Abstract
In animals, cyclin-dependent kinase inhibitors (CKIs) are important regulators of cell cycle progression. Recently, putative CKIs were also identified in plants, and in previous studies, Arabidopsis thaliana plants misexpressing CKIs were found to have reduced endoreplication levels and decreased numbers of cells consistent with a function of CKIs in blocking the G1-S cell cycle transition. Here, we demonstrate that at least one inhibitor from Arabidopsis, ICK1/KRP1, can also block entry into mitosis but allows S-phase progression causing endoreplication. Our data suggest that plant CKIs act in a concentration-dependent manner and have an important function in cell proliferation as well as in cell cycle exit and in turning from a mitotic to an endoreplicating cell cycle mode. Endoreplication is usually associated with terminal differentiation; we observed, however, that cell fate specification proceeded independently from ICK1/KRP1-induced endoreplication. Strikingly, we found that endoreplicated cells were able to reenter mitosis, emphasizing the high degree of flexibility of plant cells during development. Moreover, we show that in contrast with animal CDK inhibitors, ICK1/KRP1 can move between cells. On the one hand, this challenges plant cell cycle control with keeping CKIs locally controlled, and on the other hand this provides a possibility of linking cell cycle control in single cells with the supracellular organization of a tissue or an organ.
INTRODUCTION
During development of higher eukaryotes, many different cell types are produced, all of which can substantially differ in their cell cycle program (e.g., in the presence and length of the different cell cycle phases or in the proliferation activity) (Jakoby and Schnittger, 2004). Common cell cycle variants in both animals and plants are endocycles, in which cells replicate their DNA without undergoing a subsequent mitosis leading to polyploid cells (Edgar and Orr-Weaver, 2001). Endoreplication has been implicated in cell differentiation and cell growth, for instance, in the development of Drosophila melanogaster nurse cells, Medicago truncatula nodule cells, or Arabidopsis thaliana leaf hairs (trichomes) (Kondorosi et al., 2000; Edgar and Orr-Weaver, 2001; Schnittger and Hulskamp, 2002; Sugimoto-Shirasu and Roberts, 2003; Kondorosi and Kondorosi, 2004). In addition to cell type–specific settings, cell number and cell size are also coordinated on a supracellular level, maintaining tissue and organ growth in a highly predictable manner as well as taking environmental conditions into account (Day and Lawrence, 2000; Doonan, 2000; Potter and Xu, 2001).
The central convergence point of eukaryotic cell cycle control, where intrinsic and extrinsic cues are integrated, is a group of Ser/Thr kinases, CYCLIN DEPENDENT KINASEs (CDKs). Activated CDKs phosphorylate a plethora of proteins, resulting in the entry into a new round of DNA replication and the entry into mitosis, respectively. Recently, many putative CDK substrates have been identified (Ubersax et al., 2003). However, little is known about how CDK activity is modified for different cell cycle modes, in particular in endocycles.
One way of controlling CDK activity is mediated by CDK inhibitors (CKIs) that stochiometrically bind to CDKs and inhibit their kinase activity. In animals, two classes of inhibitors have been identified, the Inhibitor of CDK4 (INK4) class and the CDK Interacting Protein/CDK Inhibitor Protein (CIP/KIP) family.
The INK4 class comprises p15, p16, p18, and p19, which inhibit CDK4 but can also bind to CDK6. Inhibitors of the CIP/KIP family block cyclin D–, E–, and A–dependent kinases, but predominantly inhibit CDK2 activity (Pavletich, 1999; Sherr and Roberts, 1999). Besides a negative role in CDK regulation, CKIs have also been found to help assemble and stabilize a CDK4-cyclin D complex (Sherr and Roberts, 1999). It is not clear, however, whether these CDK-cyclin D-CKI complexes are active (Olashaw et al., 2004).
Putative CKIs have also been found in plants (Wang et al., 1998; De Veylder et al., 2001; Jasinski et al., 2002). In Arabidopsis, seven proteins were identified, which display homologies to the animal p27Kip1 protein and thus were named INHIBITORs/INTERACTORs OF CDK (ICKs) or KIP RELATED PROTEINS (KRPs) (Wang et al., 1998; De Veylder et al., 2001). The homology to p27Kip1, however, is restricted to ∼30 amino acids in the C terminus, and information about plant CKIs is still very limited.
In yeast two-hybrid interaction assays, ICK1/KRP1 could bind to CDKA;1 and CYCLIN D3;1, and it has been demonstrated that ICK1/KRP1 can inhibit the histone phosphorylation activity of CDKA;1 in vitro (Wang et al., 1997, 1998). In several misexpression studies, it has been found that ICK/KRPs can block endoreplication and reduce cell numbers, leading to dwarfed plants in extreme cases (Wang et al., 2000; De Veylder et al., 2001; Zhou et al., 2002; Schnittger et al., 2003). All these results are consistent with the presumed function of ICK/KRPs as inhibitors of CDKs at the G1-S transition point.
Here, we show that ICK1/KRP1 can also function outside of a G1-phase. After misexpression of ICK1/KRP1, we observed that cells skipped mitosis and underwent endoreplication. Our data suggest that ICK1/KRP1 acts in a concentration-dependent manner and only blocks G1-S transition at high concentrations, whereas at low concentrations G2-M transition is blocked. In contrast with animal CKIs, we demonstrate that Arabidopsis ICK1/KRP1 can act in a non-cell-autonomous manner with possible implications for tissue organization and organ growth. In addition, we found that cells induced by ICK1/KRP1 to endoreplicate prematurely can still adopt a new cell fate in accordance with a developmental program. Moreover, we found that endoreplicated cells even can reenter mitosis, demonstrating the high degree of flexibility of plants cells during development.
RESULTS
Trichome-Neighboring Cells in ProGL2:ICK1/KRP1-Misexpressing Plants Are Enlarged and Have an Increased DNA Content
Arabidopsis trichomes have been used as model cells to study cell cycle regulation in a developmental context (Schnittger et al., 2002a, 2002b, 2003). Trichomes are single-celled leaf hairs, which are initiated with a controlled distance to each other in the basal part of young and growing leaves. Archetypical for many differentiating cells, incipient trichomes exit a mitotic program and switch to an endoreplication mode. Concurrent with outgrowth and initiation of branches, trichomes undergo approximately four rounds of endoreplication, leading to a mature trichome with three to four branches and a DNA content of ∼32C (Marks, 1997; Hulskamp et al., 1999).
Specific misexpression of ICK1/KRP1 in trichomes using the GLABRA2 promoter (ProGL2) resulted in trichomes with reduced cell size, a decreased number of branches, and reduced endoreplication levels, and finally these trichomes underwent cell death (Figures 1A and 1B) (Schnittger et al., 2003). This effect was enhanced in plants expressing an N-terminally truncated protein version (amino acids 2 to 108 were deleted), named ICK1/KRP1109, providing in planta evidence for a previously identified negative regulatory domain in the N terminus of ICK1/KRP1 (Wang et al., 1998; Schnittger et al., 2003).
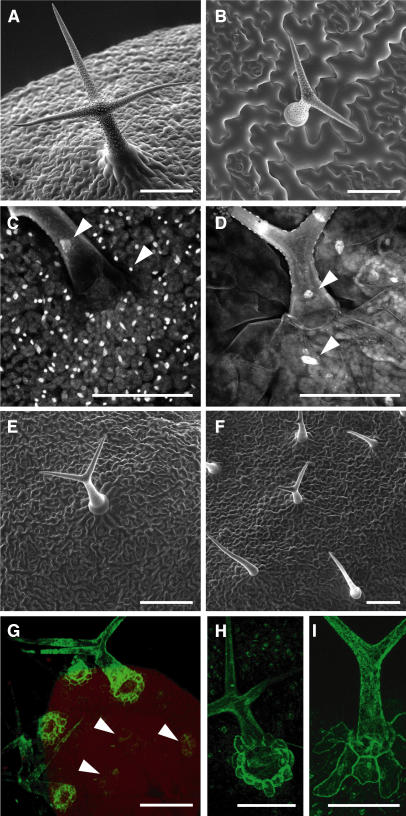
Figure 1.
Morphological Analysis.
(A) and (B) Scanning electron micrographs of mature Arabidopsis trichomes and their neighboring cells.
(A) Typical wild-type trichome with three branches surrounded by rectangular trichome-neighboring cells, which are polarized toward the trichome.
(B) Trichomes in ProGL2:ICK1/KRP1109 are smaller and develop fewer branches, whereas trichome-neighboring cells are lobed and greatly enlarged but are still oriented toward the trichome.
(C) and (D) Light micrograph of DAPI-stained trichomes and their neighboring cells; arrowheads point at trichome and trichome-neighboring cell nuclei, respectively.
(C) In the wild type, the nuclei of trichome-neighboring cells are much smaller than trichome nuclei.
(D) In ProGL2:ICK1/KRP1109 misexpressing plants, trichome-neighboring nuclei are much larger than in the wild type, whereas trichome nuclei are smaller.
(E) and (F) Scanning electron micrographs of glabra3 (E) and constitutive pathogen response5 (F) trichome mutants with reduced cell size and a decreased number of branches, yet trichome-neighboring cells are not enlarged or altered in shape.
(G) to (I) Confocal laser scanning micrograph (G) of enhancer trap line 254 showing the youngest state when GFP is detectable in the trichome-neighboring cells (arrowheads). At that time point, trichomes are already three branched. Close-up (H) of enhancer trap line 254 showing GFP fluorescence in a mature trichome and its neighboring cells. ProGL2:ICK1/KRP1109 crossed in 254 (I) showing GFP expression in the enlarged trichome-neighboring cells.
Bars = 100 μm.
Analyzing the cells surrounding a trichome on old rosette leaves of plants expressing the ProGL2:ICK1/KRP1109 construct, we made an unexpected observation: the trichome-neighboring cells were strongly enlarged (Figures 1A and 1B). Whereas wild-type trichome-neighboring cells reached in average a total surface cell area of ∼1200 μm2, on comparable leaves of ProGL2:ICK1/KRP1109 plants, trichome-neighboring cells encompassed a >10 times larger total surface area of ∼13500 μm2 (Table 1). Examining transgenic plants carrying the full-length ICK1/KRP1 misexpression construct, which showed a weaker trichome phenotype, we observed an enlargement of the trichome-neighboring cells to an average of 4800 μm2 (Table 1).
Table 1.
Total Surface Area of Trichome-Neighboring Cells
| Line | Total Surface Areaa | Σ Cells | |
|---|---|---|---|
| Landsberg erecta | 1,208 ± 493 | (1,114) | 40 |
| gl3 | 1,422 ± 789 | (1,159) | 38 |
| cpr5 | 1,103 ± 761 | (790) | 54 |
| ProGL2:KRP1*b | 4,755 ± 2,120 | (4,908) | 55 |
| ProGL2:KRP1109* | 13,459 ± 6,295 | (13,180) | 45 |
| ProGL2:YFP:KRP1* | 1,101 ± 425 | (1,154) | 46 |
| ProGL2:KRP1109:YFP* | 2,495 ± 1,253 | (2,007) | 86 |
| ProGL2:GUS:YFP:KRP1109* | 717 ± 346 | (587) | 54 |
| 254 | 675 ± 241 | (639) | 51 |
| ProUAS:YFP:KRP1109* in 254 | 738 ± 392 | (658) | 47 |
Total surface area of trichome-neighboring cells on rosette leaves was measured from at least five different plants per line; average ± sd and median in parenthesis are given in μm2.
ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
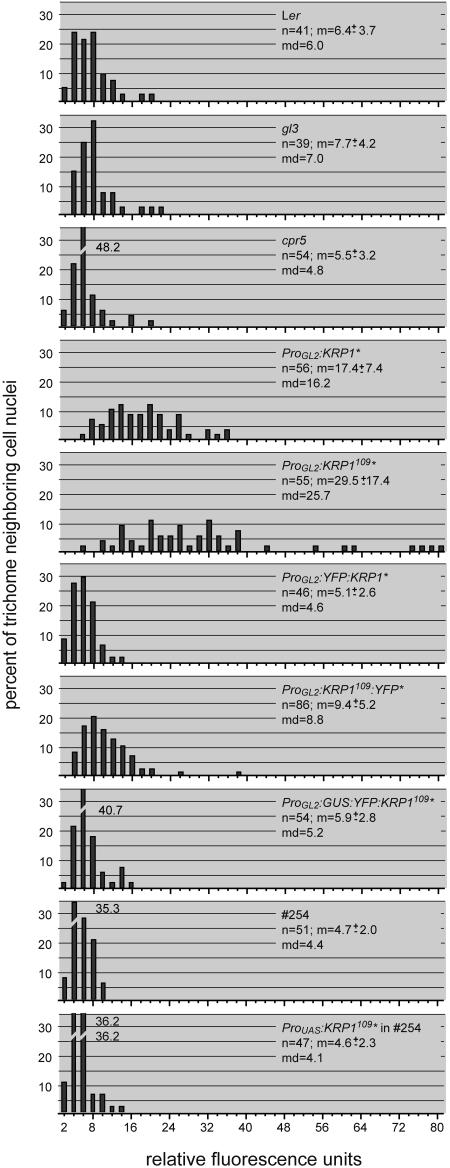
Because cell size is often correlated with the degree of cellular polyploidization, we measured the DNA content by quantifying the fluorescence of 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei (Figures 1C, 1D, and 2). We detected a strongly increased DNA content in the trichome-neighboring cells in the ICK1/KRP1-misexpressing plants, a mean of 17.4C versus 6.4C in the wild type (Figure 2). An even stronger increase in DNA levels was measured in plants expressing the truncated ICK1/KRP1109 construct with an average of 29.5C, and occasionally, extremely enlarged nuclei with up to 80C were found (Figure 2).
Figure 2.
Analysis of DNA Content.
Distributions of trichome-neighboring cell DNA contents are given in relative fluorescence units. Relative fluorescence units are calibrated with the fluorescence of guard cell nuclei of the analyzed leaves so that two relative fluorescence units represent ∼2C. The sample size (n), the mean (m) ± standard deviation, and the median (md) are given. ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
The observed cell enlargement and increase in nuclear size of the trichome-neighboring cells in the ICK1/KRP1-misexpression plants are reminiscent of a trichome developmental program. Trichome patterning is thought to involve a mutual inhibition mechanism, by which all epidermal cells compete with each other to adopt trichome cell fate (Larkin et al., 2003). Hence, we raised the hypothesis that because of a compromised and eventually dead trichome as a result of ICK1/KRP1 misexpression, the lateral inhibition is released and the trichome-neighboring cells start to develop into trichomes. To address this question, we used a very sensitive trichome-fate reporter construct, in which a fusion of a nuclear localization signal (nls) to the GREEN FLUORESCENT PROTEIN (GFP) and the β-GLUCURONIDASE (GUS) is under the control of the GL2 promoter (ProGL2:nls:GFP:GUS). Analysis of this marker line, however, revealed no expression in cells surrounding the ICK1/KRP1-misexpressing trichomes, indicating that an initiated trichome developmental program is not responsible for the observed phenotype (Figures 3A and 3I).
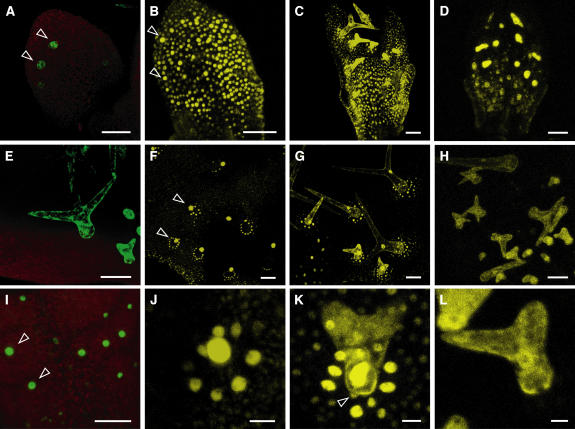
Figure 3.
Analysis of Intracellular and Intercellular Localization of ICK1/KRP1 and ICK1/KRP1109.
(A) to (L) Confocal laser scanning micrographs of rosette leaves.
(A) and (E) The expression of GFP5ER under control of the GLABRA2 promoter in young (A) and old (E) rosette leaves is only detectable in trichome and trichome precursor cells. Two young developing trichomes are indicated by arrowheads.
(B) and (F) Localization and distribution of YFP:ICK1/KRP1 fusion protein in young (B) and old (F) rosette leaves. In young leaves, the nuclei of almost all epidermal cells show a YFP:ICK1/KRP1 signal. In old leaves, the YFP:ICK1/KRP1 signal is detected in the nuclei of trichomes and in concentric rings around a trichome in the nuclei of trichome-neighboring cells. Trichomes are indicated by arrowheads.
(C) and (G) Localization and distribution of ICK1/KRP1109:YFP fusion protein in young (C) and old (G) rosette leaves. The distribution of the fusion protein of the truncated ICK1/KRP1109 has an even greater range with two to three concentric rings around a trichome.
(D) and (H) GUS:YFP:ICK1/KRP1109 protein localization is shown in young (D) and old (H) leaves. GUS:YFP:ICK1/KRP1109 can only be detected in trichome precursor cells and trichomes but not in surrounding cells.
(J) to (L) Close-up of protein localization of YFP:ICK1/KRP1 (J), ICK1/KRP1109:YFP (K), and GUS:YFP:ICK1/KRP1109 (L) within a trichome and its neighboring cells. Whereas the full-length ICK1/KRP1 fused to YFP is strictly nuclear localized (J), the truncated ICK1/KRP1109:YFP is found in the trichomes in both compartments (nucleus and cytoplasm), and the trichome-neighboring cells show a nuclear localization. Note that ICK1/KRP1109:YFP could also be found in nuclei of mesophyll cells, indicated by an arrowhead (K).
(I) Analysis of the marker line ProGL2:nls:GFP:GUS crossed in ProGL2:ICK/KRP1109. A GFP signal is only detectable in the trichomes (arrowheads) and not in the surrounding cells.
Bar = 50 μm in (A) to (I) and 10 μm in (J) to (L).
To further investigate whether the enlargement of the trichome-neighboring cells could be a response to a compromised trichome-differentiation program, we analyzed the cells surrounding a trichome in glabra 3 (gl3) and constitutive pathogen response 5 (cpr5) mutant plants. Trichomes in both mutants have reduced endoreplication levels, are smaller than wild-type trichomes, and develop mostly only two branches (Hulskamp et al., 1994; Kirik et al., 2001). In addition, similar to trichomes on ProGL2:ICK1/KRP1-expressing plants, cpr5 mutant trichomes have been reported to die. However, neither the trichome-neighboring cells in the gl3 nor in the cpr5 mutant displayed any significant difference to wild-type trichome-neighboring cells with respect to cell size and DNA content (Figures 1E, 1F, and 2, Table 1).
Taken together, these data suggest that trichome-neighboring cell enlargement and increase in DNA content is due to ICK1/KRP1 misexpression in trichomes and is not a general feature of an altered trichome development.
Intercellular Localization of ICK1/KRP1
Based on the conclusion that the phenotype of trichome-neighboring cells is specific for ICK1/KRP1 misexpression, we reasoned two different scenarios by which ICK1/KRP1 could influence the cells surrounding a trichome. First, ICK1/KRP1 might act indirectly, and its expression in trichomes would induce a non-cell-autonomous response. Alternatively, given that plant cells are symplastically connected by plasmodesmata (Ding et al., 2003; Oparka, 2004), ICK1/KRP1 itself might move into the neighboring cells.
To test the localization and mobility of ICK1/KRP1, the yellow fluorescent protein (YFP) was fused to ICK1/KRP1 and ICK1/KRP1109, and misexpression lines using the GL2 promoter were generated. Homozygous lines were created, and based on mRNA expression strength, comparable lines were chosen as reference lines for further investigations (see Supplemental Figure 1A online). All data provided in the following was obtained from the same reference line. As a control, transgenic plants expressing a cell-autonomous version of the GFP with a localization signal for the endoplasmatic reticulum (ProGL2:GFP5ER) and plants expressing an untagged YFP protein (ProGL2:YFP) were created (Siemering et al., 1996; Haseloff et al., 1997; Crawford and Zambryski, 2000).
Plants expressing the fusion proteins were first analyzed with respect to their trichome phenotype. Plants carrying an N-terminal YFP fusion to ICK1/KRP1 (ProGL2:YFP:ICK1/KRP1) displayed smaller and under-branched trichomes, which eventually died, resembling the ICK1/KRP1-misexpression phenotype (Table 2; data not shown). The expression of ICK1/KRP1 with a C-terminal fusion (ProGL2:ICK1/KRP1:YFP) did not result in a phenotype, and transgenic plants were not further analyzed. For ICK1/KRP1109, plants misexpressing both N- and C-terminal fusion proteins with YFP resembled the phenotype of ProGL2:ICK1/KRP1109 plants (Table 2). Similarly to the expression of the unfused ICK1/KRP1, we recognized that expression of fusion proteins containing the N-terminally truncated ICK1/KRP1109 resulted in a stronger trichome phenotype than the expression of fusion protein with the ICK1/KRP1 full-length version (Table 2; data not shown). Thus, although fusions in the C terminus to the full-length ICK1/KRP1 seemed to interfere with protein action, we conclude that a fusion with YFP in the other three constructs did not result in an altered ICK1/KRP1 protein activity as judged by their trichome phenotypes.
Table 2.
Trichome Branch Number
| Number of Branches in Percentage per Leafa
|
|||||
|---|---|---|---|---|---|
| Line | 1 | 2 | 3 | 4 | Σ Trichomes |
| Landsberg erecta | 0.0 ± 0.0 | 0.2 ± 0.9 | 99.6 ± 1.2 | 0.2 ± 0.8 | 477 |
| ProGL2:KRP1*b | 13.7 ± 11.1 | 65.3 ± 14.4 | 21.0 ± 14.9 | 0.0 ± 0.0 | 428 |
| ProGL2:KRP1109* | 19.5 ± 15.3 | 71.1 ± 15.1 | 9.4 ± 12.9 | 0.0 ± 0.0 | 150 |
| ProGL2:YFP:KRP1* | 13.0 ± 4.9 | 58.7 ± 10.2 | 28.3 ± 9.4 | 0.0 ± 0.0 | 488 |
| ProGL2:KRP1109:YFP* | 14.8 ± 8.0 | 65.4 ± 7.2 | 19.8 ± 7.1 | 0.0 ± 0.0 | 338 |
| ProGL2:GUS:YFP:KRP1109* | 21.2 ± 6.2 | 59.2 ± 6.0 | 19.6 ± 3.9 | 0.0 ± 0.0 | 335 |
All trichomes on rosette leaf numbers 3 and 4 were counted from at least 10 plants per line; the average ± sd is given, and the branch number with the highest percentage is shown in bold.
ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
To determine whether the fusion proteins were expressed as complete proteins, we performed protein gel blot experiments of the generated transgenic plants and probed the blots with antibodies raised against GFP, which also recognizes YFP. For plants expressing ProGL2:YFP, a strong band could be detected at the expected size of 27 kD. We found that the majority of the ICK1/KRP1109 fusion proteins can be detected at the predicted size of 37 kD (see Supplemental Figure 1B online). For the full-length version, we could not detect a band, although on RNA level the construct appeared even to be slightly stronger expressed than the truncated version (see Supplemental Figures 1A and 1B online). We and others have previously shown that a negative regulatory signal resides in the N terminus of the ICK1/KRP1 protein (Schnittger et al., 2003; Zhou et al., 2003). The limitation in detection of the full-length CKI argues that this domain might regulate the stability of ICK1/KRP1 protein. Consistently, Zhou et al. (2003) recently reported that an N-terminally truncated version was present in much higher abundance than the full-length inhibitor.
Next, the localization of the fusion proteins was analyzed by confocal laser scanning microscopy. As controls, we first analyzed the expression of two GL2 reporter lines in a wild-type background and in plants expressing ProGL2:ICK1/KRP1109. In the wild type, both GFP5ER and a nls:GFP:GUS fusion protein expressed from the GL2 promoter were only detected in trichomes and trichome precursor cells (Figures 3A and 3E; data not shown). In the F1 generation of the cross of the ProGL2:nls:GFP:GUS reporter line with the reference line expressing ProGL2:ICK1/KRP1109, the GFP signal was still restricted to trichomes and trichome precursor cells, indicating that trichome-specific expression of ICK1/KRP1109 did not alter the expression domain of the GL2 promoter (Figure 3I).
In contrast with the trichome-specific localization of the two GL2 promoter reporter lines, we could detect the ICK1/KRP1 fusion proteins also in cells around trichomes. In young leaves, ICK1/KRP1 fusion protein could be detected in many epidermal cells (Figures 3B and 3C). In older leaves, the full-length ICK1/KRP1 fused to YFP was predominantly found in one to two concentric rings around a trichome, and the truncated version ICK1/KRP1109 was detectable in three to four rings with decreasing intensity (Figures 3F and 3G). Also, we could detect a weak YFP signal in the nuclei of mesophyll cells, demonstrating that movement of ICK1/KRP1 fused to YFP is not restricted to epidermal cells but reflects rather a general feature of ICK1/KRP1-YFP fusion proteins (Figures 3K, arrowhead). Based on these localization patterns, it is conceivable that the unfused ICK1/KRP1 when expressed in trichomes will also enter the neighboring cells.
A morphological analysis of the trichome-neighboring cells revealed, however, that only plants expressing the N-terminally truncated ICK1/KRP1109 fused to YFP displayed a significant increase in trichome-neighboring cell size and DNA content with ∼2500 μm2 and 9.4C (Table 1, Figure 2; data not shown). Thus, in contrast with trichomes, the alterations of the trichome-neighboring cells were correlated with the protein size of the misexpressed ICK1/KRP1 protein (i.e., smaller proteins caused a more severe phenotype: ICK1/KRP1109 [10 kD] > ICK1/KRP1 [22 kD] > ICK1/KRP1109:YFP [37 kD] > YFP:ICK1/KRP1 [49 kD]).
To address the dynamics of the movement of ICK1/KRP1 and to test whether larger fusion proteins were less abundant in trichome-neighboring cells than smaller ICK1/KRP1 versions, we compared the fluorescence intensities of ICK1/KRP1-YFP fusions with that of free YFP. As previously reported, the YFP-related GFP is able to diffuse up to 16 cells wide in microprojectile bombardment experiments in Arabidopsis (Itaya et al., 2000). Consistently, in the generated transgenic plants expressing YFP without any localization signals from the GL2 promoter (ProGL2:YFP), YFP could be detected in trichomes and in neighboring cells (see Supplemental Figures 2A and 2B online). Determination of the fluorescence intensity of trichome-neighboring cell nuclei in comparison with trichome nuclei revealed for ICK1/KRP1109:YFP (37 kD) a similar ratio of ∼0.5 as for YFP (27 kD), whereas for the larger ICK1/KRP1 fusion (49 kD), a lower ratio of ∼0.2 was obtained (see Supplemental Figure 2C online). This is consistent with a reduced movement and, therefore, a lower concentration of increasingly larger fusion proteins in trichome-neighboring cells.
However, we could not exclude that the different ICK1/KRP1 protein versions have different molecular properties in trichome-neighboring cells versus trichomes (e.g., protein stability and/or nuclear import rate), which could influence the ratio of fluorescence intensities independent of protein size. To test more directly for a protein size–dependent movement, we created transgenic plants expressing another ICK/KRP fusion protein, in which the GUS protein was combined with YFP:ICK1/KRP1109; the size of this fusion protein is ∼105 kD. Expression of GUS:YFP:ICK1/KRP1109 from the GL2 promoter caused a significant reduction in trichome branch number similarly to the other ICK1/KRP1 protein versions, demonstrating the functionality of this fusion protein (Table 2). Confocal laser scanning microscopy revealed that GUS:YFP:ICK1/KRP1109 was restricted to trichomes (Figures 3D, 3H, and 3L), and no increase in trichome-neighboring cell size nor DNA content was observed (Figure 2,Table 1).
Taken together, we conclude that ICK1/KRP1 can act non-cell-autonomously and that the phenotype of the trichome-neighboring cells in the ICK1/KRP1 misexpression lines is due to a direct action of the CKI in the neighboring cells.
Intracellular Localization of ICK1/KRP1
In animals, the intracellular localization of the CKI p27Kip1 is strictly regulated and appears to be inherently connected with protein abundance and activity (Sherr and Roberts, 1999; Slingerland and Pagano, 2000). The general notion is that p27Kip1 exerts its inhibitory function in the nucleus and becomes degraded in the cytoplasm (Tomoda et al., 1999; Connor et al., 2003). The regulatory elements that mediate p27Kip1 localization are not conserved in plant CKIs; therefore, we were interested in the intracellular localization of ICK1/KRP1.
Whereas YFP expressed from the GL2 promoter could be detected in the nucleus and the cytoplasm, ICK1/KRP1 fused with YFP exhibited a nuclear localization (Figures 3B, 3F, and 3J; see Supplemental Figure 2B online). While this work was in progress, a similar intracellular localization of ICK1/KRP1 was reported by analyzing GFP fusions with ICK1/KRP1 (Zhou et al., 2003). Consistent with the report by Zhou and colleagues, we found that YFP fusions with the truncated ICK1/KRP1109 localized to the nucleus and the cytoplasm in trichomes (Figures 3C, 3G, and 3K; data not shown); a cytoplasmic localization was even more prominent for the GUS:YFP:ICK1/KRP1109 fusion protein (Figures 3D, 3H, and 3L).
In the trichome-neighboring cells, however, both N- and C-terminal YFP fusions with ICK1/KRP1109 could only be detected in the nucleus (Figure 3K; data not shown). On the one hand, this could indicate different cell type–dependent dynamics of the intracellular localization of ICK1/KRP1. On the other hand, it is very well possible that a cytoplasmic fraction of ICK1/KRP1109:YFP was below the detection limit because already in the much brighter stained trichomes the cytoplasmic fluorescence was weak (see Supplemental Figure 2C for a reduction of fluorescence intensities in trichome-neighboring cells).
Premature Endoreplication Does Not Interfere with the Adaptation of Cell-Specific Marker Gene Expression
In the wild type, the cells directly neighboring a trichome develop into morphologically distinct cells, called socket or support cells. Socket cells are rectangular versus the typically lobed pavement cells and are oriented in their longitudinal axis toward the trichome (Figure 1A). In addition, the expression of a few genes and enhancer trap lines has been found to discriminate socket cells from epidermal pavement cells (Molhoj et al., 2001; Vroemen et al., 2003).
Because the trichome-neighboring cells in the ICK1/KRP1-misexpressing plants were greatly enlarged and developed lobes (Figure 1B), we asked whether these cells still have socket cell fate. The analysis of two GAL4 enhancer trap lines from the Scott Poethig collection (http://enhancertraps.bio.upenn.edu/) marking trichome socket cells, 232 and 254, crossed into the reference line for ProGL2:ICK1/KRP1109 revealed expression in the cells surrounding a trichome (Figures 1G to 1I; data not shown; note also the increase in cell size and the enlarged neighboring-cell nuclei in line 254 expressing ProGL2:ICK1/KRP1109). In addition, most of the cells surrounding a trichome were still polarized toward the trichome (Figure 1B). Taken together, these data suggested that the trichome-neighboring cells in ICK1/KRP1-misexpressing plants have developed, at least to some degree, into socket cells.
Entry into an endoreplication cycle has been found to be associated with cell differentiation and the adoption of the special cell morphology occurring after cell fate specification (Nagl, 1976; Sugimoto-Shirasu and Roberts, 2003). Our data, however, implied that trichome-neighboring cells in the ICK1/KRP1-misexpressing plants become specified as socket cells independent and after the onset of an endoreplication program. To explore this hypothesis, we analyzed the cell division activity around incipient wild-type and ICK1/KRP1-misexpressing trichomes more closely. Figure 4 shows that in cells adjacent to young and growing wild-type trichomes, newly formed cell walls can be found, indicating a recent cell division (Figures 4A to 4C). By contrast, around young trichomes of ICK1/KRP1-misexpressing plants, we observed that the neighboring cells had already started to enlarge (Figures 4D to 4F). Consistent with this, we found in DAPI staining that nuclei of trichome-neighboring cells in ICK1/KRP1-misexpressing plants had already started to endoreplicate in contrast with wild-type leaves (Figures 4G and 4H).
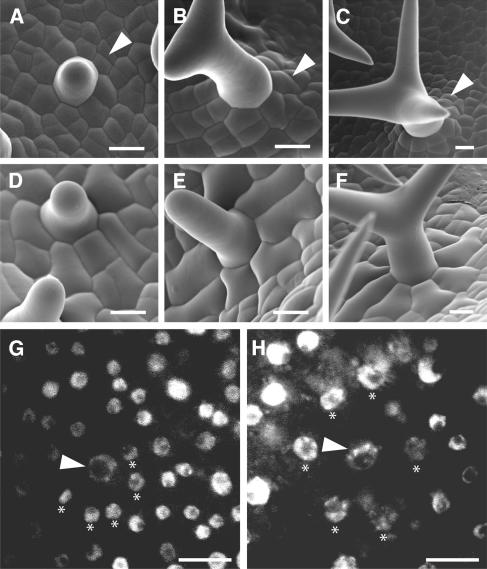
Figure 4.
Analysis of Cell Division Activity in Trichome-Neighboring Cells.
(A) to (F) Scanning electron micrographs showing the development of trichome-neighboring cells in wild-type ([A] to [C]) and ProGL2:ICK1/KRP1109 ([D] to [F]) plants. Wild-type trichome-neighboring cells divide until the centrally located trichome develops its third branch. Examples for newly formed cell walls are marked by arrowheads. By contrast, in ProGL2:ICK1/KRP1109 plants, trichome-neighboring cells enlarge and do not divide.
(G) and (H) Light micrographs of DAPI-stained trichomes with their neighboring cells in the wild type (G) and ProGL2:ICK1/KRP1109 (H) at an early stage of trichome development (corresponding to [A] and [D]). Correspondingly to the cell enlargement and the absence of cell division, trichome-neighboring cells in ProGL2:ICK1/KRP1109 plants start to endoreplicate as seen by the increased nuclear sizes of the trichome-neighboring cells in comparison with the wild type. Arrowheads point to the trichome nuclei, and the nuclei of the trichome-neighboring cells are marked by asterisks.
Bars = 10 μm.
As judged by their morphology, the dividing cells around an incipient trichome on wild-type plants have not acquired a specific fate (Figures 4A and 4B). Also, from previous studies it is known that trichomes and trichome socket cells are not of clonal origin, suggesting that socket cells become recruited by trichomes at some later stage of trichome development (Larkin et al., 1996). Consistent with this, the expression of the two socket cell markers used above only starts when the trichome is already three-branched and expanded (Figure 1G). Further evidence from the gl2 mutant supports an instruction of socket cells at a time point late during trichome development. In gl2 mutants, two classes of trichomes can be found: one class of expanding and even branching trichomes surrounded by socket cells, and the other class displays aborted trichomes, which had started to grow out but failed to expand and become arrested as young bulges (Koornneef, 1990; Rerie et al., 1994). In this latter class, stomata can be found to develop in direct contact with trichomes, suggesting that socket cells have not yet been specified. Finally, in the ICK1/KRP1-misexpressing plants, the socket cell marker became also expressed at later stages of trichome development (data not shown). Taken together, these findings suggest that in the ICK1/KRP1-misexpressing plants, endoreplication has started in the trichome-neighboring cells before these cells have been specified as socket cells; thus, we conclude that plant cells can be specified independent of an endoreplication program.
The Induction of Endocycles by ICK1/KRP1 Depends on the Cell Cycle Mode and the Developmental State
To test whether ICK1/KRP1 is generally a positive regulator of endoreplication in trichome-neighboring cells and its expression is always sufficient to promote endoreplication, we misexpressed ICK1/KRP1 at late stages of socket cell development. For that, ICK1/KRP1109 was cloned behind a UAS regulatory element and introduced into the GAL4 driver line 254 from the Scott Poethig collection by transformation (cf. Figure 1G) (http://enhancertraps.bio.upenn.edu/). Examining plants expressing pUAS:ICK1/KRP1109 in the GAL4 line 254 for a socket cell phenotype revealed neither an alteration in cell size nor in DNA content in comparison to line 254 itself or in wild-type plants (Figure 2, Table 1; see Supplemental Figure 1 online). This observation together with the finding that the trichome-neighboring cells will undergo a few cell division rounds when the GL2 promoter is already highly active (cf. Figures 3A and 4A to 4C) indicated that the induction of endocycles by ICK1/KRP1 depends on the developmental state and/or the cell cycle mode of the cells. This is also supported by the observation that in all transgenic lines generated expressing the various ICK1/KRP1 constructs in trichomes, we have never observed any indication for an increase of endoreplication levels in trichomes by ICK1/KRP1.
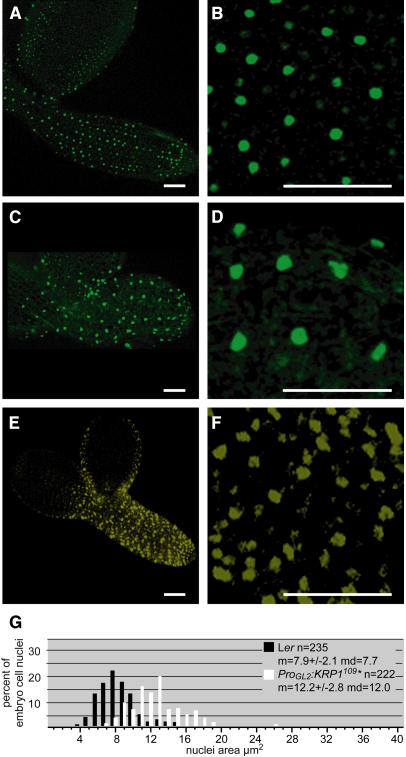
To test further whether induction of endocycles by ICK1/KRP1 depends on the cell cycle mode of the cells, we analyzed the effect of ICK1/KRP1 misexpression in other proliferating cells. For that, we made use of the observation that GL2 is also expressed during embryo development starting at heart stage and persisting until bent cotelydon stage (Lin and Schiefelbein, 2001; Costa and Dolan, 2003). Figures 5A and 5B show a torpedo stage embryo with the typical expression pattern of the GL2 promoter in approximately every second cell file in the embryonic epidermis. Expression of ICK1/KRP1 under the GL2 promoter did not alter this expression pattern, as revealed by the analysis of the GL2 promoter reporter line ProGL2:nls:GFP:GUS crossed into plants expressing ProGL2:ICK1/KRP1109 (Figures 5C and 5D). Similar to leaves, we found that ICK1/KRP1-YFP fusion proteins were found in almost all epidermal cells and also weaker in subepidermal cells, demonstrating that the movement of ICK1/KRP1 is not restricted to leaf cells (Figures 5E and 5F).
Figure 5.
Analysis of ICK1/KRP1109 Misexpression in Embryonic Epidermis Cells.
(A) to (D) Confocal laser scanning micrographs of ProGL2:nls:GFP:GUS reporter line in wild-type torpedo stage embryos (A) and ProGL2:ICK1/KRP1109 embryos (C). Close-up of hypocotyl epidermal nuclei in wild-type (B) and ProGL2:ICK1/KRP1109 (D) expressing embryos. Cells in the hypocotyl of a torpedo-stage embryo show GFP-positive nuclei in alternating cell files.
(E) and (F) Confocal laser scanning micrographs of a ProGL2:ICK1/KRP1109:YFP embryo. The YFP signal can be detected in all cell files of the hypocotyl. Bars in (A) to (F) = 50 μm.
(F) Close-up of hypocotyl epidermal nuclei.
(G) Analysis of the area of propidium iodide–stained hypocotyl nuclei of embryos of the same age for the wild type (black) and ProGL2:ICK1/KRP1109 (white), showing an enlargement of nuclear sizes in ProGL2:ICK1/KRP1109 expressing plants. The sample size (n), the mean (m) ± standard deviation, and the median (md) are given. ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
Next, we attempted to determine the DNA content of epidermal cells in embryos of plants misexpressing ICK1/KRP1. However, measurements of fluorescence intensities were compromised because of a small cell size and, therefore, a close vicinity of nuclei giving rise to high background fluorescence. Therefore, DNA levels were approximated by nuclear sizes. For that, we analyzed plants carrying a ProGL2:nls:GFP:GUS construct and compared the nuclear sizes of pGL2-positive cells in this line with ProGL2:ICK1/KRP1109 plants (Figures 5A to 5D). Nuclei in the ICK1/KRP1109-misexpressing embryos were larger than in the wild type, supporting the hypothesis that ICK1/KRP1 induced endoreplication in mitotic cells. A quantification of nuclear sizes using the DNA stain propidium iodide revealed approximately an area of 12 μm2 in ProGL2:ICK1/KRP1109 embryos, whereas in wild-type embryos, the nuclei of epidermal cells spanned an average area of ∼8 μm2 (Figure 5E).
Because of the experimental limitation of embryonic epidermis, we sought for another promoter active in dividing cells, yet not active in all mitotic cells to interfere as little as possible with plant fertility and viability. For this purpose, we used the promoter of the TOO MANY MOUTHS gene (ProTMM) (Nadeau and Sack, 2002a). TMM is expressed during early leaf development in cells of the stomatal lineage and some adjacent cells; many of these cells will undergo at least one more cell division during leaf development.
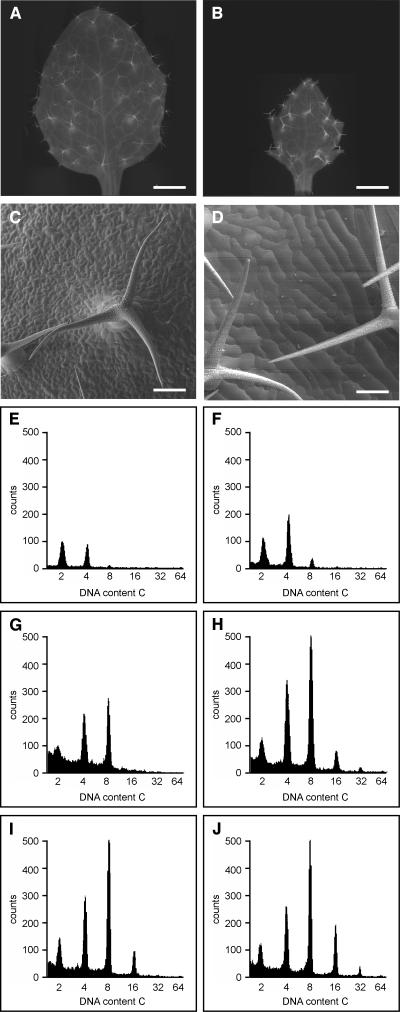
Transgenic plants misexpressing from the TMM promoter the N-terminally truncated ICK1/KRP1 version fused to YFP were generated. The phenotypes obtained were striking. Expression of ICK1/KRP1109 from the TMM promoter resulted in plants with smaller leaves with an increased degree of serration and displaying much fewer but greatly enlarged epidermal cells (Figures 6A to 6D). Also here, we found that the ICK1/KRP1-YFP fusion proteins moved between cells in the epidermis and also into the mesophyll layer, and it is likely that the strong leaf phenotype resulted from a large number of cells receiving ICK1/KRP1109 (data not shown).
Figure 6.
Analysis of ICK1/KRP1109 Misexpression in TMM-Positive Cells.
(A) to (D) Light micrographs of wild-type (A) and ProTMM:YFP:ICK1/KRP1109 (B) rosette leaves. Scanning electron micrographs of wild-type (C) and ProTMM:YFP:ICK1/KRP1109 (D) rosette leaves of the same age. Note the strong reduction in cell number and the enormous increase in cell size of all pavement cells in the transgenic line. Bars = 1 mm in (A) and (B) and 100 μm in (C) and (D).
(E) to (J) Fluorescence-activated cell sorting analysis of the first and second rosette leaves from wild-type ([E], [G], and [I]) or ProTMM:YFP:ICK1/KRP1109 ([F], [H], and [J]) plants. The 10-d-old ([E] and [F]), 15-d-old ([G] and [H]), and 20-d-old ([I] and [J]) seedlings are shown. In the transgenic line, a quantitative and a qualitative shift toward more replicated nuclei compared with the wild type is visible at all time points.
To assess whether endoreplication levels were increased, we studied the degree of polyploidization in 10-, 15-, and 20-d-old seedlings by flow cytometry (Figures 6E to 6J). At all time points, we found a quantitative as well as qualitative shift toward higher replication levels in comparison with wild-type plants. In leaves of 10-d-old ProTMM:YFP:ICK1/KRP1109 seedlings, elevated levels for 4C and 8C nuclei as well as a new, although small, 16C peak were present (Figures 6E and 6F). In 15-d-old seedlings, the 16C peak was increased and a new 32C peak appeared (Figures 6G and 6H), and in 20-d-old seedlings, a greater 16C peak and a pronounced 32C peak were detected (Figures 6I and 6J). Taken together, our data showed that ICK1/KRP1 can block cell divisions and induce endoreplication in mitotic cells.
Mode of ICK1/KRP1-Induced Endoreplication
From animals, it is known that a conversion of a mitotic cycle into an endocycle can be initiated from different phases of a mitotic cell cycle discriminating different endocycles. For instance, in the first endocycles of Drosophila nurse cells, a new G1-phase is initiated shortly after S-phase, whereas mammalian megakaryocytes progress through a G2-phase and switch to a G1-phase with the beginning of mitosis (Edgar and Orr-Weaver, 2001).
To determine how ICK1/KRP1-induced endocycles proceeded, we used a promoter reporter line for a mitotic cyclin, CYCLIN B1;2 (CYC B1;2, ProCYCB1;2:DB:GUS), which marks cells in a late G2-phase until M-phase of a cell division cycle (Schnittger et al., 2002a). Next, the number of ProCYCB1;2:DB:GUS-positive socket cells surrounding outgrowing but not yet maturated trichomes were compared in a wild-type background and in plants misexpressing ICK1/KRP1109 from the GL2 promoter. We found that wild-type as well as ProGL2:ICK1/KRP1109 plants displayed approximately the same proportion of stained cells adjacent to a trichome, 31% versus 35% (Table 3). Thus, endoreplicating trichome-neighboring cells in ICK1/KRP1 misexpressing plants still entered a G2-phase.
Table 3.
ProCYCB1;2:DB:GUS in Socket Cells of Young Trichomes
| Line | Percentage of Young Trichomes with at Least One GUS-Positive Socket Cella | n | Σ Trichomes |
|---|---|---|---|
| ProCYCB1;2:DB:GUS in Landsberg erecta | 31.3 ± 4.2 | 4 | 400 |
| ProCYCB1;2:DB:GUS in ProGL2:KRP1109*b | 34.5 ± 2.9 | 4 | 400 |
Socket cells of young trichomes (stage 2 to stage 5 according to Szymanski et al., 1998) were analyzed; average ± sd per 100 counted trichomes.
ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
We found that in the ICK1/KRP1-induced endocycles the anaphase promoting complex/cyclosome (APC/C) was active. This became evident because the GUS reporter used was fused to the N-terminal 149 amino acids of CYCLIN B1;1, including the destruction box (DB) (Schnittger et al., 2002a). Such a marker becomes degraded in late mitosis with the onset of APC/C activity, which degrades mitotic substrates as cyclins and securin and promotes by that exit from mitosis (Colon-Carmona et al., 1999). Trichome-neighboring cells in ICK1/KRP1-misexpressing plants, however, did not display a continuous staining of the DB:GUS marker, indicating a cyclic degradation of the marker.
Expression of ICK1/KRP1 Can Correct the siamese Mutant Phenotype, and Reduction versus Increase of Endoreplication Levels Correlate with ICK1/RKP1 Transcript Levels
The observation that ICK1/KRP1 could only induce endoreplication in cells with a mitotic cell cycle program and not in endoreplicating cells as trichomes or trichome-neighboring cells suggested that ICK1/KRP1 acts by blocking a mitotic activity while allowing S-phase entry rather than by actively promoting S-phase entry. This is also supported by the cyclic expression of a late G2 reporter.
It is not clear, however, why ICK1/KRP1 misexpression only in trichomes and not in proliferating cells appeared to interfere with S-phase entry. To test whether other developmental cues might be responsible for a differential response of trichome-neighboring cells versus trichomes with respect to S-phase entry, we made use of the siamese (sim) mutant. In sim mutant plants, trichomes undergo mitosis leading to clustered and multicellular trichomes with strongly reduced endoreplication levels; yet these multicellular trichomes display characteristics of typical trichomes with branch formation and papillae on the outer surface (Figure 7A) (Walker et al., 2000).
Figure 7.
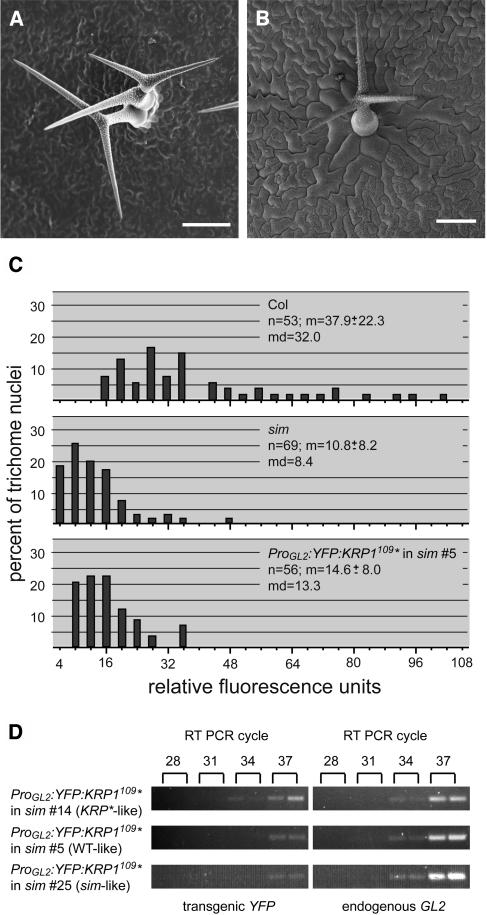
Misexpression of ICK1/KRP1109 in sim.
(A) and (B) Scanning electron micrographs of mature Arabidopsis trichomes. In (A), a typical multicellular sim trichome is shown. (B) shows a unicellular wild type–like trichome in sim mutants misexpressing ProGL2:ICK1/KRP1109 (as seen in line ProGL2:YFP:ICK1/KRP1109 in sim 5); note that trichome-neighboring cells are enlarged. Bars = 100 μm.
(C) Analysis of trichome DNA content of Columbia, sim, and ProGL2:YFP:ICK1/KRP1109 in sim line 5. Distributions of trichome DNA contents are given in relative fluorescence units. The median value of Columbia trichomes was set as 32C. From this value the respective C values of the trichome nuclei were calculated. The sample size (n), the mean (m) ± standard deviation, and the median (md) are given.
(D) Semiquantitative RT-PCR showing the relative expression strength of YFP:ICK1/KRP1109 in three independent lines misexpressing ProGL2:YFP:ICK1/KRP1109 in the sim mutant background. These lines resemble either a ICK/KRP-like, a wild type–like, or a sim-like phenotype. The expression strength was compared with the endogenous expression of GL2. The numbers at top indicate the RT-PCR cycle number. Line 14 showed the strongest, line 5 an intermediate, and line 25 the weakest transgene expression, which correlates with their phenotypes. ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
ProGL2:YFP:ICK1/KRP1 and ProGL2:YFP:ICK1/KRP1109 were introduced into sim mutant plants, and analysis of the 14 and 28 generated transgenic plants showed the following three phenotypical classes: 7% (for ProGL2:YFP:ICK1/KRP1) and 43% (for ProGL2:YFP:ICK1/KRP1109) of the plants displayed a ICK1/KRP1 misexpression-like phenotype (i.e., small trichomes with fewer branches, which eventually died); 64/40% contained almost wild type–like trichomes with none or only few clusters (Figure 7B); and 29/18% developed sim-like clustered and multicellular trichomes. Similar results were also obtained by crossing the untagged ICK1/KRP1 and ICK1KRP1109 misexpression lines into sim plants as well as by introducing the untagged version by plant transformation into sim plants (data not shown).
Next, we measured the DNA content of wild type–like sim mutant plants expressing ProGL2:YFP:ICK1/KRP1109. Although nuclei of these trichomes did not fully reach wild-type replication levels, both a quantitative and a qualitative increase in endoreplication levels was found. In sim mutants, ∼20% of the individual nuclei have a DNA content of 4C or less, and the average DNA content of all nuclei is ∼8C. By contrast, all of the trichome nuclei on plants expressing ProGL2:YFP:ICK1/KRP1109 in the sim mutant background had a DNA content of more than 4C, and the overall average DNA content was ∼13C (Figure 7C, line 15). These data showed that ICK1/KRP1 expression can at least partially rescue the sim mutant phenotype. Thus, also in a trichome environment, ICK1/KRP1 expression can induce endoreplication, suggesting that the difference between trichomes and trichome-neighboring cells is more directly associated with the execution of a mitotic program than with other developmental differences.
Furthermore, the spectrum of phenotypes obtained by expressing ICK1/KRP1 in sim mutant plants suggested that ICK1/KRP1 could act in a concentration-dependent manner. Semiquantitative RT-PCR of representative plants from the different phenotypical classes revealed that weak sim-like and wild type–like phenotypes were correlated with low expression strength of the ICK1/KRP1 construct, whereas an ICK1/KRP1-like phenotype was associated with higher expression levels of the construct (Figure 7D). Thus, our data suggest that ICK1/KRP1 supplies a mitosis-suppressing function that is compatible with an endoreplication program at a low concentration, whereas at higher levels of expression, ICK1/KPP1 blocks cell cycle progression completely.
Endoreplicated Trichome Socket Cells Reenter Mitosis
Along with maturation and differentiation, most of Arabidopsis leaf cells switch to an endoreplication cycle (Melaragno et al., 1993) (cf. Figures 6E, 6G, and 6I). Correspondingly, cell divisions become progressively restricted to the basal part of the leaf and finally stop completely (Donnelly et al., 1999).
Surprisingly, we observed that in very old leaves of ProGL2:ICK1/KRP1109 plants, the ProCYCB1;2:DB:GUS reporter was expressed again, that is in trichome socket cells, indicating that these cells again entered a G2-phase (Figure 8A). A comparison with wild-type plants carrying the ProCYCB1;2:DB:GUS transgene confirmed that in comparable stages on wild-type leaves, cell divisions have ceased with the exception of a few meristemoid cells at the leaf base. We determined the ratio of GUS-positive trichome-neighboring cells to total number of trichomes and obtained for leaves of ProGL2:ICK1/KRP1109 plants with a few meristemoid cells in a G2-phase a ratio of ∼0.024 and on somewhat older leaves without any other detectable cells in a G2-phase a ratio of ∼0.006 (Table 4). Analysis of these mature socket cells with scanning electron microscopy revealed new cell walls in very large cells (Figure 8B).
Figure 8.
Analysis of Late Cell Divisions in Endoreplicated Trichome-Neighboring Cells.
(A) Light micrograph of a whole-mount GUS staining of the reporter line ProCYCB1;2:DB:GUS in ProGL2:ICK1/KRP1109 showing GUS activity in one trichome-neighboring cell of an old rosette leaf, in which no other cell divisions are detectable.
(B) Scanning electron micrograph of trichome-neighboring cells surrounding a dead trichome of an old rosette leaf of ProGL2:ICK1/KRP1109-expressing plants. Arrowheads mark a straight wall indicative for a newly formed wall in an enlarged trichome-neighboring cell. Bars in (A) and (B) = 100 μm.
(C) to (F) Confocal laser scanning micrographs of wild-type nonendoreplicated nuclei ([C] and [E]) at different mitotic stages. (C) shows a metaphase nucleus with condensed chromosomes from a root meristem cell. (E) reflects a late anaphase/early telophase nucleus (marked by arrowheads) from a young leaf epidermal cell. (D) and (F) show mitotic figures in endoreplicated nuclei of trichome-neighboring cells in ProGL2:ICK1/KRP1109-expressing plants. Note the increased DNA content compared with the wild type. Condensed chromosomes most likely reflect metaphase (D) or late anaphase/early telophase (F). Bars = 5 μm.
Table 4.
ProCYCB1;2:DB:GUS in Socket Cells of Mature Trichomes
| Line | Stagea | GUS-Positive Socket Cells per Total Trichome Number per Leafb | Σ Leaves | Σ Trichomes |
|---|---|---|---|---|
| ProCYCB1;2:DB:GUS | GUS+ | 0.000 ± 0.000 | 9 | 326 |
| in Landsberg erecta | GUS− | 0.000 ± 0.000 | 15 | 304 |
| ProCYCB1;2:DB:GUS | GUS+ | 0.024 ± 0.035 | 16 | 508 |
| in ProGL2:KRP1109*c | GUS− | 0.006 ± 0.031 | 52 | 868 |
Stage GUS+, GUS staining in other epidermal cells besides socket cells; stage GUS−, GUS staining only in socket cells.
Mature trichomes (stage 6 according to Szymanski et al., 1998) were analyzed; average ± sd.
ICK1/KRP1 is abbreviated as KRP1 and is marked by an asterisk.
Intriguingly, at the time when the ProCYCB1;2 marker is turned on again, the majority of the trichomes on ProGL2:ICK1/KRP1109 plants are dead; in addition, this is about the time when the activity of the GL2 promoter ceases (Szymanski et al., 1998). This correlation suggested that only after the withdrawal from the ICK1/KRP1 regime trichome-neighboring cells entered mitosis.
The general notion is that cells, which have started an endoreplication program, are terminally differentiated and cannot reenter mitosis (Nagl, 1976; Melaragno et al., 1993; Edgar and Orr-Weaver, 2001). However, at the time when neighboring cells resumed cell division, all of them appeared to have undergone substantial endoreplication, suggesting that endoreplicated cells were able to reenter mitosis. To find further support for this possibility, we examined DAPI-stained leaves for the appearance of mitotic figures (Figures 8C to 8F). Figures 8D and 8F show two representative mitotic figures, most likely a metaphase (Figure 8D) and a late anaphase or telophase (Figure 8F) of trichome-neighboring cells in ICK1/KRP1-misexpressing plants. The comparison with similar mitotic stages of wild-type root meristem cells or young leaf cells, which are not polyploid (Figures 8C and 8E), revealed that mitotic figures obtained from ICK1/KRP1-misexpressing plants contained more DNA than dividing cells in the wild type (Figures 8D and 8F). This demonstrates that endoreplicated trichome-neighboring cells underwent mitosis.
As judged by the number of cell walls we identified with scanning electron microscopy, many neighboring cells reentered mitosis (Figure 8B). DAPI staining revealed that the most common nuclear type was an interphase nucleus, indicating that cell divisions did not result in abnormal mitoses or mitotic arrest but rather that mitosis of an endoreplicated cell proceeded without aberrations. Thus, we conclude that plant cells maintain the ability after going through endoreplication cycles to divide again, demonstrating a high degree of flexibility in plant development.
DISCUSSION
Here, we have shown that the Arabidopsis CKI ICK1/KRP1, besides an inhibitory role at the G1-S transition point, can block cell division and induce endoreplication. In addition, we found that ICK1/KRP1 can act non-cell-autonomously. These findings present a new view on the functions of CKIs, especially with respect to tissue organization and organ growth control in plants. Moreover, the work on ICK1/KRP1 resulted in the finding that already endoreplicated cells can adopt a certain cell fate, and finally that endoreplicated cells can reenter a mitotic cycle.
CKIs as Multiple Cell Cycle Switches
Based on this study and previous experiments, we propose that CKIs have at least three functions in plants. First, ICK/KRPs might be important regulators involved in switching from a mitotic to an endoreplicating cell cycle mode in differentiating cells. As demonstrated by misexpression in trichome-neighboring cells, embryonic epidermis cells, and ProTMM-positive cells, ICK1/KRP1 is a very potent inhibitor of entry into mitosis, whereas it allows S-phase to proceed. Such an inhibitory function might be needed in cells determined to switch to an endoreplication cycle but still containing mitotic regulators. For instance, in Medicago, mRNA of a mitotic cyclin has been detected in the zone of nitrogen-fixing nodules, in which cells will enter an endoreplication cycle (Cebolla et al., 1999). Consistently, the ICK1/KRP1 mRNA was detected in Arabidopsis in mature leaves, in which cells often endoreplicate (Ormenese et al., 2004). Lastly, the rescue of sim mutant trichomes by ICK1/KRP1 expression argues for a function of CKIs in facilitating the switch to an endoreplication cycle. Intriguingly, SIM encodes a small protein with limited homology to ICK/KRPs (J. Larkin, personal communication).
Derived from our finding that ICK/KRPs can block entry into mitosis, we postulate a second function of ICK/KRPs in dividing cells by assisting to establish a G1-phase. Licensing of origins of replication in a G1-phase requires a low CDK activity (Stern and Nurse, 1996). One way to inactivate kinase activity after a preceding mitosis is the APC/C-dependent destruction of mitotic cyclins (Peters, 1998; Harper et al., 2002). In addition, it has been shown that in Drosophila a special CDK inihibitor, ROUGHEX (RUX), binds to and inactivates mitotic CDK complexes, helping to establish a G1-phase with low CDK activity (Foley et al., 1999; Foley and Sprenger, 2001). RUX is an essential gene in Drosophila, demonstrating that there is a high demand for this inhibitory activity. Recently, for the human CKIs p21Cip1 and p27Kip1 and for the RETINOBLASTOMA protein, a similar function in controlling mitotic exit by inactivating mitotic CDK activity was found (Chibazakura et al., 2004). A function of ICK/KRPs in contributing to a G1-phase could also explain the expression of ICK/KRPs in highly proliferating cells, an observation that is so far not understood and appears even contradictory to the previously described function of ICK/KRPs as inhibitors of cell proliferation (Breuil-Broyer et al., 2004; Ormenese et al., 2004). Additional hints for a function of ICK1/KRP1 in or after mitosis come from transcriptional profiling studies of an Arabidopsis cell culture that revealed an expression peak of ICK1/KRP1 mRNA in late G2/M-phase (Menges and Murray, 2002; Menges et al., 2003). Furthermore, genes expressed in late G2-phase and mitosis often contain mitosis-specific activator elements in their promoters, for instance, the promoter of CYCB1;2 shows five elements (Ito et al., 1998; Ito, 2000). In the promoter of ICK1/KRP1, at least eight mitosis-specific activator elements can be found, supporting an expression during mitosis. However, it remains to be seen how in this scenario ICK1/KRP1 is prevented from a premature inhibition of a mitotic CDK complex.
Finally, as shown in previous experiments, misexpression of ICK/KRPs can lead to cells with a reduced DNA content (De Veylder et al., 2001; Jasinski et al., 2002; Schnittger et al., 2003). Therefore, the third function of ICK/KRPs might be to terminate/assist to terminate mitotic as well as endoreplication cycles. This is supported by the analysis of ICK1/KRP1 transcript over time. In 5-week-old Arabidopsis leaves, in which presumably all cell cycle activity has ceased, an increased level of ICK1/KRP1 transcript in comparison with CDKA;1 was found (Wang et al., 1998).
Throwing the Switch
What determines which CKI function is executed? Why does an endoreplicating cell undergo an S-phase block, whereas a proliferating cell is preferentially blocked at mitosis? It is conceivable that ICK1/KRP1 could target different CDK complexes or that it has different affinities to the various cyclin-CDK complexes in endoreplicating trichomes versus mitotic cells. Also, additional components might be present in mitotic and endoreplicating cells, respectively. Misexpression of human p21Cip1, for instance, has led to endoreplication only if the RETINOBLASTOMA protein is absent (Niculescu et al., 1998). Also, the Drosophila inhibitor RUX was found upon misexpression to block mitosis and convert the 16th embryonic cycle into an endocycle. Earlier embryonic cycles, however, were only converted when cyclin E was also absent (Vidwans et al., 2002). Thus, ICK1/KRP1 could have a cell type–specific function depending on a specific set of cell cycle regulators.
All previous data, however, were obtained from misexpression studies using strong promoters, either the GL2 promoter or the 35S promoter of Cauliflower mosaic virus (CaMV35S), precluding any analysis of CDK function at weaker concentrations. In this study, we looked at ICK1/KRP1 moving from trichomes into their neighboring cells and the comparison of fluorescence intensities of YFP-tagged ICK/KRP proteins between trichomes and their neighboring cells revealed a more than twofold difference for ICK1/KRP1109:YFP to YFP:ICK1/KRP1 between the two cell types. In addition, the GL2 promoter appears to have weaker expression in young embryos than later in trichome or root development as judged by the strength of the in situ hybridization signal and fluorescence intensity of reporter genes (Lin and Schiefelbein, 2001; Costa and Dolan, 2003). Not much is known about the relative strength of the TMM promoter, but it is presumably weaker than the CaMV35S promoter. Thus, it is possible that CKIs act as concentration-dependent switches that block entry into S-phase only at high concentrations. This is substantiated by our finding that an ICK1/KRP1 misexpression–like phenotype was found in sim mutant plants with high levels of ICK1/KRP1 expression, whereas at lower expression levels, increased endoreplication levels in comparison with the sim mutant were found. Interestingly, the study of temperature-sensitive CDK alleles in yeast has suggested that for entry into mitosis, higher levels of CDK activity are required than for entry into S-phase (MacNeill et al., 1991; Ayscough et al., 1992). One deduction from the above is that if CKIs are involved in establishing endocycles, and thus are already expressed in endoreplicating cells (e.g., trichomes), the additional expression of ICK1/KRP1 might then reach a threshold concentration of CKI, resulting in a block of S-phase entry. This could explain why among the large number of transgenic plants generated expressing various ICK/KRP versions in trichomes we have only found plants with apparently reduced endoreplication levels in trichomes.
Of course, cell type–specific action and concentration dependency of CKIs are not mutually exclusive. Also, endocycles induced in trichome-neighboring cells differed from endocycles in wild-type trichomes because in trichomes neither CYCB1;1 nor CYCB1;2 promoter activity can be recognized (Schnittger et al., 2002a).
Remarkably, the CYCB1;1 promoter reporter indicating a G2-phase did not accumulate in endoreplicating trichome-neighboring cells. This reporter carries a DB, indicating that at least some activity of the APC/C remained even though CDK activity was presumably blocked. In animals and yeast, CDK activity has been found to be needed to phosphorylate the CDC20 class of APC/C-cofactors and by that activate the APC/CCDC20 (Shteinberg et al., 1999; Kramer et al., 2000). One possibility for ICK1/KRP1-misexpressing plants could be that only the affinity to certain substrates or only certain CDKs might be blocked by ICK1/KRP1, still permitting the activation of APC/CCDC20. One candidate for a CDK that cannot be blocked by ICK/KRPs are the plant-specific B-type CDKs (Joubes et al., 2000). B-type CDKs were not found to interact with ICK/KRPs in yeast two-hybrid studies but are highly expressed during mitosis (De Veylder et al., 2001; Porceddu et al., 2001; Zhou et al., 2002).
Alternatively, a different APC/C complex also could be involved because the CDC20-dependent APC/C is active only in late mitosis (Shteinberg et al., 1999; Kramer et al., 2000). In animals, with the end of mitosis and during a G1-phase of a following cell cycle, another APC/C is assembled containing the CDH1 cofactor class (Zachariae et al., 1998). Studies from Drosophila have revealed that the APC/CCDH1 is also active in the G2-phase and needs to be inactivated before mitosis to allow accumulating mitotic cyclins (Grosskortenhaus and Sprenger, 2002). In contrast with CDC20, phosphorylation has been found to inactivate CDH1 (Kotani et al., 1999; Kramer et al., 2000). Thus, in the case of ICK1/KRP1 misexpression, another possibility is that blocked CDK activity might result in an active ACP/CCDH1. Yet, it remains to be seen whether plant APC/C is similarly regulated by CDC20 and CDH1 homologs.
Non-Cell-Autonomous Action of CKIs
To our knowledge, CKIs have not been found to function in a non-cell-autonomous manner in animals. The finding that ICK1/KRP1 can move between cells adds another level of complexity to plant development and challenges cell cycle control on a tissue and organ level. One way for plants to keep CKIs in check might be the nuclear localization and the high instability of the proteins. In contrast with YFP expressed from the GL2 promoter, full-length ICK1/KRP1 protein could not be detected on protein gel blots. For the N-terminally truncated protein ICK1/KRP1109, a band of the expected size was found. Intriguingly, whereas the full-length ICK1/KRP1 was exclusively found in the nucleus, the truncated version was also located in the cytoplasm. Similar results were recently obtained by Zhou et al. (2003) analyzing roots of plants misexpressing ICK1/KRP1 from the CaMV35S promoter. One likely possibility is that ICK1/KRP1 becomes degraded in the cytoplasm and that for this degradation a motif in the N terminus of the protein is required. In animals, p27Kip1 abundance and localization is strictly regulated (Sherr and Roberts, 1999; Slingerland and Pagano, 2000). p27Kip1 exerts its inhibitory function in the nucleus, and in many experimental systems, p27Kip1 has been found to become degraded in the cytoplasm (Tomoda et al., 1999; Connor et al., 2003). Because no sequence similarities exist between their regulatory domains, it will be interesting to see whether ICK1/KRP1 is subject to similar regulatory pathways as p27Kip1 in animals.
Besides controlling CKIs within a cell, the non-cell-autonomous action of ICK1/KRP1 offers a possibility to link decisions on a cellular level with the supracellular division and growth pattern in organs. For instance, it has been found that starting from the leaf tip, epidermal cells enter an endocycle (Melaragno et al., 1993). CKIs could help to spread the entry into an endoreplication cycle. In addition, CKIs could be involved in linking developmental programs (e.g., trichomes with trichome-neighboring cells). In contrast with other epidermal cells, we found that the level of endoreplication in trichome-neighboring cells is quite constant around 4C to 8C. Of course, this could be a feature of socket cell fate. Alternatively, this could also be an indirect effect resulting from a diffusion of CKIs from a centrally located trichome leading to a coordinated entry in and perhaps a coordinated exit from an endoreplication cycle. Analysis of trichome mutants with increased and decreased endoreplication levels might help to answer this question.
The molecular mechanism of the non-cell-autonomous action of ICK1/KRP1 remains to be analyzed in detail. Transport through plasomodesmata appears to be highly regulated, and at least for some nuclear localized proteins, a controlled transport mechanism has been found (Gallagher et al., 2004). Conversely, plasmodesmata also allow the passive diffusion of small molecules. The size exclusion limit for nontargeted symplastic movement has been estimated to be ∼60 kD in young tobacco (Nicotiana tabacum) leaves and ∼40 kD in older tobacco and Arabidopsis leaves (Oparka et al., 1999; Crawford and Zambryski, 2000; Itaya et al., 2000). Thus, on the one hand, ICK1/KRP1 is nuclear localized; on the other hand, ICK1/KRP1, even fused to YFP, might be small enough (22 and 49 kD, respectively) to diffuse between cells, whereas a GUS:YFP:ICK1/KRP1109 fusion with 105 kD was retained in trichomes. Yet a third alternative is that not the protein but the mRNA moves between cells (Ruiz-Medrano et al., 1999; Kim et al., 2001), and detailed analyses will be required in the future to understand the nature and possible function of the ICK1/KRP1 non-cell autonomy.
Endocycles and Terminal Differentiation
Endocycles are often regarded as a state of terminal differentiation because the switch to an endocycle is often associated with cell differentiation (Edgar and Orr-Weaver, 2001; Sugimoto-Shirasu and Roberts, 2003). Evident examples for this connection are Arabidopsis trichomes (Marks, 1997; Hulskamp et al., 1999), salivary gland cells in Drosophila (Smith and Orr-Weaver, 1991), and human thrombocytes (Zybina and Zybina, 1996).
Here, we have shown that endocycles might be much more dynamic and flexible than previously thought. Our first observation was that the onset of an endoreplication program still allows cells to adopt and, thus, change their fate. Our second observation was that an endoreplicated cell can reenter a mitotic cycle. Interestingly, already >50 years ago it was observed that polyploid plant and animal cells could occasionally reduce their number of chromosomes and return to a diploid chromosome set (Grell, 1946; Huskins, 1948a, 1948b). Here, we have shown that a reduction of DNA content is not limited to tetraploid cells, but even highly endoreplicated cells appeared to divide.
What causes these enlarged cells to reenter mitosis? Three possibilities are conceivable. In animals, it has been observed that binding of p27Kip1 can stabilize a cyclin D-CDK4 complex (LaBaer et al., 1997; Cheng et al., 1999; Bagui et al., 2000). Correspondingly, ICK1/KRP1 could conserve a mitotic regulator complex in the trichome-neighboring cells, and after ICK1/KRP1 is not supplied any longer by the trichome, the mitotic complex is liberated. A mitotic complex stabilized by ICK1/KRP1 might include cyclin D because in plants D-type cyclins have been found to have mitotic activities as well (Schnittger et al., 2002b; Koroleva et al., 2004).
On the other hand, a cell size checkpoint might operate and induce late cell divisions. The endoreplicated trichome-neighboring cells in ICK1/KRP1-misexpressing plants are at least for some time the largest cells found in the epidermis and likely of the entire leaf. Thus, a cell-autonomous control mechanism might be responsible for the onset of cell divisions, and after a certain size might be reached, a new cell division could be initiated.
Finally, a non-cell-autonomous control mechanism based, for instance, on stomata index also might be responsible. We noticed that new stomata complexes were formed by many cell divisions of trichome-neighboring cells (Figure 1B). Stomata density is tightly controlled on the leaf blade (Nadeau and Sack, 2002b; Bergmann, 2004). Stomata can only be generated by cell divisions; therefore, leaf growth by cell expansion in maturating leaves would lead to a dramatic decrease in stomata density. Interestingly, a few 2C cells were found in maturing leaf areas in which other cells had undergone a few rounds of endoreplication (Melaragno et al., 1993). These cells have been interpreted as a reserve for regenerating cells and also for stomata formation. In light of our findings, however, these cells might not be set aside but could be generated by divisions of endoreplicated cells.
These different possibilities remain to be tested in the future, but it has emerged that plant cell cycle control is much more flexible than anticipated, and detailed analysis of division patterns will be needed in the future to get a deeper insight into the dynamics of plant cell cycle control in the context of organ and tissue development.
METHODS
Plant Material, Growth Conditions, Plant Transformation, and Crosses of Transgenic Lines
Arabidopsis thaliana plants were grown under long-day conditions (16 h of light, 8 h of darkness) between 18 and 25°C under standard greenhouse conditions. The Arabidopsis accessions Landsberg erecta and Columbia-0 were used as wild-type controls. At least 20 transgenic plants were generated for all expression constructs. For further analysis, several representative reference lines displaying a typical phenotype and with comparable transgene expression were chosen. For all data obtained for one expression construct, the same transgenic reference line was used. For ProGL2:YFP:ICK1/KRP1, ProGL2:ICK1/KRP1109:YFP, ProGL2:YFP:ICK1/KRP1109, and ProGL2:GUS:YFP:ICK1/KRP1109, ∼40% of the transgenic plants resembled the ICK1/KRP1-misexpression phenotype. T2 plants were tested for segregation, and homozygous T3 plants were established and analyzed. Approximately 50% of the ∼100 primary transformants of ProTMM:YFP:ICK1/KRP1109 showed a phenotype. For ProTMM:YFP:ICK1/KRP1109, ProGL2:YFP, ProGL2:GFP5ER, and ProGL2:YFP:ICK1/KRP1109 in sim, plants were analyzed in the T2 generation. The ProGL2:ICK1/KRP1, ProGL2:ICK1/KRP1109, ProGL2:nls:GFP:GUS, and ProCYCB1;2:DB:GUS expression lines have been described previously (Schnittger et al., 2002a; Schnittger and Hulskamp, 2003). Plants were transformed as previously described (Schnittger et al., 2002a). Enhancer trap lines 232 and 254 were obtained from S. Poethig (http://enhancertraps.bio.upenn.edu/). For crosses between transgenic lines, homozygous T3 plants were used, and the F1 generation was analyzed.
Expression Constructs
To achieve trichome expression, a 2.1-kb HindIII-NheI fragment from the 5′ upstream region of the GL2 gene was used. For ProGL2:YFP and ProGL2:GFP5ER, the plant transformation vector pBI101.1ProGL2 (Szymanski et al., 1998) was used. In all other cases, the plant transformation vector pAM-PAT-GW was used (a kind gift from Bekir Ülker, MPIZ Cologne). To achieve expression in stomatal precursors, the plant transformation vector pAM-PAT-GWProTMM was created, which contains a 0.5-kb fragment from the 5′ upstream region of the TMM gene (Nadeau and Sack, 2002a). For socket cell expression of ICK1/KRP1109, the plant transformation vector pAM-PAT-GWProUAS, which contains five GAL4 binding domains in tandem, was used (a kind gift from Martina Pesch, University of Cologne). To generate ICK1/KRP1:YFP, YFP:ICK1/KRP1, ICK1/KRP1109:YFP, and YFP:ICK1/KRP1109 fusion constructs, gene-specific primers for the cDNAs of ICK1/KRP1 and EYFP-N1 (Clontech, Palo Alto, CA) were used and a two-step PCR was performed. The upper primers contained a BamHI and the lower primers a SacI recognition site. After the second PCR step, the obtained fragments were subcloned into pGEM-T vector (Promega, Mannheim, Germany) and sequenced, and then excised with BamHI and SacI and subcloned into a modified pENTR1A vector (Invitrogen, Karlsruhe, Germany) containing a SacI restriction site in its multiple cloning site. Thereafter, a recombination into the respective plant transformation vector was performed using Gateway LR Clonase enzyme mix (Invitrogen). To generate the GUS:YFP:ICK1/KRP1109 fusion construct, a two-step PCR was performed with primers containing attB sites using GUS and YFP:ICK1/KRP1109 cDNAs as templates. After the second PCR step, the DNA fragment was cloned into pDONR201 (Invitrogen) using Gateway BP Clonase enzyme mix and sequenced. Thereafter, a recombination into the pAM-PAT-GWProGL2 plant transformation vector was performed using Gateway LR Clonase enzyme mix (Invitrogen). For the expression construct ProGL2:YFP, the YFP cDNA from the pEYFP-N1 vector (Clontech, Palo Alto, CA) was excised with XhoI and NotI and subcloned into XhoI and NotI of pBSII-KS (Stratagene, La Jolla, CA). The YFP cDNA was then cut out with BamHI and SacI and cloned into BamHI- and SacI-digested pBI101.1ProGL2. For the expression construct ProGL2:GFP5ER, the GFP5ER cDNA was excised from pmGFP5 (a kind gift of Jim Haselhoff) with BamHI and SacI and subcloned into pUC18 digested with BamHI and SacI (pART73). Then, GFP5ER was excised from pART73 with BamHI and SacI and inserted into pBI101.ProGL2 (pART72). To express ICK1/KRP1109 in socket cells, the cDNA was amplified by PCR with gene-specific primers containing attB1 and attB2 recombination sites. The PCR fragment was then recombined into pDONR201 vector using Gateway BP Clonase enzyme mix (Invitrogen) and sequenced. To obtain the construct ProUAS:ICK1/KRP1109, an LR reaction was performed with the pAM-PAT-GWProUAS vector. Unless stated otherwise, all manipulations were performed using standard molecular methods.
RT-PCR Analysis
RNA was prepared with the RNeasy mini kit (Qiagen, Hilden, Germany). For each sample, RNA was treated with DNase I to avoid contamination with genomic DNA. Reverse transcription was performed with SuperScript II RNase H reverse transcriptase (Invitrogen) followed by PCR. For ProGL2:ICK1/KRP1, ProGL2:ICK1/KRP1109, ProGL2:YFP:ICK1/KRP1, ProGL2:ICK1/KRP1109:YFP, ProGL2:GUS:YFP:ICK1/KRP1109, and ProUAS:ICK1/KRP1109, 100 ng of total RNA were used. The upper primers were designed against the 5′ untranslated region of the GL2 gene (5′-GAGGAGAAGAGGGAAGAGATCATAA-3′), which is included in the GL2 promoter fragment, YFP (5′-GCTGACCCTGAAGTTCATCTG-3′) for ProGL2:GUS:YFP:ICK1/KRP1109, the att-recombination site of the pAM-PAT-GWProUAS construct (5′-CAAGTTTGTACAAAAAAGCAG-3′), and EF1 (5′-ATGCCCCAGGACATCGTGATTTCAT-3′). The lower primers were designed against ICK1/KRP1 (5′-TTTACCCATTCGTAACGTCCTTCTA-3′), YFP (5′-TGATATAGACGTTGTGGCTGTTG), and EF1 (5′-TTGGCGGCACCCTTAGCTGGATCA-3′). A total of 20 μL of RT-PCR products, after 25, 30, 35, and 40 cycles for EF1 or 30, 35, 40, and 45 cycles for ICK1/KRP1 and YFP, were separated on agarose gels and visualized by UV excitation of ethidium bromide–stained DNA. For ProGL2:YFP:ICK1/KRP1109 in sim lines 5, 14, and 25, 200 ng total RNA were used. The upper primer used was designed against the 5′ untranslated region of the GL2 gene. The lower primers were designed against ICK1/KRP1 or GL2 (5′-TCTTTCTCTTATTAGTGCCCTTGT-3′). A total of 20 μL of RT-PCR products, after 28, 31, 34, and 37 cycles, were separated on agarose gels. All reverse transcription expression analyses were performed in duplicate and repeated at least once in an independent experiment.
Protein Work
Fifteen- to twenty-five-day-old seedlings were harvested, directly frozen in liquid nitrogen, and homogenized in Laemmli buffer including Protease Inhibitor Cocktail (Sigma-Aldrich, Munich, Germany). The amount of protein was determined by Bradford reagent. Protein extracts from ProGL2:YFP:ICK1/KRP1, ProGL2:ICK1/KRP1109:YFP, and ProGL2:YFP misexpressing lines were applied onto a 15% SDS PAGE and blotted on Hybond ECL membrane (Amersham, Freiburg, Germany). Protein gel blots were incubated with anti-GFP IgG monoclonal antibody (1814460; Roche, Mannheim, Germany) in a 1/500 dilution followed by a goat anti-mouse IgG antibody conjugated with ImmunoPure Peroxidase (Pierce, Rockford, IL) in a 1/2000 dilution. For detection, the SuperSignal West Pico kit (Pierce) and BioMax Light films (Kodak, Stuttgart, Germany) were used. To determine an equal loading and transfer of the probes, the membrane was stained with Ponceau S. All protein gel blot analyses were performed in duplicate and repeated at least once in an independent experiment.
Microscopy
Light microscopy was performed with an Axiophot microscope (Zeiss, Heidelberg, Germany) or a Leica DM RA2 (Wetzlar, Germany) equipped with differential interference contrast (Nomarski) and epifluorescence optics. The DISKUS software package (Carl H. Hilgers-Technisches Büro, Königswinter, Germany; version 4.30.19) was used to quantify the fluorescence intensity of DAPI-stained leaves to determine nuclear and cell sizes and to measure the nuclear size of propidium iodide–stained embryos in optical sections. Cryo-scanning electron microscopy was performed as described by Rumbolz et al. (1999). Confocal laser scanning microscopy was performed with a Leica DM-Irbe or a LSM 510 META microscope (Zeiss).
Measurements of DNA Content and Nuclear Area
For DNA measurements of trichome-neighboring cells, whole-mount preparations of rosette leaves three and four were used. Leaves were first incubated in 70% ethanol overnight and then vacuum infiltrated for 30 min in DAPI solution (0.25 μg/mL in water). The DAPI intensity was quantified and the background fluorescence was substracted using the DISKUS software package (Carl H. Hilgers-Technisches Büro; version 4.30.19). For each analyzed leaf, the fluorescence intensity of 20 guard cell nuclei was also measured, and the average value was set as 2C. From this value, the respective C values of the trichome-neighboring cell nuclei were estimated.
For DNA measurements of trichomes, rosette leaves were vacuum infiltrated for 30 min in formaldehyde solution (3.7% formaldehyde in PBS + 0.1% Tween [PBST]) followed by a 2-h incubation at 4°C. Samples were washed two times for 15 min in PBST. Then, leaves were vacuum infiltrated in DAPI solution (0.25 μg/mL, 5% DMSO in PBST) for 15 min and incubated overnight in DAPI solution at 4°C; thereafter, leaves were washed two times in PBST. For measurement of DAPI intensity, see above. The median value of Columbia trichomes was set as 32C. From this value, the respective C values of the trichome nuclei were estimated. For determination of nuclear area, embryos were stained for 30 min in a propidium iodide solution (100 μg/mL in water). The area of the hypocotyl nuclei was measured using the DISKUS software package.
Fluorescent-Activated Cell Sorting Analysis
Ten to twenty leaves of rosette leaves one and two of 10-, 15- and 20-d-old seedlings were harvested and cut with a razor blade into small pieces on ice. Cold propidium iodide buffer (100 mM Tris-HCl, pH 7, 5 mM MgCl2, 85 mM NaCl, 0.1% [v/v] Triton, 50 μg/mL of propidium iodide, and 50 μg/mL of RNase A) was added to the samples. The suspension was filtered through Cell Trics filters (Partec, Münster, Germany) with 50-μm pores. Fluorescent-activated cell sorting analysis was performed with FACSCalibur (Becton Dickinson, Heidelberg, Germany). For each sample, 100,000 counts were measured. All measurements were performed in duplicate.
YFP Intensity Measurements
Confocal laser scanning micrographs were taken from young developing trichomes on young rosette leaves. The pinhole was adjusted for each scan so that the YFP intensity was not saturated. Then, the YFP intensity of the trichome nucleus and its respective neighboring cell nucleus was measured using the Zeiss software package (LSM 510 version 3.2SP2). For the transgenic lines ProGL2:YFP:ICK1/KRP1, ProGL2:ICK1/KRP1109:YFP, and ProGL2:YFP, the ratio of average YFP intensity of trichome neighboring cell nucleus to trichome nucleus was calculated. For each line, at least six independent measurements of single trichomes and two to six neighboring cells were performed.
GUS Assays
Whole-mount GUS assay was performed as described by Sessions et al. (1999).
Computer Work
Photographs were processed using Adobe Photoshop CS 8.0 and Adobe Illustrator CS 11.0 (Mountain View, CA).
Sequence data from this article are as follows: AtICK1/KRP1 has the Arabidopsis gene code At2g23430 and GenBank accession number NM_127907; AtGL2 has the Arabidopsis gene code At1g79840 and GenBank accession number NM_106633; AtTMM has the Arabidopsis gene code At1g80080 and GenBank accession number NM_106657.
Supplementary Material
Acknowledgments
We are grateful to Martina Pesch (Universität Köln, Köln, Germany) and Bekir Ülker (Max-Planck-Institut für Züchtungsforschung, Köln, Germany) for providing plant transformation vectors. We thank John Larkin (University of Louisiana, Baton Rouge, LA) for sharing unpublished results. We would like to thank Xiaoguo Zhang (University of Florida, Gainesville, FL) for his help in creating the ProTMM-expression vector. We thank Christoph Göttlinger (University of Cologne) for assistance in flow cytometry. We thank Elmon Schmelzer and Rolf-Dieter Hirtz from Central Microscopy of the Max-Planck-Institute for Plant Breeding Research in Cologne for advice and help with microscopy devices. We are grateful to Markus Dürenberger and his team from the Scanning Electron Microscopy Laboratory (University of Basel, Basel, Switzerland), especially Marcel Düggelin, for his help with the scanning electron microscopy analysis. We would like to thank Daniel Bouyer, Axel Dienemann, Niko Geldner, Viktor Kirik, Swen Schellmann, Ian Searle, Frank Sprenger, and members of the authors' laboratories for critically reading the manuscript and helpful comments. Support of the Volkswagen-Stiftung to A.S. is kindly acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Arp Schnittger (schnitt@mpiz-koeln.mpg.de).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.030486.
References
- Ayscough, K., Hayles, J., MacNeill, S.A., and Nurse, P. (1992). Cold-sensitive mutants of p34cdc2 that suppress a mitotic catastrophe phenotype in fission yeast. Mol. Gen. Genet. 232, 344–350. [DOI] [PubMed] [Google Scholar]
- Bagui, T.K., Jackson, R.J., Agrawal, D., and Pledger, W.J. (2000). Analysis of cyclin D3-cdk4 complexes in fibroblasts expressing and lacking p27(kip1) and p21(cip1). Mol. Cell. Biol. 20, 8748–8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, D.C. (2004). Integrating signals in stomatal development. Curr. Opin. Plant Biol. 7, 26–32. [DOI] [PubMed] [Google Scholar]
- Breuil-Broyer, S., Morel, P., de Almeida-Engler, J., Coustham, V., Negrutiu, I., and Trehin, C. (2004). High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J. 38, 182–192. [DOI] [PubMed] [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Olah, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18, 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, M., Olivier, P., Diehl, J.A., Fero, M., Roussel, M.F., Roberts, J.M., and Sherr, C.J. (1999). The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18, 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibazakura, T., McGrew, S.G., Cooper, J.A., Yoshikawa, H., and Roberts, J.M. (2004). Regulation of cyclin-dependent kinase activity during mitotic exit and maintenance of genome stability by p21, p27, and p107. Proc. Natl. Acad. Sci. USA 101, 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Connor, M.K., Kotchetkov, R., Cariou, S., Resch, A., Lupetti, R., Beniston, R.G., Melchior, F., Hengst, L., and Slingerland, J.M. (2003). CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol. Biol. Cell 14, 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, S., and Dolan, L. (2003). Epidermal patterning genes are active during embryogenesis in Arabidopsis. Development 130, 2893–2901. [DOI] [PubMed] [Google Scholar]
- Crawford, K.M., and Zambryski, P.C. (2000). Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10, 1032–1040. [DOI] [PubMed] [Google Scholar]
- Day, S.J., and Lawrence, P.A. (2000). Measuring dimensions: The regulation of size and shape. Development 127, 2977–2987. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inze, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Qi, Y. (2003). Symplasmic protein and RNA traffic: Regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6, 596–602. [DOI] [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Doonan, J. (2000). Social controls on cell proliferation in plants. Curr. Opin. Plant Biol. 3, 482–487. [DOI] [PubMed] [Google Scholar]
- Edgar, B.A., and Orr-Weaver, T.L. (2001). Endoreplication cell cycles: More for less. Cell 105, 297–306. [DOI] [PubMed] [Google Scholar]
- Foley, E., O'Farrell, P.H., and Sprenger, F. (1999). Rux is a cyclin-dependent kinase inhibitor (CKI) specific for mitotic cyclin-Cdk complexes. Curr. Biol. 9, 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley, E., and Sprenger, F. (2001). The cyclin-dependent kinase inhibitor Roughex is involved in mitotic exit in Drosophila. Curr. Biol. 11, 151–160. [DOI] [PubMed] [Google Scholar]
- Gallagher, K.L., Paquette, A.J., Nakajima, K., and Benfey, P.N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14, 1847–1851. [DOI] [PubMed] [Google Scholar]
- Grell, M. (1946). Cytological studies in culex I: Somatic reduction divisions. Genetics 31, 60–76. [PubMed] [Google Scholar]
- Grosskortenhaus, R., and Sprenger, F. (2002). Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev. Cell 2, 29–40. [DOI] [PubMed] [Google Scholar]
- Harper, J.W., Burton, J.L., and Solomon, M.J. (2002). The anaphase-promoting complex: It's not just for mitosis any more. Genes Dev. 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Schnittger, A., and Folkers, U. (1999). Pattern formation and cell differentiation: Trichomes in Arabidopsis as a genetic model system. Int. Rev. Cytol. 186, 147–178. [DOI] [PubMed] [Google Scholar]
- Huskins, C.L. (1948. a). Chromosome multiplication and reduction in somatic tissues. Nature 161, 80–83. [DOI] [PubMed] [Google Scholar]
- Huskins, C.L. (1948. b). Segregation and reduction in somatic tissues. J. Hered. 39, 311–325. [Google Scholar]
- Itaya, A., Liang, G., Woo, Y.-M., Nelson, R.S., and Ding, B. (2000). Nonspecific intercellular protein trafficking probed by green-fluorescent protein in plants. Protoplasma 213, 165–175. [Google Scholar]
- Ito, M. (2000). Factors controlling cyclin B expression. Plant Mol. Biol. 43, 677–690. [DOI] [PubMed] [Google Scholar]
- Ito, M., Iwase, M., Kodama, H., Lavisse, P., Komamine, A., Nishihama, R., Machida, Y., and Watanabe, A. (1998). A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M., and Schnittger, A. (2004). Cell cycle and differentiation. Curr. Opin. Plant Biol. 7, 661–669. [DOI] [PubMed] [Google Scholar]
- Jasinski, S., Riou-Khamlichi, C., Roche, O., Perennes, C., Bergounioux, C., and Glab, N. (2002). The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3;1-overexpressing plants. J. Cell Sci. 115, 973–982. [DOI] [PubMed] [Google Scholar]
- Joubes, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inze, D., Umeda, M., and Renaudi, J.P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43, 607–620. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. [DOI] [PubMed] [Google Scholar]
- Kirik, V., Bouyer, D., Schöbinger, U., Bechtold, N., Herzog, M., Bonneville, J.-M., and Hülskamp, M. (2001). CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr. Biol. 11, 1891–1895. [DOI] [PubMed] [Google Scholar]
- Kondorosi, E., and Kondorosi, A. (2004). Endoreduplication and activation of the anaphase-promoting complex during symbiotic cell development. FEBS Lett. 567, 152–157. [DOI] [PubMed] [Google Scholar]
- Kondorosi, E., Roudier, F., and Gendreau, E. (2000). Plant cell-size control: Growing by ploidy? Curr. Opin. Plant Biol. 3, 488–492. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1990). Mutations affecting the testa colour in Arabidopsis. Arabidopsis Inf. Serv. 27, 1–4. [Google Scholar]
- Koroleva, O.A., Tomlinson, M., Parinyapong, P., Sakvarelidze, L., Leader, D., Shaw, P., and Doonan, J.H. (2004). CycD1, a putative G1 cyclin from Antirrhinum majus, accelerates the cell cycle in cultured tobacco BY-2 cells by enhancing both G1/S entry and progression through S and G2 phases. Plant Cell 16, 2364–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, S., Tanaka, H., Yasuda, H., and Todokoro, K. (1999). Regulation of APC activity by phosphorylation and regulatory factors. J. Cell Biol. 146, 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kramer, E.R., Scheuringer, N., Podtelejnikov, A.V., Mann, M., and Peters, J.M. (2000). Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell 11, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer, J., Garrett, M.D., Stevenson, L.F., Slingerland, J.M., Sandhu, C., Chou, H.S., Fattaey, A., and Harlow, E. (1997). New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11, 847–862. [DOI] [PubMed] [Google Scholar]
- Larkin, J., Brown, M., and Schiefelbein, J. (2003). How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54, 403–430. [DOI] [PubMed] [Google Scholar]
- Larkin, J., Young, N., Prigge, M., and Marks, M. (1996). The control of trichome spacing and number in Arabidopsis. Development 122, 997–1005. [DOI] [PubMed] [Google Scholar]
- Lin, Y., and Schiefelbein, J. (2001). Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development 128, 3697–3705. [DOI] [PubMed] [Google Scholar]
- MacNeill, S.A., Creanor, J., and Nurse, P. (1991). Isolation, characterisation and molecular cloning of new mutant alleles of the fission yeast p34cdc2+ protein kinase gene: Identification of temperature-sensitive G2-arresting alleles. Mol. Gen. Genet. 229, 109–118. [DOI] [PubMed] [Google Scholar]
- Marks, M.D. (1997). Molecular genetic analysis of trichome development in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 137–163. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423–442. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. [DOI] [PubMed] [Google Scholar]
- Molhoj, M., Jorgensen, B., Ulvskov, P., and Borkhardt, B. (2001). Two Arabidopsis thaliana genes, KOR2 and KOR3, which encode membrane-anchored endo-1,4-beta-D-glucanases, are differentially expressed in developing leaf trichomes and their support cells. Plant Mol. Biol. 46, 263–275. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002. a). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002. b). Stomatal development in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society Plant Biologists), doi/10.1199/tab.0066, http://www.aspb.org/publications/arabidopsis/.
- Nagl, W. (1976). Zellkern und Zellzyklen: Molekularbiologie, Organisation und Entwicklungsphysiologie der Desoxyribonucleinsaeure und des Chromatins. (Stuttgart, Germany: Ulmer).
- Niculescu III, A.B., Chen, X., Smeets, M., Hengst, L., Prives, C., and Reed, S.I. (1998). Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol. Cell. Biol. 18, 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olashaw, N., Bagui, T.K., and Pledger, W.J. (2004). Cell cycle control: A complex issue. Cell Cycle 3, 263–264. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. (2004). Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 9, 33–41. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. [DOI] [PubMed] [Google Scholar]
- Ormenese, S., de Almeida Engler, J., De Groodt, R., De Veylder, L., Inze, D., and Jacqmard, A. (2004). Analysis of the spatial expression pattern of seven Kip related proteins (KRPs) in the shoot apex of Arabidopsis thaliana. Ann. Bot. (Lond.) 93, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich, N.P. (1999). Mechanisms of cyclin-dependent kinase regulation: Structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 287, 821–828. [DOI] [PubMed] [Google Scholar]
- Peters, J.M. (1998). SCF and APC: The yin and yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 10, 759–768. [DOI] [PubMed] [Google Scholar]
- Porceddu, A., Stals, H., Reichheld, J.P., Segers, G., De Veylder, L., Barroco, R.P., Casteels, P., Van Montagu, M., Inze, D., and Mironov, V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276, 36354–36360. [DOI] [PubMed] [Google Scholar]
- Potter, C.J., and Xu, T. (2001). Mechanisms of size control. Curr. Opin. Genet. Dev. 11, 279–286. [DOI] [PubMed] [Google Scholar]
- Rerie, W.G., Feldmann, K.A., and Marks, M.D. (1994). The GLABRA2 gene encodes a homeodomain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Rumbolz, J., Kassemeyer, H.-H., Steinmetz, V., Deising, H.B., Mendgen, K., Mathys, D., Wirtz, S., and Guggenheim, R. (1999). Differentiation of infection structures of the powdery mildew fungus Uncinula necator and adhesion to the host cuticle. Can. J. Bot. 78, 409–421. [Google Scholar]
- Schnittger, A., and Hulskamp, M. (2002). Trichome morphogenesis: A cell-cycle perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., and Hulskamp, M. (2003). PCD in development of plant vegetative tissue. In When Plant Cells Die, J. Gray, ed (Sheffield, UK: Academic Press), pp. 106–130.
- Schnittger, A., Schobinger, U., Bouyer, D., Weinl, C., Stierhof, Y.D., and Hulskamp, M. (2002. b). Ectopic D-type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc. Natl. Acad. Sci. USA 99, 6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger, A., Schobinger, U., Stierhof, Y.D., and Hulskamp, M. (2002. a). Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 12, 415–420. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., Weinl, C., Bouyer, D., Schobinger, U., and Hulskamp, M. (2003). Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell 15, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J., and Roberts, J.M. (1999). CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Shteinberg, M., Protopopov, Y., Listovsky, T., Brandeis, M., and Hershko, A. (1999). Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 260, 193–198. [DOI] [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Slingerland, J., and Pagano, M. (2000). Regulation of the cdk inhibitor p27 and its deregulation in cancer. J. Cell. Physiol. 183, 10–17. [DOI] [PubMed] [Google Scholar]
- Smith, A.V., and Orr-Weaver, T.L. (1991). The regulation of the cell cycle during Drosophila embryogenesis: The transition to polyteny. Development 112, 997–1008. [DOI] [PubMed] [Google Scholar]
- Stern, B., and Nurse, P. (1996). A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 12, 345–350. [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6, 544–553. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Tomoda, K., Kubota, Y., and Kato, J. (1999). Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398, 160–165. [DOI] [PubMed] [Google Scholar]
- Ubersax, J.A., Woodbury, E.L., Quang, P.N., Paraz, M., Blethrow, J.D., Shah, K., Shokat, K.M., and Morgan, D.O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425, 859–864. [DOI] [PubMed] [Google Scholar]
- Vidwans, S.J., DiGregorio, P.J., Shermoen, A.W., Foat, B., Iwasa, J., Yakubovich, N., and O'Farrell, P.H. (2002). Sister chromatids fail to separate during an induced endoreplication cycle in Drosophila embryos. Curr. Biol. 12, 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen, C.W., Mordhorst, A.P., Albrecht, C., Kwaaitaal, M.A., and de Vries, S.C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.D., Oppenheimer, D.G., Concienne, J., and Larkin, J.C. (2000). SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127, 3931–3940. [DOI] [PubMed] [Google Scholar]
- Wang, H., Fowke, L.C., and Crosby, W.L. (1997). A plant cyclin-dependent kinase inhibitor gene. Nature 386, 451–452. [DOI] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. [DOI] [PubMed] [Google Scholar]
- Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L.C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24, 613–623. [DOI] [PubMed] [Google Scholar]
- Zachariae, W., Schwab, M., Nasmyth, K., and Seufert, W. (1998). Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282, 1721–1724. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Fowke, L.C., and Wang, H. (2002). Plant CDK inhibitors: Studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparison in transgenic Arabidopsis plants. Plant Cell Rep. 20, 967–975. [Google Scholar]
- Zhou, Y., Li, G., Brandizzi, F., Fowke, L.C., and Wang, H. (2003). The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant J. 35, 476–489. [DOI] [PubMed] [Google Scholar]
- Zybina, E.V., and Zybina, T.G. (1996). Polytene chromosomes in mammalian cells. Int. Rev. Cytol. 165, 53–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.