Abstract
In plants, defensive proteins secreted to leaf aerial surfaces have not previously been considered to be a strategy of pathogen resistance, and the general occurrence of leaf surface proteins is not generally recognized. We found that leaf water washes (LWW) of the experimental plant Nicotiana tabacum tobacco introduction (TI) 1068 contained highly hydrophobic, basic proteins that inhibited spore germination and leaf infection by the oomycete pathogen Peronospora tabacina. We termed these surface-localized proteins tobacco phylloplanins, and we isolated the novel gene T-Phylloplanin (for Tobacco Phylloplanin) and its promoter from N. tabacum. Escherichia coli–expressed T-phylloplanin inhibited P. tabacina spore germination and greatly reduced leaf infection. The T-phylloplanin promoter, when fused to the reporter genes β-glucuronidase and green fluorescent protein, directed biosynthesis only in apical–tip cell clusters of short, procumbent glandular trichomes. Here, we provide evidence for a protein-based surface defense system in the plant kingdom, wherein protein biosynthesis in short, procumbent glandular trichomes allows surface secretion and deposition of defensive phylloplanins on aerial surfaces as a first-point-of-contact deterrent to pathogen establishment. As yet uncharacterized surface proteins have been detected on most plant species examined.
INTRODUCTION
Surface protection is an innate defensive strategy in which microbes are directly inhibited at their first point of host contact, usually at the boundary between the host and the external environment. Although studies of chemical-based leaf surface protection in plants have focused on secreted secondary metabolites (e.g., glandular trichome exudates), animal studies have focused on secreted surface proteins deployed at host–pathogen interfaces such as skin or intestinal epithelia (Gallo and Huttner, 1998; Schroder, 1999).
Fungi and fungi-like (e.g., oomycete) pathogens are the major causes of plant disease (Lucas, 1998), resulting in annual crop losses of ∼20% worldwide, necessitating extensive control by synthetic fungicides (Knight et al., 1997). Many of these organisms reproduce via airborne spores and transiently exploit the plant leaf surface, or phylloplane, as a starting point for host ingress. Spores of the oomycete pathogen Peronospora tabacina, the causal agent of blue mold disease on several Nicotiana species, germinate on the leaf surface by forming a germination tube and then penetrate the plant epidermal layer with an infection peg (Svircev et al., 1989). For successful phylloplane germination to occur, spores must tolerate preformed biochemicals present on the leaf surface.
Although some surface biochemicals are presumed to leach passively from the leaf interior (e.g., sugars), others are biosynthesized selectively by specialized epidermal cells for delivery to the phylloplane. Trichomes are simple or glanded epidermal appendages that occur on most plants. Glandular secreting trichomes are found on ∼30% of vascular plants (Dell and McComb, 1978; Fahn, 2000; Wagner et al., 2004), and they produce surface-accumulated exudates that usually contain hydrophobic isoprenoids and phenylpropanoids, the latter including flavonoids, phenolics, tannins, quinones, etc. (Kelsey et al., 1984; Spring, 2000). In Solanaceae plants, amphipathic sugar esters are also commonly found in glandular trichome exudates (Wagner, 1999). Such compounds have been associated with insect resistance in many plants, and pest resistance is often correlated with glandular trichome density (Thomson and Healey, 1984; Kronestedt-Robards and Robards, 1991). Two well-studied cases of glandular trichome–based insect resistance are found in the plant family Solanaceae. Sugar esters produced by tall glandular trichomes (TGSTs) of primitive tomato (Lycopersicon pennellii) and potato (Solanum berthaultii) species, and the diterpenoid cembratriene-ol produced by tobacco TGSTs, have been shown to inhibit aphid infestation (Goffreda et al., 1990; Steffens and Walters, 1990; Wang et al., 2001). Antimicrobial activities of trichome exudate compounds (particularly monoterpenoids and sesquiterpenoids) have also been reported, but these are less studied than insect resistance (Kelsey et al., 1984).
The phylloplane of the experimental plant Nicotiana tabacum tobacco introductions (TI) 1068 (Figure 1A) contains TGSTs, small, procumbent (bent to the surface) glandular secreting trichomes (SGTs), simple glandless trichomes, preformed biochemicals including cembrenoid and labdenoid diterpenes, and sugar esters, surface waxes, and some volatiles (Wagner, 1999). Diterpenoids and sugar esters, the major components of tobacco leaf exudate, are synthesized only by glandular head cells of TGSTs (Kandra and Wagner, 1988). TGST exudate accumulates under the cuticle surrounding the gland and may pass through cuticular striae to migrate down the trichome stalk to disperse widely on the leaf surface, and exudate can accumulate to ∼17% of leaf dry weight. Leaf exudate cembratriene diols (α-CBT-diols and β-CBT-diols), labdenediol, sclareol, and sucrose esters, in sufficient amounts, have been reported to inhibit P. tabacina spore germination (Cruickshank et al., 1977; Menetrez et al., 1990; Kennedy et al., 1992). However, resistance is found only in species and cultivars that accumulate high levels of these compounds or when they are applied experimentally. Leaf washing with water (Hill, 1966) and acetone (Reuveni et al., 1987) to remove cuticular components before spore inoculation was shown to increase the plant's susceptibility to P. tabacina.
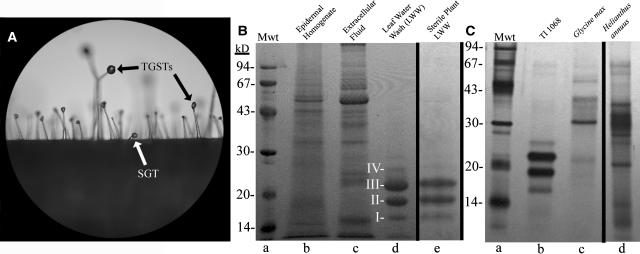
Figure 1.
Proteins Are Present on Plant Leaf Surfaces.
(A) Magnification (×40) of TI 1068 phylloplane with TGSTs and SGTs identified.
(B) Coomassie blue–stained SDS-PAGE gel of TI 1068–derived samples. Phylloplanins I to IV are identified. Loaded volumes of LWW (lane d) and sterile-grown plant LWW (lane e) represent 25-cm2 leaf surface area. Mwt (lane a) denotes protein standards.
(C) Silver-stained SDS-PAGE gel of LWW from field-grown TI 1068 (lane b; 10 cm2), G. max (lane c; 30 cm2), and H. annuus (lane d; 6 cm2). Mwt (lane a) denotes protein standards.
Although secondary metabolites in glandular trichome exudates are often associated with surface-localized insect and microbe resistance, defensive proteins have not generally been considered to be products of glanded trichomes. Secreted defensive proteins are also not known to constitute a surface defense system in plants, even though such a system exists in animals. Kowalski et al. (1992) reported that large amounts of polyphenol oxidase are produced and stored in type A (short-glanded) trichomes of primitive potato S. berthaultii. When trichomes are mechanically disrupted by insects walking on the plant surface, trichome-accumulated polyphenol oxidase and phenols are mixed to form polyphenols that immobilize aphids and reduce infestation. This mechanism appears to be restricted to type A trichomes of S. berthaultii, and polyphenol oxidase protein is not broadly dispersed on leaf surfaces. There are also a few other reports of protein enrichment in surface structures (trichomes or hydathodes) of plants, but proteins are not thought to be secreted or to be widely dispersed on the surface. Enzymes related to glutathione biosynthesis and plant metallothioneins have been reported to be enriched in trichomes, suggesting a role for trichomes in ion sequestration and removal (Wagner et al., 2004). Hydathodes, surface appendages with a direct connection to the vasculature, that primarily function in the removal of excess water (guttation) and ions, have been reported to accumulate antimicrobial, pathogenesis-related proteins (Samac and Shah, 1991), leading to the suggestion that these proteins may function to inhibit bacteria that enter open hydathode pores.
We hypothesized that plants, like animals, can produce and disperse proteins to aerial leaf surfaces and that these proteins contribute to host defense. Here, we describe the discovery of N. tabacum leaf surface proteins, termed T-phylloplanins (for tobacco phylloplanins), which are inhibitory to P. tabacina, and the recovery of the gene T-Phylloplanin. We also describe the elucidation of the T-phylloplanin promoter and provide evidence for phylloplanin biosynthesis in SGTs.
RESULTS AND DISCUSSION
SDS-PAGE analyses of leaf water wash (LWW) from greenhouse-grown TI 1068 leaves indicated the presence of four bands with molecular masses of 16 (I), 19 (II), 21 (III), and 25 (IV) kD (Figure 1B, lane d), which we collectively termed T-phylloplanins. T-phylloplanins in LWW were relatively pure and abundant, compared with proteins present in leaf epidermal cells (Figure 1B, lane b) or leaf extracellular fluid (Figure 1B, lane c), suggesting selective deployment on the phylloplane. Sterile-grown TI 1068 LWW contained T-phylloplanins (Figure 1B, lane e), indicating that these proteins were not formed by leaf surface microbes and were not induced by pathogen attack. From measurement of the protein concentration in LWW (bicinchoninic acid assay), we estimate that the phylloplane of greenhouse-grown TI 1068 leaves contains 100 to 200 ng protein/cm2 leaf surface. Field-grown TI 1068 LWW also contained T-phylloplanins, indicating that leaf surface proteins are present under natural conditions (Figure 1C, lane b), and T-phylloplanins were renewed after washing (data not shown). N. tabacum cultivars TI 1112 and TI 1406, which lack TGSTs and secretion, respectively, produce substantial T-phylloplanins (data not shown), so diterpene/sugar ester–producing TGSTs are not the site of T-phylloplanin biosynthesis. Field-grown soybean (Glycine max) and sunflower (Helianthus annuus) LWW contained varying amounts of phylloplanins (Figure 1C, lanes c and d), as did greenhouse-grown maize (Zea mays), tomato (L. esculentum), soybean, and potato (S. tuberosum) (data not shown), but these proteins were not further characterized. LWW of frozen TI 1068 leaves that were cold-brushed to completely remove TGSTs and SGTs (Wang et al., 2001) contained a similar amount of T-phylloplanins per unit surface area to that found in LWW of undisturbed leaves, indicating that T-phylloplanins are not restricted to SGTs but are rather generally dispersed on the leaf surface.
T-Phylloplanins Inhibit P. tabacina Spore Germination and Leaf Infection
P. tabacina is an oomycete pathogen that reproduces via airborne spores (Lucas, 1975), and initial host contact and spore deposition commence at the phylloplane (Svircev et al., 1989). LWW from greenhouse-grown TI 1068 plants inhibited P. tabacina spore germination (Figure 2A, gel b; mean lethal dose ∼15 to 20 ng/μL [50 spores/μL]), as did LWW from sterile-grown plants (data not shown). Protein digestion by immobilized proteinase K relieved the inhibition of spore germination (Figure 2A, gel c), indicating that proteins were necessary for inhibition. Spore germination was not affected by water incubated with immobilized proteinase K (data not shown). Also, once spore germination was initiated, addition of LWW (100 ng/μL total protein) immediately arrested germination tube growth and development (data not shown). Using gas chromatography (GC), the levels of residual exudate diterpenes found in LWW (data not shown) were less than one-tenth of the mean lethal dose reported to inhibit P. tabacina germination (Kennedy et al., 1992), and we also were unable to detect nicotine in LWW (data not shown).
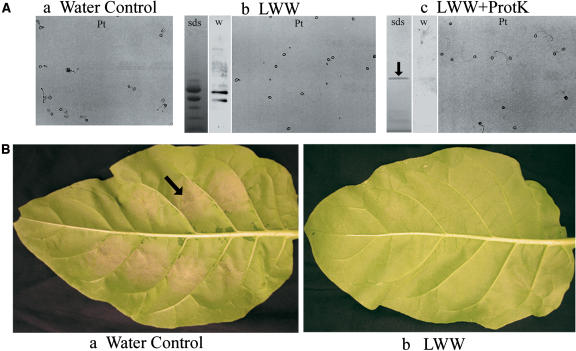
Figure 2.
Proteins in TI 1068 LWW Inhibit P. tabacina Spore Germination and Leaf Infection.
(A) P. tabacina spore germination assay (Pt), Coomassie blue–stained SDS-PAGE gel blot (sds), and protein gel blot with a 1:10,000 dilution of phylloplanin antiserum (w). Gel a, water plus spores; gel b, TI 1068 LWW (diluted to 100 ng/μL total protein) plus spores; gel c, TI 1068 LWW (100 ng/μL total protein) digested with proteinase K (ProtK) plus spores. The arrow marks residual, soluble proteinase K.
(B) P. tabacina leaf infection assay of cv Petite Havana. Photograph a, water plus spores (104 spores/mL); a sporulating lesion is indicated with the arrow. Photograph b, TI 1068 LWW (diluted to 50 ng/μL) plus spores (104 spores/mL).
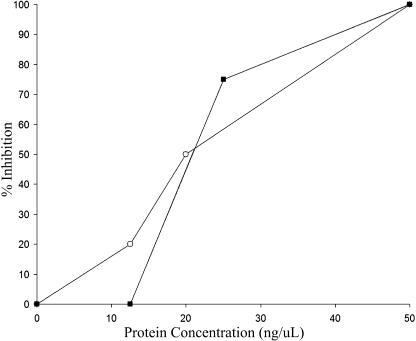
Intact N. tabacum cv Petite Havana SR1 plants, considered susceptible to P. tabacina, were infected by applying spores (50 spores/μL in 4 μL of water) to the leaf surface. After 5 d, sporulating lesions developed at sites of application (Figure 2B, photograph a). T-phylloplanins in TI 1068 LWW, when mixed with spores at total protein concentrations of 50 ng/μL or greater, inhibited leaf infection by P. tabacina (Figure 2B, photograph b). At 25 ng/μL total protein, we observed ∼75% inhibition, and no inhibition occurred with titrations of <12.5 ng/μL total protein (data not shown). Similar results were observed in three independent experiments and in identical experiments using the susceptible cv KY 14 (data not shown). Figure 3 shows the inhibitory effect of T-phylloplanins in LWW on P. tabacina spore germination and leaf infection. LWW of Petite Havana and KY 14 contain less phylloplanins I to IV than TI 1068, and unlike TI 1068 LWW, they produce low trichome exudate (data not shown). We speculate that other surface chemicals (e.g., surface lipids or TGST exudate components) may influence or accentuate phylloplanin activity, dispersion, or longevity by acting as adducts or as solubilizing agents. Thus, a combination of T-phylloplanins and high TGST exudates may provide maximal inhibition of spore germination. It is difficult to estimate the role of a single component such as T-phylloplanins in blue mold susceptibility or resistance outside of the experimental conditions used here, but we propose that T-phylloplanins are a key component.
Figure 3.
Inhibition of P. tabacina Spore Germination and Leaf Infection by T-Phylloplanins in LWW.
For both assays, the results of a single experiment that is representative of three separate experiments are shown. Open circles, spore germination; closed squares, leaf infection.
Isolation of the Novel T-Phylloplanin Gene
N. tabacum T-phylloplanins I to IV share an identical N-terminal amino acid sequence (Table 1). Internal amino acid sequences were elucidated from peptides generated by trypsin digestion of T-phylloplanins II and IV and pepsin digestion of total LWW (Table 1). Degenerate, deoxyinosine-containing primers were synthesized and used in RT-PCR with cDNA generated from N. tabacum total leaf RNA as a template, and a 332-bp fragment was amplified. RNA ligase–mediated rapid amplification of cDNA ends (RLM-RACE) was used to recover a full-length, novel N. tabacum T-Phylloplanin cDNA sequence (Figure 4; accession number AY705384) of 666 bp encoding a hydrophobic, basic (50% hydrophobicity, estimated pI of 9.3; VectorNTI; Invitrogen, Carlsbad, CA), 15.4-kD protein containing 150 amino acids. Based on the N terminus recovered from the mature T-phylloplanin (Ile-24), the first 23 amino acids constitute a signal sequence that targets the protein to the secretory pathway (TargetP version 1.0 [Emanuelsson et al., 2000]). The molecular mass of the mature protein is estimated to be ∼13 kD. We did not recover a protein of this mass from the leaf surface but instead recovered four apparent bands of higher molecular masses. Although differences in amino acid composition may account for the differences in migration, we speculate that the molecular masses of native T-phylloplanins I to IV could be increased because of the occurrence of covalent adducts with cuticular lipids or of trichome exudate diterpenes or sugar esters. These covalent adducts would be retained in SDS-PAGE, and they could increase phylloplanin solubility in TGST exudate (diterpenes and sugar esters) and aid in phylloplanin dispersion on the leaf surface. Amphipathic sugar esters (∼24% of TI 1068 weight) are known to solubilize largely hydrophobic diterpenes (∼73%) of TGST exudate. We note that highly hydrophobic, basic, saposin-like proteins of animals (see below) also display anomalous migration on SDS-PAGE (Curstedt et al., 1987), and we suggest that T-phylloplanins may behave similarly.
Table 1.
Amino Acid Sequences Recovered from T-Phylloplanin N-Terminal Analysis, Trypsin Digestion, and Pepsin Digestion
| Method | Peak (min) | T-Phylloplanin | Amino Acid Sequence | Name |
|---|---|---|---|---|
| N terminus | N/A | I | ILVPTLVST | |
| N/A | II | ILVPTLVSTHISGLVFCSV | aa-N1 | |
| N/A | III | ILVPTLVSTHISGLVFCSV | aa-N1 | |
| N/A | IV | ILVPTLVSTHISGLVFCSV (major) | aa-N1 | |
| Trypsin | 36.2 | I | ASVQLR | aa-T1 |
| 59.8 | I | ILNLNI (major) | aa-T4 | |
| CGATNVISSTIT (minor) | aa-T2 | |||
| 56.7 | III | LVVATPLSTCxATLxSVG | aa-T3 | |
| 58.7 | III | ILNLNI (major) | aa-T4 | |
| CGATxVxSSTIT (minor) | aa-T2 | |||
| Pepsin | 35 | I, II, III, IV | IRVGLAPTG | aa-P1 |
N/A, not applicable; aa, amino acid.
Figure 4.
Nucleotide and Predicted Amino Acid Sequences of T-Phylloplanin cDNA.
Nucleotides are numbered at right. Start and stop codons are underlined, and the signal sequence is shown in boldface. Segments corresponding to peptides aa-N1, aa-T1, aa-T2, aa-T3, aa-T4, and aa-P1 are marked by lines above the amino acid sequence and labeled.
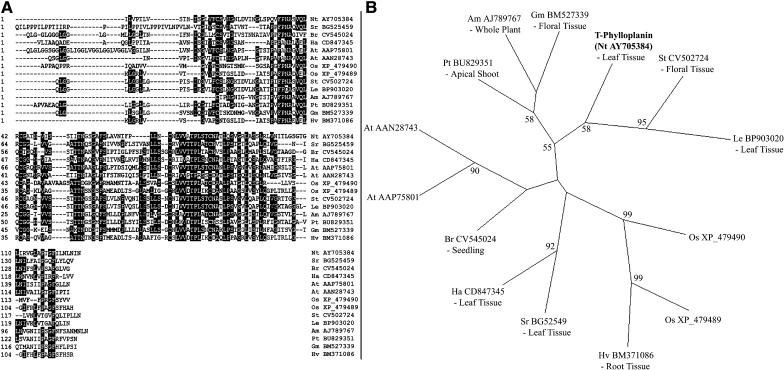
BLAST searches (Altschul et al., 1990) with the T-Phylloplanin gene sequence against the nonredundant and EST GenBank databases yielded several significant hits from Nicotiana sequences, including an amplified fragment linked polymorphism from N. tabacum (GenBank accession number AJ538724) and several EST sequences from N. tabacum and N. sylvestris. BLAST searches also indicated that homologous genes of unknown functions exist in many other plants. A ClustalW alignment (DNASTAR Software, Madison, WI) between T-phylloplanin and selected sequences giving significant tBLASTn scores from various other plant species (Figure 5A) indicated that regions of amino acid identity exist and possibly represent conserved motifs. We constructed an unrooted phylogenetic tree (Figure 5B) to show the evolutionary relationships between these sequences and to indicate the tissue localizations of ESTs. The tree indicates that T-phylloplanin groups with similar sequences from other solanaceous plants that also bear glandular secreting trichomes, and it is intriguing that the S. tuberosum gene is expressed in floral tissue (which may bear trichomes). Similar sequences from the monocots rice (Oryza sativa) and barley (Hordeum vulgare) also form a distinct group in the phylogenetic tree, with the gene from H. vulgare being expressed in root tissue. The genomic structure of T-Phylloplanin was elucidated from N. tabacum genomic DNA using a GenomeWalker kit. The gene contains two exons (175 and 278 bp) that are separated by a 508-bp intron (data not shown).
Figure 5.
Multiple Alignment and Phylogenetic Analysis of T-Phylloplanin and Similar Sequences in Other Plants.
(A) The amino acid sequence of T-phylloplanin was aligned against sequences giving significant BLAST similarity scores using the ClustalW algorithm of DNASTAR Lasergene software. Amino acids conserved between any six sequences are indicated in reverse contrast. Species identifiers are explained in (B).
(B) Unrooted phylogenetic tree showing the evolutionary relationships between the sequences in (A). Bootstrap values of >50% are given on the respective branches. The first two letters of the acronyms indicate the species (Am, Antirrhinum majus; At, Arabidopsis thaliana; Br, Brassica rapa; Gm, Glycine max; Ha, Helianthus annuus; Le, Lycopersicon esculentum; Nt, Nicotiana tabacum; Os, Oryza sativa; Pt, Populus tremuloides; Sr, Stevia rebaudiana; St, Stevia tuberosum). The GenBank accession numbers of the sequences follow the species identifiers. Tissue localizations of ESTs and cDNAs are indicated beneath the acronyms.
Escherichia coli–Expressed T-Phylloplanin Inhibits P. tabacina
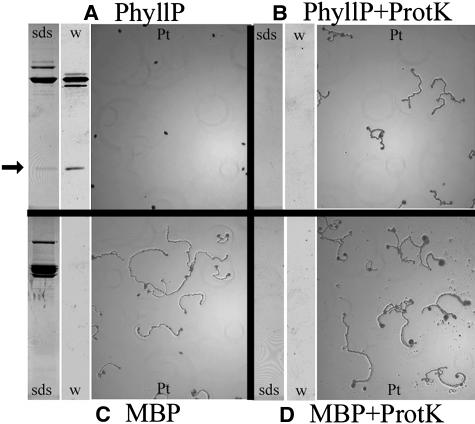
A 10.3-kD portion of the T-Phylloplanin gene (T-PhyllP) was expressed in E. coli as a fusion protein with Maltose Binding Protein (MBP). Soluble fusion protein (MBP-T-PhyllP) was purified on an amylose column, cut with the protease factor Xa to release T-PhyllP, and desalted on a 3-kD centrifugal filter. Both MBP-T-PhyllP and T-PhyllP reacted with the phylloplanin-specific antibody (Figure 6). The sample containing T-PhyllP inhibited P. tabacina spore germination at total protein concentrations of >160 ng/μL (Figure 6A). Protease digestion relieved the T-PhyllP inhibition of spore germination (Figure 6B). A control sample containing MBP alone, produced by an empty pMal-c2x vector and treated exactly as the T-PhyllP sample, had no effect on spore germination (Figure 6C), nor did protease-treated MBP (Figure 6D) at total protein concentrations of ≤500 ng/μL. We note that no inhibition of spore germination was observed with the MBP-T-PhyllP fusion protein not treated with factor Xa (data not shown). We conclude that released T-PhyllP is responsible for the observed inhibition, and because it is evident (Figure 6A, SDS gel) that released-T-PhyllP is a minor component of the sample (<10% of total protein), the inhibitory concentration of T-PhyllP is considered to be <160 ng/μL. T-PhyllP was lost when purification from MBP and factor Xa was attempted (data not shown).
Figure 6.
E. coli–Expressed TI 1068 T-Phylloplanin Inhibits P. tabacina Spore Germination.
Coomassie blue–stained SDS-PAGE gel blots (sds), protein gel blots with a 1:10,000 dilution of T-phylloplanin antiserum (w), and P. tabacina spore germination assays (Pt).
(A) E. coli–expressed MBP-PhyllP (160 ng/μL total protein) treated with factor Xa. The arrow indicates released T-PhyllP.
(B) E. coli–expressed MBP-T-PhyllP (160 ng/μL total protein) treated with factor Xa and proteinase-K (ProtK). The volume used was equivalent to that in (A).
(C) E. coli–expressed MBP (200 ng/μL total protein) treated with factor Xa.
(D) E. coli–expressed MBP (200 ng/μL total protein) treated with factor Xa and proteinase K. The volume used was equivalent to that in (C).
In leaf infection assays performed with KY 14 plants, T-PhyllP did not totally inhibit infection, but it greatly reduced necrotic leaf damage. MBP and uncut MBP-T-PhyllP fusion samples allowed successful infections (data not shown). We speculate that the lack of total inhibition with T-PhyllP may be attributable to insufficient protein concentration or the absence of another interacting protein; alternatively, we speculate that adducts with lipids or trichome exudate components are essential for a native protein–like response.
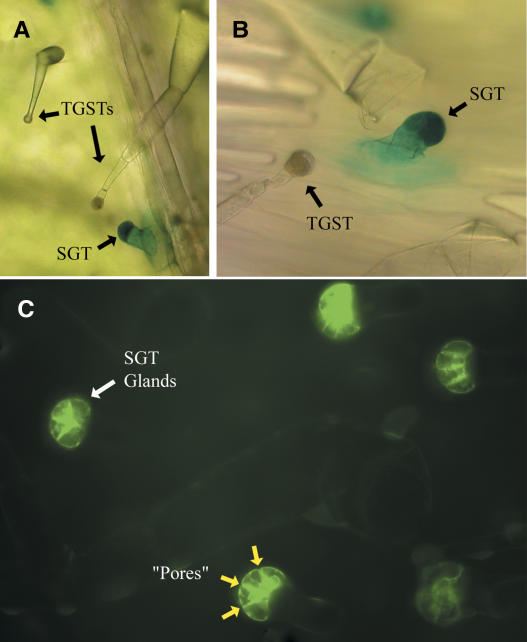
The T-Phylloplanin Promoter Region Directs Expression in Small Glandular Trichomes
We elucidated 1.8 kb of genomic DNA sequence upstream from the T-Phylloplanin transcription start site. A 1.1-kb region of this DNA, as well as the 5′ untranslated region and the T-Phylloplanin signal sequence, was fused in-frame with the reporter genes β-glucuronidase (GUS) and green fluorescent protein (GFP) and introduced into TI 1068 plants using Agrobacterium tumefaciens–mediated transformation. GUS and GFP were expressed only in SGTs (Figure 7), indicating the activity of a SGT-specific promoter. We found no evidence that GUS or GFP exits the SGTs. This is not surprising in that these reporter proteins are water soluble. TI 1068 SGTs are uniformly distributed over the leaf surface and protrude over surrounding epidermal cells (Figure 1A). We suggest that T-phylloplanins are biosynthesized locally in SGTs and are secreted to the leaf surface, where, because of their hydrophobicity and basicity, they dissolve in TGST exudate and are dispersed widely on the leaf surface during exudate flow. We note that certain animal saposin proteins are also highly hydrophobic and basic, are secreted by epithelial cells, and operate as components of innate immunity at the pulmonary air–water interface (Weaver and Conkright, 2001).
Figure 7.
The T-Phylloplanin Promoter Directs Protein Expression Only in SGTs.
(A) Magnification of a 5-bromo-4-chloro-3-indolyl-β-glucuronic acid–stained plantlet leaf from TI 1068 with GUS under the control of the T-phylloplanin promoter. TGSTs are also indicated.
(B) An X-gluc–stained SGT on a TI 1068 plantlet expressing GUS under the control of the T-phylloplanin promoter. Surface structures are indicated.
(C) Fluorescent magnification/detection of a TI 1068 plantlet with GFP under the control of the T-phylloplanin promoter. GFP was present only in SGT gland cells. The yellow arrows indicate constrictions between gland cells that we speculate may be pores to release protein to the leaf surface.
Ultrastructural studies by Akers et al. (1978) defined the subcellular structures of N. tabacum cv Xanthi SGTs and TGSTs. Glands of procumbent SGTs were observed to have approximately four cells separated by large, specifically oriented intracellular spaces that contained substantial OsO4-stained material. The nature of the accumulated substance was not defined, but we now speculate that this substance is T-phylloplanins, because the pattern of intracellular space disposition observed by Akers et al. (1978) is strikingly similar to what we have observed here using the T-phylloplanin–promoter–GFP construct (Figure 7C). Also, all tobaccos we have examined but one (smooth leaf N. glauca) produce phylloplanins. We conclude that T-phylloplanins are produced in SGT gland cells and speculate that they are secreted to gland extracellular spaces and then transferred outside of the glands through constrictions at the termini of intracellular spaces, which we speculate to be secretory pores (Figure 7C, yellow arrows) of unknown structure.
The majority of plant pathogens are fungi. When airborne spores land on a leaf surface, germination is the initial step leading to host colonization. We hypothesize that by rapidly inhibiting spore germination at the leaf surface, preformed plant proteins may suppress pathogen infection before induced defenses become functional, in a manner analogous to that of secreted surface proteins of animals. Our hypothesis is supported by our observations that surface-accumulated N. tabacum T-phylloplanins and E. coli–expressed T-PhyllP inhibit P. tabacina spore germination in vitro and limit leaf infection in situ. Our hypothesis is also supported by the observation that the T-phylloplanin promoter directs reporter gene expression specifically in SGTs, and T-phylloplanins are retained on leaves from which trichomes were completely removed by brushing of frozen tissue. From these observations, we propose that T-phylloplanins are secreted to and broadly dispersed on the leaf surface. Three observations link the gene T-Phylloplanin to T-phylloplanin proteins collected from the leaf surface. First, all amino acid sequences recovered from leaf surface T-phylloplanins I to IV are present in the predicted protein sequence from T-Phylloplanin, representing 54% of the mature protein open reading frame. Second, we provide a functional link between the gene and the proteins by replicating LWW blue mold inhibition with E. coli–expressed T-PhyllP. The T-phylloplanin promoter is a third, critical link between the gene and surface-disposed T-phylloplanins and implicates SGTs as the sites of T-phylloplanin biosynthesis and delivery to the surface.
We suggest that secreted phylloplanins (such as T-phylloplanins) may represent a novel leaf surface defense system in tobaccos, and perhaps generally in the plant kingdom, wherein protein biosynthesis in a specific trichome type allows the deposition and dispersion of phylloplanins on leaf aerial surfaces to deter pathogen establishment. Further study is needed to elucidate the mechanisms of T-phylloplanin–mediated fungal inhibition, and tandem mass spectrometry–based techniques may help identify posttranslational modifications or covalent adducts speculated to be present in T-phylloplanins I to IV. We must also regard SGTs as specialized biosynthetic structures akin to TGSTs because this study demonstrates a unique biosynthetic capability of procumbent SGTs, structures that are found on several different plants. Further study is needed to verify whether OsO4-stained material found in tobacco SGTs by Akers et al. (1978) is T-phylloplanin and to understand the details of how tobacco SGTs deliver T-phylloplanins to the leaf surface. We also must determine whether these surface-disposed protein factories can be used to enhance disease resistance and/or for molecular farming. We also recognize that T-phylloplanins are the first-studied representatives of a widespread protein family that exists in many different plants. Based on the various tissue localizations of homologous ESTs, phylloplanins may play different roles in different plants. Further study is needed to characterize phylloplanins of other plants, to determine their sites of synthesis and tissue localizations, and to elucidate their functions.
METHODS
Biological Material and Growth Conditions
Greenhouse tobacco plants (Nicotiana tabacum TI 1068, 1112, and 1406 and cv KY 14 and Petite Havana SR1 [hereafter referred to by TI number or cultivar name]) were germinated and grown in soil under natural light at 22 to 24°C with weekly fertilization (20:20:20, N:P:K). Plants were transplanted into 15-cm pots and treated with the insecticide Marathon (Olympic Horticultural Products, Mainland, PA) at 3 to 4 weeks after emergence. Field plants (TI 1068, soybean [Glycine max], and sunflower [Helianthus annuus]) were grown at a farm near Lexington, Kentucky, during the 2002 growing season.
To grow sterile TI 1068 plants, seeds were immersed in 10% (v/v) sodium hypochlorite for 10 min, rinsed briefly in 70% (v/v) ethanol, washed four times in sterile water, and germinated on MS medium (Life Technologies, Grand Island, NY) containing B5 vitamins (100 mg/L myo-inositol, 10 mg/L thiamine-HCl, and 1 mg/L each pyridoxine-HCl and nicotinic acid) in a 22°C growth chamber under fluorescent illumination (light and dark periods of 16 and 8 h daily). Individual plants were transferred to PlantCons (ICN Biomedicals, Aurora, OH) containing MS agar at 3 weeks after emergence.
Escherichia coli strain ER2508 (New England Biolabs, Beverly, MA) was stored and propagated as described by the supplier. Spores of Peronospora tabacina (isolate KY-79) were harvested from sporulating lesions on KY 14 plants as described (Reuveni et al., 1986).
Phylloplanin Collection and SDS-PAGE
Water-soluble phylloplane components were collected in LWW from mature, fully expanded leaves of all greenhouse-grown and field-grown plants by washing freshly detached leaves in 200 mL of nanopure water for 15 s (NANOpure water system D4751; Barnstead/Thermolyne, Dubuque, IA). Cut petioles or cut leaf surfaces were not exposed to wash solutions.
LWW were filtered (No. 1 filter paper; Whatman, Clifton, NJ), lyophilized to dryness, resuspended in 3 mL of sterile water, and centrifuged at 12,000g for 5 min at 21°C. The supernatants were filtered (13 mm/0.45-μm syringe filter; Corning Glass Works, Corning, NY) to exclude bacteria and fungi.
Proteins were separated by SDS-12% Gly-PAGE (Laemmli, 1970) or SDS-15% Tricine-PAGE (Judd, 1994) using a Mini-Protean II electrophoresis system (Bio-Rad, Hercules, CA), according to the manufacturer's instructions, and visualized with Coomassie Brilliant Blue R 250 or silver staining.
Protein concentration was estimated using the bicinchoninic acid assay (Pierce Chemical, Rockford, IL) with BSA as a standard. Leaf surface areas were estimated by tracing leaves onto uniform-weight paper and weighing the cutouts.
Collection of Epidermal Peels and Extracellular Fluid
Epidermal peels were prepared from greenhouse-grown TI 1068 plants as described (Kandra et al., 1990) and pulverized with liquid N2, and proteins were analyzed by SDS-PAGE. Extracellular fluid was collected using a vacuum infiltration method (Terry and Bonner, 1980) and analyzed by SDS-PAGE.
GC Analysis
Trichome exudate was collected from greenhouse-grown TI 1068 by immersing unwashed leaves for 15 s in 200 mL of acetonitrile. The wash solutions were filtered (No. 1 filter paper; Whatman) and dried, and trichome exudate was resuspended in 5 mL of acetonitrile and quantified by GC (flame ionization detection) as trimethylsilyl derivatives prepared in dimethylformamide, as described previously (Wang et al., 2001). To determine the amounts of trichome exudate biochemicals occurring in LWW, volumes equivalent to 200-cm2 leaf surface areas were transferred to glass GC vials and dried in a vacuum oven (37°C) overnight. Trichome exudate biochemicals were extracted at 21°C with methylene chloride, dried, solubilized, derivatized, and analyzed by GC. The amount of residual trichome exudate biochemicals in LWW was assessed relative to total trichome exudate on an equivalent surface area basis.
T-Phylloplanin Amino Acid Sequencing
Proteins in greenhouse-grown TI 1068 LWW were separated by SDS-PAGE, transferred to polyvinyl difluoride (Immobilon-psq; Millipore, Bedford, MA) using a Mini-Protean II electroblot apparatus (Bio-Rad), and visualized with Coomassie Brilliant Blue. T-phylloplanin bands were subjected to N-terminal sequencing using automated Edman degradation (Matsudaira, 1987) at the University of Kentucky Macromolecular Structure Analysis Facility (Lexington, KY). To recover internal amino acid sequence information, LWW from greenhouse-grown TI 1068 was separated by SDS-PAGE and stained with Coomassie Brilliant Blue, and 21- and 19-kD bands were excised and digested with trypsin. Total proteins in TI 1068 LWW were also digested with pepsin. Resulting tryptic or peptic peptides were separated by reversed-phase HPLC (Aquapore RP-300, 7-μm particle size, octyl reversed-phase column [Applied Biosystems, San Jose, CA]) and manually collected based on absorbance at 214 nm, and samples were reduced in volume under vacuum to ∼50 μL. Amino acid sequence analyses of tryptic peptides were performed as described above. For peptic peptides, similar analyses were performed at the Protein Facility of Iowa State University (Ames, IA).
Degenerate RT-PCR, RLM-RACE, and Elucidation of Genomic Structure
Total RNA was extracted from TI 1068 leaf tissue (100 mg fresh weight) with an RNeasy kit (Qiagen, Chatsworth, CA), and cDNA was synthesized from 5 μg of total RNA using an Omniscript RT kit (Qiagen). PCR was performed using PCR master mix (Promega, Madison, WI) containing 3 μL of cDNA template and 4 μM of each primer in a 50-μL volume. Successful amplification of a PCR product occurred with the primers 5′-ACWTTIGTITCIACWCATATYTCIGGICTIGTYTTTTG-3′ and 5′-AARAAICCIGTIGGIGCIARICCIACYCTAAT-3′, where I = inosine, W = A or T, Y = C or T, and R = A or G. Amplification was for 46 cycles using the following thermal profile: 95°C for 45 s, 50°C for 45 s, 72°C for 1 min, followed by a final 4-min extension at 72°C. The PCR product was size-fractionated by electrophoresis on a 1% (w/v) agarose gel, extracted using a Qiaex II kit (Qiagen), cloned into a pGem-T vector (Promega), and sequenced.
For RLM-RACE, total RNA was extracted from TI 1068 leaf tissue as described above. A GeneRacer kit (Invitrogen) containing SuperScript III was used to generate cDNAs, according to the manufacturer's instructions. Successful amplification of a 3′ RACE product occurred with the GeneRacer 3′Primer and the gene-specific primer 5′-CTCAGTCCCCAAGTTTTTCCTAATGCATCAG-3′. Successful amplification of a 5′ RACE product occurred with the GeneRacer 5′Primer and the gene-specific primer 5′-GGCCAAGAAAGTTAACTAGCTGATGCATA-3′. PCR cycling parameters were according to the GeneRacer protocol.
T-Phylloplanin genomic structure was elucidated using a GenomeWalker kit (Clontech, Palo Alto, CA), according to the manufacturer's protocol, using genomic DNA isolated from TI 1068 leaf tissue (100 mg fresh weight) with a DNeasy plant kit (Qiagen). Primary PCR was performed with a sense outer adaptor primer (AP1), provided in the kit, and the antisense T-Phylloplanin–specific primer 5′-TGGAACAAGTATGGCAAATGCAGCGGGG-3′. Primary PCR cycling parameters were 7 cycles of 25 s at 94°C and 3 min at 72°C, followed by 32 cycles of 25 s at 94°C and 3 min at 67°C, with a final extension of 7 min at 67°C. Products of primary PCR were diluted 1:25, and 1 μL was used in nested PCR with a sense inner adaptor primer (AP2), provided in the kit, and the nested antisense T-Phylloplanin–specific primer 5′-GGGGGTTGCGATTAATGCAGCCAAAAGGAAAA-3′. Nested PCR cycling parameters were 5 cycles of 25 s at 94°C and 3 min at 72°C, followed by 20 cycles of 25 s at 94°C and 3 min at 67°C, with a final extension of 7 min at 67°C. Amplified PCR products were amplified, size-fractionated by gel electrophoresis, gel-extracted, cloned into pGem-T, and sequenced.
Expression Vector Construction and Fusion Protein Purification
To overexpress the T-Phylloplanin gene in E. coli, a 10.3-kD portion of the coding sequence (His-33 to Gly-142, termed PhyllP) and the full-length mature protein-coding sequence (Ile-24 to Asn-150) were amplified incorporating XbaI and PstI restriction sites (PhyllP sense, 5′-AGCTTCTAGACATATTTCGGGGCTGGTTTT-3′; PhyllP antisense, 5′-AGCTCTGCAGTTAGCCGGTGGGGGCGAGGCC-3′; full sense, 5′-AGCTTCTAGAATACTTGTTCCAACACT-3′; full antisense, 5′-AGCTCTGCAGTTAATTGATGTTAAGA-3′; the restriction sites are underlined). The PCR products were digested with XbaI and PstI and cloned into the pMal-c2x expression vector (New England Biolabs) to create a translation fusion between the gene inserts and malE (which encodes MBP). Protein expression was induced at OD600 = 0.5 by the addition of 0.1 mM isopropyl-β-d-thiogalactoside. Cells were harvested and resuspended in column binding buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, and 1 mM EDTA) containing 1 mg/mL lysozyme. Cell lysate was centrifuged at 10,000g for 10 min, and the resulting supernatant was collected. Fusion protein was purified using amylose-mediated column chromatography (New England Biolabs) according to the manufacturer's instructions and examined by SDS-PAGE. Fractions containing purified fusion protein were pooled and concentrated to ∼1 mg/mL using a 3-kD centrifugal filter (Microsep 3K Omega; Pall Laboratories, Fort Myers, FL). Factor Xa (New England Biolabs) was added, and samples were incubated for 48 h at 21°C. Salts and buffer components were removed using a 3-kD centrifugal filter, and protein concentration was adjusted to 1 mg/mL with the addition of sterile water.
T-Phylloplanin Antibody and Protein Gel Blots
TI 1068 LWW was separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Phylloplanin III was excised and used to generate a rabbit polyclonal antibody (Strategic Biosolutions, Newark, DE). Immunodetection was performed using a 1:10,000 dilution of phylloplanin antiserum and a 1:10,000 dilution of horseradish peroxidase–coupled anti-rabbit secondary antibody (Sigma-Aldrich, St. Louis, MO).
Protease Treatment
Insoluble proteinase K affixed to acrylic beads (100 mg; P0803; Sigma-Aldrich) was placed into mini-spin filters (732-6027; Bio-Rad). The filters containing beads were placed into empty 1.5-mL Eppendorf tubes, and the filters were washed with sterile water (700 μL; 2600g for 1 min). The flow-through was discarded, and washing was repeated five times. The spin filters were transferred to empty 1.5-mL Eppendorf tubes. Samples were added to filters containing protease beads and incubated at 37°C for 4 h, with periodic inversion to mix. The tubes were then centrifuged at 2600g for 10 min, and the flow-through from each was collected and analyzed by SDS-PAGE or used in blue mold assays.
P. tabacina Spore Germination and Leaf Infection Assays
Freshly collected P. tabacina spores were mixed with various concentrations of TI 1068 LWW, proteinase K–treated TI 1068 LWW, or water incubated with proteinase K and germinated for 16 h in dark, humidified chambers as water drops (4-μL drops; 50 spores/μL) on microscope slides. The spores were then inspected visually at ×100 magnification for germination. The absence of a germination tube after 16 h indicated inhibition. Similar experiments were performed with T-PhyllP, MBP, proteinase K–treated T-PhyllP, and proteinase K–treated MBP. To assess the immediacy of germination tube arrest by LWW, spores were observed after 3 h.
For the leaf infection assay, 6-week-old, greenhouse-grown Petite Havana SR1 plants were preconditioned by incubation in a 21°C growth room (14 h of light) for 5 d. Dilution series (1, 5, 12.5, 25, 50, 75, and 100 ng protein/μL) of TI 1068 LWW were prepared and mixed with freshly collected P. tabacina spores immediately before inoculation. For each LWW dilution, 8 to 10 drops (4-μL drops; 100 spores/μL) were applied to one leaf of preconditioned plants. Plants were placed in dark, humidified chambers for 16 h to provide optimal conditions for infection and then returned to the growth room. Treated leaves were excised 5 d after inoculation, placed in dark, humidified chambers for 16 h, and then inspected for sporulation. The formation of P. tabacina sporulating lesions indicated successful leaf infection.
Elucidation of T-Phylloplanin Promoter Sequence and Activity
Genomic DNA was isolated from TI 1068 leaf tissue (100 mg fresh weight) using a DNeasy plant mini kit (Qiagen). The DNA sequence upstream of the T-Phylloplanin gene was recovered using a GenomeWalker kit (Clontech), according to the manufacturer's protocol. Briefly, ∼4 μg of genomic DNA was digested to completion (36 h) in four separate reactions with restriction enzymes that generated blunt ends (DraI, EcoRV, PvuII, and StuII). The resulting libraries were purified by phenol:chloroform extraction and precipitation. Digested genomic DNA in each library was then ligated to 5′GenomeWalker Adaptor molecules and purified again. A primary PCR for each library was performed with a sense outer adaptor primer (AP1), provided in the kit, and the antisense T-Phylloplanin–specific primer 5′-TGGAACAAGTATGGCAAATGCAGCGGGG-3′. Primary PCR cycling parameters were seven cycles of 25 s at 94°C and 3 min at 72°C, followed by 32 cycles of 25 s at 94°C and 3 min at 67°C, with a final extension of 7 min at 67°C. Products of primary PCR were diluted 1:25, and 1 μL was used in nested PCR with a sense inner adaptor primer (AP2), provided in the kit, and the nested antisense T-Phylloplanin–specific primer 5′-GGGGGTTGCGATTAATGCAGCCAAAAGGAAAA-3′. Nested PCR cycling parameters were five cycles of 25 s at 94°C and 3 min at 72°C, followed by 20 cycles of 25 s at 94°C and 3 min at 67°C, with a final extension of 7 min at 67°C. A 1.8-kb product was amplified from the StuII-based library, gel-extracted, cloned into pGem-T, and sequenced.
PCR using a T-Phylloplanin promoter–specific sense primer (5′-TGCTCCCACCACTAGAATCACCA-3′) and a T-Phylloplanin–specific antisense primer with an XbaI cut site (5′-AGCTTCTAGATGTTGGAACAAGTATGG-3′; the XbaI site is underlined) was then used to amplify the region of N. tabacum genomic DNA that included the first 25 amino acids of the T-phylloplanin protein (which included the signal sequence), the 5′ untranslated region, and a further 1.1 kb upstream. The PCR product was then cut with XbaI and HindIII (at a restriction site endogenous to the promoter) and cloned into the HindIII-XbaI sites of pBIMC (kindly provided by D. Falcone; pBIMC is a variant of pBI121 modified to include a polylinker in place of the GUS gene) to replace the Cauliflower mosaic virus 35S promoter and create the vector pBI-PhylloProm. To analyze the spatial expression of the promoter, the reporter genes GUS and sGFP (kindly provided by D. Falcone) were amplified by PCR with primers that incorporated XbaI and XhoI restriction sites (GUS sense, 5′-AGCTTCTAGAATGTTACGTCCTGTAGAAACCCCA-3′; GUS antisense, 5′-AGCTCTCGAGTCATTGTTTGCCTCCCTGCT-3′; GFP sense, 5′-AGCTTCTAGAATGGTGAGCAAGGGCGAGGA-3′; GFP antisense, 5′-AGCTCTCGAGGCTTTACTTGTACAGCTCGT-3′; the restriction sites are underlined). The PCR products were gel-extracted, cut with XbaI and XhoI, and ligated between XbaI-XhoI sites in the polylinker of pBI-PhylloProm to create in-frame fusions with the T-Phylloplanin start codon and signal sequence. These constructs were transformed into Agrobacterium tumefaciens GV3101 by triparental mating and introduced into TI 1068 using the leaf disc method (Horsch et al., 1985). Kanamycin-resistant plantlets were derived from kanamycin-resistant callus tissue and transferred to soil. Leaf discs from pBI-PhylloProm:GUS explants were stained for GUS activity by incubation with 0.1% 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Jefferson, 1987) and photographed. Leaf discs from pBI-PhylloProm:GFP explants were magnified and photographed using an Axioplan-2 imaging system (Zeiss, Jena, Germany).
Bioinformatic Analysis
Homologous open reading frames of selected cDNA or EST sequences giving significant (e-value cutoff of 10e-04) BLASTn, BLASTp, and tBLASTx (Altschul et al., 1990) scores against T-phylloplanin nucleotide and amino acid sequences were first analyzed for the presence of signal peptides using TargetP. A multiple alignment of protein sequences with the predicted signal peptides removed was performed using the ClustalW algorithm (DNASTAR). An unrooted phylogenetic tree was constructed using the maximum parsimony algorithm PROTPARS in the PHYLIP version 3.63 software package (Felsenstein, 2004), and tree robustness was estimated with 1000 bootstrapped data sets. The tree was displayed with TREEVIEW version 3.2 software (Page, 1996).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY705384.
Acknowledgments
We thank J.T. Hall and L.J. Asher for plant maintenance and technical assistance, B.C. Li for assistance with blue mold assays, D.L. Falcone for providing the pBIMC vector and reporter genes, and M. Goodin for assistance with GFP detection. This work was supported by a Kentucky Tobacco Research and Development Center grant to G.J.W. and a Jeffrey Graduate Fellowship to R.W.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: George J. Wagner (gwagner@uky.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031559.
References
- Akers, C.P., Weybrew, J.A., and Long, R.C. (1978). Ultrastructure of glandular trichomes of leaves of Nicotiana tabacum L., cv Xanthi. Am. J. Bot. 65, 282–292. [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, C.W., and Lipman, D.L. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Cruickshank, I.A.M., Perrin, D.R., and Mandryk, M. (1977). Fungitoxicity of duvatrienediols associated with the cuticular wax of tobacco leaves. Phytopathol. Z. 90, 243–249. [Google Scholar]
- Curstedt, T., Jornvall, H., Robertson, B., Bergman, T., and Berggren, P. (1987). Two hydrophobic low-molecular-mass protein-fractions of pulmonary surfactant: Characterization and biophysical activity. Eur. J. Biochem. 168, 255–262. [DOI] [PubMed] [Google Scholar]
- Dell, B., and McComb, A.J. (1978). Plant resins: Their formation, secretion, and possible functions. Adv. Bot. Res. 6, 227–316. [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fahn, A. (2000). Structure and function of secretory cells. Adv. Bot. Res. 31, 37–75. [Google Scholar]
- Felsenstein, J. (2004). PHYLIP (Phylogeny Inference Package) Version 3.6. (Seattle: University of Washington).
- Gallo, R.L., and Huttner, K.M. (1998). Antimicrobial peptides: An emerging concept in cutaneous biology. J. Invest. Dermatol. 111, 739–743. [DOI] [PubMed] [Google Scholar]
- Goffreda, J.C., Szymkowiak, E.J., Sussex, I.M., and Mutschler, M.A. (1990). Chimeric tomato plants show that aphid resistance and triacylglucose production are epidermal autonomous characters. Plant Cell 2, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A.V. (1966). Effect of inoculum spore load, length of infection period, and leaf washing on occurrence of Peronospora tabacina Adam (blue mould) of tobacco. Aust. J. Agric. Res. 17, 133–146. [Google Scholar]
- Horsch, R.B., Fry, J.E., Hoffman, N.L., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method of transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying for chimeric genes in plants: The GUS fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Judd, R.C. (1994). Electrophoresis of peptides. In Basic Protein and Peptide Protocols, J.M. Walker, ed (Totowa, NJ: Humana Press), pp. 49–57.
- Kandra, L., and Wagner, G.J. (1988). Studies of the site and mode of biosynthesis of tobacco trichome exudate components. Arch. Biochem. Biophys. 265, 425–432. [DOI] [PubMed] [Google Scholar]
- Kandra, L., Severson, R., and Wagner, G.J. (1990). Modified branched-chain amino-acid pathways give rise to acyl acids of sucrose esters exuded from tobacco leaf trichomes. Eur. J. Biochem. 188, 385–391. [DOI] [PubMed] [Google Scholar]
- Kelsey, R.G., Reynolds, G.W., and Rodriguez, E. (1984). The chemistry of biologically active constituents secreted and stored in plant glandular trichomes. In Biology and Chemistry of Plant Trichomes, E. Rodriguez, P.L. Healey, and I. Mehta, eds (New York: Plenum Press), pp. 187–240.
- Kennedy, B.S., Nielsen, M.T., Severson, R.F., Sisson, V.A., Stephenson, M.K., and Jackson, D.M. (1992). Leaf surface chemicals from Nicotiana affecting germination of Peronospora tabacina (Adam) sporangia. J. Chem. Ecol. 18, 1467–1479. [DOI] [PubMed] [Google Scholar]
- Knight, S.C., Anthony, V.M., Brady, A.M., Greenland, A.J., Heaney, S.P., Murray, D.C., Powell, K.A., Schulz, M.A., Spinks, C.A., Worthington, P.A., and Youle, D. (1997). Rationale and perspectives on the development of fungicides. Annu. Rev. Phytopathol. 35, 349–372. [DOI] [PubMed] [Google Scholar]
- Kowalski, S.P., Eannetta, N.T., Hirzel, A.T., and Steffens, J.C. (1992). Purification and characterization of polyphenol oxidase from glandular trichomes of Solanum berthaultii. Plant Physiol. 100, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronestedt-Robards, E., and Robards, A.W. (1991). Exocytosis in gland cells. Soc. Exp. Biol. Semin. Ser. 45, 199–232. [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lucas, G.B. (1975). Diseases of Tobacco. (Raleigh, NC: Biological Consulting Associates).
- Lucas, J.A. (1998). Plant Pathology and Plant Pathogens. (Oxford: Blackwell Science).
- Matsudaira, P. (1987). Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262, 10035–10038. [PubMed] [Google Scholar]
- Menetrez, M.L., Spurr, H.W., Danehower, D.A., and Lawson, D.R. (1990). Influence of tobacco leaf surface chemicals on germination of Peronospora tabacina Adam sporangia. J. Chem. Ecol. 16, 1565–1576. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Reuveni, M., Tuzun, S., Cole, J.S., Siegel, M.R., and Kuc, J. (1986). Removal of duvatrienediols from the surface of tobacco leaves increases their susceptibility to blue mold. Phytopathology 76, 1092–1098. [Google Scholar]
- Reuveni, M., Tuzun, S., Cole, J.S., Siegel, M.R., Nesmith, W.C., and Kuc, J. (1987). Removal of duvatrienediols from the surface of tobacco leaves increases their susceptibility to blue mold. Physiol. Mol. Plant Pathol. 30, 441–451. [Google Scholar]
- Samac, D.A., and Shah, D.M. (1991). Developmental and pathogen-induced activation of the Arabidopsis acidic chitinase promoter. Plant Cell 3, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, J.-M. (1999). Epithelial antimicrobial peptides: Innate local host response elements. Cell. Mol. Life Sci. 56, 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring, O. (2000). Chemotaxonomy based on metabolites from glandular trichomes. Adv. Bot. Res. 31, 153–175. [Google Scholar]
- Steffens, J.C., and Walters, D.S. (1990). Biochemical aspects of insect resistance by glandular trichomes of the Solanaceae. In Naturally Occurring Pest Bioregulators, P. Hedin, ed (Washington, DC: ACS Books) pp. 66–82.
- Svircev, A.M., McKeen, W.E., and Smith, R.J. (1989). Host-parasite relations: Morphology and ultrastructure. In Blue Mold of Tobacco, W.E. McKeen, ed (St. Paul: APS Press), pp. 43–104.
- Terry, M.E., and Bonner, B.A. (1980). An examination of centrifugation as a method of extracting an extracellular solution from peas, and its use for the study of indoleacetic acid-induced growth. Plant Physiol. 66, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, W.W., and Healey, P.L. (1984). Cellular basis of trichome secretion. In Biology and Chemistry of Plant Trichomes, E. Rodriguez, P.L. Healey, and I. Mehta, eds (New York: Plenum Press), pp. 95–112.
- Wagner, G.J. (1999). Leaf surface chemistry. In Tobacco: Production, Chemistry and Technology, D.L. Davis and M.T. Nielsen, eds (Oxford: Blackwell Science), pp. 292–303.
- Wagner, G.J., Wang, E., and Shepherd, R.W. (2004). New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. (Lond.) 93, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E., Wang, R., DeParasis, J., Loughrin, J.H., Gan, S.S., and Wagner, G.J. (2001). Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat. Biotechnol. 19, 371–374. [DOI] [PubMed] [Google Scholar]
- Weaver, T.E., and Conkright, J.J. (2001). Functions of surfactant proteins B and C. Annu. Rev. Physiol. 63, 555–578. [DOI] [PubMed] [Google Scholar]