleavage fragments present in vivo. It is probable that one or more of these factors prevented our detection of the miR165/166 targets previously reported by Floyd and Bowman (2004) in gymnosperms and lycopods, who analyzed preselected mRNAs for evidence of miRNA-mediated cleavage. It is likely that careful construction of representative cDNA libraries coupled with more extensive PCR optimization could enable the discovery of additional targets from plants that have not yet been sequenced.
If, as these data suggest, most ancient miRNAs in plants have always been regulating the same targets, it follows that the downstream molecular effects of deeply conserved miRNA circuits may also be conserved, although perhaps with differing morphological outcomes. Such highly conserved, molecularly compact developmental modules would seem to be excellent substrates for the natural selection of plant form. Although the molecular identities of the miRNAs and their targets have remained constant, it is easy to envision that small changes in the temporal, spatial, or environmental regulation of these modules over time could have had large phenotypic effects on plant morphology. It is interesting to consider the extent to which the deeply conserved miRNA–target modules may have been recruited for nonhomologous functions in different plant lineages. Understanding in molecular detail both miRNA regulation of these conserved targets and how the targets themselves cause their downstream effects in diverse model systems should significantly enhance the understanding of the molecular roots of plant morphology.
METHODS
Array Design
The At_Dm_Ce_v1 array consists of 225 C6-aminolink deoxyoligonucleotide probes spotted in quadruplicate onto Codelink slides (Amersham Biosciences, Piscataway, NJ) as described for a vertebrate miRNA array (Baskerville and Bartel, 2005). Fifty spots were designed against Arabidopsis thaliana small RNAs, 13 against predicted Oryza sativa miRNAs, 79 against Drosophila melanogaster miRNAs, and 88 against Caenorhabditis elegans miRNAs (five probes detect miRNAs conserved between C. elegans and D. melanogaster). Probes were either shortened or lengthened (using nucleotides complementary to our 5′ adapter sequence) to obtain a nearest neighbor Tm (20 nM probe concentration, 50 mM NaCl; Breslauer et al., 1986) of ∼55°C (mean = 54.79°C, sd = 1.33°C). Plant small silencing RNAs can be classified as either miRNAs or siRNAs by the structure of their parent genes and their genetic requirements for biogenesis and function (Reinhart et al., 2002; Bartel, 2004; Xie et al., 2004). The 63 plant probes corresponded to 45 miRNAs, two endogenous siRNAs, one trans-acting siRNA, and 15 as yet unclassified small RNAs (see Supplemental Table 1 online). Arabidopsis probes were from the following sources: 30 Arabidopsis miRNAs from the miRNA registry (http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml), two Arabidopsis miRNAs from the Arabidopsis Small RNA Project (ASRP; http://asrp.cgrb.oregonstate.edu/; Gustafson et al., 2005), seven unclassified small RNAs (representing three families) cloned by the ASRP that met our criteria for inclusion, five novel small RNAs found as a result of ongoing small RNA cloning in our laboratory (R. Rajagopalan and D.P. Bartel, unpublished data), two predicted family members of these five new RNAs, and four previously described endogenous siRNAs, which also appear in the ASRP database. The O. sativa probes were derived from the miRNA registry. Separate probes were made for related sequences if the two sequences differed by one or more nucleotides in the center portion of the sequence, which was defined as more than four nucleotides from both the 5′ and 3′ ends. A set of 225 reference oligonucleotides containing a sense version of each target sequence flanked by sequences representing our 5′ and 3′ adapters was also synthesized for use during hybridizations, as described (Baskerville and Bartel, 2005). Amplification and Cy5 end-labeling of this synthetic set of oligonucleotides provide a constant reference signal that allows comparison of different Cy3-labeled biological samples (Baskerville and Bartel, 2005). In this method, both the reference and experimental samples consist of single-stranded, end-labeled DNA. A complete listing of target RNAs, probes, and reference oligonucleotides is found in Supplemental Table 1 online.
RNA Sources and Extractions
C. elegans RNA was obtained from wild-type, mixed stage worms and from glp-4(bn2) worms cultured under standard conditions. Arabidopsis total RNA samples from inflorescences (stages 1 to 12), siliques (>4 d after fertilization), stems, cauline leaves, and rosette leaves were harvested from wild-type Col-0 50- to 60-d-old, long-day (16 h light/8 h dark) grown plants at 18°C. Arabidopsis root RNA samples were derived from Col-0 roots harvested from 14-d-old plants grown in constant light in liquid culture (1× MS salts + vitamins, 1% sucrose, and 5 mM Mes-KOH, pH 5.7), shaking at 60 rpm in constant light at 22°C. Short-day and constant light seedling RNA samples were taken from Col-0 10-d-old seedlings grown in soil under an 8-h-light/16-h-dark regime or under constant light at 18°C, respectively. All biological replicate samples were derived from two separate crops grown at different times under the same conditions. Nicotiana benthamiana RNA was obtained from leaves of 21- to 28-d-old plants grown under long-day conditions (16 h light/8 h dark) at 26°C. O. sativa cv indica (rice) RNA was derived from 7-d-old seedlings grown under long-day conditions on plates containing 1× MS salts + vitamins, 1% sucrose, 10 mM Mes-KOH, pH 5.7, and 0.8% bacto-agar. Triticum aestivum (wheat) total RNA was derived from wheat germ lysate prepared as described (Tang et al., 2003). Liriodendron tulipifera (tulip tree—a Magnoliid) total RNA was harvested from mature leaves of a specimen located on Cambridge Street, Cambridge, MA, in July. Pinus resinosa (red pine—a Gymnosperm) total RNA was derived from mature needles of a specimen located in John F. Kennedy Park, Cambridge, MA, in July. Ceratopteris thalictroides (water sprite—a fern) total RNA was derived from the leaves and stems of a specimen purchased from Doctors Foster and Smith (Rhinelander, WI). Selaginella uncinata (a lycopod) total RNA was derived from the leaves and stems of a specimen purchased from Plant Delights Nursery (Raleigh, NC). Polytrichum juniperinum (a moss) total RNA was derived from leafy gametophytes collected in Nickerson State Park, Brewster, MA, in October. Total RNA from all Arabidopsis, N. benthamiana, O. sativa, and T. aestivum samples was harvested as described by Mallory et al. (2001). Total RNA from all other specimens was prepared using a method for pine tree RNA isolation (Chang et al., 1993).
Array Hybridizations
Small RNAs were fractionated, sequentially ligated to 3′ and 5′ adapters, and reverse transcribed as described (Lau et al., 2001). First-stage PCR used oligonucleotides 17.92 and 17.93D (Lau et al., 2001) and proceeded until amplifications were in linear stage (as determined by visualization of products from successive cycles; typically 17 to 19 cycles). A 1/100 dilution of this reaction was used as template in a labeling PCR using oligonucleotides 5′ Cy3-labeled 17.93D and a reverse oligo containing a 20-nucleotide 5′ poly(A) tract followed by an internal 18-carbon spacer and the 17.92D sequence (17.92_c18_A20) for 10 cycles to create an asymmetric PCR product (Baskerville and Bartel, 2005). A Cy5-labeled reference library was generated by 10 cycles of PCR using 5′ Cy5-17.93D and 17.92_c18_A20 using a 45 nM pool containing equal amounts of all 225 reference oligonucleotides as template. Labeled PCR products were fractionated through a 6% denaturing polyacrylamide gel, enabling excision of the shorter Cy3- or Cy5-labeled strand. Samples were adjusted to 5 μM in water. For each hybridization, 2 μL of 5 μM Cy3-labeled sample and 2 μL of 5 μM Cy5-labeled reference was added to 20 μL of hybridization buffer (3.5× SSC, 1% [m/v] BSA, 0.1% [m/v] SDS, 93 μg/mL salmon testes DNA, 187 μg/mL Escherichia coli tRNA, and 37 μg/mL polyadenine) for a final concentration of 0.417 μM each. After heating for 4 min at 85°C, samples were applied to arrays that had been prehybridized for 45 min in 3.5× SSC, 1% (m/v) BSA, 0.1% (m/v) SDS, rinsed with deionized water, and dried. Arrays were incubated at 57° for 16 h, then washed for 5 min at 50° in 2× SSC, 0.1% SDS, followed by 10 min at room temperature in 0.1× SSC, 0.1% SDS, and 3 × 1 min at room temperature in 0.1× SSC. Arrays were then dried and scanned using the GenePix 4000B (Axon Instruments, Union City, CA) at 10 μm per pixel, line average two, and constant photomultiplier tube gains for both 635 nm and 532 nm.
miRNA Array Data Analysis
Raw data was extracted from scanned array images using GenePix Pro 5.1 (Axon Instruments). Spots with an unacceptably low signal in the reference channel (defined as less than or equal to the median background at 635 nm plus 4 standard deviations) were eliminated from analysis, as well as rare spots whose median intensities at either 532 or 635 nm were saturated. Median local background was then subtracted from median spot intensities to arrive at background-corrected median intensities in both channels for all spots. Typical global normalizations for standard two-channel arrays operate on the assumption that, on average, the total intensities of both channels should be equal (Causton et al., 2003); this assumption is clearly false for experiments comparing a constant, synthetic sample against varying biological samples. Instead, a limited global normalization was performed based upon the noncognate spot intensities: The summed median intensities in both the 635 channel (Cy5) and 532 channel (Cy3) were derived from all noncognate spots (i.e., the D. melanogaster and C. elegans spots for plant experiments) from each hybridization. The ratio of total noncognate Cy3/total noncognate Cy5 (a) was calculated for each hybridization, and a mean ratio (ā) derived from all arrays to be compared. For each array n, a normalization factor bn was derived by dividing an/ā; the final value for each spot was the ratio of background corrected median Cy3/background corrected median Cy5 divided by bn. Because of our desire to always use the same amplified, synthetic Cy5-labeled reference set for every experiment, we did not perform dye-swap experiments. Thus, although we cannot rule out small dye-specific effects, they are expected to be minimal because the dyes were introduced via end-labeled oligonucleotides rather than by direct incorporation and because our normalization procedure incorporates the nonspecific background Cy3/Cy5 ratio in its calculations. After normalization, the four replicate spots for each small RNA were averaged together. The determination of a lower limit of detection (thresholding) was also guided by the presence of the noncognate probes: Values for all noncognate spots in a given analysis were compiled into a histogram. The value at which greater than or equal to 99% of all noncognate values were lower was called the lower limit of detection in these analyses. Finally, RNAs that were called detected and whose total Cy3 + total Cy5 intensities were in below the 25th percentile of all spots in the analysis were manually reexamined and eliminated from consideration if warranted. To find small RNAs that are differentially expressed in at least one of the organs studied, the values derived from the four replicate spots on each array were first condensed to the mean. Single-factor analysis of variance was performed on the 29 small RNAs that were expressed at detectable levels in at least half of the organs tested in all biological replicates, and those with P-values < 0.01 were listed as being differentially expressed. Using the Bonferroni-Holm stepdown correction to adjust P-values for multiple comparisons, we find that eight of these (F3-3_B01-5, miR157, miR172, miR156, miR396, miR398, miR160, and miR163) have corrected P-values < 0.05, whereas six [miR167, miR169, miR394, miR158, siR480(+), and miR171] have corrected P-values between 0.05 and 0.118. Hierarchical clustering of log2 transformed values was performed with Cluster (M. Eisen, Stanford University, Stanford, CA) and visualized using Java Treeview (M. Eisen). Files containing the normalized, detected, and log2-transformed data used in the C. elegans analysis, Arabidopsis organ map, and the phylogenetic survey are available in Supplemental Tables 3 to 5 online, respectively.
mRNA Array Data Analysis
Raw data from the following triplicate experiments were downloaded from the AtGenExpress expression atlas of wild-type Arabidopsis development (The Arabidopsis Information Resource, http://www.arabidopsis.org/, accession number ME00319): ATGE_7 (green parts of seedlings, 7 d, 23°C,continuous light, soil grown), ATGE_13 (rosette leaf 4,1 cm long, 17 d, 23°C, continuous light, soil grown,), ATGE_26 (cauline leaves, 21+ d, continuous light, 23°C, soil grown), ATGE_27 (stem, 2nd internode, 21+ d, continuous light, 23°C, soil grown), ATGE_29 (shoot apex, inflorescences, 21 d, continuous light, 23°C, soil grown), ATGE_78 (siliques, with seeds, stage 5; 8 weeks, continuous light, 23°C, soil grown), and ATGE_93 (roots, 15 d, long days [16/8], 22°C, 1× MS agar with 1% sucrose). These data correspond to our miRNA array data for long-day seedlings, rosette leaves, cauline leaves, stems, inflorescences, siliques, and roots, respectively. Raw expression values from each hybridization were normalized by dividing each value by the median value of the chip and multiplying the result by 100. The resulting expression values for the miRNA targets of the differentially expressed miRNAs shown in Figure 2B, control WRKY and MADS box genes, and paralogous nontargets were retrieved. These values were normalized on a per-gene basis, such that each value was divided by the median value of that gene across all tissues examined. Expression values from calls flagged “absent” or “marginal” were excluded from the median calculation and subsequently substituted for the lowest observed “present” value for that gene in the experiments examined. The median, log2-transformed values for each gene of interest were then calculated and plotted against the medians of similarly normalized relative expression values of the cognate miRNAs. Linear correlation coefficients between relative miRNA and relative target, paralogous nontarget, and control mRNAs were calculated as by Baskerville and Bartel (2005). The targets were as defined by Jones-Rhoades and Bartel (2004), with the exception of the targets of the trans-acting siRNA designated siR480(+), for which we examined the expression levels of At5g18040 and At4g29770, which were validated by Vazquez et al. (2004); note that for three of the differentially expressed small RNAs (miR163, miR158, and the unclassified small RNA F3-3_B01-5) shown in Figure 2B, there are no conserved predicted targets. The control genes were 20 randomly selected WRKY and 15 randomly selected MADS box transcription factors that were flagged “present” in the majority of the tissues analyzed. Two different random WRKY genes and either one or two different random MADS box genes were randomly paired with the values for each of the 10 miRNAs with known targets. Gene pairings and normalized median relative expression levels for this experiment are located in Supplemental Table 8 online.
RNA Gel Blots
Approximately 25 μg of total (lanes 1 to 5 of Figure 4B) or poly(A)-depleted (lanes 6 to 9 of Figure 4B) RNA was fractionated through a 15% polyacrylamide-urea gel along with 33P end-labeled RNA standards and transferred and fixed to a nylon filter as described (Lau et al., 2001). 32P end-labeled DNA oligonucleotides antisense to the Arabidopsis miR390 (5′-GGCGCTATCCCTCCTGAGCTT-3′), Arabidopsis miR160 (5′-TGGCATACAGGGAGCCAGGCA-3′), and the U6 small nuclear RNA (snRNA) (5′-TTGCGTGTCATCCTTGCGCAGG-3′) were prepared using T4 polynucleotide kinase. Hybridizations were performed in PerfectHyb-Plus (Sigma-Aldrich, St. Louis, MO) supplemented at 100 μg/mL with denatured salmon testes DNA, at 20°C below the Tm of the probe, where Tm is defined by 4(n G+C) + 2(n A+T). Blots were washed at 50°C 2 × 10 min in 2× SSC, 0.2% SDS followed by 2 × 5 min in 0.5× SSC, 0.2% SDS, and imaged using a phosphor imager. Blots were stripped in between hybridizations by washing for 30 min, at room temperature, with 200 mL of initially boiling 0.1% SDS, and exposed for at least 16 h to verify complete removal of probe before rehybridization.
Empirical Discovery of miRNA Targets and miRNA 5′ Definition
The 3′-RACE oligonucleotides were designed for queried miRNA targets (see Supplemental Table 6 online) to be antisense to a consensus of known Arabidopsis targets and EST homologs of Arabidopsis targets containing plausible miRNA complementary sites (Jones-Rhoades and Bartel, 2004); therefore, some contained degeneracy at certain positions. Poly(A) RNA from P. resinosa, C. thalictroides, S. uncinata, and P. juniperinum was selected from total RNA using batch binding with Oligotex beads, as recommended by the manufacturer (Qiagen, Valencia, CA). cDNA libraries were constructed using an RNA-ligase mediated procedure (GeneRacer; Invitrogen, Carlsbad, CA). Two libraries were made for each sample; in the full-length library, poly(A) RNA was treated with calf intestinal phosphatase then with tobacco acid pyrophosphatase to enrich for capped messages, and in the cleavage library, these steps were omitted to enrich for miRNA-mediated cleavage products. In both cases, reverse transcription was primed with the GeneRacer oligo(dT) primer. The 3′-RACE was performed using the full-length libraries as templates, miRNA complementary site specific 3′-RACE oligonucleotides, and the GeneRacer 3′ oligo. Bands were gel-purified, cloned, and sequenced, and the sequence was then used to design a 5′-RACE oligo specific for the candidate cDNA. 5′-RACE, using cleavage libraries first nonspecifically amplified using GeneRacer 5′- and 3′-oligonucleotides, was then performed with GeneRacer 5′-nested oligo and gene-specific oligonucleotides to amplify 3′ cleavage products (Kasschau et al., 2003). Any resulting bands were gel-purified, cloned, and sequenced to determine 5′ ends. For the fern-172-1 target, a full-length cDNA was obtained from the full-length fern library using the GeneRacer 5′ oligo and the gene-specific oligo (5′-TGCGGAGCTAGTGCAGGTTCTGAAA-3′). PCR-based detection and 5′ end definitions used the primary RT-PCR of the fern small RNA library with an oligo corresponding to the 5′ adapter used during cloning (5′-ATCGTAGGCACCTGAAA-3′) and the miRNA-specific oligonucleotides (fern miR171, 5′-AGCGATATTGGCGCGGC-3′; fern miR172, 5′-GCAGCATCATCAAGA-3′) exactly as described by Lim et al. (2003). All oligonucleotide sequences used in the course of these experiments that are not listed above are listed in Supplemental Table 6 online.
Cloning and Sequencing of P. juniperinum Small RNAs
Cloning of small RNAs was performed according to Lau et al. (2001). Sequences were filtered to remove labeled marker RNAs, snRNA fragments, tRNA fragments, and rRNA fragments. Because there is no genomic sequence data currently available for P. juniperinum, it is possible that the filtered data set may contain some unrecognized portions of snRNAs, tRNAs, and rRNAs, as well as sequencing errors. The sequences and cloning frequencies of the filtered P. juniperinum small RNAs are available within Supplemental Table 7 online.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY974141 through AY974169. Microarray data from this article have been deposited with the ArrayExpress data libraries under accession numbers E-MEXP-291 through E-MEXP-293.
Supplementary Material
Acknowledgments
We thank the AtGenExpress project (coordinated by L. Nover, T. Altmann, and D. Weigel) for the availability of the expression atlas of normal Arabidopsis development. We also thank the Whitehead Institute Center for Microarray Technology (Cambridge, MA) for printing the array, S. Baskerville for advice at all stages of array design and implementation, R. Rajagopalan for sharing novel small RNA sequences, M. Jones-Rhoades for generous computational assistance and constructive comments, as well as H. Vaucheret, A. Grimson, and A. Mallory for constructive comments on this manuscript. This research was supported by a Helen Hay Whitney Foundation postdoctoral fellowship to M.J.A. and by a National Institutes of Health grant to D.P.B.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael J. Axtell (axtell@wi.mit.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032185.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Adai, A., Johnson, C., Mlotshwa, S., Archer-Evans, S., Manocha, V., Vance, V., and Sundaresan, V. (2005). Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 15, 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36, 1282–1290. [DOI] [PubMed] [Google Scholar]
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak, T., Zhang, W., Morris, Q., Blencowe, B.J., and Hughes, T.R. (2004). Probing microRNAs with microarrays: Tissue specificity and functional inference. RNA 10, 1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.C., Sieber, P., Wellmer, F., and Meyerowitz, E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15, 303–315. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Baskerville, S., and Bartel, D.P. (2005). Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and Van de Peer, Y. (2004). Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101, 11511–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. (2003). bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36. [DOI] [PubMed] [Google Scholar]
- Breslauer, K.J., Frank, R., Blocker, H., and Marky, L.A. (1986). Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton, H.C., Quackenbush, J., and Brazma, A. (2003). A Beginner's Guide to Microarray Gene Expression Data Analysis. (Oxford: Blackwell Science).
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas, D.V., and Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7, 512–520. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Floyd, S.K., and Bowman, J.L. (2004). Gene regulation: Ancient microRNA target sequences in plants. Nature 428, 485–486. [DOI] [PubMed] [Google Scholar]
- Gustafson, A.M., Allen, E., Givan, S., Smith, D., Carrington, J.C., and Kasschau, K.D. (2005). ASRP: The Arabidopsis small RNA project database. Nucleic Acids Res. 33D, D637–D640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Kim, J., Jung, J.-H., Reyes, J.L., Kim, Y.-S., Kim, S.-Y., Chung, K.-S., Kim, J.A., Lee, M., Lee, Y., Narry Kim, V., Chua, N.H., and Park, C.-M. (2005). microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky, A.M., King, K.S., Donahue, C.P., Khrapko, K., and Kosik, K.S. (2003). A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9, 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Liang, R.Q., Li, W., Li, Y., Tan, C.Y., Li, J.X., Jin, Y.X., and Ruan, K.C. (2005). An oligonucleotide microarray for microRNA expression analysis based on labeling RNA with quantum dot and nanogold probe. Nucleic Acids Res. 33, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., and Johnson, J.M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. (2003). The microRNAs of Caenorhabditis elegans. Genes Dev. 17, 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.G., et al. (2004). An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 101, 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Bartel, D.P., and Bartel, B. (2005). MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17, 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004. a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004. b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- McHale, N.A., and Koning, R.E. (2004). MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16, 1730–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Gubler, F. (2005). The Arabidopsis GAMYB-Like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska, E.A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M., and Horvitz, H.R. (2004). Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 5, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, P.T., Baldwin, D.A., Scearce, L.M., Oberholtzer, J.C., Tobias, J.W., and Mourelatos, Z. (2004). Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 1, 155–161. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., and Lohmann, J. (3April2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 10.1038/ng1543. [DOI] [PubMed]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527. [DOI] [PubMed] [Google Scholar]
- Shigyo, M., and Ito, M. (2004). Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes Evol. 214, 105–114. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Koo, S., White, N., Peralta, E., Esau, C., Dean, N.M., and Perera, R.J. (2004). Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 32, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Girke, T., Jain, P.K., and Zhu, J.-K. (2005). Cloning and characterization of microRNAs from rice. Plant Cell 17, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, J.M., Parker, J., Perou, C.M., and Hammond, S.M. (2004). A custom microarray platform for analysis of microRNA gene expression. Nat. Methods 1, 47–53. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. [DOI] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.H., and Gaasterland, T. (2004). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2004). Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45, 369–385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.