Abstract
SE-WAP41, a salt-extractable 41-kD wall-associated protein that is associated with walls of etiolated maize (Zea mays) seedlings and is recognized by an antiserum previously reported to label plasmodesmata and the Golgi, was cloned, sequenced, and found to be a class 1 reversibly glycosylated polypeptide (C1RGP). Protein gel blot analysis of cell fractions with an antiserum against recombinant SE-WAP41 showed it to be enriched in the wall fraction. RNA gel blot analysis along the mesocotyl developmental axis and during deetiolation demonstrates that high SE-WAP41 transcript levels correlate spatially and temporally with primary and secondary plasmodesmata (Pd) formation. All four of the Arabidopsis thaliana C1RGP proteins, when fused to green fluorescent protein (GFP) and transiently expressed in tobacco (Nicotiana tabacum) epidermal cells, display fluorescence patterns indicating they are Golgi- and plasmodesmal-associated proteins. Localization to the Golgi apparatus was verified by colocalization of transiently expressed AtRGP2 fused to cyan fluorescence protein together with a known Golgi marker, Golgi Nucleotide Sugar Transporter 1 fused to yellow fluorescent protein (GONST1:YFP). In transgenic tobacco, AtRGP2:GFP fluorescence is punctate, is present only in contact walls between cells, and colocalizes with aniline blue–stained callose present around Pd. In plasmolyzed cells, AtRGP2:GFP remains wall embedded, whereas GONST1:YFP cannot be found embedded in cell walls. This result implies that the targeting to Pd is not due to a default pathway for Golgi-localized fusion proteins but is specific to C1RGPs. Treatment with the Golgi disrupting drug Brefeldin A inhibits Pd labeling by AtRGP2:GFP. Integrating these data, we conclude that C1RGPs are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus.
INTRODUCTION
Plasmodesmata (Pd) (plasmodesma singular) are specialized membranous tunnels that interconnect neighboring plant cells and facilitate direct cytoplasmic cell-to-cell transport through cell walls. Transport through Pd mediates many processes, among them, information transfer to allow the coordination of development, the movement of photosynthesis products from mature to developing tissues, responses to pathogen attacks, systemic gene silencing, etc. (reviewed in Ding et al., 1999; Zambryski and Crawford, 2000; Haywood et al., 2002; Heinlein and Epel, 2004). In addition to their normal function, Pd are exploited by viruses and viroids for cell-to-cell movement and the long-distance spread of infection (reviewed in Ding et al., 1999; Beachy and Heinlein, 2000; Citovsky and Zambryski, 2000; Heinlein and Epel, 2004).
In higher plants, Pd may be monocoaxial tunnels, termed simple Pd, or they may be branched, forming a network of interconnected coaxial tunnels (Ehlers and Kollmann, 2001). In young (sink) leaves, most Pd are simple, whereas in mature (source) leaves, there are both simple and branched Pd (Oparka et al., 1999). The outer boundary of a Pd is delineated by a membrane that is continuous with the plasma membranes (PMs) of the two adjoining cells. Concentric with the outer PM is a second membrane termed desmotubule that is derived from and continuous with the endoplasmic reticulum membranes of the two cells. Between the concentric membranes is a sleeve that interconnects the cytoplasm of the neighboring cells. High-resolution electron micrographs reveal that within this sleeve are particles that have been variously interpreted as being embedded in the PM (Ding et al., 1992) and/or in the desmotubule (Botha et al., 1993) or to be present entirely within the cytoplasmic sleeve (Overall, 1999).
Contrary to the structural analysis, knowledge regarding the biochemical composition of Pd is still fragmentary, largely because of the difficulty in isolating pure Pd without subcellular membrane contaminants. Previously, we reported on the development of a procedure for isolating cell walls containing embedded Pd (Epel et al., 1996). Wall-associated proteins (WAPs) extracted from a wall fraction prepared from the mesocotyls of etiolated maize (Zea mays) seedlings were separated by SDS-PAGE, and an antiserum was prepared against a band of ∼41 kD that was highly enriched in this fraction. This antiserum, termed S-41, immunolocalized mainly to Pd and the Golgi apparatus (Epel et al., 1996). The 41-kD protein was shown to be released from the wall fraction by 3 M NaCl or by pH 11, but not by 2% Triton X-100, indicating that this protein is a peripheral membrane protein (Epel et al., 1996).
Here, we report the cloning and sequencing of the gene that encodes for the salt-extractable 41-kD protein termed SE-WAP41 that is recognized by S-41. SE-WAP41 is a member of the reversibly glycosylated polypeptides (RGPs) protein family, a family of highly conserved plant-specific proteins, which self-glycosylate in the presence of certain nucleotide sugars (Dhugga et al., 1991, 1997; Delgado et al., 1998). RGP proteins have been subdivided into two classes originally termed RGP1 and RGP2 (Langeveld et al., 2002). We will refer to RGP1 and RGP2 classes as C1RGPs and C2RGPs accordingly. SE-WAP41 is a member of C1RGPs. We show here that members of C1RGPs from Arabidopsis thaliana, when expressed as green fluorescent protein (GFP) fusions, target to the Golgi apparatus and become incorporated into Pd.
RESULTS
SE-WAP41 Is an RGP Protein
Previously, we demonstrated that S-41 immunolabeled Pd and Golgi (Epel et al., 1996). Because this antiserum was generated against a protein band from an SDS-PAGE gel, it may recognize more than one protein that could be present in the band. Furthermore, this antiserum cross-reacted with two additional proteins. A combination of experimental methods was used to molecularly characterize the protein or proteins recognized by S-41, which targets to Pd. In one approach, we employed S-41 to screen an expression library generated using mRNA obtained from meristematic tissues of 5-d-old maize seedlings. Screening ∼4.3 × 105 plaque forming units yielded 40 clones recognized by S-41. Hybridization experiments revealed that the isolated clones represent two hybridization groups, termed H1 and H2, containing 19 and 21 clones, respectively, indicating that S-41 identified two distinct proteins from the expression library.
In parallel, purified SE-WAP41, a salt-extractable 41-kD WAP, that was recognized by S-41 (Epel et al., 1996) was microsequenced, yielding a 25–amino acid sequence: NLDFLGM?RPFFQPYHLIIVQDGDP. Based on this sequence, a degenerative primer was synthesized to allow identification of isolated clones encoding the SE-WAP41 internal sequence.
The degenerative primer was used in combination with a reverse primer from the library's vector sequence in a PCR reaction to identify clones previously isolated employing the S-41 antiserum and that encode a protein that contains the sequenced peptide. Seven clones were identified, all belonging to the H1 hybridization group, establishing that this hybridization group encodes the SE-WAP41 protein.
The insert lengths of the various clones belonging to H1 were determined by an additional PCR reaction with primers complementary to vector sequences on both sides of the insert, and the clone that had the longest insert was sequenced. The open reading frame of the insert encodes a protein containing 364 amino acids with a calculated mass of 41,038 D, matching the size of the 41-kD WAP. The predicted gene product contained the internal sequence previously obtained by SE-WAP41 microsequencing, confirming that the clone encoded SE-WAP41. The sequence of the maize gene encoding SE-WAP41 protein was deposited in the EMBL and National Center for Biotechnology Information data libraries (accession number U89897). High sequence homology of SE-WAP41 to members of the RGP protein family (pfam 03214) resulted in SE-WAP41 annotation as an RGP protein.
A representative of the H2 hybridization group was sequenced and found to encode an aldolase protein (data not shown). It remains to be seen whether this aldolase protein is Pd associated or merely unspecifically identified by the S-41 antiserum. Work with clones in the H2 group was not further pursued.
Antiserum Raised against Recombinant SE-WAP41 Uniquely Identifies a 41-kD Protein That Is Highly Enriched in the Wall Fraction
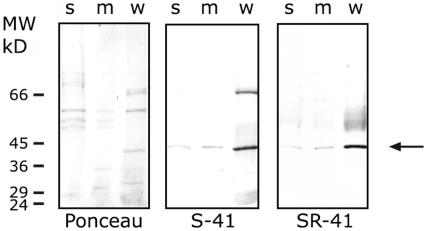
SE-WAP41 expressed in Escherichia coli as a His tag fusion was used to immunize rabbits. The antiserum obtained from one rabbit, termed SR-41, exhibited good immunogenic activity with the purified recombinant protein in protein gel blots. Protein gel blot analysis with SR-41 of proteins from the soluble, the membranal, and the wall fractions demonstrated that SE-WAP41 is highly enriched in the wall fraction that contains embedded Pd, compared with the membranal and soluble fractions (Figure 1). The membrane and soluble fractions are more weakly labeled, suggesting lower levels of SE-WAP41 association with these fractions. This protein gel blot analysis also verified that SR-41 antiserum recognizes the 41-kD protein, without recognizing the other major bands labeled by S-41 (Figure 1). Additionally, SR-41 did not recognize aldolase (data not shown).
Figure 1.
Protein Gel Blot Analysis Shows SE-WAP41 Enrichment in Wall Fraction.
Proteins extracted from the soluble (s), membranal (m), and wall (w) fractions were separated by SDS-PAGE (6 μg/lane), electroblotted to nitrocellulose, and stained with Ponceau (left), reacted with S-41 antiserum (center), or with SR-41 antiserum (right).
SE-WAP41 mRNA Expression Levels Are Highest in Meristematic and Elongating Tissues
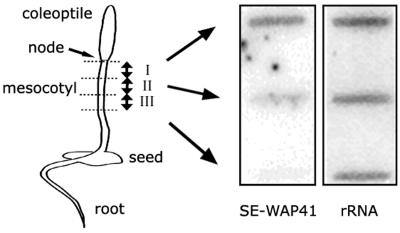
The formation of both simple primary and secondary Pd and their modification to branched Pd occurs primarily in meristematic tissues and in expanding and/or elongating tissues (Ehlers and Kollmann, 2001; Roberts and Oparka, 2003). If SE-WAP41 is a Pd-associated protein, it is expected that in etiolated maize seedlings its expression should be highest in the mesocotyl node, which is a meristem tissue, and in a 1- to 2-cm region just below the node, a region in which cells are undergoing extensive elongation. SE-WAP41 mRNA expression levels were compared in segments, each 1 cm in length, along the mesocotyl developmental gradient. RNA gel blot analysis using SE-WAP41 cDNA probe showed that SE-WAP41 transcript level was highest in the region containing the mesocotyl node and the most extensively expanding cells and decreased along the mesocotyl developmental gradient (Figure 2).
Figure 2.
RNA Gel Slot Blot Analysis of SE-WAP41 mRNA from Mesocotyl Sections of 5-d-Old Etiolated Maize Seedlings.
RNA samples were prepared from upper mesocotyl segments: section I contains mesocotyl meristem and region of most rapid cell growth; section II also contains zone of elongation; section III contains mature nondividing and nonelongating cells. The ribosomal 26S rRNA probe served as a loading control. Left, 5-d-old etiolated maize seedling; right, RNA slot blot.
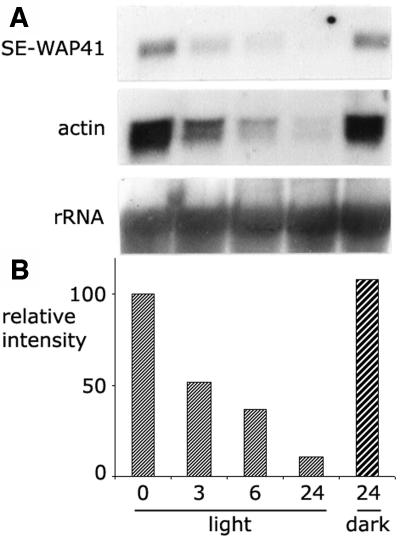
The correlation between cell division and mRNA expression level was further examined in a second experimental system. Exposure of etiolated seedlings to light brings about a cessation of cell divisions in the mesocotyl meristem (Iino, 1982; Yahalom et al., 1987). RNA gel blot analysis was used to examine the effect of light on SE-WAP41 mRNA expression levels in mesocotyls of etiolated maize seedlings. After exposure to light, SE-WAP41 mRNA levels decreased 2-fold after 3 h, 3.5-fold after 6 h, and 10-fold after 24 h (Figure 3). Actin mRNA levels displayed a similar decrease (Figure 3) indicative of cell division cessation.
Figure 3.
RNA Gel Blot Analysis Shows That SE-WAP41 mRNA Levels Decrease When Etiolated Maize Seedlings Are Exposed to Light.
(A) Five-day-old etiolated maize seedlings were exposed to white light for 3, 6, and 24 h or kept in the dark for 24 h. A blot of total RNA extracted from the upper 1 cm of mesocotyls was hybridized to cDNAs of SE-WAP41, actin, and 26S-rRNA. The ribosomal 26S rRNA probe served as a loading control. The decreased actin mRNA levels is indicative of cell division cessation.
(B) Relative SE-WAP41 mRNA levels. Values were calculated relative to 26S-rRNA intensity in each lane.
Transiently Expressed AtRGP:GFPs Associate with Pd and Golgi Vesicles
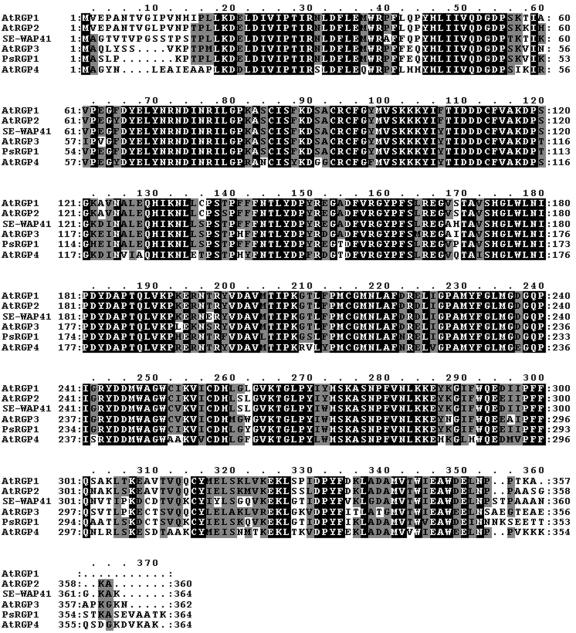
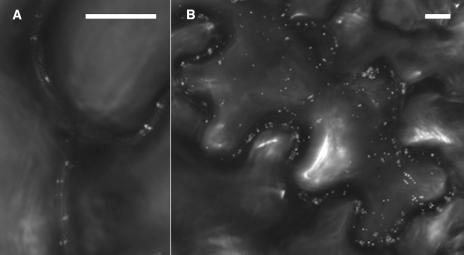
Further study was done with genes of Arabidopsis because its genome has been fully sequenced and open reading frames annotated. The Arabidopsis genome encodes four homologous genes termed AtRGP1, AtRGP2, AtRGP3, and AtRGP4, which belong to C1RGPs. These Arabidopsis proteins show high degrees of amino acid sequence identity with SE-WAP41 (Figure 4). Amino acid sequence analysis (http://www.ncbi.nlm.nih.gov/BLAST/) revealed that AtRGP2 is the closest homolog of SE-WAP41, sharing 87% amino acid sequence identity. AtRGP2 was fused to GFP to examine its intracellular localization. AtRGP2:GFP was transiently expressed in tobacco (Nicotiana tabacum) leaf epidermal cells using Agrobacterium tumefaciens leaf injection. Confocal microscopy revealed fluorescence in several distinct sites inside epidermal cells. Punctate paired fluorescent foci were detected on opposite sides of the cell wall—a pattern consistent with AtRGP2 being a plasmodesmal-associated protein (Figures 5A and 5B). Fluorescence was also observed as small fluorescence foci throughout the cell cytoplasm (Figure 5B), which moved rapidly throughout the cytoplasm and along cytoplasmic strands through the vacuole, in a manner characteristic of Golgi vesicles (see Supplemental Video 1 online). Fluorescence could also be seen in large amorphous areas inside cells (data not shown), apparently representing inclusion bodies resulting from the massive quantities of AtRGP2:GFP produced inside the cells during transient expression. Fluorescence was also detected in small areas inside cell nuclei (data not shown).
Figure 4.
SE-WAP41, AtRGP1, AtRGP2, AtRGP3, AtRGP4, and PsRGP1 Share High Amino Acid Sequence Identity.
Alignment was made with ClustalX (Thompson et al., 1997). Black boxes denote identical amino acids; gray boxes denote similar amino acids.
Figure 5.
Localization of Transiently Expressed AtRGP2:GFP in Tobacco Epidermal Cells 48 h after Infection by Agrobacterium.
(A) Punctate fluorescence can be seen along cell walls as paired fluorescence foci spanning common wall of adjacent cells.
(B) A projection of several sections showing AtRGP2:GFP both in fluorescent bodies within cell cytoplasm representing Golgi vesicles and in paired fluorescence foci spanning cell walls.
Bars = 10 μm.
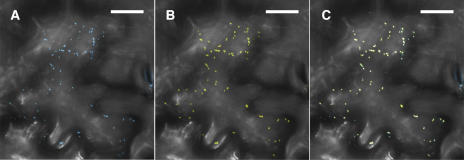
Our supposition that the AtRGP2:GFP mobile foci represent Golgi bodies is supported by the report by Dhugga et al. (1997), in which it was shown that PsRGP1, a pea (Pisum sativum) C1RGP that shares high sequence identity with SE-WAP41 and AtRGP proteins (Figure 4), immunolocalizes to the trans-Golgi apparatus. To verify that the mobile foci are indeed Golgi bodies, Golgi Nucleotide Sugar Transporter 1 fused to yellow fluorescent protein (GONST1:YFP), which has been shown to label Golgi bodies (Baldwin et al., 2001), was employed. AtRGP2 when fused to cyan fluorescent protein (AtRGP2:CFP) and coexpressed with GONST1:YFP clearly colocalize with GONST1:YFP (Figure 6).
Figure 6.
AtRGP2:CFP Colocalizes with the Golgi Apparatus Marker GONST1:YFP.
Colocalization is shown in a projection of five optical sections through a tobacco leaf epidermal cell transiently coexpressing AtRGP2:CFP and GONST1:YFP. AtRGP2:CFP fluorescence is shown in (A), GONST1:YFP fluorescence is shown in (B), and both are shown overlaid in (C). Optical sections were taken through the upper cortical cytoplasm near the cuticle where there are no Pd. Fluorescence seen in stomatal guard cells at the right edge of the photograph is autofluorescence induced by the 458-nm irradiation. Bars = 20 μm.
To examine whether plasmodesmal association is a general characteristic of C1RGPs, AtRGP1:GFP, AtRGP3:GFP, and AtRGP4:GFP were constructed and transiently expressed in tobacco leaf epidermal cells. These fusion proteins displayed fluorescence patterns identical to that presented by AtRGP2:GFP in all respects (data not shown), except that AtRGP1:GFP fluorescence levels were lower. These fluorescence patterns are consistent with AtRGP1, AtRGP3, and AtRGP4 being plasmodesmal-associated proteins. The observation that all four Arabidopsis C1RGPs appear to be plasmodesmal associated indicates that plasmodesmal association is a general characteristic of members of C1RGPs.
Stably Expressed AtRGP2:GFP Is Plasmodesmal Associated
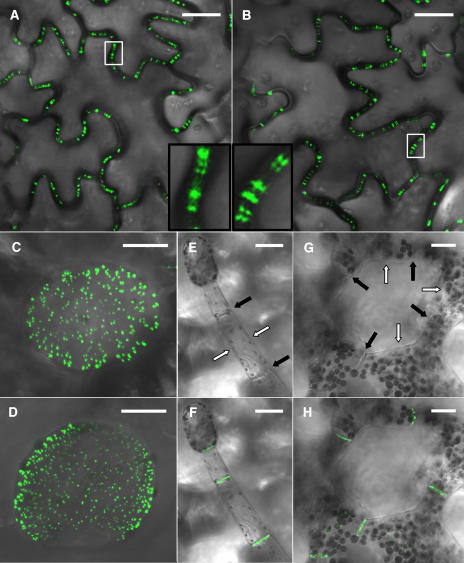
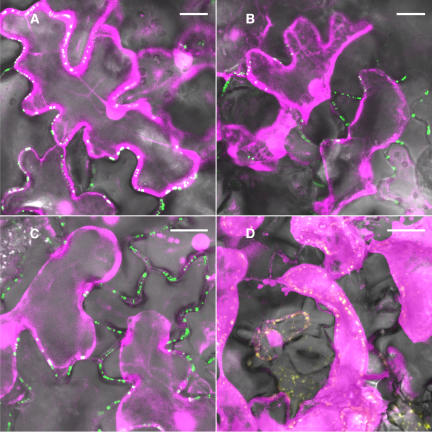
Transgenic tobacco plants constitutively expressing the AtRGP2:GFP fusion under the control of the 35S promoter of the Cauliflower mosaic virus were generated. In these plants, AtRGP2:GFP fluorescence was localized in punctate fluorescence foci inside cell walls (Figure 7A). This pattern is similar to that presented by transgenic tobacco plants constitutively expressing a GFP fusion of a known Pd marker, the movement protein of Tobacco mosaic virus (MPTMV:GFP) (Figure 7B). The cell wall at the base of a trichome is especially rich in Pd. Transgenic MPTMV:GFP expressing tobacco plants exhibit a characteristic dense punctate pattern where these Pd are fluorescently marked (Figure 7C). The same pattern is presented by the transgenic AtRGP2:GFP expressing plants (Figure 7D), supporting the conclusion that AtRGP2 localizes to Pd.
Figure 7.
Stable Expression of AtRGP2:GFP and of MPTMV:GFP in Transgenic Tobacco Plants.
Identical fluorescence patterns are presented by leaf epidermal cells in AtRGP2:GFP (A) and MPTMV:GFP (B) expressing transgenic plants: punctate fluorescence spans walls as elongated bars, or paired foci, or appears as single foci inside the cell wall. The areas inside the white boxes are enlarged in the insets in (A) and (B). The trichome-epidermis interface in AtRGP2:GFP (D) and MPTMV:GFP (C) expressing plants displays identical fluorescence patterns. In trichome (F) and spongy mesophyll cells (H) of AtRGP2:GFP expressing transgenic tobacco, fluorescence is detected only in wall areas where there is cell–cell contact and is absent from wall areas without cell–cell contact. To emphasize wall partitions, the same trichome (E) and spongy mesophyll cells (G) are shown with the fluorescence channel turned off. Black arrows indicate wall areas where there is cell–cell contact, and white arrows indicate wall areas without cell–cell contact. Bars = 20 μm.
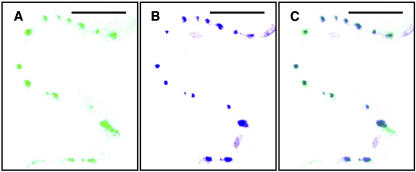
AtRGP2:GFP fluorescence was seen only in wall areas that contain Pd, where two cells are in contact, whereas wall areas devoid of Pd were unlabeled, as demonstrated by trichome cells and spongy mesophyll cells. Tobacco trichomes are hair-like structures, protruding from the epidermis surface, consisting of a single file of several cells (Figure 7E). Therefore, tobacco trichome cells have both Pd containing walls, those walls separating two cells, and lateral walls that are only in contact with the air surrounding the trichome, which are devoid of Pd. Spongy mesophyll cells (Figure 7G) likewise have regions with no cell–cell contact, where walls face an intercellular space and are devoid of Pd, and regions with cell–cell contact, where walls contain Pd. Fluorescence was detected only within cell wall regions where there is cell–cell contact, in both trichome cells (Figure 7F), and spongy mesophyll cells (Figure 7H). This differentiating labeling of walls further indicates that AtRGP2 is associated with Pd or with the wall sheath surrounding Pd. To verify that the punctate fluorescence foci inside cell walls indeed represent Pd, we employed callose staining by aniline blue. Callose has been widely used as a plasmodesmal marker (Baluska et al., 1999; Gorshkova et al., 2003; Bayer et al., 2004; and others). When leaves of AtRGP2:GFP expressing transgenic tobacco were stained by aniline blue, AtRGP2:GFP clearly colocalized with the aniline blue–stained callose (Figure 8) present around Pd. Wild-type tobacco, not expressing GFP, when stained by aniline blue and photographed with GFP conditions displayed no fluorescently labeled Pd (data not shown). Likewise, transgenic AtRGP2:GFP expressing transgenic tobacco not stained by aniline blue but photographed with aniline blue conditions also displayed no fluorescently labeled Pd (data not shown). These negative control experiments verify that colocalization is not the result of fluorescence bleedthrough between GFP and aniline blue channels.
Figure 8.
AtRGP2:GFP in Transgenic Tobacco Colocalizes with Aniline Blue–Stained Callose Present around Pd.
AtRGP2:GFP is shown in green (A), aniline blue–stained callose is shown in blue (B), and both are shown overlaid (C) in a section of cell wall between epidermal cells. Bars = 20 μm.
In mature tobacco tissue, AtRGP2:GFP fluorescence was not detected in mobile bodies possibly because of the lower steady state levels present during stable expression.
AtRGP2:GFP Remains inside Cell Walls of Plasmolyzed Cells
Plasmolysis of cells in leaves causes shrinkage of the cell protoplast and retraction of the PM from the wall, with Pd remaining embedded in the cell wall (Turner et al., 1994). Plasmolysis was induced in leaves of the transgenic AtRGP2:GFP expressing tobacco plants to learn about the association of AtRGP2 with Pd. The fluorescent dye DsRed1, a cytoplasmic marker, was used to allow better visualization of the receding cytoplasm and PM during plasmolysis. DsRed1 was transiently expressed in leaf epidermal cells of transgenic tobacco plants stably expressing AtRGP2:GFP, before they were plasmolyzed, thus marking the cytoplasm of these cells (Figure 9A). After plasmolysis, AtRGP2:GFP fluorescence clearly remained inside cell walls, in regions where the cytoplasm became disassociated from both sides of the cell wall (Figure 9B). This result demonstrates that the punctate sites are not within the cortical cytoplasm nor PM associated, but are embedded in the cell wall. This is similar to the result obtained in plasmolyzed transgenic tobacco plants stably expressing MPTMV:GFP (Figure 9C). To examine if association with Pd occurs for any stably expressed Golgi-localized GFP fusion protein, through some default pathway, GONST1:YFP expressing transgenic tobacco plants were generated. When these plants were plasmolyzed, no fluorescence remained in cell walls (Figure 9D).
Figure 9.
Both AtRGP2:GFP and MPTMV:GFP but Not GONST1:YFP Remain inside Cell Walls in Plasmolyzed Cells.
The cytoplasmic marker DsRed1 (shown in magenta) was transiently expressed in leaf epidermal cells ∼72 h before plasmolysis. The DsRed1-labeled cytoplasm can be seen to be appressed to cell walls before plasmolysis in AtRGP2:GFP expressing transgenic plants (A) and to retract from walls of plasmolyzed cells in both AtRGP2:GFP expressing transgenic plants (B) and MPTMV:GFP expressing transgenic plants (C). Both GFP fusion proteins (shown in green) remain inside cell walls in areas where the protoplast retracts from junction walls in both AtRGP2:GFP (B) and MPTMV:GFP (C) transgenic plants. By contrast, in GONST1:YFP expressing transgenic plants, GONST1:YFP (shown in yellow) does not remain inside cell walls in areas where the protoplast retracts from junction walls (D). Bars = 20 μm.
Brefeldin A Inhibits Labeling of Pd by AtRGP2:GFP
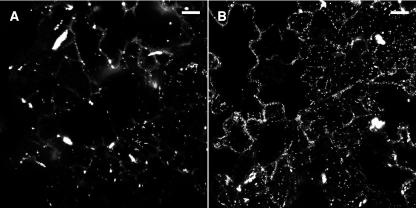
The presence of AtRGP GFP fusions in both Pd and the Golgi apparatus led us to hypothesize that C1RGPs are delivered to Pd via the Golgi apparatus. Brefeldin A (BFA), a disruptor of the Golgi apparatus (reviewed in Nebenfuhr et al., 2002), was used to examine this hypothesis. Epidermal cells of tobacco leaves treated with BFA during transient expression of AtRGP2:GFP were compared with cells not treated with BFA. BFA-treated cells displayed a decrease in the numbers of fluorescently labeled Golgi bodies (Figure 10). Fluorescently labeled foci, containing labeled Pd or clusters of Pd in pit fields that are too close together to resolve individually, were counted in five consecutive confocal sections representing the middle layer of each cell. On average, BFA-treated cells presented 24.4 ± 2.95 fluorescently labeled Pd, whereas mock-treated cells presented 77.3 ± 10.85 fluorescently labeled Pd. Thus, BFA-treated cells displayed a decrease of approximately threefold in the average number of fluorescently labeled Pd when compared with cells not treated with BFA. A paired-sample t test showed results to be significantly different at P < 0.001. The fact that the number of AtRGP2:GFP-labeled Pd was lower in the presence of the Golgi-disrupting BFA, combined with the observation that GFP fusions of AtRGPs label the Golgi apparatus, demonstrates that C1RGPs pass through the Golgi apparatus before their arrival at Pd.
Figure 10.
BFA Reduces AtRGP2:GFP Labeling of Both Golgi and Pd.
Projections of 10 optical sections through the abaxial side of tobacco leaf epidermal cells treated with BFA (A) or mock treated (B). Bars = 20 μm.
DISCUSSION
C1RGPs Are Plasmodesmal-Associated Proteins
In an earlier study, we reported on the isolation of a 41-kD protein from mesocotyl walls of etiolated maize seedlings that we proposed was a Pd-associated protein (Epel et al., 1996). The molecular identity of the plasmodesmal-associated protein, however, was not determined. In this study, we have cloned and sequenced the gene encoding this protein and have identified it as belonging to the RGP protein family pfam 03214 (the Conserved Domain Architecture Retrieval Tool). The RGP pfam 03214 is a family of highly conserved plant-specific proteins that are present in both monocotyledons (Gupta et al., 2000; Langeveld et al., 2002) and dicotyledons (Dhugga et al., 1997; Delgado et al., 1998; Bocca et al., 1999; Zhao and Liu, 2002). Members of this family are reversibly self-glycosylated in the presence of UDP-glucose, UDP-xylose, and UDP-galactose (Dhugga et al., 1991, 1997; Delgado et al., 1998); glycosylation occurs on an Arg residue (Singh et al., 1995).
Hydrophobic cluster analysis suggested that RGPs may be β-glycosyltransferases (Saxena and Brown, 1999). RGPs have been subdivided into two classes, termed here C1RGP and C2RGP. Members of C2RGP have been suggested to be similar to processive β-glycosyltransferases, which contain two conserved domains, designated domains A and B (Langeveld et al., 2002). Based on structure analysis, members of C1RGPs, to which SE-WAP41 belongs, have been suggested to be nonprocessive β-glycosyltransferases because they possess domain A but lack domain B characteristics of processive β-glycosyltransferases (Saxena and Brown, 1999). Attempts to show that RGPs function as β-glycotransferases, however, have been unsuccessful. Recent data suggest that members of this family, specifically members of C1RGPs, may function as UDP-sugar carriers, leading to the hypothesis that they function in delivering UDP-sugars from the cytoplasm to a transporter in the Golgi membrane (Faik et al., 2000; Porchia et al., 2002), where transmembrane β-glycosyltransferases function in transporting the UDP-sugars into the Golgi lumen. However, the possibility that C1RGPs may be transported by the Golgi to the Pd and become incorporated into the Pd was not taken into account.
Several lines of evidence presented here support the hypothesis that C1RGPs are plasmodesmal-associated proteins. Cell fractionation experiments showed SE-WAP41 enrichment in the Pd-containing cell wall fraction (Figure 1). Confocal microscopy reveals similar fluorescence patterns in tobacco cells expressing either AtRGP GFP fusions (Figures 5A, 5B, 7A, and 7D) or the known Pd marker MPTMV:GFP (Figures 7B and 7C). Moreover, AtRGP2:GFP fluorescence remained within the wall during plasmolysis (Figure 9B), was present only inside Pd containing cell walls, and was absent from walls devoid of Pd (Figures 7F and 7H). Colocalization of aniline blue–stained callose present around Pd with AtRGP2:GFP fluorescence foci inside cell walls (Figure 8) demonstrates that these AtRGP2:GFP foci represent Pd. The fact that GONST1:YFP, another Golgi-localized fluorescent fusion protein, does not associate with Pd when stably expressed in transgenic tobacco, as demonstrated by plasmolysis (Figure 9D), indicates that association with Pd is specific to C1RGPs and does not occur by some default pathway shared by any Golgi-localized fluorescent fusion protein. The lack of GONST1:YFP localization to Pd may be indicative of the existence of a sorting or retrieval mechanism for differential targeting to or accumulation in different compartments, the nature of which is, as yet, unclear.
Because Pd synthesis occurs mainly in meristematic and growing tissues, we would expect that C1RGP synthesis and association with Golgi would be highest in these regions. RNA gel blot analysis showed such a correlation, with higher SE-WAP41 mRNA levels measured in regions undergoing cell division and elongation (Figures 2 and 3).
The fact that all Arabidopsis C1RGP proteins appear to be plasmodesmal associated suggests that association with Pd may be a general characteristic of C1RGPs.
C1RGPs Are Transported to Pd via the Golgi Apparatus
Varied evidence suggests that the Golgi apparatus serves as the vehicle by which C1RGPs are delivered to Pd. PsRGP1 was previously immunolocalized to the trans-Golgi (Dhugga et al., 1997). Arabidopsis cell suspension fractionation data indicated that AtRGP1 is localized to the Golgi apparatus (Delgado et al., 1998). During AtRGP2:GFP transient expression, the fluorescent fusion protein is associated with mobile bodies, which are streaming within the cytoplasm (Figure 5B; see Supplemental Video 1 online). These mobile bodies were shown to be Golgi vesicles by the colocalization of AtRGP2:CFP with the known Golgi marker GONST1:YFP (Figure 6). The fact that the Golgi disrupting drug BFA greatly reduces the number of AtRGP2:GFP foci seen throughout the cytosol during transient expression (Figure 10) further supports the conclusion that these foci represent Golgi vesicles. Most importantly, labeling of Pd by transiently expressed AtRGP2:GFP was reduced by approximately threefold in the presence of the Golgi disruptor BFA, demonstrating that passage through the Golgi apparatus is required for AtGRP2:GFP targeting to Pd.
The role we propose for the Golgi apparatus as the vehicle for C1RGPs delivery to Pd is not surprising. It is known that Golgi vesicles participate in the formation of both primary and secondary Pd. During cell division, vesicles arriving from the Golgi apparatus fuse with each other to build primary Pd through the forming cell wall (Jones, 1976; Hepler, 1982; Robards and Lucas, 1990; Ehlers and Kollmann, 2001). Golgi vesicles are also actively involved in the formation of branched and secondary Pd (Ehlers and Kollmann, 2001).
The antiserum used to immunolocalize PsRGP1, unlike antiserum S-41, was reported to label only Golgi bodies but not cell walls (Dhugga et al., 1997). Because, at that time, Pd were not considered as potential RGP targets, a detailed examination of Pd for immunolabeling was not performed (K. Dhugga, personal communication). Thus, based on available information, one cannot exclude the possibility that PsRGP1, unlike C1RGPs studied by us, is not a plasmodesmal-associated protein.
Whereas fluorescent Golgi were visible during AtRGP2:GFP transient expression, in leaf cells of transgenic AtRGP2:GFP tobacco plants, they were not detected. The marked difference between the levels of AtRGP2:GFP in the Golgi and in Pd are probably due to permanent accumulation of AtRGP2:GFP in Pd, whereas in the Golgi, the protein is in transit, being shuttled by the Golgi network from the site of synthesis to its final destination in Pd. During transient expression, the cells ectopically produce very large quantities of AtRGP2:GFP in a very short time, enabling it's detection as fluorescence associated with the Golgi apparatus (Figure 5B).
Protein gel blots of different cell fractions detected C1RGPs not only in membranes and wall fractions but also in the soluble fraction (Figure 1) (Dhugga et al., 1997; Delgado et al., 1998). Immunogold electron microscopy, however, detects almost no cytosolic RGP other than that associated with Golgi (Epel et al., 1996; Dhugga et al., 1997). This, as well as the fact that high salt concentrations release SE-WAP41, led us to agree with the conclusion of Dhugga et al. (1997) that most RGP present in the cytosolic fraction was the result of RGP release from Golgi membranes or from Pd (Epel et al., 1996) during tissue homogenization and subcellular fractionation.
C1RGPs Position inside Pd: Topology and Function Intertwined
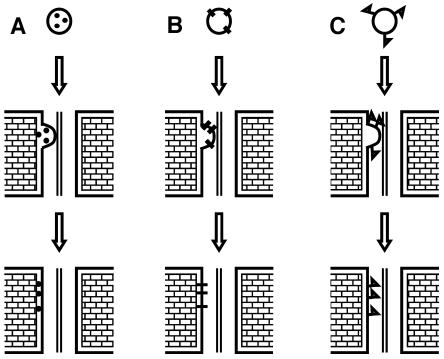
Golgi-associated proteins can either reside in the Golgi lumen, have a transmembrane region crossing the Golgi membrane, or be peripherally attached to the cytosolic side of Golgi membranes. For proteins delivered to Pd via Golgi vesicles, each of these possibilities would result in their arrival to different destinations within a Pd. When Golgi vesicles fuse with the PM, their lumen spills outwards to the cell wall, while their cytosolic side faces the cytosol. Therefore, a protein residing in the Golgi lumen would be exported to the cell wall surrounding the Pd (Figure 11A), while a protein with a transmembrane region would become an integral Pd plasma membrane protein with Golgi lumen–facing domains exposed to the apoplast (Figure 11B), and a protein peripherally associated with the cytosolic side of the Golgi membrane would reach the inner side of the PM facing the cytoplasmic sleeve of the Pd (Figure 11C). Present bioinformatic (http://smart.embl-heidelberg.de/ and http://psort.nibb.ac.jp) and biochemical data indicate that pea, maize, and Arabidopsis C1RGPs are peripherally associated with the outer Golgi membrane (Dhugga et al., 1991, 1997; Epel et al., 1996; Delgado et al., 1998). It is therefore most likely that upon fusion of the Golgi to the PM of a Pd or in the vicinity of a Pd, C1RGPs would be attached to the PM facing the cytoplasmic sleeve of the Pd (Figure 11C). The integration of C1RGPs onto the cytosolic facing side of plasmodesmal PM could be a factor in establishing the size exclusion limit for soluble proteins. Exactly how C1RGPs and/or other membrane-associated Pd proteins that are targeted to the Pd orifice by Golgi-derived vesicles, or other means, enter and line the cytoplasmic sleeve is an interesting question that will need to be addressed in future research; this may be an active directed process or simply involve diffusion of proteins or protein complexes along the Pd membranes or a mass flow of membranes as a result of the addition of membranes to the Pd membrane network.
Figure 11.
An Illustration Showing That the Nature of a Protein's Association with Golgi Determines Its Destination inside a Pd.
(A) Proteins residing in the Golgi lumen are exported to the cell wall surrounding the Pd.
(B) Proteins with transmembrane regions arrive at the PM delineating the Pd.
(C) Proteins peripherally associated with the cytosolic side of the Golgi membrane reach the inner side of the PM facing the cytoplasmic sleeve of the Pd.
Previously, it has been suggested that RGP proteins are involved in the biosynthesis of plant cell wall polysaccharides (Dhugga et al., 1991, 1997; Delgado et al., 1998; Bocca et al., 1999; Zhao and Liu, 2002). If C1RGPs face the plasmodesmal cytoplasmic sleeve, it is unlikely that they could be directly involved in cell wall polysaccharide metabolism around Pd because they would not be exposed to the cell wall nor would they be attached to the outer face of the PM. Nevertheless, C1RGPs may be indirectly involved in cell wall polysaccharide metabolism despite such a location inside Pd. This could be achieved if C1RGPs function in delivering nucleotide sugars to putative transporters residing in the plasmodesmal PM, thus allowing the export of nucleotide sugars by these putative transporters through plasmodesmal PM to cell wall domains surrounding Pd. Alternatively, C1RGPs may function as glycosyl-transport proteins, shuttling UDP-sugars to or from glycosyltransferases associated with Pd and involved in Pd modification or in the modification of cytoplasmic components in a manner that would potentiate passage or inhibit passage through the Pd.
Additional research is necessary to determine the functions performed by C1RGPs in Pd. Whatever Pd-related functions RGP proteins may perform, their association with Pd appears to be a general characteristic rather than a species-specific phenomenon. This is evident from the data regarding both SE-WAP41 of monocot origin and AtRGPs of dicot origin. GFP fusions of the four AtRGP proteins were correctly targeted to Pd when expressed in tobacco. Therefore, we conclude that both the signal responsible for C1RGPs targeting to Pd and the mechanism of their delivery to Pd are general and species independent.
METHODS
Plant Material
Zea mays cv Jubilee (Roger Brothers Company, Idaho Falls, ID) seeds were imbibed overnight and grown in moist vermiculite in the dark for 5 d at 25°C. Nicotiana tabacum cv Samson and cv Xanthi plants were grown in 10-cm pots on a mix of equal volumes of soil and vermiculite (Pecka Hipper Gan, Rehovot, Israel) at 25°C under long-day conditions (16-h-light/8-h-dark cycles).
Extraction and Microsequencing of SE-WAP41
The cell wall fraction (CW) was prepared from the first upper centimeter of mesocotyl of etiolated Z. mays seedlings as previously described (Epel et al., 1996). CW from 40 g of fresh tissue were incubated overnight at 4°C in 50 mL of SE buffer (3 M NaCl, 5.6% β-mercaptoethanol, and 50 mM Hepes, pH 7.4), and the salt-extracted (SE) proteins were separated from the CW by centrifuging at 12,000g for 5 min. The pelleted walls were than washed with equal volume (50 mL) of extraction buffer without the NaCl, the walls were removed by centrifugation as above, and the supernatants were combined. After desalting and concentration by ultrafiltration with a 100-kD transfer cutoff (Amicon; Millipore, Billerica, MA), the proteins were separated by SDS-PAGE as previously described (Epel et al., 1996). The 41-kD band, containing ∼11 μg protein, was cut from the gel. After trypsin digestion in-gel, the peptides were extracted from the gel, separated by reverse phase HPLC and microsequenced by automated Edman degradation by Eurosequence (Groningen, The Netherlands).
Isolation of SE-WAP41 from an Expression Library
A cDNA library constructed in the λZAP II (Stratagene, La Jolla, CA) with poly(A) RNA from meristematic tissues of mesocotyl and first leaf of 5-d-old etiolated maize seedlings was screened with purified S-41 polyclonal antiserum at a dilution of 1:2000 obtained as previously described (Epel et al., 1996). The S-41 antiserum employed was first purified on a Protein A column and then on a Sepharose 4B column linked to total bacterial proteins (Harlow and Lane, 1988). An alkaline phosphatase linked goat-anti-rabbit antibody (Jackson Immuno Research Laboratories, West Grove, PA), diluted 1:4000, was used to identify plaques expressing proteins recognized by S-41 by a color reaction in the presence of 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (Sambrook et al., 1989). All DNA procedures were performed by standard techniques (Sambrook et al., 1989). Phage isolates were converted to plasmid clones following the Stratagene λZAP excision protocol. Hybridization groups were determined on nitrocellulose filters containing plaques of all isolated clones by hybridization at high stringency conditions with cDNA of an arbitrarily selected clone radioactively labeled by a Random Primed DNA labeling kit (Boehringer Mannheim, Mannheim, Germany). The screening of clones by PCR was performed on isolated plaques using a degenerated forward primer 5′-TTYTTYCANCCWTAYCAYCT-3′, where Y = A or G, W = A or T, and N = A or G or C or T, and reverse primer complementary to sequence of flanking vector region.
Preparation of Antiserum against Recombinant SE-WAP41
For the expression of recombinant SE-WAP41, the cDNA was PCR amplified employing the forward primer 5′-CGAATTCATATGGCGGGCACGGTGACG-3′ and the reverse primer 5′-CCGGATCCTACTTTGGCCTTGCCATTTGC-3′, which contained flanking NdeI and BamHI sites, respectively, and the PCR product was cloned into corresponding sites of pET16 (Novagen, Darmstadt, Germany) in fusion to the polyHis. The resulting plasmid pET16:SE-WAP41 was transformed to BL21(DE3) cells, and expression of His:SE-WAP41 was induced by 2 mM isopropyl-β-d-thiogalactopyranoside in log phase cultures for 4 h. The fusion protein was affinity purified on Ni-NTA-Agarose beads (Qiagen, Hilden, Germany), released with imidazole, and then rechromatographed, and the bound protein was cleaved in column from the His tag by Factor Xa (Sigma-Aldrich, St. Louis, MO). The purified protein was mixed with complete Freund's adjuvant (Difco Laboratories, Detroit, MI), and the mixture was injected into 10 to 20 sites on the backs of 1-month-old female New Zealand rabbits. The subsequent four booster injections, spaced at a 4-week interval, were prepared with incomplete Freund's adjuvant.
Preparation of Soluble and Membranal Cellular Fractions
Cell extracts were prepared and fractionated as previously described (Yahalom et al., 1991) with modifications: the filtrates from first homogenization and after nitrogen disruption bomb were combined and centrifuged for 1 h at 90,000g at 4°C. The supernatant was collected for analysis of soluble proteins. The pellet was resuspended in buffer HM/7.5 (20 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 10 mM EGTA, 2 mM EDTA, and 1 mM phenylmethylsulphonyl fluoride), repelleted under the same conditions, and used to extract membranal proteins.
RNA Blot Analysis
Plant material from different tissues was harvested and frozen in liquid nitrogen, and total RNA was extracted using TRIzol reagent (Gibco BRL Life Technologies, Gaithersburg, MD) as described by the manufacturer. Ten micrograms of RNA were slot blotted or glycoxylated, separated in 1.4% agarose gel, and passively transferred to a Hybond-N nylon filter (Amersham, Fairfield, CT). Hybridization was performed with random primed 32P-labeled probes at 65°C for SE-WAP41 containing the whole cDNA, including 3′ and 5′ noncoding sequence, and for maize 26S-rRNA and at 55°C for actin from soybean (Glycine max) in a solution containing 0.26 M phosphate buffer, pH 7.2, 7% SDS, 1 mM EDTA, and 1% BSA. Filters were washed at 61°C, first in 0.26 M phosphate buffer, pH 7.2, containing 1% SDS, then in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.1% SDS, and finally in 1× SSC, 0.1% SDS. The 26S-rRNA gene was used as a loading control. Scanning was done with a Fuji BAS 1000 phosphor imager (Tokyo, Japan).
Construction of Plant Expression Plasmids
To create plasmids pBinGFP, pBinDsRed1, and pBinCFP, cassettes containing the 35S promoter of Cauliflower mosaic virus, omega sequence enhancer, a synthetic GFP (sGFP) gene (Chiu et al., 1996), the DsRed1 gene (Clontech Laboratories, Palo Alto, CA), or the enhanced CFP gene (Clontech Laboratories), respectively, and the nitric oxide synthase transcriptional terminator were cloned into the HindIII-EcoRI sites of pBINPLUS (van Engelen et al., 1995). To construct plasmids pBinAtRGP2:GFP and pBinAtRGP2:CFP, the AtRGP2 gene was amplified without it's stop codon by PCR performed with a 5′-TCTAGAATTCATGGTTGAGCCGGCGAATACTG-3′ forward primer containing XbaI and EcoRI sites and a 5′-TCTAGATACTGCTCTCAAGCTTTTGCCACTG-3′ reverse primer containing an XbaI site and cloned into a unique XbaI site present in pBinGFP or pBinCFP, respectively, between the omega enhancer sequence and the sGFP start codon. EcoRI digestion was used to verify AtRGP2 orientation in pBinAtRGP2:GFP and pBinAtRGP2:CFP. Plasmid pBinAtRGP3:GFP was constructed in the same way using a 5′-TCTAGAATTCATGGCGCAATTGTATTCTTCCGTG-3′ forward primer and a 5′-TCTAGAATTTTTGCCTTTTGGTGCCTCAGC-3′ reverse primer. Construction of pBinAtRGP1:GFP was identical, except AtRGP1 was amplified with a 5′-CCTAGGAATTCATGGTTGAGCCGGCGAACAC-3′ forward primer containing AvrII and EcoRI sites, and a 5′-CCTAGGAGCTTTAGTGGGTGGGTTAAGCTC-3′ reverse primer containing an AvrII site and AvrII-AtRGP1-AvrII fragment was cloned into the unique XbaI site in pBinGFP. Plasmid pBinAtRGP4:GFP was constructed in the same way as pBinAtRGP1:GFP using a 5′-CCTAGGAATTCATGGCGGGCTACAACCTCGAAGCTATC-3′ forward primer and a 5′-CCTAGGCTTGGCCTTGACATCTTTGCCATCGCTC-3′ reverse primer.
Transient Expression
Transient expression was induced using Agrobacterium tumefaciens leaf injection (Lavy et al., 2002). The abaxial side of N. tabacum cv Samson or cv Xanthi leaves was injected with Agrobacterium strain GV3101 harboring the indicated plasmids, using a 1-mL syringe without a needle. Observation of transient expression was limited to epidermal cells at the lower face of the leaves. GFP fluorescence was examined 24 to 48 h after injection. DsRed1 fluorescence was examined ∼72 h after injection.
Stable Transformations of Tobacco
To create AtRGP2:GFP or GONST1:YFP expressing transgenic plants, wild-type N. tabacum cv Samson were transformed by Agrobacterium strain GV3101 harboring pBinAtRGP2:GFP or pBinGONST1:YFP plasmids accordingly, using a modified leaf disk method (Gallois and Marinho, 1995). Leaf disks were incubated with Agrobacterium for 30 min, washed, and placed for cocultivation in MS medium containing 2 μg/mL of kinetin and 0.8 μg/mL of indoleacetic acid. The rooting medium, contrary to Gallois and Marinho (1995), did not contain hormones. Analysis was performed on leaves of T0 plants.
Callose Staining and Colocalization with AtRGP2:GFP
Callose was stained by incubating leaf segments for 40 min in a mixture of 0.1% aniline blue in double distilled water and 1 M glycine, pH 9.5, at a volume ratio of 2:3, respectively, and premixed at least 1 d before use. Colocalization was viewed with a DMRBE fluorescence light microscope (Leica, Wetzlar, Germany); aniline blue–stained callose fluorescence was viewed with a band-pass 340- to 380-nm excitation filter, an RKP 400 dichromatic mirror, and a long-pass 425-nm emission filter; AtRGP2:GFP fluorescence was viewed with a band-pass 450- to 490-nm excitation filter, an RKP 510 dichromatic mirror, and a band-pass 515- to 560-nm emission filter. GFP and aniline blue monochrome photographs were converted to magenta and yellow, respectively, using Zeiss LSM 5 image browser (Jena, Germany), and color images were inverted using Adobe Photoshop 7.0 ME (Mountain View, CA) to obtain green and blue images, respectively, on white background.
Plasmolysis
Cut leaf section from plants stably expressing AtRGP2:GFP, MPTMV:GFP, or GONST1:YFP were incubated in 30% glycerol until epidermal cells were plasmolyzed. DsRed1 was transiently expressed in the leaves by Agrobacterium leaf injection ∼72 h before they were plasmolyzed. Plasmolysis was determined by monitoring the retraction of the DsRed1 fluorescently labeled protoplast from the cell wall.
Treatment with BFA
During transient expression of AtRGP2:GFP, at ∼22 h after Agrobacterium injection, the abaxial side of tobacco leaves was injected with 50 μg/mL of BFA in 0.1% DMSO on one side of the major vein, while the opposite side of the major vein was mock injected with 0.1% DMSO. Injections were performed using 1-mL syringes without a needle, so that the intercellular spaces beneath the epidermis were completely filled. Leaf segments located at equal distances to either side of the major vein and at the same area along the baso-petal axis were cut and examined by confocal microscopy. Optical sections 0.9 μm thick through leaf epidermal cells were scanned by confocal laser scanning microscopy 2 to 5 h after injection of BFA/DMSO (24 to 27 h after Agrobacterium injection). Photographed cells did not include large cells at the base of trichomes or small cells adjacent to stomatal guard cells. The experiment was repeated twice with four leaves from four plants. Fluorescently labeled Pd were counted in 10 BFA-treated cells and 10 mock-treated cells. In each cell, the total number of fluorescently labeled Pd was counted in five consecutive optical sections representing the approximate middle layer of the cell.
Fluorescence Confocal Microscopy
Fluorescence confocal microscopy was performed using a Zeiss R510 confocal laser scanning microscope. GFP excitation was performed with an argon laser set to 488 nm, and emission was detected with a 525 nm ± 20 nm band-pass filter combination. CFP excitation was performed with an argon laser set to 458 nm, and emission was detected with a 500 nm ± 25 nm band-pass filter. When YFP was used with CFP, YFP excitation was performed with an argon laser set to 514 nm, and emission was detected with a 550 nm ± 20 nm band-pass filter combination. When YFP was used with DsRed1, YFP excitation was performed with an argon laser set to 488 nm, and emission was detected with a 525 nm ± 20 nm band-pass filter combination. The different YFP conditions were used to prevent bleedthrough between fluorescent channels. DsRed1 excitation was performed with a helium-neon laser set to 543 nm, and emission was detected with a 587.5 nm ± 27.5 nm band-pass filter. To expose spongy mesophyll for imaging, leaves were torn by hand so that the epidermis at the lower face of leaf sections was peeled off. Image analysis was performed with the Zeiss LSM-5 image browser.
Sequence data from this article appear in the EMBL/GenBank data libraries under the following accession numbers: AtRGP1 (AF013627), AtRGP2 (AF013628), AtRGP3 (AF034255), AtRGP4 (AF329280), PsRGP1 (U31565), and SE-WAP41 (U89897).
Supplementary Material
Acknowledgments
We thank G. Kotlizky for use of the maize seedlings mesocotyl expression library, C. Reichel and R.N. Beachy for use of MPTMV:GFP transgenic tobacco, I.J. Delgado and N.V. Raikhel for kindly supplying AtRGP1, AtRGP2, AtRGP3, and AtRGP4 clones, U. Hannania and A. Avni for kindly supplying pBinGFP plasmid, K. Bracha-Drori and S. Yalovsky for kindly supplying pBinGONST1:YFP plasmid, A. Levy for development of the callose staining procedure, and S. Schuster for materials. This research was supported by Resource Grant Award IS-3222-01C from the U.S.–Israel Binational Agricultural Resource and Development Fund, by the Israel Science Foundation (Grant 723/00-17.1), and by an Eshkol Doctoral Fellowship to G.S. from the Israeli Ministry of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Bernard L. Epel (blepel@post.tau.ac.il).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031823.
References
- Baldwin, T.C., Handford, M.G., Yuseff, M.I., Orellana, A., and Dupree, P. (2001). Identification and characterization of GONST1, a Golgi-localized GDP-mannose transporter in Arabidopsis. Plant Cell 13, 2283–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska, F., Samaj, J., Napier, R., and Volkmann, D. (1999). Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J. 19, 481–488. [DOI] [PubMed] [Google Scholar]
- Bayer, E., Thomas, C.L., and Maule, A.J. (2004). Plasmodesmata in Arabidopsis thaliana suspension cells. Protoplasma 223, 93–102. [DOI] [PubMed] [Google Scholar]
- Beachy, R.N., and Heinlein, M. (2000). Role of P30 in replication and spread of TMV. Traffic 1, 540–544. [DOI] [PubMed] [Google Scholar]
- Bocca, S.N., Kissen, R., Rojas-Beltran, J.A., Noel, F., Gebhardt, C., Moreno, S., du Jardin, P., and Tandecarz, J.S. (1999). Molecular cloning and characterization of the enzyme UDP-glucose: Protein transglucosylase from potato. Plant Physiol. Biochem. 37, 809–819. [DOI] [PubMed] [Google Scholar]
- Botha, C.E.J., Hartley, B.J., and Cross, R.H.M. (1993). The ultrastructure and computer-enhanced digital image-analysis of plasmodesmata at the Kranz mesophyll-bundle sheath interface of Themeda-triandra var imberbis (Retz) A. Camus in conventionally fixed leaf blades. Ann. Bot. (Lond.) 72, 255–261. [Google Scholar]
- Chiu, W.L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., and Zambryski, P. (2000). Systemic transport of RNA in plants. Trends Plant Sci. 5, 52–54. [DOI] [PubMed] [Google Scholar]
- Delgado, I.J., Wang, Z.H., de Rocher, A., Keegstra, K., and Raikhel, N.V. (1998). Cloning and characterization of AtRGP1: A reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 116, 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga, K.S., Tiwari, S.C., and Ray, P.M. (1997). A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: Purification, gene cloning, and trans-Golgi localization. Proc. Natl. Acad. Sci. USA 94, 7679–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga, K.S., Ulvskov, P., Gallagher, S.R., and Ray, P.M. (1991). Plant polypeptides reversibly glycosylated by UDP-glucose: Possible components of Golgi beta-glucan synthase in pea cells. J. Biol. Chem. 266, 21977–21984. [PubMed] [Google Scholar]
- Ding, B., Itaya, A., and Woo, Y.M. (1999). Plasmodesmata and cell-to-cell communication in plants. Int. Rev. Cytol. 190, 251–316. [Google Scholar]
- Ding, B., Turgeon, R., and Parthasarathy, M.V. (1992). Substructure of freeze-substituted plasmodesmata. Protoplasma 169, 28–41. [Google Scholar]
- Ehlers, K., and Kollmann, R. (2001). Primary and secondary plasmodesmata: Structure, origin, and functioning. Protoplasma 216, 1–30. [DOI] [PubMed] [Google Scholar]
- Epel, B.L., vanLent, J.W.M., Cohen, L., Kotlizky, G., Katz, A., and Yahalom, A. (1996). A 41 kDa protein isolated from maize mesocotyl cell walls immunolocalizes to plasmodesmata. Protoplasma 191, 70–78. [Google Scholar]
- Faik, A., Desveaux, D., and Maclachlan, G. (2000). Sugar-nucleotide-binding and autoglycosylating polypeptide(s) from nasturtium fruit: Biochemical capacities and potential functions. Biochem. J. 347, 857–864. [PMC free article] [PubMed] [Google Scholar]
- Gallois, P., and Marinho, P. (1995). Leaf disk transformation using Agrobacterium tumefaciens expression of heterologous genes in tobacco. In Plant Gene Transfer and Expression Protocols, H. Jones, ed (Totowa, NJ: Humana Press), pp. 39–48. [DOI] [PubMed]
- Gorshkova, E.N., Erokhina, T.N., Stroganova, T.A., Yelina, N.E., Zamyatnin, A.A., Kalinina, N.O., Schiemann, J., Solovyev, A.G., and Morozov, S.Y. (2003). Immunodetection and fluorescent microscopy of transgenically expressed hordeivirus TGBp3 movement protein reveals its association with endoplasmic reticulum elements in close proximity to plasmodesmata. J. Gen. Virol. 84, 985–994. [DOI] [PubMed] [Google Scholar]
- Gupta, P., Raghuvanshi, S., and Tyagi, A.K. (2000). PCR-amplification and cloning of the coding region of a cDNA for a reversibly glycosylated polypeptide from rice with possible involvement in the biosynthesis of glucans. J. Plant Biochem. Biotechnol. 9, 99–102. [Google Scholar]
- Harlow, E.D., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Haywood, V., Kragler, F., and Lucas, W.J. (2002). Plasmodesmata: Pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (suppl.), S303–S325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein, M., and Epel, B.L. (2004). Macromolecular transport and signaling through plasmodesmata. In International Review of Cytology: A Survey of Cell Biology, Vol. 235, K.W. Joen, ed (San Diego: Academic Press), pp. 93–164. [DOI] [PubMed]
- Hepler, P.K. (1982). Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma 111, 121–133. [Google Scholar]
- Iino, M. (1982). Inhibitory action of red light on the growth of the maize mesocotyl: Evaluation of the auxin hypothesis. Planta 156, 388–395. [DOI] [PubMed] [Google Scholar]
- Jones, M.G.K. (1976). The origin and development of plasmodesmata. In Intercellular Communication in Plants: Studies on Plasmodesmata, B.E.S. Gunning and A.W. Robards, eds (Heidelberg, Germany: Springer-Verlag), pp. 81–105.
- Langeveld, S.M.J., Vennik, M., Kottenhagen, M., van Wijk, R., Buijk, A., Kijne, J.W., and de Pater, S. (2002). Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Physiol. 129, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy, M., Bracha-Drori, K., Sternberg, H., and Yalovsky, S. (2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14, 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr, A., Ritzenthaler, C., and Robinson, D.G. (2002). Brefeldin A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka, K.J., Roberts, A.G., Boevink, P., Santa Cruz, S., Roberts, L., Pradel, K.S., Imlau, A., Kotlizky, G., Sauer, N., and Epel, B. (1999). Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97, 743–754. [DOI] [PubMed] [Google Scholar]
- Overall, R.L. (1999). Structure of plasmodesmata. In Plasmodesmata, Structure, Function, Role in Cell Communication, A.J. van Bel and W.J.P. van Kestern, eds (Heidelberg, Germany: Spring-Verlag), pp. 129–148.
- Porchia, A.C., Sorensen, S.O., and Scheller, H.V. (2002). Arabinoxylan biosynthesis in wheat. Characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol. 130, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, A.G., and Oparka, K.J. (2003). Plasmodesmata and the control of symplastic transport. Plant Cell Environ. 26, 103–124. [Google Scholar]
- Robards, A.W., and Lucas, W.J. (1990). Plasmodesmata. Annu. Rev. Plant Physiol. 41, 369–419. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Saxena, I.M., and Brown, R.M. (1999). Are the reversibly glycosylated polypeptides implicated in plant cell wall biosynthesis non-processive beta-glycosyltransferases? Trends Plant Sci. 4, 6–7. [DOI] [PubMed] [Google Scholar]
- Singh, D.G., Lomako, J., Lomako, W.M., Whelan, W.J., Meyer, H.E., Serwe, M., and Metzger, J.W. (1995). Beta glucosylarginine: A new glucose protein bond in a self-glucosylating protein from sweet corn. FEBS Lett. 376, 61–64. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A., Wells, B., and Roberts, K. (1994). Plasmodesmata of maize root tips: Structure and composition. J. Cell Sci. 107, 3351–3361. [DOI] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A., and Stiekema, W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Yahalom, A., Epel, B.L., Glinka, Z., Macdonald, I.R., and Gordon, D.C. (1987). A kinetic analysis of phytochrome controlled mesocotyl growth in Zea mays seedlings. Plant Physiol. 84, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalom, A., Warmbrodt, R.D., Laird, D.W., Traub, O., Revel, J.P., Willecke, K., and Epel, B.L. (1991). Maize mesocotyl plasmodesmata proteins cross-react with connexin gap junction protein antibodies. Plant Cell 3, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski, P., and Crawford, K. (2000). Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu. Rev. Cell Dev. Biol. 16, 393–421. [DOI] [PubMed] [Google Scholar]
- Zhao, G.R., and Liu, J.Y. (2002). Isolation of a cotton RGP gene: A homolog of reversibly glycosylated polypeptide highly expressed during fiber development. Biochim. Biophys. Acta 1574, 370–374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.