Abstract
The orientation of cell expansion is a process at the heart of plant morphogenesis. Cellulose microfibrils are the primary anisotropic material in the cell wall and thus are likely to be the main determinant of the orientation of cell expansion. COBRA (COB) has been identified previously as a potential regulator of cellulose biogenesis. In this study, characterization of a null allele, cob-4, establishes the key role of COB in controlling anisotropic expansion in most developing organs. Quantitative polarized-light and field-emission scanning electron microscopy reveal that loss of anisotropic expansion in cob mutants is accompanied by disorganization of the orientation of cellulose microfibrils and subsequent reduction of crystalline cellulose. Analyses of the conditional cob-1 allele suggested that COB is primarily implicated in microfibril deposition during rapid elongation. Immunodetection analysis in elongating root cells revealed that, in agreement with its substitution by a glycosylphosphatidylinositol anchor, COB was polarly targeted to both the plasma membrane and the longitudinal cell walls and was distributed in a banding pattern perpendicular to the longitudinal axis via a microtubule-dependent mechanism. Our observations suggest that COB, through its involvement in cellulose microfibril orientation, is an essential factor in highly anisotropic expansion during plant morphogenesis.
INTRODUCTION
In the absence of cell movement, plant morphogenesis is dependent on tight regulation of cell division and expansion. Plant development is characterized by complex patterns of growth that determine organ form and function. Most plant cells grow anisotropically by controlling how their extracellular matrix, the cell wall, yields to internal and isotropic turgor pressure. When the direction of maximal anisotropic expansion coincides with the longitudinal axis of the growing organ, it is called elongation. The primary cell wall combines unique properties of strength and rigidity but also of plasticity and viscosity, physical properties that derive from the nature of the different wall polymers: a load-bearing cellulose/xyloglucan array and a compression-resistant pectin gel (Carpita and Gibeaut, 1993; Roberts, 1994). Cell expansion depends directly on the local control of these structures and must combine remodeling activities to allow for slippage of existing polymers with the incorporation of newly synthesized materials (Cosgrove, 2000, 2001). The molecular mechanisms that regulate the orientation and extent of cell expansion remain poorly understood, and the identification of the key proteins involved is of paramount importance.
Among the different wall materials, cellulose microfibrils represent the most important component in the control of anisotropic expansion. Inhibition of cellulose biosynthesis with compounds such as 2,6-dichlorobenzonitrile (DCB), isoxaben, or thaxtomin (Scheible et al., 2003) results in a rapid loss of growth anisotropy. Cellulose-deficient mutants of Arabidopsis thaliana, such as rsw1 (Arioli et al., 1998; Williamson et al., 2001) and korrigan (kor/rsw2) (Nicol et al., 1998; Lane et al., 2001), also show a dramatic loss of anisotropic expansion often accompanied by cell swelling.
Cellulose microfibrils are synthesized at the plasma membrane by multimeric complexes called rosettes (Doblin et al., 2002), which are typically arranged in parallel arrays, oriented transversely to the axis of elongation. This pattern is thought to be critical for the proper orientation of cell expansion (Taiz, 1984). Biophysical, pharmacological, and genetic studies have suggested that the direction of alignment of microfibrils can be regulated at multiple levels. However, despite its importance for morphogenesis and development, the mechanism for aligning cellulose microfibrils is poorly understood.
For many years, the deposition of microfibrils has been considered to be under the control of microtubules (Green, 1962; Ledbetter and Porter, 1963; Hepler and Newcomb, 1964). Cortical microtubules have been demonstrated both genetically and chemically to regulate the directionality of cell expansion. Mutants in which the microtubule arrays are either disorganized, as in tonneau (ton) (Camilleri et al., 2002) and bot1/fra2 (Bichet et al., 2001; Burk et al., 2001; Burk and Ye, 2002), or more severely disrupted, as in mor1-1 (Whittington et al., 2001; Sugimoto et al., 2003), lose their ability to control growth anisotropy. Similar defects can be phenocopied by the application of microtubule-depolymerizing agents, such as oryzalin (Baskin et al., 1994).
Reduction in the anisotropy of expansion upon disruption of microtubules and the correlation between the alignment of cortical microtubules and cellulose microfibrils are the two principal reasons for the belief that cortical microtubules align newly synthesized microfibrils. The most popular models posit that microtubules constrain rosette movement by serving either as tracks or guard rails (Giddings and Staehelin, 1991). A functional relationship between microtubule and microfibril alignment recently received support from several studies. Gardiner et al. (2003) demonstrated, for secondary cell wall synthesis, that cortical microtubules are required to confine cellulose synthase subunits to regions of cell wall deposition during xylem development. Moreover, the characterization of the botero/fra2 mutant (deficient in a katanin p60 subunit; Burk and Ye, 2002), in which cortical microtubules and cellulose microfibrils are both disorganized, supports the existence of a functional link between the two networks, although it should be noted that the rate of cellulose synthesis is also reduced in this mutant. Recently, Baskin (2001) proposed an alternative model in which microfibrils are aligned by virtue of binding an oriented scaffold, with the orientation cues provided by either the cell wall or the cortical microtubules.
Furthermore, the idea that microtubules influence growth anisotropy by controlling microfibril alignment may be incomplete. There are actually few cases in which the alignment of cellulose has been examined carefully after microtubule disruption, and several cases in which little change in microfibril orientation occurred despite a wholesale change in microtubule orientation (Baskin, 2001). For example, recently, the mor1-1 mutant was reported to have undisturbed microfibril alignment despite significantly disrupted cortical microtubule arrays (Sugimoto et al., 2003), and recovery of well-ordered microfibrils can proceed without the oriented microfibrillar or microtubular scaffold (Himmelspach et al., 2003).
First identified in a genetic screen for regulators of cell expansion, the cobra (cob) mutant was shown to have a conditional and root-specific cell expansion defect (Benfey et al., 1993; Hauser et al., 1995). Initial characterization indicated that COB is a membrane protein required to orient cell expansion at the onset of rapid elongation in the root, a function that appeared to be associated with crystalline cellulose production (Schindelman et al., 2001). In this article, an analysis of COB expression and a newly identified null allele indicate that COB is a key regulator of diffuse anisotropic expansion throughout postembryonic development. We provide evidence that COB is anchored on the extracellular side of the plasma membrane by a glycosyl-phosphatidyl inositol (GPI) moiety and also released into the wall. We show that COB is required for the oriented deposition of cellulose microfibrils and is aligned in narrow bands perpendicular to the longitudinal axis in cells undergoing rapid elongation, a pattern that depends on, but is not closely related to, cortical microtubule organization.
RESULTS
COB Expression Is Developmentally Regulated and Linked to Anisotropic Expansion
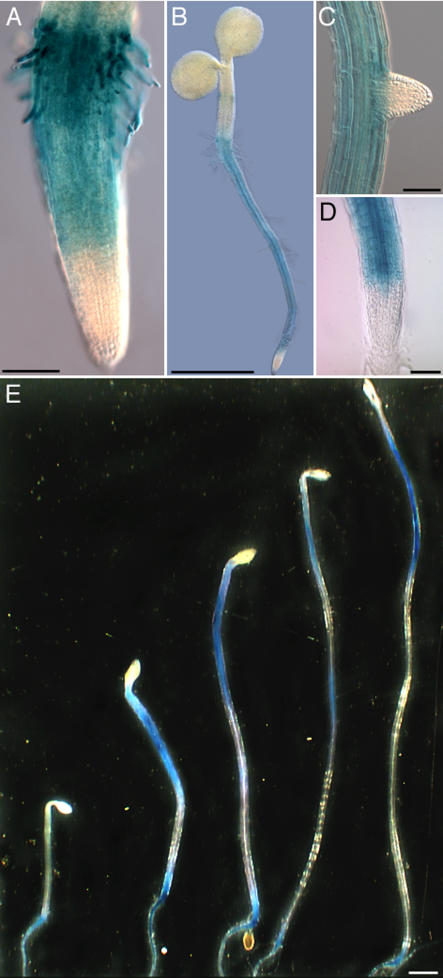
Previously, COB was shown to be expressed in most plant organs on the basis of RNA gel blots and RT-PCR analyses (Schindelman et al., 2001; Roudier et al., 2002). To specify the spatial and temporal expression pattern of COB during plant development, a β-glucuronidase (GUS) reporter fusion was generated. Approximately 2.5 kb of COB 5′ upstream sequence was fused to the uidA gene followed by 1.4 kb of COB 3′ downstream sequence, and the resulting PCOB:GUS fusion was introduced into wild-type Arabidopsis. Assaying GUS activity at different times in the plant life cycle revealed that COB was expressed only postembryonically, with its strongest expression in the root. Weaker GUS activity was detectable in growing leaves, particularly in epidermal cells, guard cells, and fully developed trichomes, as well as in the stigma and anthers of developing flowers. GUS activity was first detectable in the root tips of germinating seedlings (Figure 1A), where it showed a sharp upregulation at or near the onset of the elongation zone, and expression remained strong in the more differentiated parts of the root. At all stages, COB expression was absent in primary and lateral root meristems (Figures 1B to 1D). This expression pattern is consistent with the in situ hybridization pattern reported previously (Schindelman et al., 2001). COB expression was also observed in etiolated hypocotyls, which undergo rapid anisotropic expansion (Figure 1E). During skotomorphogenesis, the GUS staining pattern in the hypocotyls mirrored the acropetal elongation gradient (Gendreau et al., 1997; Refrégier et al., 2004), providing further evidence linking COB expression and rapid longitudinal expansion.
Figure 1.
Localization of GUS Activity under the Control of COB cis-Regulatory Regions.
(A) to (D) GUS staining in 2-d-old root (A), 7-d-old seedling (B), and 12-d-old lateral and primary roots ([C] and [D]).
(E) COB expression in etiolated hypocotyls at 3 to 7 d after germination.
Bars = 100 μm in (A), (C), and (D), 1 mm in (B), and 2.5 mm in (E).
COB Is GPI-Anchored and N-Glycosylated
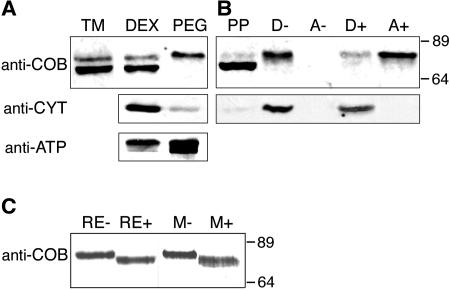
Analysis of the COB amino acid sequence predicted its substitution with a GPI anchor (Schindelman et al., 2001). To validate the glypiation of COB, we prepared total membranes from Arabidopsis callus culture and partitioned them into a dextran phase (DEX), which contains endomembranes and is depleted in plasma membrane, and a polyethylene glycol (PEG) phase, which is plasma membrane-enriched. Protein gel blot analysis of the resulting fractions using anti-COB affinity-purified antibodies demonstrated that COB was associated with the plasma membrane (Figure 2A). As expected, the endoplasmic reticulum–localized cytochrome b5 was depleted from the plasma membrane–enriched fraction, whereas the plasma membrane–localized ATPase was enriched. To determine whether COB is a GPI-anchored protein, total membranes were fractionated by Triton X-114 phase partitioning, thereby separating peripheral membrane proteins from integral and GPI-anchored proteins, which are present in the detergent phase. The detergent phase was then incubated with a phosphatidylinositol-specific phospholipase C (PI-PLC) and subjected again to phase partitioning. As shown in Figure 2B, in the absence of PI-PLC treatment, the COB protein was present in the detergent and not detectable in the aqueous phase, whereas the PI-PLC treatment of this detergent fraction resulted in the release of the COB protein from the membrane fraction into the soluble (aqueous) fraction. In a control, the endoplasmic reticulum–localized and integral membrane protein cytochrome b5 remained insensitive to the PI-PLC treatment.
Figure 2.
COB Is Plasma Membrane–Localized, GPI-Anchored, and N-Glycosylated.
(A) Protein blot probed successively with anti-COB, anti-cytochrome b5, and anti-ATPase antibodies. Total membrane proteins (TM) were phase-partitioned into endomembranes (DEX) and plasma membrane–enriched protein fractions (PEG).
(B) Protein blot probed successively with anti-COB and anti-cytochrome b5 antibodies. Proteins associated with total membranes were solubilized in Triton X-114 and phase-partitioned into an aqueous peripheral protein fraction (PP) and a detergent phase (D). The pure detergent phase was treated with PI-PLC (+) or with a buffer (−) and phase-partitioned into new detergent (D) and aqueous phases (A).
(C) Protein blot probed with anti-COB antibodies. Root extracts (RE) or microsomal membranes (M) were incubated with peptide N-glycosidase F (+) or buffer (−), and proteins were separated by SDS-PAGE.
Molecular weights are indicated at right.
COB, like many other extracellular proteins, has been predicted to be N-glycosylated (Borner et al., 2002; Roudier et al., 2002). When crude root extracts or microsomal membrane fractions were treated with peptide N-glycosidase F, the COB protein detected by immunoblot analysis displayed greater mobility compared with the undigested samples (Figure 2C). This change in protein mobility indicated that COB in its membrane-bound form had been posttranslationally modified by the addition of N-linked glycans. In the total and endomembrane fractions, the affinity-purified anti-COB antibodies also recognized a smaller protein, which partitioned as a peripheral membrane protein (Figures 2A and 2B). We hypothesize that this protein band corresponds to a cleaved and unmodified version of COB present in the endoplasmic reticulum.
COB Is Required for Anisotropic Cell Expansion throughout the Plant
The primary developmental defect of the cob alleles described previously was associated with root elongation (Benfey et al., 1993; Hauser et al., 1995; Schindelman et al., 2001). The discrepancy between the root-specific defects and the widespread expression pattern reported here could be explained either by functional redundancy with other genes in the aerial part of the plant (Roudier et al., 2002) or by a degree of functionality retained by the missense alleles whose phenotype was characterized. To distinguish between these hypotheses, we identified a putative null allele of cob in the Syngenta Arabidopsis Insertion Library (SAIL) (Sessions et al., 2002). Line 735D10 contains a T-DNA insertion in the fifth exon of COB at amino acid Met-213 and was renamed cob-4. Our analyses indicate that this mutation is recessive, monogenic, and nuclear. The BASTA resistance marker associated with the T-DNA cosegregated with the mutant phenotype, and the phenotype was complemented by expressing the COB cDNA under the 35S promoter (data not shown). No detectable signal was observed in protein gel blots of cob-4 protein extracts using anti-COB antibodies (see Methods), providing evidence that cob-4 is a null allele. cob-4 plants were sterile, as a result of the complete absence of inflorescence stem development. The phenotype of transheterozygous cob-1/cob-4 plants was very similar to that of cob-1/cob-1 homozygotes, conditional on growth rate and apparent only in the root (data not shown). This finding suggests that cob-1 is a partial loss-of-function allele of COB.
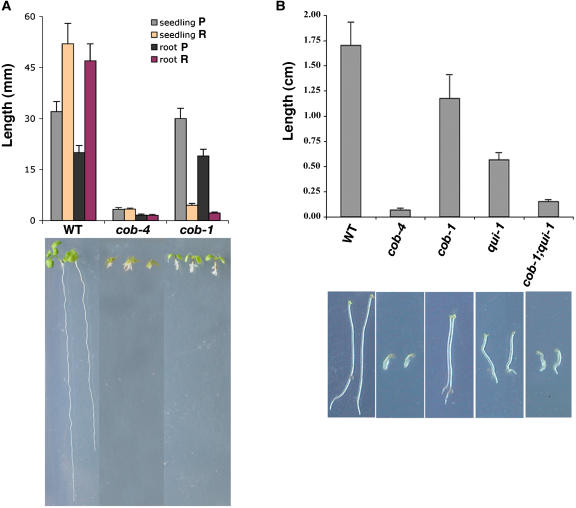
The growth of cob-4 seedlings was reduced dramatically compared with that of wild-type seedlings in all conditions tested. When grown on vertical plates, cob-4 seedlings were ∼90% shorter (measured from the shoot apical meristem to the root tip) and significantly thicker. A reduction in length was seen in all organs (Figure 3A). By contrast, cob-1 seedlings were similar to the wild type when grown on 0.3% sucrose (the permissive condition) and were ∼15% larger than cob-4 when grown on 4.5% sucrose (the restrictive condition). Dark-grown hypocotyls of cob-4 seedlings had an ∼95% reduction in length compared with the wild type (Figure 3B). This finding demonstrates that COB is also essential for the rapid elongation of the etiolated hypocotyl. By contrast, cob-1 and qui-1 (a conditional mutant allele of the cellulose synthase subunit AtCESA6) had only 25 and 67% reductions in hypocotyl length, respectively. Interestingly, the homozygous cob-1 qui-1 double mutant showed an additive phenotype, with an 85% reduction in hypocotyl length (Figure 3B).
Figure 3.
Homozygous cob-4 Plants Have a Severe Growth Defect.
(A) Length measurements of the conditional cob-1 and null cob-4 alleles under different growth conditions. Data for cob-1, cob-4, and wild-type plants grown under restrictive conditions are shown. P and R, permissive and restrictive conditions, respectively. Each bar represents the average ± sd of 15 seedlings.
(B) Reduced hypocotyl elongation in cob and qui-1 mutants as well as in the cob-1 qui-1 double mutant. Hypocotyl lengths of 7-d-old dark-grown seedlings under permissive condition are shown. Each bar represents the average ± sd of 15 hypocotyls.
Photographs of the relevant wild-type and mutant seedlings are shown below the bar graphs.
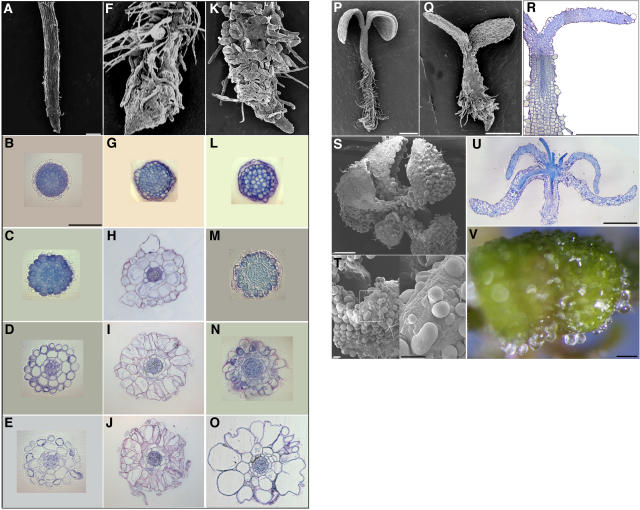
Observations of roots from 7-d-old seedlings by scanning electron microscopy revealed that cob-1 and cob-4 have similar but not identical root-swelling phenotypes (Figures 4A, 4F, and 4K). Paradoxically, in seedlings continuously grown under restrictive conditions, the meristem of cob-1 roots (77 individuals out of 80) showed abnormal radial expansion, whereas the same region of the root in cob-4 mutants showed little or no change in cell shape compared with the wild type (43 individuals out of 45). In both genotypes, root hairs emerged close to the root tip from cells that appeared to have undergone little elongation. To compare the cellular morphology in the different zones of the mutants, transverse sections were made at increasing distances from the root tip. In the root cap and quiescent center region, the mutants were similar to the wild type (Figures 4B, 4G, and 4L). Differences between the two alleles became apparent in the meristem, where cells exhibited abnormal radial expansion in cob-1 but remained fairly normal in cob-4 (Figures 4C, 4H, and 4M). Furthermore, cob-1 roots attained their maximal diameter in the early elongation zone (Figures 4D, 4I, and 4N), whereas cob-4 roots expanded radially throughout the elongation zone, ultimately reaching a diameter that was nearly twice that of the wild type. Also in the elongation zone, cells in the cob-4 epidermis took on a characteristic bulging morphology (Figure 4O). In both alleles, cell walls were occasionally broken, mostly in the epidermal layer. Cell wall breakage was rare and did not seem to derive from incomplete cell plate formation or abnormal cell divisions.
Figure 4.
cob-4 Has Dramatic Morphological Alterations in Most Organs.
Root phenotype analysis of wild-type ([A] to [E]), cob-4 ([F] to [J]), and cob-1 ([K] to [O]) plants.
(A), (F), and (K) Scanning electron micrographs of 10-d-old roots. Bar = 100 μm.
(B) to (E), (G) to (J), and (L) to (O) Light micrographs of 3-μm transverse sections in the region of the root cap-quiescent center ([B], [G], and [L]), meristem ([C], [H], and [M]), early elongation zone ([D], [I], and [N]), and late elongation zone [[E], [J], and [O]). Bar = 100 μm.
(P) to (R) Scanning electron and light micrographs of 1-week-old cob-1 (P), cob-4 (Q), and corresponding longitudinal section (R). Bars = 500 μm.
(S) to (V) Scanning electron ([S] and [U]) and light (V) micrographs of 18-d-old cob-4 aerial organs and corresponding longitudinal section (T). Bars = 500 μm in (S) and (U) and 100 μm in (T) and (V).
Whereas cob-1 aerial organs are similar to wild-type organs (Figure 4P) (Hauser et al., 1995; Schindelman et al., 2001), 7-d-old cob-4 hypocotyls and cotyledons are much smaller and thicker than those of the wild type (Figure 4Q). There is also widespread bulging of epidermal cells (Figures 4S to 4V), although the shoot apical meristem, leaf primordia, and very young leaves do not appear to be affected. Longitudinal sections of entire seedlings at different developmental stages confirmed the absence of sustained anisotropic cell expansion as well as the ballooning of epidermal cells (Figures 4R and 4U). The sections also revealed some internal expansion abnormalities in leaf mesophyll cells, which reduced or eliminated intercellular air spaces. In the absence of hypocotyl, petiole, and stem elongation, 3-week-old cob-4 plants appeared severely stunted, with thick and vitreous organs. Callus-like clusters often appeared on the adaxial side of the leaves. Together, these morphological observations highlight the essential and specific role of COB in the control of anisotropic expansion during plant morphogenesis.
COB Is Required for the Oriented Deposition of Cellulose Microfibrils during Rapid Anisotropic Expansion
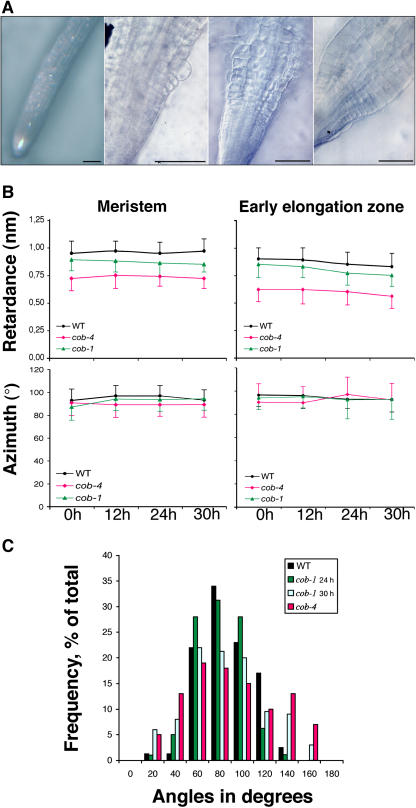
Previous work has shown that cob-1 has lower levels of crystalline cellulose when grown for 1 week under restrictive conditions, suggesting that COB is required for cellulose synthesis (Schindelman et al., 2001). To investigate this notion further, we used polarized-light microscopy and digital imaging to determine the relationship between the expansion defect observed in cob roots and the amount and degree of organization of cellulose microfibrils. The birefringent retardation (retardance) measured within a single cell wall is proportional to the amount of crystalline cellulose and its net degree of orientation (Preston, 1974). We focused our analyses on epidermal cells at the border of the meristem and elongation zones, where COB expression is sharply upregulated. At this developmental transition, postmitotic cells abruptly accelerate their elongation rate 5- to 10-fold (van der Weele et al., 2003). In the elongation zone, cellulose microfibrils (and cortical microtubules) are oriented perpendicular to the direction of rapid elongation (Sugimoto et al., 2000; Baskin et al., 2004). To follow microfibril behavior during a gradual loss of growth anisotropy resulting from COB deficiency, and to dissect primary from secondary effects triggered by the absence of functional COB, we took advantage of the conditional nature of the cob-1 mutation and performed measurements at 0, 12, 24, and 30 h after a shift from permissive to restrictive conditions, with equivalent measurements made for the wild type and cob-4. At the beginning of the condition-shift experiment (0 h), cob-1 roots had a wild-type appearance; swelling was first detectable after ∼12 h and spread during the time course, being apparent first in the elongation zone and only subsequently in the meristem (Figure 5A; see Supplemental Figure 1 online).
Figure 5.
Crystalline Cellulose Organization and Root Morphology in the cob Alleles.
(A) Time-course light micrographs of cob-1 root morphology at 0, 12, 14, and 30 h after the condition shift. Bars = 100 μm.
(B) Quantification by polarized-light microscopy of the amount of cellulose (retardance) and its net orientation (azimuth) in the walls of meristematic and elongating cells. Data from 5-d-old wild-type, cob-1, and cob-4 seedlings at 0, 12, 24, and 30 h after transfer to high-growth-rate media are shown. Each data point represents the average ± sd of at least 15 cell walls.
(C) Frequency distribution of cellulose microfibril-azimuth angles measured in the elongation zone of wild-type, cob-1, and cob-4 roots after the condition shift. F-test values (0.2 significance level) indicate that the modal distributions of azimuth angles are significantly different between the wild type and cob-4 (P = 0.081) and between cob-4 and cob-1 (24 h) (P = 0.056), whereas the wild type and cob-1 (24 h) are not significantly different (P = 0.87). At 30 h after the condition shift, the modal distributions of azimuth angles between the wild type and cob-1 (30 h) are significantly different (P = 0.129).
In the wild type, average retardance was highest in the meristem and relatively constant at different time points and slightly lower but still constant in the elongation zone (Figure 5B, top). By contrast, cells in the cob-4 meristematic and elongation zones had lower retardance than wild-type cells. At time 0, cob-1 cell walls had comparable retardance with the wild type, which did not change during the time course, although there was a nonsignificant trend toward reduced retardance in both regions (Figure 5B). These data suggest that the gradual loss of COB function did not trigger a significant reduction in the quantity of microfibrils or their net orientation in the root growth zone. For comparison, permanent loss of COB function in cob-4 roots or in cob-1 seedlings grown under restrictive conditions for several days (Schindelman et al., 2001) resulted in a significant decrease in retardance, which can be considered a secondary effect of the mutations.
To examine the orientation of the microfibrils, the average values of optical azimuth were obtained from the same cell wall regions where retardance was measured (Figure 5B, bottom). No evident difference between the average azimuth, oriented approximately transverse to the longitudinal axis, was detected in the cob alleles compared with the wild type in either root zone. However, we consistently noted a greater variability in the azimuth measurements performed on the cob alleles, especially in cob-4, in cells from the elongation zone compared with wild-type cells. The frequency distribution of azimuths for the measured cell wall regions had a sd of ∼20° for the wild type but was wider and flatter for cob-4 and cob-1 at 30 h (Figure 5C). This variability indicates that the loss of COB activity disrupts the processes that align microfibrils, even in cob-1, in which levels of cellulose were apparently maintained.
To complement the data obtained with polarized-light microscopy, which responds to the crystalline cellulose throughout the wall, we performed field emission scanning electron microscopy (FESEM) to image directly the innermost cellulose microfibrils in longitudinal walls. As shown in Figures 6A to 6D, in the successive wild-type root zones, the most recently deposited microfibrils were parallel and oriented transversely to the elongation axis, as reported previously (Sugimoto et al., 2000). The deposition patterns of cellulose microfibrils in cells within the cob-4 meristem (Figure 6F) were similar to those of the wild type (Figure 6B). By contrast, in the cob-4 elongation zone, the cell walls showed apparently randomly oriented microfibrils (Figure 6G). This disorganization of microfibrils was also visible in cells within the maturation zone (Figure 6H). FESEM analysis of cob-1 walls allowed us to image microfibrils resulting from a partial loss of COB function and in cells exhibiting less dramatic morphological alterations. Because of the difficulty of the FESEM procedure, cell walls in cob-1 roots were analyzed only at 24 h after the condition shift. At this stage, the cob-1 root was significantly swollen (Figure 6I). In the cob-1 meristem, most of the walls analyzed displayed parallel cellulose microfibrils that were oriented perpendicular to the longitudinal axis (Figures 6J and 6J′). However, in the elongation zone, microfibrils were sometimes transverse (Figure 6K), as they are at an approximately comparable region of the wild type, and sometimes disordered (Figure 6K′), as in cob-4. Farther away from the tip, cell walls in cob-1 tended to resemble those in cob-4 (Figures 6L and 6L′). In summary, 24 h after the condition shift, partial loss of COB function resulted in local disorganization of cellulose microfibrils. Together, these data demonstrate that COB is required for the proper orientation of newly synthesized cellulose microfibrils in regions that experience rapid expansion.
Figure 6.
Organization of Cellulose Microfibrils Is Altered in the Elongation Zone of cob Mutants.
FESEM images of the innermost layer of the cell wall of wild type ([A] to [D]), cob-4 ([E] to [H]), and cob-1 ([I] to [L′]) cryoplaned roots. The low-magnification (×250) images of the root indicate cells in the different root zones from which wall texture was analyzed at higher magnification (×100,000) FESEM imaging. In the meristem of the different genotypes ([B], [E], [J], and [J′]), microfibrils are approximately parallel and oriented transversely to the elongation axis. In the wild type, this typical microfibril organization pattern is maintained throughout the elongation zone ([C] and [D]). In cob-4, microfibrils in the elongation zone (G) and at the border with the maturation zone (H) are distributed randomly. Under conditions similar to a 24-h period after a shift to restrictive conditions, microfibrils in the cob-1 elongation zone ([K] and [K′]) and at the border with the maturation zone ([L] and [L′]) show a clear departure from the transverse orientation observed in the wild type. Microfibrils in cells of the cob-1 elongation zone often exhibit different net orientations in different zones of the same wall ([K] and [K′]). (J) and (J′), (K) and (K′), and (L) and (L′) represent images of two different areas in the same wall. Bars = 250 nm except in (A), (E), and (I), where they = 100 μm.
The Distribution of COB during Cell Elongation Is Dependent on Cortical Microtubule Organization
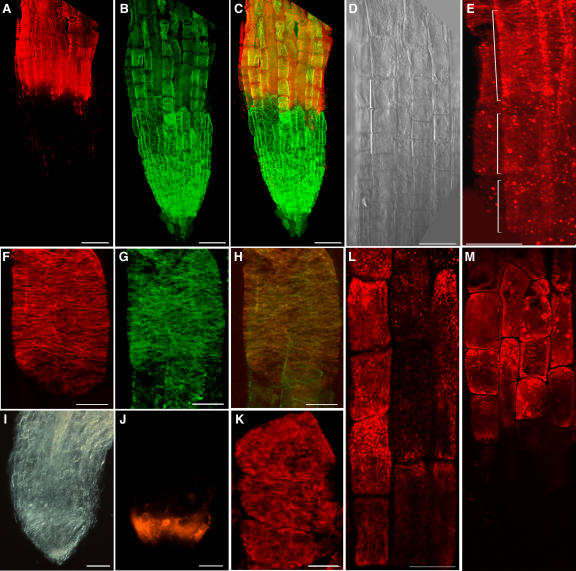
The distribution of the COB protein in elongating cells was analyzed in intact roots using whole-mount indirect immunofluorescence and confocal laser scanning microscopy. In agreement with its expression pattern, COB was detected in the elongation zone, whereas no signal above background was observed in the meristem (Figure 7A). Higher up in the elongation zone, the signal became weaker, and no significant signal was detected in the maturation zone. As expected from the absence of signal on protein gel blots, no significant signal was detected in the cob-4 mutant (data not shown). A closer examination of the wild-type root revealed that the COB protein was first detectable as a diffuse cytoplasmic and peripheral signal at the onset of the elongation zone, and soon the majority of the detected protein became organized in discrete domains on or in the vicinity of the plasma membrane (Figures 7D and 7E). In cells beginning to expand rapidly, COB patches appeared to be organized into narrow bands oriented perpendicularly to the longitudinal axis in a pattern reminiscent of cortical microtubules (Figures 7E and 7F). Double labeling of COB and cortical microtubules (Figures 7B and 7G), although confirming the similar orientation of cortical microtubules and the COB bands in elongating cells, only occasionally revealed COB-dependent fluorescence directly overlying cortical microtubules (Figures 7C and 7H). A similar transverse COB pattern was observed in cob-1 roots under both permissive and restrictive conditions (data not shown). The orientation of the cortical microtubules was not significantly affected in cob-1 (Hauser et al., 1995) or in cob-4 at the beginning of the elongation zone (data not shown).
Figure 7.
COB Immunolocalization in 5-d-Old Roots.
Whole-mount confocal scanning micrographs of wild-type roots ([A] to [E]), wild-type elongating cells ([F] to [H] and [L]), and ton2 roots ([I] to [K]). Bars = 50 μm in (A) to (E) and (I), (J), (L) and (M) and 10 μm in (F) to (H) and (K). Brackets in (D) and (E) indicate the same three elongating cells.
(A), (E), (F), and (J) to (L) Indirect fluorescence immunolocalization of COB using anti-COB antibodies.
(B) and (G) Visualization of cortical microtubules by indirect immunofluorescence.
(C) Combined images of COB (A) and microtubules (B) at the root tip.
(D) and (I) Differential interference contrast micrographs of the wild-type elongation zone and the ton2-14 root tip, respectively.
(H) Combined images of COB (F) and microtubules (G) in an elongating cell.
(L) COB staining after treatment with 10 μM oryzalin for 45 min. Note patches, compared with the normal banding pattern.
(M) COB staining after treatment with 50 μM brefeldin A for 45 min. Note intracellular accumulation of COB-containing clumps.
The relationship between the transverse COB bands and the cortical microtubule network was investigated using genetic and pharmacological approaches. Whole-mount immunolocalization using anti-COB antibodies was performed in weak and strong alleles of the ton2 mutant, which is characterized by a disturbance or loss of orientation of the cortical microtubules (without apparent depolymerization) in elongating cells (Camilleri et al., 2002). In both ton2-14 and ton2-13, COB was detected in the compacted root elongation zone (Figures 7I and 7J) and was more randomly distributed in these cells than in the wild type (Figure 7K). When wild-type roots were treated for 45 min with 10 μM oryzalin, a period in which microtubules depolymerized completely (data not shown) but cell swelling was undetectable, the COB aggregates appeared to be severely randomized (Figure 7L). These findings indicate that COB distribution at the surface of elongating cells depends on the organization of cortical microtubules.
COB Follows a Typical GPI Secretion Path and Is Predominantly Associated with the Longitudinal Cell Walls
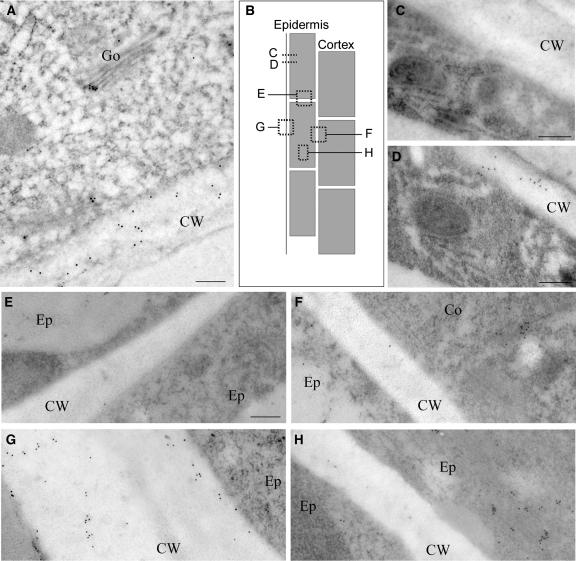
As a GPI-anchored protein, COB would be expected to follow a secretion path and reach the cell periphery through vesicular trafficking. Treatment of elongating root cells with brefeldin A (a vesicle-trafficking inhibitor) resulted in a strong decrease of the COB banding pattern (still visible as weak bands at the cell surface or as dots along the longitudinal walls) and triggered the accumulation of COB in large intracellular compartments (Figure 7M). This suggests that the polar secretion of COB is very active in root elongating cells and shows that the signal observed is a combination of COB-containing vesicles and COB at the cell surface. COB distribution at the subcellular level was further examined by transmission electron microscopy of high-pressure-frozen, immunogold-labeled material from the elongation zone of wild-type roots. At the outer face of epidermal cells (Figure 8A), COB was associated with the Golgi and abundant in the cell wall, supporting its transit along a Golgi-derived vesicle secretion pathway. Serial sectioning in the transverse plane (Figures 8C and 8D) revealed that COB was abundant in some sections and not present in others, consistent with COB's distribution to transverse bands seen in immunofluorescence preparations (Figures 7E and 7F). In contrast with its abundance at the outer epidermal cell wall, COB was present at lower abundance in other walls, particularly the radial (Figure 8E) and even the inner longitudinal (Figures 7H and 8F) walls of the epidermis. Consistent with the rapid upregulation at the onset of elongation, in the root division zone COB was not detected in the walls and was only occasionally detected close to the Golgi, in cells near the onset of the elongation zone (data not shown). In the wall sections examined, COB was present on and at various distances from the plasma membrane (Figures 8A, 8D, and 8G), suggesting that it exists in both a GPI-anchored, plasma membrane–associated form and a cleaved form.
Figure 8.
COB Subcellular Distribution in the Root Elongation Zone Revealed by 10-nm Immunogold Labeling of Sectioned High-Pressure-Frozen Material.
(A) Tangential section from the outer epidermis shows COB associated with the Golgi and in the cell wall.
(B) Scheme of the root in longitudinal view depicts the section planes from which the images in (C) to (H) were obtained.
(C) and (D) Transverse sections from the same series of the outer epidermis ∼350 nm apart. Note the absence of COB from section (C) but its relative abundance in the cytoplasm and cell wall of (D), consistent with the banding pattern observed by immunofluorescence.
(E) Transverse anticlinal section between adjacent epidermal cells shows low abundance of COB.
(F) Longitudinal periclinal section between the epidermis and the cortex shows COB in the cytoplasm but relatively little in the cell wall.
(G) Outer epidermal periclinal section shows abundant COB in both the cytoplasm and the cell wall.
(H) Longitudinal-radial section between adjacent epidermal cell files shows COB in the cytoplasm but relatively little in the cell wall.
Co, cortex; CW, cell wall; Ep, epidermis; Go, Golgi. Bars = 250 nm.
DISCUSSION
COB Is Required for Highly Anisotropic Expansion throughout Plant Development
The relationship between net cellulose orientation in the primary wall and the direction of organ growth was established decades ago (Green, 1980). Nevertheless, the mechanisms underlying the regulation of microfibril orientation in elongating cells remain largely unknown. The data presented here indicate that the GPI-anchored protein COB is an important component of these mechanisms during rapid elongation.
To dissect the molecular mechanisms underlying anisotropic expansion and to understand more precisely the function of COB, we characterized a T-DNA insertion allele, cob-4, and took advantage of the conditional nature of the previously described cob-1 allele (Schindelman et al., 2001). The extensive swelling specifically in cells that would otherwise undergo highly anisotropic growth makes cob-4 an intriguing cell expansion mutant among those characterized to date. Phenotypic analysis showed that COB is specifically involved in the control of diffuse anisotropic expansion, in agreement with its expression pattern. The observation of organized and functional meristems and normal radial patterning in cob roots suggests that cell division is affected minimally if at all in the mutants. Likewise, root hairs seem unaffected by cob mutations, and pollen development appears normal, as judged by the expected transmission ratios of the mutant alleles. These observations indicate that COB is not involved in the process of tip growth but is required specifically for the regulation of diffuse anisotropic expansion.
Recently, relative elongation rates have been measured in the Arabidopsis root at high spatial and temporal resolution (van der Weele et al., 2003). The authors characterized the growth zone as comprising two zones of roughly constant relative elongation rates separated by an abrupt transition. The apical zone (corresponding to the meristem and probably including some postmitotic cells) has a low relative rate, ∼7%/h, and the basal zone (corresponding to the canonical elongation zone) has a high rate of ∼50%/h. The extent of anisotropy increases concomitantly with the acceleration of relative elongation rates, because the tangential expansion rate actually decreases (Baskin et al., 2004). Strikingly, the position at which anisotropy increases is approximately the same as where COB expression is abruptly upregulated. This finding is consistent with our hypothesis that COB is required to achieve highly anisotropic expansion.
The expansion of leaf pavement cells has also been shown to involve two mechanistically distinct growth phases (Fu et al., 2002). Phenotypic analysis of leaf development in cob-4 suggests that COB is required primarily for the second phase, which is characterized by extensive cell growth, and not for the initial morphogenesis of the different epidermal cell types. In cob-4, the shoot apical meristem and leaf primordia appear normal, whereas in the leaf epidermis, the complete loss of COB function resulted in leaves with only small, roughly isodiametric shapes, suggesting that the ability of the cells to expand anisotropically is lost.
Cell elongation in dark-grown hypocotyls was also described recently as a biphasic growth pattern, with the growth acceleration second phase gaining ground acropetally from the base (Refrégier et al., 2004). The absence of hypocotyl elongation in cob-4 is also consistent with an important role for COB during this striking example of rapid polar growth. Thus, in the leaf, hypocotyl, and root, it appears to be the phase of maximal and developmentally regulated anisotropic expansion that requires COB function.
COB Is Required Primarily for the Orientation and Secondarily for the Synthesis of Microfibrils
The cob-4 dwarf phenotype with radially expanded cells is reminiscent of strong alleles of cellulose synthesis–deficient mutants such as rsw1 (mutant in the cellulose synthase subunit AtCESA1; Arioli et al., 1998; Williamson et al., 2001) or kor (mutant in a membrane-bound β-1,4-endoglucanase; Nicol et al., 1998; Lane et al., 2001). Likewise, cob-1 roots, grown under restrictive conditions for several days, had less cellulose than did wild-type roots, as assayed by several independent methods (Schindelman et al., 2001). Furthermore, reduced cellulose synthesis, triggered either genetically as in rsw1 or chemically with DCB, also causes the disorganization of microfibrils (Sugimoto et al., 2001), similar to that seen for cob-4. Therefore, COB may participate in cellulose synthesis, with the chaotic deposition being a consequence of decreased synthesis rates.
However, our observations suggest that COB participates directly in regulating microfibril orientation, with the decreased cellulose synthesis reflecting feedback from the disordered deposition. Within 1 d of shifting cob-1 to the restrictive condition, when swelling was first evident, FESEM images revealed that cellulose microfibrils in many cells were disorganized. On the other hand, retardance of the cell wall was reduced little if at all even after 30 h. If only the innermost cell wall layers are being disorganized, then the expected reduction in retardance may be no more than the trend seen in Figure 4. Given that swelling in cob-1 after 30 h is well advanced, the combination of polarized-light and FESEM data indicate that the loss of microfibril order is the primary defect in the mutant, with the loss of cellulose synthesis occurring secondarily.
Further linking COB function to microfibril orientation, we found that the net orientation of microfibrils within the subcellular regions of the cell wall was more variable in cob-4 and in cob-1 after 30 h under restrictive conditions. Because the retardance of the same regions was reduced not at all (or only slightly), this finding implies that the net direction of alignment of one patch of cell wall was different from that of another. Interestingly, a similar effect on microfibril alignment also occurs when roots are exposed to low concentrations of the microtubule inhibitor oryzalin (Baskin et al., 2004).
Whether the secondary loss of cellulose synthesis results in the accumulation of amorphous β-glucans by a crystallization defect, as occurs in rsw2/kor (Peng et al., 2000; His et al., 2001), remains to be determined. It is interesting that in cob-4 walls imaged with FESEM, we consistently noted the presence of globular clusters extending from microfibrillar structures, which could represent lumps of noncrystallized cellulose.
Cellular Location of COB Function
We have shown that COB is GPI-substituted and N-glycosylated. Proteomics-based approaches demonstrated that other members of the COB family are also GPI-modified (Borner et al., 2003; Elortza et al., 2003; Lalanne et al., 2004). Our immunodetection analyses revealed that in elongating root cells, COB could exist in the different forms normally taken by a GPI-modified protein. Gold label was detected in the Golgi, and transport vesicles, presumably identifying the secretory pathway, and both glycosylated and nonglycosylated forms were detected in microsomal membranes. Gold label was also detected at the plasma membrane as well as within the cell wall. GPI-anchored proteins can be released from the anchor by PI-specific phospholipases, as yet uncharacterized in plants. The presence of considerable COB within the cell wall suggests that during anisotropic expansion, the GPI anchorage is temporary, and the activity of PI-specific phospholipases may represent a way of regulating the amount of anchored COB.
This raises the question of the functional form of COB in the context of microfibril orientation. The fact that in the partial loss-of-function cob-1 allele COB expression and COB localization are not visibly affected (data not shown) indicates that the defects observed are related directly to COB's function and not to a problem of protein stability or targeting. Moreover, engineered versions of COB, either mutated at the potential GPI cleavage attachment sites (N384T, G385W, and S387P) or truncated from the probable cleavage site (Asn-384) and driven by the 35S promoter, were not able to complement the cob phenotype (0 individuals out of 16 and 15 T2 plants, respectively). Transient expression assays using green fluorescent protein (GFP) translationally fused with either the mutated or the truncated C terminus of COB showed GFP mistargeting to endocompartments, whereas the GFP fusion with a wild-type COB C terminus was localized primarily in the plasma membrane (data not shown). These observations highlight the functional significance of GPI substitution and suggest that the membrane-anchored COB or its wall form, or possibly both, represent the functional forms. In animals, GPI-anchored proteins are frequently subjected to specific targeting pathways, often proposed to involve sterol- or sphingolipid-rich microdomains of the plasma membrane (Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998; Muniz and Riezman, 2000), and similar targeting of GPI-anchored proteins may occur in plants (Borner et al., 2005). In the case of COB, the observed asymmetric targeting of this protein to the outer epidermal face opens new avenues of investigation to understand how plant cells take advantage of glypiation to effect specific localization.
Association between COB and Microtubules
In elongating epidermal cells, the COB protein is distributed mainly near the cell surface in transverse bands that parallel cortical microtubules. Based on the transmission electron microscopy data, it appears that these bands reflect clustering within the cell wall at variable distances from the plasma membrane. Unfortunately, microtubules were only rarely encountered in the cryofixed, freeze-substituted material; thus, it could not be determined with confidence whether there is any positional relation between microtubules and the COB-positive clusters.
When microtubule organization is genetically or pharmacologically challenged, the COB banding pattern becomes disrupted and COB-labeled patches become more prominent in the cytosol, implying that microtubules are required, directly or indirectly, for COB localization. Because the banding is disrupted even after a relatively brief oryzalin treatment, it appears that at least some of the label seen in the confocal images arises from cytoplasmic material in addition to plasma membrane and cell wall–bound material, as shown by the partial loss of the banded localization of COB after brief treatment with brefeldin A.
Although the mechanism leading to a banded appearance of COB remains to be established, the microtubule dependence of COB localization provides an intriguing link between the function of the protein and cellulose orientation. A recently proposed model postulates that nascent cellulose microfibrils are oriented by binding to a scaffold of cell wall polysaccharides or plasma membrane proteins, which, in turn, are oriented according to positional information provided by either cortical microtubules or the cell wall (Baskin, 2001). The COB protein, with its particular surface localization and distribution during rapid elongation, is a candidate for one of these scaffolding proteins.
At present, these scaffolds are purely hypothetical, and recent experiments combining the mor1-1 mutant, in which microtubules are disrupted, and DCB treatment, which disorganizes microfibrils, suggest that neither preexisting cell wall templates nor microtubules are required for microfibril alignment (Himmelspach et al., 2003). This, along with other observations (Wiedemeier et al., 2002; Sugimoto et al., 2003), has led to a new model whereby microtubules influence growth anisotropy by regulating the relative length of microfibrils (Wasteneys, 2004). According to this model, the microtubule-dependent patterning of COB could support the formation and extension of relatively long microfibrils, either by preventing the breakage of newly formed microfibrils or by regulating the activity of synthase complexes (Wasteneys, 2004). The potential cellulose binding capacity of COB (Roudier et al., 2002) and the phenotypic alterations reported here fit well with this latest model. Loss of microfibril orientation in the cob mutants would reflect a degraded orienting mechanism, possibly associated with a reduced degree of polymerization. In this scenario, the oriented pattern of COB at the cell surface may reflect a mechanism for the patterned delivery of cell wall precursors rather than for direct microfibril alignment. Despite the present ambiguity about the connection between microtubule and microfibril alignments, the experiments reported here establish COB as a pivotal player in the regulation of the anisotropic expansion of higher plant cells.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown as described previously (Benfey et al., 1993). All of the mutant lines used in this work are in the Columbia background. The SAIL line 735D10 (Syngenta, San Diego, CA), which contains a T-DNA insertion in the fifth exon of the COB gene, was backcrossed twice, propagated as a heterozygote, and renamed cob-4.
GUS Reporter Construct
To generate the PCOB:GUS reporter construct, a −2509 to +45 fragment containing the entire 5′ flanking region of COB and a 1443-bp fragment of the 3′ end of COB were amplified by PCR from genomic DNA using the respective primer pairs 5′-TTACTAAAACAGCACTAGCCAGC-3′/5′-CGGATCCCGATGCCATGGACAATTTGGAGACGATGGAGGTGG-3′ and 5′-ATTTGCGGCCGCTCGGATTTACGGTTTTGCCACTGG-3′/5′-ACGGATGGAAATTGGACTGAAATGC-3′, subcloned as HindIII/BamHI and NotI fragments, respectively, and sequenced. The uidA gene was cloned in frame between the two COB genomic sequences as a NcoI/BamHI fragment. The entire construct was then excised as a SacI fragment and cloned into the plant transformation vector pBIN19. The resulting vector was transformed into the Arabidopsis accession Col-0 by the floral-dipping method (Clough and Bent, 1998). GUS staining was performed on T2 plants as described by Malamy and Benfey (1997).
Light and Scanning Electron Microscopy and Sectioning
For scanning electron microscopy, specimens were either fixed in 2.5% glutaraldehyde, dehydrated through an ethanol series, and critical point dried in liquid CO2 or cryofixed in an Oxford CT1500 cryochamber (Oxford Instruments, Oxon, UK). After sputter coating with gold, samples were visualized with an Amray FE-1850 scanning electron microscope (KLA-Tencor, San Jose, CA) operated at 5 kV. For sectioning, fixed plantlets and roots were embedded in Historesin (Technovit 7100; Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. Three-micrometer plastic sections were stained with toluidine blue (0.5% [w/v] in 0.1 M phosphate buffer, pH 7.2) to visualize the cell wall. Light micrographs were taken on a DMRXA2 microscope (Leica, Wetzlar, Germany).
Immunolocalization and Confocal Microscopy
Anti-COB polyclonal IgY antibodies were produced by GenWay Biotech (San Diego, CA) against amino acids 147 to 297. Antibodies were raised in Rhode Island Red hens, and total IgY was isolated from egg yolk by standard methods. COB-specific IgY was further purified through affinity chromatography using recombinant COB fusion protein produced in Escherichia coli as ligand. Whole-mount immunolocalizations were performed as described by Friml et al. (2003) except that incubations were done either overnight at 4°C or for 3 h at room temperature. For oryzalin and brefeldin A treatments, seedlings were incubated in 1 mL of liquid growth medium (0.5× MS medium [Sigma-Aldrich, St. Louis, MO] and 1% sucrose, pH 5.7) containing 10 μM oryzalin (100 mM stock in ethanol; Duchefa, Haarlem, The Netherlands) or 50 μM brefeldin A (100 mM stock in DMSO; Molecular Probes, Eugene, OR) and equal amounts of solvent for control. Washing was done twice for 10 min, and incubation was stopped by fixation. Antibodies used and final concentrations (v/v) were as follows: affinity-purified anti-COB IgY antibodies, 1:200; affinity-purified rabbit anti-chicken IgY Alexa Fluor 647–conjugated secondary antibodies (Genway Biotech, San Diego, CA), 1:200; anti-α-tubulin antibodies (Sigma-Aldrich), 1:1000; and fluorescein isothiocyanate–conjugated anti-mouse IgG secondary antibodies (1:200). Fluorescent images were collected on a confocal laser scanning microscope (LSM 510 system; Carl Zeiss MicroImaging, Jena, Germany). Each image shown represents either a single focal plan or a projection of individual images taken as a Z-series. Images acquired using Zeiss software were imported into Adobe Photoshop (Mountain View, CA) for cropping, contrast adjustment, and assembly.
Biochemical Fractionation, PI-PLC Sensitivity, and N-Glycosylation Test
Total membrane isolation and Triton X-114 phase separation were performed as described by Borner et al. (2003). Dextran-PEG partitioning was performed as described by Sherrier et al. (1999). For PI-PLC treatment, the washed Triton X-114 phase was split into two aliquots. PI-PLC (Sigma-Aldrich) was added to one aliquot, and both were incubated at 37°C for 1 h (with intermittent mixing). BSA was added as carrier protein to the aqueous phases. Proteins were precipitated using cold acetone and subsequently subjected to SDS-PAGE and protein gel blot analysis. Anti-cytochrome b5 and anti-ATPase antibodies were a kind gift of J. Napier (Rothamsted Research, Harpenden, UK) and M. Boutry (Université Catholique de Louvain, Louvain-La-Neuve, Belgium), respectively. To determine whether COB was N-glycosylated, roots of 1-week-old seedlings were ground and microsomes were prepared as described by Jinn et al. (2000), then they were resuspended in the denaturing buffer supplied with the peptide N-glycosidase F and treated according to the manufacturer's instructions (New England Biolabs, Beverly, MA).
Polarized-Light Microscopy and FESEM
Birefringent retardation of root cell walls was measured on 2-μm longitudinal plastic sections using a polarized-light microscope (Jenapol; Zeiss) equipped for circularly polarized-light quantitative digital imaging (LC Pol Scope; Cambridge Research Instruments, Cambridge, UK). Longitudinal cell walls were assayed that were parallel to the plane of the section in areas devoid of any visible heterogeneity. Retardance and optical azimuth were calculated from the intensity measurements by the Pol Scope software, which implements the algorithm described by Oldenbourg and Mei (1995). We did not observe a significant difference in retardance between the epidermis and the cortex layer for the three genotypes tested; data from both cell types were combined to generate the graphs. For each time point, five root longitudinal sections were examined and at least three measurements were performed per section and root zone. Samples were prepared and longitudinal walls were analyzed by FESEM as described by Himmelspach et al. (2003) using a Hitachi 4500 field emission scanning electron microscope (Tokyo, Japan). For each genotype, three roots and at least three different cells for each root zone were analyzed.
Transmission Electron Microscopy
Roots from wild-type (Col-0) seedlings grown, as described by Sugimoto et al. (2000), for 5 d under constant light (80 μmol·m−2·s−1) at 21°C were cut into 1.5-mm lengths with a razor blade while submerged in hexadecane. Cut root tissue was transferred into flat specimen holders filled with hexadecane and cryofixed in a high-pressure freezing unit (Bal-Tech HPM010; Balzers, Lichtenstein). For LR White (London Resin Company, London, UK) embedding, frozen samples were put into cryogenic vials containing freeze substitution media (0.25% glutaraldehyde and 0.1% uranyl acetate in acetone) and placed into a dry ice–acetone bath as described (Rensing et al., 2002). This bath was placed at −20°C for 7 d and warmed to 4°C for 4 h, then brought to room temperature. The tissue was rinsed with fresh acetone and infiltrated with LR White embedding resin according to the following schedule: drop by drop for 3 d; then 50, 75, and 85% (v/v) LR White:acetone, each step for 12 h; and finally 100% LR White resin for 36 h with resin exchanged every 12 h. Polymerization was achieved at 60°C for 24 h. Thin sections (<70 nm) were cut with an Ultracut E ultramicrotome (Leica) and mounted on Formvar-coated 200-mesh nickel grids.
For immunogold labeling, all incubations of sections were performed by placing the grids on droplets of solution section-side-down at room temperature. Thin sections mounted on grids were incubated for 10 min on the droplets of 10 mM NH4Cl in TBS (10 mM Tris and 0.25 M NaCl, pH 7) to block free aldehydes. Nonspecific antibody binding sites on the sections were blocked by incubating the sections for 30 min in 5% nonfat dried milk in TBS. The sections were then incubated for 1 h with chicken anti-COB antibody diluted 1:100 in 0.5% nonfat dried milk in TBS. After rinsing in TBS five times, the sections were incubated for 1 h with goat anti-chicken IgG conjugated to 10-nm colloidal gold (Aurion, Wageningen, The Netherlands) diluted 1:100 in 0.5% nonfat dried milk in TBS. The sections were then rinsed in TBS five times, in distilled water twice, and air dried. Grids were stained in 2% aqueous uranyl acetate for 30 min and in lead citrate for 5 min and then observed using a Hitachi H7600 transmission electron microscope.
Supplementary Material
Acknowledgments
We thank I. Tan (New York University) for expert technical assistance with scanning electron microscopy. Casey J. Roehrig is thanked for her skilled technical assistance. M. Pastuglia (Institut National de la Recherche Agronomique, Versailles) is thanked for the gift of ton2-14 and ton2-13 seeds. We thank H. Höfte and K. Birnbaum for critical reading of the manuscript. Polarized-light microscopy done in the Baskin laboratory was supported by a grant to T.I.B. from the U.S. Department of Energy (Award 94ER20146), which does not constitute endorsement by that department of the views expressed herein. FESEM and transmission electron microscopy done in the Wasteneys laboratory was supported by grants to G.O.W. from the Australian Research Council (DP0208872) and the Natural Sciences and Engineering Research Council of Canada (DP298264-04), respectively. This material is based on work supported by the National Science Foundation under Grant 0209754 to P.N.B.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Philip N. Benfey (philip.benfey@duke.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031732.
References
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Beemster, G.T.S., Judy-March, J.E., and Marga, F. (2004). Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135, 2279–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, T.I., Wilson, J.E., Cork, A., and Williamson, R.E. (1994). Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant Cell Physiol. 35, 935–942. [PubMed] [Google Scholar]
- Benfey, P.N., Linstead, P.J., Roberts, K., Schiefelbein, J.W., Hauser, M.T., and Aeschbacher, R.A. (1993). Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 119, 57–70. [DOI] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Borner, G.H., Lilley, K.S., Stevens, T.J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: A proteomic and genomic analysis. Plant Physiol. 132, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Stevens, T.J., Arkin, I.T., and Dupree, P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., Macaskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis: Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Camilleri, C., Azimzadeh, J., Pastuglia, M., Bellini, C., Grandjean, O., and Bouchez, D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14, 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2000). Loosening of plant cell walls by expansins. Nature 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2001). Wall structure and wall loosening: A look backwards and forwards. Plant Physiol. 125, 131–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin, M.S., Kurek, I., Jacob-Wilk, D., and Delmer, D.P. (2002). Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiol. 43, 1407–1420. [DOI] [PubMed] [Google Scholar]
- Elortza, F., Nuhse, T.S., Foster, L.J., Stensballe, A., Peck, S.C., and Jensen, O.N. (2003). Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics 2, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Friedrichson, T., and Kurzchalia, T.V. (1998). Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394, 802–805. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Mayer, U., Palme, K., and Muster, G. (2003). Automated whole mount localisation techniques for plant seedlings. Plant J. 34, 115–124. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J.C., Taylor, N.G., and Turner, S.R. (2003). Control of cellulose synthase complex localization in developing xylem. Plant Cell 15, 1740–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Höfte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (San Diego: Academic Press), pp. 85–99.
- Green, P.B. (1962). Mechanism for plant cellular morphogenesis. Science 138, 1404–1405. [DOI] [PubMed] [Google Scholar]
- Green, P.B. (1980). Organogenesis: A biophysical view. Annu. Rev. Plant Physiol. 31, 51–82. [Google Scholar]
- Hauser, M.T., Morikami, A., and Benfey, P.N. (1995). Conditional root expansion mutants of Arabidopsis. Development 121, 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P.K., and Newcomb, E.H. (1964). Microtubules and fibrils in the cytoplasm of coleus cells undergoing secondary wall deposition. J. Cell Biol. 20, 529–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelspach, R., Williamson, R.E., and Wasteneys, G.O. (2003). Cellulose microfibril alignment recovers from DCB-induced disruption despite microtubule disorganization. Plant J. 36, 565–575. [DOI] [PubMed] [Google Scholar]
- His, I., Driouich, A., Nicol, F., Jauneau, A., and Höfte, H. (2001). Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-beta-glucanase. Planta 212, 348–358. [DOI] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., and Walker, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. [PMC free article] [PubMed] [Google Scholar]
- Lalanne, E., Honys, D., Johnson, A., Borner, G.H., Lilley, K.S., Dupree, P., Grossniklaus, U., and Twell, D. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, D.R., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter, M.C., and Porter, K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J. 12, 957–963. [DOI] [PubMed] [Google Scholar]
- Muniz, M., and Riezman, H. (2000). Intracellular transport of GPI-anchored proteins. EMBO J. 19, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Höfte, H. (1998). A plasma membrane-bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenbourg, R., and Mei, G. (1995). New polarized light microscope with precision universal compensator. J. Microsc. 180, 140–147. [DOI] [PubMed] [Google Scholar]
- Peng, L., Hocart, C.H., Redmond, J.W., and Williamson, R.E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. [DOI] [PubMed] [Google Scholar]
- Preston, R.D. (1974). The Physiological Biology of Plant Cell Walls. (London: Chapman and Hall), pp. 75–108.
- Refrégier, G., Pelletier, S., Jaillard, D., and Höfte, H. (2004). Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 135, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing, K.H., Samuels, A.L., and Savidge, R.A. (2002). Ultrastructure of vascular cambial cell cytokinesis in pine seedlings preserved by cryofixation and substitution. Protoplasma 220, 39–49. [DOI] [PubMed] [Google Scholar]
- Roberts, K. (1994). The plant extracellular matrix in a new expansive mood. Curr. Opin. Cell Biol. 6, 688–694. [DOI] [PubMed] [Google Scholar]
- Roudier, F., Schindelman, G., DeSalle, R., and Benfey, P.N. (2002). The COBRA family of putative GPI-anchored proteins in Arabidopsis: A new fellowship in expansion. Plant Physiol. 130, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R., Fry, B., Kochevenko, A., Schindelasch, D., Zimmerli, L., Somerville, S., Loria, R., and Somerville, C.R. (2003). An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell 15, 1781–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Himmelspach, R., Williamson, R.E., and Wasteneys, G.O. (2003). Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15, 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2001). Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: Microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215, 172–183. [DOI] [PubMed] [Google Scholar]
- Taiz, L. (1984). Plant cell expansion: Regulation of cell wall mechanical properties. Annu. Rev. Plant Physiol. 35, 585–657. [Google Scholar]
- van der Weele, C.M., Jiang, H.S., Palaniappan, K.K., Ivanov, V.B., Palaniappan, K., and Baskin, T.I. (2003). A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth: Roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol. 132, 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, R., and Mayor, S. (1998). GPI-anchored proteins are organized in submicron domains at the cell surface. Nature 394, 798–801. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2004). Progress in understanding the role of microtubules in plant cells. Curr. Opin. Plant Biol. 7, 651–660. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Wiedemeier, A.M., Judy-March, J.E., Hocart, C.H., Wasteneys, G.O., Williamson, R.E., and Baskin, T.I. (2002). Mutant alleles of Arabidopsis RADIALLY SWOLLEN 4 and 7 reduce growth anisotropy without altering the transverse orientation of cortical microtubules or cellulose microfibrils. Development 129, 4821–4830. [DOI] [PubMed] [Google Scholar]
- Williamson, R.E., Burn, J.E., Birch, R., Baskin, T.I., Arioli, T., Betzner, A.S., and Cork, A. (2001). Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215, 116–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.